Abstract
Non-coding RNAs are a complex class of nucleic acids, with growing evidence supporting regulatory roles in gene expression. Here we identify a non-coding RNA located head-to-head with the gene encoding the Glioma-associated oncogene 1 (GLI1), a transcriptional effector of multiple cancer-associated signaling pathways. The expression of this three-exon GLI1 antisense (GLI1AS) RNA in cancer cells was concordant with GLI1 levels. siRNAs knockdown of GLI1AS up-regulated GLI1 and increased cellular proliferation and tumor growth in a xenograft model system. Conversely, GLI1AS overexpression decreased the levels of GLI1, its target genes PTCH1 and PTCH2, and cellular proliferation. Additionally, we demonstrate that GLI1 knockdown reduced GLI1AS, while GLI1 overexpression increased GLI1AS, supporting the role of GLI1AS as a target gene of the GLI1 transcription factor. Activation of TGFβ and Hedgehog signaling, two known regulators of GLI1 expression, conferred a concordant up-regulation of GLI1 and GLI1AS in cancer cells. Finally, analysis of the mechanism underlying the interplay between GLI1 and GLI1AS indicates that the non-coding RNA elicits a local alteration of chromatin structure by increasing the silencing mark H3K27me3 and decreasing the recruitment of RNA polymerase II to this locus. Taken together, the data demonstrate the existence of a novel non-coding RNA-based negative feedback loop controlling GLI1 levels, thus expanding the repertoire of mechanisms regulating the expression of this oncogenic transcription factor.
Keywords: non-coding RNA, gene expression, transcription, chromatin, GLI1, cancer
INTRODUCTION
Antisense transcription is a common phenomenon in the human genome (Conley and Jordan, 2012; Magistri et al., 2012; Morris and Vogt, 2010). Up to 70% of the transcriptome has antisense partners modulating the transcription rate, mRNA stability, transport or translation efficiency of the sense gene. Both sense and antisense RNAs can encode proteins or be non-protein coding transcripts; however, experimental evidence indicates that antisense transcripts are generally non-protein coding RNAs (Faghihi and Wahlestedt, 2009; Katayama et al., 2005). To date, a number of functionally-characterized human non-coding antisense transcripts have been reported (Carrieri et al., 2012; Ebralidze et al., 2008; Faghihi et al., 2008; Holdt et al., 2013; Johnsson et al., 2013; Li et al., 2012a; Morris et al., 2008; Onoguchi et al., 2012; Pandey et al., 2008; Prensner et al., 2013; Rinn et al., 2007; Yu et al., 2008; Zhu et al., 2012). Their role in regulating the expression of protein coding genes is a well-established concept and has become a subject of increasing interest (Faghihi and Wahlestedt, 2009; Mattick and Makunin, 2006; Ozsolak et al., 2010; Werner et al., 2009), however, the detailed mechanism underlying the interplay between the sense and antisense remains in part elusive.
Here using a combination of in vitro and in vivo assays, we have identified a novel sense-antisense pair controlling the expression of the transcription factor GLI1, a known effector of oncogenic pathways. In different malignancies the pro-tumoral function of GLI1 is associated with its increase expression. Thus, understanding the mechanisms influencing gene expression of GLI1 are particularly relevant, as these may represent additional means of constraining the oncogenic capacity of GLI1 (Nilsson et al., 2000). We identify a non-coding GLI1 RNA (GLI1AS), originating from the antisense strand of the human GLI1 gene, which elicits negative feedback on GLI1 expression via local chromatin remodeling. These findings may allow the development of novel strategies, based on epigenetic modulation, which could achieve an effective reduction of the capacity of GLI1 to act as an oncogene.
MATERIALS AND METHODS
Cell lines and culture
The alveolar rhabdomyosarcoma RMS13 and the pancreatic carcinoma PANC1 cell lines were purchased from ATCC (Manassas, VA). The embryonal rhabdomyosarcoma cells lines used were Rh36, a kind gift from P. Houghton (St. Jude Children’s Research Hospital, Memphis, TN), CCA a kind gift from P.L. Lollini (University of Bologna, Italy), and RD, purchased from ATCC (Manassas, VA). The Daoy medulloblastoma cell line was a kind gift of F. Aberger (Univesity of Salzburg, Austria).
RMS13 and Rh36 cells were cultured in RPMI-1640 Medium + 10% fetal bovine serum (FBS), Daoy cells in EMEM + 10% FBS. CCA, RD and PANC1 cells were cultured in DMEM supplemented with L-glutamine and 10% FBS. All cell lines were maintained in a 5% CO2 humidified incubator.
PANC1 cells were treated with 5 ng/ml of TGFβ1 recombinant ligand (R&D Systems, Minneapolis, MN) in serum free media and collected 24 h after treatment. Daoy cells were treated with 200nM SAG in 0.5% FBS and harvested after 48 h. cDNA from the PC3 and 22Rv1 prostatic carcinoma, PANC1 pancreatic carcinoma, A549 lung adenocarcinoma and AGS gastric adenocarcinoma was generously provided by Dr. Matthias Lauth, Karolinska Institutet.
Patient specimens
Human basal cell carcinoma biopsy and normal skin specimens were collected at the Dermatology and Venerology Unit, Karolinska University Hospital, Stockholm, Sweden. The clinical diagnosis was made by a dermatologist and was confirmed by histopathological evaluation. The studies were approved by the Regional Committee of Ethics.
The human breast cancer samples collection was approved by the ethic institutional review board for "Biobanking and use of human tissue for experimental studies" of the Pathology Services of the Azienda Ospedaliera Città della Salute e della Scienza di Torino or the Ethical Committee of the Universidad del Rosario. Written informed consent was obtained from all patients for their tissue to be used in research. The samples were collected from residual tissue, that is tissue not used for diagnostic and therapeutic purposes.
RNA isolation
Nuclear and cytoplasmic RNAs were isolated using the Paris kit (Ambion Life Technologies) following the manufacturer’s instructions. Total RNA from cells was prepared with the RNeasy kit (Qiagen, Hamburg, Germany) or TRIzol reagent (Invitrogen) followed by cDNA synthesis with p(dT)15 primer (Roche) or random N6 primers (New England Biolabs) and Superscript II (Invitrogen).
Transfection of cell lines with siRNA
siRNAs targeting human GLI1 and GLI1AS (Table 2 and Figure 1A), were purchased from Sigma-Aldrich. All siRNAs were designed using the BLOCK-iT™ RNAi Designer (Invitrogen) and the siDESIGN Center (Dharmacon) software tools except si1G, which had been designed by Sigma-Aldrich. As control, a non-targeting siRNA was used (MISSION® siRNA Universal Negative Control, Sigma-Aldrich). Cells were plated in 60 mm dishes at 30 to 50% confluency, and transfections were performed with Lipofectamine 2000 (Invitrogen). Transfection efficiencies were confirmed by siGLO (Green Transfection Indicator, Dharmacon).
Table 2.
siRNAs targeting GLI1 (G) and GLI1AS (AS)
| siRNA NAME | SEQUENCE, SENSE STRAND |
|---|---|
| si1G | 5’ GUCAUAGUCACGCCUCGAAdTdT |
| si2G | 5’ GCCCAGAUGAAUCACCAAAdTdT |
| si3G | 5’ CUCGCGAUGCACAUCUCCAdTdT |
| si1AS | 5’ GCUGUUGGCCUCACCCUUUdTdT |
| si2AS | 5’ GGUGAGGAGUCUAUGGAGCdTdT |
| si3AS | 5’ GCCGUUCUCACACAUGACAdTdT |
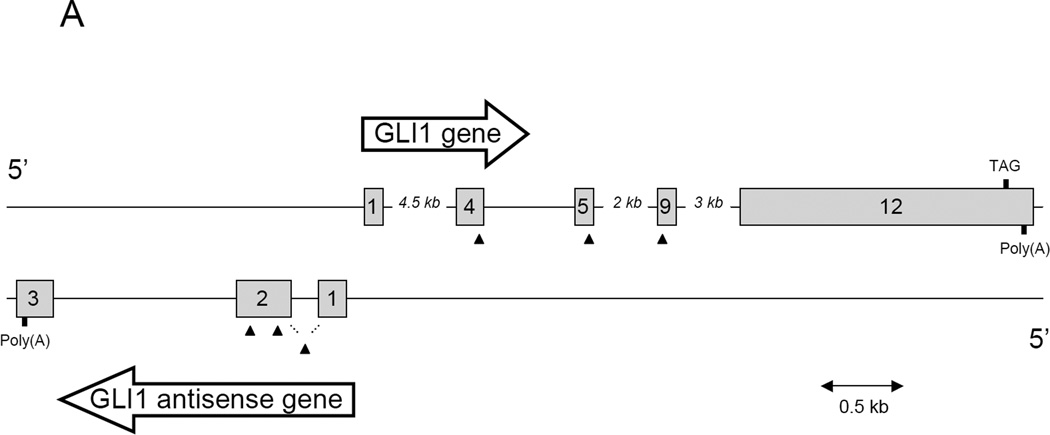
Figure 1. Genomic organization and expression of GLI1 and GLI1AS.
A. Schematic representation of the GLI1AS/GLI1 genomic region. GLI1AS, transcribed from the same region in chromosome 12q13.2 as GLI1 but from the opposite strand, is composed of three exons without any long open reading frame. Exons are numbered and depicted as grey rectangles. Arrowheads show the position of the siRNAs used, with the translation termination TAG codon of GLI1 and the poly(A) signals also indicated.
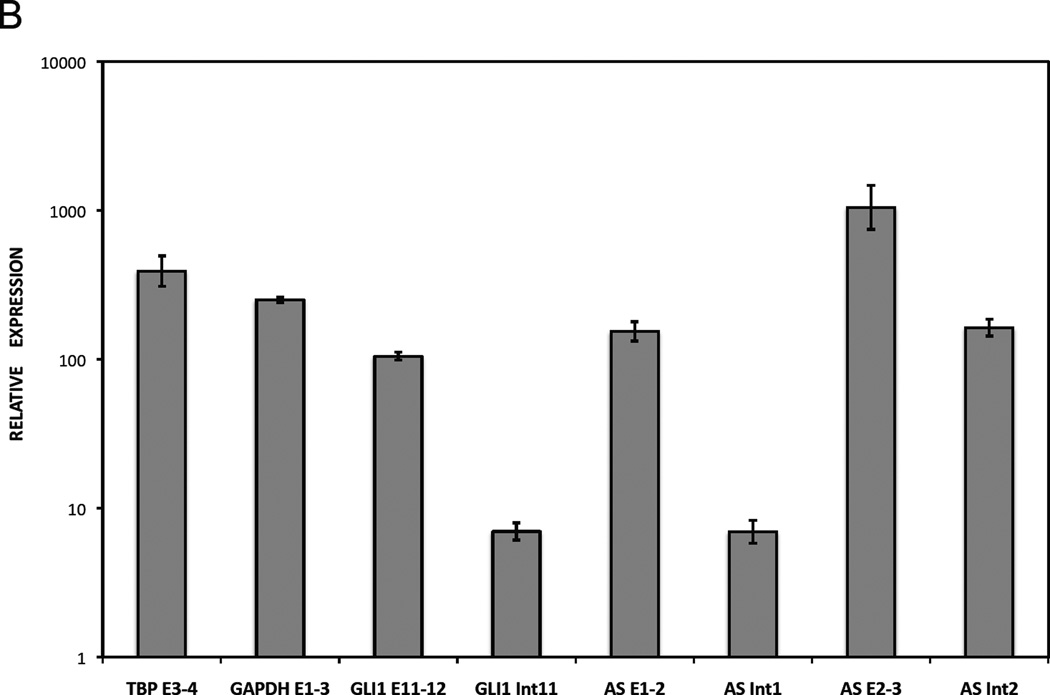
B. Polyadenylation of GLI1AS in RMS13 cells. Total RNA was subjected to cDNA synthesis in the presence or absence of oligo(dT) primers followed by real-time PCR analysis. The amplicons used (Table 1, Supplementary figure 4) were, TPB exon 3 to exon 4 (TBP E3–4), GAPDH exon 1 to exon 3 (GAPDH E1–3), GLI1 exon 11 to exon 12 (GLI1 E11–12), GLI1 exon 11 to intron 11 (GLI1 Int11), GLI1AS exon 1 to exon 2 (AS E1–2), GLI1AS intron 1 to exon 2 (AS Int1), GLI1AS exon 2 to exon 3 (AS E2–3) and GLI1AS exon 2 to intron 2 (AS Int2). Data from triplicate experiments for each amplicon are represented as relative expression (2−ΔCt values), calculated by subtracting the Ct value in the absence of oligo(dT) from the Ct value in the presence of oligo(dT) (ΔCt). Note that in the presence of oligo(dT) primers the increased expression of the GLI1AS exonic amplicons is comparable to that of the TBP E3–4, GAPDH E1–3 or GLI1 E11–12 amplicons (polyadenylated mRNAs). On the other hand, the intronic amplicons reveal a lower increase in expression (unprocessed RNAs). Error bars indicate the standard deviation.
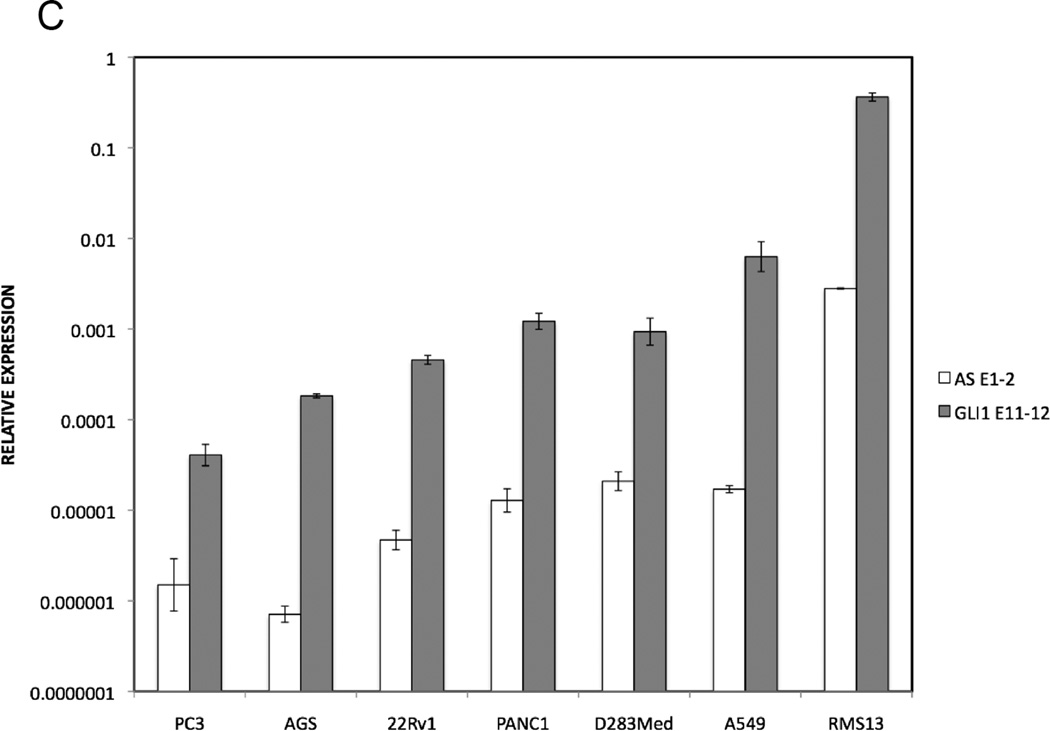
C. Expression of GLI1AS and GLI1 in cancer cell lines. Real-time RT/PCR analysis of the expression of GLI1AS and GLI1 in PC3 and 22Rv1 prostatic carcinoma, PANC1 pancreatic carcinoma, A549 lung adenocarcinoma, AGS gastric adenocarcinoma, D283Med medulloblastoma, and RMS13 rhabdomyosarcoma cell lines. Data are represented as relative expression (2−ΔCt values), calculated by subtracting the Ct value of the housekeeping gene ACTB from the Ct value of the GLI1 and GLI1AS transcripts (ΔCt). The amplicons used (Table 1, Supplementary figure 4) were, GLI1 exon 11 to exon 12 (GLI1 E11–12) and GLI1AS exon 1 to exon 2 (AS E1–2). Error bars indicate the standard deviation.
