Abstract
Objective
To determine to what extent oligoclonal band (OCB) specificities are clonally interrelated and to what degree they are associated with corresponding B-cell responses in the peripheral blood of multiple sclerosis (MS) patients.
Methods
Mass-spectrometric proteomic analysis of isoelectric focused (IEF) CSF IgG was used in combination with next-generation deep-immune repertoire sequencing of peripheral blood (PB) and CSF IgG heavy chain variable regions (IgG-VH) from MS patients.
Results
We find evidence for ongoing stimulation and maturation to antibody expressing B cells to occur primarily inside the CNS compartment. B cells participating in OCB production can also be identified in peripheral blood; these cells appear to migrate across the blood-brain barrier (BBB) and may also undergo further antigen-stimulation in the periphery. In individual patients different bands comprising OCB are clonally related.
Interpretation
Our data provide a high-resolution molecular analysis of OCB and strongly support the concept that OCB are not merely the terminal result of a targeted immune response in MS but represent a component of active B cell immunity that is dynamically supported on both sides of the blood-brain barrier.
INTRODUCTION
The presence of soluble clonal IgG, also referred to as “oligoclonal bands” (OCB), in the cerebrospinal fluid (CSF) represents a central immunodiagnostic feature for MS, detected in more than 95 % of patients1. OCB result from intrathecal antigen-driven immune responses2, 3 against as yet unknown target antigens. Typically visualized as discrete bands on isoelectric focusing gel electrophoresis, OCB have been associated with a more rapid conversion from clinically isolated syndrome (CIS) to clinically definite MS4–6, evidence that they may reflect more active CNS-directed autoimmunity or otherwise contribute to tissue damage. Under treatment with natalizumab, a peripherally–acting anti-VLA4 monoclonal antibody that blocks immune cell migration into the CNS and effectively reduces MS disease activity, CSF IgG levels can decrease and OCB disappear7, 8. These changes are associated with a decrease in intrathecal lymphocytes7, 9, suggesting that an ongoing exchange of immune cells between the peripheral blood and the CNS is required to maintain intrathecal B cell stimulation and OCB.
Although oligoclonal CSF IgG have been shown to be products of intrathecal plasma cells and B cells3, 10, essentially nothing is known about how B-cells and/or plasma cells migrate to the CNS compartment and establish immunologically active sites in MS brain. It is likely that antigen-directed affinity maturation and terminal differentiation of B cells contributing to OCB occurs within perivascular infiltrates and meningeal lymphoid-like follicles in the CNS11, 12. Attempts to define the antigenic specificity of B cell immunity in MS have yielded mixed and partially conflicting results. While some studies demonstrated binding of intrathecally produced IgG and/or OCB to myelin13, myelin proteins14, 15, and Epstein-Barr virus antigens16, others have reported an absence of reactivity to myelin antigens17.
Our own work suggests that IgG expressing B cells and/or plasma cells participate in an ongoing exchange across the blood-brain barrier (BBB), a process operative during clinically active and also seemingly quiescent phases of the disease18. It is not known if B cells or plasma cells that express Ig corresponding to OCBs are also present in the peripheral blood where they may contribute to CNS targeted immune responses. Bystander activation in the periphery has been suggested as one mechanism by which proinflammatory B cells might support CNS directed T cell mediated autoimmunity in MS19; however, there is currently no evidence in MS linking antigen-driven immunity in the periphery to targeted humoral immune responses in the CNS compartment.
We performed extensive mass-spectrometric proteomic analysis of CSF OCB from 5 MS patients; previously described IgG heavy chain variable region (IgG-VH) sequence datasets obtained by deep-immune repertoire sequencing of peripheral blood (PB) and CSF were used as reference to match proteomic data with IgG-VH transcripts18. Our findings suggest that ongoing antigenic stimulation and maturation of B cells to antibody expressing plasma cells and plasmablasts occurs mostly inside the CNS compartment. More important, we show that B cells involved in OCB expression may also egress into the peripheral blood where they may undergo further affinity maturation.
MATERIALS AND METHODS
Patients and Samples
CSF samples from five patients who provided informed consent and who met diagnostic criteria for MS20 were available for our studies (Table 1; Table S1 for additional clinical information). All patients displayed multiple OCB in their CSF that were not detectable in their serum. Immediately after CSF collection via lumbar puncture, CSF was centrifuged and supernatants stored separately from cell pellets at −80°C; peripheral blood (PB) was obtained during the same patient visit. All 5 patients have been previously reported18. IgG heavy (H) chain variable region (IgG-VH) transcriptomes were generated from CSF cells and PB mononuclear cells (PBMC) using deep immune repertoire sequencing (DIRS). OCB IgG proteomes were analyzed by mass-spectrometry using IgG isolated from CSF supernatants and separated by isoelectric focusing (IEF). The study was approved by the UCSF Committee on Human Research.
Table 1. Patient characteristics.
Shown are age (years), sex (F/M), treatment (Tx; IVMP, intravenous methylprednisolone; GA, glatiramer acetate); CSF volume obtained for this study, CSF white cell count (WBC), presence of OCB as determined by a clinical laboratory (+ if ≥ 2 CSF-restricted IgG bands); and numbers of IgG-VH sequences per patient contained in our IgG-VH reference database. Refer to Table S1 for additional clinical information. The CSF/PB VHref-CSF represents IgG-VH sequences that are restricted to the CSF or belong to bi-compartmental clusters of IgG-VH sequences; VHref-PB contains IgG-VH sequences from peripheral blood without a link to the CSF compartment (see Methods).
| Patient ID | Age | Sex | CSF vol (ml) | WBC | OCB | VHref-CSF | VHref-PB |
|---|---|---|---|---|---|---|---|
| MS-1 | 29 | F | 8 | 1 | + | 294 | 135,346 |
| MS-2 | 37 | F | 10 | 1 | + | 419 | 60,609 |
| MS-3 | 39 | F | 10 | ND | + | 1,025 | 127,973 |
| MS-4 | 39 | F | 10 | 3 | + | 61 | 78,136 |
| MS-5 | 22 | M | 8 | 4 | + | 1,723 | 50,207 |
Deep immune repertoire sequencing (DIRS)
In brief, RNA from the CSF cell pellet or PBMC (1×106) was isolated and reverse transcribed using the SMARTer™ RACE cDNA Amplification Kit (Clontech). cDNA was amplified using the SMARTer™ RACE universal 5’ primer mix and an IgG isotype specific 3’ primer (5’-GGG AAG ACS GAT GGG CCC TTG GTG G-3’) to allow for unbiased, immunoglobulin heavy chain variable germline gene (IGHV)-independent amplification of IgG-VH. IgG-VH transcript libraries were prepared using GS FLX Titanium kits (Lib-L chemistry, 454 Sequencing, Roche). PCR amplification was performed at 95°C for 5 minutes; 95°C for 30 seconds, 65°C for 30 seconds, 72°C for 1 minute for 33 cycles; and 72°C for 7 minutes. IgG-VH PCR products were purified using AMPure XP beads (Beckman Coulter Genomics) and quantified using a Quant-iT™ PicoGreen dsDNA kit (Invitrogen). Purified PCR products were diluted to 1×109 molecules/µl and subjected to emulsion PCR and unidirectional sequencing (from the IgG isotype-specific primer end) using the GS FLX Titanium Lib-L chemistry (Roche).
