Abstract
Various types of neurons and glia are generated following a precise spatial and temporal order during neurogenesis. The mechanisms that control this sequential generation of neuronal and glial cell types from the same progenitor population are not well understood. Growth differentiation factor 11 (Gdf11) belongs to the TGF-β family of proteins and is expressed transiently in newly born neurons adjacent to the progenitor domain in the developing spinal cord. We examined the phenotypes of Gdf11−/− mouse embryos and found that without Gdf11, neuronal differentiation in the spinal cord progresses at a slower rate. Higher progenitor proliferation rate, along with a delay in gliogenesis, is also observed in Gdf11−/− spinal cord but only after the peak of Gdf11 expression, indicating that Gdf11 can cause long-lasting changes in progenitor properties. These changes can be preserved in vitro, as neurospheres derived from Gdf11−/− and wild-type littermates at a stage after, but not before the onset of Gdf11 expression, exhibit differences in proliferation and differentiation potential. Moreover, these changes in progenitor properties can be induced in vitro by the addition of Gdf11. We also demonstrate that the effects of Gdf11 on progenitor cells are associated with its ability to upregulate p57 Kip2 and p27 Kip1 while downregulating Pax6 expression. These results support a model in which Gdf11 secreted by newly born neurons in the developing spinal cord facilitates the temporal progression of neurogenesis by acting as a positive feedback signal on the progenitor cells to promote cell cycle exit and decrease proliferation ability, thus changing their differentiation potential.
Introduction
Distinct types of neurons and glia are generated from progenitor cells following a precise spatial and temporal order during development of the nervous system. The spatial identity of a neuron is determined by the position of its progenitor cell in the neural tube and the signals that it receives along the dorsoventral (D-V) and rostrocaudal (R-C) axes (for review, see Lumsden and Krumlauf, 1996; Diez del Corral and Storey, 2004; Wilson and Maden, 2005). The neuronal temporal identity is influenced by both progenitor intrinsic properties and environmental cues, as demonstrated by cortical progenitor transplantation experiments performed in ferrets (McConnell and Kaznowski, 1991; Frantz and McConnell, 1996; Desai and McConnell, 2000). In addition, in vitro experiments performed with cultured cortical progenitor cells further indicated that the mechanisms governing the temporal order of neurogenesis are encoded within the progenitors and their immediate progeny (Qian et al., 2000; Shen et al., 2006).
In the developing spinal cord, sequential generation of neurons and glia has been observed in different progenitor domains along the D-V axis. At the onset of neurogenesis, ventrally located motor neurons (MNs) are the first type of neurons generated in the spinal cord, and various subtypes of MNs are born from the MN progenitor domain (pMN) at different times during development. For example, MNs destined for the medial portion of the lateral motor column (LMCm) are generated before MNs populating the lateral portion of the lateral motor column (LMCl) (Hollyday and Hamburger, 1977). After the production of MNs, progenitors in the pMN domain will then give rise to oligodendrocyte precursor cells (OLPs) (Kessaris et al., 2001; Novitch et al., 2001; Zhou et al., 2001). The dorsal spinal progenitors first produce six different types of early born dorsal interneurons (dI1–6) that migrate into the deep layers of the dorsal horn, followed by two types of late born dorsal interneurons (dILA and dILB) that migrate into the superficial dorsal horn (Matise, 2002; Helms and Johnson, 2003). Following the cessation of neuronal differentiation, dorsal progenitors also give rise to OLPs in the dorsal spinal cord (Cai et al., 2005). However, the mechanisms that control the timing of these transitions are not well understood.
Growth and differentiation factor 11 (Gdf11) belongs to the TGF-β family (Gamer et al., 1999). Gdf11 exerts its function by interacting with activin type IIA and IIB receptors (Oh et al., 2002) and type I receptor Alk5 (Andersson et al., 2006) to induce phosphorylation of Smad2/3. We have demonstrated that a Gdf11 signal originating from the node/tail bud region is essential for proper R-C patterning of the developing spinal cord (Liu, 2006). In addition, a different function of Gdf11 in controlling progenitor proliferation and/or differentiation has been demonstrated in olfactory epithelia and the retina (Wu et al., 2003; Kim et al., 2005).
