Abstract
Mitochondria have roles in many cellular processes, from energy metabolism and calcium homeostasis to control of cellular lifespan and programmed cell death. These processes affect and are affected by the redox status of and ATP production by mitochondria. Here, we describe the use of two ratiometric, genetically encoded biosensors that can detect mitochondrial redox state and ATP levels at subcellular resolution in living yeast cells. Mitochondrial redox state is measured using redox-sensitive Green Fluorescent Protein (roGFP) that is targeted to the mitochondrial matrix. MitoroGFP contains cysteines at positions 147 and 204 of GFP, which undergo reversible and environment-dependent oxidation and reduction, which in turn alter the excitation spectrum of the protein. MitGO-ATeam is a Förster resonance energy transfer (FRET) probe in which the ε subunit of the FoF1-ATP synthase is sandwiched between FRET donor and acceptor fluorescent proteins. Binding of ATP to the ε subunit results in conformation changes in the protein that bring the FRET donor and acceptor in close proximity and allow for fluorescence resonance energy transfer from the donor to acceptor.
Keywords: roGFP, GO-ATeam, FRET, ROS
INTRODUCTION
Mitochondria are essential organelles for ATP production, biosynthesis of amino acids, fatty acids, heme, iron sulfur clusters and pyrimidines. Mitochondria also play pivotal roles in calcium homeostasis, and in regulation of apoptosis.1 Increasing evidence links mitochondria to aging and age-related diseases including Parkinson's disease, Alzheimer's disease, amyotrophic lateral sclerosis, and Huntington's disease.2 While individuals live their entire lives with mutations in mitochondrial proteins that are associated with neurodegenerative diseases, the disease symptoms occur only later in life. This indicates that changes occur in mitochondria with age that allow disease pathology to emerge. Indeed, mitochondrial fitness is correlated with overall cell health and lifespan in yeast and mammalian cells.3,4 Here, we describe how to use genetically encoded, ratiometric fluorescent biosensors to assess two critical features of mitochondria in living yeast cells: redox state and ATP levels.
Mitochondrial function in aerobic energy mobilization is well established. Mitochondrial redox state is a product of reducing and oxidizing species in the organelle, including NAD+/NADH, FAD/FADH2, NADP+/NADPH, glutathione/glutathione disulfide (GSH/GSSG) and reactive oxygen species (ROS). Uncoupling mitochondria or hypoxia affects mitochondrial respiratory activity and alters the ratio of NAD+ to NADH in the organelle. ROS, which are produced from inefficient electron transfer between complexes of the electron transport chain in the inner mitochondrial membrane, as well as from the deamination of amines via monoamine oxidase in the outer mitochondrial membrane5, damage lipids, proteins and nucleic acids and have been linked to aging and age-associated neurodegenerative diseases6,7,8. ROS also play a role in signal transduction in mitochondria, through oxidation of GSH. For example, NADH dehydrogenase not only contributes to ROS production but also is regulated through interactions with the glutathione pool9,10. α-Ketoglutarate dehydrogenase and aconitase, components of the TCA cycle, exhibit reduced activity in oxidizing environments11,12. Indeed, redox-dependent regulation of aconitase activity is conserved from bacteria to mammals13,14. Thus, monitoring the redox state and ATP levels of mitochondria is crucial to understanding their function and role in disease pathology.
Biochemical methods have been used to assess the redox state or ATP levels of whole cells or isolated mitochondria. Widely used methods to assess the redox state of whole cells or isolated mitochondria are based on measuring the levels of the redox pair GSH/GSSG15. The luciferin-luciferase system is commonly used to measure mitochondrial ATP levels in either permeabilized whole cells or isolated mitochondria.16,17,18,19,20 In this assay, luciferase binds to ATP and catalyzes the oxidation of and chemiluminescence from luciferin.21 The intensity of the emitted light is proportional to the amount of ATP in the reaction mixture.22
These methods have revealed fundamental information regarding mitochondrial function, including the finding that patients with neurodegenerative diseases, such as Alzheimer's disease, have abnormally low ATP levels.23 However, they cannot be used to image living, intact cells. Moreover, methods based on whole-cell analysis provide an average of redox state or ATP levels in all compartments of the cell. Measurements in isolated organelles are potentially problematic because mitochondrial redox state or ATP levels may change during subcellular fractionation. Finally, recent studies from our laboratory and others indicate that mitochondria within individual cells are heterogeneous in function, which in turn affects the lifespan of mother and daughter cells.3 Thus, there is a need to measure mitochondrial ATP levels and redox state in living cells with subcellular resolution.
The biosensors for mitochondrial function described here are both based on GFP. Redox-sensitive GFP (roGFP)24,25 is a GFP variant in which surface-exposed cysteines are added to the molecule. roGFP, like wild-type GFP, has two excitation peaks (at ~400 nm and ~480 nm) and one emission peak at ~510 nm. Oxidation of the cysteine residues in roGFP results in an increase in excitation at ~400 nm. Reduction of those cysteines favors excitation at ~480 nm. Thus, the ratio of 510 nm emission upon excitation of roGFP at 480 and 400 nm reveals the relative amount of reduced and oxidized roGFP, which reflects the redox state of the fluorophore's environment.
Two versions of roGFP are widely used: roGFP1 and roGFP2. Both contain the same cysteine insertions. roGFP1 is based on wild-type GFP and roGFP2 is based on S65T GFP, which has more efficient excitation at 480 nm and less efficient excitation at 400 nm compared to wtGFP24. roGFP1 is less pH sensitive than roGFP2 and its dynamic range extends further into the reduced range. Thus, roGFP1 may be more useful for monitoring more reducing compartments such as mitochondria or the cytosol, and compartments with variable pH, such as endosomes. roGFP2 offers brighter signal and, in some studies, a greater dynamic range than roGFP124,26. Studies in Arabidopsis thaliana indicate that the time required for response to changes in redox state is similar for both sensors (t1/2 for oxidation, 65 and 95 sec and t1/2 for reduction, 272 and 206 sec, for roGFP1 and roGFP2, respectively).26
MitGO-ATeam2 is a minimally invasive, reliable sensor that measures mitochondrial ATP in the budding yeast Saccharomyces cerevisiae. GO-ATeam is a Förster resonance energy transfer (FRET) probe that consists of the ε subunit of the FoF1-ATP synthase sandwiched between FRET donor and acceptor fluorescent proteins (GFP and orange fluorescent protein (OFP), respectively).27 Binding of ATP to the ε subunit results in conformational changes in the protein that bring the FRET donor in close proximity to the acceptor and allow for energy transfer from donor to acceptor. There are two variants of GO-ATeam, GO-ATeam1 and GO-ATeam2. GO-ATeam2 has a higher affinity for MgATP than GO-ATeam1, making it more suitable for measuring the typically lower [ATP] in mitochondria compared to the cytosol.27
To probe mitochondrial redox state, we constructed a fusion protein (mito-roGFP1) consisting of roGFP1 fused to the ATP9 leader sequence and expressed from a centromere-based (low copy number) yeast expression plasmid under control of the strong glyceraldehyde-3-phosphate dehydrogenase (GPD) promoter (p416GPD, Addgene). We used roGFP1 to probe the redox status of mitochondria in the context of aging of the model fungus Saccharomyces cerevisiae1. We find that roGFP1 can detect changes in mitochondrial redox state that occur during aging and in response to nutrient availability but has no apparent detrimental effect on yeast cells. We also see variability in the redox state of mitochondria within individual living yeast cells, a finding that underscores the importance of a biosensor with subcellular spatial resolution.
