Abstract
Shotgun proteomics experiments integrate a complex sequence of processes, any of which can introduce variability. Quality metrics computed from LC-MS/MS data have relied upon identifying MS/MS scans, but a new mode for the QuaMeter software produces metrics that are independent of identifications. Rather than evaluating each metric independently, we have created a robust multivariate statistical toolkit that accommodates the correlation structure of these metrics and allows for hierarchical relationships among data sets. The framework enables visualization and structural assessment of variability. Study 1 for the Clinical Proteomics Technology Assessment for Cancer (CPTAC), which analyzed three replicates of two common samples at each of two time points among 23 mass spectrometers in nine laboratories, provided the data to demonstrate this framework, and CPTAC Study 5 provided data from complex lysates under Standard Operating Procedures (SOPs) to complement these findings. Identification-independent quality metrics enabled the differentiation of sites and run-times through robust principal components analysis and subsequent factor analysis. Dissimilarity metrics revealed outliers in performance, and a nested ANOVA model revealed the extent to which all metrics or individual metrics were impacted by mass spectrometer and run time. Study 5 data revealed that even when SOPs have been applied, instrument-dependent variability remains prominent, although it may be reduced, while within-site variability is reduced significantly. Finally, identification-independent quality metrics were shown to be predictive of identification sensitivity in these data sets. QuaMeter and the associated multivariate framework are available from http://fenchurch.mc.vanderbilt.edu and http://homepages.uc.edu/~wang2x7/, respectively.
INTRODUCTION
The diverse methods, instrument platforms, and bioinformatics used in shotgun proteomics frequently inhibit the reproduction of experiments among different laboratories1–3. Most reproducibility analyses of the data produced in these experiments have constrained themselves to the peptide and protein identifications the experiments produced4,5. More recently, bioinformatics teams have produced software tools for generating quality metrics that leverage identifications6–8.
The NCI Clinical Proteomic Technology Assessment for Cancer (CPTAC) was designed to characterize proteomic methods for their use in clinical samples. The first study conducted by CPTAC (2006-2007) was intended to provide a baseline set of data from common samples to evaluate the variability of LC-MS/MS data collections prior to the use of Standard Operating Procedures (SOPs) for CPTAC joint studies. While later studies from CPTAC used more complex samples (such as yeast lysates9 or blood plasma10), CPTAC Study 1 employed a defined mixture of twenty proteins. Later studies emphasized Thermo LTQ and Orbitrap instruments for proteomic inventories4 and AB Sciex QTRAP instruments for targeted quantitation10, but Study 1 was conducted on all instruments that CPTAC principal investigators had identified as available for experimentation in their proposals. The availability of raw data from 16 electrospray-mass spectrometers in nine distinct instrument models enables a unique vantage for evaluating variability in these platforms. CPTAC Study 5, on the other hand, limited its focus to Thermo LTQ and Orbitrap instruments but demonstrated the variability reduction of an SOP, coupled with more complex samples.
Variability in a complex technology such as shotgun proteomics may be characterized by multiple metrics, and sets of metrics imply the need for a multivariate approach to monitoring and evaluation. Such an approach can model the correlations between features and control the overall probability of falsely signaling an outlier when one is not present (the usual α-level in univariate hypothesis testing). It is hard to evaluate this probability from a large number of individual metrics if the correlation among the features is extensive11. To effectively evaluate and monitor experiments, one should jointly analyze the array of metrics. If a more diverse set of features can be characterized by metrics, a more comprehensive appraisal of the system becomes possible, so looking beyond identifications to include signal intensity and chromatography is essential6. Summarizing among metrics presents opportunities for diagnosing the mechanism causing variability.
In this study, we use multidimensional performance metrics generated by QuaMeter to evaluate the data from CPTAC Studies 1 and 5. CPTAC Study 1 sought to characterize the performance of a broad collection of mass spectrometers prior to any cross-site coordination of instrument protocols. The data obtained in this large scale study have not been rigorously analyzed until now. The results from this study are of particular interest from the perspective of quality control (QC). We show how robust multivariate statistical methods can produce insights on the cross-platform, laboratory, sample, and experimental variability assessment of large-scale experiments by effective data visualization and modeling. The methods developed here can be applied in a broad range of quality control studies with multidimensional performance metrics.
EXPERIMENTAL SECTION
Creation of NCI-20 reference material
An aqueous mixture of twenty purified human proteins (referred to as the “NCI-20”) was produced by NIST. The twenty proteins were chosen based on several criteria including the commercial availability of highly-purified or recombinant preparations, the identity of the protein as either “classical” plasma proteins or potential plasma biomarkers of cancer12, and the availability of commercial immunoassays for the chosen protein. The concentrations of the twenty human proteins in the NCI-20 mixture spanned a range similar to that of proteins in human plasma, specifically from 5 g/L to 5 ng/L. The target concentration of most of the proteins in the NCI-20 mixture (with the exception of human albumin) reflects the clinically-relevant concentration range in human plasma, as reported in the literature13.
To prepare the NCI-20 mixture, stock solutions of each commercial protein preparation were prepared in 25 mmol/L phosphate buffered saline, pH 7.4, containing 5 mmol/L acetyl tryptophan and 4 mmol/L sodium azide. Table 1 in the Supporting Information of the CPTAC Repeatability article lists the commercial sources of the proteins in the NCI-20 mixture4. A portion of the NCI-20 mixture was aliquotted (600 × 125 μL aliquots) into polypropylene microcentrifuge tubes and stored at −80 °C. These aliquots were labeled as “Sample 1A” for the CPTAC interlaboratory study. Another portion of the NCI-20 mixture was denatured in RapiGest SF (Waters, Millford MA), reduced by dithiothreitol, alkylated with iodoacetamide, and proteolytically digested with immobilized trypsin (Thermo Pierce, Rockford IL) to prepare “Sample 1B” for the study. A portion of this digest was aliquotted (600 × 125 μL) into polypropylene microcentrifuge tubes and stored at −80 °C.
Harvesting data from CPTAC
The CPTAC program conducted a series of multi-site experiments that employed data-dependent technologies:
Study 1: diverse instruments analyzing NCI-20 without SOP
Study 2: LTQs and Orbitraps analyzing NCI-20 under SOP 1.0
Study 3: LTQs and Orbitraps analyzing yeast and NCI-20 under SOP 2.0
Study 5: LTQs and Orbitraps analyzing yeast and BSA-spiked yeast under SOP 2.1
Study 6: LTQs and Orbitraps analyzing yeast and UPS1-spiked yeast under SOP 2.2
Study 8: LTQs and Orbitraps analyzing yeast in two concentrations without SOP
Study 1 was unusual for its inclusion of a broad range of instrument types and replication of its experiments to span at least two weeks, and yet these data have never been evaluated in the peer-reviewed literature. Study 5 is valuable for producing six replicates of both complex and defined samples under SOP for six different instruments. The two experiments were selected because they provided sufficient replication for characterizing the participating instruments, and they provide significant contrast due to the included sample types and the different levels of methodology control.
Overview of CPTAC Study 1
Study 1 was designed as a first group experiment for the CPTAC network. The study evaluated the variability of replicate data sets for NCI-20 samples that were digested at the individual sites (1A) or centrally at NIST (1B). It attempted three aims in assessing clinical proteomics technologies. The first was to benchmark proteomic identification for a defined mixture across a diverse set of platforms and laboratories. The second evaluated the week-to-week reproducibility associated with MS/MS instrumentation. The third aim sought to evaluate the variability introduced by de-centralizing the initial step of protein digestion by trypsin; if on-site digestion led to less comparable results among sites, centralized digestion would be justified for shared program experiments. Three vials of sample 1A and sample 1B were sent to each lab in each shipment, with one week intervening between two shipments. The date on which the first LC-MS/MS was conducted for each instrument was defined as day one. The expected output from each lab would include six LC-MS/MS analyses on day one (split evenly between 1A and 1B samples) and another six LC-MS/MS analyses on day eight. A total of 17 electrospray tandem mass spectrometers of seven different models took part in the study starting in November, 2006, with an additional four MALDI tandem mass spectrometers and two MALDI peptide mass fingerprinting instruments rounding out the study. Table S1 in the Supporting Information provides a list of mass spectrometers included in Study 1. Table S2 provides the historical context for proteomic instruments spanning the 1990s and the first decade of the 2000s.
