Abstract
Background
Chronic mucocutaneous candidiasis (CMC) and hyper IgE syndrome (HIES) are primary immunodeficiencies mainly caused by mutations in STAT1 and STAT3, respectively. CMC and HIES patients have an increased risk for skin and mucosal infections with fungal pathogens and Staphylococcus aureus. However, it is unknown whether the genetic defects in these patients also affect the skin and mucosal microbiome, which in turn may influence host defense mechanisms.
Methods
The skin and oral microbiome of CMC and HIES patients was compared to that of healthy controls at five body sites using 16S rRNA sequencing. The influence of skin colonizers on the immune response was investigated using in-vitro experiments.
Results
The microbiome of CMC and HIES patients contained more Gram-negative bacteria, especially Acinetobacter spp, and less of the normal Corynebacterium spp., compared to healthy controls. Exposure of human primary leukocytes to Acinetobacter suppressed the cytokine response to C. albicans and S. aureus, while the normal Corynebacteria did not suppress cytokine responses.
Discussion
These results demonstrate that central mediators of immune responses like STAT1 and STAT3 not only directly influence immune responses, but also result in changes of the skin microbiome that in turn can amplify the defective immune response against fungal and microbial pathogens.
Keywords: Microbiome, immunodeficiency, Chronic Mucocutaneous Candidiasis, Hyper IgE Syndrome
Introduction
Chronic mucocutaneous candidiasis (CMC) and hyper IgE syndrome (HIES) are primary immunodeficiencies characterized by defective mucosal host defense mechanisms, due to impaired Th1 and Th17 responses. Currently it is not known how these immune defects influence the regular colonizers on the skin and mucosa of these patients. In this study we investigated the interaction between the host genotype, microbiome and host immunity.
CMC patients display chronic fungal infections of the skin, nails and oropharynx [1,2]. CMC is classified into autosomal recessive (AR) CMC, autosomal recessive autoimmune polyendocrinopathy candidiasis ectodermal dystrophy (APECED), and autosomal dominant (AD) CMC. It has been demonstrated that APECED is caused by mutations in the autoimmune regulator (AIRE) gene [3,4], leading to the production of autoantibodies against IL-17 and IL-22 [5,6]. Recently, we and others have demonstrated that AD-CMC is mainly caused by dominant negative mutations in the signal transduced and activator of transcription (STAT) 1 gene [7–10]. These mutations lead to decreased Th1 and Th17 responses, explaining the increased susceptibility to fungal infections and lung pathology seen in these patients [2,11,12].
HIES was first described in 1966 as Job’s syndrome [4,13,14]. HIES is a heterogeneous disease, but the main characteristics include increased serum IgE levels, eczematoid dermatitis, recurrent skin en pulmonary infections with predominantly Staphylococcus aureus, and mucocutaneous candidiasis. Interestingly, AD-HIES is mainly caused by dominant negative mutations in the STAT3 gene [6,15–17]. Similar to AD-CMC, these mutations decrease production of Th1 and Th17 cytokines [8–10,18–22], explaining the increased susceptibility to infections with Staphylococcus and Candida species.
Since HIES and CMC are well defined primary immunodeficiencies with similar immune defects in mucosal and skin immunity, one may hypothesize that the mutations in STAT1 and STAT3 could also influence the microorganisms that colonize the skin and oral mucosa, the main sites affected by these diseases. Kong et al. previously demonstrated that patients with atopic dermatitis lesions carry a statistically significant higher portion of S. aureus compared to healthy controls, which may have contributed to the pathogenesis of the disease [23,24]. Since colonizing microorganisms are known to modulate the host immune response, we consequently hypothesize that a potentially skewed microbiome on the skin or mucosa of CMC and HIES patients may further increase the imbalance in the host immune response, augmenting the immune defects.
From recent human microbiome studies, the microbial communities on the skin are now more comprehensively characterized than before, particularly for healthy people [25–27]. In summary, the human skin can be divided intro three different habitat types: dry, moist and sebaceous areas; the dry skin areas are the most diverse, and contain various mixtures of predominantly Actinobacteria, Proteobacteria, Firmicutes and Bacteroidetes. Staphylococcus and Corynebacterium are the most common organisms on moist skin, and the Propionibacterium is the most abundant phylum present on sebaceous skin areas [28,29].
No information currently exists on how host genotype specifically affects the skin microbiome in humans, whereas several strong interactions have now been described between the host genotype and the gut microbiome [30]. For example, mice with different genetic backgrounds display quantitative differences in their gut microbiome [31,32]. Khachatryan et al. demonstrated that mutations in the MEFV gene, leading to familial Mediterranean fever (FMF), are associated with a decreased number and diversity of bacteria in the gut. Furthermore, during disease flares there is a shift in the bacterial populations within the Bacteroidetes, Firmicutes and Proteobacteria [33]. Li et al. demonstrated that in patients with inflammatory bowel disease there is a correlation between NOD2 genotype and overall microbial composition of the ilium [1]. TLR5−/− mice develop hallmark features of metabolic syndrome, and this is correlated to an altered gut microbiome. Interestingly, the altered microbiome in its turn can induce features of metabolic syndrome [3].
Clearly, genetic variation in the host genome can strongly influence the gut microbiome. Unfortunately, there is not a lot known about the interaction between the skin microbiome and the host immune system. To our knowledge the only paper published on this subject was about research conducted in mice. In 2012, Naik et al. demonstrated that unlike the gut microbiome, the skin microbiome has a great influence on immunity against cutaneous pathogens [5].
In the current study, the skin and oral microbiome of patients with CMC and HIES was compared to that of healthy controls. We demonstrate that the microbiome of CMC and HIES patients contains more Gram-negative bacteria and less of the normal Corynebacterium spp. compared to that of healthy controls, and that this shift has the capacity to impair the host defense against Candida and Staphylococci.