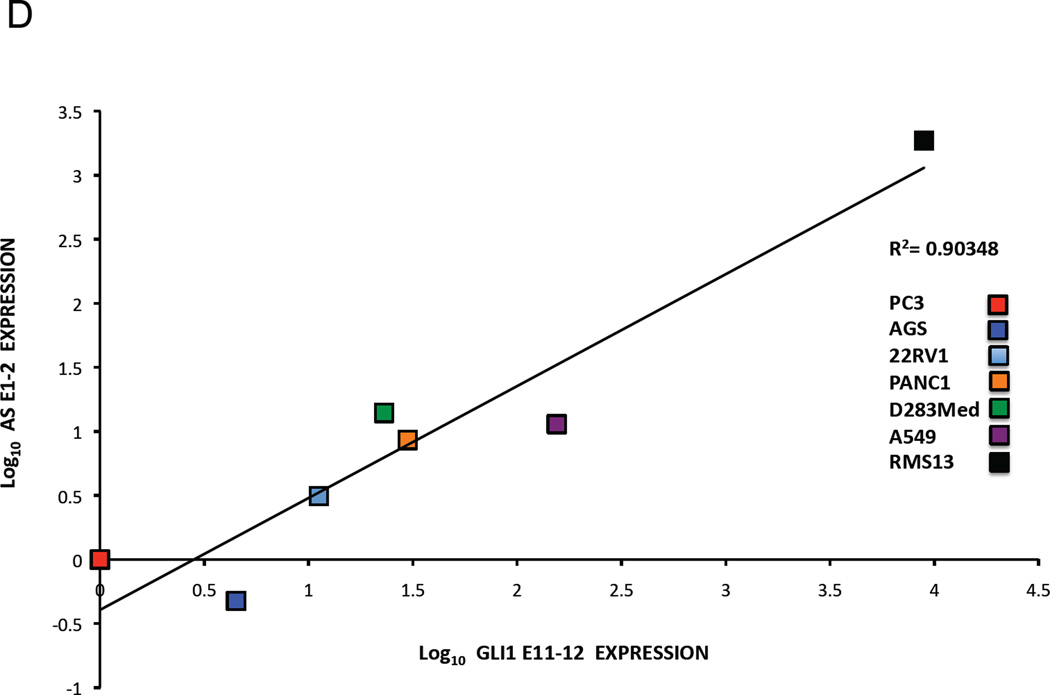
D. Co-relation of GLI1AS with GLI1 expression in cancer cell lines. The expression of GLI1AS and GLI1 in the seven cancer cell lines determined above was normalized to the expression in the PC3 prostatic carcinoma cell line, by subtracting the ΔCt in this cell line from the ΔCt in all cell lines, resulting in ΔΔCt values. Data are presented as the Log10 of the 2−ΔΔCt values. A positive correlation was observed between GLI1 (GLI1 E11–12) and GLI1AS (AS E1–2) expression, R2 = 0.90348.
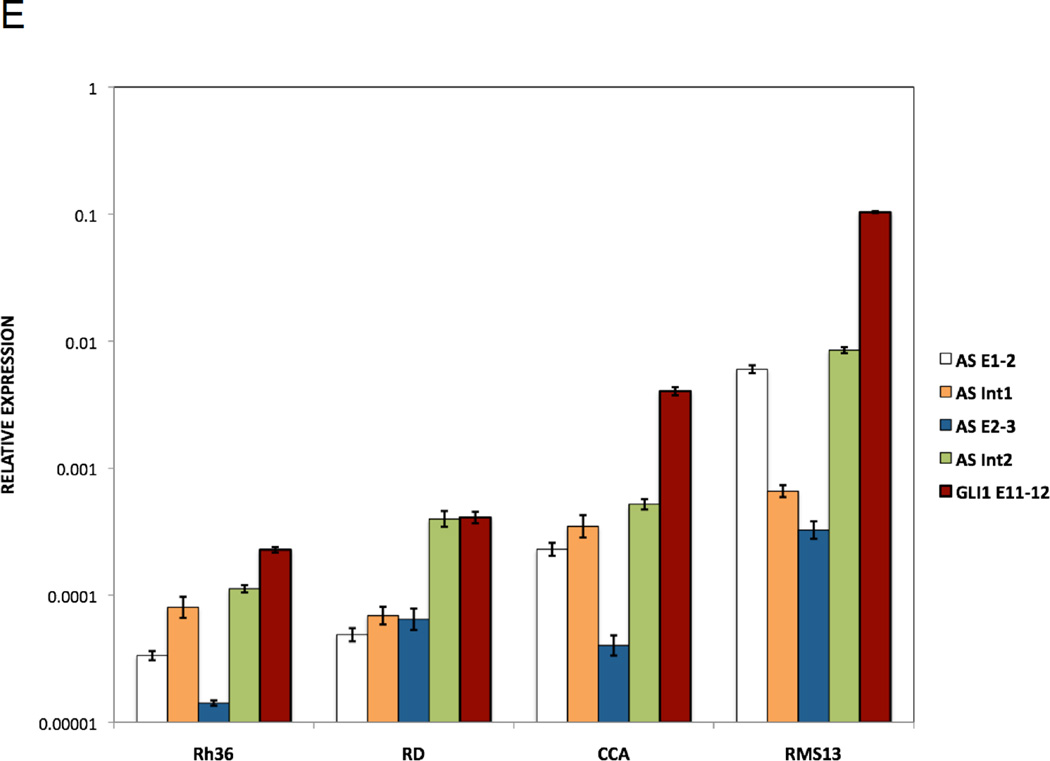
E. Spliced versus unspliced GLI1AS expression. Real-time RT/PCR analysis of the expression of GLI1AS and GLI1 in the Rh36, RD, CCA and RMS13 rhabdomyosarcoma cells lines. Data are represented as relative expression (2−ΔCt values), calculated by subtracting the Ct value of the housekeeping gene GAPDH from the Ct value of the GLI1 and GLI1AS transcripts (ΔCt). Error bars indicate the standard deviation. Note the higher expression of intron 2 retained (AS Int2) relative to intron 2 spliced (AS E2–3) GLI1AS transcripts in all cell lines analyzed.
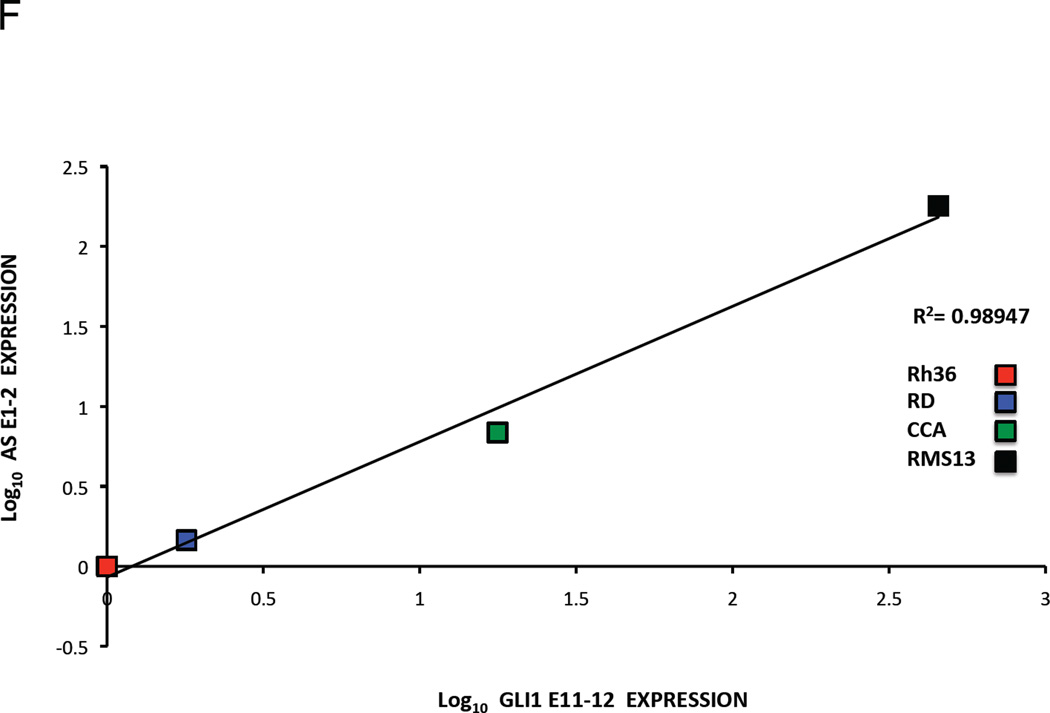
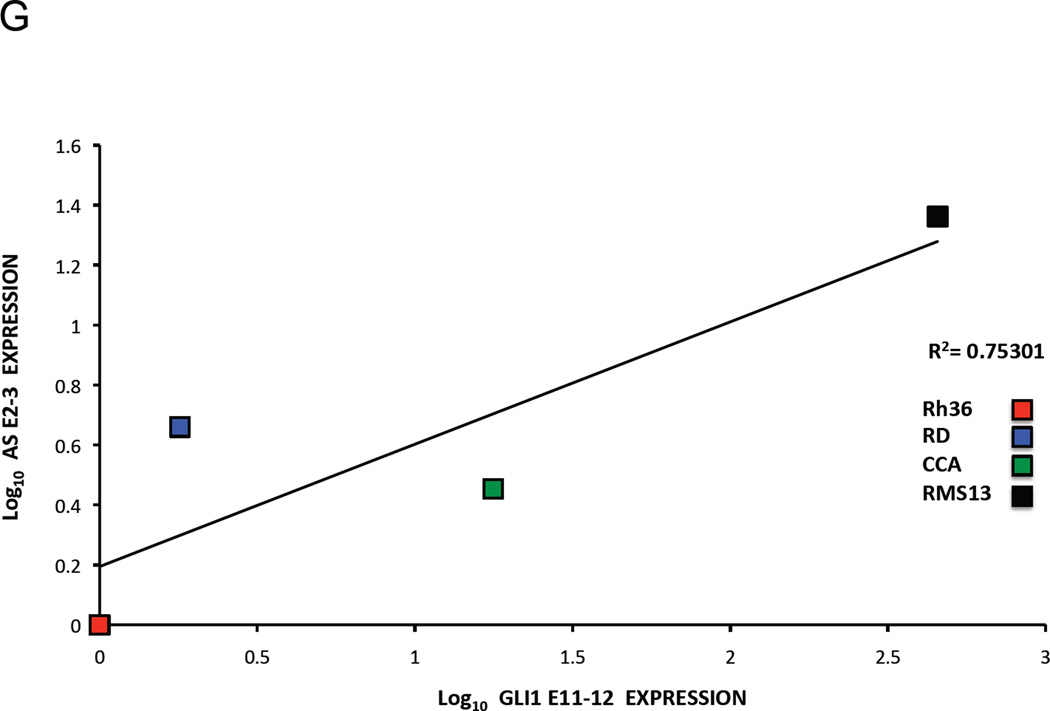
F and G. Co-relation of GLI1AS with GLI1 expression in rhabdomyosarcoma cell lines. The expression of GLI1AS and GLI1 in the four rhabdomyosarcoma cell lines determined above was normalized to the expression in the Rh36 cell line, by subtracting the ΔCt in this cell line from the ΔCt in all cell lines, resulting in ΔΔCt values. Data are presented as the Log10 of the 2−ΔΔCt values. A positive correlation was observed between GLI1 and GLI1AS, using amplicons GLI1 E11–12 and AS E1–2, R2 = 0.98947 (F) and amplicons GLI1 E11–12 and AS E2–3, R2 = 0.75301 (G).
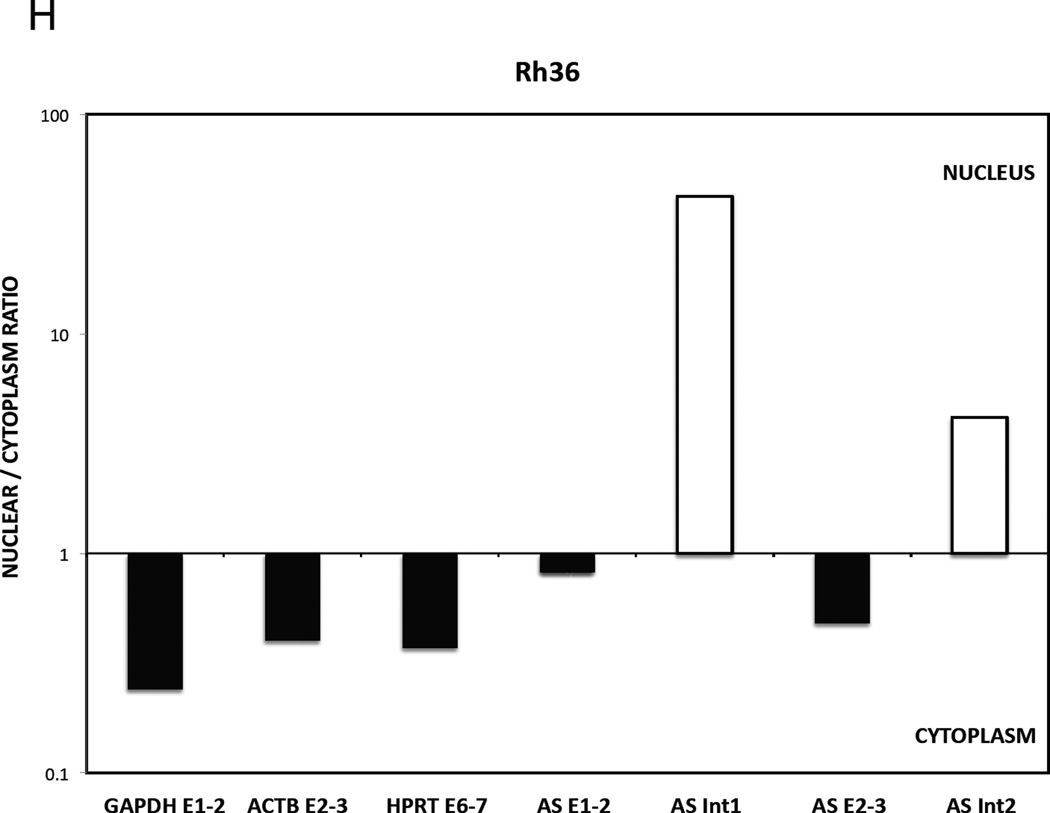
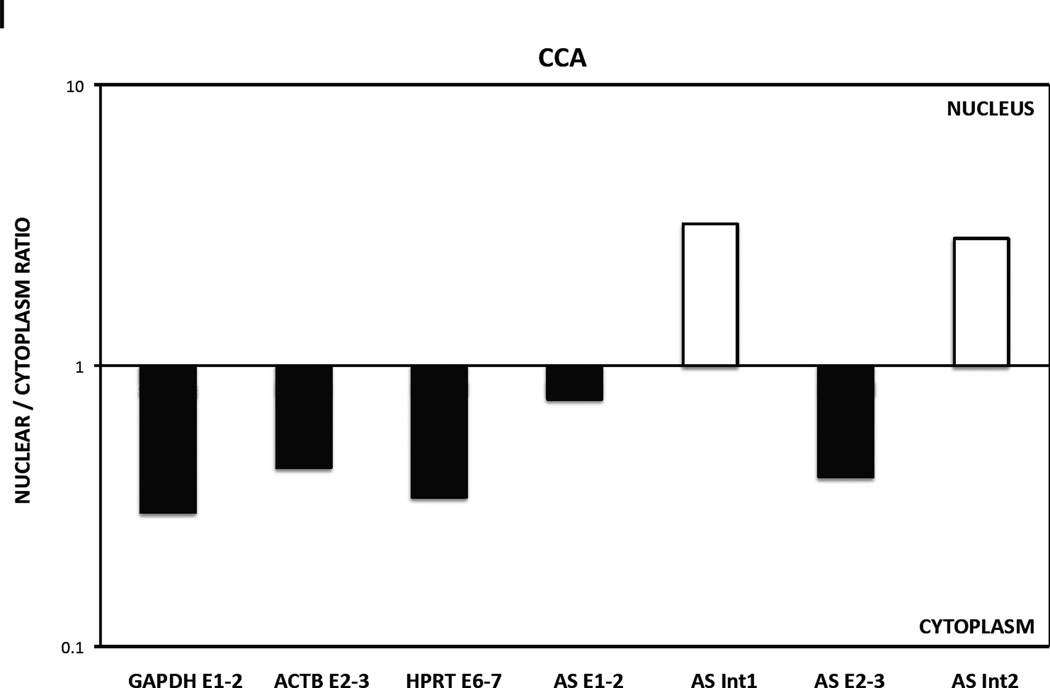
H and I. Nuclear versus cytoplasmic distribution of GLI1AS. Real-time RT/PCR analysis of the expression of GLI1AS in nuclear and cytoplasmic fractions of Rh36 (H) and CCA (I) cells. Data are represented as nuclear/cytoplasmic ratios, calculated by determining the 2−ΔCt values in the nucleus and the cytoplasm, with the ΔCt obtained by subtracting the Ct value of GAPDH in the cytoplasmic fraction from all Ct values. Note that for both cell lines the spliced forms of GLI1AS are preferentially retained in the cytoplasm (AS E1–2 and AS E2–3), similar to the exonic amplicons, GAPDH E1–3, HPRT E6–7 and ACTB E2–3, of the housekeeping genes used, while the opposite is true for the unspliced forms of GLI1AS (AS Int1 and AS Int2).
