Abstract
Alterations in the tumor suppressor p53 have been reported in tumor-associated stromal cells; however, the consequence of these alterations has not been elucidated. We investigated p53 status and responses to p53-activating drugs using tumor-associated stromal cells from A375 melanoma and PC3 prostate carcinoma xenografts, and a spontaneous prostate tumor model (TRAMP). P53-accumulation after treatment with different p53-activating drugs was diminished in tumor-associated stromal cells compared to normal stromal cells. Tumor-associated stromal cells were also less sensitive to p53-activating drugs – this effect could be recapitulated in normal stromal cells by p53-knockdown. Unlike normal stromal cells, tumor stromal cells failed to arrest in G2 after etoposide treatment, failed to up-regulate p53-inducible genes, and failed to undergo apoptosis after treatment with vincristine. The lower levels of p53 in tumor stromal cells accompanied abnormal karyotypes and multiple centrosomes. Impaired p53 function in tumor stroma might be related to genomic instability and could enable stromal cell survival in the destabilizing tumor microenvironment.
Keywords: Cancer, Tumor, Tumor microenvironment, Tumor stroma, p53, Drug resistance
Introduction
Both cancer and healing wounds are characterized by increased proliferation and infiltration of endothelial cells, inflammatory cells, and fibroblasts (Dvorak, 1986). These infiltrating stromal cells (SC) may by tissue-resident and/or bone marrow-derived and together constitute the stromal microenvironment. The function of SC in tumors and physiological wound healing is to secrete matrix proteins and factors involved in tissue remodeling, and to secrete chemotactic factors for inflammatory cells and endothelial cells. Thus, both tissue injury and cancer resemble an “activated” state whereby the host's response is designed to heal the affected tissue (Coussens & Werb, 2002).
The tumor microenvironment consists of tumor cells, SC, and the matrix proteins, growth factors, and cytokines they produce (Liotta & Kohn, 2001). Fibroblasts comprise the majority of the tumor stroma, and their role in tumors is long-recognized (Seemayer et al., 1979). For example, fibroblasts are responsible for the synthesis of fibronectin and types I, III, and V collagens that make up basement membranes; they also secrete factors such as TGFβ that support non autonomous tumor epithelial growth (Bhowmick et al., 2004; Tlsty, 2001). Functionally and phenotypically distinct myofibroblasts are also observed in most tumors (Sappino et al., 1988). Myofibroblasts express alpha smooth muscle actin (α SMA) which enhances their contractility and motility (Hinz et al., 2007) and they over-express stromal-derived factor (SDF-1) which mobilizes bone marrow-derived endothelial progenitor cells (Orimo et al., 2005).
Due to genomic instability, cancer cells are typically mutable and develop drug resistance; thus, targeting SC in the tumor microenvironment may be a viable approach for helping to eliminate solid tumors (e.g. anti-angiogenesis). However, it has recently become clear that like tumor cells, tumor-associated stromal cell may be characterized by genomic instability and p53 mutations (Hill et al., 2005; Kurose et al., 2002). For example, numerous genetic amplifications and deletions were detected in murine stromal DNA isolated from implanted tumor xenografts (Pelham et al., 2006). Furthermore, cytogenetic (Allinen et al., 2004; Fukino et al., 2004; Hida et al., 2004; Moinfar et al., 2000) and epigenetic (Kaplan et al., 2005) alterations have been described in tumor stroma and in tissue adjacent to carcinoma (Deng et al., 1994). The possibility that cytogenetic alterations in tumor SC including p53 mutations might accompany changes in p53 function have not been addressed.
In cancer cells, the principal cause of resistance to chemotherapeutic drugs is chromosomal instability accompanied by deletion of or mutations in p53; however, both drug resistance and increased sensitivity in a p53 null background have been noted (Bunz et al., 1999; Burdelya et al., 2006). P53 is mutated or lost in about half of all cancers probably as a consequence of selection pressure for p53 mutations which enable tumor cell survival (Gorgoulis et al., 2005). The importance of functional p53 in tumors has been formally proven by reintroduction of wild-type p53 in tumor cells in vivo (Martins et al., 2006; Ventura et al., 2007). However, it has been noted that re-imposition of p53 function was quickly mitigated by p53 inactivation and the emergence of p53-resistant tumors (Martins et al., 2006). Despite these detailed studies investigating p53 function in tumor cells, no studies to date have examined the relationship between p53 status and sensitivity to cytotoxic, DNA damaging agents in tumor-associated SC.
In this report, we show tumor SC, in contrast to normal SC, fail to undergo growth arrest and apoptosis after treatment with etoposide or vincristine, respectively. Tumor SC show diminished p53 expression, and p53 fails to or only marginally accumulates after treatment with p53-activating stimuli. Tumor SC also have multiple centrosomes and are aneuploid – both of which are readouts of abnormal p53 function. Together, these results indicate that, similar to tumor cells, alterations in p53 function and decreased sensitivuty to commonly used p53-activating chemotherapeutic drugs is a feature of tumor-associated SC.
Materials and Methods
Cell lines and media
All primary cells were cultured in EGM2-MV medium (Cambrex Bioscience, Rockland, ME) and were maintained in an atmosphere of 5 % CO2 at 37 C. PC3MLN4 prostate carcinoma cells were grown in HAMS with 10% FBS and A375SM melanoma cells were grown in MEM with 10% FBS. Tumor cells were grown under 10 % CO2. Normal and tumor SC were obtained from isolated endothelial cultures overtaken by rapidly growing fibroblast-like cells. This was a common occurrence in our hands, and was unavoidable without strict monitoring of the pure endothelial cultures (Hida et al., 2004).
Mice
All animal procedures were performed in compliance with Boston Children's Hospital Guidelines and were approved by the Institutional Animal Use and Care Committee. The procedures for cell isolation for A375 tumor xenografts were previously described (Hida et al., 2004). For the PC3MLN4 prostate carcinoma, one million cells were injected subcutaneously into the dorsal lateral flank of eight week old nu/nu mice. When tumors reached 1 cm3 (approximately 4-6 weeks post implantation), tumors were harvested as described by Hida et al. Tumors from five mice were combined for the cell isolation. TRAMP mice were genotyped at four weeks of age (Transnetyx, Cordova, TN) and tumors were harvested when mice reached 20-22 weeks.
Antibodies and reagents
The mouse monoclonal p53 and rabbit polyclonal pSER15 and pSER20 p53 antibodies were purchased from Cell Signaling Technologies (Danvers, MA). β actin, caldesmon, α SMA, and FSP-1 antibodies were from Sigma-Aldrich (St. Louis, MO). The CD105 antibody was from BD Pharmingen (San Jose, CA). Rabbit polyclonal pericentrin antibody was from Covance (Berkeley, CA). Etoposide and vincristine were dissolved in DMSO and were purchased from Sigma-Aldrich.
Drug treatment
To analyze p53 expression, cells were seeded at a density of 200,000 cells/10 cm2 and left overnight. The next day, the indicated drug was added, and cells were incubated between eight and 24 hours. Cell lysates were prepared in RIPA buffer and subjected to western blotting according to standard methods. Nuclear fractions were prepared using the NE-PER kit according to the manufacturer (Pierce, Rockford, IL). For viability studies, cells were seeded at 2500 cells/well in a 96-well plate. The next day, the indicated concentration of each drug was added, and the cells were incubated for an additional 72 hours. Cell counts were determined by dispersing the cells in trypsin and counting using a Coulter counter (Beckman Coulter, Model Z1).