MitGO-ATeam2 is a variant of GO-ATeam2, which has the mitochondrial signal sequence of cytochrome c oxidase subunit VIIIA inserted at the amino terminus of GO-ATeam2.27 We modified the mitGO-ATeam2 probe (kindly provided by the laboratory of H. Noji, Institute of Scientific and Industrial Research, Osaka University, Japan) for use in yeast by subcloning it, via Xba1 and HindIII sites, into the yeast expression vector pAG415GPD-ccdB (Addgene, Cambridge, MA, USA), which is a low-copy plasmid containing the strong constitutive GPD promoter. We expressed mitGO-ATeam2 in budding yeast, and find, by counterstaining with the DNA-binding dye DAPI, that it localizes exclusively to mitochondria, where it serves as an effective probe to measure physiological changes in mitochondrial ATP levels.
roGFP and GO-ATeam are both genetically encoded. As a result, they can be introduced and stably maintained in intact cells, and provide information on redox state or ATP levels in individual, living cells. Moreover, both biosensors monitor changes in redox state or ATP levels that occur under physiological conditions.28 Both probes are also ratiometric. As a result, measurements made with these probes are not affected by changes in biosensor concentration or sample illumination or thickness. Finally, both biosensors provide subcellular spatial resolution. Indeed, roGFP has been targeted to mitochondria, ER, endosomes and peroxisomes24, and can detect changes in redox state of each of these organelles, largely independent of pH.
PROCEDURE
1. Transformation of yeast cells with the biosensors
-
1.1
Transform the desired yeast strain with plasmid bearing mito-roGFP or mitGO-ATeam2 using the lithium acetate method27.
-
1.2
To confirm transformation with the plasmid-borne biosensor and to prevent loss of the plasmid, select and maintain transformants on the appropriate selective synthetic complete medium (SC-Ura for mito-roGFP, or SC-Leu for mitGo-ATeam2). If the fluorescent probe has been subcloned in a different plasmid than those described here, use the appropriate selective medium. Visualize transformants by fluorescence microscopy to confirm that they express the fluorescent biosensor and exhibit normal mitochondrial morphology.
2. Growth of cells and preparation for imaging
Cell function and response to drug treatment are highly dependent on cell density and metabolic activity. Best results are obtained when cells are actively dividing (mid-log phase, ~0.5 – 1 × 107 cells/ml). The most reliable way to generate midlog phase cultures of consistent density is to inoculate from a stationary-phase pre-culture.
-
2.1
Prepare a stationary-phase pre-culture: pick a single colony of transformed cells from a plate and inoculate selective liquid media (5 ml in a 50 ml conical-bottom tube). Grow at 30°C with shaking at 225 rpm until the optical density of the culture at 600 nm (OD600) has reached a plateau (24–48 hr).
-
2.2
Prepare a mid-log phase culture for observation: use the appropriate volume of pre-culture to inoculate 5 ml of selective media in a 50 ml conical-bottom tube. YPD media is autofluorescent and should not be used for these studies. Use synthetic complete, glucose based, dropout (SC-Ura) media. Grow cells at 30°C, shaking at 225 rpm, for 4–16 hr, until they reach mid-log phase (~0.5 – 1 × 107 cells/ml). Cell density can be determined by measuring OD600; calibrate the spectrophotometer to determine the correct OD600 reading. On our Beckman DU530 (Beckman Coulter, Indianapolis, IN), mid-log phase corresponds to an OD600 of 0.1–0.3.
Mito-roGFP1 senses fluctuations in the organelle in response to metabolic changes. For example, in this assay mitochondrial redox state changes when yeast grow on fermentable carbon sources (e.g. glucose, as in SC media) versus non-fermentable carbon sources (e.g. glycerol, as in SGlyc media), and even in different batches of the same media. Therefore, use the same batch of media for all experiments.
-
2.3
Cells are ready for concentration (see step 2.4) and imaging if no treatment is being performed. If cells are being treated, incubate cells with the appropriate treatment and continue to the next step.
-
2.4
Concentrate 1 ml of culture by centrifugation at 6,000 × g for 15 sec and resuspension of the cell pellet in 20 μl of media. These conditions maximize the number of distinguishable cells in the field of view.
-
2.5
Apply 2 μl of the resuspended cells to a slide. Cover with a coverslip (No. 1.5, preferably high-performance 170±5 μm thickness), and seal the edges of the coverslip with clear nail polish or valap (see Reagents). To seal with valap, melt a small amount on a metal spatula by holding it over a Bunsen burner flame, then spread a small amount along the edges of the coverslip.
-
2.6
Maintain cells at 30°C during imaging. An objective heater on the 100× oil immersion lens used for imaging works well for this application. Under these conditions, mitochondrial morphology, redox state and ATP levels remain unchanged during imaging for 10–15 min
3. Imaging setup
3.1 Setup for imaging mito-roGFP1 on a wide-field fluorescence microscope
The steps here are tailored to the AxioObserver.Z1 microscope equipped with a Colibri LED excitation source, a wide-field Orca ER camera and Axiovision acquisition software. Photobleaching of both channels and photoconversion of oxidized mito-roGFP1 is reduced significantly using LED illumination compared to mercury arc lamp illumination (see below).