Overview of CPTAC Study 5
Study 5 was designed as a multi-site test for a long-gradient SOP and for gauging the impacts of a spiked protein on the CPTAC yeast reference material9. SOP v2.1 defined a 184-minute data-dependent method for Thermo LTQ and Orbitrap instruments (see Supporting Information Table 2 in the CPTAC Repeatability article4). The samples included the digested NCI-20 (labeled “1B”), the yeast reference material (“3A”), and the yeast reference material with bovine serum albumin spiked at 10 fmol/μL in 60 ng/μL yeast lysate (“3B”). The run order for the experiment included the largest number of replicates for any CPTAC study, repeating twice the block of these samples: 1B, 3A, 3A, 3A, 1B, 3B, 3B, 3B, followed by an additional 1B. In total, the 1B sample should have been analyzed by at least five experiments (more if the blocks were separated by a gap), while the 3A and 3B samples yielded three early and three late replicates each. Data were collected on three LTQ and three Orbitrap mass spectrometers between October of 2007 and January of 2008. Raw data files for Studies 1 and 5 can be found at the CPTAC Public Portal: https://cptac-data-portal.georgetown.edu/cptacPublic/.
Quality Metric Generation
QuaMeter software7 is a tool built on the ProteoWizard library14 to produce quality metrics from LC-MS/MS data. In its original release, QuaMeter generated metrics styled after those of Rudnick et al6, incorporating peptide identifications along with raw data. The metrics generated by this tool, however, depended significantly upon which MS/MS scans were identified, reducing their utility in LC-MS/MS experiments with diminished identifications. For this research, a special “IDFree” mode was added to QuaMeter to produce metrics that are independent of identification success rates for MS/MS scans.
QuaMeter IDFree metrics separate into the following categories: XIC (extracted ion chromatograms), RT (retention times), MS1 (mass spectrometry), and MS2 (tandem mass spectrometry). The full list of 45 metrics is supplied in Table S3 of Supporting Information; in the analyses that follow, some metrics were omitted due to low variance or high correlation. Many of the metrics are subdivided into three or four quartiles in order to approximate the distribution of a variable for an experiment, and peak intensities are generally evaluated in logarithmic space in order to flatten large fold changes. Because the software does not make use of identification data, it emphasizes the set of precursor ions that are associated with the widest XICs; this set is found by sorting all precursor ion XIC values by full width at half maximum intensity (FWHM) and then accepting the smallest set that accounts for half of the FWHM sum.
The four RT-MSMS-Qx metrics provide a simple example of the quartiles in action. MS/MS scans could be acquired uniformly across the retention time for an LC-MS/MS experiment or more frequently during the most common elution times. In the uniform acquisition case, each of these metrics would be 0.25, implying that each quartile of MS/MS acquisition times lasted one quarter of the total retention time duration. If MS/MS acquisition rates are higher in the middle of an LC gradient, however, the Q2 and Q3 metrics will be lower than for Q1 and Q4. The MS2-Freq-Max metric, on the other hand, reports only the highest rate of MS/MS acquisition (in Hz) sustained for a full minute of the LC-MS/MS experiment.
The total intensity of MS signals can vary considerably from scan to scan, and the three MS1-TIC-Change-Qx metrics monitor this stability. The property in question is the log fold change of the total MS1 signal in one MS scan versus the total MS1 signal in the next one; a very high or very low log fold change could indicate electrospray instability. MS1-TIC-Change-Q4 compares the highest of these log fold changes to the one at the 75%ile, while -Q3 evaluates the 75%ile against the median, and the -Q2 evaluates the median against the 25%ile. Simple values like MS1-Count or MS2-Count, which report the numbers of these scans acquired in the experiment, are also provided.
Because QuaMeter needs access only to raw data, it allows a very rapid assessment of experiments. Typical run times for Thermo Orbitrap Velos raw data files are less than five minutes per LC-MS/MS experiment, largely consumed by the extraction of ion chromatograms. As a result, QuaMeter is well-positioned to give immediate feedback in support of go/no-go decisions with major experiments.
Robust Hierarchical Multivariate Statistical Toolkit
The performance evaluation in CPTAC Studies 1 and 5 features multi-level comparisons: across instrument types, across mass spectrometers, across samples, and among LC-MS/MS experiments. While metrics that evaluate this chain of complex activities have been designed6,7, there are few quantitative analysis methods able to jointly analyze these metrics and take advantage of the multivariate nature of the data. Here, we describe a series of multivariate statistical tools that can be used to visualize, explore and test rigorously the quality metrics obtained through identification-independent QuaMeter quality metrics. Multivariate statistical techniques are essential in performance evaluation of any shotgun proteomic data set, as the experiment routinely contains a chain of complex processes and the performance metrics comprise an integrated group of measures on these processes. Assessing experimental reliability and repeatability based on individual metrics is possible, but one can achieve greater sensitivity through combinations of metrics15. Multivariate statistical methods anticipate interdependence of the measurements and provide a deeper and more complete evaluation of the experimental workflow. When outliers may be present, the use of robust models helps to protect against bias, and operating without a benchmark profile is necessary when only a few experiments are available for a given instrument.
This is especially true for the shotgun proteomics experiments in CPTAC Study 1, where no cross-site protocols were implemented. The identification performance among mass spectrometers varied across orders of magnitude. Even for the same mass spectrometer, experimental methods yielded considerable changes in performance. When a benchmark profile is available, it is natural to compare each experiment with the benchmark profile for performance evaluation, quality control, and outlier detection. Hotelling's T2 and QC chart are widely implemented for quality control16. In a pilot study like CPTAC Study 1, however, evaluation has to be carried out without a benchmark. With the large variability among mass spectrometers, many assumptions used in the common multivariate methods are not appropriate. Based on these considerations, we employed robust principal component analysis. In evaluating the mass spectrometers and batch effects, we measured deviation using multivariate median and L1 distance instead of mean and Euclidean distance. These methods resist undue influence from highly variant individual mass spectrometers or flawed individual LC-MS/MS experiments. It can also be extended to detect outliers. The simultaneous evaluation of the full set of metrics may eventually pave the way for technicians to identify which element of the platform led to unusual behavior for an experiment, an essential feature for QC.
Data Visualization and Explorative Analysis
If we have only one or two metrics to measure the performance for a shotgun proteomics experiment, it is straightforward for us to visualize all the data (by a scatter plot, for example) and to do an explorative comparison among all experiments. The visualization and interpretation becomes challenging when the set of metrics expands; for example, QuaMeter provides more than 40 identification-independent metrics to measure the mass spectrometer performance in a single experiment. Robust principal component analysis is a suitable tool to use as the first step in exploratory data analysis. Ringnér provided an introduction of PCA for biological high dimensional data17.
One purpose of the PCA analysis is to replace the multidimensional correlated metrics by a much smaller number of uncorrelated components which contain most of the information in the original data set. This dimension reduction makes it easier to understand the data since it is much easier to interpret two or three uncorrelated components than a set of 40 with embedded patterns of interrelationships. The amount of information contained in the transformed metrics (PCs) is measured by the amount of variance that is accounted for by each of the newly constructed metrics. A two-dimension PC plot reduces and visualizes the data while retaining the most interesting features and maximal variability from the original data. It visualizes the performance of all the experiments based on the first two PCs (as if we had only two metrics from the beginning). Of course, some information is lost by the reduction from more than 40 metrics to only 2 components. Here, we only use the first two PCs as an example to show how the PCA is helpful for us to recognize some key features in the data, such as clusters, outliers, and deviation. A systematic examination of different combinations for components would be needed to comprehensively evaluate the data structure.