Fluorescence Activated Cell Sorting (FACS)
FACS for cell characterization was carried out on live cells used between six and ten passages. Propidium iodide and annexin V staining were done according to the manufacturer's instructions (BD Pharmingen). Cells were analyzed on a BD FACSCalibur System.
Immunofluorescence
Cells were seeded at a density of 10,000 cells/well in gelatin coated 8-well chamber slides. Confluent cells were washed twice with PBS, and then fixed with ice cold 100 % methanol at -20 C for 20 minutes. The fixed cells were rinsed briefly with PBS and then blocked for one hour at room temperature with PBS containing 5 % BSA. After blocking, antibodies were added overnight at 4 C in a humidified chamber. The next day, cells were rinsed with PBS, and then blocked again for 30 minutes at room temperature. Secondary antibodies were added and the cells were incubated an additional hour at room temperature protected from light. Finally, cells were washed with PBS and then mounted using Gel Mount (Biomeda, Foster City, CA) containing 0.4 μg/mL 4′, 6-diamidino-2-phenylindole (DAPI).
P53 sequencing
cDNA was prepared from normal and tumor SC by reverse transcription. Primers specific for the mouse p53 coding regions were used to amplify a 1,173 base pair fragment which was sub-cloned into the PCR-2 vector (Invitrogen, Carlsbad, CA). Plasmid DNA was cloned in Top10 cells, purified, and sequenced using the same primers for PCR amplification. Sequences were analyzed using Chromas software.
Karyotyping
Karyotyping was carried out by the Brigham and Women's Cytogenetics Core Facility, Boston, MA.
siRNA
Mouse siRNA for p53 was purchased from Dharmacon (Lafayette, CO). Cells were transfected using Silentfect transfection reagent according to the manufacturer's instructions (BioRad, Hercules, CA).
Results
Stromal cell characterization
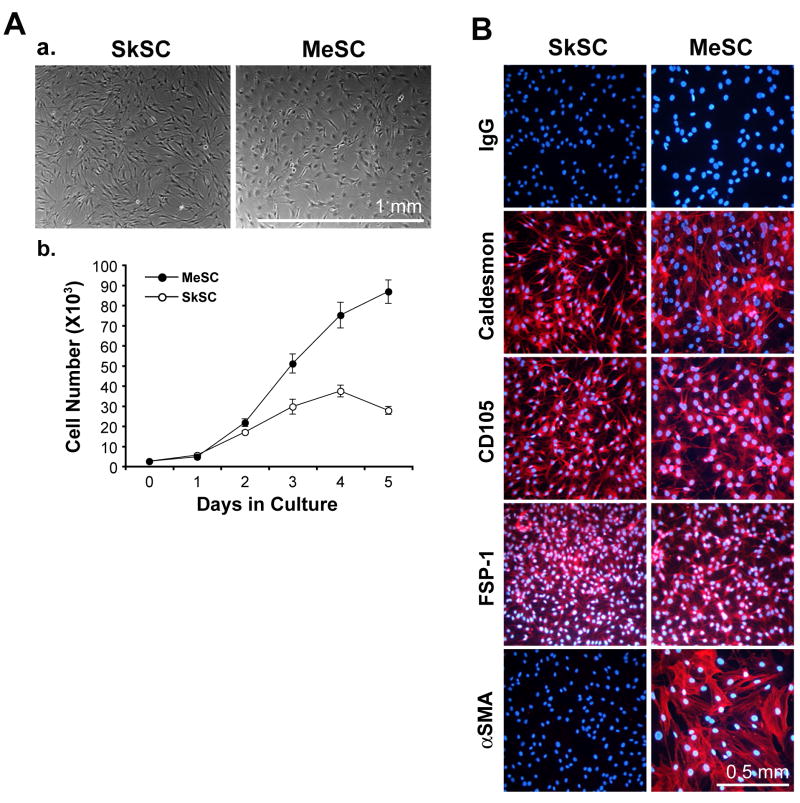
Previously, our laboratory identified cytogenetic alterations in tumor-specific endothelial cells (Hida et al., 2004). In the present study, we found that isolates of endothelial cells from A375 melanoma were frequently overtaken by fibroblast-like, stromal cells (MeSC) which grew on average 2-3 times the rate of normal SC from mouse skin (SkSC) (figure 1A). In contrast to tumor endothelial cells described by Hida et al., these cells were negative for the EC markers CD31 and VEGFR2 (data not shown), but positive for caldesmon, CD105, and fibroblast-specific protein (FSP-1) (figure 1B). α SMA expression was restricted to the SC from melanoma, possibly owing to their “activated” or myofibroblast-like phenotype observed in most tumors (Orimo et al., 2005). Based on morphology and marker expression, the isolated SC used in this study were identified as fibroblasts or myofibroblasts.
Figure 1. Stromal cell characterization.
(A) Morphology of SC from normal mouse skin or subcutaneous xenografts of A375 melanoma (a) and comparison of their growth properties in culture (b). (B) By immunofluorescence, SC stained uniformly positive for caldesmon, CD105, and FSP-1. α SMA was expressed only in the tumor SC, indicative of a myofibroblast-like phenotype.
P53 function in normal and tumor stromal cells
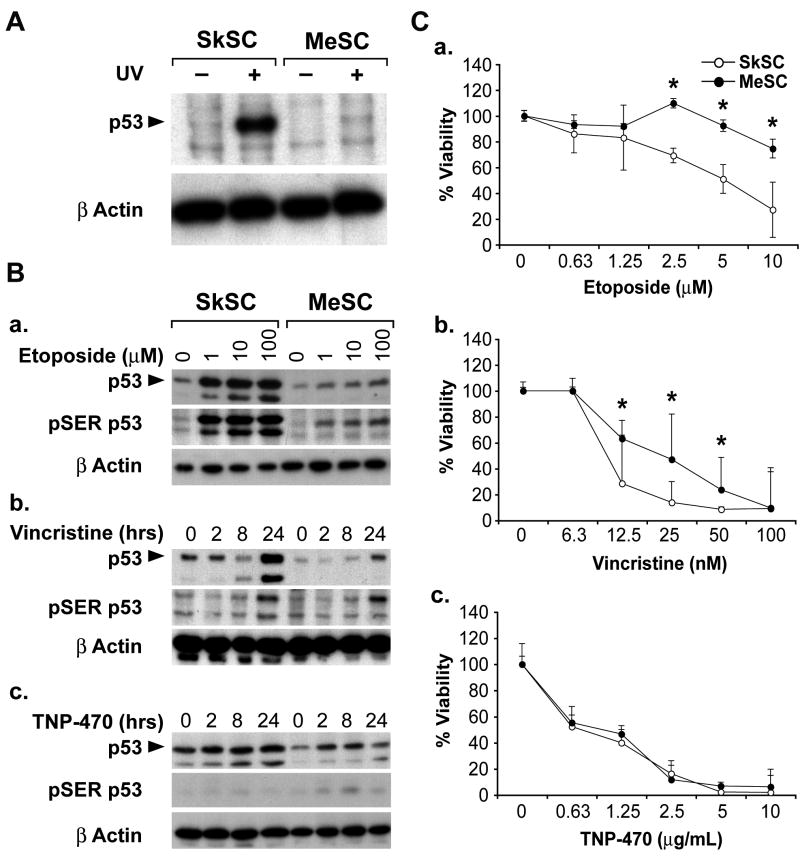
UV light causes DNA damage which is repaired via a p53-dependent mechanism (Nelson & Kastan, 1994). As expected, p53 levels were increased in normal SkSC after UV treatment (figure 2A). Strikingly, p53 only marginally accumulated in MeSC after treatment with UV light. Next, the p53 responses in SkSC or MeSC following treatment with different p53-activating drugs were determined. While etoposide caused an approximate five-fold increase in p53 and SER15-phosphorylated p53 in SkSC at all doses, only a two-fold increase in p53 was evident in MeSC (figure 2B). Time course experiments using vincristine produced similar results, with a maximum 7-8 fold increase in p53 levels in SkSC after drug treatment, but only a 1-2 fold increase in MeSC. TNP-470, a compound not known as a p53-inducer, did not cause a remarkable up-regulation of p53 or p53 phosphorylation at any time point in both SC.