Materials and Methods
Patients and healthy controls
In January 2012, 4 CMC patients, 7 HIES patients, and 10 age-matched healthy controls were included in this study. Both HIES and CMC are very rare primary immunodeficiencies, with only a few hundred patients known worldwide. The hands, feet, trunk, mouth and if present the skin lesion, were swapped aseptically using Catch-All™ Sample Collection Swab, moistened with sterile SCF-1 solution (50 mM Tris buffer [pH 8], 1 mM EDTA, and 0.5% Tween-20). Immediately after swabbing, each swab was swirled in a 2 ml Collection Tube containing 300 µl MicroBead Solution. The samples were stored at −20°C until further processing. 9 healthy controls, 4 CMC and 2 HIES patients also donated blood for in-vitro immunological studies. The study was approved by the Ethical Committee of the Radboud University Nijmegen, and the participants gave written informed consent. The study was performed in accordance with the declaration of Helsinki. The patients were asked to stop the use of any prophylactic antibiotics one week before the initiation of the study. Of the seven patients using antibiotics, three used flucloxacilline for the prophylaxis of skin Staphylococcus infections, and the other patients used prohylaxis with different antibiotics for respiratory infections. In one of the CMC patients we were not able to stop antibiotic therapy (cotrimoxazol, ganciclovir) due to a chronic pulmonary infection and CMV infection associated with low CD4 counts.
Microbial DNA isolation
Microbial DNA was isolated using the UltraClean™ Microbial DNA Isolation Kit (MO BIO laboratories, Uden, The Netherlands), according to instruction of the manufacturer. The presence of microbial DNA was confirmed using a PCR to detect 16S rRNA gene. The following page-purified primers were used: F338: 5’-act-cct-acg-gga-ggc-agc-ag-3’ (V3) and 1061R: 5’-crr-cac-gag-ctg-acg-ac-3’ (V6) (Biolegio, Nijmegen, The Netherland). The PCR reaction was performed using KOD DNA polymerase (EMD Millipore, Amsterdam, The Netherlands). The PCR reaction was as follows: 2 minutes 95°C, followed by 30 cycles of 20 seconds 95°C; 10 seconds 55°C; 15 seconds 70°C, followed by 10 minutes 70°C. 5 µL of PCR product was checked on agarose gel.
16S rRNA gene profiling and sequencing
The 16S rRNA gene dataset consists of Illumina MiSeq paired-end sequences targeting the V4 variable region. Genomic DNA was subjected to 16S amplifications using the 515 Fwd and 806 Rev primers as previously described in detail [7], with the following modifications to the previously described protocol: (1) only one PCR amplification was performed, instead of three; (2) only 30 PCR cycles, instead of 35; (3) an extension time of 5 minutes, instead of 1.5 to reduce chimera formation; (4) a size selection was applied to retrieve amplicons between 375 bp – 425bp using SAGE pippin prep, in order to reduce a-specific amplification products derived from host DNA, and (5) the amplicons were sequenced for 175 bases in each direction, using custom designed primers that are complimentary to the V4 amplification primers. Sequencing was performed according to the manufacturer’s specifications.
Processing 16S rRNA gene sequence data
Sequences were processed using a pipeline implemented in QIIME 1.4.0 [11] as pick_reference_otus_through_otu_table.py, which picks OTUs by mapping the sequences against a subset of the latest Greengenes database (version Oct 2012) [13] filtered at 97% identity, resulting in a table of counts by OTU and sample, with a representative sequence for each OTU. Classification was used as provided through Greengenes. The resulting OTU tables were checked for mislabeling [15] and contamination [18], and further microbial community analysis and visualizations. Statistical association analysis was done both with a univariate approach using LEfSe [23,34], and in a multivariate approach as described previously [25,27]. LEfSe is a non-parametric factorial Kruskal-Wallis (KW) sum-rank test to detect organisms with significant differential abundance between sample sets; biological significance is subsequently investigated using a set of pairwise tests among subclasses using the (unpaired) Wilcoxon rank-sum test. As a last step, LEfSe uses Linear Discriminant Analysis to estimate the effect size of each differentially abundant feature.
Isolation of PBMC
Peripheral blood mononuclear cells (PBMCs) were obtained by density centrifugation of diluted blood (1 part blood to 1 part pyrogen-free saline) over Ficoll-Paque (Pharmacia Biotech, Uppsala, Sweden). PBMCs were washed twice in saline and suspended in culture medium (RPMI 1640 Dutch modifications (Sigma-Aldrich, Zwijndrecht, The Netherlands) supplemented with 1% gentamicin, 1% L-glutamine and 1% pyruvate (Life Technologies, Nieuwekerk, the Netherlands). The cells were counted in a hemocytometer, and their number was adjusted to 5 × 106 cells/mL.
Microorganisms
C. albicans ATCC MYA-3573 (UC 820) as grown overnight in Sabouraud broth at 37°C, cells were harvested by centrifugation, washed twice, and resuspended in culture medium. C. albicans was heat-killed for one hour at 100 °C. Staphylococcus aureus (American type culture collection) 25923 was heat-killed for 30 minutes at 100 °C. Acinetobacter baumannii (ATCC-15308) and Corynebacterium jeikeium (ATCC-BAA-949) were obtained from American Type Culture Collection, grown overnight, harvested by centrifugation, washed twice, and resuspended in culture medium.
In-vitro stimulation assays
5 × 105 PBMCs in a volume of 100 µL per well were pre-stimulated for 24 hours at 37°C in round-bottom 96-wells plates (Greiner, Nurnberg, Germany), with Corynebacterium jeikeium, Acinetobacter baumannii or Staphylococcus aureus (1×106/mL). Cells were restimulated with C. albicans (1×106/mL) or S. aureus (1×107/mL) for 24 hours or 7 days. When cells were stimulated for 7 days this was done in the presence of 10% human pooled serum. Experiments were performed in duplicate. After 1 or 7 days of incubation, duplicate supernatants were pooled and stored at −20°C until assayed.
