Abstract
Histone variants are key players in shaping chromatin structure, and, thus, in regulating fundamental cellular processes such as chromosome segregation and gene expression. Emerging evidence points towards a role for histone variants in contributing to tumor progression, and, recently, the first cancer-associated mutation in a histone variant-encoding gene was reported. In addition, genetic alterations of the histone chaperones that specifically regulate chromatin incorporation of histone variants are rapidly being uncovered in numerous cancers. Collectively, these findings implicate histone variants as potential drivers of cancer initiation and/or progression, and, therefore, targeting histone deposition or the chromatin remodeling machinery may be of therapeutic value. Here, we review the mammalian histone variants of the H2A and H3 families in their respective cellular functions, and their involvement in tumor biology.
Keywords: Histone variants, Histone chaperones, Histone post-translational modifications, Cancer, Chromatin remodeler
Introduction
DNA-templated processes, such as DNA replication and repair, gene expression, and chromosome segregation, must contend with the chromatin template. Chromatin, the physiological form of the eukaryotic genome, is a polymer of DNA and protein, consisting mostly of histone proteins. The nucleosome is the basic repeating unit of chromatin, a macromolecule of 146 bp of DNA wrapped around an octamer of histones: two of each canonical histones H2A, H2B, H3, and H4 or variants thereof [1, 2]. Chromatin structure is highly dynamic and can be modulated through a number of different mechanisms, including nucleosome remodeling, histone Post-Translational Modification (PTMs), and, relevant to this review, the incorporation and exchange of canonical histones with histone variants.
Histone variants are most prominent from the H2A and H3 families, and linker histone H1 family, and, in general, represent a small portion of the total cellular histone pool [3, 4]. As histone variants contain sequence and structural variations of the canonical histones, they may be regarded as “mutant” histone proteins that replace their conventional counterparts within the nucleosome [3, 4]. Due to these differences, histone variants may also be subject to distinct PTMs that, in turn, recruit specific chromatin-related factors to further modify the chromatin template [3]. Histone variants can have temporal and tissue-specific expression, and their incorporation into the nucleosome requires the assistance of additional factors, known as histone chaperones. Chaperones can function in coordination with, or are sometimes themselves, chromatin remodeling factors [5–7].
It has become clear in recent years that histone variants confer unique functions within the chromatin template, and, in turn, increase chromatin complexity. In particular, histone variants contain a unique ability to regulate key cellular and developmental processes, and, when deregulated, may contribute to cancer initiation and progression. Indeed, a growing body of evidence links histone variants to cancer biology. For example, the expression level of particular variants correlates with tumor malignancy in a number of different tumor types, and, thus, histone variants may be utilized as prognostic indicators in cancer (described below in detail) [8–10]. Recent insights have shed light on the mechanistic role of histone variants in cancer progression [11–14] and, remarkably, the first histone variant mutations have just been uncovered in cancer [15, 16]. Furthermore, mutations and altered expression of chromatin remodelers, some of which are directly responsible for histone variant deposition, have also been reported [17–20]. Collectively, these observations point towards a critical, yet unappreciated, role for histone variants and their chaperones in cancer biology.
In this review, we first aim to provide the reader with an overview of histone variants, with a focus on the mammalian H2A and H3 families and their chaperones. Next, we highlight the roles of histone variants and their chaperones in tumor biology and discuss the diverse tumor types they influence. Throughout the review, we will use the recently established phylogeny-based nomenclature proposed for histone variants [21].
H2A variants
The H2A family is the largest family of variants among the core histones (i.e. 19 in humans). Most of these variants encode canonical H2A, but the others are considered “atypical” variants, namely H2A.X, H2A.Z, macroH2A (mH2A), H2A.B (Barr body-deficient), and H2A.J, as well as isoforms and splice variants thereof [22]. Compared to other histone variant families, which generally differ slightly from their canonical counterparts in amino acid sequence, the H2A atypical variants are notably divergent from canonical H2A. Thus, the H2A variant family may be the most structurally diverse family, and these structural differences result in a multitude of biological functions (see Table 1). The H2A family has been the focus of extensive research in recent years, and here we will focus on H2A.Z, H2A.X, and mH2A, which to date have been identified to exert a role in human cancer.
Table 1.
Deposition, localization, and functional properties of histone variants implicated in human cancer
| Variant | Function | Chaperones and regulatory factors |
Distribution patterns | Deposition | Mutant phenotype |
|---|---|---|---|---|---|
| H2A variant | |||||
| H2A.Z (H2A.Z.1, H2A.Z.2 and H2A.Z.2.2) |
Transcriptional regulation (mostly activation) Genome stability and chromosome segregation Telomere stability (S. cerevisiae) Others |
SRCAP complex p400 complex (including Tip60) INO80 (negative regulator, S. cerevisiae) |
TSSs, enhancers, insulators Pericentric chromatin Subtelomeric regions (S. cerevisiae) |
Replication-independent |
T.thermophila, D. melanogaster, X. Laevis, M. musculus: lethal (H2A.Z.1) S. cerevisiae, S. pombe: transcriptional defects, chromosmome loss (H2A.Z.1) |
| H2A.X | DSB repair factor recruitmenta Meiotic gene silencinga (M. musculus) |
Tip60 (D.melanogaster) FACT Nucleolin |
Genome-wide DSB sitesa |
Replication-independent DSB-induced (IRIF)a |
M. musculus: chromosomal aberrations |
| mH2A (mH2A.1 and mH2A.2) |
Transcriptional regulation (mostly repression) | ATRX (negative regulator) | Upstream and downstream of TSS Telomeres SAHFs Xi |
Replication-independent |
D. rerio: brain developmental defects (mH2A.2) M. musculus: female liver steatosis (mH2A.1) |
| H3 variant | |||||
| CENP-A | Centromere structure and function Kinetochore assembly (chromosome segregation) |
HJURP Mis18 complex |
Centromeres | Replication-independent (M/G1) | M. musculus: lethal |
| H3.3 | Gene activation Telomere homeostasis Epigenetic memory (X. laevis) Spermatogenesis (D. melanogaster) |
ATRX-Daxx HIRA DEK |
Euchromatin (active genes) Promoters and regulatory sequences Telomeric repeats Pericentric heterochromatin |
Replication-independent |
D. melanogaster, T. thermophila: sterility M. musculus (hypomorphic allele): lethality or fertility defects |
All data refer to mammalian histone variants, unless otherwise specified
γ-H2A.X
H2A.Z
H2A.Z structure and function
First described in the 1980s [23–25], H2A.Z is highly conserved throughout eukaryotic evolution, with a sequence conservation of ~90 %. Together with its low sequence identity to the canonical H2A (~60 %), a unique and necessary function for H2A.Z is likely [26]. While deletion of H2A.Z in simple eukaryotes such as fission and budding yeast is not lethal [27, 28], this variant is essential for viability in other model organisms, such as Tetrahymena thermophila [29], Drosophila melanogaster [30], Xenopus laevis [31], and Mus musculus [32].
Here, we point out that what is commonly referred to as H2A.Z is one of two isoforms, namely H2A.Z.1. Recently, a second isoform, H2A.Z.2, has been identified [33–35]. Encoding a second H2A.Z gene appears to be vertebrate-specific, and the two isoforms arose from a common origin in early chordate evolution [36]. They are the products of two non-allelic genes, namely H2AFZ (H2A.Z.1) and H2AFV (H2A.Z.2), and are expressed across a wide variety of human tissues [34]. While differing by only three amino acids at the protein level, H2A.Z.1 and H2A.Z.2 are encoded by unique nucleotide sequences [34, 35]. Knockout studies suggest that the genes are not redundant, and point toward a possible functional diversification of H2A.Z.1 and H2A.Z.2 [32, 36]. Supporting this view, the bromodomain-containing protein Brd2 was recently identified as an H2A.Z.1-enriched binding factor, suggesting the potential for isoform-specifc effects on gene expression [37]. However, our understanding of isoform-specific functions remains unclear and technically limited due to the lack of isoform-specific antibodies. Further increasing the complexity of H2A.Z biology, a third isoform, H2A.Z.2.2 (a splice variant of H2A.Z.2), was recently uncovered [38, 39]. Due to its shorter and distinct C-terminus, H2A.Z.2.2 is loosely associated with chromatin and forms the least stable nucleosome described thus far [38, 39]. In this review, we will mostly focus on H2A.Z.1, as it is well studied to date. However, we note that the studies described in the following paragraphs cannot necessarily distinguish between the isoforms, particularly those using antibody-based approaches.
The structure of H2A.Z.1-containing nucleosomes was resolved more than 10 years ago, and, despite the sequence divergence, it is surprisingly similar to nucleosomes containing canonical H2A [40]. However, a structural comparison of H2A.Z.1 and H2A revealed regions where the two diverge, including the L1 loop and the C-terminal docking domain. The L1 loop was originally thought to impair the formation of H2A.Z.1-H2A dimers [40]; however, it was later shown that heterotypic nucleosomes can form, both in vitro and in vivo [41, 42]. While questioned whether such heterotypic nucleosomes were stable or just an intermediate in a stepwise pathway to exchange H2A-with H2A.Z-containing nucleosomes, recent data suggest these heterotypic nucleosomes may be regulated in a cell cycle-dependent manner [43]. The docking domain comprises the interaction surface between the (H2A–H2B) dimer and the (H3–H4)2 tetramer, and is likely to influence the stability of H2A.Z.1-containing nucleosomes, although this is subject to debate [40, 44] and reviewed in [22].
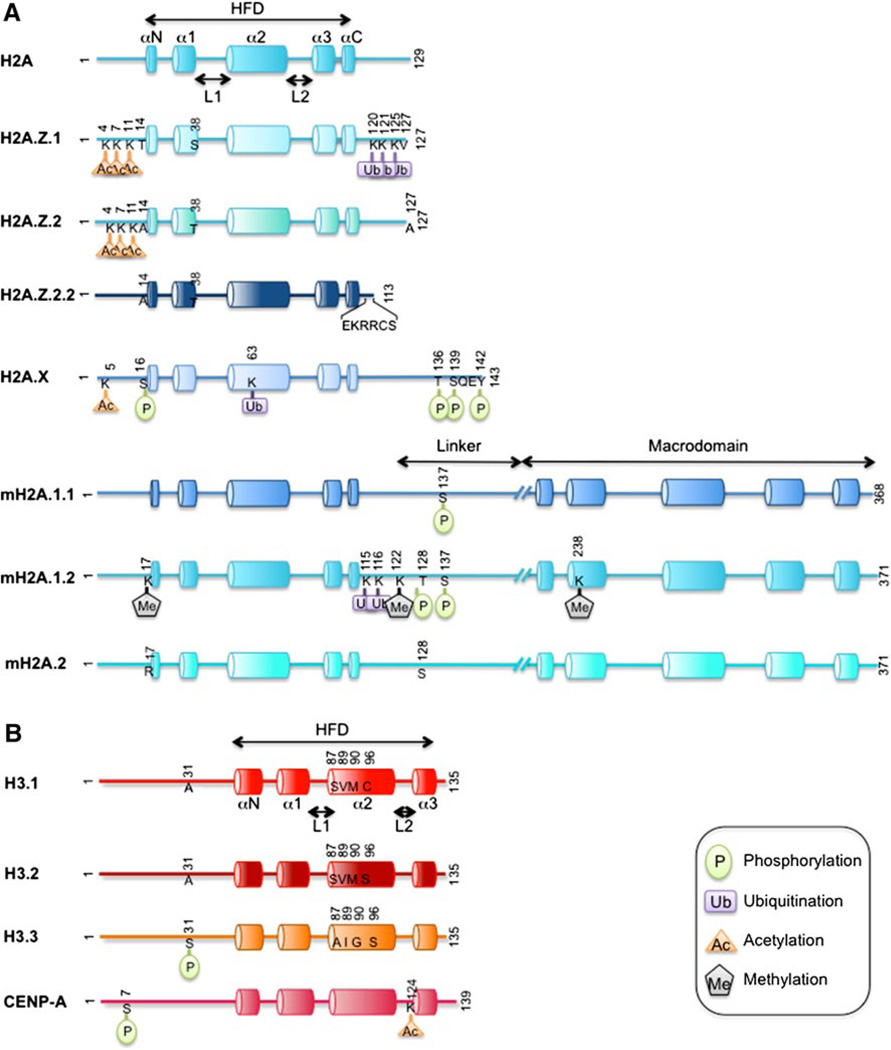
Similar to canonical histones, H2A.Zs are subject to PTMs (Fig. 1). Multiple lysine residues in the N-terminal tail are acetylated in organisms from yeast to human [45–48]. Generally, the hyperacetylated form of H2A.Z localizes at the transcriptional start site (TSS) of active genes, consistent with a less stable nucleosome and an open chromatin conformation which is suitable for gene expression. In addition, Immunofluorescence (IF) analyses detected the C-terminus of H2A.Z to be monoubiquitinated at the inactive X chromosome of female mammals [49] (Fig. 1). Therefore, PTMs of H2A.Z may help to reconcile the apparently conflicting associations of H2A.Z with both active and repressed transcription (see below). Of note, both H2A.Z.1 and H2A.Z.2 are acetylated on the same N-terminal lysines, while ubiquitylation on H2A.Z.2 has not been reported thus far [34] (Fig. 1).
Fig. 1.
Schematic representation of the mammalian H2A and H3 families of histone variants with a role in human cancer, and relative PTMs. a H2A variants (blue) and b H3 variants (red) are drawn relative to their peptide length. Protein sequences that are highly divergent between the canonical histones and their variants are depicted in different color shades. Specific amino acids are depicted where key differences are functionally relevant (e.g., H3 variants’ chaperone binding specificity), or when they are found to be post-translationally modified (PTMs are indicated by symbols as shown in the legend). Cylinders depict alpha-helical structures; other features discussed in the text noted by black arrows (see text for details)
H2A.Z is constitutively expressed throughout the cell cycle and is incorporated into chromatin in a replication-independent manner [50, 51]. Once incorporated into chromatin, H2A.Z exerts pleiotropic effects. For example, H2A.Z influences a plethora of different cellular processes and events, such as transcriptional regulation, epigenetic memory [52], heterochromatin boundaries [53], genome stability and chromosome segregation [54–57], and integrity of telomeres [58], further reviewed elsewhere [4, 59–61].
Although recently described as a player in reorganizing chromatin architecture at sites of DNA double-strand breaks (DSBs) [62], the best characterized role of H2A.Z is in transcription, which was initially suggested by Allis and colleagues [24], who showed that H2A.Z resides exclusively in the transcriptionally active macronucleus in T. thermophila. With the advent of high-throughput techniques, such as ChIP-chip and ChIP-seq, evidence has accumulated for a role of H2A.Z in transcription. H2A.Z is enriched at gene promoters in yeast [63–66] and in higher eukaryotes [67, 68], as well as on other regulatory regions, such as insulators and enhancers [61, 67, 68]. While H2A.Z generally has a positive effect on gene expression, it also negatively regulates transcription at specific gene targets [69] (see below). As mentioned before, such controversy can potentially be reconciled at least in part by different PTMs of H2A.Z [45, 46, 48, 49].
The non-random distribution of H2A.Z along chromosomes is mediated by the concerted action of both positive and negative regulators of H2A.Z deposition. H2A.Z is incorporated into chromatin by ATP-dependent chromatin remodeling complexes, including the Swr1 complex in yeast [70–72], or by its orthologs p400 [73] and SRCAP (Snf2-Related CBP (CREB-binding Protein) activator protein [74, 75]) in mammals. A number of “canonical” histone chaperones such as NAP-1 (Nucleosome Assembly Protein-1) and FACT (FAcilitates Chromatin Transcription) have also been identified to interact with H2A.Z in vivo [72, 76, 77]; however, since they are not required for in vitro histone exchange, their effective impact on H2A.Z deposition remains ambiguous. Evidence points towards a context-dependent action of p400 and/or SRCAP (i.e. gene-, cell type-, and developmentally-specific), and the possibility of functional redundancy cannot be ruled out. The intriguing possibility that p400 and SRCAP show a differential specificity for the two H2A.Z isoforms has been addressed but appears not to be the case [35]. Interestingly, recent studies have shed light on the process of H2A.Z eviction from nucleosomes. In particular, Peterson and colleagues found that the INO80 complex evicts H2A.Z in yeast [78], but whether this process also takes place in higher eukaryotes remains to be determined.
Overall, it is reasonable to consider that what dictates the final readout of gene expression is not merely the presence/absence of H2A.Z itself (or its post-translationally modified forms). It may also involve a combinatorial effect of H2A.Z, its targeting factors, transcription factors that it may work cooperatively with, PTMs on other histones, and the combination with other histone variants (e.g., H3.3) within H2A.Z-containing nucleosomes (see below) [79–81].
H2A.Z: an oncogenic histone variant
The first hints of H2A.Z involvement in tumor malignancy come from a number of microarray studies aimed at identifying the gene expression signatures of human cancer. These studies detected increased levels of H2A.Z in colorectal, breast, lung, and bladder cancers [8, 82–84] (Table 2). In addition, several members of the SRCAP and p400 complexes have been implicated in cancer through similar microarray-based studies in cell lines or in patient biopsies [84–89] (Table 3). Accordingly, we reported that global levels of H2A.Z are increased in metastatic melanoma cell lines as compared to primary lines [12].
Table 2.
H2A and H3 families of histone variants implicated in human cancer
| Variant | Cancer type | Dysfunction | Proposed mechanism in promoting cancer | References |
|---|---|---|---|---|
| H2A.Z (H2A.Z.1) | Colorectal cancer | Overexpression | N.D. | [82] |
| Undifferentiated cancers | Overexpression | N.D. | [83] | |
| Metastatic breast carcinoma | Overexpression | N.D. | [84] | |
| Primary breast cancer | Overexpression | ER alpha- and Myc-dependent upregulation | [8] | |
| Breast cancer (MCF7 cell line) | Overexpression | Increased proliferation | [14] | |
| Breast cancer | Overexpression | H2A.Z recruited at promoters of ERalpha target genes | [17] | |
| Melanoma | Overexpression | N.D. | [12] | |
| Prostate cancer (LNCaP cell line) | Overexpression | Myc-mediated H2AFZ upregulation | [91] | |
| Prostate cancer (LNCap xenograft) | N.D. | H2A.Zub evicted from PSA promoter/enhancer upon activation | [90] | |
| Prostate cancer (LNCaP cell line) | N.D. | H2A.Zac associated with oncogene activation, unmodified H2A.Z with tumor-suppressor silencing | [48] | |
| H2A.X | B-CLL and T-PLL | Translocations and deletions chr 11q23 | Increased genome instability | [130–132] |
| Head and neck squamous cell carcinoma | Gene deletion | Increased genome instability | [133] | |
| Non-Hodgkin lymphoma | Gene mutation | Increased genome instability | [135] | |
| Gastrointestinal stromal tumor | Upregulation | Promote apoptosis upon the treatment with a kinase inhibitor | [138] | |
| Breast cancer | Gene deletion | Increased genome instability | [134] | |
| mH2A.1 | Lung cancer | Reduced protein levels/splicing defects | Suppression of cell proliferation via reduced PARP-1 | [10, 13] |
| Breast cancer | Reduced protein levels/splicing defects | N.D. | [10, 13] | |
| Melanoma | Transcriptional downregulation | Upregulation of CDK8 | [12] | |
| Testicular, bladder, ovarian, cervical, endometrial cancers | Splicing defects | N.D. | [13] | |
| Colon cancer | Reduced protein levels and splicing defects | N.D. | [9, 13] | |
| mH2A.2 | Lung | Reduced protein levels | N.D. | [10] |
| Breast | Reduced protein levels | N.D. | [10] | |
| Melanoma | Transcriptional downregulation | Upregulation of CDK8 | [12] | |
| CENP-A | Colorectal cancer | Overexpression | Aneuploidy | [222] |
| Invasive testicular germ cell tumors | Overexpression | N.D. | [224] | |
| HCC | Overexpression | Deregulation of cell cycle and apoptotic genes | [226] | |
| Breast cancer | Overexpression | N.D. | [225] | |
| Lung adenocarcinoma | Overexpression | N.D. | [223] | |
| H3.3 | Carcinoma of the esophagus | Overexpression | N.D. | [262] |
| GBM | Mutation (K27M, G34V/R) | N.D. | [15] | |
| Pediatric DIPG | Mutation (K27M, G34V/R) | Inhibition of EZH2 activity, reduced H3K27me3 | [16, 263] |
Table 3.