Figure 2. P53 function in tumor stromal cells.
(A) Western blotting of whole cell lysates from UV-treated SkSC and MeSC. Cells were treated with 100mJ/cm2 for five minutes and lysates were prepared six hours later. (B) Western blotting of whole cell lysates from eight hour etoposide-treated (a), one nM vincristine-treated (b), or 10 μg/mL TNP-470-treated cells (c). The same blots were stripped and re-probed with rabbit polyclonal pSER15 p53 antibodies and then mouse monoclonal β actin antibodies. (C) Dose response curves for etoposide (a), vincristine (b), and TNP-470 (c). Cells were plated in triplicate and treated with each drug for 72 hours before dispersing in trypsin and counting. An (*) indicates results are statistically significant (p< 0.05) by student's t-test.
MeSC are less sensitive to p53-activating drugs
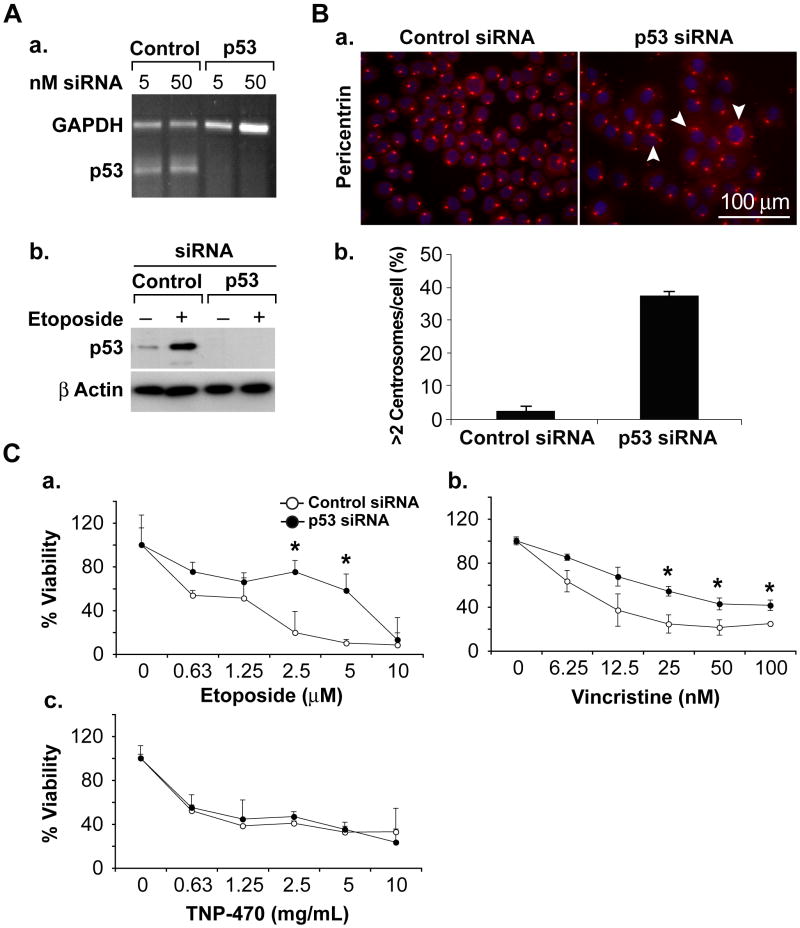
We compared the viability of SkSC and MeSC following treatment with each p53-activating drug. Although both cell types were growth inhibited by etoposide and vincristine, MeSC were consistently less sensitive when compared to SkSC. In contrast, no difference in viability was seen in SkSC or MeSC treated with TNP-470 which also did not up-regulate p53 (figure 2C). To confirm a p53-specific effect, we knocked down p53 in SkSC using p53 siRNA. P53 knockdown was complete at the RNA and protein levels (figure 3A). As a readout of p53 function, we measured centrosome numbers in SkSC after p53 knockdown (Bennett et al., 2004). P53 knockdown in SkSC resulted in multiple centrosomes per cell, consistent with p53's role in regulating centrosome duplication (figure 3B). Furthermore, the dose-response curves for etoposide and vincristine showed that p53 knockdown rendered SkSC less sensitive to each drug (figure 3C). No change in sensitivity to TNP-470 was observed, consistent with TNP-470's failure to activate p53.
Figure 3. P53 knockdown in SkSC decreases sensitivity to etoposide and vincristine.
(A) P53 knockdown was complete at the RNA (a) and protein levels (b) using siRNA. (B) P53 knockdown resulted in multiple, less uniform centrosomes (a) and about 30 % of the cells had an abnormal > 2 pericentrin signals per cell. (C) Cell numbers determined in p53 knockdown SkSC after a 72 hour treatment with etoposide (a), vincristine (b), and TNP-470 (c). Cells were plated in triplicate and treated with each drug for 72 hours before dispersing in trypsin and counting. An (*) indicates results are statistically significant (p< 0.05) by student's t-test.