To maximize signal and resolution, use the highest numerical aperture possible in the objective, and the lowest magnification that provides sufficient spatial resolution. In addition, for mito-roGFP imaging, verify that the objective transmits well at 365 nm. The 100×/1.3NA EC PlanNeofluar objective (Zeiss) works well for this application.
Configure the acquisition software to capture the oxidized and reduced mito-roGFP1 species. We use the following conditions.
-
3.1.1
Configure the channel for oxidized mito-roGFP to use excitation at 365 nm (100% LED power) and an emission filter suitable for GFP, such as the 38 HE filter set (Zeiss), with the included excitation filter removed from the cube. Removal of the excitation filter allows excitation at both 365 nm and 470 nm without the need to switch filters, thus increasing achievable time resolution.
-
3.1.2
Configure the channel for reduced mito-roGFP to use excitation at 470 nm (100% LED power) and, as mentioned above, the same emission filter cube used for the oxidized mito-roGFP.
Set the camera to 1×1 binning to optimize spatial resolution.
-
3.1.3
Set software to acquire a z stack consisting of 11 slices with 0.5 μm spacing, collecting both channels at each z position. This mode of acquisition is slower than acquiring each z stack in turn, but it prevents artifacts arising from mitochondrial motion between acquisition of the oxidized and reduced channels.
-
3.1.4
Image several cells to determine an appropriate exposure time, producing a strong but not saturating signal. It is important to maintain the ratio of exposure times for oxidized and reduced mito-roGFP1 for all experiments. For example, if the exposure times for oxidized and reduced mito-roGFP1 are 300 and 100 ms, respectively, then the exposure time for oxidized mito-roGFP1 should be 3 times that for reduced mito-roGFP for all experiments.
3.2 Setup for imaging mitGO-ATeam2 on a spectral confocal microscope
To quantify ATP levels using mitGO-ATeam2, the fluorescence from GFP (emission maximum 510 nm) must be distinguished from that from OFP (emission maximum 560 nm). There are fluorescence filter sets that resolve the fluorescence emitted from these fluorophores. However, we find that a spectral detector, available on many laser scanning confocal microscopes, works best for this application. A spectral detector separates the fluorescence emission into many components (typically 32 or more) according to wavelength. Several adjacent wavelength bands can be combined into one image channel. The wavelengths to be combined are selected empirically so as to maximize signal from GFP and OFP while avoiding crossover of signal that would confound the FRET measurement. Use the highest numerical aperture lens available. We use a 100×/1.49 or 60×/1.49 Apo-TIRF lens. We use the Nikon A1R confocal microscope running NIS Elements software. Configure the acquisition software to capture the ATP-bound and ATP-unbound mitGO-ATeam2 species. We use the following conditions.
-
3.2.1
Set pinhole to 1.0 Airy units (AU), which on our system corresponds to a z resolution of about 0.42 μm.
-
3.2.2
Set scan zoom to approach the Nyquist sampling limit, which will maximize spatial information in the image. On our system the pixel size is 0.12 μm.
-
3.2.3
Because mitochondria may move during imaging, it can be helpful to increase imaging speed by cropping the field to 512 × 256 pixels. The total time required to image one frame in our system is 1.0 s, including a 2-fold scan average. Depending on desired spatial resolution, the field can be further cropped to increase time resolution.
-
3.2.4
Excite at 488 nm, and collect emission from 500 – 520 nm for GFP, and from 550 – 580 nm for OFP. Actual optimal values may vary with characteristics of the imaging system.
-
3.2.5
The optimal laser power for our system is between 6.0 and 6.4%. The optimal laser power will vary for each microscope system, but will ideally be as low as possible while still producing an interpretable image. Use an internal or external power meter to monitor changes in the laser power, which occur normally over time in any optical system.
-
3.2.6
Adjust detector gain and illumination light intensity to maximize the detected range of pixel values but avoid saturating the signal, for as many cells as possible. Do not analyze any cells containing more than 1% saturated pixels. Image all samples using the same objective, laser power, scan zoom, pixel size, gain, and offset.
4. Image acquisition
-
4.1
Locate the focal plane of cells of interest using transmitted light to minimize bleaching the fluorescent probe.
-
4.2
After locating one or more cells of interest, collect a z-series through the whole depth of a typical cell (approx. 7 μm) using a step size of 0.5 μm.
-
4.3
Image other cells of interest on the slide, but do not image a single slide for longer than 15 min. After 15 min on the slide, cells lose viability.
5. Analysis
Mitochondrial ATP level is determined by measuring the ratio of the mitGO-ATeam2 emission at 560 nm to that at 510 nm.27 The redox state of the organelle is measured as the reduced to oxidized (R/O) ratio of mito-roGFP; i.e., emission at 510 nm upon excitation at 365 nm divided by emission at 510 nm upon excitation at 470 nm. Before calculating the ratio, we subtract background and determine a threshold value for pixels belonging to the fluorescent mitochondria.
Public domain (e.g. ImageJ30) or commercially available (e.g. Volocity, Perkin-Elmer) software can be used for analysis of mito-roGFP1 or mitGO-ATeam2. Depending on the software used for image acquisition and for analysis, the images may first need to be converted to another format, such as TIFF, before opening them in the analysis software. If images are converted, it is essential to verify that pixel values are not changed during the conversion. Analysis of mito-roGFP1 data, using both programs is described below. Program menus and options to select within each menu are highlighted in bold italics.
5.1 ImageJ analysis
-
5.1.1
Open images and change type to 32 bit: Image→ Type → 32 bit.
-
5.1.2
Draw a region of interest (ROI) in an area where there are no cells. Calculate the mean intensity in this ROI: Analyze → Measure.
-
5.1.3
Subtract the calculated mean background from the stack: Process → Math → Subtract.
-
5.1.4
Using the subtracted z-stack, find the middle slice and threshold on mitochondria: Image → Adjust → Threshold and click Apply on the Threshold window. Apply to all slices in the stack. Check Set background pixels to NaN.
-
5.1.5
Create the ratio z stack: Process → Image Calculator Divide the reduced stack by the oxidized z stack for mito-roGFP1 analysis. Divide the 560 nm (ATP bound) image by the 510 nm (ATP unbound) image for mitGO-ATeam2 analysis.
-
5.1.6
Draw an ROI around the area of interest. Choose Analyze → Tools → ROI Manager, and click Add to record the ROI. Multiple regions may be stored in the manager. In ROI Manager, select all ROIs, then choose More → Multi-Measure to measure all stack slices. Export data to a spreadsheet for analysis.