IgG-VH repertoire sequence analysis
For all IgG-VH sequencing reads, IGHV and immunoglobulin heavy chain joining (IGHJ) germline gene usage, and IgG heavy chain CDR3 region (H-CDR3) were determined using VDJFasta software as previously described21; in addition to unique sequence identifiers resulting from DIRS, each sequence was tagged with a patient-specific identifier and the compartment of origin (CSF or PBMC). Reads not passing quality criteria (uniquely classified IGHV, IGHJ and H-CDR3) were filtered from subsequent analysis. An average of 16,892 (± 10,248, SD) CSF IgG-VH and 318,130 (± 114,984, SD) PBMC IgG-VH were generated from each sample (CSF IgG-VH per patient: MS-1: 5,661; MS-2: 20,752; MS-3: 25,658; MS-4: 6,164; MS-5: 26,226) (PBMC IgG-VH per patient: MS-1: 507,411; MS-2: 235,515; MS-3: 243,301; MS-4: 347,616; MS-5: 256,809). Only sequences with contiguous reading frames from H-CDR1 to the joining region were used to generate our reference databases (see below). Clusters of clonally related IgG-VH sequences were identified using a distance metric approach as previously described (Levenshtein distance of 0 or 1 between their H-CDR3 amino acid sequences22).
Our study was designed 1) to identify proteomic IgG signatures in CSF (OCB peptides) that would allow us to determine whether OCB are exclusively produced by CSF B cells or can also be linked to peripheral B cell repertoires, and 2) to determine whether separate IgG bands (OCB) on IEF electrophoresis might contain clonally related IgG. Availability of DIRS data permitted clustering of IgG-VH into groups of clonally related sequences that were derived exclusively from CSF, exclusively from PB, or from both compartments. To match IgG-VH transcriptomic with proteomic data, two IgG-VH sequence databases containing sequences that resulted from DIRS of CSF and PBMC of all 5 patients were generated:
-
-
VHref-CSF contained a pool of 3,522 IgG-VH sequences from all 5 patients reported here that were either derived from CSF-restricted IgG-VH sequence clusters, or found in PB and clonally related to CSF-derived IgG-VH. In VHref-CSF 2,308 IgG-VH derived from CSF, and 1,214 from PB. VHref-CSF permitted identification of OCB peptides that are clearly linked to the CSF compartment, either because they are directly found in the CSF IgG-VH transcriptome, or because they map to PB IgG-VH sequences that are clonally related to CSF IgG-VH.
-
-
VHref-PB contained a pool of 452,270 PBMC IgG-VH transcripts from all 5 patients reported here. None of the IgG-VH in VHref-PB have clonally related sequences among CSF IgG-VH. VHref-PB permitted identification of OCB peptides mapping to IgG-VH sequences contained in PB-restricted clusters, i.e. for which no clonally related IgG-VH could be found in the CSF-associated IgG-VH transcriptome from the same patient.
Isolation of IgG from CSF, isoelectric focusing, and mass spectrometry
4–6 ml of cell-free CSF supernatant was incubated overnight with Protein G sepharose 4 Fast Flow beads (GE healthcare) in the presence of protease inhibitors and IgG eluted using glycine buffer at pH2.8. The eluate was immediately neutralized using Tris buffer and processed for isoelectric focusing using the ReadyPrep 2D Cleanup kit (Biorad) according to manufacturers instructions. Eluates resuspended in rehydration buffer (Biorad) were loaded onto pH 3–10, 11 cm IPG strips (Biorad) and rehydrated for 16 hours. Isoelectric focusing (IEF) on IPG strips was performed according to manufacturer’s instructions. At the completion of the IEF run, the IPG strips were fixed in trichloroacetic acid (TCA) (Sigma) and sulfosalicylic acid (Sigma) for 1 hour and stained with colloidal Blue staining kit (Invitrogen) for 1 hour. Discrete bands representing clonal IgG were excised from the IPG strips and processed for mass spectrometry. Established methods of tryptic in-gel digestion were used for processing of IPG strip slices containing OCB (see Supplemental Methods for details).
IgG-VH proteome/transcriptome comparisons
Masses of tryptic OCB peptides as determined by tandem mass-spectrometry (see Supplemental Methods) were compared to in silico trypsin-digested IgG-VH contained in the reference databases (VHref-CSF, VHref-PB). Due to VHref-CSF/PB only containing variable region (IgG-VH) sequences, OCB peptides mapping to IgG constant regions were not included in our analyses. We applied a multi-step approach to determine whether OCB peptides were 1) patient-specific, 2) mapped to IgG-VH clusters that support a B cell exchange between CSF and PB, and 3) map to IgG-VH clusters represented by separate IgG bands on IEF electrophoresis: First, we identified “patient-specific” OCB peptides that were found exclusively in the same patient’s CSF proteome and IgG-VH transcriptome represented by VHref-CSF. For example, a CSF OCB peptide derived from patient MS-1 was considered patient-specific if it mapped exclusively to patient MS-1 IgG-VH transcripts in VHref-CSF; patient-specific OCB peptides from patient MS-2 had to map to MS-2 IgG-VH transcripts, MS-3 OCB peptides had to map to MS-3 IgG-VH transcripts, etc., to be considered “patient-specific”. Second, we analyzed all OCB peptides to determine whether they mapped uniquely to a single patient’s PBMC IgG-VH in VHref-PB but not in VHref-CSF; only one peptide was found meeting these criteria and was also considered “patient-specific”. Third, OCB peptides were assigned to clusters of clonally related IgG-VH sequences (see next paragraph). Only patient-specific peptides with full sequence confirmation by tandem mass-spectrometry (see Supplemental Methods) were used for further analysis. All patient-specific OCB peptides were analyzed via IgBlast (http://www.ncbi.nlm.nih.gov/igblast/) to determine similarity with known IGHV germline gene sequences.
OCB-associated IgG-VH clusters and lineages
To determine whether peptides mapped to IgG-VH expressed by clonally related B cells exclusively found in the CSF, or PB, or in both compartments, OCB peptides were first aligned to IgG-VH sequences contained in our databases (VHref-CSF/VHref-PB) and all IgG-VH transcripts containing exact OCB peptide sequence matches were retrieved from our databases. Then, IgG-VH sequences with identical IGHV and IGHJ usage, and highly similar/identical H-CDR3 amino acid sequence were retrieved from our IgG-VH databases to generate groups (clusters) of clonally related IgG-VH sequences representative of CSF OCB-derived peptides.