P53 localizes to the nucleus in MeSC but the total cellular p53 pool is reduced
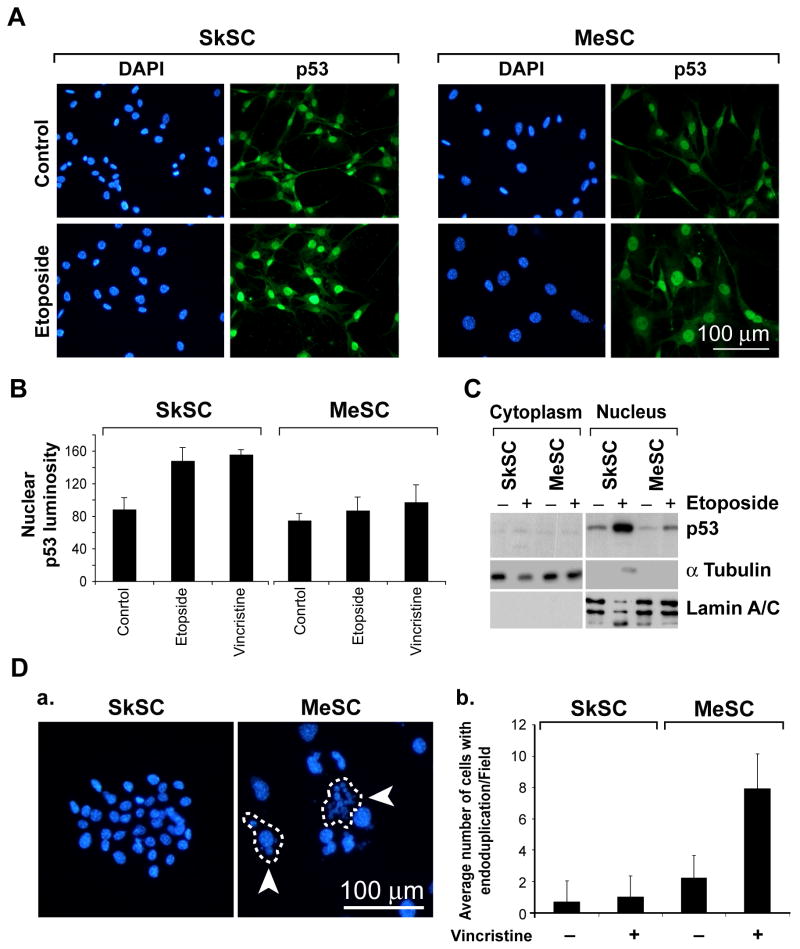
Because p53 mutations may result in its sequestration in cytoplasm or nucleus, we determined the sub-cellular localization of p53 by immunofluorescence and cell fractionation studies. By immunofluorescence, p53 localized predominately to the nucleus in both SkSC and MeSC in untreated cells and cells treated with etoposide (figure 4A). However, when the results were quantified, nuclear p53 levels in SkSC treated with etoposide or vincristine were approximately two fold higher compared to MeSC (figure 4B). Western blotting of purified nuclear and cytosolic extracts of etoposide-treated cells confirmed that p53 was predominately localized to the nucleus, where it was strikingly increased in SkSC, but not MeSC (figure 4C). Of note, almost 10 times as many cells with endoduplicated nuclei (nuclear division without cell division) were detected in MeSC compared to SkSC after vincristine treatment (figure 4D). Endoduplication is commonly observed in p53-null mouse embryonic fibroblasts treated with microtubule inhibitors and occurs due to checkpoint failure in the absence of normal p53 function (Lanni & Jacks, 1998).
Figure 4. P53 localizes to the nucleus in SkSC and MeSC but the total cellular p53 pool is reduced.
(A) P53 immunofluorescence in SkSC and MeSC treated with etoposide (10 μM, eight hours). Note the predominant nuclear staining for p53 in both cell types, although the p53 signal in MeSC is diminished. (B) The luminosity for the nuclear p53 signal in treated and untreated cells was measured in 10 cells from three random fields and plotted. (C) Western blots of purified nuclear and cytoplasmic fractions from etoposide-treated cells (10 μM, eight hours). As loading controls, blots were stripped and re-probed with γ tubulin (cytoplasm) and lamin A/C (nucleus). (D) Vincristine-treated cells (1 nM, 24 hours) were methanol-fixed and the nuclei stained with DAPI. The circled cells are single cells and the arrows point to endoduplicated nuclei. Cells in ten random fields were counted, and the average number of cells with multiple nuclei (> 3/cell) was plotted.
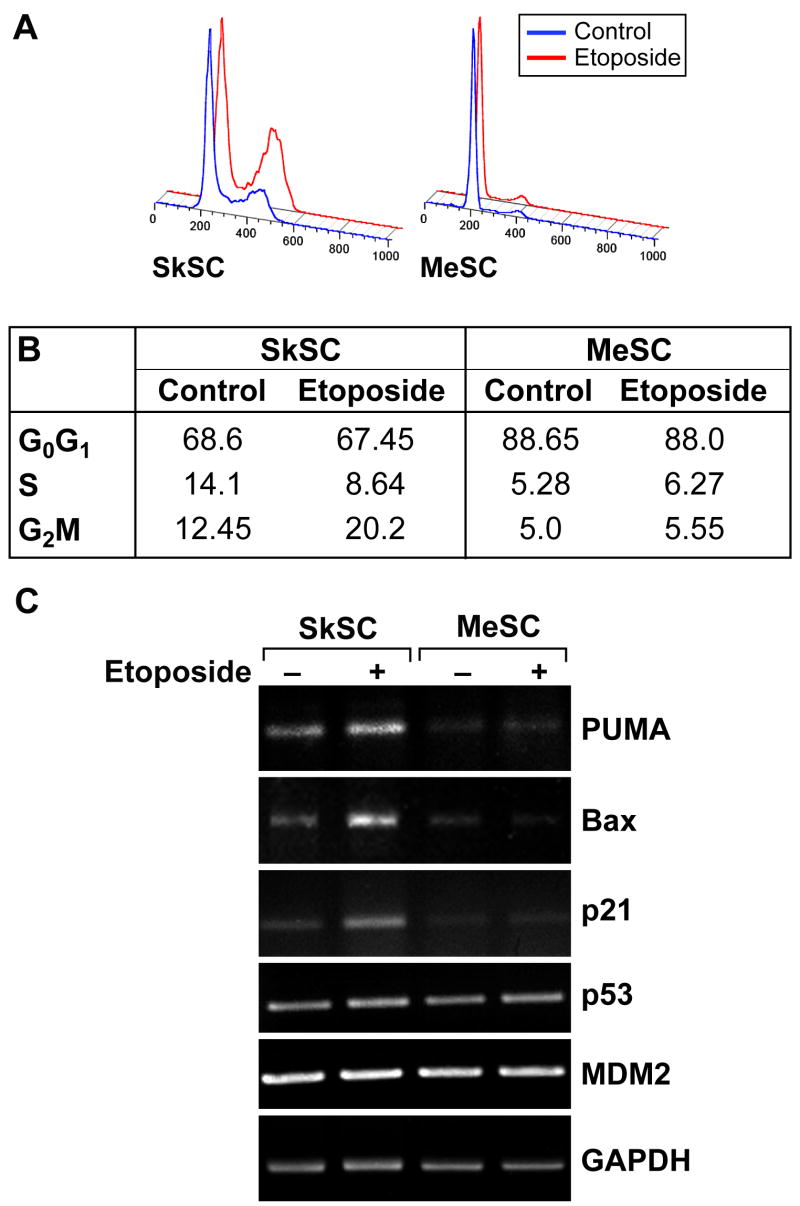
MeSC fail to arrest in G2 after etoposide treatment
Because the growth inhibitory effects of etoposide are mainly due to G2 arrest, cell cycle status in etoposide-treated cells was determined by propidium iodide (PI) staining followed by FACS. In SkSC treated with etoposide, the number of cells in G2 almost doubled compared to untreated cells. In contrast, MeSC failed to accumulate in G2 (figure 5A and 5B). By semi-quantitative RT-PCR, the expression of p53-inducible genes related to cell cycle arrest and apoptosis including PUMA, Bax, and p21 were up-regulated in SkSC, but not in MeSC after etoposide treatment (figure 5C). Notably, the constitutive levels of these genes were also qualitatively lower in MeSC relative to SkSC. No differences in p53 or the negative regulator of p53 stability, MDM2, were detected. Taken together, these results were consistent with a failed induction of p53 and p53-inducible genes related to apoptosis or cell cycle arrest in MeSC after etoposide treatment.