Expression constructs and protein detection
PCR primers were designed to amplify, a 579 bases 5’ segment of GLI1AS, a 407 bases 3’ segment of GLI1AS and a full-length GLI1AS from cDNA (Supplementary figure 1A), and the products were cloned into the KpnI and BamHI sites of the pCMV5 vector. The GLI1 expression construct (Flag-tagged) has been described previously (Shimokawa et al., 2008). The constructs were used to transfect Rh36 cells as described before (Tostar et al., 2012). Western analysis was performed using a GLI1 rabbit polyclonal antibody (Cell Signaling) and a Vinculin mouse monoclonal antibody (Sigma-Aldrich) as previously (Tostar et al., 2012).
RACE analysis
RACE was performed with the GeneRacer kit (Invitrogen) as described in the users’ manual. Total RNA from CCA, Rh36 and RMS13 cell lines was enzymatically treated with CIP (Calf Intestinal Phosphatase) to remove the 5’ phosphates of truncated mRNAs and with TAP (Tobacco Acid Pyrophosphatase) to remove the 5’ cap structure, both followed by ethanol extraction and phenol RNA precipitation. Subsequently, the GeneRacer RNA oligo was ligated to the 5’ exposed phosphate of the RNA using T4 RNA ligase. The ligated RNA was then reversed transcribed into first-strand cDNA using Superscript III and the GeneRacer OligodT primer, creating known priming sites at both the 5’ and 3’ ends. Gene specific primers (Supplementary table) were designed preferably at exon junctions and obtained from Sigma-Aldrich. Subsequently, initial touchdown and nested PCR were performed as recommended by the manufacturer. The RACE PCR products were analyzed on 1–1.5% agarose gels, followed by band excision, DNA extraction and sequencing.
Real-time PCR
Real-time PCR was carried out with Power SYBR Green (Applied Biosystems, Foster City, CA) on a 7500 fast real-time PCR system (Applied Biosystems) or with 1X IQ SYBR Green Supermix (Bio-Rad) on a C1000 Thermal Cycler (Bio-Rad) with primers designed to detect the spliced and unspliced GLI1 and GLI1AS RNAs, PTCH1, PTCH2, ADAR2 and INHBE (Table 1, Supplementary figure 4). All amplifications were run at least in triplicate and the fold change was normalized to the expression of one of the following housekeeping genes, ACTB, GAPDH, TBP or RPLPO. These were chosen based on the least variation in the samples analyzed. The relative expression was determined by the ΔCt method. To assess the subcellular distribution of GLI1AS equal amounts of RNA from the nuclear and cytoplasmic fractions were reverse transcribed and analyzed in comparison to the expression of the three housekeeping genes ACTB, GAPDH and HPRT. Data are represented as ratios (R) of the relative expression between the nucleus and the cytoplasm (R= 2−ΔCt nucleus / 2−ΔCt cytoplasm). The ΔCt values were calculated by subtracting from the Ct values the Ct value of GAPDH in the cytoplasmic fraction.
Table 1.
Primers for real-time PCR
| PRIMER NAME | SEQUENCE |
|---|---|
| TBP E3 forward | 5’ GCCAGCTTCGGAGAGTTCTGGGATT |
| TBP E4 reverse | 5’ CGGGCACGAAGTGCAATGGTCTTTA |
| GAPDH E1 forward | 5’ CTTCGCTCTCTGCTCCTCCTGTTCG |
| GAPDH E3 reverse | 5’ ACCAGGCGCCCAATACGACCAAAT |
| ACTB E2 forward | 5’ CAAGGCCAACCGCGAGAAGATGAC |
| ACTB E3 reverse | 5’ GCCAGAGGCGTACAGGGATAGCACA |
| RPLPO E6 forward | 5’ CCTTCTCCTTTGGGCTGGTCATCCA |
| RPLPO E7 reverse | 5’ CAGACACTGGCAACATTGCGGACAC |
| HPRT E6 forward | 5’ TGCAGACTTTGCTTTCCTTGGTCAGG |
| HPRT E7 reverse | 5’ CCAACACTTCGTGGGGTCCTTTTCA |
| GLI1 E11 forward | 5’ CAGCTACATCAACTCCGGCCAATAGGG |
| GLI1 E12 reverse | 5’ TGCTGCGGCGTTCAAGAGAGACTG |
| GLI1 E11 forward | 5’ GACCAGCTACATCAACTCCGGCCAAT |
| GLI1 Int 11 reverse | 5’ TCCTCCTCTCAACCTCAGGGCAGGT |
| GLI1 E1 forward | 5’ GAGCCCAGCGCCCAGACAGA |
| GLI1 Int 1 reverse | 5’ CTGGGATGAGTCCCTAAGAAGC |
| GLI1 Int 3 forward | 5’ GTGCATGGAGAGACATGCCCCTTT |
| GLI1 E4 reverse | 5’ GGCATCCGACAGAGGTGAGATGGAC |
| AS E1 forward | 5’ GAACACGCTGCTGTTGGCCTCAC |
| AS E2 reverse | 5’ ACCTCGTGAACCACACCCTGCAC |
| AS Int 1 forward | 5’ AGGGGGAGAAGTTCAACCGAGAACG |
| AS E2 reverse | 5’ CATCCAAAGGGTGAGGCCTGCAG |
| AS E2 forward | 5’ GGGGAACTGAAGGAGGAGCCCAGTT |
| AS E3 reverse | 5’ TGACCTCTTGATCCACCCGACTTGG |
| AS E2 forward | 5’ CCCACTTAAAAGCCCAGGAGGTGGA |
| AS Int 2 reverse | 5’ CTGCCCCTCCCTTATATCCGGTCAC |
| PTCH1 forward | 5’ AATGGGTCCACGACAAAGCCGACTA |
| PTCH1 reverse | 5’ TCCCGCAAGCCGTTGAGGTAGAAAG |
| PTCH2 forward | 5’ TCTTTCTGGGACTGTTGGCCTTTGG |
| PTCH2 reverse | 5’ CCTCCCCCAGCTTCTCCTTGGTGTA |
| INHBE forward | 5’ CCGGAGACTACAGCCAGGGAGTGTG |
| INHBE reverse | 5’ GTACAGGTGGTGGGACCGAGGAGTG |
| INHBE E1 forward | 5’ TCAGCCTTCCTGAGTCCCAGAC |
| INHBE E1 reverse | 5’ AATGAGGGCACAGTGACAGCAG |
| INHBE Int 1 forward | 5’ GCAGGGCTTGGGGAGTCTCAGAGG |
| INHBE Int 1 reverse | 5’ TTCCCTGTCTGCTCTTTGCCTGTTG |
| ADAR2 forward | 5’ ACCGCAGGTTTTAGCTGACGCTGT |
| ADAR2 reverse | 5’ CCAGCCAGCACTTTTCTGCGAGCGT |
Cell proliferation - EdU incorporation assay
Cells were seeded in 60 mm dishes, treated with siRNA for 48 hours, followed by a 4-hours 10 µM EdU (5-ethynyl-2’deoxyuridine) incubation. EdU were detected by fluorescent-azide coupling reaction (Click-iT, Invitrogen, Eugene, OR), and subsequently, 10 000 cells were counted on a FACS calibur machine (BD Biosciences, Stockholm, Sweden) to determine the percentage of cells in the population that are in the S-phase.
Gating was performed to eliminate aggregated cells, and non-stained cells were used to define on the FACS plot the area with the cells that have incorporated EdU.
Chick chorioallantoic membrane (CAM) assay
Fertilized chicken eggs purchased from Ova Production AB (Vittinge, Sweden) were placed in a 60% humidified, 37°C incubator without CO2. On day 10, a square window was opened and transfected cells, re-suspended in 30µl of growing medium (around one million cells per embryo), were applied on top to the CAM as described previously (Brooks et al., 1994). The eggs were sealed and returned to the incubator for an additional 7 days. On day 17, the tumors were collected, weighed and measured. All tumors were fixed in 4% paraformaldehyde, embedded in paraffin and processed for sectioning. Additionally, the tumors were stained with hematoxylin and eosin and further analyzed by standard light microscopy in order to ensure the presence of tumor tissue.
Chromatin immunoprecipitation (ChIP) assays
The ChIP analysis was carried out by using the SimpleChIP® Enzymatic Chromatin IP Kit (Cell Signaling). Briefly, cells were cross-linked in 1% formaldehyde, quenched with 1XGlycine and lysed according to the manufacturer's recommendations. Subsequently, the samples were treated with Micrococcal Nuclease to digest DNA at an average length of 150–900 bp, then subjected to a water bath sonicator (Bioruptor) for breaking the nuclear membrane and finally incubated overnight at 4°C with the appropriate antibody. ChIP Grade Protein G Magnetic Beads (Cell Signaling) were used to pull down the antibody and the associated chromatin. The antibody bound chromatin was eluted in 1XChIP elution buffer (Cell Signaling) followed by reverse cross-linking and protease K treatment. DNA was purified with ‘DNA purification spin columns’ supplied in the kit. The following antibodies were used: Normal Rabbit IgG (#2729, Cell Signaling) as negative control, Histone H3 (D2B12) XP® rabbit mAb (ChIP formulated) (#4620, Cell Signaling) as positive control, Tri-Methyl-Histone H3 (Lys27) Antibody (#9756, Cell Signaling), Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb (#9751, Cell Signaling) and Rpb1 CTD (4H8) Mouse mAb (#2629, Cell Signaling). The immunoprecipitated DNA was analyzed by real-time PCR using primer sets for the INHBE, GLI1AS and GLI1 genes (Table 1, Supplementary figure 4). A serial dilution of the 2% input chromatin DNA (1:5, 1:25, 1:125) was used to create a standard curve and determine the efficiency of amplification.
RESULTS
Identification of a non-coding RNA, antisense to the GLI1 gene, GLI1AS
In silico analysis of EST databases identified a transcript, GLI1AS, flanking the GLI1 promoter and encoded on the opposite (antisense) strand. Rapid Amplification of cDNA Ends (RACE) in Rh36, CCA and RMS13 cancer cells determined the sequence of GLI1AS RNA, which initiates 155 nucleotides upstream of the major transcription start site of the GLI1 gene (Liu et al., 1998) in an opposite orientation (Figure 1A, Supplementary figure 1). The 5’ and 3’ RACE analysis revealed that the 885-nucleotide, three-exon GLI1AS RNA is 5’ capped and 3’ poly(A) tailed but without any long open reading frame (GenBank accession number JX675466). Polyadenylation of GLI1AS was also confirmed by cDNA synthesis in the presence or absence of oligo(dT) primers in RMS13 cells (Figure 1B). This would be in-line with data from the FANTOM5 ZENBU gLyphs Genome Browser (http://fantom.gsc.riken.jp/5/suppl/Kanamori-Katayama_et_al_2011), indicating a number of transcription initiation sites at the 5’ end of the GLI1 gene both in the sense and antisense orientation, with some having the potential for transcriptional overlap (Supplementary figure 2). Finally, evidence for the presence of active transcription at the GLI1AS/GLI1 genomic region was gathered from the UCSC Genome Browser (Supplementary figure 3).
GLI1AS and GLI1 have a concordant expression in tumors
Expression analysis of GLI1AS in the following cancer cell lines: PC3, 22Rv1 PANC1, A549, AGS, D283Med, RMS13, RD, Rh36, and CCA show a remarkable co-regulation between GLI1 and GLI1AS. (Figure 1C and D). Further analysis using exonic and intronic PCR primer sets (Table 1, Supplementary figure 4) defined the splicing pattern of GLI1AS in RMS13, RD, Rh36, and CCA cells. Transcripts retaining GLI1AS intron 2 were more abundant than transcripts with the intron being removed in all cell lines analyzed. A similar but less pronounced pattern was also observed for intron 1 in RD, Rh36, and CCA but not RMS13 cells (Figure 1E). Additionally, a correlation between the expression of GLI1AS, interrogated by spliced exons 1 and 2 or 2 and 3, and GLI1 was detected (Figure 1F and G). These observations are reminiscent of the finding that sense/antisense transcript pairs often reveal a concordant regulation and a lower expression of the antisense transcript (Faghihi et al., 2008; Faghihi and Wahlestedt, 2009; Katayama et al., 2005).
To gain further insight into a possible biological function of GLI1AS we also determined its subcellular localization. Through nuclear/cytoplasmic fractionation of Rh36 and CCA cells, followed by RNA isolation and real-time RT/PCR, we found that the unspliced GLI1AS RNAs are preferentially retained in the nucleus, while the spliced GLI1AS RNAs transported to the cytoplasm (Figure 1H and I). A similar pattern was also observed for RMS13 cells (Supplementary figure 5).