In this case, robust PCA18 was applied to all of the Sample 1A and 1B experiments collectively in CPTAC Study 1 and Sample 3A and 3B in CPTAC Study 5. The following metrics were excluded from PCA because of insufficient variability among sites in Study 1: RT-Duration, MS2-PrecZ*, XIC-FWHM-Q1, and XIC-FWHM-Q3. Two additional metrics (RT.MSMS.Q4 and MS2.Count) in Study 5 were removed because of the high correlation with RT.MSMS.Q2 (Pearson correlation >0.99). Multivariate analysis loses very little information from this removal because one can predict the values for these stripped variables almost perfectly from the retained information.
Factor analysis provides another means for dimensional reduction. The technique evaluates the relationship between unobservable, latent variables (factors) and the observable, manifest variables (QuaMeter metrics). It describes the covariance relationships among observed metrics in terms of a few underlying, but unobservable, quantities called factors. Unlike principal components analysis, however, factor analysis emphasizes the relationship among a small set of metrics associated with each factor. The factor analysis is carried out on the sample correlation matrix estimated using robust methods. The number of factors may be chosen to provide a reasonable amount of explanatory power for the total variability in the original data. The maximum likelihood method is used to estimate the loading matrix and other parameters in the factor model. To facilitate the selection of important metrics, we employ the varimax criterion19 to disperse the squares of loadings as much as possible. Factor analysis should reveal combinations of key metrics that can be used to monitor performance.
Dissimilarity
The dissimilarity between two experiments is measured by the Euclidean distance between the robust PCA coordinates for each LC-MS/MS experiment. Euclidean distance is an appropriate metric because dissimilarity is a comparison of only two experiments. Thus, any abnormal experiment will influence only the dissimilarity measures that include that experiment. Mathematically, the dissimilarity between two p-dimensional coordinates x1 and x2 is
The larger the dissimilarity values are, the less similar the two experimental runs. This measure of dissimilarity can be further developed into statistical distance when stable estimation of correlation is available.
This distance measure is designed to be automatically outlier-proof as pairwise comparison does not require a benchmark profile. Any abnormal experiments (outliers) can be easily identified by their large distances from other experiments, while clusters of experiments can be identified by small dissimilarity values among a set. These pairwise comparisons also facilitate examination of key factors, such as run order, digestion methods and mass spectrometers. When a benchmark is needed in the analysis, such as in ANOVA, careful consideration is needed in the selection of the benchmark. One key issue is that the benchmark cannot be very sensitive to any outlying experimental run. This is why we employed L1 distance rather than Euclidean distances in the nested ANOVA below.
Nested ANOVA Model
The ANOVA model decomposes the total variability among the metrics into different sources. It is applied here to study the mass spectrometer and batch effects. Experiments in Study 1 were designed to be carried out in two-week intervals. The actual time was frequently longer and more scattered. In Study 5, 3A and 3B samples were both separated to two batches. To examine the mass spectrometer and batch effects, we calculated the multivariate median for all experiments and the L1-distance of each experiment from the multivariate median. If all experiments produced similar data, we would expect their vectors of metrics to randomly distribute around a central vector of metrics (the median vector). Any non-random pattern in the performance of experiments could impose a pattern on the distances of the vector metrics of affected runs from the center. For example, if the performance depends on the individual mass spectrometers, we would expect the metric vectors to cluster together for different mass spectrometers. We choose to use L1-distance as it is less influenced by experiments with idiosyncratic performance, and combines metric distances for a simple univariate ANOVA analysis. L1 distance can also provide information on outliers and clusters, though it is less sensitive than the Euclidean pairwise comparison detailed above. The inability to take into account the direction of change is a drawback of using L1 distance. As a result, the analysis may not be able to differentiate effects that contribute equal distances but in opposite directions, producing a false negative. Multivariate ANOVA analysis is a possible way to overcome this no-direction problem; however, with a large set of metrics to examine simultaneously, it is computationally and methodologically more challenging, and the estimates may involve a large magnitude of uncertainty.
As the experiments were carried out during different time spans for different mass spectrometers, the batches are nested within each mass spectrometer. A nested ANOVA model was developed as follows20:
where Mass Spectrometeri represents the effect of the ith mass spectrometer, Batchj(Mass Spectrometeri) represents the jth batch effect on the ith mass spectrometer and Residualsijk is the residual for the kth experiment in the jth batch on the ith mass spectrometer. In this ANOVA model, both the mass spectrometer and the batch effects were assumed to be random. The estimated variance of these random effects can evaluate which factor contributes the most variability in experimental performance. A statistical significance test can also be carried out to test if mass spectrometer and batch effects are significant. The ANOVA model could be extended to include more factors such as instrument types and the sample types.
Peptide Identification
Tandem mass spectra from each Study 1 mass spectrometer were identified using spectral library search. The Pepitome algorithm21 compared spectra to the NIST human ion trap library, using a 1.5 m/z precursor tolerance for average masses and a 1.25 m/z precursor tolerance for monoisotopic masses. Each peptide in the library was included in normal and scrambled form to allow for decoy measurement22. Fragments in all cases were allowed to vary from their expected positions by up to 0.5 m/z; while these settings for precursor and fragment tolerances may not have been as tight as optimal for each instrument, the much smaller search space of spectral libraries protected against significant reductions in identification efficiency. Peptides were allowed to be semi-tryptic or modified if the spectrum library contained those possibilities. Identifications were filtered and assembled in IDPicker 3.0, build 520, using a 0.02 PSM q-value threshold23 and requiring ten spectra to be observed for each protein. These settings resulted in a 4.11% protein FDR, with 373 distinct proteins identified (picking up extra proteins from carryover, in some cases) that spanned 3266 distinct peptides and 232,351 identified spectra. A total of 1040 distinct peptides and 217,157 spectra (93.6%) were accounted for by the NCI-20 proteins. Only the hits to the following RefSeq entries were accepted as legitimate for further analysis: NP_000468.1 (albumin), NP_001054.1 (serotransferrin), NP_000499.1, NP_005132.2, NP_000500.2 (fibrinogen alpha, beta, and gamma, respectively), and NP_004039.1 (beta-2-microglobulin). A spreadsheet of hits for each experiment in terms of distinct peptides, identified spectra, and matches (sub-variants of peptides by precursor charge or PTM-state) is available in Supporting Information.
Data from Study 5 were also identified by Pepitome. The search for samples 3A and 3B included the NIST ion trap libraries for yeast and BSA, mapped to the UniProt reference proteome set for yeast plus the sequence for bovine serum albumin. The search for sample 1B employed the same library as in Study 1. When Orbitraps were able to provide confident monoisotopic measurements, a precursor mass tolerance of 10 ppm was applied; otherwise, a 1.5 m/z precursor mass tolerance was applied. For samples 3A/3B, IDPicker 3.0 employed a 0.01 PSM q-value threshold and required six spectra per protein, yielding a 2.55% protein FDR. To achieve a protein FDR under 5% for the sample 1B report allowed a 0.05 PSM FDR threshold and required three spectra per protein for a 3.64% protein FDR. Only the hits to NCI-20 proteins and bovine trypsin were included in identification counts; when all data were included for sample 1B, a total of 79 human protein groups were detected, along with three decoys and trypsin, though spectral counts were far lower for proteins not found in the NCI20. A spreadsheet of the NCI20 identifications resulting from each experiment is available in Supporting Information.
RESULTS AND DISCUSSION
The reproducibility of LC-MS/MS experiments has been a controversial subject. Most analyses of this topic, however, have limited themselves to the peptide and protein identifications produced from these data sets. By using the “IDFree” metrics produced by QuaMeter to characterize data, however, this study examines the signals recorded in LC-MS/MS rather than the derived identifications. QuaMeter produced metrics for sixteen of the seventeen electrospray instruments for Study 1 (ProteoWizard14 did not support centroiding algorithms for Waters instruments). Because all six mass spectrometers in Study 5 were Thermo instruments, QuaMeter was able to produce metrics successfully from all experiments. This analysis begins with dimensionality reduction and data visualization, then quantifies the relationship between experiments, and finally builds a hierarchical model to test the impact of multiple factors on experimental outcomes. At the end, the correlation between ID-free and ID-based evaluations is discussed.