Cytokine assays
The concentration of IL-1β, IL-17, IL-22 and TNF-α (R&D Systems, Abingdon, UK) and IFNy (Sanquin, Amsterdam, the Netherlands) was measured in cell culture supernatants using enzyme-linked immuno sorbent assay (ELISA), according to the instructions of the manufacturer. These particular cytokines were measured because they play an important role in the host immune response against C. albicans [35] and S. aureus [36].
Immunological statistical analysis
The differences between groups were analyzed using the Wilcoxon signed rank test. All statistical analyses were 2-sided, and differences were considered statistically significant if p≤ 0.05 (*) or p ≤0.01 (**).
Results
Patient and control characteristics
The microbiome structure of 11 immunodeficient patients (4 CMC and 7 HIES patients) was obtained from several skin and oral locations during their regular visit in the outpatient clinic (January 2012). In addition, 10 non-affected healthy individuals served as controls. Controls were matched on age, but not on gender as it was previously shown that this is not associated with substantial variation in the human micriobiome [2,27]. At the time of sampling, 4 out of 4 CMC patients, and 1 out of 7 HIES patients had an active skin lesion, which was also sampled (Table 1). DNA was extracted from skin and oral samples, and the 16S rRNA gene was amplified and sequenced on the Illumina MiSeq platform. The resulting sequences were then processed with QIIME taking a reference-based approach (see Methods). At the end of this process, the median sequencing depth was 6,389 reads per sample.
Table 1.
Baseline characteristics of participants
| Characteristic | CMC patients |
HIES patients |
Age-matched healthy controls |
|---|---|---|---|
| Number | 4 | 7 | 10 |
| Gender (male:female) | 2:2 | 5:2 | 8:2 |
| Age (mean ± SD) | 46 ± 16 | 46 ± 5 | 47 ± 10 |
| Active lesion | 4 (100%) | 2 (29%) | - |
| Genetic defect | STAT1 | STAT3 | - |
Sample diversity
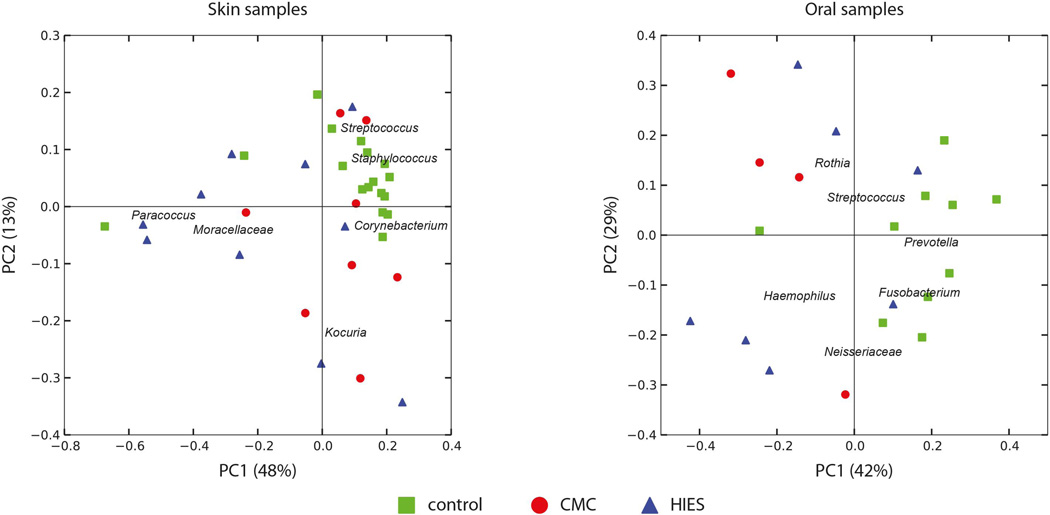
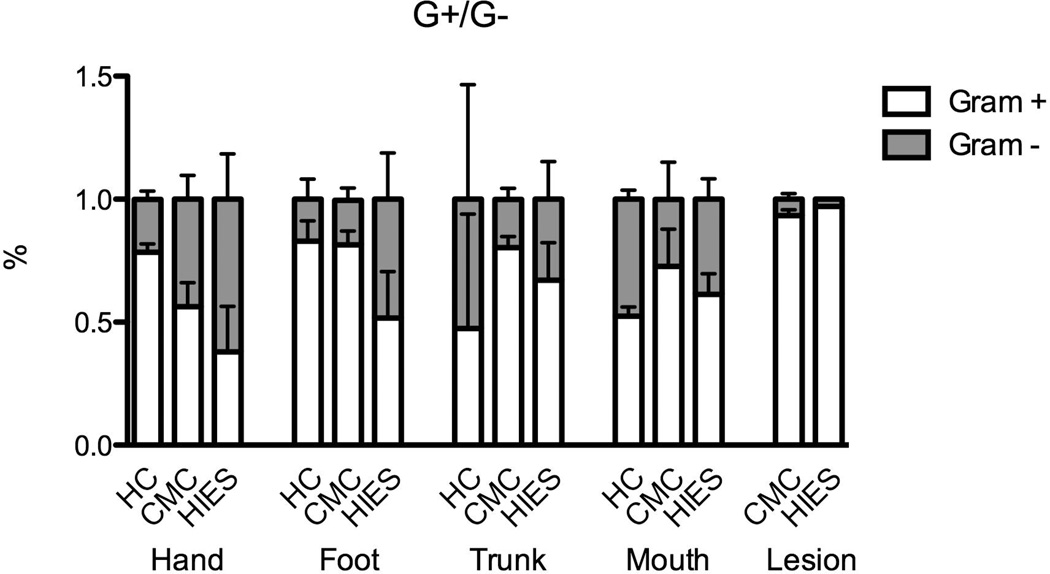
We compared the microbial community profiles among subjects to relate disease phenotype to variation in microbiome structure (Figure 1). Oral and skin samples were analyzed separately, as between-site variation in the human microbiome is greater than between-subject variation within-site [4,27]. The skin microbiome composition was very similar among healthy controls and was dominated by Staphylococcus, Streptococcus and Corynebacterium as expected. In contrast, in HIES and CMC patients there was more variation in microbiome structure, which was characterized by an increase of Moracellaceae (mainly Actinobacter) (Figure 1A). The oral samples of healthy controls are largely defined by the presence of Prevotella and Fusobacterium, with an effect of disease on oral composition in a non-unidirectional way (Figure 1B). In general, both CMC and HIES patients have relatively little Gram-positive bacteria on their hands and feet compared to healthy controls (Figure 2). The relative abundance of phyla per individual is displayed in Supplemental Figure 1.