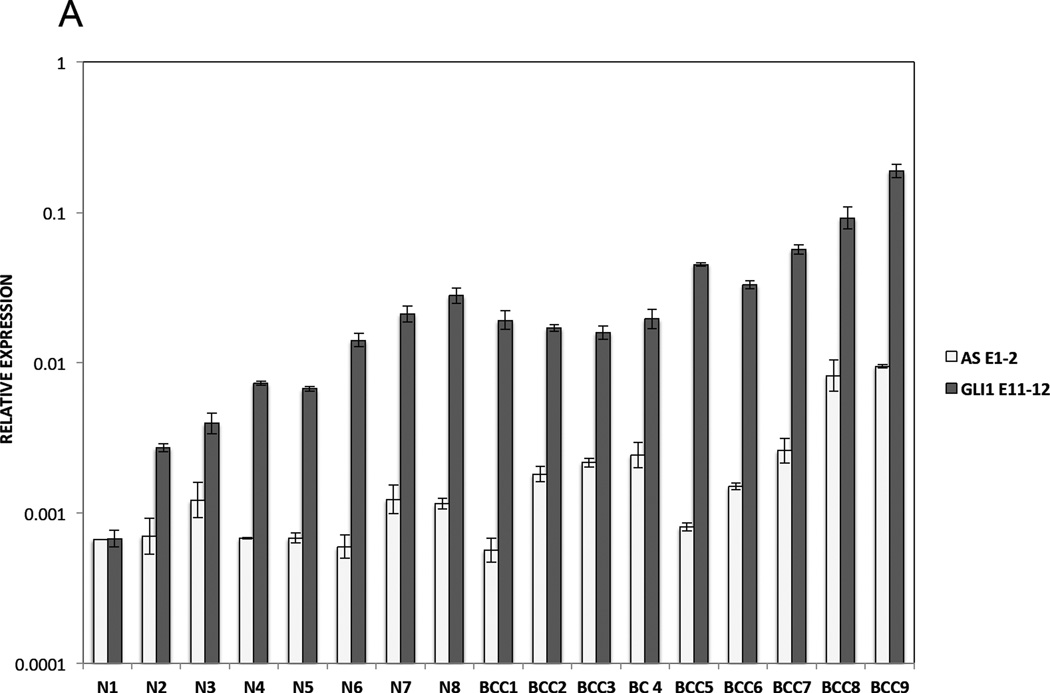
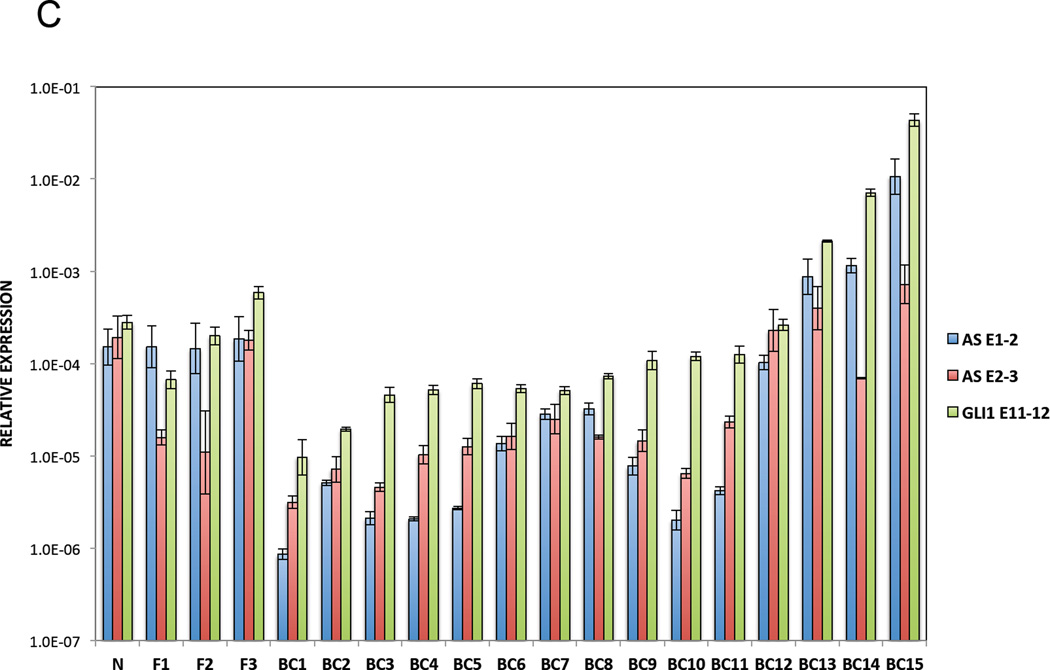
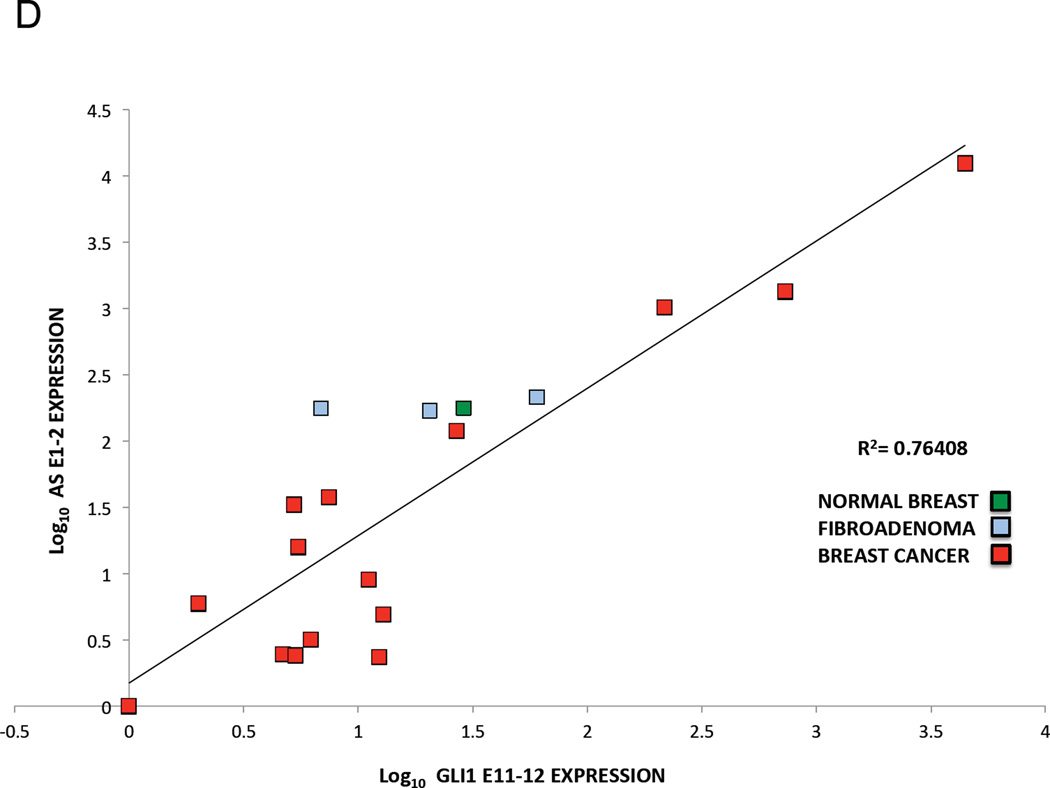
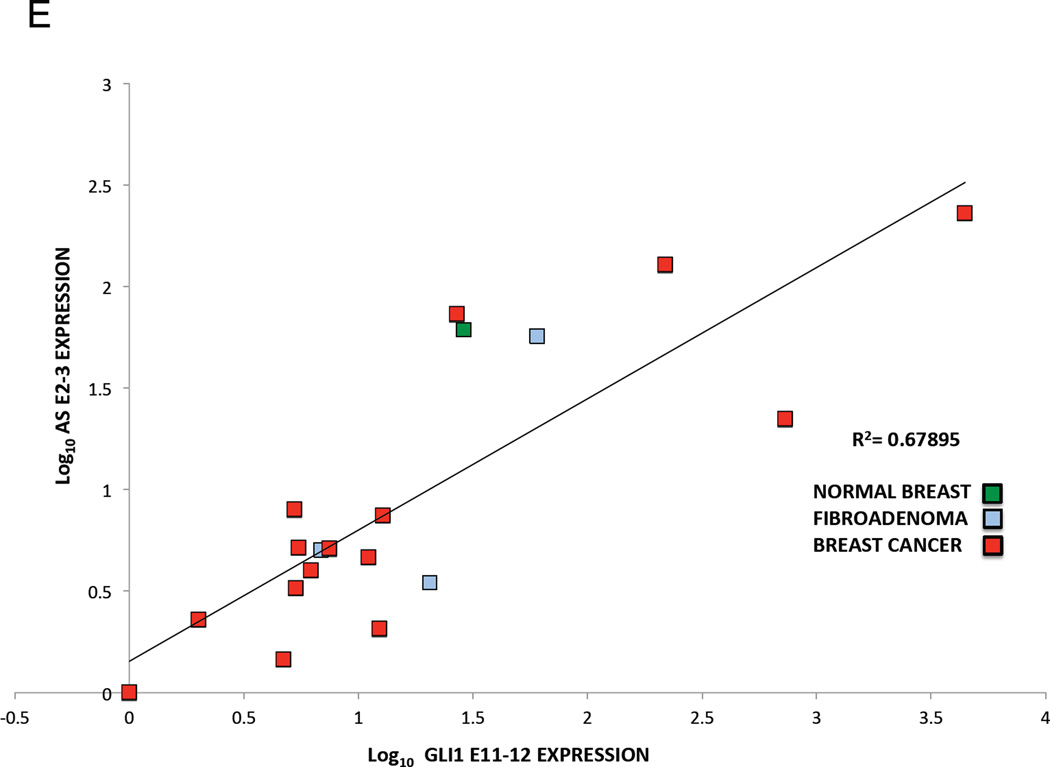
Finally, we determine the translational significance of these findings by examining GLI1AS expression in a panel of basal cell carcinomas (BCCs), a tumor type characterized by increased GLI1 levels (Gailani et al., 1996). GLI1AS levels were lower than GLI1 and more pronounced in the nine BCC compared to the eight normal skin samples analyzed, showing an apparent co-regulation with GLI1 expression, similarly to what was previously seen in the cancer cell lines (Figure 2A and B,). Moreover, the levels of GLI1 and GLI1AS were analyzed in breast cancer, a tumor type where GLI1 expression is thought to participate in the pathogenesis of this malignancy (Li et al., 2012b). A well co-related pattern of GLI1 and GLI1AS expression in the 15 tumor samples, the three benign breast fibroadenomas and the normal breast analyzed was observed (Figure 2C, D and E).
Figure 2. Expression of GLI1AS in basal cell carcinoma and breast cancer. Co-regulation with GLI1.
A. Real-time RT/PCR analysis of the expression of GLI1AS and GLI1 in basal cell carcinoma and normal skin. Data are represented as relative expression (2−ΔCt values), calculated by subtracting the mean Ct value of the housekeeping gene RPLPO from the Ct value of the GLI1 and GLI1AS transcripts (ΔCt).
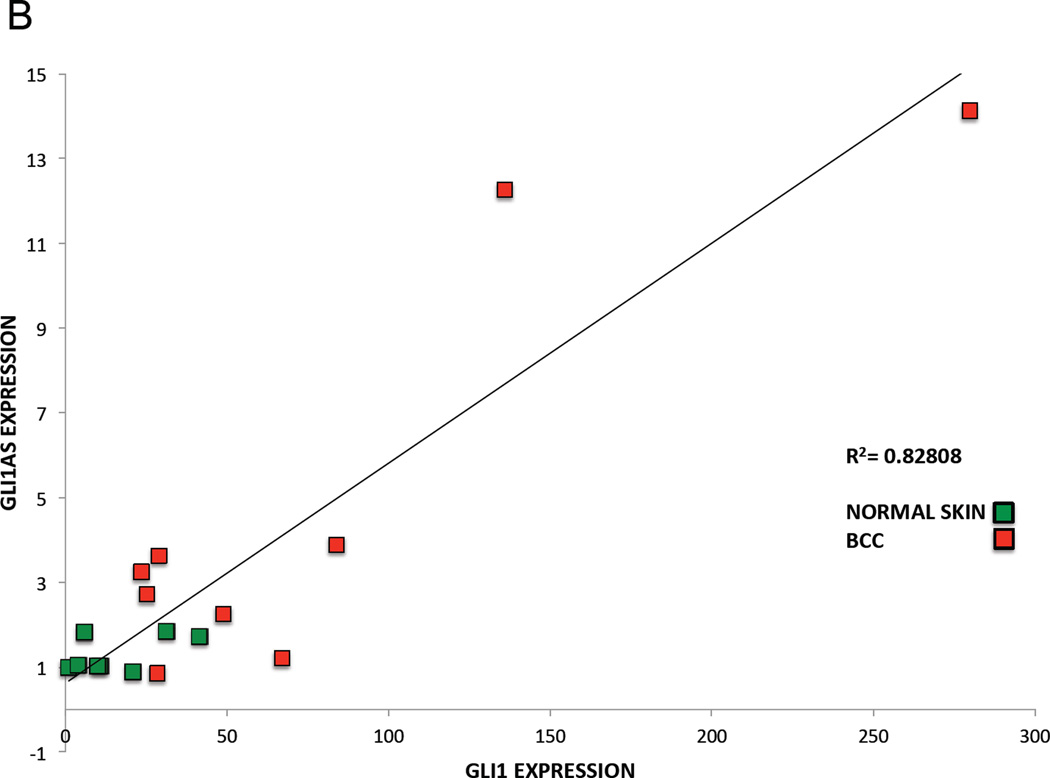
B. Co-relation of GLI1AS with GLI1 expression in basal cell carcinoma and normal skin. The expression of GLI1AS and GLI1 in the eight normal skin and the nine basal cell carcinoma samples determined above was normalized to the expression in the skin sample with lowest GLI1 expression, by subtracting the ΔCt value of this sample from the ΔCt in all samples, resulting in ΔΔCt values. Data are presented as normalized relative expression (2−ΔΔCt values). A positive correlation was observed between GLI1 and GLI1AS expression, R2 = 0.82808 (Pearson correlation = 0.91, P<0.05).
C. Real-time RT/PCR analysis of the expression of GLI1AS and GLI1 in breast cancer, benign fibroadenoma and normal breast samples. Data are represented as relative expression (2−ΔCt values), calculated by subtracting the Ct value of the housekeeping gene ACTB from the Ct value of the GLI1 and GLI1AS (AS E1–2 and AS E2–3) transcripts (ΔCt).
D, E. Co-relation of GLI1AS with GLI1 expression in breast cancer. The expression of GLI1AS and GLI1 in the 15 breast tumor samples, the three benign fibroadenomas and the normal breast sample determined above was normalized to the expression of the least expressing sample (BC1), by subtracting the ΔCt in BC1 from the ΔCt in all samples, resulting in ΔΔCt values. Data are presented as the Log10 of the 2−ΔΔCt values. A positive correlation was observed between GLI1 (GLI1 E11–12) and GLI1AS (AS E1–2, D or AS E2–3, E) expression, R2 = 0.76408 (Pearson correlation = 0.874, P<0.05) or 0.67895 (Pearson correlation = 0,824, P<0.05), respectively.
Opposing regulatory effects of GLI1AS on GLI1 versus GLI1 on GLI1AS
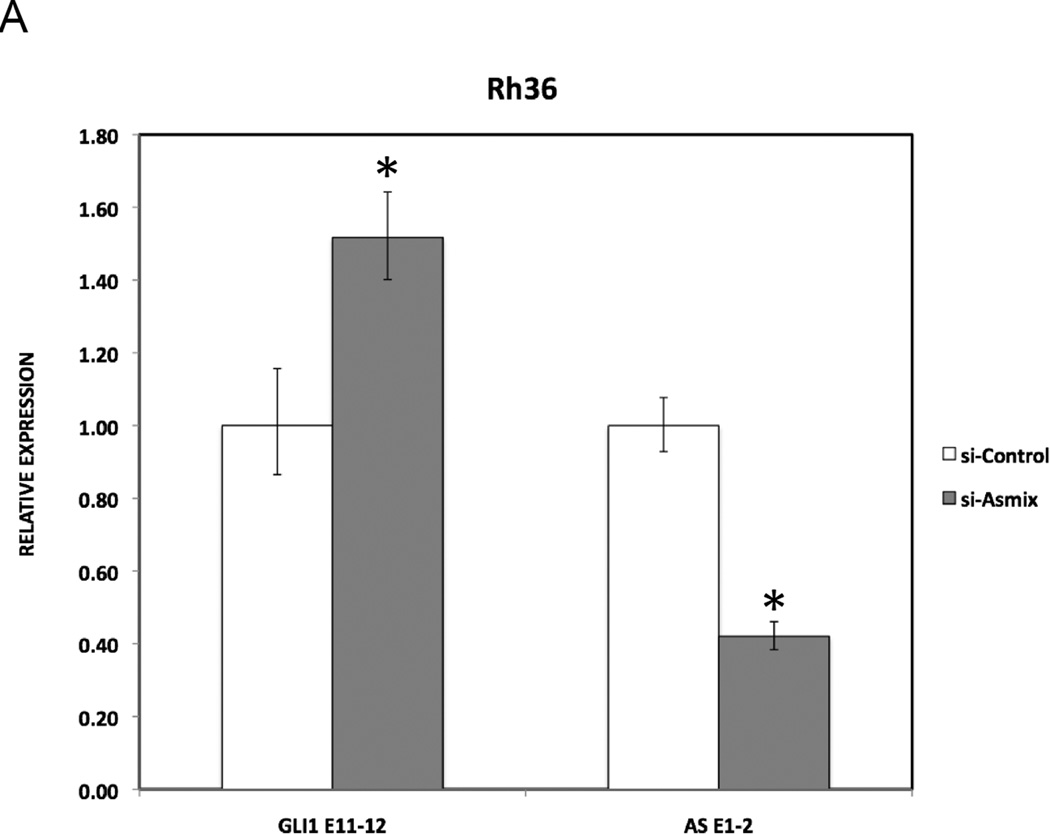
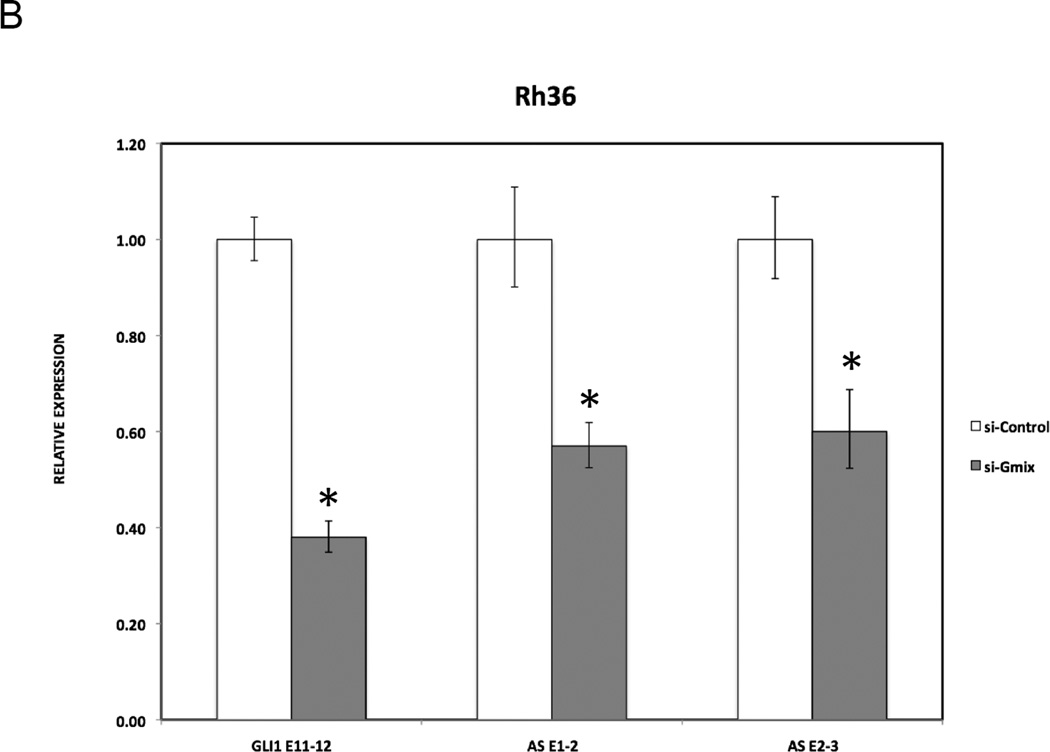
In order to investigate whether the GLI1AS transcript regulates the expression of GLI1, three short interfering RNA (siRNA) molecules targeting GLI1AS were designed (Figure 1A, Table 2) and used as a mixture (si-ASmix). Transfection of the si-ASmix into Rh36 cells resulted in a significant GLI1AS knockdown, accompanied by an increase in GLI1expression (Figure 3A). Similar results were found in CCA cells (Supplementary figure 6). Interestingly, transfection of three siRNAs targeting GLI1 (Figure 1A, Table 2) (si-Gmix) in the Rh36 and CCA cells resulted in a decrease of GLI1AS levels (Figure 3B and Supplementary figure 6). Thus, depletion of one member of the GLI1 / GLI1AS pair has opposing effects on the other partner. GLI1 knockdown reduces GLI1AS but GLI1AS knockdown increases GLI1.