Dimensionality Reduction and Data Visualization
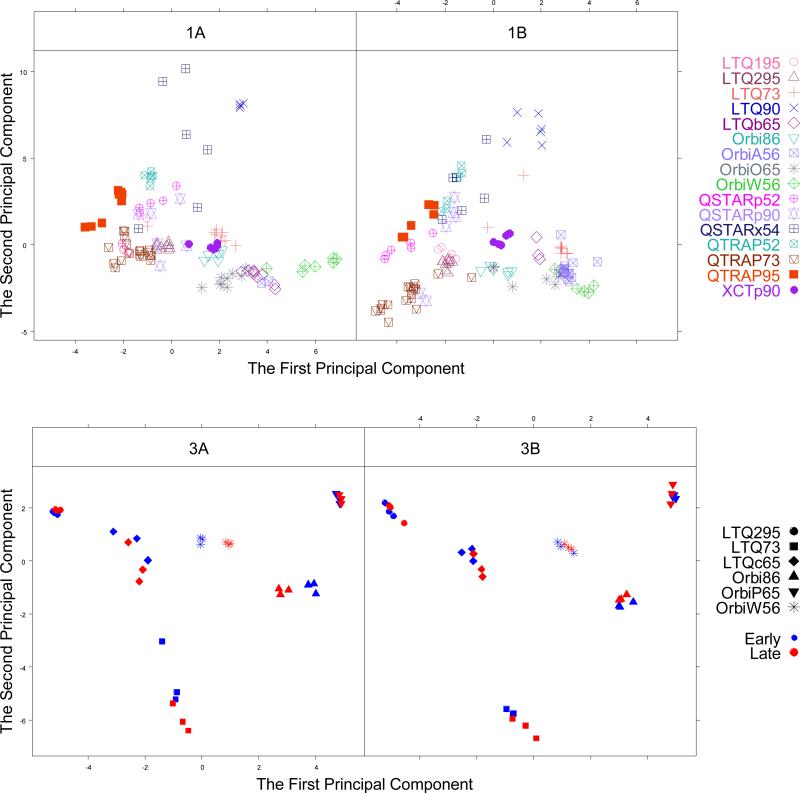
Univariate analysis would look at each QuaMeter metric in isolation to find differences between experiments, but multivariate analysis is able to combine the information of multiple metrics, recognizing correlations among them. Principal Components Analysis (PCA) is a widely used dimensionality reduction method that summarizes multidimensional inputs to a set of uncorrelated components, sorting them by the fraction of variance accounted for by each. The first two components of the robust PCA (PC1 and PC2) for the QuaMeter IDFree metrics from Studies 1 and 5 are visualized in Figure 1.
Figure 1.
The plot of the first two principal components. The upper two panels are for the 16 electrospray mass spectrometers from CPTAC Study 1. The left panel is for Sample 1A (digested on-site) and the right panel is for Sample 1B (centrally digested at NIST). The lower two panels are for the 6 mass spectrometers from CPTAC Study 5. The left panel is for Sample 3A (the yeast reference material) and the right panel is for Sample 3B (yeast reference material spiked with bovine serum albumin). The first three experiments are labeled as blue (“early”) and the last three experiments are labeled as red (“late”).
The PCA plot yields a good snapshot for data exploration. In general, the experiments for a particular mass spectrometer group together. This grouping can reflect the similarity of metrics that are characteristic of the instrument type where the same method is applied. That being said, different users of the same model of instrument may be quite separate in the plot; the five LTQ laboratories in Study 1 range considerably on the first two principal components. Because PCA was conducted jointly for lab digestion protocols (1A) and for centrally digested samples (1B) in Study 1 and the yeast (3A) and the spiked yeast sample (3B) in Study 5, comparing placements between plots for a given site is possible. While it is tempting to say that instruments like QSTARx54 were more variable than QTRAP52 on the plots for Study 1 because of how their symbols distribute, one needs to take the other principal components into account before framing this interpretation.
For Study 1 the first two principal components account for 22% and 19% of the variability in the QuaMeter metrics, respectively. For Study 5, these proportions are 42% and 23%. PC1 and PC2 in study 5 account for a larger proportion of metric variability than in Study 1, which may be impacted by the lack of an SOP guiding Study 1. Since the first two metrics account for only part of the total variance, the PCA plot can be deceptive; experiments that appear close together on the plot might look quite distant when a third or fourth dimension is taken into account. The third principal component, in this case, would describe an additional 10% of total variability in Study 1 and 20% of total variability in Study 5. As Ringnér suggests17, it is critical to systematically check different combinations of components when visualizing data by PCA.
Seeing the same symbol for each experiment flattens the available information to mask important attributes. Clearly one should expect that laboratories that use the same kinds of instruments or separations are likely to produce more similar results than those using different instruments or separations, a fact not represented by each site having an independent symbol. When data are produced in rapid succession (perhaps even without interleaved blanks), one can expect them to be more similar than when they are produced with more than a month of intervening time (See results in Nested ANOVA section). Incorporating factors of this type can yield a more much subtle analysis of variability for a multi-instrument study of this type.
To understand the underlying process that influences experimental performance, we also carried out exploratory factor analysis. In Study 1, the six-factor model represented more than 60% of the total variability. Almost all variability (95%) was accounted for by a six-factor model in Study 5. Examination of the loading matrix in Table S4 of Supporting Information may help to understand the key aspects that differentiated mass spectrometer performance. In Study 1, the factor accounting for the greatest overall variability (18%) was associated with intense precursor ions (XIC-Height-Q2, -Q3, -Q4), TIC concentration in the middle two quartiles of retention time (RT-TIC-Q2, -Q3), and a high rate of MS/MS acquisition (MS2-Count, MS2-Freq-Max). The second factor, accounting for an additional 10.6% of variability, favored wider peaks (XIC-FWHM.Q2) and MS scans that were heavily populated with ions for fragmentation (MS1-Density-Q1, -Q2). While all of these elements may seem equally associated with experiments producing large numbers of identifications, they varied separately among different experiments, allowing them to be separated to different factors. In Study 5, the first factor accounts for more than 40% of the total variability. Its features include narrow chromatographic peaks (XIC-FWHM-Q2), good contrast in peak heights (heightened XIC-Height-Q3) without extremes (lower XIC-Height-Q4), continued MS/MS acquisition during late chromatography (lower RT-MSMS-Q1, Q2, and Q3) and greater contrasts in MS total ion current (higher MS1.TIC.Q3, Q4), and sparse MS scans (low MS1-Density) accompanied by MS/MS scans populated with many peaks (high MS2-Density). The contrasts in metrics produced from both studies demonstrate that that correlation structure among metrics can differ significantly under different experimental protocols. No single combination of factors would be appropriate for all possible purposes to which these quality metrics may be applied.
Dissimilarity Evaluation
When QuaMeter produces its metrics for a given experiment, it reduces it to a vector of numbers, each representing a metric value. That vector can be thought of as a coordinate in a multidimensional space. PCA transforms from one set of dimensions to another, but each point in the new space continues to represent an individual LC-MS/MS experiment. The distance metrics described in Methods find the distance between a pair of those points just as the Pythagorean Theorem finds the length of the hypotenuse in a right triangle.