Figure 1. Covariance of skin and oral microbial community structure with disease subtype in immunodeficient patients.
The variation in microbial diversity of (A) 35 skin and (B) 21 oral samples from healthy controls (HC, green squares), chronic mucocutaneous candidiasis (CMC, red circles), and hyper IgE syndrome (HIES, blue triangles) patients. The principle coordinates plot uses the weighted UniFrac beta-diversity metric, with the covariance of selected microbial taxa overlaid as a biplot. Organisms characteristic for the healthy skin samples are Staphylococcus and Corynebacterium spp., with Moracellaceae spp. driving the different community in disease.
Figure 2. CMC and HIES patients are colonized by relatively little gram-positive bacteria.
Mean percentage of Gram-positive and Gram-negative bacteria per body site, per group. Bars represent mean + SEM from 1–10 subjects.
Assessing significant covariation of microbiome structure with disease
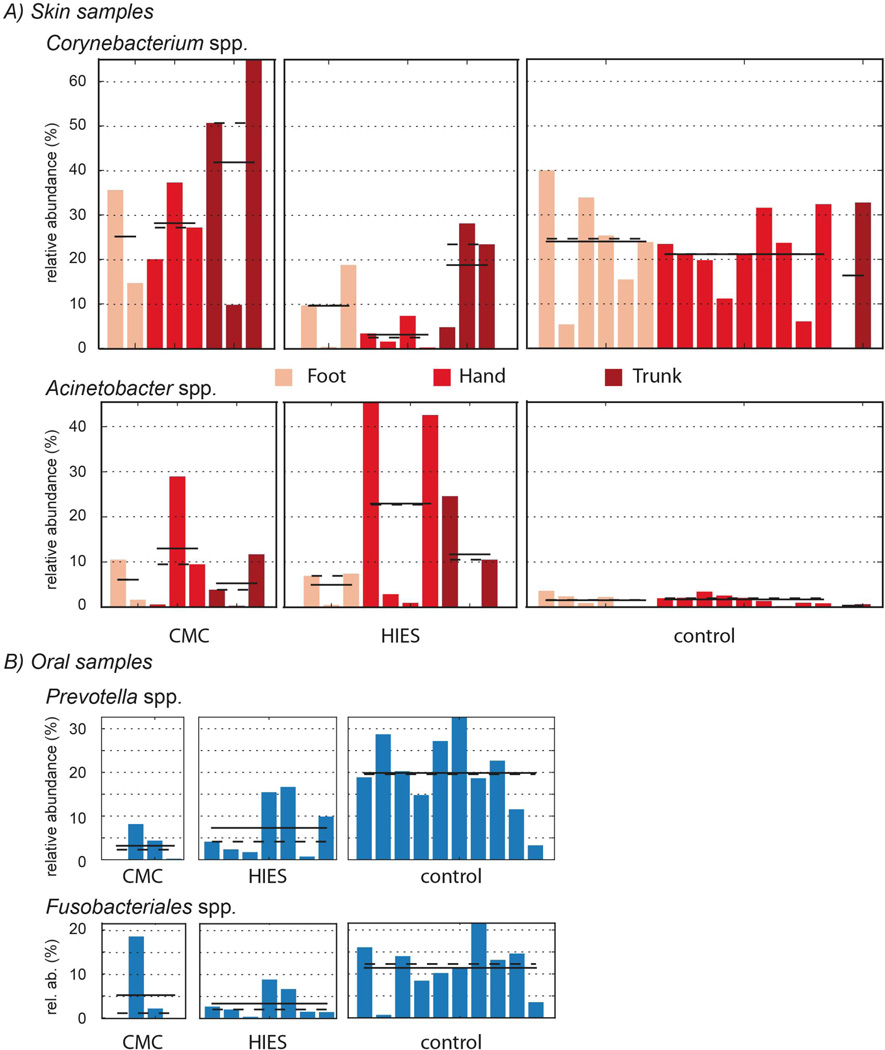
Next, we looked in more detail at the differences between healthy controls and immunodeficient patients using univariate association tests taking into account disease phenotype, and body site for skin samples. The results found in skin samples were verified using multivariate tests (Figure 3, Supplementary Table 1). Using the univariate LEfSe test for microbial biomarkers, we observed lower levels of Corynebacterium on the hands and feet of HIES patients. On the other hand, the levels of Acinetobacter were strongly increased in both HIES and CMC patients (Figure 3A). The relative abundance of Prevotella and Fusobacteria, which are key species in oral samples of healthy controls, was lower in both CMC and HIES patients (Figure 3B, see Methods). The composition of the microbiome in skin lesions varied substantially between immunocompromised individuals, but in general contained a lot of Fusobacteria and Firmucutes (data not shown). There was no significant association between microbiome composition and gender (data not shown).
Figure 3. Selected associations of microbial clade abundance with body site and disease subtype.
A subset of taxa that significantly differ in relative abundance as determined by LEfSe between disease subtypes at skin (A) or oral body sites (B). Subclasses of disease subtype are separated, with specific body sites differentially colored. The mean and median relative abundance of the taxa are indicated with a solid and dashed lines, respectively (LEfSe * = p < 0.05).