Figure 3. GLI1AS knockdown increases GLI1 expression in the Rh36 cell line.
A. Real-time RT/PCR analysis of the expression of GLI1 and GLI1AS transcripts in Rh36 cells, following knockdown of GLI1AS by a 48-hr treatment with si-ASmix, respectively. Data are represented as relative expression (2−ΔΔCt values), calculated by subtracting the Ct value of the housekeeping gene TBP from the Ct value of the GLI1 and GLI1AS transcripts (ΔCt), and normalized to the ΔCt value obtained by a control siRNA (si-Control), by subtracting this control ΔCt value from the ΔCt values of the si-Gmix and si-ASmix samples (ΔΔCt). Error bars indicate the standard deviation. *, Statistical significant, P<0.01 compared to control, calculated by the Student’s t-test.
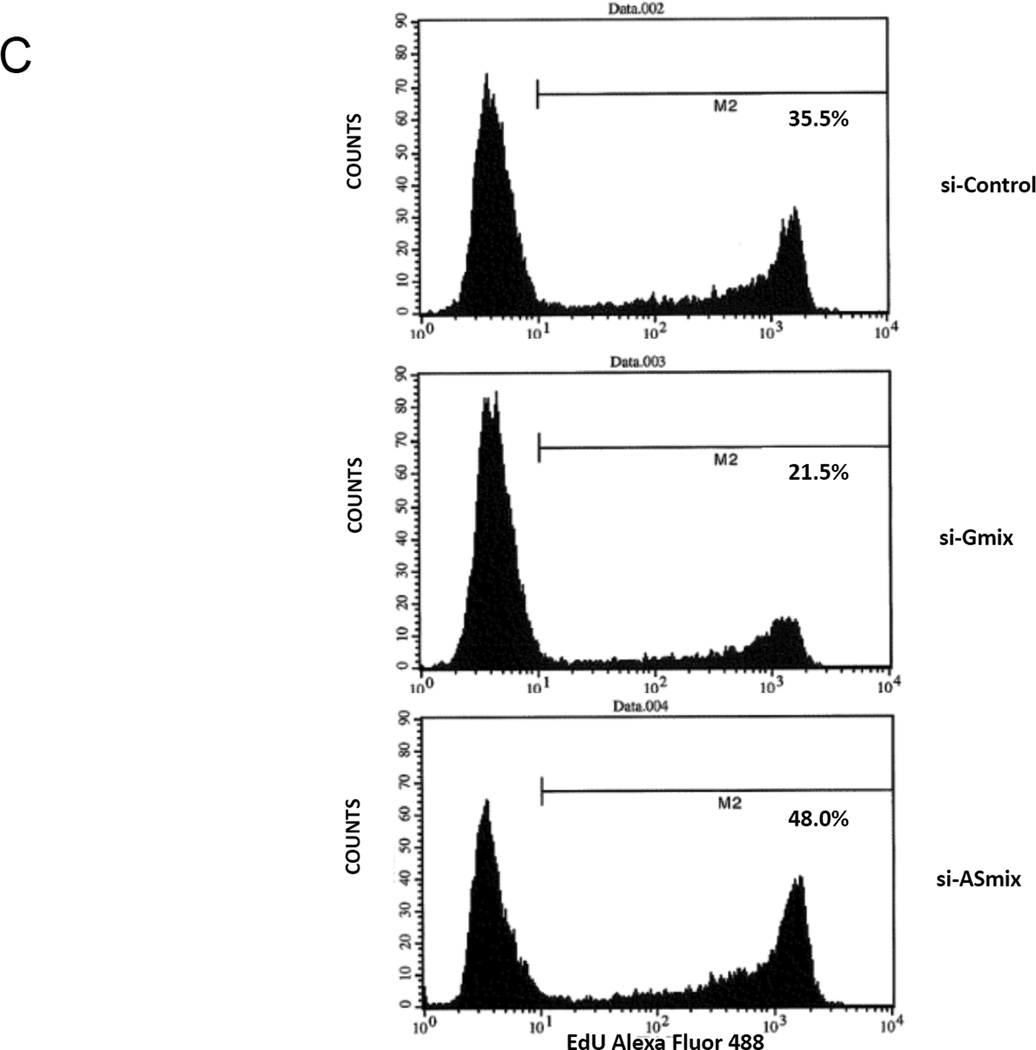
B. The expression of GLI1 and GLI1AS transcripts in Rh36 cells, following knockdown of GLI1 by a 48-hr treatment with si-Gmix was analyzed as in (A). Note that siRNA treatment effectively reduced expression of the targeted genes. Additionally, the si-ASmix increased the GLI1 mRNA (GLI1 E11–12) while the si-Gmix conferred a decrease in GLI1AS (AS E1–2 and AS E2–3).
C. Rh36 cells, cultured for 48 hours after transfection with si-Control, si-Gmix or si-ASmix, were subjected to the EdU incorporation assay for 4 hours. The percentage of cells labeled with Alexa Fluor 488 azide and detected by flow cytometry is shown. Note that treatment with the si-Gmix reduces proliferation, while treatment the si-ASmix increases proliferation, in-line with the changes in GLI1 mRNA levels.
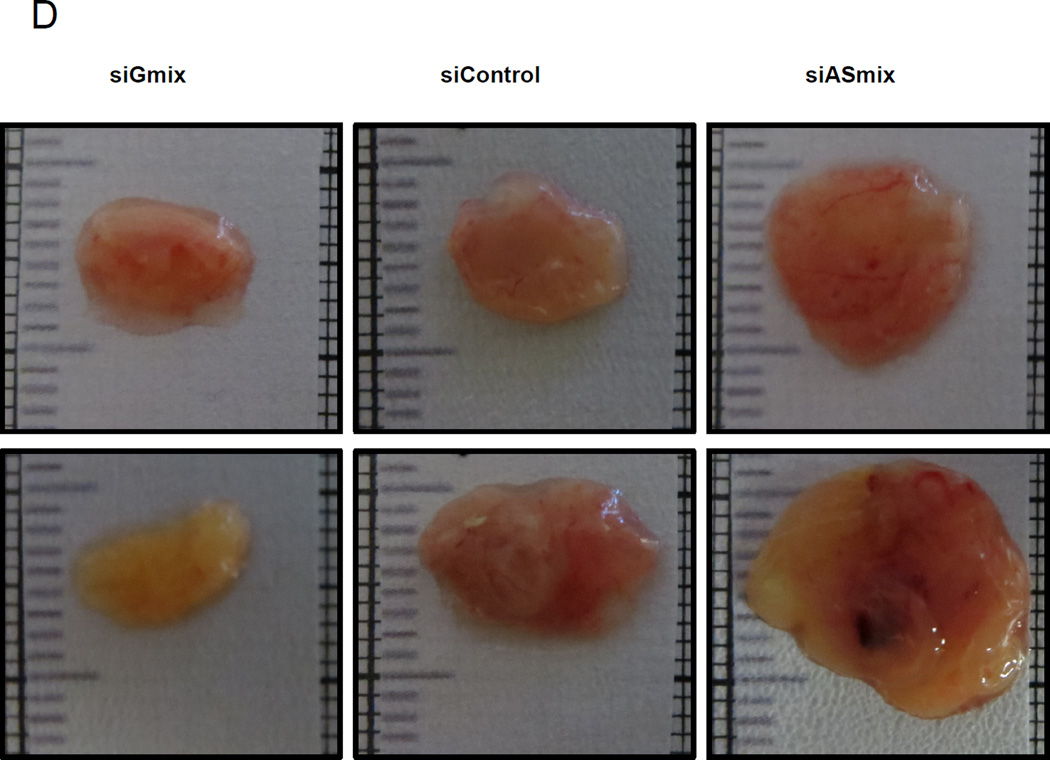
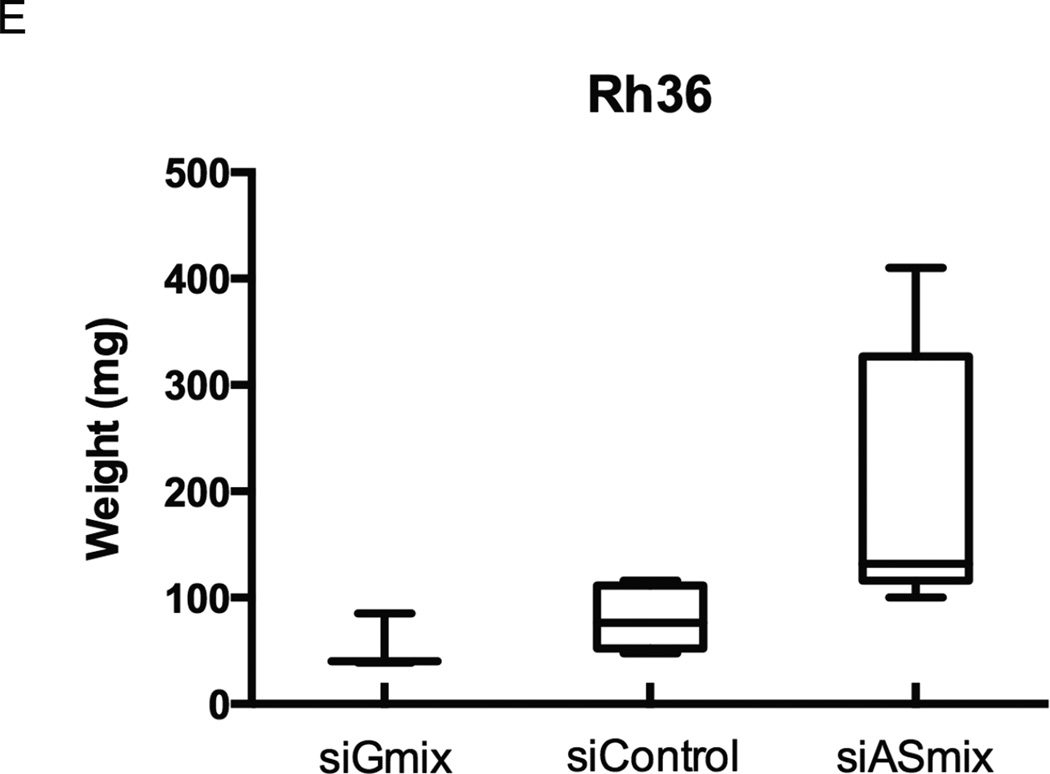
D, E. Rh36 cells, cultured for 48 hours after transfection with si-Control, si-Gmix or si-ASmix were introduced onto the chorioallantoic membrane of fertilized chicken eggs (CAM assay) and tumors were collected after 10 days. Two representative tumors from each experimental treatment are shown D. Tumor weights (in mg), from three independent experiments, are represented as box plots, with the box denoting median ± interquartile range (IQR) and the whiskers the range from minimum to maximum E.
To evaluate the biological implications of the impact of GLI1AS on GLI1 expression, the siRNA-treated Rh36 cells, a line in which proliferation is dependent on GLI1 (Tostar et al., 2010), were further analyzed. EdU incorporation assay showed a decrease in proliferation by the si-Gmix and an increase by the si-ASmix, in-line with the changes of the GLI1 mRNA (Figure 3C).
To examine whether the endogenous modulation of GLI1 and GLI1AS levels in Rh36 cells has an impact on tumor growth, the CAM xenograft model was used. Treatment of Rh36 cells with the si-Gmix decreased their capacity to form tumors in this model, On the other hand an increased tumor weight was observed following treatment with the si-ASmix (Figure 3D and E).
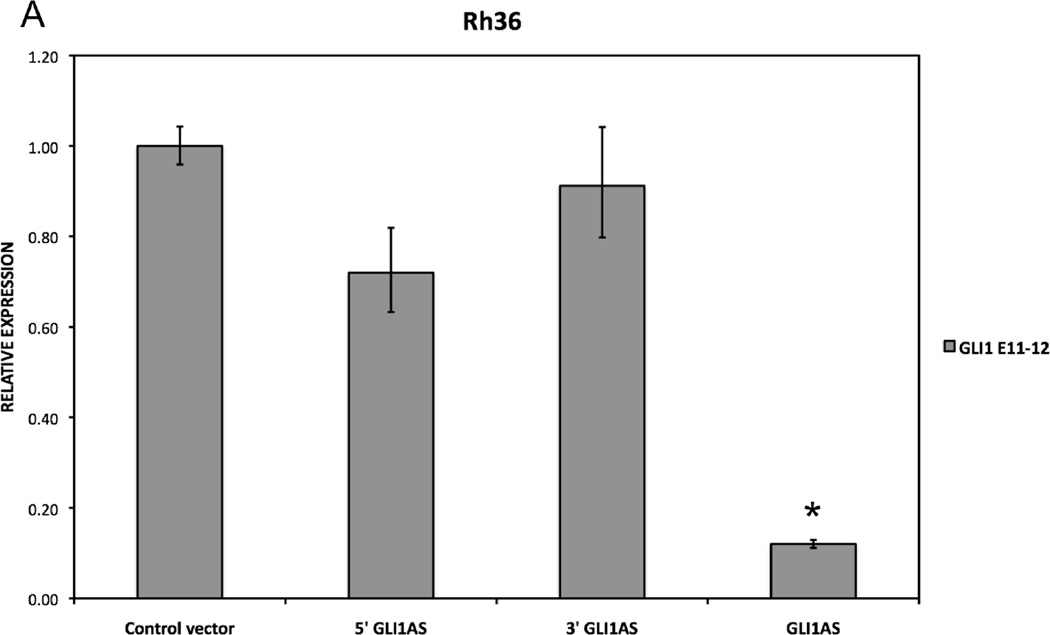
To determine the impact of increased levels of the GLI1AS RNA on GLI1, expression constructs of a full-length GLI1AS, a 5’ segment of GLI1AS and a 3’ segment of GLI1AS in the pCMV5 vector were generated as described in supplementary figure 1A. Interestingly, the 5’ and the 3’ GLI1AS constructs did not elicited major changes in the GLI1 mRNA levels but the full-length GLI1AS construct conferred an almost 10-fold reduction of the GLI1 mRNA (Figure 4A). Additionally, the full-length but not the 5’ or the 3’ constructs decreased the expression of the GLI1 protein (Figure 4B) and down-regulated well-established GLI1 target genes, PTCH1 and PTCH2 (Shimokawa et al., 2008) (Figure 4C). These findings suggest that the complete GLI1AS RNA sequence/structure is needed to elicit regulatory effects on GLI1. Additionally, retention of the intron 1 sequence in the 5’ construct did not confer major changes on its limited capacity to modulate GLI1 levels / GLI1 target genes (Supplementary figure 7).