Figure 5. MeSC fail to arrest in G2 after etoposide treatment.
(A) Cells treated with one μM etoposide were analyzed by FACS 24 hours later. Note the larger G2 peak on the histogram in etoposide-treated SkSC but not in MeSC. (B) DNA histograms were analyzed and the results from two experiments were averaged and are shown in the figure (numbers are percent). (C) Semi-quantitative RT-PCR analysis for p53-indicible genes in SC treated with one μM etoposide for 24 hours.
MeSC fail to undergo apoptosis after vincristine treatment
Dual staining using PI and annexin V (AV) in vincristine-treated cells indicated an increase in AV+/PI+ cells in SkSC, but not MeSC (figure 6A). The number of early apoptotic AV+ cells increased six times above untreated cells in SkSC but only two times in MeSC (figure 6B). Thus, the decreased sensitivity to vincristine in MeSC relative to SkSC was most likely due to decreased p53-dependent apoptosis.
Figure 6. MeSC fail to undergo apoptosis after vincristine treatment.
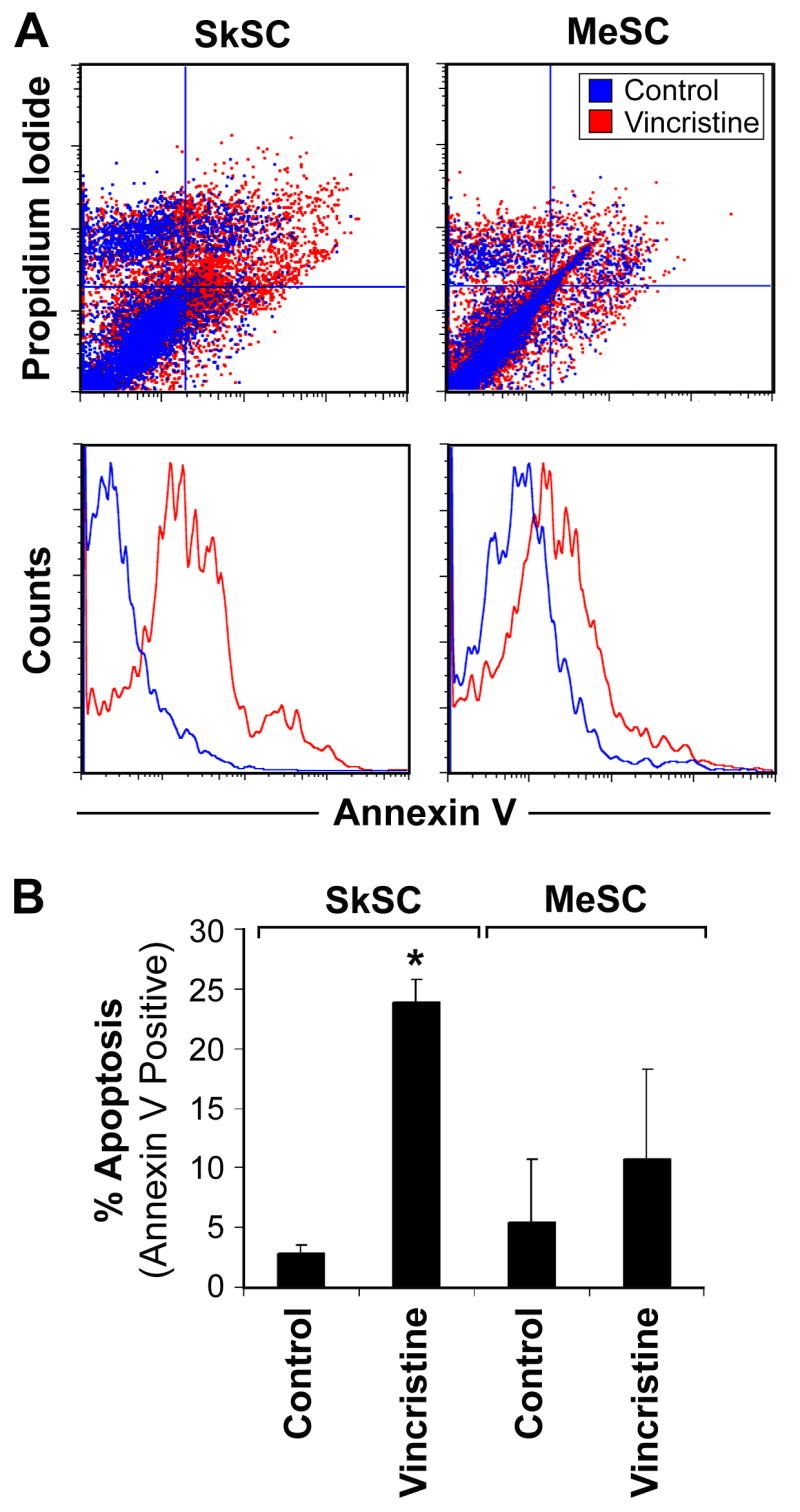
(A) Cells treated with one nM vincristine for 24 hours were double stained with PI and AV and analyzed by FACS. An increase in PI+/AV+ and PI-/AV+ cells was detected in vincristine-treated SkSC compared to MeSC. (B) The average numbers of early apoptotic AV+ cells from two experiments were plotted. An (*) indicates results are statistically significant (p< 0.05) by student's t-test.