CMC/HIES-related skin colonizers impair innate immune host responses
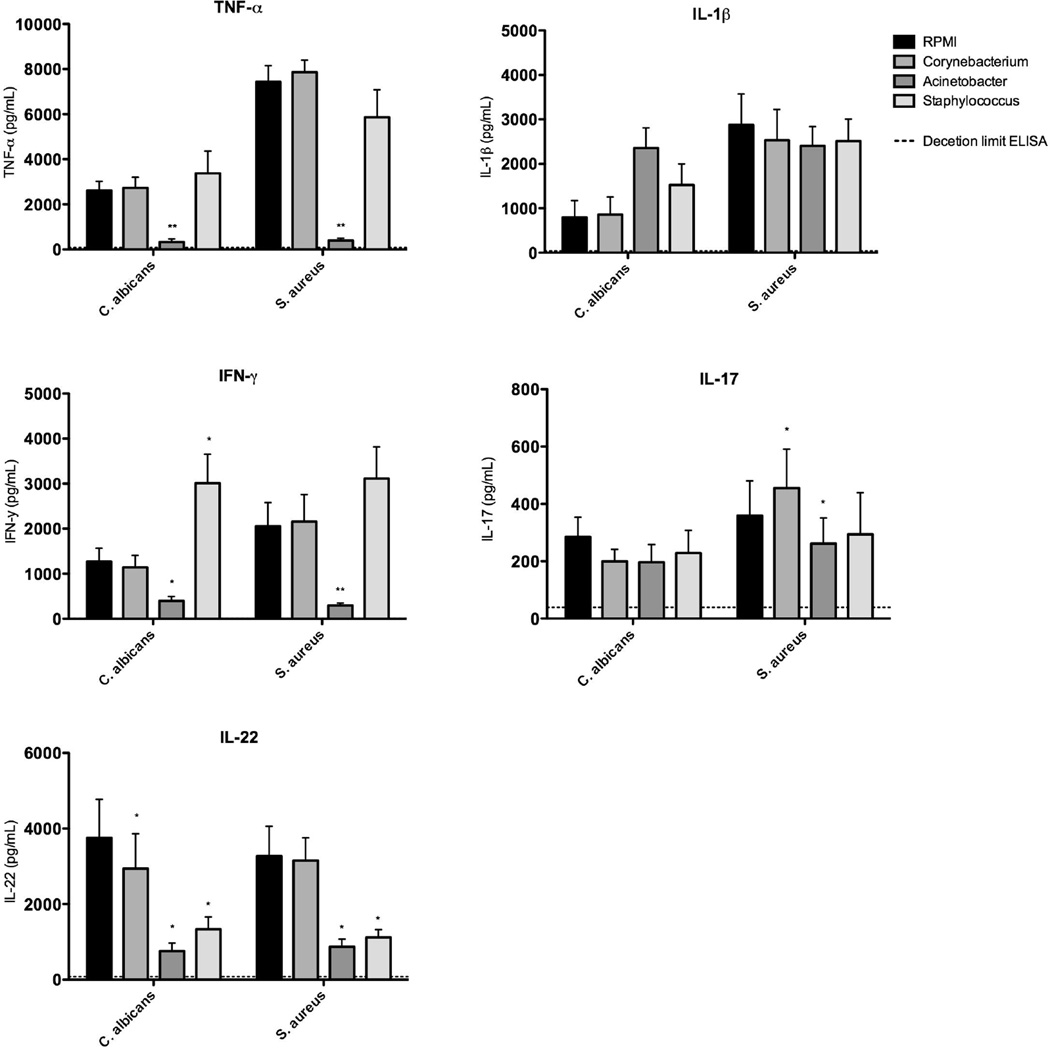
To investigate whether microorganisms abundantly found on the skin of CMC and HIES patients can modify host immune responses to the pathogens encountered in these diseases, PBMC of healthy volunteers were exposed either to C. jeikeium (normal colonizer), A. baumannii (HIES/CMC colonizer) or S. aureus (CMC colonizer). After 24h of incubation, the cells were restimulated with C. albicans or S. aureus. Exposure of PBMCs to Corynebacteria normally found in the human skin microbiota did not affect Candida and Staphylococcus-induced cytokine responses. In contrast, exposure of immune cells to Acinetobacter found on the skin of HIES and CMC patients strongly inhibited the normal TNF, IFNy and IL-22 responses of human PBMCs, while the IL-1β and IL-17 responses remained relatively unaffected (Figure 4). A similar inhibitory effect on TNF and IFNy production by Acinetobacter was found in PBMCs isolated from CMC and HIES patients (data not shown), while PBMC of CMC and HIES patients already had absent IL-17 and IL-22 production due to their genetic defect [6,22,37].
Figure 4. Acinetobacter suppress typical immune responses.
PBMC of healthy volunteers were prestimulated for 24 hours with RPMI, Corynebacterium, Acinetobacter or Staphylococcus, and stimulated with C. albicans or S. aureus for 1 or 7 days. Cytokine concentrations were measured in cell culture supernatants using ELISA. Bars represent mean + SEM of 10 healthy volunteers (Wilcoxon * = p < 0.05, ** = p < 0.01).
Discussion
In this study, we characterized the microbiome of CMC and HIES patients and compared it to that of healthy controls. The skin and oral microbiota of our healthy controls was dominated by typical human-associated phyla, mainly the Firmicutes and Proteobacteria, as expected [8–10,27]. In contrast, patients with HIES displayed significantly decreased colonization by Corynebacteria, while Acinetobacter was more prevalent in both CMC and HIES patients. Additionally, exposure of healthy PMBCs to Acinetobacter decreased subsequent cytokine production in response to either C. albicans or S. aureus exposure.
This is one of the first studies on the microbiome of patients with a primary immunodeficiency. Because of their specific defects in cellular immunity, the microbiota of patients with STAT1 and STAT3 mutations provide a unique opportunity to define the role of Th17 immunity for the composition of the normal flora of an individual. One important observation of the present study was the identification of an overrepresentation of Gram-negative bacteria (particularly Acinetobacter spp) in the skin microbiome of patients with STAT1/STAT3 defects, whereas the presence of the normal Corynebacteria was decreased. This is interesting in that it mirrors the findings of previous work in patients with multifactorial diseases like psoriasis [2,12,38], chronic obstructive pulmonary disease (COPD) [4,14,39], or AD [6,16,17,24]. During AD disease flares the amount of Corynebacterium is decreased [8–10,19–22,24], similar to CMC and HIES patients.