Chaperones of H2A and H3 histone variants and their proposed mechanisms in human cancer
| Chaperone | Cancer type | Dysfunction | Proposed mechanism in promoting cancer | References | Variant |
|---|---|---|---|---|---|
| SRCAP | Ovarian cancer | Overexpression | N.D. | [89] | H2A.Z |
| Metastatic breast carcinoma | Overexpression | N.D. | [84] | ||
| Thyroid carcinoma | Overexpression | N.D. | [85] | ||
| Prostate cancer | Overexpression | N.D. | [86] | ||
| Prostate cancer | N.D. | Deregulation of cell proliferation | [20] | ||
| Tip60 | Colon carcinomas | Downregulation | N.D. | [88] | |
| Lung carcinomas | Downregulation | N.D. | [88] | ||
| Breast carcinoma | Downregulation | N.D. | [87] | ||
| p400 | Breast cancer | Overexpression | Activation of ERα target genes | [17] | |
| HJURP | Lung cancer | Overexpression | Aneuploidy | [227] | CENP-A |
| Breast cancer | Overexpression | Aneuploidy | [228] | ||
| HIRA | Endothelial cells | Upregulation | Promotion of tumor neovascularization | [264] | H3.3 |
| Daxx | GBM | Mutation | N.D. | [15] | |
| PanNET | Mutation | Loss of function | [18, 19] | ||
| ATRX | ATMDS | Mutation | Deregulation of α-globin expression | [266] | H3.3 (mH2A?) |
| GBM | Mutation | Loss of nuclear localization | [15] | ||
| Pediatric DIPG | Mutation | Loss of function | [16] | ||
| Other cancers of the CNS | Mutation | Loss of nuclear localization | [279] | ||
| Neuroblastoma | Mutation/deletion | Loss of function | [274] | ||
| PanNET | Mutation/deletion | Loss of function | [18, 19] |
A direct role for H2A.Z in cancer was found in hormone-dependent breast [8, 14, 17] and prostate cancer [20, 48, 90, 91], where an effect on cellular proliferation has been demonstrated (discussed below; see Tables 2, 3). Zucchi et al. [84] first reported an upregulation of H2AFZ and SRCAP in invasive and metastatic breast cancer compared to normal mammary epithelium. Furthermore, immunostaining and tissue microarray screening revealed that H2A.Z.1 is overexpressed in primary breast tumor samples from over 700 patients. This elevated level of H2A.Z.1 expression is significantly associated with metastasis to lymph nodes and with shorter overall patient survival [8, 14].
A recent genome-wide ChIP-chip approach of Estrogen Receptor α (ERα) and Myc binding sites, combined with estrogen-stimulated gene expression arrays, has identified the mechanism of H2A.Z.1 induction in breast cancer cells: in response to estrogen, ERα activates the proto-oncogene c-myc, which is recruited to the H2AFZ promoter and stimulates H2AFZ transcription [8]. In turn, H2A.Z positively regulate Myc expression, creating a positive feedback loop important for regulating ERα signaling both in normal and in pathological conditions [14]. Collectively, these data support a link between H2A.Z.1 and ERα-dependent breast tumor proliferation. Evidence that H2A.Z.1 may be a causal factor of breast cancer came from elegant functional studies by Gaudreau’s laboratory [14, 17]. By selectively depleting cellular levels of H2A.Z.1 or p400, H2A.Z.1 was shown to be essential for ER signaling, as were members of the p400 complex. Indeed, loss of H2A.Z or p400 led to a severe defect in estrogen signaling, and, in turn, estrogen-driven cell proliferation [17]. Conversely, ectopic expression of H2A.Z increased proliferation in the hormone-dependent MCF-7 cell line, particularly when estrogen levels were low [14]. In addition, by ChIP-chip analysis, H2A.Z.1 was reported to be specifically and cyclically recruited to promoters of ERα target genes upon estrogen stimulation, while its presence at their distal enhancers was independent of gene induction [17].
The role exerted by H2A.Z in regulating Androgen Receptor (AR)-mediated transcription of Prostate-Specific Antigen (PSA) and prostate cancer development was also reported recently. These studies accounted for H2A.Z PTMs, its chromatin deposition complexes and, importantly, the two H2A.Z isoforms [20, 48, 90, 91]. In a genome-wide study aimed at characterizing the epigenetic landscape of normal prostate epithelial cells and prostate cancer, H2A.Z was reported to be associated with both active and inactive promoters, while its acetylated form was present at promoters of actively transcribed genes, mutually exclusive of repressive marks such as DNA methylation and H3K27me3. Acetylated H2A.Z is associated with oncogene activation, and with the deacetylated form with silencing of tumor-suppressor-genes in prostate cancer cells [48]. Of note, H2A.Z.1 but not H2A.Z.2 expression is specifically upregulated in response to androgen treatment, and this occurs at least in part through increased binding of Myc to the H2AFZ promoter, as observed for estrogen-stimulated H2AFZ expression in breast cancer cells [8, 91].
Seemingly paradoxical observations have been accumulating on how H2A.Z regulates the expression of PSA [20, 90, 91]. Unlike what has been observed at ER targets [17], the levels of H2A.Z, and of its acetylated and ubiquitylated forms, decrease at PSA promoter/enhancer upon androgen treatment and concomitant with PSA induction. This suggests that H2A.Z poises the PSA gene for activation, but is evicted once the hormone is present [91]. Since a decreased expression of PSA is observed upon knockdown of SRCAP [20], this chaperone may also be required for the priming function of H2A.Z at the PSA gene. Collectively, these findings have been conducted in prostate cancer cell lines, and whether H2A.Z effectively correlates with prostate cancer progression and drives prostate cancer growth requires further investigation.
H2A.Z has also been implicated in cellular senescence— an irreversible loss of proliferative capacity. Senescence is triggered by progressive telomere shortening, activated oncogenes (Oncogene-Induced Senescence; OIS), and other forms of cellular stress. Importantly, senescence bypass appears to be an important step in the development of cancer (for a recent review, see [92]). First, Chan et al. [93] showed that p400 negatively affects the induction of senescence by decreasing p21 expression, and later Gevry et al. [69] demonstrated that this effect is mediated by deposition of H2A.Z by p400 at the p21 promoter. Indeed, depletion of both H2A.Z and p400 induced premature senescence in human fibroblasts. Thus, H2A.Z is a negative regulator of p21 expression and suppresses the senescence response [69]. Although intriguing, the role of H2A.Z in driving cancer through senescence bypass remains to be tested. In addition, our understanding of H2A.Z’s role in transcriptional repression remains limited.
Collectively, these findings strongly suggest that H2A.Z is an oncogene. We envision that what drives cancer initiation and/or progression is an increased incorporation of H2A.Z into chromatin, which can result from either an upregulation of H2A.Z transcription per se, and/or via changes in expression or activity of its chromatin deposition complexes. Given the association of H2A.Z in transcriptional activity, we speculate that H2A.Z fine tunes gene expression or modulates additional regulatory or non-coding regions of the genome that are relevant to disease progression. However, as mentioned previously, H2A.Z also participates in orchestrating DSBs repair [62], maintaining the integrity of telomeres [58], genome stability, and chromosome segregation [54, 56, 57], which are all candidate mechanisms underlying tumorigenesis. Thus, we anticipate a mechanistically complex role for H2A.Z in driving tumor progression.
H2A.X
H2 A.X structure and function
Like H2A.Z, H2A.X was first identified in human cells in the 1980s [25]. H2A.X is highly conserved and present in all eukaryotes, although at variable levels. For instance, while H2A.X represents roughly the total H2A pool in budding yeast, in mammals it only constitutes 2–25 % of the H2A pool, depending on the cell line or tissue [94]. Unlike H2A.Z, which diverged from H2A only once, it seems that H2A.X had multiple evolutionary origins [26]. H2A.X is typically expressed throughout the cell cycle, and it is ubiquitously incorporated throughout the genome during DNA replication in physiological conditions. H2A.X is probably best known for its well-studied phosphorylation site, S139ph, with critical roles in the DNA Damage Response (DDR), such that H2A.X has been coined as the “histone guardian” of the genome [94, 95] (see below). Consistent with this, H2A.X-deficient mice are viable, but radiation-sensitive, growth-retarded, and immune-deficient [96–98], exhibiting repair defects and chromosomal instability.
H2A.X is highly similar to the canonical H2A in the Histone Fold Domain (HFD); however, its unique feature is a longer C-terminal extension containing a serine–glutamine (SQ) motif followed by an acidic and hydrophobic residue [SQ(E/D)ϕ]. Of note, this motif is conserved in all species with regard to its sequence and position relative to the C-terminus (Fig. 1). H2A.X is subject to a number of different PTMs, among which the S139 phosphorylation (S139ph) in the conserved SQ(E/D)ϕ motif is extensively characterized. S139 is rapidly phosphorylated upon DNA damage and is localized to DNA DSBs (Figs. 1, 2e). This form is referred to as γ-H2A.X, since it was originally identified in mammalian cells when treated with gamma irradiation [94, 99]. Of note, the canonical H2A in mammals does not contain S139, and therefore only H2A.X can be phosphorylated at this site. The phosphorylation appears within minutes of DNA damage, and in mammals it spreads over a 2-Mb domain surrounding the DSB [99]. H2A.X S139ph is carried out by members of the Phospho-Inositide 3-Kinase-related protein Kinase (PIKK) family (ATM, ATR, and DNA-PK) [100, 101], and, accordingly, it has been reported that mice deficient for ATM are severely deficient for γ-H2A.X foci formation [102].
Fig. 2.
Localization of H2A variants by immunofluorescence. a mH2A.1 localizes to the Xi, note also staining throughout the nucleus. b mH2A.1 localizes to SAHFs in cells induced to senesce by oncogenic RAS signaling, note also the presence of Xi. c Phosphorylated mH2A.1 on S137 (S137ph) is excluded from the Xi and has a distinct nuclear localization pattern compared to mH2A.1. d mH2A.2, like mH2A.1, localizes to the Xi and throughout the nucleus. e y-H2A.X (S139ph) localizes to IRIF upon DNA damage induced by γ-irradiation. All imaging performed in IMR-90 female primary lung fibroblasts at a 40X magnification. Histones or histone modification antibodies were labeled with Alexa Fluor 488 (green) and DAPI (blue) was used to visualize the nucleus
Upon DNA damage, repair proteins, histone modifying enzymes, and chromatin remodeling complexes accumulate in subnuclear foci called IRradiation-Induced Foci (IRIF) [103] (Fig. 2e). The impairment in IRIF formation is observed upon H2A.X deletion or S139 mutation [104], suggesting that S139ph might itself be recognized by one or more DDR proteins, serving as a signal for recruitment and assembly at sites of DNA damage. However, γ-H2A.X is not required for the initial formation of IRIF in vivo, but is rather essential for their retention [97]. Recent elegant live-cell imaging studies have been instrumental in devising a model for the kinetics of recruitment and accumulation of DDR players at DSB sites [105]. A detailed description of the DDR response is beyond the aim of this review, and we refer the reader to a number of comprehensive recent reviews [106, 107].
Aside from its role in DDR, γ-H2A.X is required for orchestrating chromatin remodeling in meiotic silencing of unpaired chromosomes in male mice [108], and, in particular, of sex chromosomes during meiosis [109]. Here, γ-H2A.X was shown to precede the deposition of another histone variant, H3.3 [110].
Besides S139, other residues such as serine 16, threonine 136, and tyrosine 142 are phosphorylated on H2A.X [111–113] (Fig. 1), conferring variable effects on nucleosome structure and ultimately on cellular biology. Interestingly, Y142 was shown to be constitutively phosphorylated in physiological conditions, and is dephosphorylated by specific phosphatases upon DNA damage [112, 114, 115]. In addition to phosphorylation, H2A.X is modified by ubiquitylation [116–121] and acetylation [122] (Fig. 1). Mutagenesis experiments of the residues subject to PTMs highlighted the importance of the crosstalk between H2A.X PTMs in the regulation of DDR. The emerging scenario is that the extent of a single PTM, as well as a balance between the different PTMs, is critical to fine-tune the outcome of the cellular response to DNA damage. For further reading on the role of H2A.X PTMs in orchestrating the DDR, we refer the reader to the original works cited above or to other excellent reviews [123, 124].
Regarding its incorporation into nucleosomes, the Tip60 HAT (Histone Acetyl Transferase) complex was first reported as a major exchange activity in Drosophila [125]. Later, an approach aimed at identifying factors that can regulate H2A.X exchange in human cells uncovered the histone chaperone FACT (for a recent review, see [126]). Furthermore, FACT-mediated H2A.X deposition is facilitated by S139ph, suggesting that it acts in response to DNA damage [127]. However, the machinery required for H2A.X turnover still remains unclear, as additional chaperones including nucleolin have also been reported [128].
H2 A.X: a genomic caretaker
DSBs are lesions with the potential to induce genomic instability and gene mutation. Thus, given the critical function of γ-H2A.X in orchestrating the cellular response to DSBs, it is not surprising that H2A.X has become a key player in tumor biology (Table 2). Even though the H2A.X null mice are not cancer prone, deficiency of histone H2A.X (and H2A.X haplo-insufficiency) results in increased genomic instability and cancer incidence in the context of p53 deficiency [104, 129]. H2A.X−/−p53−/− and H2A.X−/+p53−/− mice develop T and B lymphomas and solid tumors exhibiting dramatic genomic instability [104, 129].
Deletion of band 11q23—where H2AFX maps—has been detected at a particularly high frequency in several human cancers, including hematological malignancies such as B cell chronic leukemia (B-CLL) and T cell prolymphocytic leukemia (T-PLL). In B-CLL, it is associated with rapid disease progression and poor survival [130–132]. Notably, alteration in H2AFX copy number has also been described in solid tumors such as head and neck squamous cell carcinoma and breast cancer [133, 134]. In all cases, chromosomal instability was observed. Interestingly, not only deletions but also Single Nucleotide Polymorphisms (SNPs) of H2AFX have recently been reported to play a role in lymphoma susceptibility and development [135]. Overall, these findings strongly suggest that H2A.X might contribute to cancer initiation and progression, and support the idea that it could be a genome caretaker and a tumor suppressor in certain genetic contexts.
In addition to its role in tumorigenesis, γ-H2A.X has been used both as a diagnostic tool and as an indicator of efficiency of treatments based on DNA damaging agents used in both chemotherapy and radiotherapy [136–138]. Finally, γ-H2A.X may be used as a biomarker to quantify the genotoxic potential of novel anticancer compounds in both cultured cells and in animal models.
mH2A
mH2A structure and function
Among the histone variants, mH2A may be the most structurally distinct, due to a large and evolutionarily ancient macro domain of ~30 kDa at its C-terminal end [26, 139] (Fig. 1). This domain results in a histone approximately three times the size of canonical H2A. The N-terminal histone domain shares only ~65 % sequence identity with canonical H2A, and is connected to the macro domain by a basic linker sequence, which resembles a histone tail [140–142].
To date, three isoforms of mH2A have been reported in mammals. Two different genes, namely H2AFY and H2AFY2, encode, respectively, for mH2A.1 and mH2A.2 [141, 143]. mH2A.1.1 and mH2A.1.2 are splicing variants encoded by H2AFY, which differ only in a single exon (~30 amino acids) in the macro domain [143]. Although subtle, this differential exon allows mH2A.1.1, but not mH2A.1.2, to bind NAD+ metabolites. However, the functional consequences of this have yet to be fully explored in vivo [144, 145].
Pehrson and Fried [142] identified mH2A.1 in 1992, and soon after its initial identification, it gained acceptance as a repressive histone. Both mH2A.1 and mH2A.2 were reported to coat the transcriptionally inactive X (Xi) chromosome in female mammalian cells by IF [146] (Fig. 2a, d), and more recently by ChIP-seq [147] (recently reviewed in [148]). However, the contribution of mH2A to silencing of the X chromosome is perhaps best described as a complementary, but not an essential, silencing mechanism [149, 150], as mH2A.1 knockout female mice are viable and undergo normal X inactivation [147, 151, 152].
Additional observations have strengthened the notion of mH2A as a transcriptional repressor. It is associated with regions of condensed chromatin such as Senescence-Associated Heterochromatic Foci (SAHFs) [153] (Fig. 2b), inactive alleles of imprinted genes, and silenced endogenous retroviruses [154, 155]. Furthermore, analyses of mH2A.1 and mH2A.2 genomic occupancy have recently yielded interesting insights into this variant, showing that it is also present at autosomal genes, and hence likely involved in genome-wide transcriptional regulation [147, 156–159]. In particular, mH2A localizes to subtelomeric genes [159], cell–cell signaling genes [157], and pluripotency-related and developmental-specific genes [156, 157, 159], generally— but not always—preventing their expression [157]. Although this might argue for a functional role of mH2A in development and differentiation, this hypothesis remains questionable based on the lack of strong phenotypes reported from mH2A knockout mice [147, 151, 152]. Of the two knockout studies performed, only one reported a mild, gender-specific phenotype—female mH2A.1 null mice had higher incidence of liver steatosis [151].
Crystal studies of mH2A.1 by Luger and colleagues suggested that subtle alterations of mH2A.1’s L1 loop, compared to that of canonical H2A, result in a nucleosome with less flexibility, and potentially contribute to the repressive nature of mH2A [160] (Fig. 3). While these studies were performed using only the histone domain of mH2A, [160], it has been speculated that the macro domain protrudes from the nucleosome core, thereby inhibiting the association of certain factors by steric hindrance, and/or by promoting a specific and context-dependent association with unique factors [140, 160]. To date, several proteins have indeed been identified to interact with the macro domain, sometimes in an isoform-specific manner, and at least a few of such interactions create a repressive chromatin environment (reviewed in [140]). Supporting the steric hindrance model, studies have provided evidence that (1) Transcription Factors (TF) bind less effectively to mH2A.1 containing nucleosomes, and (2) SWI/SNF remodeling activity and/or binding to mH2A nucleosomes is less optimal than that of canonical nucleosomes. As a consequence, transcriptional initiation is hindered [161–164]. Furthermore, while the HFD of mH2A is able to pack DNA very similarly to canonical H2A [165], the linker region stabilizes the wrapping of DNA around the histone core by interacting with DNA around the nucleosomal entry/exit site, and this potentially contributes to the more condensed nature of mH2A-containing chromatin [166]. Taken together, the in vitro biochemical data provide mechanistic evidence for the role of mH2A as a transcriptional repressor.