To understand the relationship of OCB peptides with IgG-VH lineage development during affinity-maturation, we generated lineage trees (examples see Figures 4 to 6) of clustered IgG-VH sequences containing OCB peptide sequences. IgG-VH sequences with a contiguous reading frame spanning at least from the 5’ end of H-CDR1 to the 3’ end of H-CDR3 were aligned using ClustalW 2.123. Putative germline sequences including the V-D-J junction were determined using SoDA24 and used as tree-root for lineage tree calculations. IgTree software25 (kindly provided by Dr. Ramit Mehr, Bar-Ilan University, Ramat-Gan, Israel) was used to place IgG-VH sequences along lineage trees. Lineage trees were displayed using the hierarchic (for clusters up to 50 nodes) or organic (for clusters >50 nodes) layout in Cytoscape (Version 2.8.326); in Figures 4 to 6, IgG-VH exclusively found in CSF are labeled blue, those in PB are red, and those present in both compartments are green. Putative germline sequences are black and represent the root of each lineage tree. Unknown intermediates calculated by IgTree are beige. The size of tree nodes is proportional to the number of identical sequences contained within each node. IgG-VH amino acid alignments in were generated using the MAFFT algorithm27 within Jalview28 software.
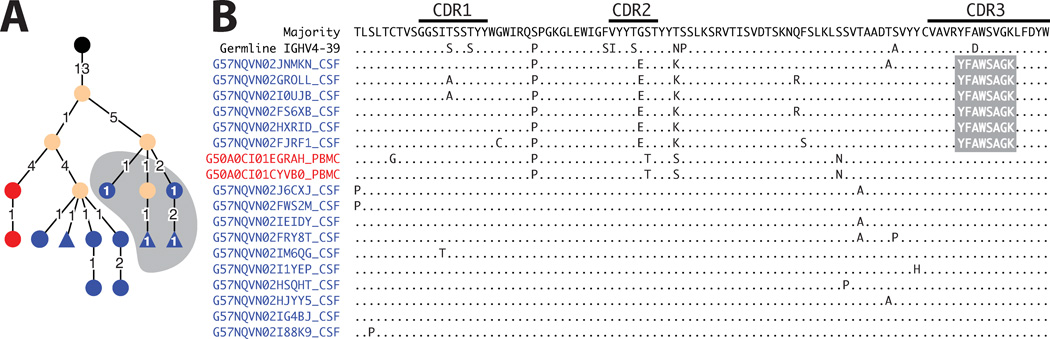
Figure 4. Bi-compartmental B cell lineage contributing to OCB production in patient MS-2.
Shown are the lineage tree (hierarchic layout; A) and alignment (B) of IgG-VH aminoacid sequences from CSF and PBMC. Lineage trees are calculated using nucleotide sequences IgTree software and displayed using Cytoscape (see Methods). In the lineage tree, each round node represents at least one unique IgG-VH sequence ranging from at least the 5’ end of H-CDR1 to the 3’ end of H-CDR3; larger nodes represent up to hundreds of identical sequences. CSF derived IgG-VH are represented by blue nodes, PB derived IgG-VH are red, and identical sequences found in both compartments are green. Numbers between nodes are numbers of nucleotide mutations; unlabeled connections between nodes represent single nucleotide mutations. Putative germline sequences were determined using SoDA24 and are labeled black; hypothetical intermediates calculated by IgTree are beige. Numbers on lines between nodes (edges) represent mutational steps (nucleotides) between nodes. Triangular nodes contain 2 or more singleton sequences in leaves. The OCB peptide (YFAWSAGK) identified from IEF band “D” (Figure 1) maps to the H-CDR3 and was only found in a CSF-restricted sub-lineage. Additional sub-lineages in PB and CSF suggest B cell affinity maturation and plasma cell maturation (OCB production) to occur in parallel in both compartments.
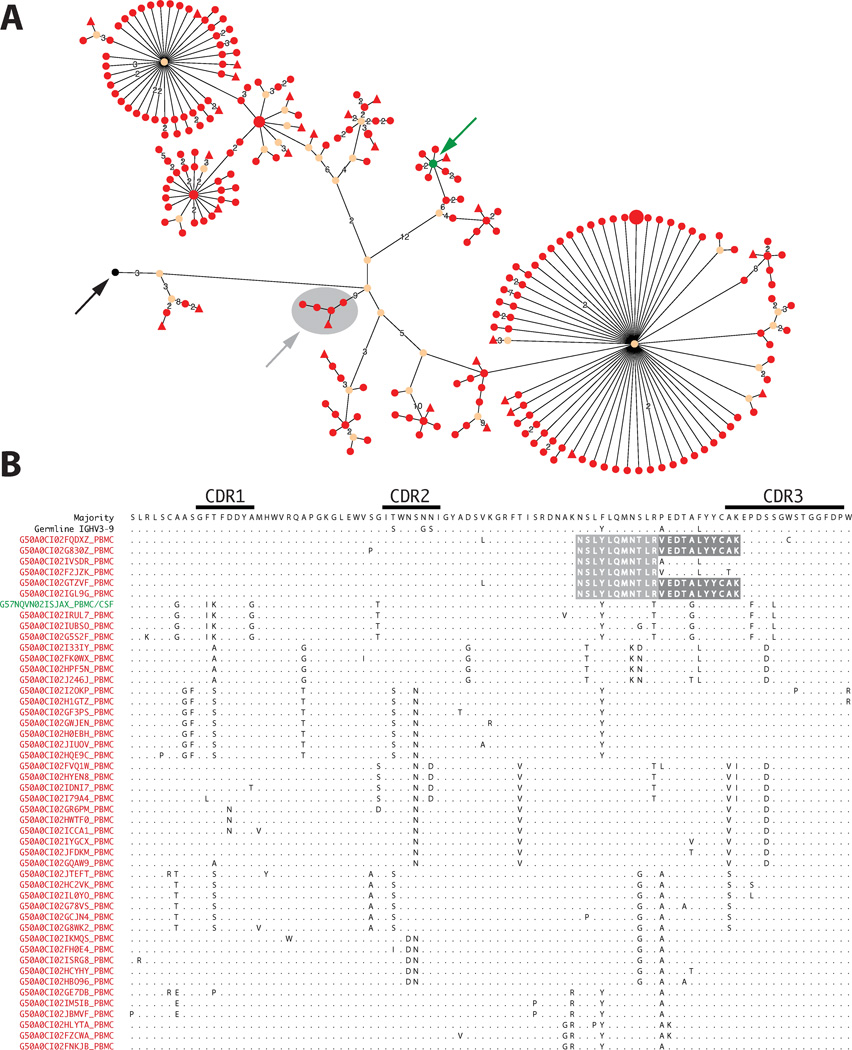
Figure 6. B cell clusters participating in OCB production undergo immune stimulation in the periphery.