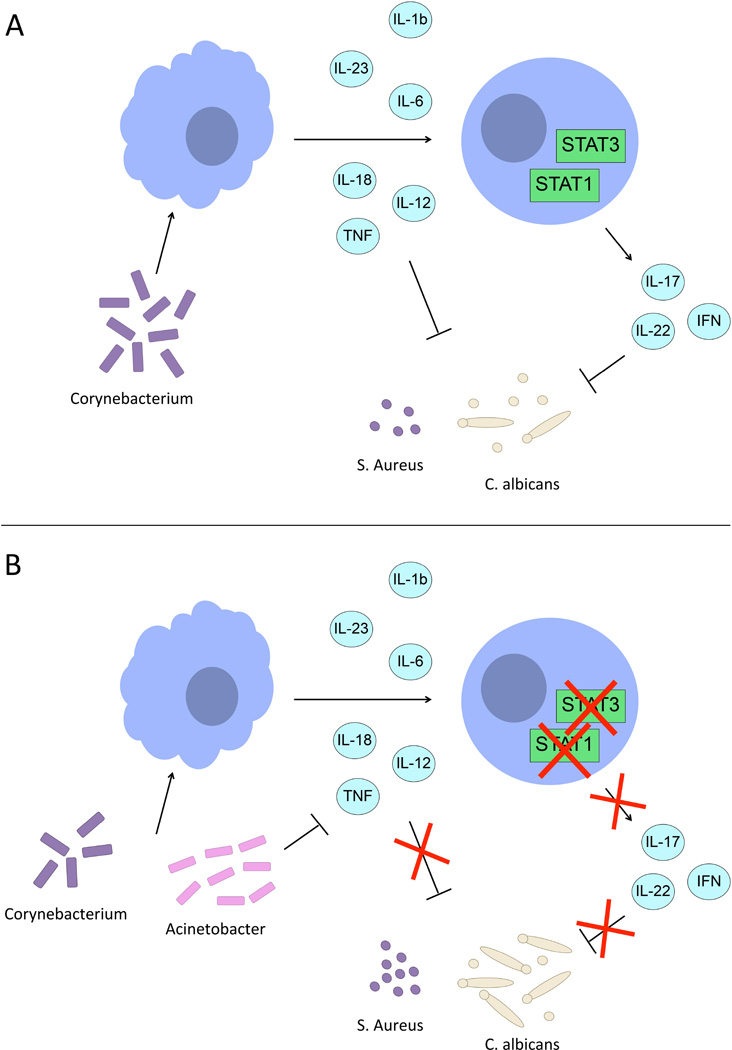
The functional consequence for the immune system of the shift from normal to Gram-negative flora in the CMC and HIES patients was investigated by exposing human primary leukocytes to either Corynebacterium or Acinetobacter, before their stimulation with C. albicans or S. aureus. They are two of the relevant pathogens for these diseases, as CMC and HIES patients often suffer from skin infections with fungal pathogens (especially dermatophytes) and HIES patients from skin abscesses with S. aureus [24,40]. While Corynebacteria did not impair antifungal and antistaphylococcal responses, exposure of human PBMCs to Acinetobacter significantly inhibited their capacity to produce proinflammatory cytokines such as TNF or IFNy when stimulated with C. albicans or S. aureus. Both cytokines are crucial for host defense against these two pathogens. Thus, while CMC and HIES patients have IL-17 immune defects mediated by mutations in Stat1 or Stat3, the microbiome skewed towards a Gram-negative microflora amplifies the susceptibility to infection by inhibiting TNF and IFNy responses (Figure 5). Recently, the role of skin microbiota for T-cell responses has been demonstrated to involve IL-1R-dependent innate immune signals in mice [5,26,27]. Our data confirm and extend for the first time these observations to humans, in which the shifted microbiota of patients with immunodeficiencies has the capacity to impair normal cellular immune responses.
Figure 5. Acinetobacter suppresses TNF-α, IFNy and IL-22.
Proposed model of typical interactions between the microbiome and the immune system in healthy individuals (A) and in STAT1/STAT3 immunodeficient patients (B).
Remarkably, the presence of Staphylococcus was not significantly increased across all HIES patients, despite its regular role in skin abscesses [14,28,29]. This could be related to the prophylaxis with narrow-spectrum antibiotics specifically directed towards S. aureus, such as flucloxacilline [30,41], that have been used previously by some of the patients. Although the HIES patients stopped the antibiotics before the collection of the microbiome samples, we cannot exclude that the long-term use of these regimens may be the cause of the low colonization with staphylococci. It has been previously demonstrated that S. aureus can secrete chemotaxis inhibitory protein of staphylococci (CHIPS), which binds to the formyl peptide receptor and C5a on neutrophils, thereby interfering with neutrophil chemotaxis [31,32,42]. Interestingly, neutrophils are a critical component of innate immunity with respect to C. albicans infections. Therefore, the excess of S. aureus found in CMC patients might be another factor explaining their reduced resistance to C. albicans infections. It might be interesting to learn whether the skin microbiome of these patients is also affected with respect to fungal colonization. However, better tools to detect fungal species still need to be developed [33,43].
The present study has also a few limitations. Firstly, we do not have direct information on the actual bacterial loads, which might play an important role in the emergence of infections, as we only measured the relative abundance of different bacterial 16S genes. Kembel et al. demonstrated that correcting for an estimated amount of 16S gene copy number for all species can alter the community structure [44]. Because of the above mentioned reasons, the bacterial numbers used in the stimulation experiments might not fully reflect the in-vivo situation, but they nevertheless enabled us to study the immunomodulatory effect of the bacteria identified in the CMC/HIES microbiome. Secondly, these data are based on a relatively small cohort containing only 4 CMC and 7 HIES patients. CMC and HIES are both very rare diseases, making it impossible for us to include more patients at this time. However, all these patients are very homogeneous in terms of their immune defects, and the clear changes in microbiome suggest that the shifts identified are real. Nevertheless, larger studies to validate the data presented here are warranted. Thirdly, some of the CMC and HIES patients were treated with antibiotics and antifungals in the period preceding the study, although with one exception of a CMC patient they temporarily stopped them prior to sampling. In addition, the patients recruited in this study were not hospitalized (with the exception of the same patient being continuously treated), precluding the possibility of hospital-acquired microorganisms. Antibiotic use has been associated with decreased bacterial diversity, and the prior antibiotic regimens used by some of the patients directed against fungi and staphylococci may have shifted the microbiome towards a Gram-negative bacteria-dominated colonization. This does not however influence the conclusion that CMC and HIES patients are colonized with relatively many Gram-negative bacteria, and that they can influence normal innate immune responses.