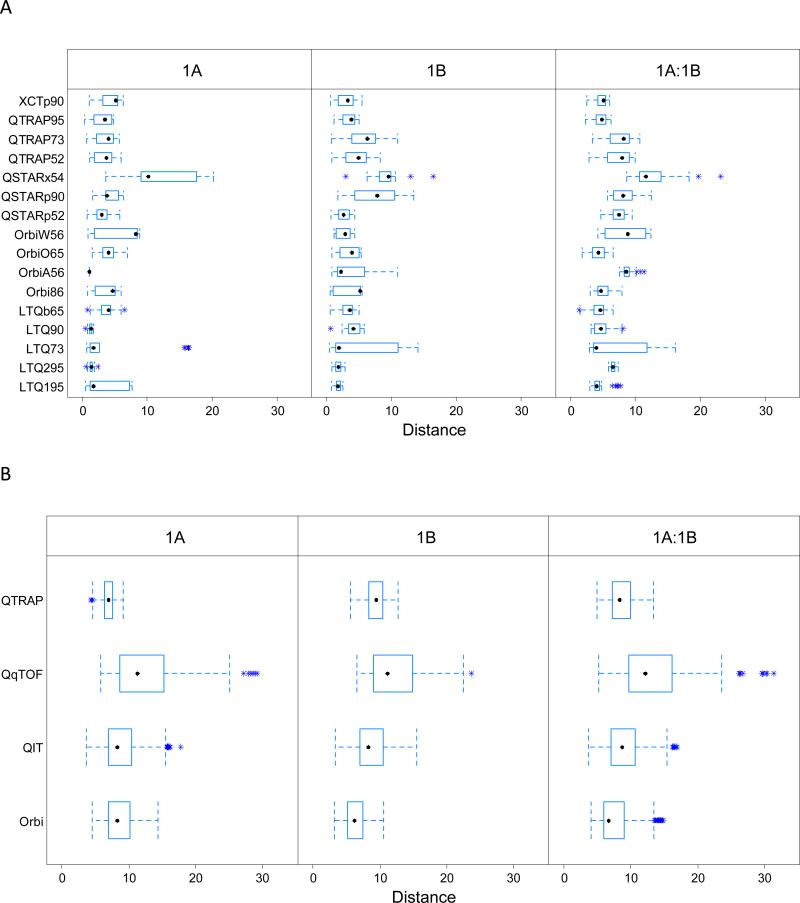
Relationships between pairs of LC-MS/MS experiments can take place at several levels. In the case of Study 1, when a particular mass spectrometer produced multiple replicates for sample 1A (digested on-site) and for sample 1B (digested centrally), the data for those experiments were likely to yield high similarity since the same operator employed the same mass spectrometer. That said, comparisons for a given sample type will also share digestion technique and may reduce variability further. These comparisons can be found in Figure 2A. At a higher level, one can group together the data from all instruments of a particular type, in this case generalized to QTRAP, QIT (Quadrupole Ion Trap), Orbi, and QqTOF (Quadrupole-collision cell-Time-of-Flight). These comparisons, within and between the two sample types, appear in Figure 2B. In cases where an individual laboratory employed instruments of different types, comparisons between experiments from the two instruments could determine the extent to which common operation imposed greater similarity (Figure S1A in Supporting Information). Finally, mass spectrometers of different types employed by different laboratories might be assumed to produce the largest degree of expected difference in performance (Figure S1B in Supporting Information). In each case, the three panels separate all possible pairs of sample 1A experiments, all possible pairs of sample 1B experiments, and all possible pairs of samples 1A and 1B experiments.
Figure 2.
Dissimilarity measures. Each panel evaluates the distance between metrics in PC space for every possible pair of files. 2A: Study 1 experiments from the same mass spectrometer (in the same lab); 2B: Study 1 experiments from the same type of instruments but different laboratories; 2C: Study 5 experiments from the same mass spectrometer (in the same lab); 2D: Study 5 experiments from the same type of instruments but different laboratories. Note that the x-axis scale differs between Study 1 and Study 5.
Several conclusions emerge from the Study 1 dissimilarity analysis. When experiments are compared within the 1A or 1B sample type, median dissimilarity values are approximately half the values seen when a 1A experiment is compared to a 1B experiment (see Table S5A for summary statistics). This demonstrates that trypsin digestion protocols can contribute substantial variability even when the starting protein mix is identical; alternatively, this result may suggest that shipping samples as peptides induces different effects than shipping samples as proteins. For some instruments, outliers are obvious in this analysis. For LTQ73, the “sample_1A205_03” experiment was very distant from every other produced by this instrument. One can also observe that some instruments are more inherently variable than others. QSTARx54 produced substantially higher mean distances than others. When classes of instruments are considered rather than individual instruments (Figure 2B), the AB SCIEX QTRAP 4000s produced relatively small distances from one experiment to the next; the three QTRAPs were operated under nearly identical methods from lab to lab.
Figures 2C and 2D show the dissimilarity analysis on sample 3A and 3B in Study 5. There are no abnormal runs based on the distance measures. Compared with that of Study 1, a striking decrease in the dissimilarity for experiments on the same spectrometer was observed (2C). Table S5B lists the medians and interquartile ranges from QIT and Orbitrap instruments in Study 1 as well as those from Study 5 within and across mass spectrometers. The consistency among experimental runs and higher similarity suggests that the implementation of an SOP reduced the level and spread of the dissimilarity between experiments. The distance for experiments from different mass spectrometers of the same instrumental type was also reduced, but with a much smaller magnitude. As shown in Table S5B, while the level of the dissimilarity does not change greatly compared with QIT and Orbi instruments in Study 1, the variability of the distance is reduced from 3.5~3.8 to 1~2. These results show that the implementation of SOP can greatly increase the reproducibility and repeatability of the experiments within and across laboratories, even though a large amount of variability is still retained for different mass spectrometers.
Mass Spectrometer and Batch Effects
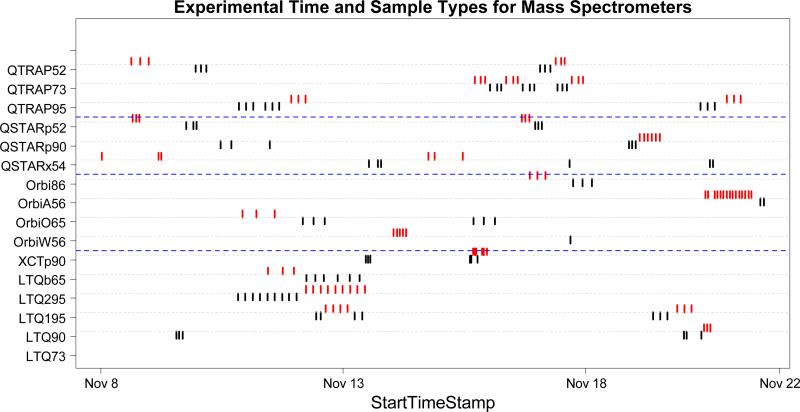
Statistical testing on the mass spectrometers and batch effects was carried out using a nested ANOVA model. In Study 1, we choose the first three and the last three experiments for each mass spectrometer to produce a balanced design with two batches labeled “early” and “late.” By this criterion, a total of 90 experiments from 15 mass spectrometers were selected for Study 1. The experimental design of Study 5 allows for the inclusion of all runs on Sample 3A and 3B in the analysis from 6 mass spectrometers. A snapshot of Study 1 is shown in Figure 3, displaying all experiments that were completed within 15 days of November 6, 2006, the starting date of the study. As shown in Figure 3, almost every laboratory produced data for 1A consecutively and data for 1B consecutively. As a result, potential batch effects were confounded with sample effects (i.e. if the platforms were varying in performance through time, it might easily appear as if it were varying in response to different samples). To avoid the possible confounding problem, only data from sample 1B were selected for ANOVA analysis. Consequently, we were not able to characterize the effect of digestion method on variability in Study 1. Figure S2 in the Supporting Information shows the time points for each of the experiments from the 6 different mass spectrometers in Study 5. The yeast only sample (3A, red) and the yeast spiked with BSA sample (3B, blue) were analyzed in two batches of three consecutive experiments. The sample effect is again confounded with the batch effect. Thus, ANOVA was performed separately on Samples 3A and 3B.
Figure 3.
Each dot represents a CPTAC Study 1 experiment that was conducted within 15 days of the starting date on November 6, 2006. Black dots represent sample 1A, while red dots represent sample 1B.