Lineage tree (organic layout, A) and alignment of representative IgG-VH amino acid sequences (B) from patient MS-5 are shown. Germline node (black arrow), node comprised of CSF and PBMC IgG-VH (green arrow), and PB-derived sub-cluster identified by OCB peptide-searches (gray arrow) are indicated. In the alignment, matched OCB peptides are shaded in gray; IgG-VH is derived from IGHV3–9 germline segment. Only representative IgG-VH are shown. Organic layout was chosen over hierarchic layout because the latter generates very extensive horizontal images. For additional information please refer to legends of Figures 4 and 5.
RESULTS
Oligoclonal band peptides map to IgG-VH transcripts from CSF and/or PB
As expected, isoelectric focusing (IEF) of purified patient CSF IgG revealed OCB in all 5 CSF samples (Figure 1). Tryptic digestion of OCB, tandem mass-spectrometry, and matching of peptide masses to VHref databases yielded combined 385 OCB peptides from the 5 patients reported here (Table S2). About 1/3 (n=135) of all identified OCB peptides were determined as not patient-specific and were excluded from further analysis. 249 OCB peptides were patient-specific in VHref-CSF, 79 of which were fully sequence-confirmed and used for further analyses (Tables 2, S2, S3); 1 additional sequence-confirmed and entirely H-CDR3 derived OCB peptide from patient MS-1 CSF could be specifically mapped to that patient’s IgG-VH transcriptome in VHref-PB but not in VHref-CSF (Table S3, gray shading). Together, 80 patient-specific OCB peptides were identified and used for further analysis.
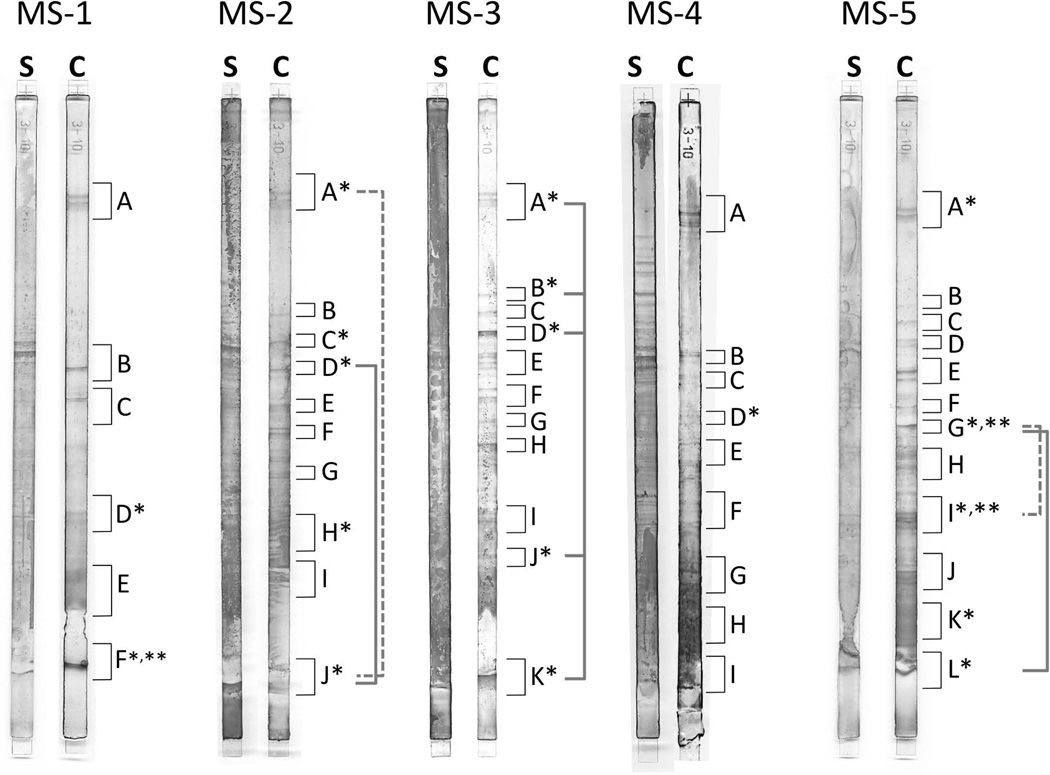
Figure 1. OCB represent diverse and partially related IgG populations.
Shown are isoelectric focused CSF and serum IgG from patients MS-1 to MS-5. IEF gels were stained and processed for mass-spectrometry as described in Methods; analyzed gel slices are labeled with letters. (*) Gel slices yielding patient-specific OCB bands. (**) Gel slices yielding peptides linking to PBMC-only clusters or sub-clusters. IEF bands yielding IgG OCB peptides belonging to the same cluster are connected by gray brackets; dashed and solid lines were used for better visual separation.
Table 2.
OCB Peptides and associated IgG-VH clusters. Shown are numbers of OCB peptides that could be uniquely assigned to a specific patient in our IgG-VH reference database and associated IgG-VH clusters. CSF restricted IgG-VH clusters are those in which OCB peptides mapped to IgG-VH sequence clusters that were exclusively found in the CSF.
| Patient ID |
OCB peptides | Associated IgG-VH clusters | CSF-restricted IgG-VH clusters |
|---|---|---|---|
| MS-1 | 7 | 6 | 5 |
| MS-2 | 20 | 14 | 13 |
| MS-3 | 38 | 20 | 20 |
| MS-4 | 3 | 2 | 2 |
| MS-5 | 12 | 10 | 4 |
General OCB peptide characteristics
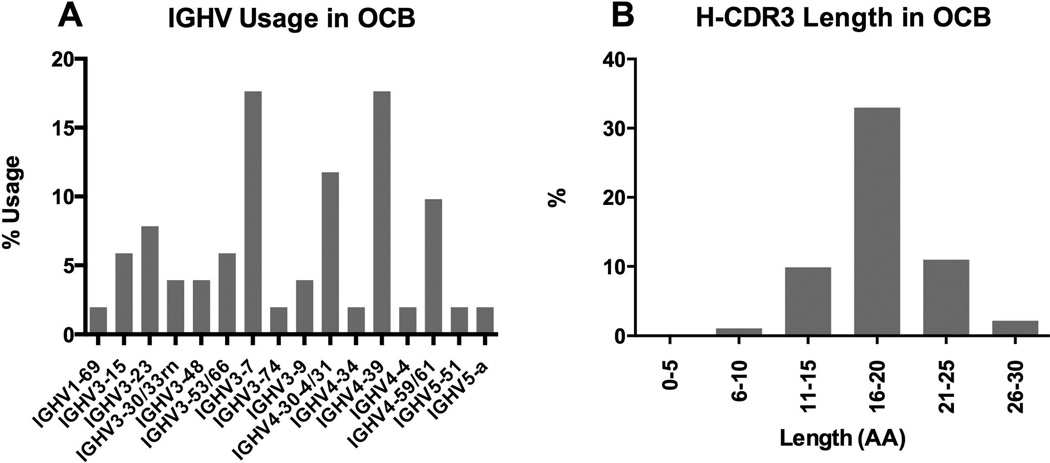
The 80 patient-specific OCB peptides were derived from 18 IEF gel slices (Figure 1). OCB peptide lengths ranged from 8 to 27 amino acids; peptide coverage per IgG-VH sequence ranged from 1 to 4; an overview of OCB peptides mapping to IgG-VH sequences is shown in Figure S1. IGHV usage among all OCB peptides reported here and associated IgG-VH is shown in Figure 2A; IGHV3-7 (17.7%), IGHV4-30-4/31 (11.8%), IGHV4-39 (17.7%), and IGHV4-59/61 (9.8%) were most frequently found; H-CDR3 length of peptide-associated IgG-VH (Figure 2B) ranged from 10 to 27 AA with an average H-CDR3 length of 18.1 ± 3.5 (SD) AA as expected for a typical normal IgG population.