Fig. 3.
The L1 loops of mH2A.1 and canonical H2A are structurally distinct. a, c Crystal structure of nucleosomes containing canonical H2A (a, red) or mH2A.1 (c, blue) by in silico homology models. H2B, H3 and H4 are in light blue and DNA is in beige. b Superimposition of H2A and mH2A.1 LI loops illustrates a closer organization of the two mH2A.1 molecules compared to those of canonical H2A [160]
Until recently, the mechanisms and factors involved in mH2A chromatin incorporation were unknown. Our group identified the SWI/SNF helicase ATRX (Alpha Thalassemia/MR, X-linked) to interact with all three isoforms of mH2A in their chromatin-free state [159]. In contrast to its role in H3.3 deposition [167–169] (see below), our observations suggest that ATRX is a negative regulator of mH2A chromatin association. Loss of ATRX results in the increased deposition of mH2A at the α-globin gene cluster, concomitant with the loss of α-globin gene expression that is often observed in ATRX syndrome patients [159]. Together with the reported role of INO80 in the regulation of H2A.Z [78], this is another example of negative regulation of chromatin deposition, which may include nucleosome eviction or inhibition of deposition. Therefore, a dynamic balance between histone deposition and eviction/inhibition is likely required to maintain proper chromatin states and, in turn, a specific biological readout. However, despite these findings, the mechanisms by which mH2A is actively incorporated at distinct genomic loci still remain unclear.
To date, a handful of mH2A PTMs have been identified (primarily through mass spectrometry analyses), and their functional roles are largely unknown. These include ubiquitylation of mH2A.1.2 at lysines 115 and 116 (K115, K116) [170, 171], methylation of mH2A1.2 at lysines 17, 122 and 238 (K17, K122, K238) [170], phosphorylation of mH2A.1.2 at threonine 128 (T128) [170, 172], and phosphorylation of both mH2A.1.1 and 1.2 at serine 137 (S137) [172] (Figs. 1, 2c). Some of these PTMs orchestrate the association of mH2A with the inactive X. Indeed, the ubiquitylation of mH2A.1, mediated by CULLIN3/SPOP ubiquitin ligase complex [173, 174], is important for association with the Xi. In contrast, the S137 phosphorylated form of mH2A.1 is excluded from the Xi and co-localizes with RNA polymerase II, suggestive of a role in gene activation [172] (Fig. 2c). Furthermore, while S137ph is present throughout the cell cycle, it is enriched during mitosis and is Cyclin-Dependent Kinases (CDK)-dependent. While it remains to be tested, this finding implicates mH2A in cell cycle regulation.
The role of mH2A at the Xi has been extensively dissected, and while not involved in the initial stages of silencing, mH2A may be a key player in reinforcing silent chromatin states [173, 175]. This is consistent with the finding that mH2A levels in chromatin increase during mouse Embryonic Stem Cell (mESC) differentiation and in mouse development [158, 176, 177]. Recent studies have investigated the role of mH2A isoforms in the context of mESC differentiation via RNA interference approaches. Consistent with the mouse knockout studies, Tanasijevic and Rasmussen [177] found that knockdown of mH2A does not perturb X inactivation and cells effectively differentiate towards multiple lineages. On the other hand, Buschbeck and colleagues [178] found that knockdown of mH2A inhibits proper differentiation via incomplete inactivation of pluripotency genes and reduced activation of lineage-specific genes. Therefore, the role of mH2A in differentiation remains unclear, and we anticipate that studies in the context of genetically deficient mice will shed light on this interesting biology.
Finally, while mH2A isoforms have not been shown to play a significant role in development, these histone variants can act as barriers in the reprogramming of differentiated cells into their pluripotent counterparts. Via Somatic Cell Nuclear Transfer (SCNT) and TF-based reprogramming (induced Pluripotent Stem Cells; iPSCs), the enhanced induction of pluripotency was observed in the absence of mH2A isoforms [158, 179, 180]. As mH2A.1 and mH2A.2 are enriched at pluripotency genes in differentiated cells, mH2A isoforms act as an ‘epigenetic barrier’ that needs to be overcome (potentially via their active eviction from chromatin) in order to induce such genes during reprogramming [158, 180]. This suggests that mH2A inhibits cellular reprogramming and plasticity, and may therefore also present a barrier to tumor formation (see below).
mH2A: a pleiotropic tumor suppressor
In recent years, the number of studies addressing the role of mH2A in cancer initiation and progression has increased considerably (Table 2). As a whole, these data suggest a role for particular mH2A isoforms as tumor suppressors, whose expression is reduced (compared to normal tissues and/or early cancer stages) in several tumor types, including melanoma, lung, testicular, bladder, colon, ovarian, breast, cervical, and endometrial cancers [9, 10, 12, 13, 181]. However, mH2A isoforms vary in their functional contribution to different tumors, as do the mechanisms of mH2A downregulation, suggesting cancer- and cell type-specific roles for mH2A.
Initial studies suggested that mH2A isoforms act as a biomarker for disease. In patient samples of non-small cell lung cancer, decreased levels of mH2A.1.1 and mH2A.2 (and to a lesser extent mH2A.1.2) were shown to inversely correlate with cell proliferation and with disease-free survival [10]. The same inverse correlation between mH2A.1.1 expression and proliferation has recently been reported in colon cancer [9]. Interestingly, mH2A.1.1 levels are significantly reduced in a wider panel of cancer types, and the mechanism has been attributed to splicing of the mH2A.1 transcript [13]. In particular, QKI (QuaKIng), a factor that regulates the alternative splicing of mH2A.1 pre-mRNA, is transcriptionally downregulated in certain cancer types. This results in increased splicing and expression of the mH2A.1.2 transcript, with a concomitant downregulation of mH2A.1.1. This switch from mH2A.1.1 to mH2A.1.2 affects cell proliferation in various cancer cell lines [13]. Recently, the RNA helicases Ddx17 and Ddx5 were also identified in the regulation of mH2A1 alternative splicing in breast cancer [11].
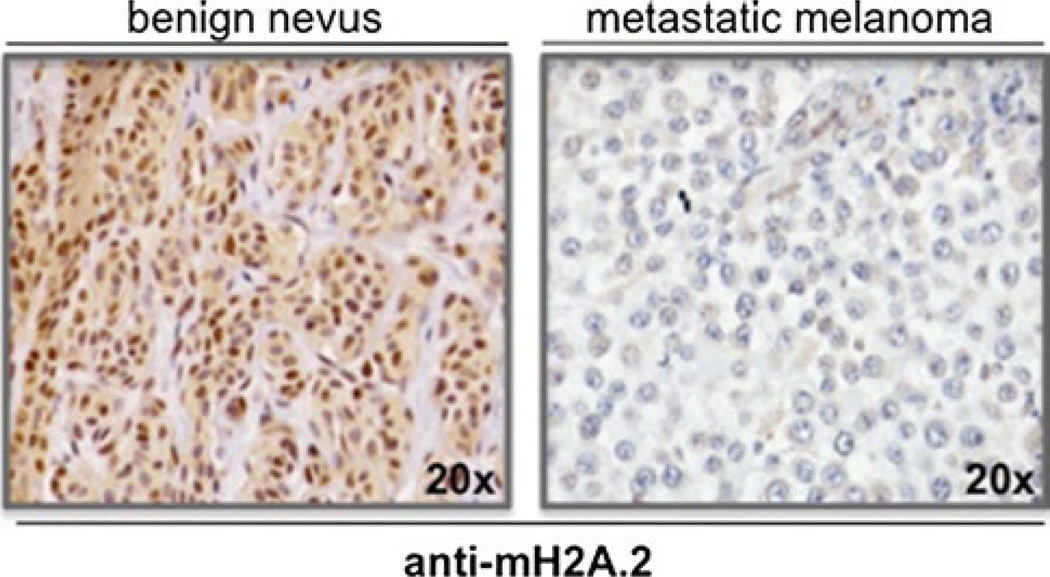
Our group reported that downregulation of mH2A isoforms is regulated by transcriptional loss of both mH2A.1 and mH2A.2 in malignant melanoma [12]. In the case of mH2A.2, silencing was through promoter DNA hypermethylation. The loss of mH2A isoforms significantly correlates with melanoma malignancy, as mH2A.1 and mH2A.2 are downregulated in metastatic cell lines and lesions from metastatic patients as compared to primary tumors and benign nevi (Fig. 4). Furthermore, with loss- and gain-of-function studies in vitro and in vivo, we showed that mH2A loss drives melanoma progression, at least in part, through deregulation of CDK8 (Cyclin-Dependent-Kinase 8), which has been implicated as an oncogene in colorectal carcinoma [12, 182]. Collectively, these studies implicate mH2A as a driver in cancer largely due to its effect on cell proliferation [9, 10, 12, 13] and on tumor invasiveness [11, 12].
Fig. 4.
mH2A.2 loss correlates with melanoma malignancy. Immunostaining of mH2A.2 in benign nevi and metastatic melanoma specimens (representative samples shown at 20X magnification). mH2A.2 is visualized using DAB (brown) and nuclei by haematoxylin (blue). Note loss of staining in metastatic melanoma cells
mH2A has also been described as a marker of senescence [153], and, as previously discussed in this review, OIS is considered a barrier to malignant transformation [92]. Interestingly, all three mH2A isoforms are enriched in SAHFs [153]. Furthermore, using an oncogenic K-RasV12 OIS mouse model [183], Sporn and colleagues [10] found that mH2A1.1 levels were higher in pre-malignant lung adenomas compared to malignant lung adenocarcinomas. Given that pre-malignant adenomas were also found to express high levels of senescence markers such as β-galactosidase and p16 compared to malignant adenocarcinomas [184], these findings reinforce the notion that mH2A loss may contribute to tumor progression through bypass of cellular senescence. In addition, human nevi display some features of OIS, most likely triggered by the activating BRAF V600E mutation [185]. Nevi remain growth-arrested for decades, but can eventually develop into melanomas. It remains unknown if/how benign nevus cells escape OIS; however, we speculate that loss of mH2A may contribute to senescence bypass and, thus, melanoma progression.
Finally, as mentioned earlier, we recently reported ATRX to be a negative regulator of mH2A deposition [159]. Numerous recent reports have described loss-of-function mutations and deletions in this chromatin remodeler in a variety of tumor types (see “H3.3” below, and Table 3). Thus, while the link between ATRX and mH2A in cancer remains unclear, the possibility of mH2A deregulation through ATRX mutation is very enticing and awaits further investigation.
H3 variants
The H3 variants have been of increasing interest in recent years, with numerous studies aimed at deciphering their function(s), chromatin deposition machinery, and genome-wide occupancy patterns, as well as the discovery of novel family members. To date, eight histone H3 variants (H3.1, H3.2, H3.3, H3t (H3.4), H3.5, H3.X, H3.Y, and CENP-A) have been reported in humans [186]. H3.1 and H3.2 are generally the most abundant H3 proteins in the cell and are often referred to as “canonical” H3. While some H3s are ubiquitously expressed, others are expressed in a tissue- or organism-specific fashion. For example, H3.1t and H3.5 appear to be testis-specific [187, 188], while H3.X and H3.Y are primate-specific [189]. Like the H2A variants, the H3 variant members differ in their primary sequence, chromatin localization, deposition timing and machinery, and importantly their functions within the chromatin template (Table 1). The H3 histones are extensively decorated by PTMs, many of which have been well studied. These can occur in the context of various cellular processes such as transcription, mitosis, meiosis and heterochromatin formation. The reader is referred to other reviews for an in-depth summary of H3 PTMs [190, 191]. For the purpose of this review, we will discuss the centromeric histone H3 variant CENP-A (CENtromere Protein-A) and H3.3, both of which have been reported to play a role in cancer.
CENP-A
CENP-A structure and function
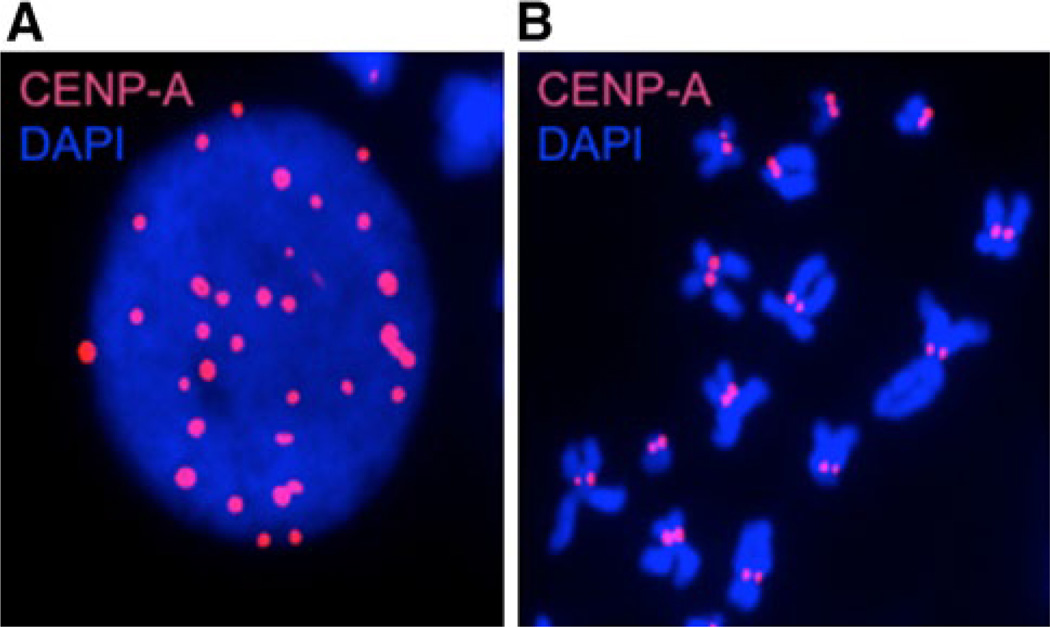
The centromere is known as the chromosomal locus that ensures fidelity in genome transmission at cell division. The H3 variant CENP-A is a highly specialized histone variant which localizes to the centromere in all eukaryotes (Figs. 1, 5). As the centromere-specific variant (Fig. 5), CENP-A has two major functions. First, it is necessary and sufficient for centromere formation and maintenance [192, 193], and second, it forms the platform onto which the kinetochore assembles, mediating chromosome segregation [194–196]. While CENP-A is present at a very low stoichiometry in the cell compared to canonical H3, CENPA is an essential gene demonstrated by CENPA null mice, which fail to survive beyond 6.5 dpc due to severe mitotic defects [197]. It has been proposed that CENP-A is an epigenetic mark that is propagated in each cell division to maintain centromere identity [198]. This is supported by the observation that all active centromeres contain CENP-A independent of the underlying DNA sequence, while inactivated centromeres do not [196].
Fig. 5.
Localization of CENP-A by immunofluorescence. a CENP-A staining (red) in interphase nucleus, foci represent centromeres. b CENP-A staining (red) of metaphase spreads. CENP-A localizes to the kinetochore of each sister chromatid. All imaging performed in nuclei isolated from a lymphoblast cell line at 100X magnification. DNA is stained with DAPI (blue)
CENP-A is a highly divergent H3 family member, with only ~60 % similarity to histone H3 within the HFD, and a distinct N-terminal tail [199]. While the global structures of H3- and CENP-A-containing nucleosomes are quite similar [200], two regions of CENP-A differ from canonical H3. These include the αN helix and the L1 loop, as highlighted in Fig. 6. The αN helix of CENP-A is at least one helical turn shorter than H3 with a structurally disordered region N-terminal to this helix, while the L1 loop has two additional residues [186] (Fig. 6). Both these differences are likely to contribute to the unique centromeric chromatin architecture [186, 192, 198] and to the specific binding of other centromeric proteins [195, 201, 202]. DNA is wrapped around CENP-A nucleosomes in a conventional left-handed manner, but, unlike H3, only the central 121 bp of DNA are visible in the crystal structure of in vitro-reconstituted CENP-A nucleosomes. This might indicate a more flexible DNA conformation at the entry and exit regions, resulting in more closely compacted nucleosomes and, thus, condensed chromatin arrays relative to the canonical H3 [186, 198, 201, 203].
Fig. 6.
CENP-A- and H3-containing nucleosomes are structurally distinct. a Crystal structure of nucleosomes containing canonical H3 (green) or CENP-A (orange) by in silico homology models are super-imposed. Boxes highlight differences in their L1 loop structures (b) and in the organization of the N-terminal helix with regard to nucleosomal DNA (c). H2A, H2B, and H4 are in blue and DNA is in beige. b The L1 loop of CENP-A contains two additional amino acids and protrudes further from the nucleosome than that of canonical H3. c The N-terminal helix of CENP-A is shorter than the corresponding region in H3, and the DNA at this region (dashed line) may be more flexible in CENP-A nucleosomes
In vitro studies of recombinant nucleosomes have established that CENP-A forms an octameric nucleosome containing two copies of CENP-A [186, 198, 203]. However, studies from various eukaryotic species have put forth competing models for the structure of the CENP-A-containing particles in vivo [192]. One of the most intriguing is the hemisome model, containing only one copy of each histone CENP-A, H4, H2A, and H2B [204, 205]. A recent study from our group showed that the predominant form of CENP-A particles at functional centromeres in asynchronous human cells is an octamer with loose super helical termini [280]. Studies using synchronized cells have suggested that CENP-A-containing particles are cell cycle-regulated, existing as octamers in S phase and hemisomes during the other stages of the cell cycle [206, 207]. This would potentially have ramifications for centromere function throughout the cell cycle, in particular for the inheritance of centromeric domains after replication.
The mechanisms of how newly synthesized CENP-A is targeted, deposited, and maintained at the centromere have long been a mystery. However, recent studies have begun to unravel these mechanisms. CENP-A expression begins late in S phase and reaches maximum levels in late G2, suggestive that it may be driven by a cell cycle-regulated promoter element [208]. Using a SNAP tag approach, which can distinguish between old and newly synthesized histones, [209], Jansen et al. [210] demonstrated that the loading of newly synthesized CENP-A, at least in metazoans, occurs independently of DNA replication, during a short window in late mitosis/early G1. In order to identify the CENP-A deposition machinery, a large-scale purification of CENP-A-interacting proteins in a chromatin-free context was performed. As a result, HJURP (Holliday Junction Recognition Protein) was identified as a CENP-A chaperone [211, 212]. Functional studies demonstrated that loss of HJURP led to a dramatic reduction in CENP-A at centromeres and impaired deposition of newly synthesized CENP-A, resulting in mitotic defects [211].