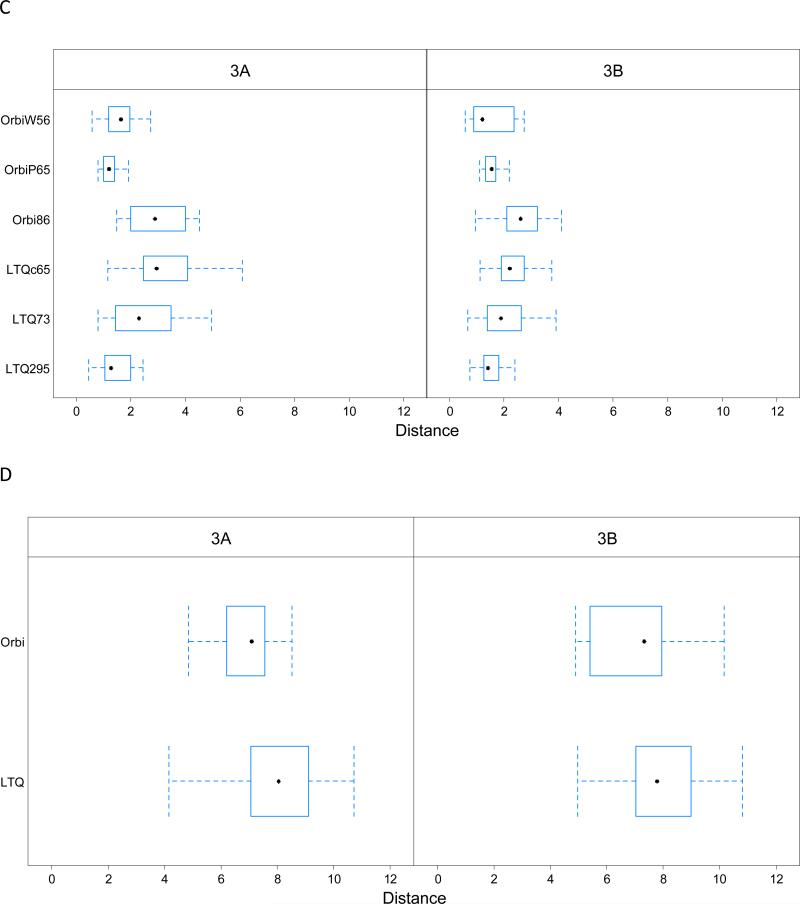
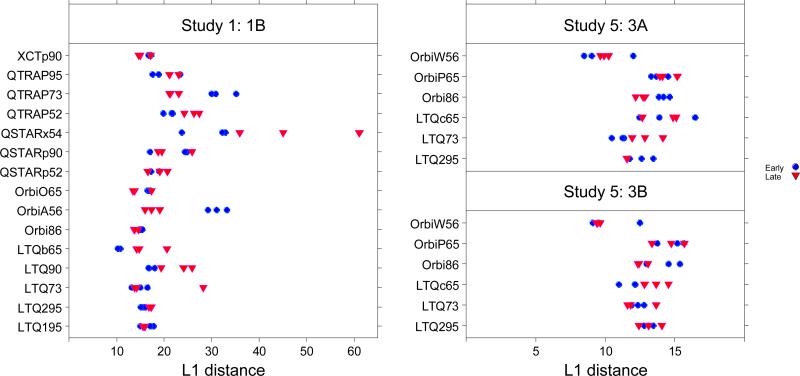
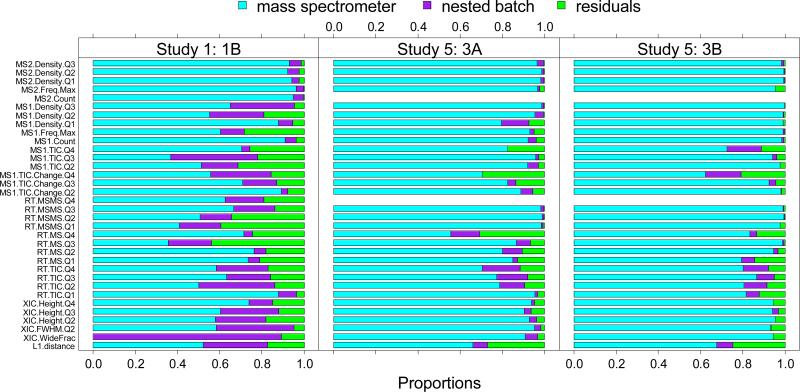
Figure 4 shows the typical L1-distances produced by different mass spectrometers in Study 1 and Study 5. The blue dots represent early experiments (first three), while the red triangles represent late experiments (last three). The distances vary across mass spectrometers and batches. In Study 1, the ANOVA on the multivariate L1 distance confirmed that both the mass spectrometer and the batch effects were significant with p-values <1E-06. The mass spectrometer accounted for 52.3% of variability (see bottom bar of Figure 5), and the batch nested within each mass spectrometer accounted for 30.3% of variability, with the remaining 17.4% unexplained. Results from Study 5 also showed strong mass spectrometer effects (p-value<0.0001) but no significant batch effect within mass spectrometers (p-value>0.10). Proportionally, the mass spectrometer accounted for most of the variability (66% in 3A and 68% in 3B), with residuals representing a much smaller fraction (27% in 3A and 25% in 3B). The upper bars of Figure 5 show the extent of impact for mass spectrometers and for nested batch on each of the metrics individually, for finer resolution. Significant associations between these sources and individual metrics are shown in Table S6 in Supporting Information.
Figure 4.
The L1-distance. Left Panel: the L1 distance for 90 experiments within 70 days used in ANOVA model for the mass spectrometer and batch effect study for Study 1; Right Panel (upper): L1 distance for Sample 3A in Study 5; Right Panel (lower): L1 distance for Sample 3B in Study 5. Distances observed for Study 1 ranged much higher than for Study 5.
Figure 5.
Proportion of variability accounted by mass spectrometer effect, nested batch effect and random errors (residuals) estimated using the nested ANOVA model on each individual metric. Left: CPTAC Study 1 (within 70 days); middle: Sample 3A CPTAC Study 5; right: Sample 3B CPTAC Study 5. The final row of the graph reflects the combination of all metrics.
Much less variability separated early and late batches for each mass spectrometer in Study 5 than in Study 1. This reduced batch effect may reflect the imposition of an SOP or may reflect the shorter time between batches. However, repeating the same sample in a consecutive series is still not good practice, even within a short span of time duration and SOP in place. Artifactual differences may arise due to the run order of experiments (see figure 10 of the CPTAC Repeatability article4). An ANOVA model on each site with batch as a factor showed that in Sample 3A, Orbi86 yielded a significant difference in performance between its two batches (p-value= 0.00612), and LTQ73 also exhibited a difference (p-value= 0.0425). In Sample 3B, LTQc65 produced a p-value of 0.03, which also demonstrates a potential batch effect. Since only a few hours separated early from late batches, the ability to find significant differences in identification-independent metrics is somewhat surprising.
Correlating Identification-free quality metrics to identifications
Identifications of tandem mass spectra are the most common way that data quality is evaluated in proteomics. For Study 1, three values were produced for each file: the number of MS/MS scans that were confidently identified to the known content of NCI-20, the number of matches (peptide variants in precursor charge and PTMs) that were detected, and the number of distinct peptide sequences that were observed. Unsurprisingly, these three measures exhibit a high degree of correlation. In Study 1, a Pearson correlation of 0.99 showed strong correlation between peptides and matches, peptides and spectra produced a value of 0.84, and matches and spectra yielded a correlation of 0.85. The values were even higher when Sample 1B was evaluated in isolation. For Study 5, the correlations among the distinct peptides, distinct matches and the filtered spectra were almost perfectly linear (>0.989). For subsequent analysis, only distinct peptide sequences were considered, since this value reasonably well reflected the information content of an identification experiment.