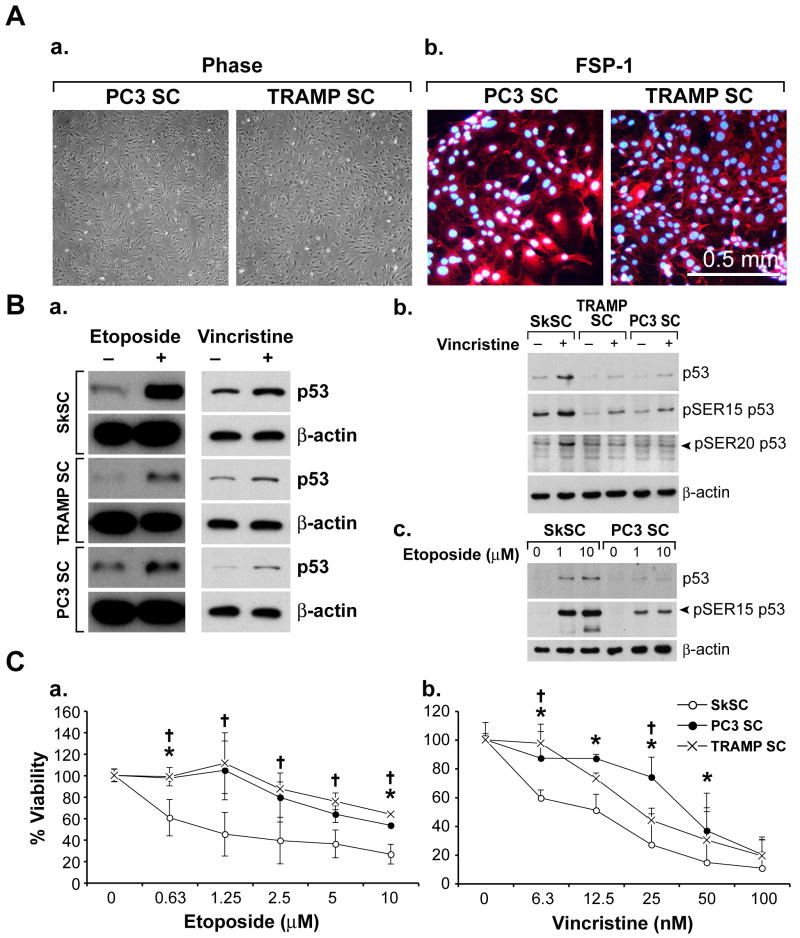
P53 function is impaired in stromal cells from PC3 and TRAMP prostate tumors
To determine if SC from different tumors also showed diminished p53 function, we isolated SC from PC3 xenografts and spontaneous prostate tumors in TRAMP mice. Both PC3SC and TRAMPSC had a fibroblast-like morphology, were negative for EC markers (data not shown) and were positive for FSP-1, indicating they were tumor-associated fibroblasts, similar to SC from A375 melanoma (figure 7A). For all tumor xenografts used in this study, diphtheria toxin (DT) was used to eliminate human tumor cells from cultures (Arbiser et al., 1999). DT could not be used in the TRAMP model because these are spontaneous tumors with no human component. However, TRAMPSC did not express epithelial-specific E-cadherin by FACS or immunofluorescence, indicating absence of contaminating tumor cells (data not shown). Similar to MeSC, both PC3SC and TRAMPSC showed only modest increases in p53 after treatment with etoposide or vincristine, while p53 levels in SkSC were markedly up-regulated (figure 7B). Both vincristine and etoposide treatment increased SER15 phosphorylation in both normal and tumor SC, although the levels were reduced in tumor SC presumably due to the decrease in total p53 levels. However, while SER20 was phosphorylated in normal SC after vincristine treatment, no increase in SER20 phosphorylation was evident in tumor SC. No SER20 phosphorylation was detected in SkSC or PC3SC after treatment with etoposide (data not shown). Similar to MeSC, and in good accord with the diminished p53 levels, TRAMPSC and PC3SC were less sensitive to etoposide and vincristine compared to SkSC (figure 7C). These results suggest that tumor-associated stromal cells from different tumor types show diminished p53 protein levels and impaired p53 function.
Figure 7. P53 function is impaired in stromal cells from PC3 and TRAMP prostate tumors.
(A) Both PC3SC and TRAMPSC had a fibroblast-like morphology (a) and uniformly expressed FSP-1 (b). (B) After treatment with etoposide (10 μM, eight hours) or vincristine (1 nM, 24 hours) p53 accumulation was diminished in TRAMPSC and PC3SC compared to normal SkSC (a). Western blotting for pSER15 and pSER20 after vincristine (b) (1nM, 24 hours) or etoposide (eight hours) treatment (c). Blots were striped and re-probed with p53 or β actin antibodies. (C) Viabilities of TRAMPSC and PC3SC after treatment with etoposide (a) or vincristine (b). Cells were plated in triplicate and treated with each drug for 72 hours before dispersing in trypsin and counting. An (*) indicates results are statistically significant (p< 0.05) by student's t-test.
Tumor stromal cells have multiple centrosomes and aneuploid karyotypes
Hida et al. recently reported abnormal centrosomes and abnormal karyotypes in tumor-specific endothelial cells (Hida et al., 2004). About 30 % of MeSC, TRAMPSC, and PC3SC also had abnormal, multiple centrosomes by pericentrin staining (figure 8A). Karyotypes on the isolated cells showed that all tumor SC had a heterogeneous aneuploid chromosomes, whereas SkSC were normal (figure 8B). Taken together, SC from different types of tumors (melanoma versus prostate) and different models (xenograft versus spontaneous) are characterized by aneuploid karyotypes and multiple centrosomes.
Figure 8. Tumor stromal cells have multiple centrosomes and aneuploid karyotypes.
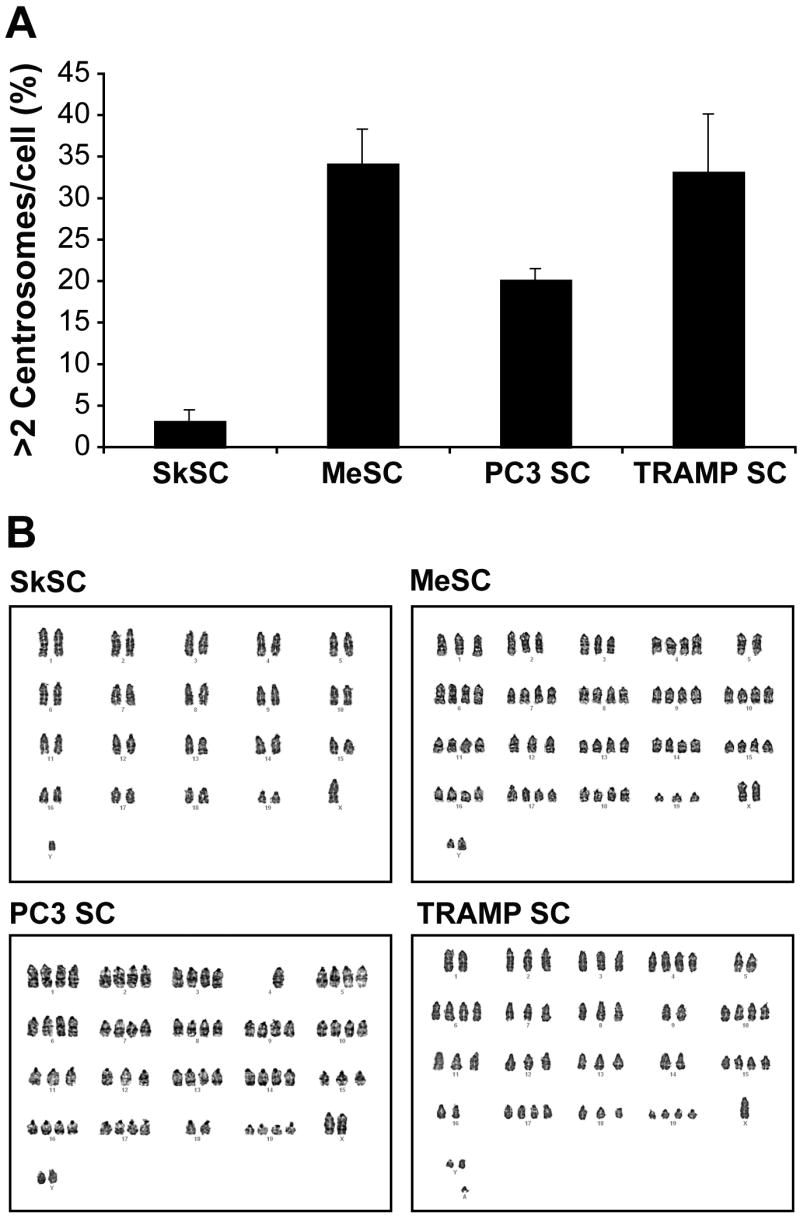
(A) Centrosomes were decorated with pericentrin antibodies and counted. One hundred cells were scored and averaged from three different fields. (B) Karyotypes for SkSC, MeSC, PC3SC, and TRAMPSC showing aneuploidy in all tumor SC, while SkSC were normal.