Recognition and targeting of CENP-A to the centromere occurs via the CENP-A Targeting Domain (CATD) [212]. An H3 chimera containing the CATD was shown to be delivered to the centromere [213], and to co-purify with HJURP from mammalian cells [212]. Moreover, the CATD is also important for the assembly of chromatin at centromeres and for the rigid intranucleosomal dynamics specifically required to generate the unique centromeric chromatin architecture [214]. Taken together, these data suggest that HJURP is not only a histone chaperone but may also be important for stabilizing the CENP–A/H4 complex [201].
In addition, the Mis18 complex was shown to play a critical role in centromere targeting of CENP-A [215]. Although the mechanism remains obscure, Mis18 deficiency results in mislocalization of CENP-A, which, in turn, leads to early embryonic lethality in mice [216]. It has recently been proposed that the CENP-A assembly machinery is responsible for the cell cycle-regulated incorporation of CENP-A. Indeed, while components of the Mis18 complex are present throughout most of the cell cycle, their activity seems to be tightly controlled by CDK1 and CDK2, thus coordinating the timing of DNA replication, cell division, and subsequent centromere maturation [217].
In mammalian cells, CENP-A is phosphorylated on serine 7 (S7) by Aurora B kinase during prophase, and then dephosphorylated during anaphase [218] (Fig. 1). An S7 CENP-A mutant is still targeted to the centromere; however, it shows defects in kinetochore function and chromosome alignments at both prometaphase and cytokenesis [219, 220]. In addition, lysine 124, which is located at the interface between the HFD and DNA, was recently reported to be acetylated at the G1/S transition, potentially causing structural alterations in the CENP-A-containing nucleosome [206].
While CENP-A is recognized as both an epigenetic mark maintaining centromere identity and a platform onto which the kinetochore is formed, controversy still exists in regards to the in vivo composition of CENP-A-containing particles. We look forward to future studies that elucidate the unique characteristics of the centromeric chromatin environment, which will also be critical for understanding its role in cancer (see below).
CENP-A: genome stability and cancer
Chromosomal aneuploidy is a common hallmark of human solid tumors, resulting mainly from chromosome mis-segregation during mitosis. Given the critical role of CENP-A in shaping the centromere/kinetochore structures, it is not surprising that its deregulation might lead to chromosomal instability and ultimately to cancer (Table 2). Defects in CENP-A functions can be caused by alterations in expression levels, which in turn alters the stoichiometry between CENP-A and HJURP [221] leading to CENP-A mis-targeting [222]. Accumulating evidence indicates that CENP-A is upregulated in a number of cancers, such as colorectal cancer [222], lung adenocarcinoma [223], invasive testicular germ cell tumors [224], breast cancer [225], and HepatoCellular Carcinoma (HCC) [226] (Table 2). HJURP is also overexpressed in certain cancers, including lung [227] and breast cancers [228] (Table 3). In breast cancer, e.g., increased HJURP and CENP-A mRNA levels are significantly associated with decreased survival rate [228].
Besides the deregulation of centromeres, additional mechanisms might be involved in CENP-A-mediated tumor formation. Indeed, by manipulating its expression in a human HCC cell line, CENP-A was shown to promote cell proliferation and to inhibit apoptosis through modulating the expression of many cell cycle and apoptotic genes [229]. Thus, we speculate that overexpression of CENP-A might lead to its mis-localization along chromosome arms, which in turn alters cellular gene expression and/or other chromosomal dynamics. However, the mechanisms underlying increased levels of CENP-A in cancer cells remain to be determined, as do the functional readouts of such upregulation.
H3.3
H3.3 structure and function
H3.3 is highly conserved in evolution, from yeast (where it is the only non-centromeric H3 variant) to human [26]. H3.3 is encoded by two genes; H3F3A on chromosome 1 and H3F3B on chromosome 17. The H3.3 genes code for an identical H3.3 protein, but harbor different untranslated regions, and thus their expression levels vary with cell type, tissue, and stage of development [230–232]. Deletion of the H3F3A gene results in phenotypes that range from lethality in the mouse to sterility in organisms such as D. melanogaster and T. thermophila [233–235]. Mice harboring a hypomorphic H3F3A allele exhibited partial neonatal lethality and significant defects in adult fertility [236]. The latter phenotype is likely due to H3.3’s role in chromosome segregation during spermatogenesis [110, 237, 238].
H3.3 differs from canonical H3.1 and H3.2 by five and four amino acids, respectively (Fig. 1). Four of these residues cluster in the interface between H3.3 and H4. This region is accessible to regulatory factors and mediates the specific interaction with distinct histone chaperones (discussed further below) [186, 239–241]. The other unique residue is the N-terminal serine 31, which is phosphorylated during mitosis. This phosphorylation confers a unique PTM to H3.3, potentially differentiating it from canonical H3 in regard to cell division dynamics [242, 243]. Besides phosphorylation, H3.3 may be subject to a distinct PTM pattern as compared to canonical to H3.1 and H3.2; however, an extensive characterizations of the PTMs of histone H3s is beyond the scope of this review [243, 244].
While the crystal structure analyses of nucleosomes containing H3.3 revealed a very similar structure as canonical H3-containing nucleosomes, those with H3.3 are less stable [186]. In particular, they dissociate from H2A–H2B dimers at lower salt concentrations [79], and intriguingly H3.3/H2A.Z-containing nucleosomes are more unstable than H3.3/H2A. Indeed, H3.3/H2A.Z nucleosomes are present at promoters and enhancers of highly expressed genes, potentially creating a “variant” chromatin environment accessible to TFs [80]. This suggests that the combination of different histone variants in the same nucleosome allows for increased complexity or fine-tuning of transcriptional regulation [79, 80].
Consistent with destabilized nucleosomes, H3.3 has been described as a replacement histone associated with transcriptionally active chromatin [245, 246]. Deposition of H3.3 occurs mainly at promoters and gene bodies of actively transcribed genes, and at regulatory sites of both active and inactive genes [80, 168, 247, 248]. ChIP–chip and ChIP-seq approaches in mammalian and chicken cells revealed that H3.3 is incorporated throughout genes and upstream regulatory regions upon induction of transcription and is therefore also associated with transcriptional elongation [80, 248, 249].
Interestingly, recent studies suggest that, besides associating with transcriptionally competent chromatin, H3.3 is also incorporated into heterochromatic regions of the genome, including telomeres and pericentric regions (discussed below) [167–169, 250]. Furthermore, H3.3 has been implicated as a ‘placeholder H3′ for CENP-A at centromeres, which occurs upon CENP-A dilution during DNA replication [251]. H3.3 was also proposed to be involved in the epigenetic maintenance of chromatin states (‘epigenetic memory’) in Xenopus nuclear transplantation studies [252]. Indeed, H3.3/H4 and not H3.1/H4 tetramers have been observed to split during DNA replication, indicating a potential mode for transmitting epigenetic marks [253, 254]. Taken together, these results suggest that H3.3 has numerous and context-dependent functions.
Unlike canonical H3.1 and H3.2, which are expressed during S phase and incorporated into chromatin in a replication-dependent fashion, H3.3 is constitutively expressed and deposited into chromatin independently of DNA replication. Interestingly, swapping any of the four unique residues of the HFD in H3.1 to their counterparts in H3.3 (Fig. 1) in Drosophila allows H3.1 incorporation into chromatin outside of S phase [255]. This strongly suggests that specific chaperone interactions are dedicated to the deposition of each H3 variant [239, 240].
Multiple chaperones have been implicated in the replication-independent chromatin deposition of H3.3. These include HIRA (HIstone cell cycle Regulation-defective homolog A), DEK, ATRX, and Daxx (death domain-associated protein) [167–169, 256, 257]. HIRA was described to deposit H3.3 throughout the cell cycle across the genome [241, 257]. Consistent with this finding, deposition of H3.3 at promoters and in the body of active genes is greatly impaired upon HIRA loss in ESC [168], and HIRA knockout mice display embryonic lethality [258]. However, it has recently been reported that ATRX, and its co-factor Daxx [259, 260], regulate H3.3 deposition at non-coding genomic loci, including telomeres and pericentric heterochromatin [167–169]. This finding clearly suggests that the deposition of H3.3 is mediated by distinct factors at specific genomic regions. While the functional significance of H3.3 deposition at heterochromatic regions remains to be explored, loss of H3.3 or ATRX results in decreased transcription of pericentric repeats, suggesting its involvement in active transcription at this region [167]. Of note, two groups recently resolved the crystal structure of H3.3-containing nucleosomes in complex with Daxx [239, 240]. By elegant residue swapping experiments, these reports demonstrated that unique residues of H3.3 are critical in mediating the Daxx interaction, thereby substantiating the importance of these residues in the differential regulation of H3 variants [239, 240] (Fig. 1).
Our group showed that ATRX also acts to negatively regulate mH2A association with chromatin, however, in a Daxx-independent fashion [159]. Thus, while distinct factors are required for site-specific deposition of H3.3, they may also regulate the incorporation of other histone variants to create a unique chromatin environment. Furthermore, because Daxx interacts with both ATRX and DEK [256, 259, 260], we can envision that Daxx gives ATRX specificity towards H3.3 over mH2A, and in turn ATRX or DEK differentially regulate H3.3 deposition at distinct genomic regions. We also note that these chaperones have post-translationally modified forms, which may also regulate their activity and specificity [256, 261]. In summary, the H3.3 histone deposition machinery is highly complex, and we look forward to future studies that facilitate a deeper understanding of these chaperones and H3.3 genome-wide chromatin deposition.
H3.3: a histone variant mutated in cancer
H3.3 and its chaperones have clearly been implicated in cancer. While H3.3 has been reported to be overexpressed in human tumors [262], the advent of high throughput sequencing has uncovered mutations in the H3F3A gene in an unbiased manner. Two recent exome sequencing studies surprisingly uncovered H3.3 mutations in pediatric Glio-Blastoma Multiforme (GBM) [15, 16] and Diffuse Intrinsic Pontine Gliomas (DIPGs) [16]. Of note, this is the first evidence of a histone variant-encoding gene mutated in human cancer. Missense mutations in the H3F3A gene were observed in 31 % of GBMs leading to amino acid substitutions of two critical residues within the histone tail (K27M and G34V/R), which are either subject to PTMs themselves (H3.3K27) or lie in very close proximity to other modified residues (H3.3K36) [15]. Both these residues play a key role in gene expression programs.
In a further study aimed at identifying somatic mutations in DIPG and non-brainstem pediatric glioblastomas (non-BS-PGs), 78 % of DIPGs and 22 % of non-BS-PGs harbored a K27M substitution in H3F3A or in the HISTH3.1 gene encoding H3.1. An additional 14 % of the non-BS-PGs were found to have a G34R substitution in the H3F3A gene [16]. Notably, these heterozygous mutations occur only in the H3F3A gene, although both H3F3A and H3F3B encode the same H3.3 protein. Because the mutations always encode the same amino acid substitutions, it suggests a gain-of-function phenotype [15]. In fact, the K27M mutation has recently been reported to directly interfere with EZH2 activity, resulting in reduced global levels of H3K27me3 [263].
Intriguingly, the H3.3 chaperones have also emerged as critical players in human cancer. For example, in response to angiogenic signals, HIRA is induced in endothelial cells leading to incorporation of acetylated H3.3K56 (a marker of active transcription) in the pro-angiogenic VEGFR1 (Vascular Endothelial Growth Factor Receptor 1) gene, and thus promotes neovascularization of the tumor [264]. Akin to the unbiased high throughput sequencing studies mentioned above, ATRX and/or Daxx mutations (and deletions) have been revealed in a variety of tumor types including pancreatic NeuroEndocrine Tumors (panNETs) [19], neuroblastoma [265], and GBM [15, 18], as well as the rare Alpha-Thalassemia MyeloDysplasia Syndrome (ATMDS) [266]. Intriguingly, most of these tumor types arise from neural crest-derived cells, suggesting a common mechanism underlying formation of these tumor types [267].
The PanNET exome study revealed that 43 % of the tumors harbored inactivating mutations in either the ATRX or Daxx genes, and appear to be independent of each other. Interestingly, the ATRX and Daxx mutations were all inframe deletions or nonsense mutations, suggesting a tumor suppressor role [19]. PanNET tumors harboring ATRX or Daxx mutations displayed Alternative Lengthening of Telomeres (ALT) and had lost the nuclear expression of either Daxx or ATRX, suggestive of a role exerted by ATRX in telomeric maintenance [18]. A high correlation between ATRX mutations and ALT was also found in Central Nervous System (CNS) tumors, and in known ALT-positive cell lines [15, 18, 265, 267]. We can envision a scenario where ATRX and Daxx mutations impair the chromatin state at telomeres potentially through deregulation of H3.3 and mH2A, leading to telomere destabilization and facilitating ALT. Yet, at the same time, not all ATRX mutant tumors exhibit ALT [16], and thus the ATRX/ALT connection at this point is merely correlative and not causal [268, 269].
The study by Schwartzentruber et al. showed that somatic loss-of-function mutations of ATRX and Daxx in GBM were frequently coupled with H3.3 mutations in the same tumors; 100 % of H3.3 G34 V /R mutations also carry an ATRX mutation. Some H3.3/ATRX-mutated GBM samples (exclusively pediatric and young adults GBMs) exhibited ALT and changes in expression profiling of brain-specific developmental genes, consistent with a dual role of H3.3 in shaping telomeric chromatin and in regulating gene expression [15, 270]. Similarly, the H3.3 K27 M mutation found in DIPG is associated with ATRX mutations, mainly in older children [16].
Whole-genome sequencing on primary neuroblastomas identified structural variants and large deletions in ATRX that led to loss of mRNA expression [265]. Interestingly, none of these patients had Daxx or H3.3 mutations, indicating that ATRX alone could be potential driver of neuroblastoma.
Collectively, it is now evident that H3.3 regulation is perturbed in cancer. Whether it is through H3.3 mutations directly or mutations in its chaperones, the challenge is now to understand the consequences for tumor cells. We anticipate that deciphering the alterations at the chromatin level by genome-wide analyses of histone variant deposition in the context of these tumors will help to elucidate the underlying biology. Given the implication of ATRX both in telomere homeostasis and in regulation of H3.3 and mH2A association with chromatin, it is now of interest to determine how loss of functional ATRX might contribute to tumorigenesis via telomere dysfunction, or other as yet unidentified cellular functions [269].
Conclusions and perspectives
Research in recent years has focused heavily on the H2A and H3 histone variant families, leading to identification of the machinery responsible for their chromatin deposition, consequences of their incorporation, biochemical properties of variant-incorporated nucleosomes, and biological functions, as well as their genome-wide localization patterns in both physiological and disease states. However, some fundamental questions in regard to histone biology still remain unanswered, as discussed throughout the review. As new technologies arise, coupled to creative and novel experimental approaches, some of these long-standing questions will be resolved.
Both H2A and H3 histone variants have been broadly shown to be key, and often essential, players in orchestrating cell differentiation and organism development [32, 176, 177, 197, 236, 271]. Due to our better understanding of cancer biology, the similarities to developmental biology are becoming evident. It is therefore not completely unexpected that the role histone variants play in cancer progression may be cell type- and context-dependent.
The last few years have witnessed an explosion of studies focused on the role of histone variants in cancer. Moreover, exome sequencing has recently been utilized to uncover somatic mutations in patient samples, and these studies have unraveled alterations in histone chaperones, chromatin remodeling complexes, and histones themselves [15, 16, 18, 19, 265, 266]. However, as a field, we have just scratched the surface in understanding the role of histone variants in cancer initiation and/or progression. Furthermore, we point out that, while some histone variants were discovered decades ago [23, 25, 142], novel variants and isoforms thereof have only recently been characterized [38, 81, 189]. However, while little is known about their biology, studies detecting their expression in human tumors are strongly suggestive of a critical, yet unknown, role in cancer development and progression; for example, newly identified H3 variants H3.X and H3.Y [189]. Thus, we believe there is a vast and intriguing biology of histone variants in cancer still awaiting discovery.
Interestingly, histone variants and histone chaperones implicated in cancer can now be ascribed to several classical types of genes associated with cancer. For example, by promoting cell proliferation, H2A.Z and SRCAP are suggested to behave like oncogenes [14, 20]. On the other hand, mH2A might be assigned to the class of tumor suppressor genes [9–13], having an inhibitory effect on growth and metastasis, while H2A.X and CENP-A display features of caretakers, which act to prevent genomic instability [133–135, 222].
We note here that epigenetic mechanisms other than histone variant incorporation/exchange have also been implicated in human cancer, i.e. DNA methylation and histone PTMs [272, 273]. In order to gain a broader picture of the contribution of epigenetics to cancer in general, studying the crosstalk between histone variants and other epigenetic mechanisms is essential. For example, a handful of studies have addressed the anti-correlation of H2A.Z deposition and DNA methylation, while others implicate a positive correlation between certain histone variants with histone PTMs [15, 16, 48, 274].
Given their fundamental role in shaping chromatin structure, histone variants (and their chaperones) likely constitute important biomarkers for diagnosis or prognosis and may even represent therapeutic targets in cancer in patients. To date, studies have demonstrated the potential prognostic utility of histone variants for multiple cancers including those of lung, breast, and colon [8–10]. Cellular levels of histone variants may also predict responses to certain chemotherapeutic agents [136, 137], serving as predictive biomarkers that could inform clinical decisions regarding courses of therapy. The main challenge now is to mechanistically understand how histone variant deregulation can contribute to cancer development and progression, and ultimately to developing novel therapeutics. Owing to the fact that histone incorporation into chromatin can potentially be reversed with small molecules that would block the relevant histone-to-histone chaperone interaction, or those that inhibit the activity of chaperones themselves, histone variant biology holds incredible translational potential. Based on the recent success of ‘epigenetic drugs’ [275–278], we anticipate and are hopeful that additional compounds, including those that regulate histone variants, will be developed and find their way to the clinic.
Acknowledgments
The authors thank Zulekha Qadeer and Alexandre Gaspar-Maia for critically reading this manuscript. The authors thank Matthew O’Connell for microscopy assistance and Pablo DeIoannes for assistance with in silico homology modeling. This work is supported by an NCI T32-CA078207 to L.F.D, a Melanoma Research Development Award (Department of Dermatology, Mount Sinai) to C.V., and The Ellison Medical Foundation New Scholar Award, Association for International Cancer Research, Hirschl/Weill-Caulier Research Award and NCI/NIH R01CA154683 to E.B.
Abbreviations
- ChIP
Chromatin ImmunoPrecipitation
- DDR
DNA Damage Response
- DSB
Double-Strand Break
- ESC
Embryonic Stem Cell
- HFD
Histone Fold Domain
- IF
ImmunoFluorescence
- OIS
Oncogene-Induced Senescence
- PTM
Post-Translational Modification
- TF
Transcription Factor
- TSS
Transcription Start Site
Contributor Information
Chiara Vardabasso, Department of Oncological Sciences, Icahn School of Medicine at Mount Sinai, 1425 Madison Avenue, New York, NY 10029, USA; Department of Dermatology, Icahn School of Medicine at Mount Sinai, 1425 Madison Avenue, New York, NY 10029, USA.