Figure 2.
IGHV usage (A) and H-CDR3 length distribution (B) of IgG-VH clusters associated with OCB peptides.
Oligoclonal band peptides reflect molecular diversity of IgG-VH
We were interested in understanding the molecular diversity associated with patient-specific OCB peptides reported here. For this purpose we selected IgG-VH that contained OCB peptide sequences from our VHref datasets and generated clusters of OCB-associated, clonally related IgG-VH. The resulting clusters are reflective of mutational activity in the IgG-VH region and clustered IgG-VH can be used for phylogenetic lineage analysis. The 80 patient-specific OCB peptides could be assigned to 52 different clusters of clonally related IgG-VH (Table 2). Among the described peptides, 57 differed from IGHV germline sequences indicating presence of somatic mutations; 18 OCB peptides did not yield germline IGHV matches in IgBlast, generally because these sequences corresponded in large part to non-germline encoded H-CDR3 region and/or adjacent IGHJ sequence (Table S3); 5 OCB peptides were identical to germline IGHV but mapped exclusively to IgG-VH from patient MS-2 or MS-5 in VHref-CSF (Table S3). In general, OCB-peptides that map largely or entirely to H-CDR3 sequences, or include mutations, are the most reliable tools to clearly assign peptides to a specific IgG-VH cluster. Germline gene-identical OCB-peptides (n=5, Table S3) were included in our analyses if they were patient-specific, and within a patient also exclusively mapped to a single cluster of clonally related IgG-VH.
In one patient (MS-3) we identified a number of highly similar OCB peptides incorporating AA mutations, demonstrating that progression of somatic hypermutation can be represented in OCB (Figure 3). Numerous OCB peptides mapped to clusters of related IgG-VH sequences that were represented in >1 band on the IEF gels (Figure 1, gray brackets; Table S3), indicating presence of related IgG in spatially separated OCB.
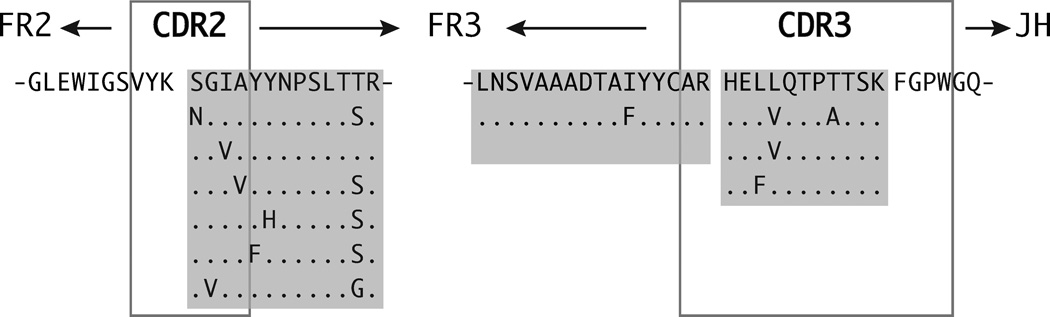
Figure 3. OCB peptides reveal signs of affinity-maturation.
Shown is an IGHV4-39 derived, truncated IgG-VH with OCB peptides identified by mass-spectrometry in IEF gel slices “A”, “B”, “D”, “J”, and “K” (Figure 1, Table S3) in patient MS-3 CSF. Shaded in light gray are aligned AA sequences of identified OCB peptides reflective of ongoing affinity-maturation in a B cell cluster contributing to OCB production in patient MS-3. H-CDR2 and H-CDR-3 are in grey frames. Dots (․) indicate identity with the consensus sequence (top).
OCB-producing B cell clusters participate in active immunity on both sides of the BBB
To address the question whether OCB are exclusively associated with intrathecal immune responses, or whether B cells in the periphery may also express the same or clonally related immunoglobulin genes, we analyzed the compartmental (i.e. CNS vs. periphery) contributions in OCB-associated IgG-VH clusters. We found the majority of patient-specific OCB peptides to be associated with IgG-VH transcripts observed exclusively in the CSF (Tables 2 and S3); this finding supports well-accepted knowledge, that significant activation of CNS-restricted B cell responses, including affinity maturation of Ig genes and terminal differentiation to antibody-secreting B cells, occurs inside the CNS in MS patients18, 29–31.
However, we also found a number of OCB peptides mapping to bi-compartmental clusters composed of CSF and/or PB IgG-VH transcripts from patients MS-1, MS-2 and MS-5 (Figures 4, 5A–D, 6): In patient MS-2 with clinically quiescent MS we identified an OCB peptide from IEF band “D” (Figure 1, MS-2) mapping to the H-CDR3 region (detected peptide: YFAWSAGK; corresponding H-CDR3: CVAVRYFAWSAGKLFDYW) of a bi-compartmental IgG-VH cluster (Figure 4) utilizing IGHV4-39 and IGHJ4 germline segments. Alignment and lineage analysis of IgG-VH sequences belonging to this cluster resulted in a lineage tree with three distinctive branches; the identified H-CDR3 OCB peptide mapped to a CSF-sublineage, in the PB and remaining CSF-sublineage this sequence portion displayed a single AA replacement from “V” to “A” at amino acid position 6 of the described peptide (Figure 4).