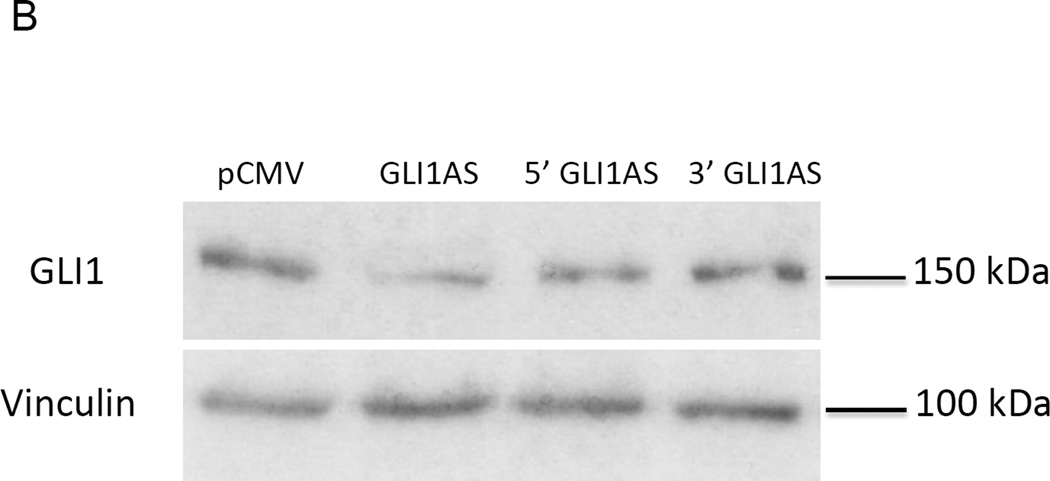
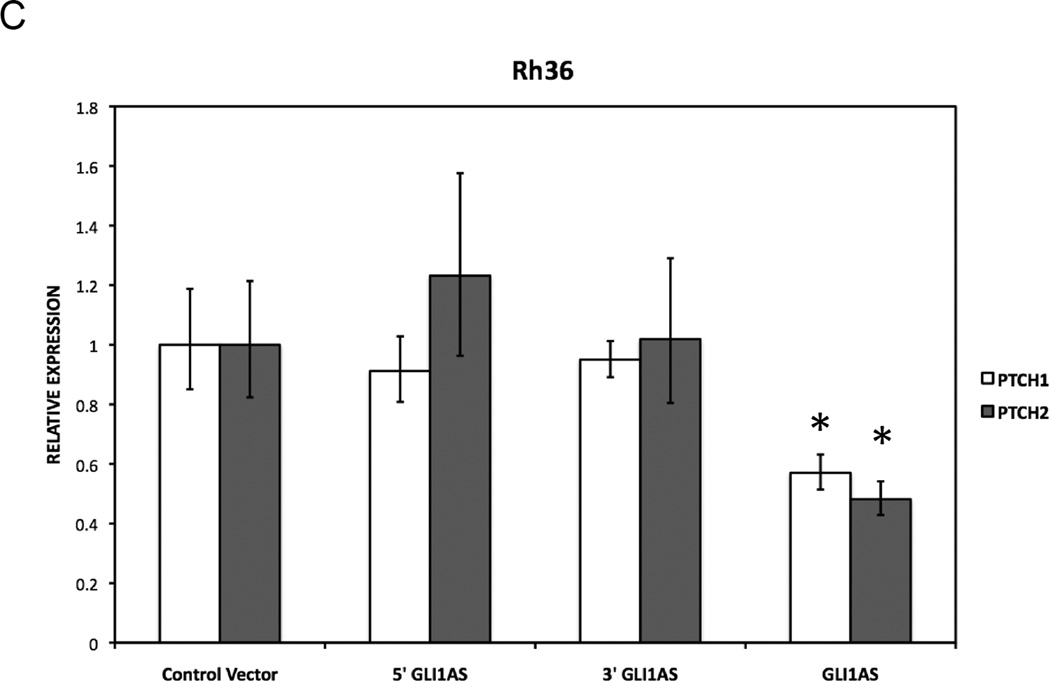
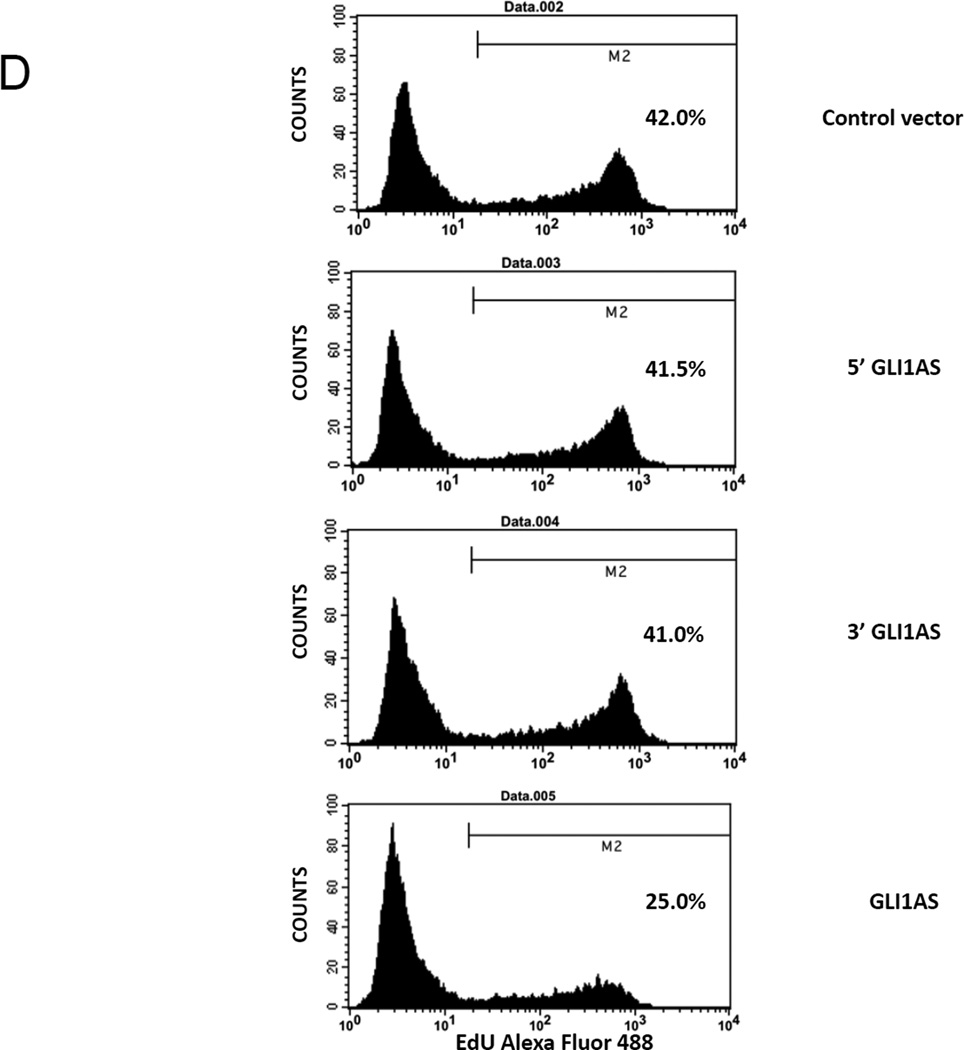
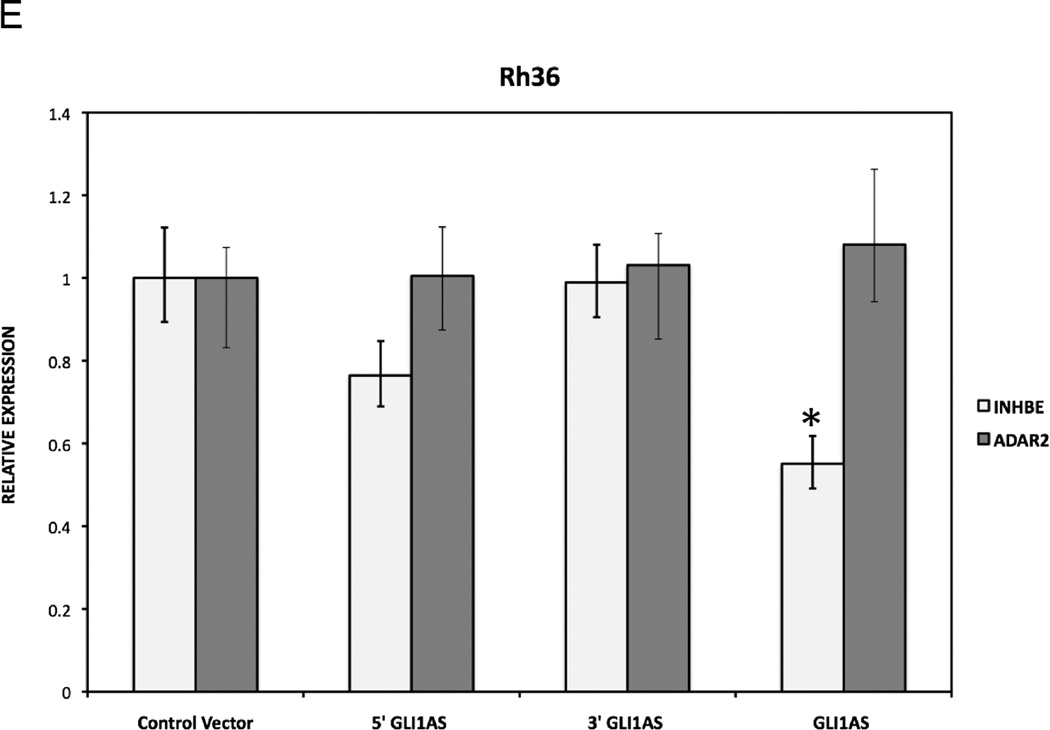
Figure 4. GLI1AS decreases GLI1 expression, while GLI1 increased GLI1AS expression.
A. Real-time RT/PCR analysis of the expression of GLI1 in Rh36 cells, 48 hrs after transfection of pCMV5 expression constructs for GLI1AS, a 579 bases 5’ segment of GLI1AS (5’GLI1AS) or a 407 bases 3’ segment of GLI1AS (3’GLI1AS). Data are represented as relative expression (2−ΔΔCt values), calculated by subtracting the Ct value of the housekeeping gene TBP from the Ct value of the GLI1 mRNA (ΔCt), and normalized to the ΔCt value obtained following transfection of the pCMV5 control vector, by subtracting this control ΔCt value from the ΔCt values of all samples (ΔΔCt). Error bars indicate the standard deviation. *, Statistical significant, P<0.01 compared to control, calculated by the Student’s t-test. Note that overexpression of GLI1AS effectively reduced GLI1 expression, while this is not the case when either the 5’ half or the 3’ half of GLI1AS are overexpressed.
B. Western blot analysis of GLI1 protein expression in Rh36 cells, following transfection of pCMV5 expression constructs for GLI1AS, 5’GLI1AS or 3’GLI1AS.
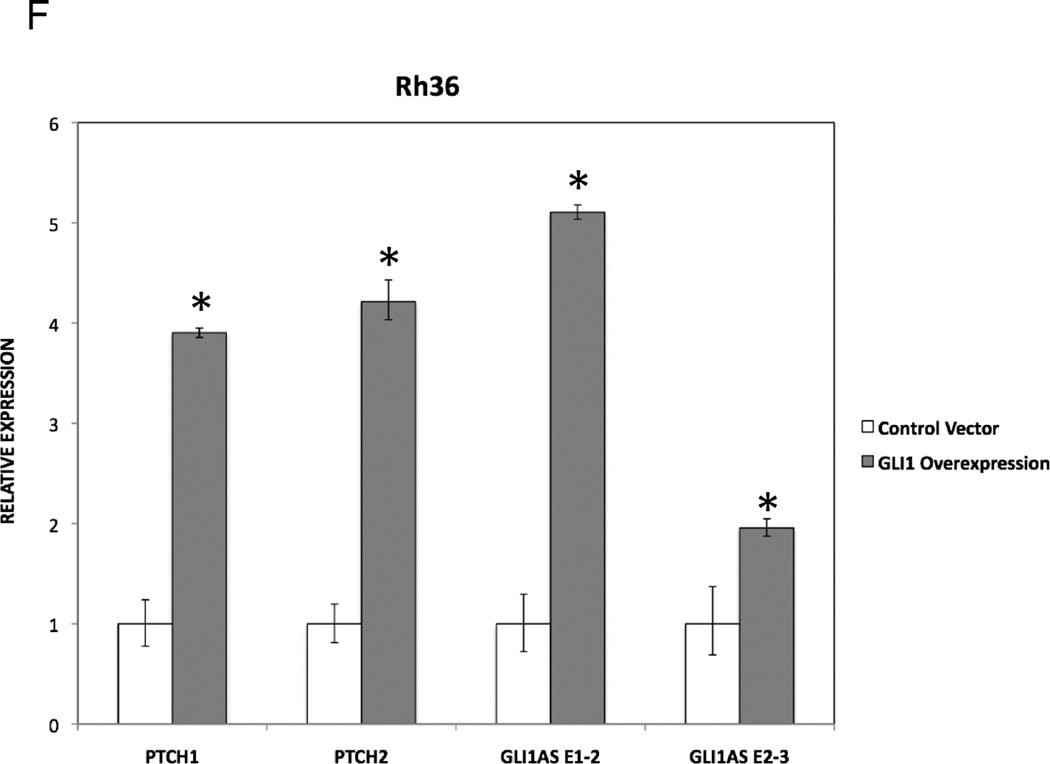
C. The expression of the typical GLI1 target genes PTCH1 and PTCH2 in Rh36 cells, following transfection of pCMV5 expression constructs for GLI1AS, 5’GLI1AS or 3’GLI1AS, is analyzed as in (A). *, Statistical significant, P<0.05 compared to control, calculated by the Student’s t-test.
D. Rh36 cells 48 hrs after transfection with pCMV5 control vector, or expression constructs for GLI1AS, the 5’ half of GLI1AS or the 3’ half of GLI1AS were subjected to the EdU incorporation assay for 4 hours. The percentage of cells labeled with Alexa Fluor 488 azide and detected by flow cytometry is shown. Note that overexpression of GLI1AS confers an effective reduction of proliferation, while this is not the case when either the 5’ half or the 3’ half of GLI1AS are overexpressed.
E. The expression of INHBE, a gene from the GLI1AS locus, and of the unrelated gene ADAR2 in Rh36 cells, following transfection of pCMV5 expression constructs for GLI1AS, 5’GLI1AS or 3’GLI1AS, is analyzed as in (A). *, Statistical significant, P<0.01 compared to control, calculated by the Student’s t-test.
F. The RNA levels of GLI1AS (AS E1–2 and AS E2–3), PTCH1 and PTCH2 in Rh36 cells following transfection of a pCMV5 expression construct for GLI1 (Materials and Methods) is analyzed as in (A). *, Statistical significant, P<0.01 compared to control, calculated by the Student’s t-test.
To determine whether the modulation of GLI1 levels by GLI1AS overexpression elicits regulatory effects at the cellular level, an analysis of the Rh36 proliferative capacity was performed. As anticipated, GLI1AS overexpression conferred a major reduction in Rh36 cellular proliferation (Figure 4D). Worth noting is that overexpression of either the 5’ GLI1AS or the 3’ GLI1AS construct did not alter the proliferative capacity of the transfected Rh36 cells.
GLI1AS modifies the chromatin landscaping and reduces the recruitment of RNA polymerase II at the GLI1/GLI1AS locus
To examine whether the repressive effects of GLI1AS on GLI1 expression are gene specific, the mRNA levels of INHBE, a gene positioned tail-to-tail to GLI1AS (Supplementary figures 3 and 4) and of an unrelated gene on chromosome 21q22.3, ADAR2, were also analyzed. ADAR2 levels were not changed by GLI1AS overexpression, however, INHBE expression was reduced, albeit not to the same extent as to that seen for GLI1 (Figure 4E).
Interestingly, transfection of a GLI1 expression construct in Rh36 cells resulted in an up-regulation of GLI1AS, similar to other typical GLI1 target genes PTCH1 and PTCH2 (Figure 4F). Of note, this treatment did not increase the INHBE levels (Supplementary figure 8). This finding rules out the possibility that INHBE is a GLI1 target gene. Consequently, the observed INHBE down-regulation by GLI1AS overexpression (Figure 4E) is not an indirect result of the reduced GLI1 levels, rather an effect of the antisense RNA on the organization of the chromatin locus.