Dan Hasson, Department of Oncological Sciences, Icahn School of Medicine at Mount Sinai, 1425 Madison Avenue, New York, NY 10029, USA; Graduate School of Biomedical Sciences, Icahn School of Medicine at Mount Sinai, 1425 Madison Avenue, New York, NY 10029, USA.
Kajan Ratnakumar, Department of Oncological Sciences, Icahn School of Medicine at Mount Sinai, 1425 Madison Avenue, New York, NY 10029, USA; Graduate School of Biomedical Sciences, Icahn School of Medicine at Mount Sinai, 1425 Madison Avenue, New York, NY 10029, USA.
Chi-Yeh Chung, Department of Oncological Sciences, Icahn School of Medicine at Mount Sinai, 1425 Madison Avenue, New York, NY 10029, USA; Graduate School of Biomedical Sciences, Icahn School of Medicine at Mount Sinai, 1425 Madison Avenue, New York, NY 10029, USA.
Luis F. Duarte, Department of Oncological Sciences, Icahn School of Medicine at Mount Sinai, 1425 Madison Avenue, New York, NY 10029, USA Department of Dermatology, Icahn School of Medicine at Mount Sinai, 1425 Madison Avenue, New York, NY 10029, USA; Graduate School of Biomedical Sciences, Icahn School of Medicine at Mount Sinai, 1425 Madison Avenue, New York, NY 10029, USA.
Emily Bernstein, Email: emily.bernstein@mssm.edu, Department of Oncological Sciences, Icahn School of Medicine at Mount Sinai, 1425 Madison Avenue, New York, NY 10029, USA; Department of Dermatology, Icahn School of Medicine at Mount Sinai, 1425 Madison Avenue, New York, NY 10029, USA.
References
- 1.Andrews AJ, Luger K. Nucleosome structure(s) and stability: variations on a theme. Annu Rev Biophys. 2011;40:99–117. doi: 10.1146/annurev-biophys-042910-155329. [DOI] [PubMed] [Google Scholar]
- 2.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein E, Hake SB. The nucleosome: a little variation goes a long way. Biochem Cell Biol. 2006;84:505–517. doi: 10.1139/o06-085. [DOI] [PubMed] [Google Scholar]
- 4.Talbert PB, Henikoff S. Histone variants—ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol. 2010;11:264–275. doi: 10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- 5.Das C, Tyler JK, Churchill ME. The histone shuffle: histone chaperones in an energetic dance. Trends Biochem Sci. 2010;35:476–489. doi: 10.1016/j.tibs.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hondele M, Ladurner AG. The chaperone-histone partnership: for the greater good of histone traffic and chromatin plasticity. Curr Opin Struct Biol. 2011;21:698–708. doi: 10.1016/j.sbi.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Park YJ, Luger K. Histone chaperones in nucleosome eviction and histone exchange. Curr Opin Struct Biol. 2008;18:282–289. doi: 10.1016/j.sbi.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hua S, Kallen CB, Dhar R, Baquero MT, Mason CE, Russell BA, Shah PK, Liu J, Khramtsov A, Tretiakova MS, Krausz TN, Olopade OI, Rimm DL, White KP. Genomic analysis of estrogen cascade reveals histone variant H2A.Z associated with breast cancer progression. Mol Syst Biol. 2008;4:188. doi: 10.1038/msb.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sporn JC, Jung B. Differential regulation and predictive potential of macroH2A1 isoforms in colon cancer. Am J Pathol. 2012;180:2516–2526. doi: 10.1016/j.ajpath.2012.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sporn JC, Kustatscher G, Hothorn T, Collado M, Serrano M, Muley T, Schnabel P, Ladurner AG. Histone macroH2A isoforms predict the risk of lung cancer recurrence. Oncogene. 2009;28:3423–3428. doi: 10.1038/onc.2009.26. [DOI] [PubMed] [Google Scholar]
- 11.Dardenne E, Pierredon S, Driouch K, Gratadou L, Lacroix-Triki M, Espinoza MP, Zonta E, Germann S, Mortada H, Villemin JP, Dutertre M, Lidereau R, Vagner S, Auboeuf D. Splicing switch of an epigenetic regulator by RNA helicases promotes tumor-cell invasiveness. Nat Struct Mol Biol. 2012;19:1139–1146. doi: 10.1038/nsmb.2390. [DOI] [PubMed] [Google Scholar]
- 12.Kapoor A, Goldberg MS, Cumberland LK, Ratnakumar K, Segura MF, Emanuel PO, Menendez S, Vardabasso C, Leroy G, Vidal CI, Polsky D, Osman I, Garcia BA, Hernando E, Bernstein E. The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature. 2010;468:1105–1109. doi: 10.1038/nature09590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Novikov L, Park JW, Chen H, Klerman H, Jalloh AS, Gamble MJ. QKI-mediated alternative splicing of the histone variant macroH2A1 regulates cancer cell proliferation. Mol Cell Biol. 2011;31:4244–4255. doi: 10.1128/MCB.05244-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Svotelis A, Gevry N, Grondin G, Gaudreau L. H2A.Z overexpression promotes cellular proliferation of breast cancer cells. Cell Cycle. 2009;9:364–370. doi: 10.4161/cc.9.2.10465. [DOI] [PubMed] [Google Scholar]
- 15.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, Hovestadt V, Albrecht S, Kool M, Nantel A, Konermann C, Lindroth A, Jäger N, Rausch T, Ryzhova M, Korbel JO, Hielscher T, Hauser P, Garami M, Klekner A, Bognar L, Ebinger M, Schuhmann MU, Scheurlen W, Pekrun A, Frühwald MC, Roggendorf W, Kramm C, Dürken M, Atkinson J, Lepage P, Montpetit A, Zakrzewska M, Zakrzewski K, Liberski PP, Dong Z, Siegel P, Kulozik AE, Zapatka M, Guha A, Malkin D, Felsberg J, Reifenberger G, von Deimling A, Ichimura K, Collins VP, Witt H, Milde T, Witt O, Zhang C, Castelo-Branco P, Lichter P, Faury D, Tabori U, Plass C, Majewski J, Pfister SM, Jabado N. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 16.Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, Qu C, Ding L, Huether R, Parker M, Zhang J, Gajjar A, Dyer MA, Mullighan CG, Gilbertson RJ, Mardis ER, Wilson RK, Downing JR, Ellison DW, Zhang J, Baker SJ St. Jude Children’s research hospital—Washington University Pediatric Cancer Genome Project. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44:251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gevry N, Hardy S, Jacques PE, Laflamme L, Svotelis A, Robert F, Gaudreau L. Histone H2A.Z is essential for estrogen receptor signaling. Genes Dev. 2009;23:1522–1533. doi: 10.1101/gad.1787109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heaphy CM, de Wilde RF, Jiao Y, Klein AP, Edil BH, Shi C, Bettegowda C, Rodriguez FJ, Eberhart CG, Hebbar S, Offerhaus GJ, McLendon R, Rasheed BA, He Y, Yan H, Bigner DD, Oba-Shinjo SM, Marie SK, Riggins GJ, Kinzler KW, Vogelstein B, Hruban RH, Maitra A, Papadopoulos N, Meeker AK. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333:425. doi: 10.1126/science.1207313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, Schulick RD, Tang LH, Wolfgang CL, Choti MA, Velculescu VE, Diaz LA, Jr, Vogelstein B, Kinzler KW, Hruban RH, Papadopoulos N. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331:1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slupianek A, Yerrum S, Safadi FF, Monroy MA. The chromatin remodeling factor SRCAP modulates expression of prostate specific antigen and cellular proliferation in prostate cancer cells. J Cell Physiol. 2010;224:369–375. doi: 10.1002/jcp.22132. [DOI] [PubMed] [Google Scholar]
- 21.Talbert PB, Ahmad K, Almouzni G, Ausio J, Berger F, Bhalla PL, Bonner WM, Cande WZ, Chadwick BP, Chan SW, Cross GA, Cui L, Dimitrov SI, Doenecke D, Eirin-López JM, Gorovsky MA, Hake SB, Hamkalo BA, Holec S, Jacobsen SE, Kamieniarz K, Khochbin S, Ladurner AG, Landsman D, Latham JA, Loppin B, Malik HS, Marzluff WF, Pehrson JR, Postberg J, Schneider R, Singh MB, Smith MM, Thompson E, Torres-Padilla ME, Tremethick DJ, Turner BM, Waterborg JH, Wollmann H, Yelagandula R, Zhu B, Henikoff S. A unified phylogeny-based nomenclature for histone variants. Epigenetics Chromatin. 2012;5:7. doi: 10.1186/1756-8935-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonisch C, Hake SB. Histone H2A variants in nucleosomes and chromatin: more or less stable? Nucleic Acids Res. 2012;40:10719–10741. doi: 10.1093/nar/gks865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allis CD, Glover CV, Bowen JK, Gorovsky MA. Histone variants specific to the transcriptionally active, amitotically dividing macronucleus of the unicellular eukaryote, Tetrahymena thermophila. Cell. 1980;20:609–617. doi: 10.1016/0092-8674(80)90307-4. [DOI] [PubMed] [Google Scholar]
- 24.Allis CD, Richman R, Gorovsky MA, Ziegler YS, Touchstone B, Bradley WA, Cook RG. Hv1 is an evolutionarily conserved H2A variant that is preferentially associated with active genes. J Biol Chem. 1986;261:1941–1948. [PubMed] [Google Scholar]
- 25.W est MH, Bonner WM. Histone 2A, a heteromorphous family of eight protein species. Biochemistry. 1980;19:3238–3245. doi: 10.1021/bi00555a022. [DOI] [PubMed] [Google Scholar]
- 26.Malik HS, Henikoff S. Phylogenomics of the nucleosome. Nat Struct Biol. 2003;10:882–891. doi: 10.1038/nsb996. [DOI] [PubMed] [Google Scholar]
- 27.Carr AM, Dorrington SM, Hindley J, Phear GA, Aves SJ, Nurse P. Analysis of a histone H2A variant from fission yeast: evidence for a role in chromosome stability. Mol Gen Genet. 1994;245:628–635. doi: 10.1007/BF00282226. [DOI] [PubMed] [Google Scholar]
- 28.Jackson JD, Gorovsky MA. Histone H2A.Z has a conserved function that is distinct from that of the major H2A sequence variants. Nucleic Acids Res. 2000;28:3811–3816. doi: 10.1093/nar/28.19.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X, Li B, Gorovsky MA. Essential and non-essential histone H2A variants in Tetrahymena thermophila. Mol Cell Biol. 1996;16:4305–4311. doi: 10.1128/mcb.16.8.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Daal A, Elgin SC. A histone variant, H2AvD, is essential in Drosophila melanogaster. Mol Biol Cell. 1992;3:593–602. doi: 10.1091/mbc.3.6.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iouzalen N, Moreau J, Mechali M. H2A.ZI, a new variant histone expressed during Xenopus early development exhibits several distinct features from the core histone H2A. Nucleic Acids Res. 1996;24:3947–3952. doi: 10.1093/nar/24.20.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Faast R, Thonglairoam V, Schulz TC, Beall J, Wells JR, Taylor H, Matthaei K, Rathjen PD, Tremethick DJ, Lyons I. Histone variant H2A.Z is required for early mammalian development. Curr Biol. 2001;11:1183–1187. doi: 10.1016/s0960-9822(01)00329-3. [DOI] [PubMed] [Google Scholar]
- 33.Coon JJ, Ueberheide B, Syka JE, Dryhurst DD, Ausio J, Shabanowitz J, Hunt DF. Protein identification using sequential ion/ion reactions and tandem mass spectrometry. Proc Natl Acad Sci USA. 2005;102:9463–9468. doi: 10.1073/pnas.0503189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dryhurst D, Ishibashi T, Rose KL, Eirin-Lopez JM, McDonald D, Silva-Moreno B, Veldhoen N, Helbing CC, Hendzel MJ, Shabanowitz J, Hunt DF, Ausió J. Characterization of the histone H2A.Z-1 and H2A.Z-2 isoforms in vertebrates. BMC Biol. 2009;7:86. doi: 10.1186/1741-7007-7-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsuda R, Hori T, Kitamura H, Takeuchi K, Fukagawa T, Harata M. Identification and characterization of the two isoforms of the vertebrate H2A.Z histone variant. Nucleic Acids Res. 2010;38:4263–4273. doi: 10.1093/nar/gkq171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.E irin-Lopez JM, Gonzalez-Romero R, Dryhurst D, Ishibashi T, Ausio J. The evolutionary differentiation of two histone H2A.Z variants in chordates (H2A.Z-1 and H2A.Z-2) is mediated by a stepwise mutation process that affects three amino acid residues. BMC Evol Biol. 2009;9:31. doi: 10.1186/1471-2148-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Draker R, Ng MK, Sarcinella E, Ignatchenko V, Kislinger T, Cheung P. A combination of H2A.Z and H4 acetylation recruits Brd2 to chromatin during transcriptional activation. PLoS Genet. 2012;8:e1003047. doi: 10.1371/journal.pgen.1003047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonisch C, Schneider K, Punzeler S, Wiedemann SM, Bielmeier C, Bocola M, Eberl HC, Kuegel W, Neumann J, Kremmer E, Leonhardt H, Mann M, Michaelis J, Schermelleh L, Hake SB. H2A.Z.2.2 is an alternatively spliced histone H2A.Z variant that causes severe nucleosome destabilization. Nucleic Acids Res. 2012;40:5951–5964. doi: 10.1093/nar/gks267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.W ratting D, Thistlethwaite A, Harris M, Zeef LA, Millar CB. A conserved function for the H2A.Z C terminus. J Biol Chem. 2012;287:19148–19157. doi: 10.1074/jbc.M111.317990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suto RK, Clarkson MJ, Tremethick DJ, Luger K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat Struct Biol. 2000;7:1121–1124. doi: 10.1038/81971. [DOI] [PubMed] [Google Scholar]
- 41.Luk E, Ranjan A, Fitzgerald PC, Mizuguchi G, Huang Y, Wei D, Wu C. Stepwise histone replacement by SWR1 requires dual activation with histone H2A.Z and canonical nucleosome. Cell. 2010;143:725–736. doi: 10.1016/j.cell.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.W eber CM, Henikoff JG, Henikoff S. H2A.Z nucleosomes enriched over active genes are homotypic. Nat Struct Mol Biol. 2010;17:1500–1507. doi: 10.1038/nsmb.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nekrasov M, Amrichova J, Parker BJ, Soboleva TA, Jack C, Williams R, Huttley GA, Tremethick DJ. Histone H2A.Z inheritance during the cell cycle and its impact on promoter organization and dynamics. Nat Struct Mol Biol. 2012;19:1076–1083. doi: 10.1038/nsmb.2424. [DOI] [PubMed] [Google Scholar]
- 44.Park YJ, Dyer PN, Tremethick DJ, Luger K. A new fluorescence resonance energy transfer approach demonstrates that the histone variant H2AZ stabilizes the histone octamer within the nucleosome. J Biol Chem. 2004;279:24274–24282. doi: 10.1074/jbc.M313152200. [DOI] [PubMed] [Google Scholar]
- 45.Bruce K, Myers FA, Mantouvalou E, Lefevre P, Greaves I, Bonifer C, Tremethick DJ, Thorne AW, Crane-Robinson C. The replacement histone H2A.Z in a hyperacetylated form is a feature of active genes in the chicken. Nucleic Acids Res. 2005;33:5633–5639. doi: 10.1093/nar/gki874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Millar CB, Xu F, Zhang K, Grunstein M. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 2006;20:711–722. doi: 10.1101/gad.1395506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ren Q, Gorovsky MA. Histone H2A.Z acetylation modulates an essential charge patch. Mol Cell. 2001;7:1329–1335. doi: 10.1016/s1097-2765(01)00269-6. [DOI] [PubMed] [Google Scholar]
- 48.V aldes-Mora F, Song JZ, Statham AL, Strbenac D, Robinson MD, Nair SS, Patterson KI, Tremethick DJ, Stirzaker C, Clark SJ. Acetylation of H2A.Z is a key epigenetic modification associated with gene deregulation and epigenetic remodeling in cancer. Genome Res. 2011;22:307–321. doi: 10.1101/gr.118919.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sarcinella E, Zuzarte PC, Lau PN, Draker R, Cheung P. Monoubiquitylation of H2A.Z distinguishes its association with euchromatin or facultative heterochromatin. Mol Cell Biol. 2007;27:6457–6468. doi: 10.1128/MCB.00241-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jackson V, Chalkley R. Histone synthesis and deposition in the G1 and S phases of hepatoma tissue culture cells. Biochemistry. 1985;24:6921–6930. doi: 10.1021/bi00345a026. [DOI] [PubMed] [Google Scholar]
- 51.Wu RS, Tsai S, Bonner WM. Patterns of histone variant synthesis can distinguish G0 from G1 cells. Cell. 1982;31:367–374. doi: 10.1016/0092-8674(82)90130-1. [DOI] [PubMed] [Google Scholar]
- 52.Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J, Brickner JH. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 2007;5:e81. doi: 10.1371/journal.pbio.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meneghini MD, Wu M, Madhani HD. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112:725–736. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- 54.Greaves IK, Rangasamy D, Ridgway P, Tremethick DJ. H2A.Z contributes to the unique 3D structure of the centromere. Proc Natl Acad Sci USA. 2007;104:525–530. doi: 10.1073/pnas.0607870104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kelly TK, Miranda TB, Liang G, Berman BP, Lin JC, Tanay A, Jones PA. H2A.Z maintenance during mitosis reveals nucleosome shifting on mitotically silenced genes. Mol Cell. 2010;39:901–911. doi: 10.1016/j.molcel.2010.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rangasamy D, Berven L, Ridgway P, Tremethick DJ. Pericentric heterochromatin becomes enriched with H2A.Z during early mammalian development. EMBO J. 2003;22:1599–1607. doi: 10.1093/emboj/cdg160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rangasamy D, Greaves I, Tremethick DJ. RNA interference demonstrates a novel role for H2A.Z in chromosome segregation. Nat Struct Mol Biol. 2004;11:650–655. doi: 10.1038/nsmb786. [DOI] [PubMed] [Google Scholar]
- 58.Shia WJ, Li B, Workman JL. SAS-mediated acetylation of histone H4 Lys 16 is required for H2A.Z incorporation at subtelomeric regions in Saccharomyces cerevisiae. Genes Dev. 2006;20:2507–2512. doi: 10.1101/gad.1439206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marques M, Laflamme L, Gervais AL, Gaudreau L. Reconciling the positive and negative roles of histone H2A.Z in gene transcription. Epigenetics. 2010;5:267–272. doi: 10.4161/epi.5.4.11520. [DOI] [PubMed] [Google Scholar]
- 60.Svotelis A, Gevry N, Gaudreau L. Regulation of gene expression and cellular proliferation by histone H2A.Z. Biochem Cell Biol. 2009;87:179–188. doi: 10.1139/O08-138. [DOI] [PubMed] [Google Scholar]