5.2 Volocity analysis
-
5.2.1
Import images into a Volocity library and create an image sequence with 2 channels.
-
5.2.2
Draw a region of interest (ROI) in an area where there are no cells. Choose: Tools → Ratio.
-
5.2.3
Use Volocity to calculate the background: Get From ROI.
-
5.2.4
Adjust the threshold to include mitochondrial structures.
-
5.2.5
Check the option to apply a rainbow LUT to the ratio channel. The intensity-modulated channel may produce a less noisy image for presentation, but it should not be used for quantitation of average ratio.
-
5.2.6
Select the Measurement tab, select the ratio channel and draw an ROI around the area of interest. Measure the ratio channel, excluding zero values. Multiple regions may be selected and measured at the same time. Export data to a spreadsheet for analysis.
RESULTS
Measuring mitochondrial redox state with mito-roGFP
Here, we show that mito-roGFP1 has the dynamic range to detect changes in mitochondrial redox state from fully oxidized to reduced in living yeast cells, without affecting yeast cell growth or mitochondrial morphology. First, we find cells expressing mitochondria-targeted GFP and roGFP1 grow at normal rates (Fig. 1A). The maximum growth rate, as measured by maximum slope of the growth curve during log-phase growth and the time to reach maximum growth rate, is similar in cells expressing mito-GFP and mito-roGFP1. Furthermore, mitochondria in yeast expressing mito-roGFP1 exhibit wild-type morphology (Fig. 1B). Specifically, they are tubular, align along the mother-bud axis, and accumulate at the tips of mother and daughter cells. Since mitochondrial dysfunction often induces fragmentation, this normal morphology supports the idea that mito-roGFP1 does not perturb mitochondrial function.
Fig. 1.
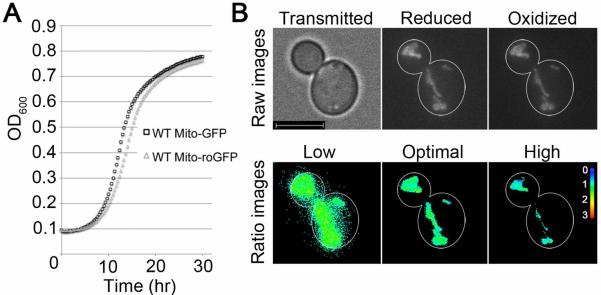
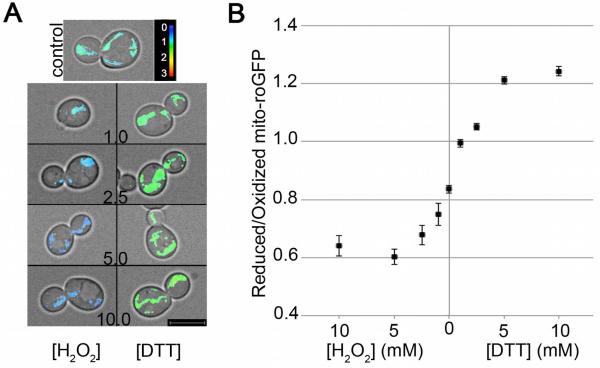
Mito-roGFP1 detects mitochondrial redox state without affecting cell growth rates or mitochondrial morphology. (A) Growth of yeast that express mitochondria-targeted GFP or ro-GFP1 was measured as change in optical density at 600 nm as a function of time of growth in SC-Ura liquid medium at 30°C. (B) Upper panels: Normal mitochondrial morphology and localization channel in raw images. Lower panels: Ratio images show the reduced channel divided by the oxidized channel (color reference at right). The images show the effect of threshold choice on results. The optimal threshold includes mitochondrial structures and excludes background
To assess the dynamic range of mito-roGFP1, we treated mid-log phase wild-type yeast cells with hydrogen peroxide (H2O2) and dithiothreitol (DTT), respectively, and measured the mean mitochondrial R/O ratio to assess mitochondrial redox state (Fig. 2). Titration with H2O2 or DTT from 0 to 10 mM results in a dose-dependent change in mean cellular R/O ratio with a measured range from 0.6 (under oxidizing conditions) to 1.23 (under reducing conditions). Thus, mito-roGFP1 is an effective biosensor for analysis of mitochondrial redox state in living cells.
Fig. 2.
Mito-roGFP1 detects changes in mitochondrial redox state in response to treatment with hydrogen peroxide or dithiothreitol. (A–B) Mid-log phase yeast cells expressing mito-roGFP1 were incubated in SC-Ura media with no treatment (0) or with the indicated concentrations of H2O2 or DTT for 20 min at 30°C. (A) Maximum intensity projections of the R/O mito-roGFP1 ratio images are superimposed on transmitted light images, which show cell outlines. The color reference for mito-roGFP1 is shown at upper right. Bar: 5 μM. (B) Quantitation of R/O mito-roGFP1 ratio under each experimental condition. Error bars are standard error of the mean. Results were obtained from at least 20 cells under each condition.
Mito-roGFP1 also offers subcellular resolution of mitochondrial redox state. This subcellular resolution reveals that mitochondria within individual yeast cells differ in relative redox state. If mitochondria within a single yeast cell are functionally distinct, it is possible that they are passively or actively segregated (Fig. 3). Such segregation could contribute to mother-daughter age asymmetry and the rejuvenation of daughter cells. This finding underscores the need for a biosensor with subcellular resolution of mitochondrial redox state.
Fig. 3.
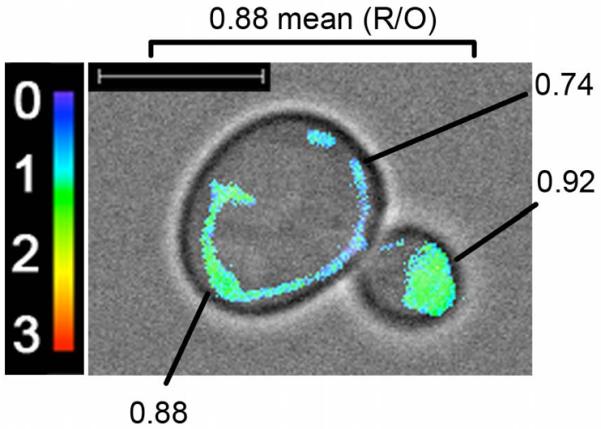
Mito-roGFP1 offers subcellular resolution of mitochondrial redox state. A maximum intensity projection of the R/O mito-roGFP1 ratio channel is superimposed on a transmitted-light image. The color reference for mito-roGFP1 is shown at upper right. Bar: 5 μM. This particular cell has a mean R/O mito-roGFP1 ratio of 0.88. However, individual mitochondria within this cell show different redox states. The numbers shown are the calculated R/O mito-roGFP1 ratio for different mitochondrial regions.