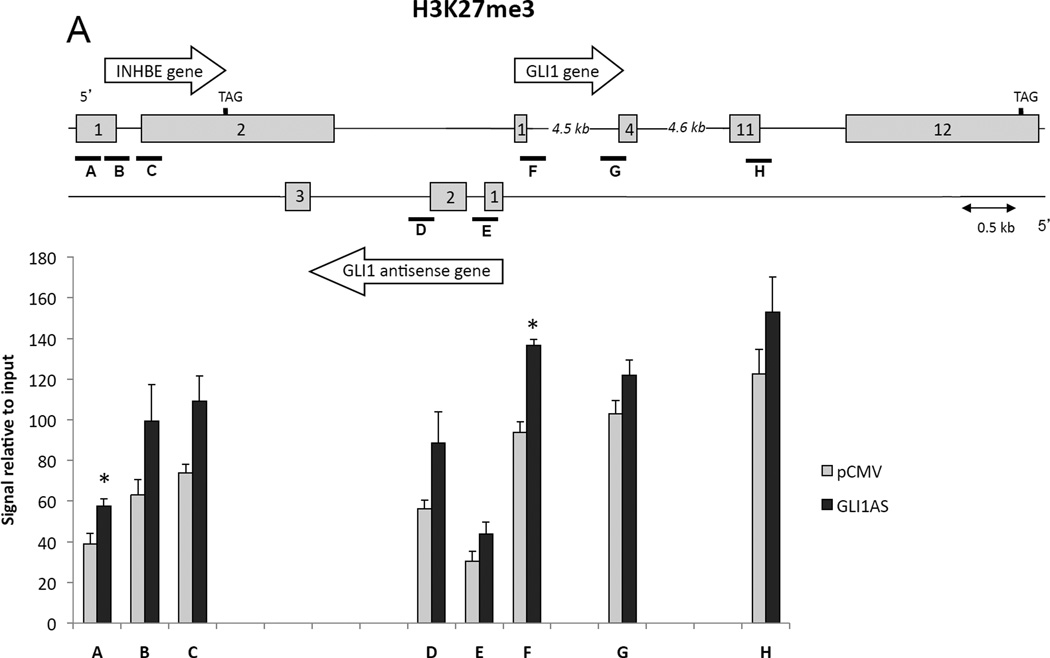
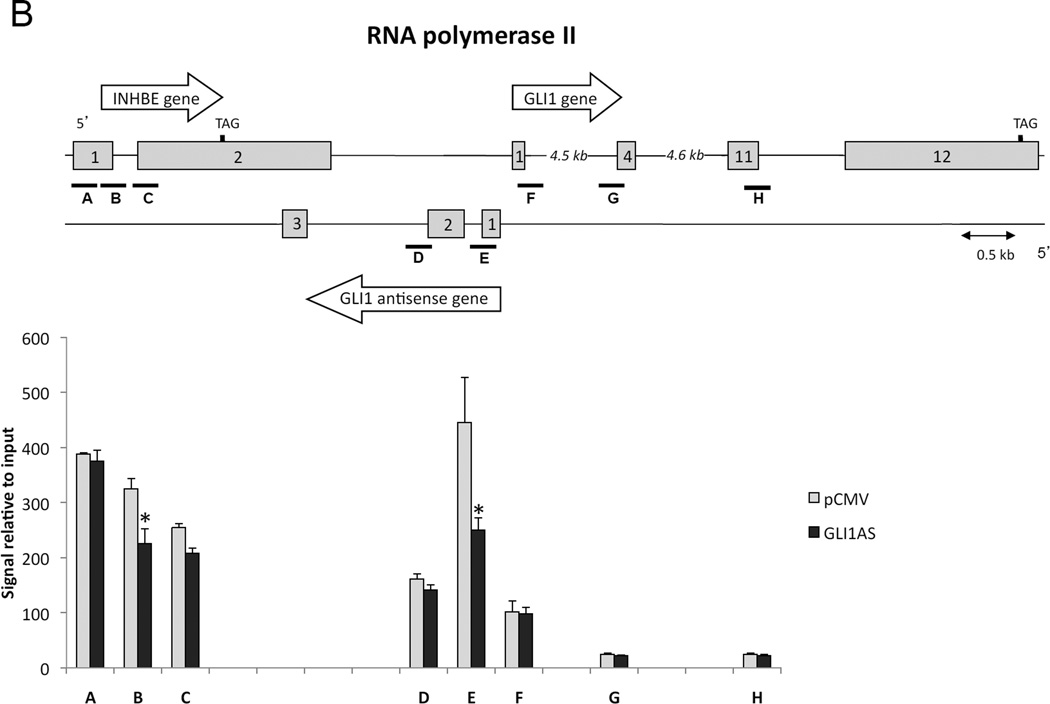
To address the role of epigenetic modifications in eliciting the GLI1 and INHBE down-regulation by GLI1AS overexpression, chromatin immunoprecipitation assays on transfected Rh36 cells were performed. The H3K27me3 signal, a mark of repressive chromatin, was increased throughout the INHBE / GLI1AS / GLI1 genomic regions, being most pronounced at the 5’ end of the INHBE and GLI1 genes, at interrogated segments A and F (Figure 5A). Surprisingly, the H3K4me3 signal was also increased up to the exon 1 of GLI1, with the major differences observed at the 5’ end the INHBE, GLI1AS and GLI1 genes, segments A, E, and F (Supplementary figure 9). This may relate to the observations that this histone mark, generally considered to associate with active transcription, has also been linked to repression of gene expression in the context of antisense transcription in yeast (Berretta et al., 2008; Pinskaya et al., 2009). Additionally, the recruitment of RNA polymerase II showed a statistical significant decrease at segments B and E. (Figure 5B). Thus, GLI1AS acts as an epigenetic modifier that represses gene expression at its locus.
Figure 5. GLI1AS acts as a chromatin remodeler at the INHBE / GLI1AS / GLI1 locus.
Chromatin immunoprecipitation assays for H3K27me3 (A), and RNA polymerase II (B) in Rh36 cells 48 hrs after transfection of a pCMV5 expression construct for GLI1AS. For real-time PCR analysis, 8 primers sets (A to H) spanning the INHBE / GLI1AS / GLI1 locus (Table 1, Supplementary figure 4) were used. Below the schematic diagram of the locus is a graph showing the immunoprecipitated signals relative to input control DNA for each interrogated segment. *, Statistical significant, P<0.01 compared to control, calculated by the Student’s t-test.
GLI1AS expression is regulated by Hedgehog and TGFβ signaling, two known inducers of GLI1 expression
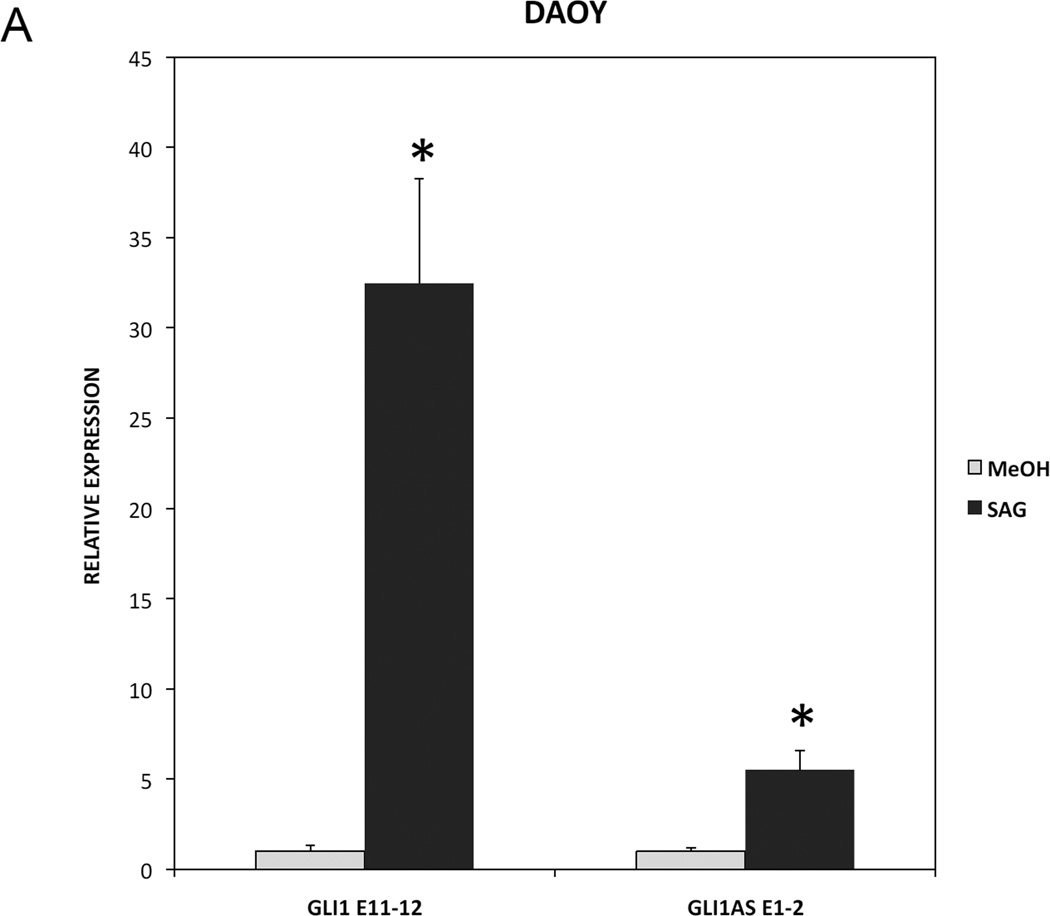
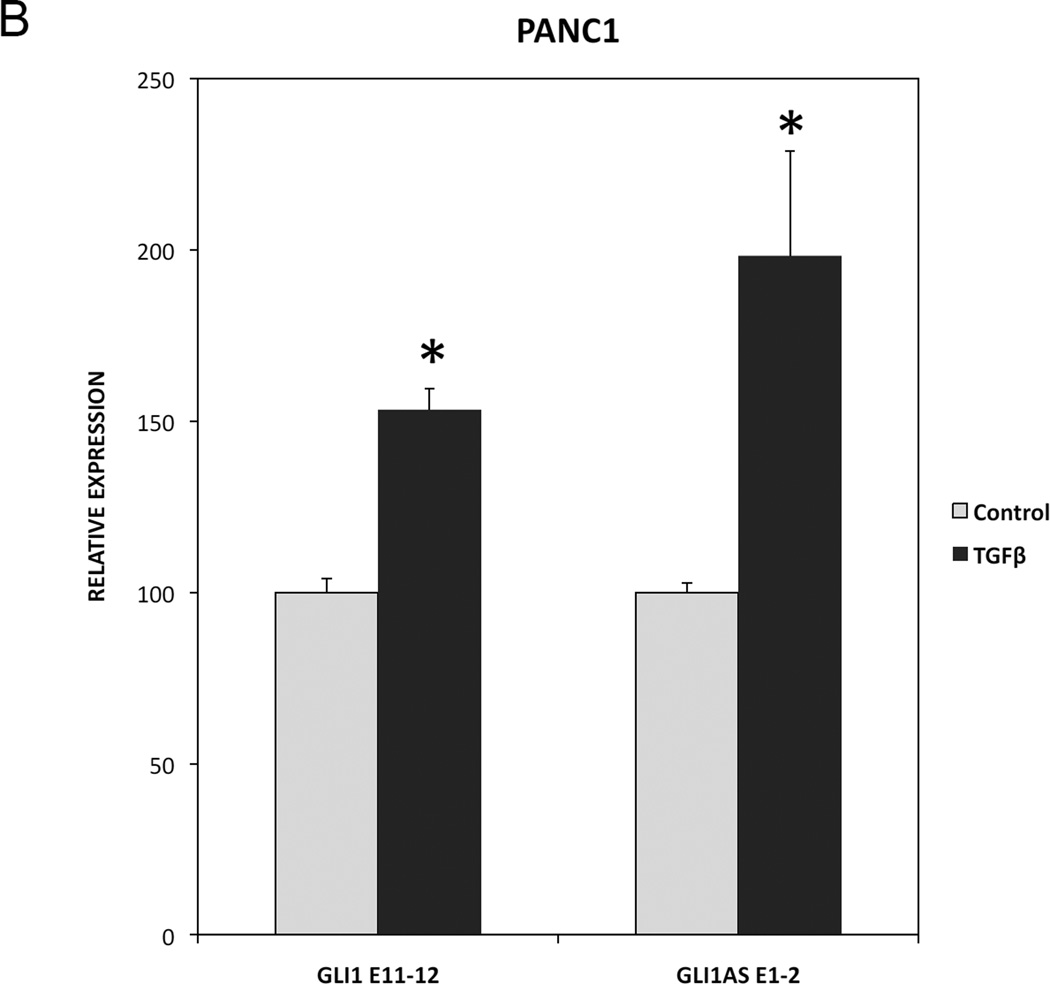
To determine whether GLI1AS has a role in signaling-dependent regulation of GLI1, Daoy and PANC1 cell lines were used. Activation of Hedgehog signaling by the Smoothened agonist SAG in Daoy cells increased not only GLI1 but also GLI1AS expression. Similarly, treatment of PANC1 cells with TGFβ increased not only GLI1 but also GLI1AS expression (Figure 6A and 6B). Thus, a concordant up-regulation of GLI1 and GLI1AS is elicited by activation of Hedgehog or TGFβ signaling.
Figure 6. Signaling-dependent regulation of GLI1AS.
A. Real-time RT/PCR analysis of the expression of GLI1 and GLI1AS in Daoy cells, following a 48-hr treatment with the Hedgehog signaling inducer SAG. Data are represented as relative expression (2−ΔCt values), calculated by subtracting the Ct value of the housekeeping gene TBP from the Ct value of the GLI1 and GLI1AS transcripts (ΔCt), and normalized to the ΔCt value obtained without treatment. Error bars indicate the standard deviation. *, Statistical significant, P<0.01 compared to control, calculated by the Student’s t-test.
B. Real-time RT/PCR analysis of the expression of GLI1 and GLI1AS in PANC1 cells, following a 24-hr treatment with the TGFβ1. Data are represented as relative expression (2−ΔΔCt values), calculated by subtracting the Ct value of the housekeeping gene ACTB from the Ct value of the GLI1 and GLI1AS transcripts (ΔCt), and normalized to the ΔCt value obtained without treatment. Error bars indicate the standard error of the mean of three experiments. *, Statistical significant, P<0.01 compared to control, calculated by the Student’s t-test.
DISCUSSION
In this study, we have characterized an RNA transcript from the antisense strand of the GLI1 gene, termed GLI1AS, with no potential to code for a long protein, which acts as a negative regulator of GLI1 expression. Moreover, GLI1, a known transcription factor, also influences GLI1AS expression but in a positive direction. GLI1AS is a polyadenylated transcript, with all intron–exon boundaries following the GU/AG consensus (Figure 1B, Supplementary figure 1). Interestingly, the pattern of expression of the spliced and unspliced GLI1AS transcripts in the rhabdomyosarcoma cells lines analyzed highlights the predominance of intron-containing RNA forms. Transcripts retaining intron 2 are consistently more abundant than transcripts with this intron excised, and to a lesser extent this is also observed for intron 1 in all cell lines but RMS13. Additionally, splicing of exon 1 to exon 2 is more prevalent than exon 2 to exon 3 splicing in all cell lines but RD (Figure 1E). Moreover, the spliced forms of GLI1AS are preferentially exported to the cytoplasm and the unspliced forms retained in the nucleus (Figure 1H and I). This finding, in addition to the evidence of capping and polyadenylation, suggest that GLI1AS is processed similarly to a typical mRNA, even though it lacks the potential to code for a long protein. However, the fact that the unspliced, nuclear localized GLI1AS forms are more abundant than the cytoplasmic, spliced forms (Figure 1E) contrasts GLI1AS with typical mRNAs.
Our data also show that GLI1AS expression is positively co-related with that of GLI1 in cancer cell lines (Figure 1C and D) and in biopsies of basal cell carcinomas and breast tumors (Figure 2). GLI1 mRNA expression levels were higher than GLI1AS across all samples examined, consistent with the results reported for most antisense transcripts with regulatory roles on the corresponding sense gene (Fish et al., 2007; Katayama et al., 2005; Oeder et al., 2007).
Interestingly, transfection of Rh36 and CCA cells with a mixture of three distinct siRNAs targeting regions of the GLI1 mRNA resulted in a reduction of not only the targeted GLI1 mRNA, but also the GLI1AS transcript, arguing that GLI1AS is a target gene of the GLI1 transcription factor (Figure 3B, Supplementary figure 6). This is further supported by the GLI1AS up-regulation elicited by GLI1 overexpression (Figure 4F). On the other hand, knockdown of GLI1AS increased GLI1 levels (Figure 3A, Supplementary figure 6), and the proliferation / tumor formation capacity of the Rh36 cell line (Figure 3C, D and E). Importantly, overexpression of GLI1AS in Rh36 cells resulted in decreased GLI1 mRNA and protein levels, decreased expression of the GLI1 target genes PTCH1 and PTCH2 and concomitant reduction of cellular proliferation (Figures 4A, B, C and D). Moreover, the expression of the INHBE gene, which is tail-to-tail with GLI1AS, was also reduced, while the unrelated ADAR2 gene was unaffected (Figure 4E). It is worth noting that these effects were observed only when the complete GLI1AS RNA was introduced into the Rh36 cells. Transfection of either a 5’ or a 3’ GLI1AS segment, or the intron 1 containing 5’ GLI1AS segment (Figure 4D, Supplementary Figure 7), did not elicit similar changes in gene expression/cellular proliferation.
Chromatin immunoprecipitation assays support the notion that GLI1AS acts as an epigenetic modifier, which elicits repressive histone marks at the INHBE / GLI1AS / GLI1 locus, and down-regulates the expression of its neighboring genes (Figure 5, Supplementary figure 9). Although H3K4m3, which is increased by GLI1AS, is generally thought to be an active chromatin mark, there is recent evidence suggesting that it may also have repressive functions (Wang et al., 2011; Zhou and Zhou, 2011). These observations are in-line with the claims that antisense transcripts can provide the means for local spreading of regulatory signals via histone modifications (Wei et al., 2011; Xu et al., 2009).
Additionally, the observed co-relation of GLI1 and GLI1AS RNA levels in a variety of cancer cell lines and in tumor samples is supportive of feedback mechanisms that keep the concordant expression of this transcript pair tightly regulated. Our experimental data demonstrate that GLI1 positively regulates GLI1AS, while GLI1AS negatively regulates GLI1. Consequently, these GLI1 / GLI1AS interactions represent a regulatory loop, which keeps the coordinate expression of the two transcripts. This bears similarities to the known feedback mechanism of Hedgehog signaling, which is elicited by the PTCH1 receptor, a target gene that also acts as a negative regulator of the pathway (Marigo and Tabin, 1996).