- 61.Zlatanova J, Thakar A. H2A.Z: view from the top. Structure. 2008;16:166–179. doi: 10.1016/j.str.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 62.Xu Y, Ayrapetov MK, Xu C, Gursoy-Yuzugullu O, Hu Y, Price BD. Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Mol Cell. 2012;48:723–733. doi: 10.1016/j.molcel.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guillemette B, Bataille AR, Gevry N, Adam M, Blanchette M, Robert F, Gaudreau L. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 2005;3:e384. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li B, Pattenden SG, Lee D, Gutierrez J, Chen J, Seidel C, Gerton J, Workman JL. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc Natl Acad Sci USA. 2005;102:18385–18390. doi: 10.1073/pnas.0507975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang H, Roberts DN, Cairns BR. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–231. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 68.Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gevry N, Chan HM, Laflamme L, Livingston DM, Gaudreau L. p21 transcription is regulated by differential localization of histone H2A.Z. Genes Dev. 2007;21:1869–1881. doi: 10.1101/gad.1545707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kobor MS, Venkatasubrahmanyam S, Meneghini MD, Gin JW, Jennings JL, Link AJ, Madhani HD, Rine J. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2004;2:E131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Krogan NJ, Keogh MC, Datta N, Sawa C, Ryan OW, Ding H, Haw RA, Pootoolal J, Tong A, Canadien V, Richards DP, Wu X, Emili A, Hughes TR, Buratowski S, Greenblatt JF. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- 72.Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 73.Fuchs M, Gerber J, Drapkin R, Sif S, Ikura T, Ogryzko V, Lane WS, Nakatani Y, Livingston DM. The p400 complex is an essential E1A transformation target. Cell. 2001;106:297–307. doi: 10.1016/s0092-8674(01)00450-0. [DOI] [PubMed] [Google Scholar]
- 74.Cai Y, Jin J, Florens L, Swanson SK, Kusch T, Li B, Workman JL, Washburn MP, Conaway RC, Conaway JW. The mammalian YL1 protein is a shared subunit of the TRRAP/ TIP60 histone acetyltransferase and SRCAP complexes. J Biol Chem. 2005;280:13665–13670. doi: 10.1074/jbc.M500001200. [DOI] [PubMed] [Google Scholar]
- 75.Ruhl DD, Jin J, Cai Y, Swanson S, Florens L, Washburn MP, Conaway RC, Conaway JW, Chrivia JC. Purification of a human SRCAP complex that remodels chromatin by incorporating the histone variant H2A.Z into nucleosomes. Biochemistry. 2006;45:5671–5677. doi: 10.1021/bi060043d. [DOI] [PubMed] [Google Scholar]
- 76.Luk E, Vu ND, Patteson K, Mizuguchi G, Wu WH, Ranjan A, Backus J, Sen S, Lewis M, Bai Y, Wu C. Chz1, a nuclear chaperone for histone H2AZ. Mol Cell. 2007;25:357–368. doi: 10.1016/j.molcel.2006.12.015. [DOI] [PubMed] [Google Scholar]
- 77.Mahapatra S, Dewari PS, Bhardwaj A, Bhargava P. Yeast H2A.Z, FACT complex and RSC regulate transcription of tRNA gene through differential dynamics of flanking nucleosomes. Nucleic Acids Res. 2011;39:4023–4034. doi: 10.1093/nar/gkq1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Papamichos-Chronakis M, Watanabe S, Rando OJ, Peterson CL. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell. 2011;144:200–213. doi: 10.1016/j.cell.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jin C, Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21:1519–1529. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G. H3.3/H2A.Z double variant-containing nucleosomes mark “nucleosome-free regions” of active promoters and other regulatory regions. Nat Genet. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Soboleva TA, Nekrasov M, Pahwa A, Williams R, Huttley GA, Tremethick DJ. A unique H2A histone variant occupies the transcriptional start site of active genes. Nat Struct Mol Biol. 2012;19:25–30. doi: 10.1038/nsmb.2161. [DOI] [PubMed] [Google Scholar]
- 82.Dunican DS, McWilliam P, Tighe O, Parle-McDermott A, Croke DT. Gene expression differences between the microsatellite instability (MIN) and chromosomal instability (CIN) phenotypes in colorectal cancer revealed by high-density cDNA array hybridization. Oncogene. 2002;21:3253–3257. doi: 10.1038/sj.onc.1205431. [DOI] [PubMed] [Google Scholar]
- 83.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. Largescale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc Natl Acad Sci USA. 2004;101:9309–9314. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zucchi I, Mento E, Kuznetsov VA, Scotti M, Valsecchi V, Simionati B, Vicinanza E, Valle G, Pilotti S, Reinbold R, Vezzoni P, Albertini A, Dulbecco R. Gene expression profiles of epithelial cells microscopically isolated from a breast-invasive ductal carcinoma and a nodal metastasis. Proc Natl Acad Sci USA. 2004;101:18147–18152. doi: 10.1073/pnas.0408260101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Arnaldi LA, Borra RC, Maciel RM, Cerutti JM. Gene expression profiles reveal that DCN, DIO1, and DIO2 are underexpressed in benign and malignant thyroid tumors. Thyroid. 2005;15:210–221. doi: 10.1089/thy.2005.15.210. [DOI] [PubMed] [Google Scholar]
- 86.Bianco-Miotto T, Chiam K, Buchanan G, Jindal S, Day TK, Thomas M, Pickering MA, O’Loughlin MA, Ryan NK, Raymond WA, Horvath LG, Kench JG, Stricker PD, Marshall VR, Sutherland RL, Henshall SM, Gerald WL, Scher HI, Risbridger GP, Clements JA, Butler LM, Tilley WD, Horsfall DJ, Ricciardelli C Australian Prostate Cancer BioResource. Global levels of specific histone modifications and an epigenetic gene signature predict prostate cancer progression and development. Cancer Epidemiol Biomarkers Prev. 2010;19:2611–2622. doi: 10.1158/1055-9965.EPI-10-0555. [DOI] [PubMed] [Google Scholar]
- 87.Gorrini C, Squatrito M, Luise C, Syed N, Perna D, Wark L, Martinato F, Sardella D, Verrecchia A, Bennett S, Confalonieri S, Cesaroni M, Marchesi F, Gasco M, Scanziani E, Capra M, Mai S, Nuciforo P, Crook T, Lough J, Amati B. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature. 2007;448:1063–1067. doi: 10.1038/nature06055. [DOI] [PubMed] [Google Scholar]
- 88.Me LL, Vidal F, Gallardo D, Diaz-Fuertes M, Rojo F, Cuatrecasas M, Lopez-Vicente L, Kondoh H, Blanco C, Carnero A, Ramón y Cajal S. New p53 related genes in human tumors: significant downregulation in colon and lung carcinomas. Oncol Rep. 2006;16:603–608. doi: 10.3892/or.16.3.603. [DOI] [PubMed] [Google Scholar]
- 89.Santin AD, Zhan F, Bellone S, Palmieri M, Cane S, Bignotti E, Anfossi S, Gokden M, Dunn D, Roman JJ, O’Brien TJ, Tian E, Cannon MJ, Shaughnessy J, Jr, Pecorelli S. Gene expression profiles in primary ovarian serous papillary tumors and normal ovarian epithelium: identification of candidate molecular markers for ovarian cancer diagnosis and therapy. Int J Cancer. 2004;112:14–25. doi: 10.1002/ijc.20408. [DOI] [PubMed] [Google Scholar]
- 90.Draker R, Sarcinella E, Cheung P. USP10 deubiquitylates the histone variant H2A.Z and both are required for androgen receptor-mediated gene activation. Nucleic Acids Res. 2011;39:3529–3542. doi: 10.1093/nar/gkq1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dryhurst D, McMullen B, Fazli L, Rennie PS, Ausio J. Histone H2A.Z prepares the prostate specific antigen (PSA) gene for androgen receptor-mediated transcription and is upregulated in a model of prostate cancer progression. Cancer Lett. 2011;315:38–47. doi: 10.1016/j.canlet.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 92.Kuilman T, Michaloglou C, Mooi WJ, Peeper DS. The essence of senescence. Genes Dev. 2010;24:2463–2479. doi: 10.1101/gad.1971610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chan HM, Narita M, Lowe SW, Livingston DM. The p400 E1A-associated protein is a novel component of the p53 - ->p21 senescence pathway. Genes Dev. 2005;19:196–201. doi: 10.1101/gad.1280205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 95.Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A. H2AX: the histone guardian of the genome. DNA Repair (Amst) 2004;3:959–967. doi: 10.1016/j.dnarep.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 96.Bassing CH, Chua KF, Sekiguchi J, Suh H, Whitlow SR, Fleming JC, Monroe BC, Ciccone DN, Yan C, Vlasakova K, Livingston DM, Ferguson DO, Scully R, Alt FW. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc Natl Acad Sci USA. 2002;99:8173–8178. doi: 10.1073/pnas.122228699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, Reina-San-Martin B, Coppola V, Meffre E, Difilippantonio MJ, Redon C, Pilch DR, Olaru A, Eckhaus M, Camerini-Otero RD, Tessarollo L, Livak F, Manova K, Bonner WM, Nussenzweig MC, Nussenzweig A. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Petersen S, Casellas R, Reina-San-Martin B, Chen HT, Difilippantonio MJ, Wilson PC, Hanitsch L, Celeste A, Muramatsu M, Pilch DR, Redon C, Ried T, Bonner WM, Honjo T, Nussenzweig MC, Nussenzweig A. AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching. Nature. 2001;414:660–665. doi: 10.1038/414660a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Stiff T, O’Driscoll M, Rief N, Iwabuchi K, Lobrich M, Jeggo PA. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 2004;64:2390–2396. doi: 10.1158/0008-5472.can-03-3207. [DOI] [PubMed] [Google Scholar]
- 101.Stucki M, Jackson SP. GammaH2AX and MDC1: anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair (Amst) 2006;5:534–543. doi: 10.1016/j.dnarep.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 102.Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem. 2001;276:42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- 103.Fernandez-Capetillo O, Celeste A, Nussenzweig A. Focusing on foci: H2AX and the recruitment of DNA-damage response factors. Cell Cycle. 2003;2:426–427. [PubMed] [Google Scholar]
- 104.Celeste A, Difilippantonio S, Difilippantonio MJ, Fernandez-Capetillo O, Pilch DR, Sedelnikova OA, Eckhaus M, Ried T, Bonner WM, Nussenzweig A. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell. 2003;114:371–383. doi: 10.1016/s0092-8674(03)00567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Soutoglou E, Misteli T. Activation of the cellular DNA damage response in the absence of DNA lesions. Science. 2008;320:1507–1510. doi: 10.1126/science.1159051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lukas J, Lukas C, Bartek J. More than just a focus: the chromatin response to DNA damage and its role in genome integrity maintenance. Nat Cell Biol. 2011;13:1161–1169. doi: 10.1038/ncb2344. [DOI] [PubMed] [Google Scholar]
- 107.van Attikum H, Gasser SM. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 2009;19:207–217. doi: 10.1016/j.tcb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 108.Turner JM, Mahadevaiah SK, Fernandez-Capetillo O, Nussenzweig A, Xu X, Deng CX, Burgoyne PS. Silencing of unsynapsed meiotic chromosomes in the mouse. Nat Genet. 2005;37:41–47. doi: 10.1038/ng1484. [DOI] [PubMed] [Google Scholar]
- 109.Fernandez-Capetillo O, Mahadevaiah SK, Celeste A, Romanienko PJ, Camerini-Otero RD, Bonner WM, Manova K, Burgoyne P, Nussenzweig A. H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Dev Cell. 2003;4:497–508. doi: 10.1016/s1534-5807(03)00093-5. [DOI] [PubMed] [Google Scholar]
- 110.van der Heijden GW, Derijck AA, Posfai E, Giele M, Pelczar P, Ramos L, Wansink DG, van der Vlag J, Peters AH, de Boer P. Chromosome-wide nucleosome replacement and H3.3 incorporation during mammalian meiotic sex chromosome inactivation. Nat Genet. 2007;39:251–258. doi: 10.1038/ng1949. [DOI] [PubMed] [Google Scholar]
- 111.Li A, Yu Y, Lee SC, Ishibashi T, Lees-Miller SP, Ausio J. Phosphorylation of histone H2A.X by DNA-dependent protein kinase is not affected by core histone acetylation, but it alters nucleosome stability and histone H1 binding. J Biol Chem. 2010;285:17778–17788. doi: 10.1074/jbc.M110.116426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xiao A, Li H, Shechter D, Ahn SH, Fabrizio LA, Erdjument-Bromage H, Ishibe-Murakami S, Wang B, Tempst P, Hofmann K, Patel DJ, Elledge SJ, Allis CD. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature. 2009;457:57–62. doi: 10.1038/nature07668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhu F, Zykova TA, Peng C, Zhang J, Cho YY, Zheng D, Yao K, Ma WY, Lau AT, Bode AM, Dong Z. Phosphorylation of H2AX at Ser139 and a new phosphorylation site Ser16 by RSK2 decreases H2AX ubiquitination and inhibits cell transformation. Cancer Res. 2011;71:393–403. doi: 10.1158/0008-5472.CAN-10-2012. [DOI] [PubMed] [Google Scholar]
- 114.Cook PJ, Ju BG, Telese F, Wang X, Glass CK, Rosenfeld MG. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature. 2009;458:591–596. doi: 10.1038/nature07849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Krishnan N, Jeong DG, Jung SK, Ryu SE, Xiao A, Allis CD, Kim SJ, Tonks NK. Dephosphorylation of the C-terminal tyrosyl residue of the DNA damage-related histone H2A.X is mediated by the protein phosphatase eyes absent. J Biol Chem. 2009;284:16066–16070. doi: 10.1074/jbc.C900032200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, Lukas J, Lukas C. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 117.Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, Pelletier L, Jackson SP, Durocher D. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 120.Pan MR, Peng G, Hung WC, Lin SY. Monoubiquitination of H2AX protein regulates DNA damage response signaling. J Biol Chem. 2011;286:28599–28607. doi: 10.1074/jbc.M111.256297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhao GY, Sonoda E, Barber LJ, Oka H, Murakawa Y, Yamada K, Ikura T, Wang X, Kobayashi M, Yamamoto K, Boulton SJ, Takeda S. A critical role for the ubiquitin-conjugating enzyme Ubc13 in initiating homologous recombination. Mol Cell. 2007;25:663–675. doi: 10.1016/j.molcel.2007.01.029. [DOI] [PubMed] [Google Scholar]
- 122.Ikura T, Tashiro S, Kakino A, Shima H, Jacob N, Amunugama R, Yoder K, Izumi S, Kuraoka I, Tanaka K, Kimura H, Ikura M, Nishikubo S, Ito T, Muto A, Miyagawa K, Takeda S, Fishel R, Igarashi K, Kamiya K. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol Cell Biol. 2007;27:7028–7040. doi: 10.1128/MCB.00579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pinto DM, Flaus A. Structure and function of histone H2AX. Subcell Biochem. 2009;50:55–78. doi: 10.1007/978-90-481-3471-7_4. [DOI] [PubMed] [Google Scholar]
- 124.Rossetto D, Avvakumov N, Cote J. Histone phosphorylation: a chromatin modification involved in diverse nuclear events. Epigenetics. 2012;7:1098–1108. doi: 10.4161/epi.21975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kusch T, Florens L, Macdonald WH, Swanson SK, Glaser RL, Yates JR, 3rd, Abmayr SM, Washburn MP, Workman JL. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- 126.W inkler DD, Luger K. The histone chaperone FACT: structural insights and mechanisms for nucleosome reorganization. J Biol Chem. 2011;286:18369–18374. doi: 10.1074/jbc.R110.180778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Heo K, Kim H, Choi SH, Choi J, Kim K, Gu J, Lieber MR, Yang AS, An W. FACT-mediated exchange of histone variant H2AX regulated by phosphorylation of H2AX and ADP-ribosylation of Spt16. Mol Cell. 2008;30:86–97. doi: 10.1016/j.molcel.2008.02.029. [DOI] [PubMed] [Google Scholar]
- 128.Kobayashi J, Fujimoto H, Sato J, Hayashi I, Burma S, Matsuura S, Chen DJ, Komatsu K. Nucleolin participates in DNA double-strand break-induced damage response through MDC1-dependent pathway. PLoS ONE. 2012;7:e49245. doi: 10.1371/journal.pone.0049245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bassing CH, Suh H, Ferguson DO, Chua KF, Manis J, Eckersdorff M, Gleason M, Bronson R, Lee C, Alt FW. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell. 2003;114:359–370. doi: 10.1016/s0092-8674(03)00566-x. [DOI] [PubMed] [Google Scholar]
- 130.Monni O, Knuutila S. 11q deletions in hematological malignancies. Leuk Lymphoma. 2001;40:259–266. doi: 10.3109/10428190109057924. [DOI] [PubMed] [Google Scholar]
- 131.Stankovic T, Stewart GS, Byrd P, Fegan C, Moss PA, Taylor AM. ATM mutations in sporadic lymphoid tumours. Leuk Lymphoma. 2002;43:1563–1571. doi: 10.1080/1042819021000002884. [DOI] [PubMed] [Google Scholar]
- 132.Thirman MJ, Gill HJ, Burnett RC, Mbangkollo D, McCabe NR, Kobayashi H, Ziemin-van der Poel S, Kaneko Y, Morgan R, Sandberg AA, Chaganti RSK, Larson RA, Le Beau MM, Diaz MO, Rowley JD. Rearrangement of the MLL gene in acute lymphoblastic and acute myeloid leukemias with 11q23 chromosomal translocations. N Engl J Med. 1993;329:909–914. doi: 10.1056/NEJM199309233291302. [DOI] [PubMed] [Google Scholar]
- 133.Parikh RA, White JS, Huang X, Schoppy DW, Baysal BE, Baskaran R, Bakkenist CJ, Saunders WS, Hsu LC, Romkes M, et al. Loss of distal 11q is associated with DNA repair deficiency and reduced sensitivity to ionizing radiation in head and neck squamous cell carcinoma. Genes Chromosomes Cancer. 2007;46:761–775. doi: 10.1002/gcc.20462. [DOI] [PubMed] [Google Scholar]
- 134.Srivastava N, Gochhait S, Gupta P, Bamezai RN. Copy number alterations of the H2AFX gene in sporadic breast cancer patients. Cancer Genet Cytogenet. 2008;180:121–128. doi: 10.1016/j.cancergencyto.2007.09.024. [DOI] [PubMed] [Google Scholar]
- 135.Novik KL, Spinelli JJ, Macarthur AC, Shumansky K, Sipahimalani P, Leach S, Lai A, Connors JM, Gascoyne RD, Gallagher RP, Brooks-Wilson AR. Genetic variation in H2AFX contributes to risk of non-Hodgkin lymphoma. Cancer Epidemiol Biomarkers Prev. 2007;16:1098–1106. doi: 10.1158/1055-9965.EPI-06-0639. [DOI] [PubMed] [Google Scholar]