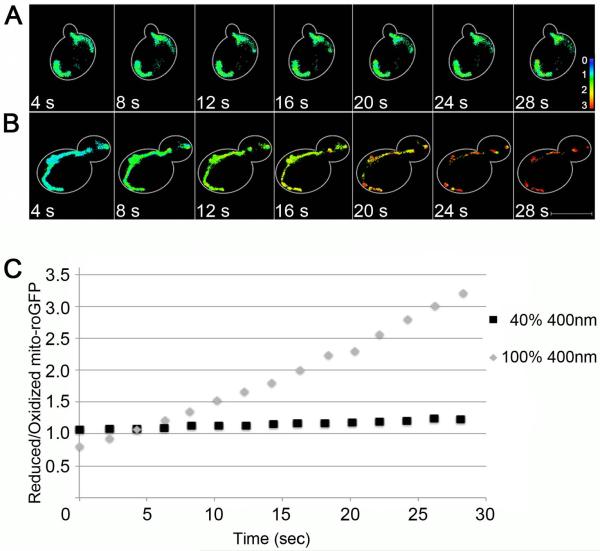
It has long been known31 that light can induce changes in the GFP chromophore. Upon exposure to high intensity 400 nm light, roGFP1 undergoes photoconversion to a species with a different emission spectrum26. When photoconversion occurs, there is a decrease in green emission from oxidized mito-roGFP1 and an increase in green emission upon excitation of reduced mito-roGFP1, which alters the ratio of R/O mito-roGFP1 (Fig. 4B). Using low-intensity excitation (e.g. illumination at 40% using the 400 nm LED in the Colibri light source), there is no significant photoconversion during the period analyzed (Fig. 4C). The preferred way to reduce photoconversion is to excite the oxidized form of roGFP at 365 nm (see Discussion).
Fig. 4.
High-intensity excitation leads to photoconversion and altered R/O mito-roGFP1 ratios. Mid-log phase yeast expressing mito-roGFP1 were illuminated with 400 nm light at 40% (A) or 100% (B) power from a LED light source, and the fluorescence from reduced and oxidized mito-roGFP1 was captured every 4 sec. Images shown are maximum projections in which colors represent the R/O ratio of mito-roGFP1 (see color scale in lower right). Bar: 5 μM. (C) The R/O ratio of mito-roGFP1 remained constant over the imaging period upon illumination at 40% LED power. However, upon illumination at 100% LED power, we observe a time-dependent change in R/O ratio of mito-roGFP1, which reflects photoswitching of the fluorophore. Black squares, 40% LED power; gray diamonds, 100% LED power
Measuring mitochondrial ATP with mitGO-ATeam2
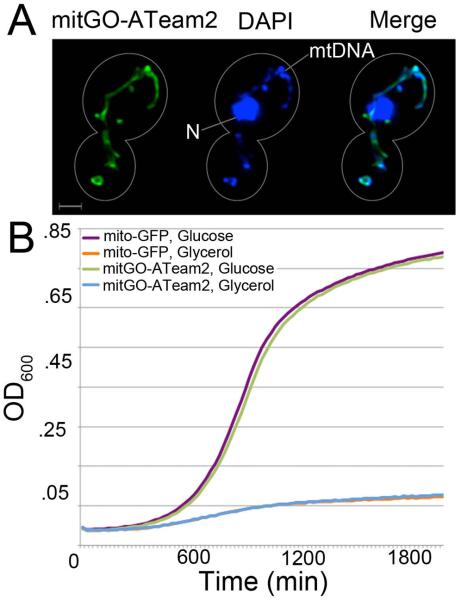
MitGO-ATeam2 localizes to mitochondria when expressed in mammalian cells.27 We find that mitGO-ATeam2 colocalizes with DAPI-stained mitochondrial DNA in yeast, and is found in tubular structures typical of wild-type yeast mitochondria (Fig. 5A). Thus, we verified that the mitochondrial targeting sequence from the mammalian cytochrome c oxidase subunit VIIIA localizes the probe to mitochondria in yeast without disrupting mitochondrial morphology. Additionally, we find that the growth rate of cells expressing mitGO-ATeam2 is similar to that of cells expressing mitochondria-targeted GFP in both glucose and glycerol media (Fig. 5B). Thus, expression of mitGO-ATeam2 does not appear deleterious to the cell or mitochondria.
Fig. 5.
MitGO-ATeam2 localizes to mitochondria in yeast and does not affect yeast cell growth rates. (A) Yeast cells expressing mitGO-ATeam2 were grown to mid-log phase, fixed using paraformaldehyde and stained with the DNA-binding dye DAPI, as described previously.19 Images shown are maximum-intensity projections of deconvolved z-series. For simplicity, the mitGO-ATeam2 images were obtained using a conventional GFP filter, so they represent only the population unbound to ATP. Cell outlines are shown in white. N: nuclear DNA. mtDNA: mitochondrial DNA. Bar: 1 μm. (B) Growth curves of wild-type yeast cells expressing mitochondria-targeted GFP or mitGO-ATeam2 in glucose-based (SC) and glycerol-based (SGlyc) liquid media at 30°C. This data is the average of quadruplicates for each strain at each time point, and is representative of two independent experiments.
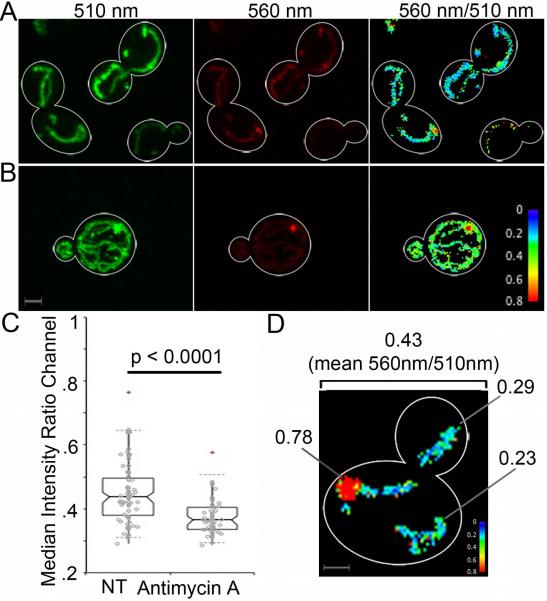
Next, we tested whether the mitGO-ATeam2 FRET responds to changes in mitochondrial ATP levels. To do so, we treated cells with antimycin A, an agent that binds to cytochrome c reductase, inhibits oxidation of ubiquinol in the electron transport chain, disrupts the proton gradient, and inhibits mitochondrial ATP production.20 The median FRET ratio decreases in cells treated with Antimycin A (Fig. 6C).