In conclusion, the assessment of the skin and oral microbiome of CMC and HIES patients with STAT1/STAT3 mutations and Th17 defects shows a shift of the microbiota from normal microorganisms such as Corynebacteria and Actinomycetaceae to a Gram-negative colonization with genera such as Acinetobacter. Importantly, Gram-negative colonization is capable of inhibiting immune responses to Candida and S. aureus, likely contributing to the increased susceptibility of the patients to these infections. These data in patients with immunodeficiencies proof that the microbiome can influence host defense and could open the possibility of microbiome-based adjuvant therapy in patients with immune defects.
Acknowledgements
This work was supported by an ERC Consolidator Grant from the European Research Council [310372 to M.G.N.]; by the National Institutes of Health [NIH 1R01HG005969 to C.H.]; and by a grant from the Crohn’s and Colitis Foundation of America [to R.J.X, D.G., and C.H.].
Footnotes
The authors declare no conflict of interest.
References
- 1.Li E, Hamm CM, Gulati AS, Sartor RB, Chen H, Wu X, et al. Inflammatory Bowel Diseases Phenotype, C. difficile and NOD2 Genotype Are Associated with Shifts in Human Ileum Associated Microbial Composition. PLoS ONE. 2012 Jun 13;7:e26284. doi: 10.1371/journal.pone.0026284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kirkpatrick CH. Chronic mucocutaneous candidiasis. J Am Acad Dermatol. 1994 Sep 11;31:S14–S17. doi: 10.1016/s0190-9622(08)81260-1. [DOI] [PubMed] [Google Scholar]
- 3.Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, et al. Metabolic Syndrome and Altered Gut Microbiota in Mice Lacking Toll-Like Receptor 5. Science. 2010 Apr 8;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aaltonen J, Björses P, Perheentupa J, Horelli Kuitunen N, Palotie A, Peltonen L, et al. An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. Nat Genet. 1997 Dec;17:399–403. doi: 10.1038/ng1297-399. [DOI] [PubMed] [Google Scholar]
- 5.Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, et al. Compartmentalized Control of Skin Immunity by Resident Commensals. Science. 2012 Aug 30;337:1115–1119. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Puel A, Döffinger R, Natividad A, Chrabieh M, Barcenas-Morales G, Picard C, et al. Autoantibodies against IL-17A, IL-17F, and IL-22 in patients with chronic mucocutaneous candidiasis and autoimmune polyendocrine syndrome type I. Journal of Experimental Medicine. 2010 Feb 15;207:291–297. doi: 10.1084/jem.20091983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012 Mar 8;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van de Veerdonk FL, Plantinga TS, Hoischen A, Smeekens SP, Joosten LAB, Gilissen C, et al. STAT1 mutations in autosomal dominant chronic mucocutaneous candidiasis. N Engl J Med. 2011 Jul 7;365:54–61. doi: 10.1056/NEJMoa1100102. [DOI] [PubMed] [Google Scholar]
- 9.Liu L, Okada S, Kong X-F, Kreins AY, Cypowyj S, Abhyankar A, et al. Gain-of-function human STAT1 mutations impair IL-17 immunity and underlie chronic mucocutaneous candidiasis. Journal of Experimental Medicine. 2011 Aug 1;208:1635–1648. doi: 10.1084/jem.20110958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takezaki S, Yamada M, Kato M, Park M-J, Maruyama K, Yamazaki Y, et al. Chronic mucocutaneous candidiasis caused by a gain-of-function mutation in the STAT1 DNA-binding domain. J Immunol. 2012 Aug 1;189:1521–1526. doi: 10.4049/jimmunol.1200926. [DOI] [PubMed] [Google Scholar]
- 11.Kuczynski J, Lauber CL, Walters WA, Parfrey LW, Clemente JC, Gevers D, et al. Experimental and analytical tools for studying the human microbiome. Nature Reviews Genetics. 2011 Dec 16;13:47–58. doi: 10.1038/nrg3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smeekens SP, Plantinga TS, van de Veerdonk FL, Heinhuis B, Hoischen A, Joosten LAB, et al. STAT1 hyperphosphorylation and defective IL12R/IL23R signaling underlie defective immunity in autosomal dominant chronic mucocutaneous candidiasis. PLoS ONE. 2011 Dec;6:e29248. doi: 10.1371/journal.pone.0029248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a Chimera-Checked 16S rRNA Gene Database and Workbench Compatible with ARB. Appl Environ Microbiol. 2006 Jul;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis SD, Schaller J, Wedgwood RJ. Job's Syndrome. Recurrent, ‘cold’, staphylococcal abscesses. Lancet. 1966 May 7;1:1013–1015. doi: 10.1016/s0140-6736(66)90119-x. [DOI] [PubMed] [Google Scholar]
- 15.Knights D, Kuczynski J, Koren O, Ley RE, Field D, Knight R, et al. Supervised classification of microbiota mitigates mislabeling errors. ISME J. 2010 Oct 7;5:570–573. doi: 10.1038/ismej.2010.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holland SM, DeLeo FR, Elloumi HZ, Hsu AP, Uzel G, Brodsky N, et al. STAT3 mutations in the hyper-IgE syndrome. N Engl J Med. 2007 Oct 18;357:1608–1619. doi: 10.1056/NEJMoa073687. [DOI] [PubMed] [Google Scholar]
- 17.Minegishi Y, Saito M, Tsuchiya S, Tsuge I, Takada H, Hara T, et al. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature. 2007 Aug 30;448:1058–1062. doi: 10.1038/nature06096. [DOI] [PubMed] [Google Scholar]