In this study, we used both in vivo and in vitro approaches to examine Gdf11 function in spinal cord neurogenesis. We provide evidence to support a model in which Gdf11 signals produced by newly differentiated neurons act as a feedback signal on the adjacent progenitors to promote cell cycle exit, inhibit proliferation, and thus facilitate temporal progression of neurogenesis in the developing spinal cord.
Materials and Methods
Mouse breeding and embryo collection
Gdf11+/− mice (McPherron et al., 1999) were maintained in a mixed 129/SvJ and C57BL/6 background and housed in a barrier facility under a 12 h light/12 h dark cycle. Noon of the day that the vaginal plug was present was considered embryonic day 0.5 (e0.5). Bromodeoxyuridine (BrdU; 100 μg/g body weight, Sigma) was injected intraperitoneally into pregnant females, and embryos were harvested 30 min or 24 h later. All protocols for animal use were approved by the Institutional Animal Care and Use Committee of the University of Virginia (Charlottesville, VA) and were in accordance with National Institutes of Health guidelines.
In situ hybridization. In situ hybridization was performed using mouse Gdf11 probes (McPherron et al., 1999) as described previously (http://physio.ucsf.edu/rubenstein/protocols/PDFs/Jessell_ISH.pdf). Briefly, e10 – e13.5 embryos were harvested, fixed with 4% paraformaldehyde in 0.1 M phosphate buffer (PB) (pH 7.4) at 4°C overnight, washed three times with PBS, equilibrated with 30% sucrose in 0.1 M PB, and then embedded in OCT compound and sectioned at 15 μm. Images were taken with an Olympus BX60 microscope and a Magnafire CCD camera.
Immunohistochemistry and cell count
Mouse embryos were harvested at e9.5–e15.5, fixed for 2 h, and processed as described above. Serial sections through brachial (forelimb) level spinal cord, a level that was least affected by Gdf11 function in R-C patterning, were collected at 10 μm, and images were acquired using a Nikon C-1 confocal microscope. Antibodies used are as follows: rat anti-BrdU, rabbit anti-Ki67, rabbit anti-Sox10 (Abcam); rabbit anti-Pax2 (Zymed); rabbit anti-microtubule-associated protein 2 (MAP2), rabbit anti-cleaved caspase3 (Cell Signaling Technology); goat anti-p57 kip2(Santa Cruz Biotechnology); mouse anti-P27 kip1, rabbit anti-phospho-histone H3 (PH3), rabbit antiglial fibrillary acidic protein (GFAP), rabbit anti-Olig1, rabbit anti-Olig2, mouse anti-myelin basic protein (MBP), rabbit anti-Sox2 (Millipore Bioscience Research Reagents Biotechnology); mouse anti-green fluorescent protein (GFP) (Invitrogen); mouse anti-BrdU, mouse anti-HB9, mouse anti-Lhx1, mouse anti-Lhx3, mouse anti-Pax6, mouse anti-Pax7 (Developmental Studies Hybridoma Bank, Department of Biology, University of Iowa, Iowa City, IA). Additional antibodies were kindly provided by Dr. T. Jessell at Columbia University, New York, NY (guinea pig anti-Lmx1b, rabbit anti-Lhx1, and rabbit anti-Isl1), and Dr. J. Dasen, New York University, New York, NY (rabbit anti-FoxP1). Cell nuclei were counterstained with To-pro-3 (To-pro3) (Invitrogen), and cell counts were obtained from 3–7 pairs of Gdf11−/− and wild-type (WT) littermates for each embryonic stage. Brachial level spinal cord tissues were sectioned at 10 μm thickness and collected serially. One of every 10 sections (100 μm apart) were counted for e10.5 embryos (five or six sections per embryo), and one of every 20 sections (200 μm apart) were counted for e11.5–e13.5 embryos (4 – 6 sections/embryo). For FoxP1/Isl1/Lhx3 cell counts, adjacent sections were stained with Sox2/ Topro3 to obtain progenitor and total cell numbers. Cell numbers and percentages are presented as mean ±SEM per section. Statistical analyses were performed using SigmaPlot Software.