Fig. 6.
MitGO-ATeam2 measures changes in ATP levels in yeast mitochondria at subcellular and suborganellar resolution. (A–B) Emission of mitGO-ATeam2 from GFP (510 nm) and OFP (560 nm), and quantitation of the 560/510 nm emission ratio of yeast cells grown overnight in glucose-based (SC) (A) or glycerol-based (SGlyc) (B) media. The color reference for the 560/510 ratio channel is shown at lower right. Bar: 1 μm. (C) Quantitation of the 560/510 ratio for cells propagated in SGlyc media with or without antimycin A (2 μg/ml) for 1 hr at 30°C. The decrease in ATP levels upon antimycin A treatment is statistically significant (Kruskal-Wallis significance test). (D) 560 nm/510 nm ratio of mitGO-ATeam2 of a midlog phase wild-type cell propagated on SC media. The color reference for the 560/510 ratio channel is shown at lower right. The cell outline is shown in white. The mean 560/510 nm ratio for the entire cell is 0.43. The numbers shown are the 560/510 nm ratios of specific regions within mitochondria. Bar: 1 μm.
Given evidence that mitGO-ATeam2 is an effective biosensor for mitochondrial ATP, we used this probe to monitor mitochondrial ATP levels in yeast that were propagated using a fermentable or non-fermentable carbon source (Fig. 6A,B). The reduction in the total fluorescent area in glucose-grown cells confirmed that glucose repression results in a decrease in mitochondrial abundance. We note cell-to-cell variation in the level of mitGO-ATeam2. Interestingly, our preliminary data indicates that there may be differences in ATP levels in different mitochondria within the same cell (Fig. 6D).
DISCUSSION
Here, we describe methods to use mito-roGFP1 and mitGO-ATeam2 as biosensors to assess mitochondrial redox state and ATP levels in living yeast cells. We find that expression of plasmid-borne mito-roGFP1 or mitGO-ATeam results in quantitative targeting to mitochondria, without any obvious effect on mitochondrial morphology or distribution or on cellular growth rates.3 Mito-roGFP1 can detect changes in mitochondrial redox state from highly oxidized to highly reduced states. Similarly, mitGO-ATeam can measure changes in mitochondrial ATP levels that occur in yeast undergoing respiration-driven growth and yeast treated with a respiration inhibitor. Moreover, since both biosensors offer subcellular resolution, they can detect heterogeneity in the redox state or ATP levels of mitochondria within individual cells. Finally, since both biosensors are ratiometric probes, they are not affected by changes in concentration or variability in sample thickness or illumination. These probes provide a minimally invasive approach to studying mitochondrial function with subcellular resolution in living cells.
To successfully apply this method, images should be acquired with the maximum possible signal-to-noise ratio. An important first step is to examine cells from 10–20 transformant colonies under the microscope and select those with robust fluorescence and normal mitochondrial morphology and growth rates. Next, imaging conditions should be optimized to produce high, but not saturating, intensities without causing photodamage to the fluorescent protein or the cells. A few cells in a population (<1 %) may have unusually high or low overall fluorescence due to variation in plasmid copy number; these can be disregarded in favor of capturing interpretable images of the majority of cells.
While the benefits of mito-roGFP are clear, it is also important to be aware of potential pitfalls. We detect differences in mitochondrial redox potential not just with carbon source-induced differences in cell metabolism, but also upon propagation of yeast in different batches of the same medium. Thus, care must be taken when selecting cell growth media for mitochondrial redox measurements. Moreover, although mito-roGFP offers unprecedented spatial resolution of mitochondrial redox state, temporal resolution of roGFP is not sufficient to detect bursts of ROS that are associated with ROS signal transduction31. Finally, care must be taken when choosing the excitation wavelength and intensity for the oxidized form of roGFP. Illumination at 400 nm optimally excites the oxidized species of roGFP. However, it also excites reduced roGFP to a greater extent compared to excitation at 365 nm. This leads to reduced sensitivity in measuring mitochondrial redox state. Furthermore, exposure of mito-roGFP1 to high-intensity illumination at 400 nm leads to photoconversion of the protein to a species with altered spectral properties. We have not seen photoconversion upon high-intensity illumination at 365 nm. Therefore, we recommend excitation using 365 nm if possible.
MitGO-ATeam2 also has limitations. For example, since mitGO-ATeam2 is itself a protein it can be damaged by cellular insults including reactive oxygen species, which may impair its biosensor activity. Additionally, GFP and OFP emission wavelengths (510 and 560 nm) must be resolved for FRET analysis, which may be difficult on conventional fluorescence microscopes.
Unlike in vitro enzyme assays, these live-cell assays report relative changes in mitochondrial ATP or redox state, and do not measure the absolute concentration of ATP or reduced cysteine. Although a standard curve could theoretically be generated by measuring the response of sensor proteins in solution, this might not apply to the different molecular environment within mitochondria. Therefore, these assays provide a means of comparing cells under different conditions. In addition, imaging conditions vary, so the exact ratio values acquired on one imaging setup may not be replicated on another.
These techniques for ratio imaging can be adapted for other ratiometric indicators, including other roGFP or GO-ATeam isoforms and other functional probes. The choice of wide-field or confocal microscopy will depend on the probe and the spatial resolution required. The wide-field method was used here for mito-roGFP because it allows excitation at 365 nm, which improves sensitivity and stability of the sensor. The confocal method with a spectral detector, as used here for mitGO-Ateam, provides a convenient way to eliminate emission crossover by adjusting the spectral detection windows. However, it is possible to perform similar experiments in wide-field microscopy by using custom filters or eliminating crossover computationally.
The most common difficulty in this type of imaging experiment is insufficient signal. Imaging hardware (especially the objective lens and detector) is critical for collecting enough signal to interpret the data.