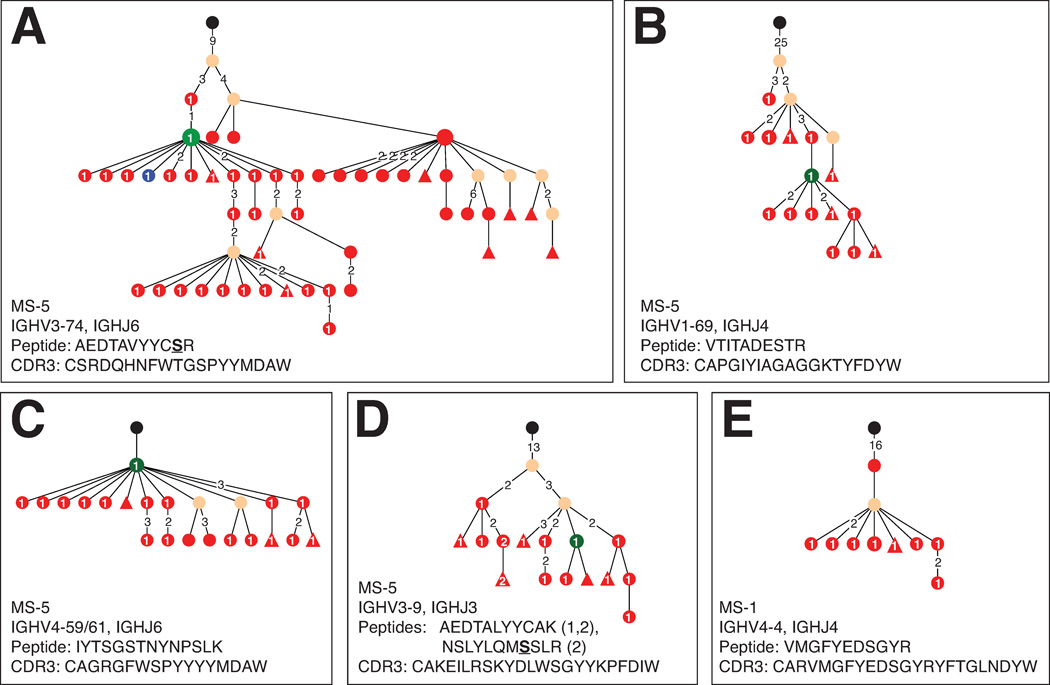
Figure 5. B cells involved in OCB-production are present in the peripheral blood.
Shown are examples of bi-compartmental IgG-VH lineages (hierarchic layout) from patient MS-5 (A–D), and a IgG-VH lineage from patient MS-1 in which OCB peptides mapped to PBMC Ig sequences exclusively (E). CSF derived IgG-VH are represented by blue nodes, PB derived IgG-VH are red, and identical sequences found in both compartments are green. Numbers between nodes are numbers of nucleotide mutations; unlabeled connections between nodes represent single nucleotide mutations. Numbers in nodes (white) are numbers of OCB peptides mapping to a specific node. Triangular nodes contain 2 or more singleton sequences in leaves. IGHV and IGHJ usage, matched peptide, and corresponding representative CDR3 are indicated per lineage. Amino acids differing from germline segments are underlined; in panel E the detected peptide is entirely located in the non-germline encoded H-CDR3.
The largest number of OCB peptides mapping to bi-compartmental IgG-VH clusters was found in patient MS-5 with active MS and a gadolinium contrast-enhancing lesion on MRI one week prior to LP. In this patient, we identified 12 patient-specific OCB peptides, 8 of which mapped to bi-compartmental IgG-VH clusters (Table S3; examples see Figures 5A–D and 6). Interestingly, 2 OCB peptides from patient MS-5 mapped exclusively to PB IgG-VH sequences in a bi-compartmental IgG-VH cluster (Figure 6). In addition, we identified a single OCB peptide from patient MS-1 (detected peptide: VMGFYEDSGYR; corresponding H-CDR3: CARVMGFYEDSGYRYFTGLNDYW; IGHV4-4, IGHJ4) that did not match any CSF-restricted or bi-compartmental IgG-VH clusters; instead, this peptide mapped to an IgG-VH cluster that exclusively comprised of PB-derived Ig transcripts found in VHref-PB (Figure 5 E).
DISCUSSSION
Oligoclonal bands (OCB) are produced by clonally expanded, terminally differentiated B cells within the CNS compartment; they are believed to mark a highly targeted immune response against a specific target antigen, or antigens that await unequivocal identification; OCB are one of the strongest indicators that an antigen-driven humoral immune process participates in MS pathology. Unfortunately, OCB cannot be readily sequenced and isolated proteomic analysis of OCB is unreliable due to amino acid changes introduced in the Ig variable region during affinity maturation. Combining transcriptomic data of Ig genes involved in OCB expression with OCB proteomic analysis by mass-spectrometry was previously applied to demonstrate that OCB are produced by intrathecal plasma cells and B cells2, 3. Using next generation deep immune repertoire sequencing, we recently demonstrated in MS and other neurological disorders that B cell repertoires on both sides of the blood brain barrier are connected18. The availability of soluble CSF IgG and comprehensive CSF and PB IgG-VH immune repertoires from 5 representative MS patients now enabled us to formally demonstrate that B cells participating in OCB production in the CSF can also be found in the periphery.
It is well accepted that OCB are produced by CSF B cells3, 10; our finding that most OCB peptides map to CSF-derived IgG-VH sequences underscores the importance of such intrathecal antigen-driven immune responses. However, it is not known if clusters of related B cells participating in OCB production are also present in the periphery; our studies did indeed reveal such a connection in three of five patients. Two patients with clinically quiescent MS displayed single OCB-producing B cell clusters that potentially participated in exchange across the BBB. The largest number of bi-compartmental IgG-VH clusters was identified in patient MS-5, an individual with active MS and a contrast-enhancing lesion on brain MRI one week prior to LP. This finding suggests enhanced migration of B cells across the BBB during active disease. Our data does not permit a clear statement regarding directionality of B cell exchange across the BBB. However, B cells that participate in OCB production likely represent a well-established and temporally invariable intrathecal immune response32. Hence, our findings in patient MS-5 provide circumstantial evidence that during active MS B cells may egress from the CNS to the periphery. Based on our findings that some CSF-associated B cell clusters apparently undergo extensive somatic-hypermutation (i.e. affinity-maturation) in the periphery, it is also possible that B cells that have already participated in intrathecal immune-stimulation and OCB production can egress from the CNS and undergo antigen-directed restimulation in the periphery. Clonal B cell reservoirs may be a source of replenishment for OCB-producing B cells; such reservoirs may be located within the CSF, in meningeal lymphoid follicle-like structures11, intraparenchymal B cell infiltrates, or the periphery.
Placing OCB peptides along lineage trees has helped to clarify the relationship between OCB production and B cell affinity maturation. We found a number of instances in which OCB peptides could be clearly assigned to an IgG-VH cluster but within this cluster mapped to a distinct sub-lineage, an observation that pertains to both, CSF-restricted, and bi-compartmental IgG-VH clusters. Thus, it is possible that clonal B cell expansion and plasma cell differentiation necessary for OCB production is a function of dynamic B cell immunity rather than its final result. Within permissive inflammatory environments, plasma cell maturation and affinity-maturation likely occur in parallel resulting in antibody populations derived from clonally related B cells that may contain VH regions reflective of different stages of affinity maturation.
An important question awaiting elucidation is whether OCB contribute to tissue damage in MS. B cell depleting therapy with the anti-CD20 targeting monoclonal antibodies rituximab and ocrelizumab was recently found to rapidly and effectively suppress inflammatory disease activity in MS33–35. Interestingly, in one report OCB were unchanged 24 weeks after rituximab therapy despite a reduction in intrathecal B cell numbers36, to some degree suggesting an inferior role of OCB in MS immune pathology. Conversely, there is evidence that limiting migration of lymphocytes across the BBB by natalizumab (anti-VLA4 antibody) significantly reduces intrathecal antibody production7, 8. Our data presented here, supports the concept that disease-relevant B cell immune responses are active on both sides of the BBB, a connection that could be effectively interrupted by natalizumab.