- 18.Knights D, Kuczynski J, Charlson ES, Zaneveld J, Mozer MC, Collman RG, et al. Bayesian community-wide culture-independent microbial source tracking. Nat Meth. 2011 Jul 17;8:761–763. doi: 10.1038/nmeth.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Renner ED, Rylaarsdam S, Aňover-Sombke S, Rack AL, Reichenbach J, Carey JC, et al. Novel signal transducer and activator of transcription 3 (STAT3) mutations, reduced TH17 cell numbers, and variably defective STAT3 phosphorylation in hyper-IgE syndrome. Journal of Allergy and Clinical Immunology. 2008 Jul;122:181–187. doi: 10.1016/j.jaci.2008.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Beaucoudrey L, Puel A, Filipe-Santos O, Cobat A, Ghandil P, Chrabieh M, et al. Mutations in STAT3 and IL12RB1 impair the development of human IL-17-producing T cells. The Journal of Experimental Medicine. 2008 Jul 7;205:1543–1550. doi: 10.1084/jem.20080321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma CS, Chew GYJ, Simpson N, Priyadarshi A, Wong M, Grimbacher B, et al. Deficiency of Th17 cells in hyper IgE syndrome due to mutations in STAT3. Journal of Experimental Medicine. 2008 Jul 7;205:1551–1557. doi: 10.1084/jem.20080218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milner JD, Brenchley JM, Laurence A, Freeman AF, Hill BJ, Elias KM, et al. Impaired T(H)17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature. 2008 Mar 12;452:773–776. doi: 10.1038/nature06764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biology. 2011 Nov 6;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Research. 2012 May 1;22:850–859. doi: 10.1101/gr.131029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biology. 2012 Sep 26;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011 Mar 16;9:244–253. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Consortium THMP. Structure, function and diversity of the healthy human microbiome. Nature. 2012 Jun 13;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, et al. Topographical and Temporal Diversity of the Human Skin Microbiome. Science. 2009 May 28;324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial Community Variation in Human Body Habitats Across Space and Time. Science. 2009 Dec 17;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. 2011 Mar 16;9:279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- 31.McKnite AM, Perez-Munoz ME, Lu L, Williams EG, Brewer S, Andreux PNLA, et al. Murine Gut Microbiota Is Defined by Host Genetics and Modulates Variation of Metabolic Traits. PLoS ONE. 2012 Jun 11;7:e39191. doi: 10.1371/journal.pone.0039191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benson AK, Kelly SA, Leggea R, Ma F, Low SJ, Kim J, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci USA. 2010 Nov 2;107:18933–18938. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khachatryan ZA, Ktsoyan ZA, Manukyan GP, Kelly D, Ghazaryan KA, Aminov RI. Predominant Role of Host Genetics in Controlling the Composition of Gut Microbiota. PLoS ONE. 2008 Aug 26;3:e3064. doi: 10.1371/journal.pone.0003064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LEfSe. [ http://huttenhower.sph.harvard.edu/lefse/]. [Google Scholar]
- 35.Gaffen SL, Hernandez-Santos N, Peterson AC. IL-17 signaling in host defense against Candida albicans. Immunol Res. 2011 Jun 30;50:181–187. doi: 10.1007/s12026-011-8226-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krishna S, Miller LS. Innate and adaptive immune responses against Staphylococcus aureus skin infections. Semin Immunopathol. 2011 Nov 6;34:261–280. doi: 10.1007/s00281-011-0292-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eyerich K, Foerster S, Rombold S, Seidl H-P, Behrendt H, Hofmann H, et al. Patients with Chronic Mucocutaneous Candidiasis Exhibit Reduced Production of Th17-Associated Cytokines IL-17 and IL-22. Journal of Investigative Dermatology. 2008 Jul 10;128:2640–2645. doi: 10.1038/jid.2008.139. [DOI] [PubMed] [Google Scholar]
- 38.Gao Z, Tseng C-H, Strober BE, Pei Z, Blaser MJ. Substantial Alterations of the Cutaneous Bacterial Biota in Psoriatic Lesions. PLoS ONE. 2008 Jul 23;3:e2719. doi: 10.1371/journal.pone.0002719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cabrera-Rubio R, Garcia-Núñez M, Setó L, Antó JM, Moya A, Monsó E, et al. Microbiome Diversity in the Bronchial Tracts of Patients with Chronic Obstructive Pulmonary Disease. J Clin Microbiol. 2012 Aug;50:3562–3568. doi: 10.1128/JCM.00767-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buckley RH. The Hyper-IgE Syndrome. CRIAI. 2001;20:139–154. doi: 10.1385/CRIAI:20:1:139. [DOI] [PubMed] [Google Scholar]
- 41.Dethlefsen Les, Huse S, Sogin ML, Relman DA. The Pervasive Effects of an Antibiotic on the Human Gut Microbiota, as Revealed by Deep 16S rRNA Sequencing. PLOS Biology. 2008 Nov 18;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Haas CJC. Chemotaxis Inhibitory Protein of Staphylococcus aureus, a Bacterial Antiinflammatory Agent. Journal of Experimental Medicine. 2004 Feb 23;199:687–695. doi: 10.1084/jem.20031636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grice EA, Segre JA. The Human Microbiome: Our Second Genome. Annu Rev Genomics Hum Genet. 2012 Sep;13:151–170. doi: 10.1146/annurev-genom-090711-163814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kembel SW, Wu M, Eisen JA, Green JL. Incorporating 16S Gene Copy Number Information Improves Estimates of Microbial Diversity and Abundance. PLoS Comput Biol. 2012 Oct 25;8:e1002743. doi: 10.1371/journal.pcbi.1002743. [DOI] [PMC free article] [PubMed] [Google Scholar]