The thresholding steps have great influence on the results, as the inclusion of background pixels can dampen any trends in the ratios obtained. Automatic methods are preferable, but due to limited signal, manual thresholding is often the best available method. If manual thresholds are used, the criteria used should be articulated as well as possible, and analysis may need to be performed blind or by different experimenters to demonstrate reproducibility. Control experiments with antimycin A (for mitGO-Ateam2) and H2O2 or DTT (for mito-roGFP) can be used to confirm the predicted changes in fluorescence ratios.
Mito-roGFP and mitGO-ATeam2 are non-invasive, ratiometric probes for monitoring mitochondrial fitness in living yeast cells. The ratiometric sensors used here were targeted to mitochondria by fusion of mitochondria-specific signal sequences. Other cellular compartments could be studied with the addition of appropriate targeting sequences to the original sensors. In addition, it is possible to combine either of these sensors with complementary fluorescent markers (e.g. for organelles or signaling factors) or sensors (e.g. for calcium) to simultaneously measure multiple processes in living cells.
REAGENTS:
| Name | Company or ingredients | Catalog Number | Comments |
|---|---|---|---|
|
| |||
| Antimycin A | Sigma-Aldrich (St. Louis, MO) | 1397-94-0 | Dissolved in ethanol to a 2 mg/mL stock solution. |
|
| |||
| SGIyc (synthetic glycerol-based) yeast growth medium | 0.67% Yeast nitrogen base without amino acids | N/A | Dissolve in H2O. Adjust pH to 5.5 with NaHCO3. Autoclave. |
| 3% Glycerol | |||
| *omit for SGIyc-Ura | 0.05% Glucose | ||
| **omit for SGIyc-Leu | 2 mg/mL adenine | ||
| 2 mg/mL uracil* | |||
| 1 mg/mL L-arginine | |||
| 1 mg/mL L-histidine | |||
| 1 mg/mL L-leucine** | |||
| 3 mg/mL L-lysine | |||
| 2 mg/mL L-methionine | |||
| 4 mg/mL L-phenylalanine | |||
| 2 mg/mL L-tryptophan | |||
| 3 mg/mL L-tyrosine | |||
|
| |||
| SC (synthetic complete, glucose-based) yeast growth medium | 0.67% Yeast nitrogen base without amino acids | N/A | Dissolve in H2O. Adjust pH to 5.5 with NaHCO3. Autoclave. |
| 3% Glucose | |||
| *omit for SGIyc-Ura | 2 mg/mL adenine | ||
| **omit for SGIyc-Leu | 2 mg/mL uracil* | ||
| 1 mg/mL L-arginine | |||
| 1 mg/mL L-histidine | |||
| 1 mg/mL L-leucine** | |||
| 3 mg/mL L-lysine | |||
| 2 mg/mL L-methionine | |||
| 4 mg/mL L-phenylalanine | |||
| 2 mg/mL L-tryptophan | |||
| 3 mg/mL L-tyrosine | |||
|
| |||
| Valap | Vaseline petroleum jelly, hard paraffin, lanolin | N/A | Combine ingredients in a 1:1:1 (w:w:w) ratio. Melt by submerging in a 70°C H2O bath. Aliquot into glass petri dishes. Store at room temperature. |
EQUIPMENT AND SOFTWARE:
| Name | Company | Catalog Number | Comments |
|---|---|---|---|
| Precleaned Gold Seal Rite-on Micro Slides | Thomas Scientific (Swedesboro, NJ) | 3050 | Size: 25 × 75 mm; Thickness: 0.93 to 1.05 mm |
| High-performance coverslips, No. 1.5, 18×18 mm | Zeiss (Thornwood, NY) | 474030-9000-000 | These are less variable in thickness (170±5 μm) than standard coverslips, reducing spherical aberration and improving 3D imaging performance |
| Fisherbrand Microscope Cover Glass, No. 1.5 | Fisher Scientific (Pittsburgh, PA) | 12-545E | Size: 22 × 22 mm, No. 1.5 thickness (170 μm) |
| A1 laser scanning confocal microscope with spectral detector and 100x/1.49NAApo-TIRF objective | Nikon (Melville, NY) | N/A | |
| AxioObserver.Z1 microscope equipped with a 100x/1.3NA EC Plan-Neofluar objective (Zeiss) and Orca ER cooled CCD camera (Hamamatsu) and controlled by Axiovision software | Zeiss (Thornwood, NY); Hamamatsu (Hamamatsu City, Japan) | ||
| Volocity 3D Image Analysis software | Perkin Elmer (Waltham, MA) | N/A | Restoration module for deconvolution; Quantitation module for ratio calculation and measurement |
| ImageJ software | National Institutes of Health (Bethesda, MD) | http://rsb.info.nih.gov/ij/ |
ACKNOWLEDGMENTS
This work was supported by awards from HHMI 56006760 to JDV, the National Institutes of Health (NIH) (2 TL1 RR 24158-6) to DMAW, and from the Ellison Medical Foundation (AG-SS-2465) and the NIH (GM45735, GM45735S1 and GM096445) to LP. GM45735S1 was issued from the NIH under the American Recovery and Reinvestment Act of 2009. The microscopes used for these studies were supported in part through a NIH / NCI grant (5 P30 CA13696).
Footnotes
DISCLOSURES: No conflicts of interest declared.