Antigens recognized by OCB or intrathecal B cells remain unknown, despite extensive efforts by many groups, and despite growing evidence that these immune responses must be driven by specific immunogenic stimuli. We find that clonally related IgG-VH sequences are associated with OCB that are spatially separated on IEF gels. Thus, immune responses against target epitopes could be represented by numerous OCB; in other words, the presence of numerous OCB does not necessarily constitute an intrathecal immune response against multiple target epitopes. Our finding that B cell clusters producing OCB in the CNS also undergo SHM in the periphery strongly suggests the presence of identical or highly similar antigens on both sides of the blood-brain barrier. Recently, antibodies against the potassium channel protein KIR4.1 have been described in almost 50% or MS patients’ serum37 and injection of purified KIR4.1 antibodies into the CSF compartment of mice was shown to induce CNS tissue damage. The initial trigger of KIR4.1 directed autoimmunity is unknown; however, molecular mimicry between peripheral and CNS self or foreign antigens could be an important concept supporting bi-compartmental B cell immunity in MS. The pathological relevance of immune responses against myelin-oligodendrocyte glycoprotein (MOG) and myelin basic protein (MBP) have been controversially discussed in MS; mimicry between MOG and the milk protein butyrophilin has been demonstrated for both antibodies and T cells in MS38, 39. MBP specific T cell clones have also been found to react to viral peptides40.
We provide evidence for a deep connection between OCB and B-cell clusters in patients with MS, and we demonstrate for the first time, that “OCB” (i.e. antibodies represented by OCB) can theoretically be produced in the periphery. In single individuals, multiple different bands are comprised of closely related protein sequences corresponding to clonally expanded B-cells present in the nervous system and also in the periphery. Remarkably, some B-cell clusters or subclusters corresponding to OCB are observed only in the peripheral blood. Perhaps sampling at an earlier or different time or at a significantly greater sequencing depth would have identified the B cells corresponding to these OCB in the CSF, or alternatively some OCB could be the product of B cells present in the meninges or brain parenchyma but not in CSF. Whatever the explanation, the finding that some OCB specificities are connected only to peripheral blood B-cells indicates that disease-relevant B-cells circulate between the CNS and peripheral compartments. Equally important, this finding suggests that the periphery represents a site of persistence and also likely of activation of pathogenic B-cells in MS. Lastly, our findings add support to the concept that molecular sequencing of BCR could be used as a diagnostic and monitoring tool for longitudinal tracking of disease activity in MS18.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to the individuals who agreed to serve as subjects for this study. We dedicate this work to the memory of our friend and colleague David R. Cox, MD, PhD, whose contributions to the concepts and technologies reported here enabled these studies; we will always be inspired by David’s lifelong dedication to biomedical research as a means to prevent suffering and improve human health. We thank Ramit Mehr, Bar-Ilan University, Ramat-Gan Israel for providing IgTree, and Richard Niles for help with mass-spectrometry experiments and data analyses. We thank Tracy Kuo, Marina Sirota, Steven Pitts, Shobha Potluri and Dave Shelton for helpful discussions and suggestions. These studies were supported by grants from Pfizer Inc., The Nancy Davis Foundation, Small Ventures USA, The Brass Family Foundation, the NMSS (RG-4868), and the NIH (K02NS072288). H.C.v.B. was also supported by an endowment from the Rachleff Family Foundation. Mass spectrometry analysis was performed by the UCSF Sandler-Moore Mass Spectrometry Core Facility, which acknowledges support from the Sandler Family Foundation, the Gordon and Betty Moore Foundation, and NIH/NCI Cancer Center Support Grant P30 CA082103.
REFERENCES
- 1.Walsh MJ, Tourtellotte WW, Roman J, Dreyer W. Immunoglobulin G, A, and M--clonal restriction in multiple sclerosis cerebrospinal fluid and serum--analysis by two-dimensional electrophoresis. Clin Immunol Immunopathol. 1985;35:313–327. doi: 10.1016/0090-1229(85)90092-3. [DOI] [PubMed] [Google Scholar]
- 2.Obermeier B, Lovato L, Mentele R, et al. Related B cell clones that populate the CSF and CNS of patients with multiple sclerosis produce CSF immunoglobulin. Journal of neuroimmunology. 2011;233:245–248. doi: 10.1016/j.jneuroim.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Obermeier B, Mentele R, Malotka J, et al. Matching of oligoclonal immunoglobulin transcriptomes and proteomes of cerebrospinal fluid in multiple sclerosis. Nat Med. 2008;14:688–693. doi: 10.1038/nm1714. [DOI] [PubMed] [Google Scholar]
- 4.Avasarala JR, Cross AH, Trotter JL. Oligoclonal band number as a marker for prognosis in multiple sclerosis. Arch Neurol. 2001;58:2044–2045. doi: 10.1001/archneur.58.12.2044. [DOI] [PubMed] [Google Scholar]
- 5.Joseph FG, Hirst CL, Pickersgill TP, et al. CSF oligoclonal band status informs prognosis in multiple sclerosis: a case control study of 100 patients. J Neurol Neurosurg Psychiatry. 2009;80:292–296. doi: 10.1136/jnnp.2008.150896. [DOI] [PubMed] [Google Scholar]
- 6.Tintore M, Rovira A, Rio J, et al. Do oligoclonal bands add information to MRI in first attacks of multiple sclerosis? Neurology. 2008;70:1079–1083. doi: 10.1212/01.wnl.0000280576.73609.c6. [DOI] [PubMed] [Google Scholar]
- 7.Villar LM, Garcia-Sanchez MI, Costa-Frossard L, et al. Immunological markers of optimal response to natalizumab in multiple sclerosis. Arch Neurol. 2012;69:191–197. doi: 10.1001/archneurol.2011.971. [DOI] [PubMed] [Google Scholar]
- 8.Harrer A, Tumani H, Niendorf S, et al. Cerebrospinal fluid parameters of B cell-related activity in patients with active disease during natalizumab therapy. Mult Scler. 2013;19:1209–1212. doi: 10.1177/1352458512463483. [DOI] [PubMed] [Google Scholar]
- 9.Stuve O, Marra CM, Jerome KR, et al. Immune surveillance in multiple sclerosis patients treated with natalizumab. Ann Neurol. 2006;59:743–747. doi: 10.1002/ana.20858. [DOI] [PubMed] [Google Scholar]