Discussion
Our study provides the first evidence for impaired p53 function in tumor-associated SC. All tumor SC examined were characterized by diminished p53 protein levels, decreased sensitivity to cytotoxic drugs, and genomic instability indicated by multiple centrosomes and aneuploid karyotypes. While the nature of the p53 defect in tumor SC is not yet clear, impaired p53 function in tumor stroma could enable SC survival in the tumor microenvironment and contribute to genomic instability.
Host SC can constitute a significant percentage of the total tumor bulk as shown in GFP-SCID mice implanted with tumor xenografts (Udagawa et al., 2006). Tumor-associated fibroblasts or mesenchymal-like cells comprise the majority of the tumor stroma. Collectively, these cells might arise from tissue-resident activated fibroblasts, tumor epithelial cells undergoing epithelial-to-mesenchymal transition, resident stem or mesenchymal-like cells, or bone-marrow derived progenitors. Recently, mesenchymal stem cells localized to breast carcinoma were shown to increase metastatic potency (Karnoub et al., 2007). Thus, by providing scaffolds for tumor cells and other SC, and by producing matrix, growth factors, and chemotactic factors for inflammatory cells and vascular progenitors, tumor SC can enable tumor growth and possibly metastasis (Bhowmick et al., 2004; Olumi et al., 1999). Targeting tumor SC may therefore be a viable approach for eliminating solid tumors (Hofmeister et al., 2008).
Recently, our laboratory reported that endothelial cells from human-to-mouse xenografts were aneuploid with centrosome abnormalities 4-6 weeks after implantation (Hida et al., 2004). Additional studies have confirmed both cytogenetic and epigenetic alterations in tumor-associated SC and tissue adjacent to carcinoma (Deng et al., 1994; Moinfar et al., 2000; Streubel et al., 2004). The mechanism(s) of these cytogenetic changes in tumor stroma are not yet clear. One possibility is that SC and tumor cells may fuse, as hypothesized in cases of allelic imbalance and genomic instability in human breast stroma (Weber et al., 2006). However, we found no evidence of human DNA in the karyotypes of murine tumor SC from human-to-mouse xenografts. Selection pressure for SC with diminished p53 function, and thus a survival advantage in the tumor microenvironment is also a possibility. For example, non-autonomous oncogenic stress in tumor epithelium may result in the selection of SC with LOH at the p53 locus (Hill et al., 2005; Kiaris et al., 2005; Lu et al., 2001). It is likely that impaired p53 function in tumor SC might enable the propagation of cells with damaged DNA due to checkpoint failure and lead to chromosomal instability.
A potential caveat related to targeting tumor SC is the loss of function of tumor suppressor genes in the stromal compartment. Hill et al. described widespread p53 loss in tumor mesenchyme after 20-25 weeks in a prostate cancer model (Hill et al., 2005). We evaluated p53 function and performed karyotypes in TRAMPSC at 22 weeks following tumor initiation and in SC from tumor xenografts at 4-6 weeks post-implantation. In either case, alterations in p53 function, diminished p53 protein levels, and abnormal karyotypes were evident, irrespective of time or tumor model. At present, we have been unable to detect mutations in the p53 coding regions in SC, nor could we detect p53 LOH by real-time PCR using genomic DNA (data not shown). Although real-time PCR is a sensitive and well-established method to detect LOH, heterogeneity in p53 status in the cultured tumor SC could confound these results. Therefore, LOH may only be evident on a cell-to-cell basis. Further studies will be needed to address the mechanism of impaired p53 function in tumor SC in our in vitro system, and whether tumor SC can acquire drug resistance in vivo.
It is also possible that alterations in p53-interacting proteins including the kinases responsible for p53 phosphorylation could contribute to impaired p53 function in tumor SC. For example, DNA damage imparts a well-characterized phosphorylation of SER15 and SER20 in the p53 transcriptional activation domain. Phosphorylation of these residues is thought to inhibit p53's interaction with MDM2 and increase its stability (Appella & Anderson, 2001). While lower levels of p53 SER15 phosphorylation were evident in tumor SC in this study, this was most likely due to the decrease in total levels of the p53 protein. On the other hand, SER20 did not appear to be phosphorylated in tumor SC relative to normal SC after vincristine treatment. It remains possible that in tumor SC, alterations in pathways secondary to p53'stability, rather than direct alterations in p53, could impart a destabilizing effect on p53 leading to its degradation.
The idea that tumor SC can contribute to tumor growth and perhaps metastasis is an emerging concept in cancer biology (Karnoub et al., 2007). Though alterations in p53 in tumor SC have been shown previously in vivo, we show that diminished p53 function accompanies genomic instability and decreased sensitivity to cytotoxic drugs.
Acknowledgments
ACD wishes to thank Leonora DeBella and the American Cancer society for supporting his research with a post doctoral fellowship. We thank Drs. Akiko Mammoto and Elisa Boscolo for the lamin A/C and tubulin antibodies. We thank Dr. Dipak Panigrahy for the TNP-470. We thank Kristin Johnson for her excellent assistance with preparing the figures for this manuscript.