REFERENCES
- 1.Rizzuto R, De Stefani D, Raffaello A, Mammucari C. Mitochondria as sensors and regulators of calcium signalling. Nature reviews. Molecular cell biology. 2012;13(9):566–78. doi: 10.1038/nrm3412. [DOI] [PubMed] [Google Scholar]
- 2.Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148(6):1145–59. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McFaline-Figueroa JR, Vevea J, et al. Mitochondrial quality control during inheritance is associated with lifespan and mother-daughter age asymmetry in budding yeast. Aging Cell. 2011;10(5):885–895. doi: 10.1111/j.1474-9726.2011.00731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamilton ML, Van Remmen H, et al. Does oxidative damage to DNA increase with age? Proc Natl Acad Sci USA. 2001;98:10469–10474. doi: 10.1073/pnas.171202698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cadenas E, Davies KJ. Mitochondrial free radical generation, oxidative stress, and aging. Free radical biology & medicine. 2000;29(3–4):222. doi: 10.1016/s0891-5849(00)00317-8. [DOI] [PubMed] [Google Scholar]
- 6.Mammucari C, Rizzuto R. Signaling pathways in mitochondrial dysfunction and aging. Mechanisms of ageing and development. 2010;131(7–8):536–43. doi: 10.1016/j.mad.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443(7113):787–95. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 8.Schon EA, Przedborski S. Mitochondria: the next (neurode)generation. Neuron. 2011;70(6):1033–53. doi: 10.1016/j.neuron.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor ER, Hurrell F, Shannon RJ, Lin T-K, Hirst J, Murphy MP. Reversible glutathionylation of complex I increases mitochondrial superoxide formation. The Journal of biological chemistry. 2003;278(22):19603–10. doi: 10.1074/jbc.M209359200. [DOI] [PubMed] [Google Scholar]
- 10.Beer SM, Taylor ER, et al. Glutaredoxin 2 catalyzes the reversible oxidation and glutathionylation of mitochondrial membrane thiol proteins: implications for mitochondrial redox regulation and antioxidant defense. The Journal of biological chemistry. 2004;279(46):47939–51. doi: 10.1074/jbc.M408011200. [DOI] [PubMed] [Google Scholar]
- 11.Tretter L, Adam-Vizi V. Inhibition of Krebs cycle enzymes by hydrogen peroxide: A key role of [alpha]-ketoglutarate dehydrogenase in limiting NADH production under oxidative stress. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2000;20(24):8972–9. doi: 10.1523/JNEUROSCI.20-24-08972.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nulton-Persson a C., Szweda LI. Modulation of mitochondrial function by hydrogen peroxide. The Journal of biological chemistry. 2001;276(26):23357–61. doi: 10.1074/jbc.M100320200. [DOI] [PubMed] [Google Scholar]
- 13.Gardner PR, Fridovich I. Effect of glutathione on aconitase in Escherichia coli. Archives of biochemistry and biophysics. 1993;301(1):98–102. doi: 10.1006/abbi.1993.1120. [DOI] [PubMed] [Google Scholar]
- 14.Gardner PR, Raineri I, Epstein LB, White CW. Superoxide radical and iron modulate aconitase activity in mammalian cells. The Journal of biological chemistry. 1995;270(22):13399–405. doi: 10.1074/jbc.270.22.13399. [DOI] [PubMed] [Google Scholar]
- 15.Schafer F, Buettner G. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radical Biology and Medicine. 2001;30(11):1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 16.Maechler P, Wang H, Wolheim CB. Continuous monitoring of ATP levels in living insulin secreting cells expressing cytosolic firefly luciferase. FEBS Lett. 1998;422:328–332. doi: 10.1016/s0014-5793(97)01618-9. [DOI] [PubMed] [Google Scholar]
- 17.Ouhabi R, Boue-Grabot M, Mazat JP. Mitochondrial ATP synthesis in permeabilized cells: Assessment of the ATP/O values in situ. Anal. Biochem. 1998;263:169–175. doi: 10.1006/abio.1998.2776. [DOI] [PubMed] [Google Scholar]
- 18.Strehler BL, Totter JR. Firefly luminescence in the study of energy transfer mechanism in substrates and enzymes determinations. Arch. Biochem. Biophys. 1952;40:28–41. doi: 10.1016/0003-9861(52)90070-2. [DOI] [PubMed] [Google Scholar]
- 19.Lemasters JJ, Backenbrock CR. Adenosine triphosphate: Continuous measurement in mitochondrial suspension by firefly luciferase luminescence. Biochem. Biophys. Res. Commun. 1973;55:1262–1270. doi: 10.1016/s0006-291x(73)80030-0. [DOI] [PubMed] [Google Scholar]
- 20.Wibom R, Lundin A, Hultman E. A sensitive method for measuring ATP formation in rat muscle mitochondria. Scand. J. Clin. Lab. Invest. 1990;50:143–152. doi: 10.1080/00365519009089146. [DOI] [PubMed] [Google Scholar]
- 21.Manfredi, Giovanni, Spinazzola, Checcarelli, Nicoletta, Naini Ali. Assay of Mitochondrial ATP Synthesis in Animal Cells. In: Pon Liza A., Schon Eric A., editors. Methods in Cell Biology, Mitochondria. Vol. 65. Academic Press; New York: 2001. pp. 133–145. [DOI] [PubMed] [Google Scholar]
- 22.DeLuca M. Firefly luciferase. Adv. Enzymol. Relat. Areas Mol. Biol. 1976;44:37–68. doi: 10.1002/9780470122891.ch2. [DOI] [PubMed] [Google Scholar]
- 23.Atamna Hani, Frey, William H., II Mechanisms of mitochondrial dysfunction and energy deficiency in Alzheimer's disease. Mitochondrion. 2007;7:297–310. doi: 10.1016/j.mito.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 24.Hanson GT, Aggeler R, et al. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J. Biol. Chem. 2004;279(13):13044–53. doi: 10.1074/jbc.M312846200. [DOI] [PubMed] [Google Scholar]
- 25.Dooley CT, Dore TM, Hanson GT, Jackson C, Remington SJ, Tsien RY. Imaging dynamic redox changes in mammalian cells with green fluorescent protein indicators. J. Biol. Chem. 2004;279(21):22284–93. doi: 10.1074/jbc.M312847200. [DOI] [PubMed] [Google Scholar]
- 26.Schwarzländer M, Fricker MD, et al. Confocal imaging of glutathione redox potential in living plant cells. Journal of microscopy. 2008;231(2):299–316. doi: 10.1111/j.1365-2818.2008.02030.x. [DOI] [PubMed] [Google Scholar]
- 27.Nakano M, Imamura H, Nagai T, Noji H. Ca2+ regulation of mitochondrial ATP synthesis visualized at the single cell level. ACS Chem Biol. 2011;6(7):709–715. doi: 10.1021/cb100313n. [DOI] [PubMed] [Google Scholar]
- 28.Berg J, Hung YP, Yellen G. A genetically encoded fluorescent reporter of ATP:ADP Ratio. Nature Methods. 2009;6(2):161–166. doi: 10.1038/nmeth.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods in enzymology. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 30.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science (New York, N.Y.) 2002;297(5588):1873–7. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- 32.Dairaku N, Kato K, et al. Oligomycin and antimycin A prevent nitric oxide-induced apoptosis by blocking cytochrome c leakage. J Lab Clin Med. 2004;143(3):141–153. doi: 10.1016/j.lab.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 33.Cannon MB, Remington SJ. Re-engineering redox-sensitive green fluorescent protein for improved response rate. 2006. pp. 45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]