Isolation, passaging, differentiation, and Gdf11 treatment of neurosphere culture
Neural progenitors were isolated from brachial level spinal cord at e9.5 and e12.5. The e9.5 spinal cord was incubated in 1 mg/ml Dispase (Roche Diagnostics)/HBSS (Invitrogen) for 15 min at 37°C and then mechanically dissociated with 200 μl pipettes. The e12.5 spinal cord was incubated in DMEM containing 5 U/ml papain and 1 mM N-acetyl-L-cysteine (Sigma) for 30 min at 37°C and then dissociated using a fire-polished glass pipette. Cells from individual embryos were seeded separately at 105cells/ml in NSC medium (DMEM/F12 containing 2 mM L-glutamine, B-27 [without retinoic acid (RA)] and N2 (Invitrogen) with 0.2% heparin, 20 μg/ml insulin (Sigma), 10 ng/ml epidermal growth factor, and 10 ng/ml basic fibroblast growth factor (PeproTech) to form neurospheres (Medrano et al., 2009). These cells were designated as passage 0 (P0).
Depending on the genotype, later passages of neurospheres (P1–P3) usually take ~ 4 – 8 d to grow in culture. For passaging, neurospheres were treated with TrypLE Express (Invitrogen) for 10 min, triturated into a single cell suspension, and then seeded at 1.5 ×104 cells/ml. For Gdf11 treatment, dissociated cells from P0 e9.5 WT, e12.5 WT, and e12.5 Gdf11−/− neurospheres were seeded in NSC medium containing 10 ng/ml recombinant human Gdf11 (R&D Systems) at P1. To determine the growth rate, neurospheres were dissociated and their cell numbers counted at the end of each passage. Growth was plotted as cumulative population doublings (PDs) (PD = log2 (N2/N1), N1 = number of cells initially seeded for each passage, N2 = number of cells harvested at the end of each passage) versus total days in culture (Baxter et al., 2004). GraphPad Prism5 was used for generating the growth curve and statistical analyses. For BrdU incorporation experiments, P3 neurospheres at day 4 were treated with 10 μM BrdU for 30 min before collection. For differentiation, P3 neurospheres at day 4 were dissociated and plated on poly-L-ornithine-treated glass coverslips at 2 × 105 cells/well with Neurobasal medium containing 2 mM L-glutamine and B27 (with RA) (Invitrogen).
Cell counts were obtained from neurospheres derived from more than three pairs of Gdf11−/− and WT littermates at e9.5 and e12.5. Three neurospheres per embryo were counted for BrdU and PH3 experiments, and four microscopic fields (600 × 600 μm) per embryo were counted for progenitor cell differentiation experiments. Cell numbers and percentages are presented as mean ± SEM per microscopic field.
To verify that endogenous Gdf11 is not expressed in neurospheres under our culture conditions, total RNA was extracted from P3 neurospheres derived from e12.5 WT and Gdf11−/− spinal cords at day 4 using TRIzol reagent (Invitrogen) and reverse-transcribed using SuperScript III RT-PCR kit (Invitrogen). Gdf11 transcripts were detected using an exon 2 primer, 5′-TGCCCCGCCCAGCCACAGTCTAC3-′ and an exon 3 primer, 5′-CCCAGCCAAAAGCCTCAAAGTCCA-3′, with 40 cycles of amplification. Glyceraldehyde 3-phosphate dehydrogenase transcripts were detected using the forward primer 5′-AAATGGTGAAGGTC-GGTGTGAACG3-′ and the reverse primer 5′-AGTGATGGCATGG-ACTGTGGTCAT3-′.
Western analyses
Forelimb level spinal cords were isolated from seven pairs of e11.5 Gdf11−/− embryos and control littermates, and total protein was extracted as described previously (Liu, 2006). Restore Western blot stripping buffer (Pierce/Thermo Fisher Scientific) was used to strip blots between different antibodies, and SuperSignal West Femto maximum sensitivity substrate (Pierce/Thermo Fisher Scientific) was used for signal detection. Quantification was performed using ImageQuant 5.2.