- 10.von B dingen HC, Gulati M, Kuenzle S, et al. Clonally expanded plasma cells in the cerebrospinal fluid of patients with central nervous system autoimmune demyelination produce "oligoclonal bands". J Neuroimmunol. 2010;218:134–139. doi: 10.1016/j.jneuroim.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 11.Serafini B, Rosicarelli B, Magliozzi R, et al. Detection of ectopic B-cell follicles with germinal centers in the meninges of patients with secondary progressive multiple sclerosis. Brain Pathol. 2004;14:164–174. doi: 10.1111/j.1750-3639.2004.tb00049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prineas JW. Multiple sclerosis: presence of lymphatic capillaries and lymphoid tissue in the brain and spinal cord. Science. 1979;203:1123–1125. doi: 10.1126/science.424741. [DOI] [PubMed] [Google Scholar]
- 13.von Büdingen HC, Harrer MD, Kuenzle S, et al. Clonally expanded plasma cells in the cerebrospinal fluid of MS patients produce myelin-specific antibodies. Eur J Immunol. 2008;38:2014–2023. doi: 10.1002/eji.200737784. [DOI] [PubMed] [Google Scholar]
- 14.Cruz M, Olsson T, Ernerudh J, et al. Immunoblot detection of oligoclonal anti-myelin basic protein IgG antibodies in cerebrospinal fluid in multiple sclerosis. Neurology. 1987;37:1515–1519. doi: 10.1212/wnl.37.9.1515. [DOI] [PubMed] [Google Scholar]
- 15.O'Connor KC, Appel H, Bregoli L, et al. Antibodies from inflamed central nervous system tissue recognize myelin oligodendrocyte glycoprotein. J Immunol. 2005;175:1974–1982. doi: 10.4049/jimmunol.175.3.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cepok S, Zhou D, Srivastava R, et al. Identification of Epstein-Barr virus proteins as putative targets of the immune response in multiple sclerosis. J Clin Invest. 2005;115:1352–1360. doi: 10.1172/JCI23661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Owens GP, Bennett JL, Lassmann H, et al. Antibodies produced by clonally expanded plasma cells in multiple sclerosis cerebrospinal fluid. Ann Neurol. 2009;65:639–649. doi: 10.1002/ana.21641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.von Büdingen HC, Kuo TC, Sirota M, et al. B cell exchange across the blood-brain barrier in multiple sclerosis. J Clin Invest. 2012;122:4533–4543. doi: 10.1172/JCI63842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bar-Or A, Fawaz L, Fan B, et al. Abnormal B-cell cytokine responses a trigger of T-cell-mediated disease in MS? Ann Neurol. 2010;67:452–461. doi: 10.1002/ana.21939. [DOI] [PubMed] [Google Scholar]
- 20.Polman CH, Reingold SC, Banwell B, et al. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann Neurol. 2011;69:292–302. doi: 10.1002/ana.22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glanville J, Zhai W, Berka J, et al. Precise determination of the diversity of a combinatorial antibody library gives insight into the human immunoglobulin repertoire. Proc Natl Acad Sci U S A. 2009;106:20216–20221. doi: 10.1073/pnas.0909775106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levenshtein VI. Binary codes capable of correcting deletions, insertions, and reversals. Soviet Physics Dokl. 1965;10:707–710. [Google Scholar]
- 23.Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 24.Volpe JM, Cowell LG, Kepler TB. SoDA: implementation of a 3D alignment algorithm for inference of antigen receptor recombinations. Bioinformatics. 2006;22:438–444. doi: 10.1093/bioinformatics/btk004. [DOI] [PubMed] [Google Scholar]
- 25.Barak M, Zuckerman NS, Edelman H, et al. IgTree: creating Immunoglobulin variable region gene lineage trees. J Immunol Methods. 2008;338:67–74. doi: 10.1016/j.jim.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 26.Smoot ME, Ono K, Ruscheinski J, et al. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 28.Waterhouse AM, Procter JB, Martin DM, et al. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qin Y, Duquette P, Zhang Y, et al. Clonal expansion and somatic hypermutation of V(H) genes of B cells from cerebrospinal fluid in multiple sclerosis. J Clin Invest. 1998;102:1045–1050. doi: 10.1172/JCI3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Owens GP, Ritchie AM, Burgoon MP, et al. Single-cell repertoire analysis demonstrates that clonal expansion is a prominent feature of the B cell response in multiple sclerosis cerebrospinal fluid. J Immunol. 2003;171:2725–2733. doi: 10.4049/jimmunol.171.5.2725. [DOI] [PubMed] [Google Scholar]
- 31.Baranzini SE, Jeong MC, Butunoi C, et al. B cell repertoire diversity and clonal expansion in multiple sclerosis brain lesions. J Immunol. 1999;163:5133–5144. [PubMed] [Google Scholar]
- 32.Walsh MJ, Tourtellotte WW. Temporal invariance and clonal uniformity of brain and cerebrospinal IgG, IgA, and IgM in multiple sclerosis. J Exp Med. 1986;163:41–53. doi: 10.1084/jem.163.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bar-Or A, Calabresi PA, Arnold D, et al. Rituximab in relapsing-remitting multiple sclerosis: a 72-week, open-label, phase I trial. Ann Neurol. 2008;63:395–400. doi: 10.1002/ana.21363. [DOI] [PubMed] [Google Scholar]
- 34.Hauser SL, Waubant E, Arnold DL, et al. B-cell depletion with rituximab in relapsing-remitting multiple sclerosis. N Engl J Med. 2008;358:676–688. doi: 10.1056/NEJMoa0706383. [DOI] [PubMed] [Google Scholar]
- 35.Kappos L, Li D, Calabresi PA, et al. Ocrelizumab in relapsing-remitting multiple sclerosis: a phase 2, randomised, placebo-controlled, multicentre trial. Lancet. 2011;378:1779–1787. doi: 10.1016/S0140-6736(11)61649-8. [DOI] [PubMed] [Google Scholar]
- 36.Cross AH, Stark JL, Lauber J, et al. Rituximab reduces B cells and T cells in cerebrospinal fluid of multiple sclerosis patients. J Neuroimmunol. 2006;180:63–70. doi: 10.1016/j.jneuroim.2006.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Srivastava R, Aslam M, Kalluri SR, et al. Potassium channel KIR4.1 as an immune target in multiple sclerosis. N Engl J Med. 2012;367:115–123. doi: 10.1056/NEJMoa1110740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stefferl A, Schubart A, Storch M, et al. Butyrophilin, a milk protein, modulates the encephalitogenic T cell response to myelin oligodendrocyte glycoprotein in experimental autoimmune encephalomyelitis. J Immunol. 2000;165:2859–2865. doi: 10.4049/jimmunol.165.5.2859. [DOI] [PubMed] [Google Scholar]
- 39.Guggenmos J, Schubart AS, Ogg S, et al. Antibody cross-reactivity between myelin oligodendrocyte glycoprotein and the milk protein butyrophilin in multiple sclerosis. J Immunol. 2004;172:661–668. doi: 10.4049/jimmunol.172.1.661. [DOI] [PubMed] [Google Scholar]
- 40.Wucherpfennig KW, Strominger JL. Molecular mimicry in T cell-mediated autoimmunity: viral peptides activate human T cell clones specific for myelin basic protein. Cell. 1995;80:695–705. doi: 10.1016/0092-8674(95)90348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.