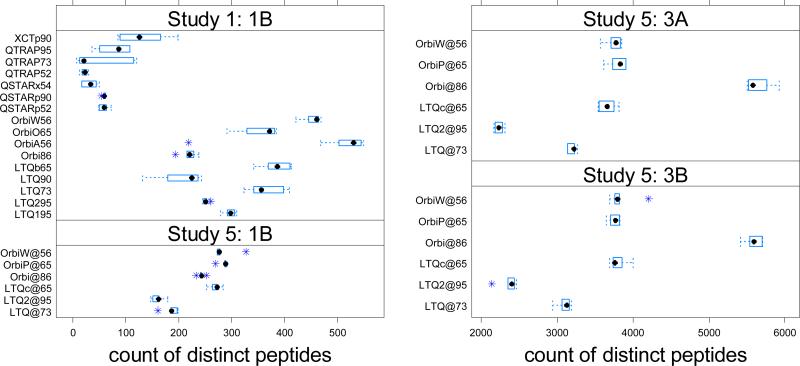
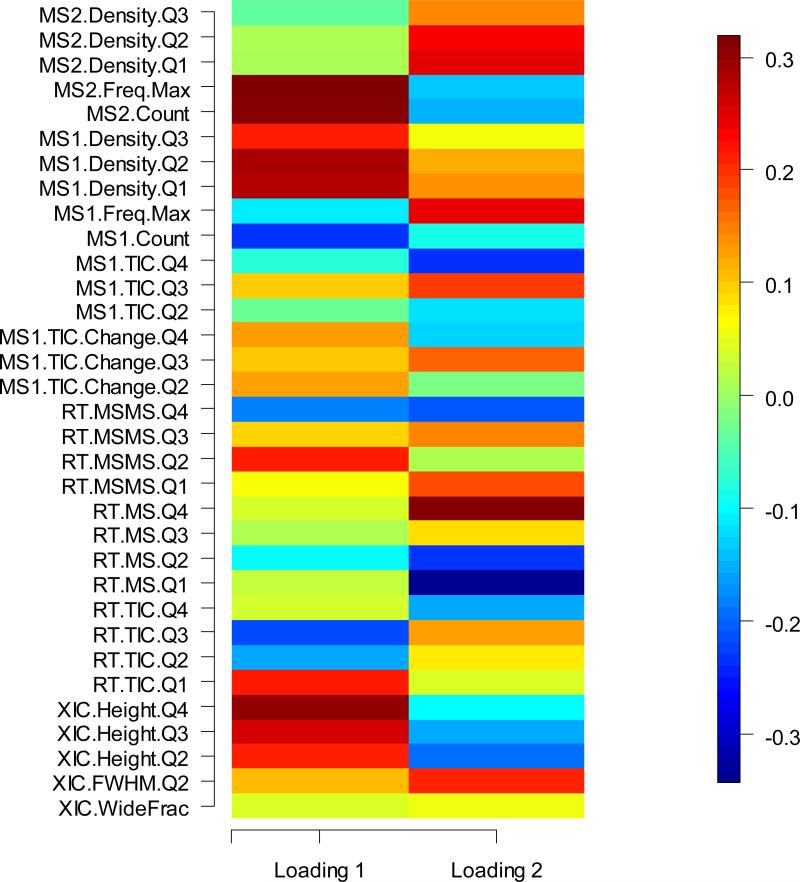
The numbers of distinct peptides were clearly associated with the type of mass spectrometers employed in Study 1 (see Figure 6). A regression was framed using an indicator variable value of ‘1’ for Thermo instruments and ‘0’ for non-Thermo instruments, using the indicator plus the principal component values (PC1 and PC2) to explain the expected number of distinct peptides identified. The indicator variable was highly significant, with a p-value of 2E-16. The PC1 score correlated significantly with the type of instruments. A one-way ANOVA analysis revealed that around 43% variability in PC1 was caused by the type of instrument in Study 1. However, even after the instrument manufacturer was taken into account, the first two principal components were significant. Looking across the entirety of the data set, the first principal component (PC1) was significantly positive (p-value = 0.000425) while the second component (PC2) was significantly negative (p-value = 0.000223). The loadings of the first two components were listed in Figure 7. The loadings ranged from 0.3 to -0.3. The first PC, which accounted for the most variability in the original metrics data, was a linear combination of all 33 metrics, with heavier weights on XIC.Height.Qx, RT.TIC.Q1, RT.TIC.Q3 (in the opposite orientation), RT.MSMS.Q2, MS1.count, MS1.Density.Qx, MS2.Count and MS2.Freq.Max. An examination of the 1B sample only obtained comparable results, though the PC effects were weaker.
Figure 6.
The number of distinct peptides identified in each experiment bore a clear relationship to the type of instrument employed. While in each case instrument vendor libraries performed operations such as charge state determination and peak picking, the peak lists for identification may not have been optimal for a variety of reasons. For this informatics workflow in Study 1 (the upper panel on the left), the Orbitrap and LTQ-class instruments from Thermo identified a greater diversity of peptides than did the instruments from other manufacturers. Study 5 results (Sample 1B, the lower panel on the left; Sample 3A: the upper panel on the right; Sample 3B: the lower panel on the right) show that the Orbitrap has a larger number of peptides identified than LTQ. Orbi@86 yielded very high sensitivity for sample 3A and 3B, with OrbiP@65, OrbiW@56, and LTQc@65 trailing behind, but still yielding more diverse peptide collections than the other LTQs.
Figure 7.
The loadings for the first two PCs for CPTAC Study 1 (Sample 1A and 1B conjointly)
A similar multiple linear regression model can be constructed with the QuaMeter metrics directly (along with the Thermo indicator variable) rather than through the principal components derived from them. This analysis for Study 1 shows a significantly positive relationship (p-value less than 0.05) for the XIC.WideFrac and MS1.TIC.Change.Q3 metrics, while producing p-values below 0.001 for positive relationships with the Thermo indicator variable and the log of MS2.Count. A significant negative relationship (p-value less than 0.05) was observed for MS1.TIC.Change.Q2, MS1.TIC.Q3, MS1.TIC.Q4, and MS1.Density.Q3. In analytical chemistry terms, this would translate to getting high identification rates when peak width is distributed among many different precursor ions, with changing signal seen in more than half of the MS1 scans and a copious number of MS/MS acquisitions. Lower identification rates, on the other hand, correspond to changing signal in less than half of the MS1 scans and seeing lots of signal in MS scans throughout the LC gradient.
CONCLUSION
This study demonstrates robust multivariate statistical methods to assess mass spectrometer and batch effects for proteomics using a collection of identification-free metrics. The adoption of multivariate statistics makes it possible to jointly model multiple aspects of an LC-MS/MS experiment, easing the visualization, comparison and analysis of variance in experimental performance. Most of the methods presented here aim to detect outlying experimental runs and identify the main sources of experimental variability, which is crucial for any QC study. Each method has its own niche. Robust PCA reduces and visualizes multivariate data. The dissimilarity measure is designed to be robust and can be easily applied to experimental profiles with any sample size and scope. The nested ANOVA, coupled with L1-distance, provides statistically rigorous analysis of the potential sources of experimental variability. The evaluation demonstrated that ID-free metrics are predictive of identification success, though ID-free metrics also provide information for more comprehensive analysis.
This research emphasized multi-site studies, but the general type of analysis is applicable in a wide variety of contexts. When data collection for a project has spanned multiple weeks (or been stopped and then restarted), researchers need to know if batch effects render the early experiments incomparable to late experiments. When an instrument has undergone service, technicians need to know whether its performance has shifted substantially to know whether preceding experiments should be re-run. This strategy should be very useful for recognizing when a subset of experiments in a large set comprise outliers due to unusual instrument conditions. In all cases, biological mass spectrometry is most useful when data differ due to biological effects rather than technical variation; these metrics and statistical models enable researchers to recognize when the less desirable outcome has occurred.
Because Study 5 was generated under the control of an SOP, one might expect that different mass spectrometers would exhibit far greater similarity than was seen in Study 1. This is confirmed by the dissimilarity analysis. The dissimilarity analysis confirms that the SOP and short time frame significantly improved the within mass spectrometer variability and also reduced the variability among mass spectrometers to a limited extent. While principal components were related to identification success after controlling for instrument type in Study 1 data, the same could not be said of Study 5. However, ANOVA showed that the across mass spectrometer effects are still a significant factor in experimental variation when one looks beyond identifications. ANOVA for Study 1 demonstrated that the greater the time elapsing between experiments, the greater the experimental variability apparent in the QuaMeter metrics. Had sample run orders been randomized in this study, the effects of centralized vs. on-site digestion might have been disentangled from batch effects.
In the evaluation of both Study 1 and Study 5, contextual data were not available to establish a “stable profile.” As a result, these data cannot be evaluated against the normal range of variation for these experiments. Even without such a profile, dissimilarity analysis can still help to identify outlying experiments. Future work will develop quality control models for dynamic performance monitoring and will also allow for systematic incorporation of the insights of laboratory technicians.