References
- Allinen M, Beroukhim R, Cai L, Brennan C, Lahti-Domenici J, Huang H, Porter D, Hu M, Chin L, Richardson A, Schnitt S, Sellers WR, Polyak K. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6:17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Appella E, Anderson CW. Post-translational modifications and activation of p53 by genotoxic stresses. Eur J Biochem. 2001;268:2764–72. doi: 10.1046/j.1432-1327.2001.02225.x. [DOI] [PubMed] [Google Scholar]
- Arbiser JL, Raab G, Rohan RM, Paul S, Hirschi K, Flynn E, Price ER, Fisher DE, Cohen C, Klagsbrun M. Isolation of mouse stromal cells associated with a human tumor using differential diphtheria toxin sensitivity. Am J Pathol. 1999;155:723–9. doi: 10.1016/S0002-9440(10)65171-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RA, Izumi H, Fukasawa K. Induction of centrosome amplification and chromosome instability in p53-null cells by transient exposure to subtoxic levels of S-phase-targeting anticancer drugs. Oncogene. 2004;23:6823–9. doi: 10.1038/sj.onc.1207561. [DOI] [PubMed] [Google Scholar]
- Bhowmick NA, Neilson EG, Moses HL. Stromal fibroblasts in cancer initiation and progression. Nature. 2004;432:332–7. doi: 10.1038/nature03096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunz F, Hwang PM, Torrance C, Waldman T, Zhang Y, Dillehay L, Williams J, Lengauer C, Kinzler KW, Vogelstein B. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J Clin Invest. 1999;104:263–9. doi: 10.1172/JCI6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdelya LG, Komarova EA, Hill JE, Browder T, Tararova ND, Mavrakis L, Dicorleto PE, Folkman J, Gudkov AV. Inhibition of p53 Response in Tumor Stroma Improves Efficacy of Anticancer Treatment by Increasing Antiangiogenic Effects of Chemotherapy and Radiotherapy in Mice. Cancer Res. 2006;66:9356–61. doi: 10.1158/0008-5472.CAN-06-1223. [DOI] [PubMed] [Google Scholar]
- Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–7. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng G, Chen LC, Schott DR, Thor A, Bhargava V, Ljung BM, Chew K, Smith HS. Loss of heterozygosity and p53 gene mutations in breast cancer. Cancer Res. 1994;54:499–505. [PubMed] [Google Scholar]
- Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N Engl J Med. 1986;315:1650–9. doi: 10.1056/NEJM198612253152606. [DOI] [PubMed] [Google Scholar]
- Fukino K, Shen L, Matsumoto S, Morrison CD, Mutter GL, Eng C. Combined total genome loss of heterozygosity scan of breast cancer stroma and epithelium reveals multiplicity of stromal targets. Cancer Res. 2004;64:7231–6. doi: 10.1158/0008-5472.CAN-04-2866. [DOI] [PubMed] [Google Scholar]
- Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA, Jr, Kastrinakis NG, Levy B, Kletsas D, Yoneta A, Herlyn M, Kittas C, Halazonetis TD. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–13. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- Hida K, Hida Y, Amin DN, Flint AF, Panigrahy D, Morton CC, Klagsbrun M. Tumor-associated endothelial cells with cytogenetic abnormalities. Cancer Res. 2004;64:8249–55. doi: 10.1158/0008-5472.CAN-04-1567. [DOI] [PubMed] [Google Scholar]
- Hill R, Song Y, Cardiff RD, Van Dyke T. Selective evolution of stromal mesenchyme with p53 loss in response to epithelial tumorigenesis. Cell. 2005;123:1001–11. doi: 10.1016/j.cell.2005.09.030. [DOI] [PubMed] [Google Scholar]
- Hinz B, Phan SH, Thannickal VJ, Galli A, Bochaton-Piallat ML, Gabbiani G. The myofibroblast: one function, multiple origins. Am J Pathol. 2007;170:1807–16. doi: 10.2353/ajpath.2007.070112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmeister V, Schrama D, Becker JC. Anti-cancer therapies targeting the tumor stroma. Cancer Immunol Immunother. 2008;57:1–17. doi: 10.1007/s00262-007-0365-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan RN, Riba RD, Zacharoulis S, Bramley AH, Vincent L, Costa C, MacDonald DD, Jin DK, Shido K, Kerns SA, Zhu Z, Hicklin D, Wu Y, Port JL, Altorki N, Port ER, Ruggero D, Shmelkov SV, Jensen KK, Rafii S, Lyden D. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature. 2005;438:820–7. doi: 10.1038/nature04186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW, Richardson AL, Polyak K, Tubo R, Weinberg RA. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–63. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- Kiaris H, Chatzistamou I, Trimis G, Frangou-Plemmenou M, Pafiti-Kondi A, Kalofoutis A. Evidence for nonautonomous effect of p53 tumor suppressor in carcinogenesis. Cancer Res. 2005;65:1627–30. doi: 10.1158/0008-5472.CAN-04-3791. [DOI] [PubMed] [Google Scholar]
- Kurose K, Gilley K, Matsumoto S, Watson PH, Zhou XP, Eng C. Frequent somatic mutations in PTEN and TP53 are mutually exclusive in the stroma of breast carcinomas. Nat Genet. 2002;32:355–7. doi: 10.1038/ng1013. [DOI] [PubMed] [Google Scholar]
- Lanni JS, Jacks T. Characterization of the p53-dependent postmitotic checkpoint following spindle disruption. Mol Cell Biol. 1998;18:1055–64. doi: 10.1128/mcb.18.2.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liotta LA, Kohn EC. The microenvironment of the tumour-host interface. Nature. 2001;411:375–9. doi: 10.1038/35077241. [DOI] [PubMed] [Google Scholar]
- Lu X, Magrane G, Yin C, Louis DN, Gray J, Van Dyke T. Selective inactivation of p53 facilitates mouse epithelial tumor progression without chromosomal instability. Mol Cell Biol. 2001;21:6017–30. doi: 10.1128/MCB.21.17.6017-6030.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell. 2006;127:1323–34. doi: 10.1016/j.cell.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Moinfar F, Man YG, Arnould L, Bratthauer GL, Ratschek M, Tavassoli FA. Concurrent and independent genetic alterations in the stromal and epithelial cells of mammary carcinoma: implications for tumorigenesis. Cancer Res. 2000;60:2562–6. [PubMed] [Google Scholar]
- Nelson WG, Kastan MB. DNA strand breaks: the DNA template alterations that trigger p53-dependent DNA damage response pathways. Mol Cell Biol. 1994;14:1815–23. doi: 10.1128/mcb.14.3.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olumi AF, Grossfeld GD, Hayward SW, Carroll PR, Tlsty TD, Cunha GR. Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Res. 1999;59:5002–11. doi: 10.1186/bcr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R, Carey VJ, Richardson AL, Weinberg RA. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–48. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- Patocs A, Zhang L, Xu Y, Weber F, Caldes T, Mutter GL, Platzer P, Eng C. Breast-cancer stromal cells with TP53 mutations and nodal metastases. N Engl J Med. 2007;357:2543–51. doi: 10.1056/NEJMoa071825. [DOI] [PubMed] [Google Scholar]
- Pelham RJ, Rodgers L, Hall I, Lucito R, Nguyen KC, Navin N, Hicks J, Mu D, Powers S, Wigler M, Botstein D. Identification of alterations in DNA copy number in host stromal cells during tumor progression. Proc Natl Acad Sci U S A. 2006;103:19848–53. doi: 10.1073/pnas.0609635104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sappino AP, Skalli O, Jackson B, Schurch W, Gabbiani G. Smooth-muscle differentiation in stromal cells of malignant and non-malignant breast tissues. Int J Cancer. 1988;41:707–12. doi: 10.1002/ijc.2910410512. [DOI] [PubMed] [Google Scholar]
- Seemayer TA, Lagace R, Schurch W, Tremblay G. Myofibroblasts in the stroma of invasive and metastatic carcinoma: a possible host response to neoplasia. Am J Surg Pathol. 1979;3:525–33. doi: 10.1097/00000478-197912000-00005. [DOI] [PubMed] [Google Scholar]
- Streubel B, Chott A, Huber D, Exner M, Jager U, Wagner O, Schwarzinger I. Lymphoma-specific genetic aberrations in microvascular endothelial cells in B-cell lymphomas. N Engl J Med. 2004;351:250–9. doi: 10.1056/NEJMoa033153. [DOI] [PubMed] [Google Scholar]
- Tlsty TD. Stromal cells can contribute oncogenic signals. Semin Cancer Biol. 2001;11:97–104. doi: 10.1006/scbi.2000.0361. [DOI] [PubMed] [Google Scholar]
- Udagawa T, Puder M, Wood M, Schaefer BC, D'Amato RJ. Analysis of tumor-associated stromal cells using SCID GFP transgenic mice: contribution of local and bone marrow-derived host cells. Faseb J. 2006;20:95–102. doi: 10.1096/fj.04-3669com. [DOI] [PubMed] [Google Scholar]
- Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–5. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- Weber F, Shen L, Fukino K, Patocs A, Mutter GL, Caldes T, Eng C. Total-genome analysis of BRCA1/2-related invasive carcinomas of the breast identifies tumor stroma as potential landscaper for neoplastic initiation. Am J Hum Genet. 2006;78:961–72. doi: 10.1086/504090. [DOI] [PMC free article] [PubMed] [Google Scholar]