- 136.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y. GammaH2AX and cancer. Nat Rev Cancer. 2008;8:957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ivashkevich A, Redon CE, Nakamura AJ, Martin RF, Martin OA. Use of the gamma-H2AX assay to monitor DNA damage and repair in translational cancer research. Cancer Lett. 2011;327:123–133. doi: 10.1016/j.canlet.2011.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Liu Y, Tseng M, Perdreau SA, Rossi F, Antonescu C, Besmer P, Fletcher JA, Duensing S, Duensing A. Histone H2AX is a mediator of gastrointestinal stromal tumor cell apoptosis following treatment with imatinib mesylate. Cancer Res. 2007;67:2685–2692. doi: 10.1158/0008-5472.CAN-06-3497. [DOI] [PubMed] [Google Scholar]
- 139.Han W, Li X, Fu X. The macro domain protein family: structure, functions, and their potential therapeutic implications. Mutat Res. 2011;727:86–103. doi: 10.1016/j.mrrev.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Buschbeck M, Di Croce L. Approaching the molecular and physiological function of macroH2A variants. Epigenetics. 2010;5:118–123. doi: 10.4161/epi.5.2.11076. [DOI] [PubMed] [Google Scholar]
- 141.Costanzi C, Pehrson JR. MACROH2A2, a new member of the MARCOH2A core histone family. J Biol Chem. 2001;276:21776–21784. doi: 10.1074/jbc.M010919200. [DOI] [PubMed] [Google Scholar]
- 142.Pehrson JR, Fried VA. MacroH2A, a core histone containing a large nonhistone region. Science. 1992;257:1398–1400. doi: 10.1126/science.1529340. [DOI] [PubMed] [Google Scholar]
- 143.Pehrson JR, Costanzi C, Dharia C. Developmental and tissue expression patterns of histone macroH2A1 subtypes. J Cell Biochem. 1997;65:107–113. doi: 10.1002/(sici)1097-4644(199704)65:1<107::aid-jcb11>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 144.Gamble MJ, Kraus WL. Multiple facets of the unique histone variant macroH2A: from genomics to cell biology. Cell Cycle. 2010;9:2568–2574. doi: 10.4161/cc.9.13.12144. [DOI] [PubMed] [Google Scholar]
- 145.Kustatscher G, Hothorn M, Pugieux C, Scheffzek K, Ladurner AG. Splicing regulates NAD metabolite binding to histone macroH2A. Nat Struct Mol Biol. 2005;12:624–625. doi: 10.1038/nsmb956. [DOI] [PubMed] [Google Scholar]
- 146.Costanzi C, Pehrson JR. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature. 1998;393:599–601. doi: 10.1038/31275. [DOI] [PubMed] [Google Scholar]
- 147.Changolkar LN, Singh G, Cui K, Berletch JB, Zhao K, Disteche CM, Pehrson JR. Genome-wide distribution of macroH2A1 histone variants in mouse liver chromatin. Mol Cell Biol. 2010;30:5473–5483. doi: 10.1128/MCB.00518-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nora EP, Heard E. Chromatin structure and nuclear organization dynamics during X-chromosome inactivation. Cold Spring Harb Symp Quant Biol. 2011;75:333–344. doi: 10.1101/sqb.2010.75.032. [DOI] [PubMed] [Google Scholar]
- 149.Csankovszki G, Panning B, Bates B, Pehrson JR, Jaenisch R. Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation. Nat Genet. 1999;22:323–324. doi: 10.1038/11887. [DOI] [PubMed] [Google Scholar]
- 150.Rasmussen TP, Mastrangelo MA, Eden A, Pehrson JR, Jaenisch R. Dynamic relocalization of histone macroH2A1 from centrosomes to inactive X chromosomes during X inactivation. J Cell Biol. 2000;150:1189–1198. doi: 10.1083/jcb.150.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Boulard M, Storck S, Cong R, Pinto R, Delage H, Bouvet P. Histone variant macroH2A1 deletion in mice causes female-specific steatosis. Epigenetics Chromatin. 2010;3:8. doi: 10.1186/1756-8935-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Changolkar LN, Costanzi C, Leu NA, Chen D, McLaughlin KJ, Pehrson JR. Developmental changes in histone macroH2A1-mediated gene regulation. Mol Cell Biol. 2007;27:2758–2764. doi: 10.1128/MCB.02334-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhang R, Poustovoitov MV, Ye X, Santos HA, Chen W, Daganzo SM, Erzberger JP, Serebriiskii IG, Canutescu AA, Dunbrack RL, Pehrson JR, Berger JM, Kaufman PD, Adams PD. Formation of macroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev Cell. 2005;8:19–30. doi: 10.1016/j.devcel.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 154.Changolkar LN, Singh G, Pehrson JR. MacroH2A1-dependent silencing of endogenous murine leukemia viruses. Mol Cell Biol. 2008;28:2059–2065. doi: 10.1128/MCB.01362-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Choo JH, Kim JD, Chung JH, Stubbs L, Kim J. Allele-specific deposition of macroH2A1 in imprinting control regions. Hum Mol Genet. 2006;15:717–724. doi: 10.1093/hmg/ddi485. [DOI] [PubMed] [Google Scholar]
- 156.Buschbeck M, Uribesalgo I, Wibowo I, Rue P, Martin D, Gutierrez A, Morey L, Guigo R, Lopez-Schier H, Di Croce L. The histone variant macroH2A is an epigenetic regulator of key developmental genes. Nat Struct Mol Biol. 2009;16:1074–1079. doi: 10.1038/nsmb.1665. [DOI] [PubMed] [Google Scholar]
- 157.Gamble MJ, Frizzell KM, Yang C, Krishnakumar R, Kraus WL. The histone variant macroH2A1 marks repressed autosomal chromatin, but protects a subset of its target genes from silencing. Genes Dev. 2010;24:21–32. doi: 10.1101/gad.1876110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gaspar-Maia A, Qadeer ZA, Hasson D, Ratnakumar K, Adrian Leu N, Leroy G, Liu S, Costanzi C, Valle-Garcia D, Schaniel C, Lemischka I, Garcia B, Pehrson JR, Bernstein E. MacroH2A histone variants act as a barrier upon reprogramming towards pluripotency. Nat Commun. 2013;4:1565. doi: 10.1038/ncomms2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ratnakumar K, Duarte LF, Leroy G, Hasson D, Smeets D, Vardabasso C, Bonisch C, Zeng T, Xiang B, Zhang DY, Li H, Wang X, Hake SB, Schermelleh L, Garcia BA, Bernstein E. ATRX-mediated chromatin association of histone variant macroH2A1 regulates alpha-globin expression. Genes Dev. 2012;26:433–438. doi: 10.1101/gad.179416.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Chakravarthy S, Gundimella SK, Caron C, Perche PY, Pehrson JR, Khochbin S, Luger K. Structural characterization of the histone variant macroH2A. Mol Cell Biol. 2005;25:7616–7624. doi: 10.1128/MCB.25.17.7616-7624.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Angelov D, Molla A, Perche PY, Hans F, Cote J, Khochbin S, Bouvet P, Dimitrov S. The histone variant macroH2A interferes with transcription factor binding and SWI/SNF nucleosome remodeling. Mol Cell. 2003;11:1033–1041. doi: 10.1016/s1097-2765(03)00100-x. [DOI] [PubMed] [Google Scholar]
- 162.Angelov D, Verdel A, An W, Bondarenko V, Hans F, Doyen CM, Studitsky VM, Hamiche A, Roeder RG, Bouvet P, Dimitrov S. SWI/SNF remodeling and p300-dependent transcription of histone variant H2ABbd nucleosomal arrays. EMBO J. 2004;23:3815–3824. doi: 10.1038/sj.emboj.7600400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chang EY, Ferreira H, Somers J, Nusinow DA, Owen-Hughes T, Narlikar GJ. MacroH2A allows ATP-dependent chromatin remodeling by SWI/SNF and ACF complexes but specifically reduces recruitment of SWI/SNF. Biochemistry. 2008;47:13726–13732. doi: 10.1021/bi8016944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Doyen CM, An W, Angelov D, Bondarenko V, Mietton F, Studitsky VM, Hamiche A, Roeder RG, Bouvet P, Dimitrov S. Mechanism of polymerase II transcription repression by the histone variant macroH2A. Mol Cell Biol. 2006;26:1156–1164. doi: 10.1128/MCB.26.3.1156-1164.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Changolkar LN, Pehrson JR. Reconstitution of nucleosomes with histone macroH2A1.2. Biochemistry. 2002;41:179–184. doi: 10.1021/bi0157417. [DOI] [PubMed] [Google Scholar]
- 166.Chakravarthy S, Patel A, Bowman GD. The basic linker of macroH2A stabilizes DNA at the entry/exit site of the nucleosome. Nucleic Acids Res. 2012;40:8285–8295. doi: 10.1093/nar/gks645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010;24:1253–1265. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, Wen D, Chapgier A, DeKelver RC, Miller JC, Lee YL, Boydston EA, Holmes MC, Gregory PD, Greally JM, Rafii S, Yang C, Scambler PJ, Garrick D, Gibbons RJ, Higgs DR, Cristea IM, Urnov FD, Zheng D, Allis CD. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lewis PW, Elsaesser SJ, Noh K-M, Stadler SC, Allis CD. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc Natl Acad Sci USA. 2010;107:14075–14080. doi: 10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chu F, Nusinow DA, Chalkley RJ, Plath K, Panning B, Burlingame AL. Mapping post-translational modifications of the histone variant MacroH2A1 using tandem mass spectrometry. Mol Cell Proteomics. 2006;5:194–203. doi: 10.1074/mcp.M500285-MCP200. [DOI] [PubMed] [Google Scholar]
- 171.Ogawa Y, Ono T, Wakata Y, Okawa K, Tagami H, Shibahara KI. Histone variant macroH2A1.2 is mono-ubiquitinated at its histone domain. Biochem Biophys Res Commun. 2005;336:204–209. doi: 10.1016/j.bbrc.2005.08.046. [DOI] [PubMed] [Google Scholar]
- 172.Bernstein E, Muratore-Schroeder TL, Diaz RL, Chow JC, Changolkar LN, Shabanowitz J, Heard E, Pehrson JR, Hunt DF, Allis CD. A phosphorylated subpopulation of the histone variant macroH2A1 is excluded from the inactive X chromosome and enriched during mitosis. Proc Natl Acad Sci USA. 2008;105:1533–1538. doi: 10.1073/pnas.0711632105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hernandez-Munoz I, Lund AH, van der Stoop P, Boutsma E, Muijrers I, Verhoeven E, Nusinow DA, Panning B, Marahrens Y, van Lohuizen M. Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc Natl Acad Sci USA. 2005;102:7635–7640. doi: 10.1073/pnas.0408918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Takahashi I, Kameoka Y, Hashimoto K. MacroH2A1.2 binds the nuclear protein Spop. Biochim Biophys Acta. 2002;1591:63–68. doi: 10.1016/s0167-4889(02)00249-5. [DOI] [PubMed] [Google Scholar]
- 175.Mermoud JE, Costanzi C, Pehrson JR, Brockdorff N. Histone macroH2A1.2 relocates to the inactive X chromosome after initiation and propagation of X-inactivation. J Cell Biol. 1999;147:1399–1408. doi: 10.1083/jcb.147.7.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Dai B, Rasmussen TP. Global epiproteomic signatures distinguish embryonic stem cells from differentiated cells. Stem Cells. 2007;25:2567–2574. doi: 10.1634/stemcells.2007-0131. [DOI] [PubMed] [Google Scholar]
- 177.Tanasijevic B, Rasmussen TP. X chromosome inactivation and differentiation occur readily in ES cells doubly-deficient for macroH2A1 and macroH2A2. PLoS ONE. 2011;6:e21512. doi: 10.1371/journal.pone.0021512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Creppe C, Janich P, Cantarino N, Noguera M, Valero V, Musulen E, Douet J, Posavec M, Martin-Caballero J, Sumoy L, Di Croce L, Benitah SA, Buschbeck M. MacroH2A1 regulates the balance between self-renewal and differentiation commitment in embryonic and adult stem cells. Mol Cell Biol. 2012;32:1442–1452. doi: 10.1128/MCB.06323-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Pasque V, Gillich A, Garrett N, Gurdon JB. Histone variant macroH2A confers resistance to nuclear reprogramming. EMBO J. 2011;30:2373–2387. doi: 10.1038/emboj.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Pasque V, Radzisheuskaya A, Gillich A, Halley-Stott RP, Panamarova M, Zernicka-Goetz M, Surani MA, Silva JC. Histone variant macroH2A marks embryonic differentiation in vivo and acts as an epigenetic barrier to induced pluripotency. J Cell Sci. 2012;125(Pt 24):6094–6104. doi: 10.1242/jcs.113019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Li X, Kuang J, Shen Y, Majer MM, Nelson CC, Parsawar K, Heichman KA, Kuwada SK. The atypical histone macroH2A1.2 interacts with HER-2 protein in cancer cells. J Biol Chem. 2012;287:23171–23183. doi: 10.1074/jbc.M112.379412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Firestein R, Bass AJ, Kim SY, Dunn IF, Silver SJ, Guney I, Freed E, Ligon AH, Vena N, Ogino S, Chheda MG, Tamayo P, Finn S, Shrestha Y, Boehm JS, Jain S, Bojarski E, Mermel C, Barretina J, Chan JA, Baselga J, Tabernero J, Root DE, Fuchs CS, Loda M, Shivdasani RA, Meyerson M, Hahn WC. CDK8 is a colorectal cancer oncogene that regulates betacatenin activity. Nature. 2008;455:547–551. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Guerra C, Mijimolle N, Dhawahir A, Dubus P, Barradas M, Serrano M, Campuzano V, Barbacid M. Tumor induction by an endogenous K-ras oncogene is highly dependent on cellular context. Cancer Cell. 2003;4:111–120. doi: 10.1016/s1535-6108(03)00191-0. [DOI] [PubMed] [Google Scholar]
- 184.Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, Benguría A, Zaballos A, Flores JM, Barbacid M, Beach D, Serrano M. Tumour biology: senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- 185.Michaloglou C, Vredeveld LC, Mooi WJ, Peeper DS. BRAF(E600) in benign and malignant human tumours. Oncogene. 2008;27:877–895. doi: 10.1038/sj.onc.1210704. [DOI] [PubMed] [Google Scholar]
- 186.Tachiwana H, Osakabe A, Shiga T, Miya Y, Kimura H, Kagawa W, Kurumizaka H. Structures of human nucleosomes containing major histone H3 variants. Acta Crystallogr. 2011;67:578–583. doi: 10.1107/S0907444911014818. [DOI] [PubMed] [Google Scholar]
- 187.Schenk R, Jenke A, Zilbauer M, Wirth S, Postberg J. H3.5 is a novel hominid-specific histone H3 variant that is specifically expressed in the seminiferous tubules of human testes. Chromosoma. 2011;120:275–285. doi: 10.1007/s00412-011-0310-4. [DOI] [PubMed] [Google Scholar]
- 188.W itt O, Albig W, Doenecke D. Testis-specific expression of a novel human H3 histone gene. Exp Cell Res. 1996;229:301–306. doi: 10.1006/excr.1996.0375. [DOI] [PubMed] [Google Scholar]
- 189.W iedemann SM, Mildner SN, Bonisch C, Israel L, Maiser A, Matheisl S, Straub T, Merkl R, Leonhardt H, Kremmer E, Schermelleh L, Hake SB. Identification and characterization of two novel primate-specific histone H3 variants, H3.X and H3.Y. J Cell Biol. 2010;190:777–791. doi: 10.1083/jcb.201002043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 191.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Black BE, Cleveland DW. Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell. 2011;144:471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.W arburton PE. Chromosomal dynamics of human neocentromere formation. Chromosome Res. 2004;12:617–626. doi: 10.1023/B:CHRO.0000036585.44138.4b. [DOI] [PubMed] [Google Scholar]
- 194.Barnhart MC, Kuich PHJL, Stellfox ME, Ward JA, Bassett EA, Black BE, Foltz DR. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J Cell Biol. 2011;194:229–243. doi: 10.1083/jcb.201012017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR, 3rd, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 196.Warburton PE. Epigenetic analysis of kinetochore assembly on variant human centromeres. Trends Genet. 2001;17:243–247. doi: 10.1016/s0168-9525(01)02283-1. [DOI] [PubMed] [Google Scholar]
- 197.Howman EV, Fowler KJ, Newson AJ, Redward S, MacDonald AC, Kalitsis P, Choo KH. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc Natl Acad Sci USA. 2000;97:1148–1153. doi: 10.1073/pnas.97.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Sekulic N, Bassett EA, Rogers DJ, Black BE. The structure of (CENP-A-H4)2 reveals physical features that mark centromeres. Nature. 2010;467:347–351. doi: 10.1038/nature09323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Sullivan KF, Hechenberger M, Masri K. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J Cell Biol. 1994;127:581–592. doi: 10.1083/jcb.127.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Tachiwana H, Kagawa W, Shiga T, Osakabe A, Miya Y, Saito K, Hayashi-Takanaka Y, Oda T, Sato M, Park S-Y, Kimura H, Kurumizaka H. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011;476:232–235. doi: 10.1038/nature10258. [DOI] [PubMed] [Google Scholar]
- 201.Bassett EA, DeNizio J, Barnhart-Dailey MC, Panchenko T, Sekulic N, Rogers DJ, Foltz DR, Black BE. HJURP uses distinct CENP-A surfaces to recognize and to stabilize CENPA/ histone H4 for centromere assembly. Dev Cell. 2012;22:749–762. doi: 10.1016/j.devcel.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Carroll CW, Silva MC, Godek KM, Jansen LE, Straight AF. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat Cell Biol. 2009;11:896–902. doi: 10.1038/ncb1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Panchenko T, Sorensen TC, Woodcock CL, Kan Z-Y, Wood S, Resch MG, Luger K, Englander SW, Hansen JC, Black BE. Replacement of histone H3 with CENP-A directs global nucleosome array condensation and loosening of nucleosome superhelical termini. Proc Natl Acad Sci USA. 2011;108:16588–16593. doi: 10.1073/pnas.1113621108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Dalal Y, Wang H, Lindsay S, Henikoff S. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 2007;5:e218. doi: 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Dimitriadis EK, Weber C, Gill RK, Diekmann S, Dalal Y. Tetrameric organization of vertebrate centromeric nucleosomes. Proc Natl Acad Sci USA. 2010;107:20317–20322. doi: 10.1073/pnas.1009563107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Bui M, Dimitriadis EK, Hoischen C, An E, Quenet D, Giebe S, Nita-Lazar A, Diekmann S, Dalal Y. Cell-cycle-dependent structural transitions in the human CENP-A nucleosome in vivo. Cell. 2012;150:317–326. doi: 10.1016/j.cell.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Shivaraju M, Unruh JR, Slaughter BD, Mattingly M, Berman J, Gerton JL. Cell-cycle-coupled structural oscillation of centromeric nucleosomes in yeast. Cell. 2012;150:304–316. doi: 10.1016/j.cell.2012.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Shelby RD, Vafa O, Sullivan KF. Assembly of CENPA into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J Cell Biol. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat Biotechnol. 2003;21:86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- 210.Jansen LE, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- 212.Foltz DR, Jansen LET, Bailey AO, Yates Iii JR, Bassett EA, Wood S, Black BE, Cleveland DW. Centromere-specific assembly of CENP-A nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Black BE, Jansen LET, Maddox PS, Foltz DR, Desai AB, Shah JV, Cleveland DW. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENPA targeting domain. Mol Cell. 2007;25:309–322. doi: 10.1016/j.molcel.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 214.Black BE, Brock MA, Bedard S, Woods VL, Jr, Cleveland DW. An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. Proc Natl Acad Sci USA. 2007;104:5008–5013. doi: 10.1073/pnas.0700390104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev Cell. 2007;12:17–30. doi: 10.1016/j.devcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 216.Kim IS, Lee M, Park KC, Jeon Y, Park JH, Hwang EJ, Jeon TI, Ko S, Lee H, Baek SH, Kim KI. Roles of Mis18alpha in epigenetic regulation of centromeric chromatin and CENP-A loading. Mol Cell. 2012;46:260–273. doi: 10.1016/j.molcel.2012.03.021. [DOI] [PubMed] [Google Scholar]