Worth noting is that in the chimpanzee (Pan troglodytes) and the Sumatran orangutan (Pongo abelii) the GLI1AS sequence is highly conserved, with all splice sites of the two introns being retained. In the more evolutionary distant northern white-cheeked gibbon (Nomascus leucogenys) the donor splice site of intron 2 is shifted from GT to AT arguing against intron removal. In the further distant white-tufted-ear marmoset (Callithrix jacchus) the acceptor splice sites of both intron 1 and intron 2 are not conserved, also arguing against intron removal. This would suggest that the splicing of the GLI1AS exons represents a recent evolutionary novelty and is in agreement with the fact that most of exon 3 consists of Alu sequences (Supplementary Figure 1A). Additionally, comparison with the mouse genome revealed conservation within the exon 1 sequence that is reduced towards the 3’ end sequences. However, efforts to detect transcripts from this genomic region in mouse tissues and embryos were negative in our hands (unpublished observations), consistently with the data of the UCSC Genome Browser. These observations are in-line with the recent claims of rapid evolutionary turnover of long non-coding RNAs (Kutter et al., 2012).
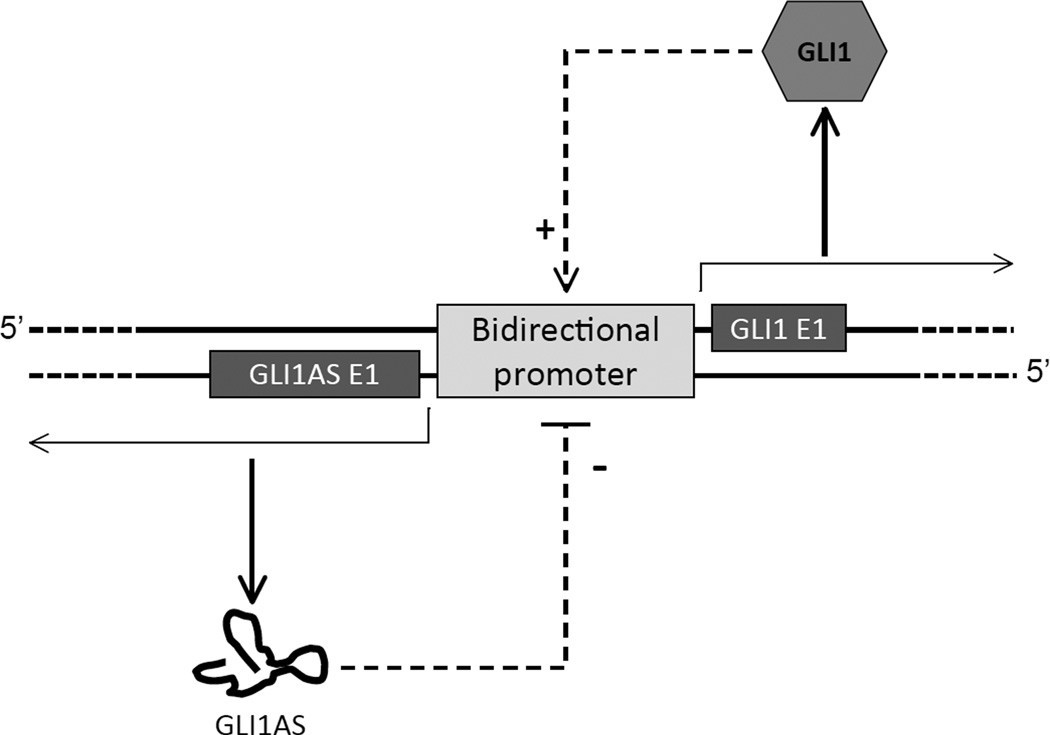
Moreover and in agreement with the finding that promoters can generate transcripts in both directions (Ntini et al., 2013), GLI1AS is likely to be the product of a GLI1 bidirectional promoter, as the physical distance between the 5’ end of the GLI1 and GLI1AS genes is short (Supplementary Figure 1B), and GLI1 up-regulates not only GLI1AS but also its own gene expression (Regl et al., 2002; Shimokawa et al., 2013; Shimokawa et al., 2008) (Figure 4F). This would suggest that antisense transcripts from bidirectional promoters can be exapted to acquire a biological role, exemplified in this case by the negative feedback of GLI1AS on GLI1 gene expression, and depicted in the model of Figure 7. This model is consistent with the findings that Hedgehog and TGFβ signaling activation up-regulates not only GLI1 but also GLI1AS expression.
Figure 7. Schematic representation of the proposed mechanism for the interplay of the GLI1AS and GLI1 regulatory effects.
The GLI1 protein, acting as a transcription factor, up-regulates it own promoter. This concomitantly increases transcription from both DNA strands, resulting not only in a GLI1 positive feedback but also in a negative feedback, which is mediated by the GLI1AS non-coding RNA.
The function of most non-coding antisense transcripts remains unknown. A certain proportion of these may constitute transcriptional noise; however, there is a growing number of examples with a regulatory impact that is physiologically significant. Consequently, the results presented herein, revealing a negative feedback of GLI1AS on GLI1 expression, with concomitant effects on cellular proliferation, provide evidence for an additional non-coding antisense RNA that is biologically relevant.
In summary, our data highlight complex regulatory mechanisms acting on the oncogenic GLI1 transcription factor, which are elicited by processes that include not only alternative splicing (Cao et al., 2012) or RNA editing (Shimokawa et al., 2013) but also chromatin remodeling through non-coding RNA.
Supplementary Material
A novel negative feedback loop on Hedgehog signaling is demonstrated
The mechanism involves a non-coding RNA antisense to the GLI1 gene, GLI1AS
GLI1AS is shown to be a target gene of the GLI1 transcription factor
GLI1AS represses gene expression at the GLI1 / GLI1AS locus
GLI1AS acts as an epigenetic modifier eliciting repressive chromatin marks
ACKNOWLEDGMENT
This study was supported by the Swedish Childhood Cancer Foundation, the Swedish Cancer Fund, the Swedish Research Council and the AFA Insurance. VEV, YD and EL were funded by scholarships from the ERACOL program of the European Union, the China Scholarship Council and the Erasmus Scholarship Program. MEFZ and MGFB were supported by NCI CA165076 and Mayo Clinic Cancer Center. The authors thank R. Toftgård, C. Finta, R. Kuiper and Å. Bergström for helpful discussions and technical support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF CONFLICTING INTERESTS
The authors declare that they have no conflict of interest.
REFERENCES
- Berretta J, Pinskaya M, Morillon A. A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S. cerevisiae. Genes & development. 2008;22:615–626. doi: 10.1101/gad.458008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks PC, Montgomery AM, Rosenfeld M, Reisfeld RA, Hu T, Klier G, Cheresh DA. Integrin alpha v beta 3 antagonists promote tumor regression by inducing apoptosis of angiogenic blood vessels. Cell. 1994;79:1157–1164. doi: 10.1016/0092-8674(94)90007-8. [DOI] [PubMed] [Google Scholar]
- Cao X, Geradts J, Dewhirst MW, Lo HW. Upregulation of VEGF-A and CD24 gene expression by the tGLI1 transcription factor contributes to the aggressive behavior of breast cancer cells. Oncogene. 2012;31:104–115. doi: 10.1038/onc.2011.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C, Forrest AR, Carninci P, Biffo S, Stupka E, Gustincich S. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- Conley AB, Jordan IK. Epigenetic regulation of human cis-natural antisense transcripts. Nucleic acids research. 2012;40:1438–1445. doi: 10.1093/nar/gkr1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebralidze AK, Guibal FC, Steidl U, Zhang P, Lee S, Bartholdy B, Jorda MA, Petkova V, Rosenbauer F, Huang G, Dayaram T, Klupp J, O'Brien KB, Will B, Hoogenkamp M, Borden KL, Bonifer C, Tenen DG. PU.1 expression is modulated by the balance of functional sense and antisense RNAs regulated by a shared cis-regulatory element. Genes & development. 2008;22:2085–2092. doi: 10.1101/gad.1654808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G, 3rd, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nature medicine. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nature reviews. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish JE, Matouk CC, Yeboah E, Bevan SC, Khan M, Patil K, Ohh M, Marsden PA. Hypoxia-inducible expression of a natural cis-antisense transcript inhibits endothelial nitric-oxide synthase. The Journal of biological chemistry. 2007;282:15652–15666. doi: 10.1074/jbc.M608318200. [DOI] [PubMed] [Google Scholar]
- Gailani MR, Ståhle-Bäckdahl M, Leffell DJ, Glynn M, Zaphiropoulos PG, Pressman C, Undén AB, Dean M, Brash DE, Bale AE, Toftgård R. The role of the human homologue of Drosophila patched in sporadic basal cell carcinomas. Nature genetics. 1996;14:78–81. doi: 10.1038/ng0996-78. [DOI] [PubMed] [Google Scholar]
- Holdt LM, Hoffmann S, Sass K, Langenberger D, Scholz M, Krohn K, Finstermeier K, Stahringer A, Wilfert W, Beutner F, Gielen S, Schuler G, Gabel G, Bergert H, Bechmann I, Stadler PF, Thiery J, Teupser D. Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks. PLoS genetics. 2013;9:e1003588. doi: 10.1371/journal.pgen.1003588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson P, Ackley A, Vidarsdottir L, Lui WO, Corcoran M, Grandér D, Morris KV. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nature structural & molecular biology. 2013;20:440–446. doi: 10.1038/nsmb.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA, Lipovich L, Batalov S, Engstrom PG, Mizuno Y, Faghihi MA, Sandelin A, Chalk AM, Mottagui-Tabar S, Liang Z, Lenhard B, Wahlestedt C. Science. Vol. 309. New York, N.Y: 2005. Antisense transcription in the mammalian transcriptome; pp. 1564–1566. [DOI] [PubMed] [Google Scholar]
- Kutter C, Watt S, Stefflova K, Wilson MD, Goncalves A, Ponting CP, Odom DT, Marques AC. Rapid turnover of long noncoding RNAs and the evolution of gene expression. PLoS genetics. 2012;8:e1002841. doi: 10.1371/journal.pgen.1002841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Wen S, Guo X, Bai B, Gong Z, Liu X, Wang Y, Zhou Y, Chen X, Liu L, Chen R. The novel long non-coding RNA CRG regulates Drosophila locomotor behavior. Nucleic acids research. 2012a;40:11714–11727. doi: 10.1093/nar/gks943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Gao HF, Wang Y, Liu F, Tian XF, Zhang Y. Overexpression of Gli1 in cancer interstitial tissues predicts early relapse after radical operation of breast cancer. Chinese journal of cancer research = Chung-kuo yen cheng yen chiu. 2012b;24:263–274. doi: 10.3978/j.issn.1000-9604.2012.10.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CZ, Yang JT, Yoon JW, Villavicencio E, Pfendler K, Walterhouse D, Iannaccone P. Characterization of the promoter region and genomic organization of GLI a member of the Sonic hedgehog-Patched signaling pathway. Gene. 1998;209:1–11. doi: 10.1016/s0378-1119(97)00668-9. [DOI] [PubMed] [Google Scholar]
- Magistri M, Faghihi MA, St Laurent G, 3rd, Wahlestedt C. Regulation of chromatin structure by long noncoding RNAs: focus on natural antisense transcripts. Trends Genet. 2012;28:389–396. doi: 10.1016/j.tig.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marigo V, Tabin CJ. Regulation of patched by sonic hedgehog in the developing neural tube. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:9346–9351. doi: 10.1073/pnas.93.18.9346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV. Non-coding RNA. Human molecular genetics. 2006;15(Spec No 1):R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- Morris KV, Santoso S, Turner AM, Pastori C, Hawkins PG. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS genetics. 2008;4:e1000258. doi: 10.1371/journal.pgen.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KV, Vogt PK. Cell cycle. Vol. 9. Georgetown, Tex: 2010. Long antisense non-coding RNAs and their role in transcription and oncogenesis; pp. 2544–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson M, Unden AB, Krause D, Malmqwist U, Raza K, Zaphiropoulos PG, Toftgård R. Induction of basal cell carcinomas and trichoepitheliomas in mice overexpressing GLI-1. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:3438–3443. doi: 10.1073/pnas.050467397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntini E, Jarvelin AI, Bornholdt J, Chen Y, Boyd M, Jorgensen M, Andersson R, Hoof I, Schein A, Andersen PR, Andersen PK, Preker P, Valen E, Zhao X, Pelechano V, Steinmetz LM, Sandelin A, Jensen TH. Polyadenylation site-induced decay of upstream transcripts enforces promoter directionality. Nature structural & molecular biology. 2013;20:923–928. doi: 10.1038/nsmb.2640. [DOI] [PubMed] [Google Scholar]
- Oeder S, Mages J, Flicek P, Lang R. Uncovering information on expression of natural antisense transcripts in Affymetrix MOE430 datasets. BMC genomics. 2007;8:200. doi: 10.1186/1471-2164-8-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onoguchi M, Hirabayashi Y, Koseki H, Gotoh Y. A noncoding RNA regulates the neurogenin1 gene locus during mouse neocortical development. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:16939–16944. doi: 10.1073/pnas.1202956109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Kapranov P, Foissac S, Kim SW, Fishilevich E, Monaghan AP, John B, Milos PM. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell. 2010;143:1018–1029. doi: 10.1016/j.cell.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Molecular cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Pinskaya M, Gourvennec S, Morillon A. H3 lysine 4 di- and tri-methylation deposited by cryptic transcription attenuates promoter activation. The EMBO journal. 2009;28:1697–1707. doi: 10.1038/emboj.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L, Vergara IA, Davicioni E, Erho N, Ghadessi M, Jenkins RB, Triche TJ, Malik R, Bedenis R, McGregor N, Ma T, Chen W, Han S, Jing X, Cao X, Wang X, Chandler B, Yan W, Siddiqui J, Kunju LP, Dhanasekaran SM, Pienta KJ, Feng FY, Chinnaiyan AM. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nature genetics. 2013 doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regl G, Neill GW, Eichberger T, Kasper M, Ikram MS, Koller J, Hintner H, Quinn AG, Frischauf AM, Aberger F. Human GLI2 and GLI1 are part of a positive feedback mechanism in Basal Cell Carcinoma. Oncogene. 2002;21:5529–5539. doi: 10.1038/sj.onc.1205748. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimokawa T, Rahman MF, Tostar U, Sonkoly E, Ståhle M, Pivarcsi A, Palaniswamy R, Zaphiropoulos PG. RNA editing of the GLI1 transcription factor modulates the output of Hedgehog signaling. RNA biology. 2013;10:321–333. doi: 10.4161/rna.23343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimokawa T, Tostar U, Lauth M, Palaniswamy R, Kasper M, Toftgard R, Zaphiropoulos PG. Novel human glioma-associated oncogene 1 (GLI1) splice variants reveal distinct mechanisms in the terminal transduction of the hedgehog signal. The Journal of biological chemistry. 2008;283:14345–14354. doi: 10.1074/jbc.M800299200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tostar U, Finta C, Rahman MF, Shimokawa T, Zaphiropoulos PG. Novel mechanism of action on Hedgehog signaling by a suppressor of fused carboxy terminal variant. PloS one. 2012;7:e37761. doi: 10.1371/journal.pone.0037761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tostar U, Toftgård R, Zaphiropoulos PG, Shimokawa T. Reduction of human embryonal rhabdomyosarcoma tumor growth by inhibition of the hedgehog signaling pathway. Genes & cancer. 2010;1:941–951. doi: 10.1177/1947601910385449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SS, Zhou BO, Zhou JQ. Histone H3 lysine 4 hypermethylation prevents aberrant nucleosome remodeling at the PHO5 promoter. Molecular and cellular biology. 2011;31:3171–3181. doi: 10.1128/MCB.05017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Pelechano V, Jarvelin AI, Steinmetz LM. Functional consequences of bidirectional promoters. Trends Genet. 2011;27:267–276. doi: 10.1016/j.tig.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner A, Carlile M, Swan D. What do natural antisense transcripts regulate? RNA biology. 2009;6:43–48. doi: 10.4161/rna.6.1.7568. [DOI] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BO, Zhou JQ. Recent transcription-induced histone H3 lysine 4 (H3K4) methylation inhibits gene reactivation. The Journal of biological chemistry. 2011;286:34770–34776. doi: 10.1074/jbc.M111.273128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Gao X, He Y, Zhao H, Yu Q, Jiang D, Zhang P, Ma X, Huang H, Dong D, Wan J, Gu Z, Jiang X, Yu L, Gao Y. An Insertion/Deletion Polymorphism within RERT-lncRNA Modulates Hepatocellular Carcinoma Risk. Cancer research. 2012;72:6163–6172. doi: 10.1158/0008-5472.CAN-12-0010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.