Supplementary Material
ACKNOWLEDGMENT
XW was supported by an interagency agreement between the NIH/NCI and the National Institute of Statistical Sciences. MCC and DLT were supported by NIH/NCI U24 CA159988. LJVM was supported by NIH/NCI U24 CA126479. DMB and SES were supported by an interagency agreement between the NIH/NCI and the National Institute of Standards and Technology.
Footnotes
Disclaimer: Certain commercial equipment, instruments, or materials are identified in this paper in order to specify the experimental procedure adequately. Such identification is not intended to imply recommendation or endorsement by the National Institute of Standards and Technology, nor is it intended to imply that the materials or equipment identified are necessarily the best available for the purpose.
References
- 1.Bell AW, Deutsch EW, Au CE, Kearney RE, Beavis R, Sechi S, Nilsson T, Bergeron JJM, HUPO Test Sample Working Group Nat. Methods. 2009;6:423–430. doi: 10.1038/nmeth.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klie S, Martens L, Vizcaíno JA, Côté R, Jones P, Apweiler R, Hinneburg A, Hermjakob H. J. Proteome Res. 2008;7:182–191. doi: 10.1021/pr070461k. [DOI] [PubMed] [Google Scholar]
- 3.Mann M. Nat. Methods. 2009;6:717–719. doi: 10.1038/nmeth1009-717. [DOI] [PubMed] [Google Scholar]
- 4.Tabb DL, Vega-Montoto L, Rudnick PA, Variyath AM, Ham A-JL, Bunk DM, Kilpatrick LE, Billheimer DD, Blackman RK, Cardasis HL, Carr SA, Clauser KR, Jaffe JD, Kowalski KA, Neubert TA, Regnier FE, Schilling B, Tegeler TJ, Wang M, Wang P, Whiteaker JR, Zimmerman LJ, Fisher SJ, Gibson BW, Kinsinger CR, Mesri M, Rodriguez H, Stein SE, Tempst P, Paulovich AG, Liebler DC, Spiegelman CJ. Proteome Res. 2010;9:761–776. doi: 10.1021/pr9006365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Resing KA, Meyer-Arendt K, Mendoza AM, Aveline-Wolf LD, Jonscher KR, Pierce KG, Old WM, Cheung HT, Russell S, Wattawa JL, Goehle GR, Knight RD, Ahn NG. Anal. Chem. 2004;76:3556–3568. doi: 10.1021/ac035229m. [DOI] [PubMed] [Google Scholar]
- 6.Rudnick PA, Clauser KR, Kilpatrick LE, Tchekhovskoi DV, Neta P, Blonder N, Billheimer DD, Blackman RK, Bunk DM, Cardasis HL, Ham A-JL, Jaffe JD, Kinsinger CR, Mesri M, Neubert TA, Schilling B, Tabb DL, Tegeler TJ, Vega-Montoto L, Variyath AM, Wang M, Wang P, Whiteaker JR, Zimmerman LJ, Carr SA, Fisher SJ, Gibson BW, Paulovich AG, Regnier FE, Rodriguez H, Spiegelman C, Tempst P, Liebler DC, Stein SE. Mol. Cell. Proteomics MCP. 2010;9:225–241. doi: 10.1074/mcp.M900223-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma Z-Q, Polzin KO, Dasari S, Chambers MC, Schilling B, Gibson BW, Tran BQ, Vega-Montoto L, Liebler DC, Tabb DL. Anal. Chem. 2012;84:5845–5850. doi: 10.1021/ac300629p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pichler P, Mazanek M, Dusberger F, Weilnböck L, Huber CG, Stingl C, Luider TM, Straube WL, Köcher T, Mechtler K. J. Proteome Res. 2012;11:5540–5547. doi: 10.1021/pr300163u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paulovich AG, Billheimer D, Ham A-JL, Vega-Montoto L, Rudnick PA, Tabb DL, Wang P, Blackman RK, Bunk DM, Cardasis HL, Clauser KR, Kinsinger CR, Schilling B, Tegeler TJ, Variyath AM, Wang M, Whiteaker JR, Zimmerman LJ, Fenyo D, Carr SA, Fisher SJ, Gibson BW, Mesri M, Neubert TA, Regnier FE, Rodriguez H, Spiegelman C, Stein SE, Tempst P, Liebler DC. Mol. Cell. Proteomics MCP. 2010;9:242–254. doi: 10.1074/mcp.M900222-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Addona TA, Abbatiello SE, Schilling B, Skates SJ, Mani DR, Bunk DM, Spiegelman CH, Zimmerman LJ, Ham A-JL, Keshishian H, Hall SC, Allen S, Blackman RK, Borchers CH, Buck C, Cardasis HL, Cusack MP, Dodder NG, Gibson BW, Held JM, Hiltke T, Jackson A, Johansen EB, Kinsinger CR, Li J, Mesri M, Neubert TA, Niles RK, Pulsipher TC, Ransohoff D, Rodriguez H, Rudnick PA, Smith D, Tabb DL, Tegeler TJ, Variyath AM, Vega-Montoto LJ, Wahlander A, Waldemarson S, Wang M, Whiteaker JR, Zhao L, Anderson NL, Fisher SJ, Liebler DC, Paulovich AG, Regnier FE, Tempst P, Carr SA. Nat. Biotechnol. 2009;27:633–641. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson RA, Wichern DW. Applied multivariate statistical analysis. 6th ed. Pearson Prentice Hall; Upper Saddle River, N.J: 2007. [Google Scholar]
- 12.Polanski M, Anderson NL. Biomark. Insights. 2007;1:1–48. [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson NL, Anderson NG. Mol. Cell. Proteomics MCP. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 14.Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S, Gatto L, Fischer B, Pratt B, Egertson J, Hoff K, Kessner D, Tasman N, Shulman N, Frewen B, Baker TA, Brusniak M-Y, Paulse C, Creasy D, Flashner L, Kani K, Moulding C, Seymour SL, Nuwaysir LM, Lefebvre B, Kuhlmann F, Roark J, Rainer P, Detlev S, Hemenway T, Huhmer A, Langridge J, Connolly B, Chadick T, Holly K, Eckels J, Deutsch EW, Moritz RL, Katz JE, Agus DB, MacCoss M, Tabb DL, Mallick P. Nat. Biotechnol. 2012;30:918–920. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor RM, Dance J, Taylor RJ, Prince JT. Bioinforma. Oxf. Engl. 2013 doi: 10.1093/bioinformatics/btt510. [DOI] [PubMed] [Google Scholar]
- 16.Xiong H, Yu LX, Qu H. AAPS PharmSciTech. 2013;14:802–810. doi: 10.1208/s12249-013-9966-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ringnér M. Nat. Biotechnol. 2008;26:303–304. doi: 10.1038/nbt0308-303. [DOI] [PubMed] [Google Scholar]
- 18.Hubert M, Engelen S. Bioinforma. Oxf. Engl. 2004;20:1728–1736. doi: 10.1093/bioinformatics/bth158. [DOI] [PubMed] [Google Scholar]
- 19.Kaiser HF. Psychometrika. 1958;23:187–200. [Google Scholar]
- 20.Karp NA, Spencer M, Lindsay H, O'Dell K, Lilley KS. J. Proteome Res. 2005;4:1867–1871. doi: 10.1021/pr050084g. [DOI] [PubMed] [Google Scholar]
- 21.Dasari S, Chambers MC, Martinez MA, Carpenter KL, Ham A-JL, Vega-Montoto LJ, Tabb DL. J. Proteome Res. 2012;11:1686–1695. doi: 10.1021/pr200874e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lam H, Deutsch EW, Aebersold R. J. Proteome Res. 2010;9:605–610. doi: 10.1021/pr900947u. [DOI] [PubMed] [Google Scholar]
- 23.Holman JD, Ma Z-Q, Tabb DL. Curr. Protoc. Bioinforma. Ed. Board Andreas Baxevanis Al. 2012 Chapter 13, Unit13.17. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.