- 217.Silva MC, Bodor DL, Stellfox ME, Martins NM, Hochegger H, Foltz DR, Jansen LE. Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Dev Cell. 2011;22:52–63. doi: 10.1016/j.devcel.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 218.Zeitlin SG, Shelby RD, Sullivan KF. CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J Cell Biol. 2001;155:1147–1158. doi: 10.1083/jcb.200108125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Kunitoku N, Sasayama T, Marumoto T, Zhang D, Honda S, Kobayashi O, Hatakeyama K, Ushio Y, Saya H, Hirota T. CENP-A phosphorylation by Aurora-A in prophase is required for enrichment of Aurora-B at inner centromeres and for kinetochore function. Dev Cell. 2003;5:853–864. doi: 10.1016/s1534-5807(03)00364-2. [DOI] [PubMed] [Google Scholar]
- 220.Zeitlin SG, Barber CM, Allis CD, Sullivan KF. Differential regulation of CENP-A and histone H3 phosphorylation in G2/M. J Cell Sci. 2001;114:653–661. doi: 10.1242/jcs.114.4.653. [DOI] [PubMed] [Google Scholar]
- 221.Mishra PK, Au WC, Choy JS, Kuich PH, Baker RE, Foltz DR, Basrai MA. Misregulation of Scm3p/HJURP causes chromosome instability in Saccharomyces cerevisiae and human cells. PLoS Genet. 2011;7:e1002303. doi: 10.1371/journal.pgen.1002303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Tomonaga T, Matsushita K, Yamaguchi S, Oohashi T, Shimada H, Ochiai T, Yoda K, Nomura F. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 2003;63:3511–3516. [PubMed] [Google Scholar]
- 223.Wu Q, Qian YM, Zhao XL, Wang SM, Feng XJ, Chen XF, Zhang SH. Expression and prognostic significance of centromere protein A in human lung adenocarcinoma. Lung Cancer. 2012;77:407–414. doi: 10.1016/j.lungcan.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 224.Biermann K, Heukamp LC, Steger K, Zhou H, Franke FE, Guetgemann I, Sonnack V, Brehm R, Berg J, Bastian PJ, Müller SC, Wang-Eckert L, Schorle H, Büttner R. Gene expression profiling identifies new biological markers of neoplastic germ cells. Anticancer Res. 2007;27:3091–3100. [PubMed] [Google Scholar]
- 225.McGovern SL, Qi Y, Pusztai L, Symmans WF, Buchholz TA. Centromere protein-A, an essential centromere protein, is a prognostic marker for relapse in estrogen receptor-positive breast cancer. Breast Cancer Res. 2012;14:R72. doi: 10.1186/bcr3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Li YMLX, Cao XZ, Wang L, Zhu MH. Expression of centromere protein A in hepatocellular carcinoma. Zhonghua Bing Li Xue Za Zhi. 2007;36:175–178. [PubMed] [Google Scholar]
- 227.Kato T, Sato N, Hayama S, Yamabuki T, Ito T, Miyamoto M, Kondo S, Nakamura Y, Daigo Y. Activation of Holliday junction recognizing protein involved in the chromosomal stability and immortality of cancer cells. Cancer Res. 2007;67:8544–8553. doi: 10.1158/0008-5472.CAN-07-1307. [DOI] [PubMed] [Google Scholar]
- 228.Hu Z, Huang G, Sadanandam A, Gu S, Lenburg M, Pai M, Bayani N, Blakely E, Gray J, Mao J-H. The expression level of HJURP has an independent prognostic impact and predicts the sensitivity to radiotherapy in breast cancer. Breast Cancer Res. 2010;12:R18. doi: 10.1186/bcr2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Li Y, Zhu Z, Zhang S, Yu D, Yu H, Liu L, Cao X, Wang L, Gao H, Zhu M. ShRNA-targeted centromere protein A inhibits hepatocellular carcinoma growth. PLoS ONE. 2011;6:e17794. doi: 10.1371/journal.pone.0017794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Albig W, Bramlage B, Gruber K, Klobeck HG, Kunz J, Doenecke D. The human replacement histone H3.3B gene (H3F3B) Genomics. 1995;30:264–272. doi: 10.1006/geno.1995.9878. [DOI] [PubMed] [Google Scholar]
- 231.Frank D, Doenecke D, Albig W. Differential expression of human replacement and cell cycle dependent H3 histone genes. Gene. 2003;312:135–143. doi: 10.1016/s0378-1119(03)00609-7. [DOI] [PubMed] [Google Scholar]
- 232.W ells D, Hoffman D, Kedes L. Unusual structure, evolutionary conservation of non-coding sequences and numerous pseudogenes characterize the human H3.3 histone multigene family. Nucleic Acids Res. 1987;15:2871–2889. doi: 10.1093/nar/15.7.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Cui B, Liu Y, Gorovsky MA. Deposition and function of histone H3 variants in Tetrahymena thermophila. Mol Cell Biol. 2006;26:7719–7730. doi: 10.1128/MCB.01139-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Hodl M, Basler K. Transcription in the absence of histone H3.3. Curr Biol. 2009;19:1221–1226. doi: 10.1016/j.cub.2009.05.048. [DOI] [PubMed] [Google Scholar]
- 235.Sakai A, Schwartz BE, Goldstein S, Ahmad K. Transcriptional and developmental functions of the H3.3 histone variant in Drosophila. Curr Biol. 2009;19:1816–1820. doi: 10.1016/j.cub.2009.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Couldrey C, Carlton MBL, Nolan PM, Colledge WH, Evans MJ. A retroviral gene trap insertion into the histone 3.3A gene causes partial neonatal lethality, stunted growth, neuromuscular deficits and male sub-fertility in transgenic mice. Hum Mol Genet. 1999;8:2489–2495. doi: 10.1093/hmg/8.13.2489. [DOI] [PubMed] [Google Scholar]
- 237.Banaszynski LA, Allis CD, Lewis PW. Histone variants in metazoan development. Dev Cell. 2010;19:662–674. doi: 10.1016/j.devcel.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Rogers RS, Inselman A, Handel MA, Matunis MJ. SUMO modified proteins localize to the XY body of pachytene spermatocytes. Chromosoma. 2004;113:233–243. doi: 10.1007/s00412-004-0311-7. [DOI] [PubMed] [Google Scholar]
- 239.E lsasser SJ, Huang H, Lewis PW, Chin JW, Allis CD, Patel DJ. DAXX envelops a histone H3.3-H4 dimer for H3.3-specific recognition. Nature. 2012;491:560–565. doi: 10.1038/nature11608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Liu CP, Xiong C, Wang M, Yu Z, Yang N, Chen P, Zhang Z, Li G, Xu RM. Structure of the variant histone H3.3-H4 heterodimer in complex with its chaperone DAXX. Nat Struct Mol Biol. 2012;19:1287–1292. doi: 10.1038/nsmb.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 242.Hake SB, Garcia BA, Duncan EM, Kauer M, Dellaire G, Shabanowitz J, Bazett-Jones DP, Allis CD, Hunt DF. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J Biol Chem. 2006;281:559–568. doi: 10.1074/jbc.M509266200. [DOI] [PubMed] [Google Scholar]
- 243.Loyola A, Almouzni Gv. Marking histone H3 variants: how when and why? Trends Biochem Sci. 2007;32:425–433. doi: 10.1016/j.tibs.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 244.Hake SB, Allis CD. Histone H3 variants and their potential role in indexing mammalian genomes: the “H3 barcode hypothesis”. Proc Natl Acad Sci USA. 2006;103:6428–6435. doi: 10.1073/pnas.0600803103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.E lsaesser SJ, Goldberg AD, Allis CD. New functions for an old variant: no substitute for histone H3.3. Curr Opin Genet Dev. 2010;20:110–117. doi: 10.1016/j.gde.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Szenker E, Ray-Gallet D, Almouzni G. The double face of the histone variant H3.3. Cell Res. 2011;21:421–434. doi: 10.1038/cr.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Chow C-M, Georgiou A, Szutorisz H, Maia e Silva A, Pombo A, Barahona I, Dargelos E, Canzonetta C, Dillon N. Variant histone H3.3 marks promoters of transcriptionally active genes during mammalian cell division. EMBO Rep. 2005;6:354–360. doi: 10.1038/sj.embor.7400366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Jin C, Felsenfeld G. Distribution of histone H3.3 in hematopoietic cell lineages. Proc Natl Acad Sci USA. 2006;103:574–579. doi: 10.1073/pnas.0509974103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.W irbelauer C, Bell O, Schübeler D. Variant histone H3.3 is deposited at sites of nucleosomal displacement throughout transcribed genes while active histone modifications show a promoter-proximal bias. Genes Develop. 2005;19:1761–1766. doi: 10.1101/gad.347705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Zhang R, Liu ST, Chen W, Bonner M, Pehrson J, Yen TJ, Adams PD. HP1 proteins are essential for a dynamic nuclear response that rescues the function of perturbed heterochromatin in primary human cells. Mol Cell Biol. 2007;27:949–962. doi: 10.1128/MCB.01639-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Dunleavy EM, Almouzni Gv, Karpen GH. H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G1 phase. Nucleus. 2011;2:146–157. doi: 10.4161/nucl.2.2.15211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Ng RK, Gurdon JB. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat Cell Biol. 2008;10:102–109. doi: 10.1038/ncb1674. [DOI] [PubMed] [Google Scholar]
- 253.Das C, Tyler JK. Histone exchange and histone modifications during transcription and aging. Biochim Biophys Acta. 2011;1819:332–342. doi: 10.1016/j.bbagrm.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Xu M, Long C, Chen X, Huang C, Chen S, Zhu B. Partitioning of histone H3–H4 tetramers during DNA replicationdependent chromatin assembly. Science. 2010;328:94–98. doi: 10.1126/science.1178994. [DOI] [PubMed] [Google Scholar]
- 255.Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 256.Hollenbach AD, McPherson CJ, Mientjes EJ, Iyengar R, Grosveld G. Daxx and histone deacetylase II associate with chromatin through an interaction with core histones and the chromatin-associated protein Dek. J Cell Sci. 2002;115:3319–3330. doi: 10.1242/jcs.115.16.3319. [DOI] [PubMed] [Google Scholar]
- 257.Ray-Gallet D, Woolfe A, Vassias I, Pellentz C, Lacoste N, Puri A, Schultz DC, Pchelintsev NA, Adams PD, Jansen LE, Almouzni G. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol Cell. 2011;44:928–941. doi: 10.1016/j.molcel.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 258.Roberts C, Sutherland HF, Farmer H, Kimber W, Halford S, Carey A, Brickman JM, Wynshaw-Boris A, Scambler PJ. Targeted mutagenesis of the Hira gene results in gastrulation defects and patterning abnormalities of mesoendodermal derivatives prior to early embryonic lethality. Mol Cell Biol. 2002;22:2318–2328. doi: 10.1128/MCB.22.7.2318-2328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Tang J, Wu S, Liu H, Stratt R, Barak OG, Shiekhattar R, Picketts DJ, Yang X. A novel transcription regulatory complex containing death domain-associated protein and the ATR-X syndrome protein. J Biol Chem. 2004;279:20369–20377. doi: 10.1074/jbc.M401321200. [DOI] [PubMed] [Google Scholar]
- 260.Xue Y, Gibbons R, Yan Z, Yang D, McDowell TL, Sechi S, Qin J, Zhou S, Higgs D, Wang W. The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc Natl Acad Sci USA. 2003;100:10635–10640. doi: 10.1073/pnas.1937626100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Berube NG, Smeenk CA, Picketts DJ. Cell cycledependent phosphorylation of the ATRX protein correlates with changes in nuclear matrix and chromatin association. Hum Mol Genet. 2000;9:539–547. doi: 10.1093/hmg/9.4.539. [DOI] [PubMed] [Google Scholar]
- 262.Graber MW, Schweinfest CW, Reed CE, Papas TS, Baron PL. Isolation of differentially expressed genes in carcinoma of the esophagus. Ann Surg Oncol. 1996;3:192–197. doi: 10.1007/BF02305800. [DOI] [PubMed] [Google Scholar]
- 263.Lewis PW, Müller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, Allis CD. Inhibition of PRC2 Activity by a Gain-of-Function H3 Mutation Found in Pediatric Glioblastoma. Science. 2013 doi: 10.1126/science.1232245. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Dutta D, Ray S, Home P, Saha B, Wang S, Sheibani N, Tawfik O, Cheng N, Paul S. Regulation of angiogenesis by histone chaperone HIRA-mediated incorporation of lysine 56-acetylated histone H3.3 at chromatin domains of endothelial genes. J Biol Chem. 2010;285:41567–41577. doi: 10.1074/jbc.M110.190025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I, Hamdi M, van Nes J, Westerman BA, van Arkel J, Ebus ME, Haneveld F, Lakeman A, Schild L, Molenaar P, Stroeken P, van Noesel MM, Ora I, Santo EE, Caron HN, Westerhout EM, Versteeg R. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–593. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- 266.Gibbons RJ, Pellagatti A, Garrick D, Wood WG, Malik N, Ayyub H, Langford C, Boultwood J, Wainscoat JS, Higgs DR. Identification of acquired somatic mutations in the gene encoding chromatin-remodeling factor ATRX in the alpha-thalassemia myelodysplasia syndrome (ATMDS) Nat Genet. 2003;34:446–449. doi: 10.1038/ng1213. [DOI] [PubMed] [Google Scholar]
- 267.Friedmann-Morvinski D, Bushong EA, Ke E, Soda Y, Marumoto T, Singer O, Ellisman MH, Verma IM. Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science. 2012;338:1080–1084. doi: 10.1126/science.1226929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Lovejoy CA, Li W, Reisenweber S, Thongthip S, Bruno J, de Lange T, De S, Petrini JH, Sung PA, Jasin M, Rosenbluh J, Zwang Y, Weir BA, Hatton C, Ivanova E, Macconaill L, Hanna M, Hahn WC, Lue NF, Reddel RR, Jiao Y, Kinzler K, Vogelstein B, Papadopoulos N, Meeker AK ALT Starr Cancer Consortium. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet. 2012;8:e1002772. doi: 10.1371/journal.pgen.1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Ratnakumar K, Bernstein E. ATRX: the case of a peculiar chromatin remodeler. Epigenetics. 2012;8:3–9. doi: 10.4161/epi.23271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C, Pfaff E, Tonjes M, Sill M, Bender S, Kool M, Zapatka M, Becker N, Zucknick M, Hielscher T, Liu XY, Fontebasso AM, Ryzhova M, Albrecht S, Jacob K, Wolter M, Ebinger M, Schuhmann MU, van Meter T, Frühwald MC, Hauch H, Pekrun A, Radlwimmer B, Niehues T, von Komorowski G, Dürken M, Kulozik AE, Madden J, Donson A, Foreman NK, Drissi R, Fouladi M, Scheurlen W, von Deimling A, Monoranu C, Roggendorf W, Herold-Mende C, Unterberg A, Kramm CM, Felsberg J, Hartmann C, Wiestler B, Wick W, Milde T, Witt O, Lindroth AM, Schwartzentruber J, Faury D, Fleming A, Zakrzewska M, Liberski PP, Zakrzewski K, Hauser P, Garami M, Klekner A, Bognar L, Morrissy S, Cavalli F, Taylor MD, van Sluis P, Koster J, Versteeg R, Volckmann R, Mikkelsen T, Aldape K, Reifenberger G, Collins VP, Majewski J, Korshunov A, Lichter P, Plass C, Jabado N, Pfister SM. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22:425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 271.Creyghton MP, Markoulaki S, Levine SS, Hanna J, Lodato MA, Sha K, Young RA, Jaenisch R, Boyer LA. H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell. 2008;135:649–661. doi: 10.1016/j.cell.2008.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Fullgrabe J, Kavanagh E, Joseph B. Histone onco-modifications. Oncogene. 2011;30:3391–3403. doi: 10.1038/onc.2011.121. [DOI] [PubMed] [Google Scholar]
- 273.Heyn H, Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nat Rev Genet. 2012;13:679–692. doi: 10.1038/nrg3270. [DOI] [PubMed] [Google Scholar]
- 274.Conerly ML, Teves SS, Diolaiti D, Ulrich M, Eisenman RN, Henikoff S. Changes in H2A.Z occupancy and DNA methylation during B-cell lymphomagenesis. Genome Res. 2010;20:1383–1390. doi: 10.1101/gr.106542.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat Biotechnol. 2007;25:84–90. doi: 10.1038/nbt1272. [DOI] [PubMed] [Google Scholar]
- 276.Marks PA, Xu WS. Histone deacetylase inhibitors: potential in cancer therapy. J Cell Biochem. 2009;107:600–608. doi: 10.1002/jcb.22185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Muller S, Filippakopoulos P, Knapp S. Bromodomains as therapeutic targets. Expert Rev Mol Med. 2011;13:e29. doi: 10.1017/S1462399411001992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Xu F, Li X. The role of histone methyltransferase EZH2 in myelodysplastic syndromes. Expert Rev Hematol. 2012;5:177–185. doi: 10.1586/ehm.12.5. [DOI] [PubMed] [Google Scholar]
- 279.Jiao Y, Killela PJ, Reitman ZJ, Rasheed AB, Heaphy CM, de Wilde RF, Rodriguez FJ, Rosemberg S, Oba-Shinjo SM, Nagahashi Marie SK, Bettegowda C, Agrawal N, Lipp E, Pirozzi C, Lopez G, He Y, Friedman H, Friedman AH, Riggins GJ, Holdhoff M, Burger P, McLendon R, Bigner DD, Vogelstein B, Meeker AK, Kinzler KW, Papadopoulos N, Diaz LA, Yan H. Frequent ATRX, CIC and FUBP1 mutations refine the classification of malignant gliomas. Oncotarget. 2012;3:709–722. doi: 10.18632/oncotarget.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Hasson D, Panchenko T, Salimian KJ, Salman MU, Sekulic N, Alonso A, Warburton PE, Black BE. The octamer is the major form of CENP-A nucleosomes at human centromeres. Nat Struct Mol Biol. 2013 doi: 10.1038/nsmb.2562. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]