In ovo electroporation. To examine the effect of Gdf11 on progenitor marker expression, electroporation was performed on somite 11–16 embryos using the pNes-Gdf11 plasmid at 2 mg/ml as described previously (Liu, 2006). Observation under a fluorescence microscope (Nikon SMZ1500) was made the day after electroporation, and only embryos with visible GFP expression were harvested for analysis. A total of 16 embryos were analyzed at HH (Hamburger and Hamilton, 1951) stage 15–17, and a total of 9 embryos were analyzed at HH stage 21–22. To examine the effects of Gdf11 and Pax6 on OLP differentiation, electroporation was performed on HH stage 13–14 embryos (before the onset of neuronal differentiation and the expression of endogenous Gdf11) with pNes-Gdf11, pRFPRNAi-Pax6A, or pRFPRNAi-Pax6B plasmid (Das et al., 2006) (all at 0.5 mg/ml). Embryos were harvested at e6 (Gdf11: three embryos; Pax6i: seven embryos) or at e7 (Gdf11: 13 embryos; Pax6i: 9 embryos), and serial sections (10 μm thickness) through brachial level spinal cord were collected for analyses. Olig2+ cells outside the progenitor domain were counted from eight pNes-Gdf11electroporated embryos collected at e7 (14 –29 sections/embryo, 80 μm-160 μm between each section), and cell numbers are presented as mean ±95% confidence to exhibit the degree of variability.
Results
Gdf11 is expressed transiently in newly born neurons in the developing spinal cord
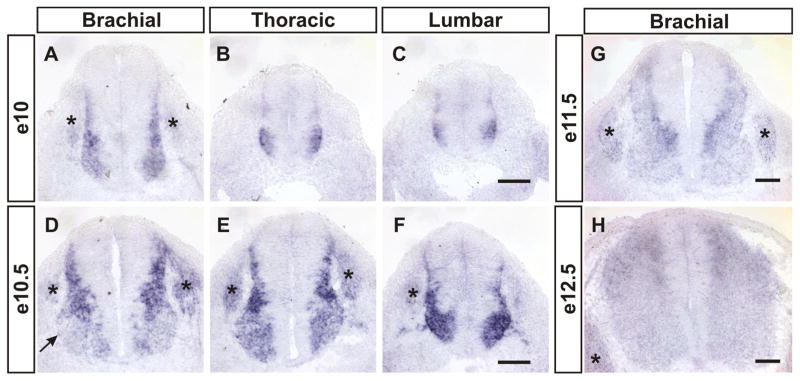
During our study of Gdf11 function in R-C patterning in the developing spinal cord, we also noticed the presence of enlarged progenitor domains along the length of spinal cord and clustered cells trapped in the dorsal midline at the lumbar levels of Gdf11−/− embryos (supplemental Fig. 1, available at www.jneurosci.org as supplemental material), indicating that Gdf11 could have a function in regulating progenitor proliferation and/or neuronal differentiation. To evaluate this possibility, we first performed in situ hybridization to examine Gdf11 expression in mouse embryonic spinal cord at different stages. Gdf11 expression is detected in the tail bud and caudal neural plate region at approximately e9.5, but not in the neural tube (Gamer et al., 1999; McPherron et al., 1999; Nakashima et al., 1999; Liu, 2006; and data not shown). By e10, Gdf11 expression is observed in cells located adjacent to the progenitor domain along the length of the spinal cord (Fig. 1A–C). A similar expression pattern persists into e10.5 (Fig. 1D–F), with the exception that the level of Gdf11 expression is downregulated in the ventral horn, where mature MNs are located (Fig. 1D). This expression pattern is similar to that of NeuroM, a basic helix-loop-helix (bHLH) transcription factor that is expressed in postmitotic premigratory cells located adjacent to the progenitor domain in the spinal cord (Roztocil et al., 1997), suggesting that Gdf11 is expressed in newly born neurons. By e11.5, lower levels of Gdf11 expression remain in the cells next to the progenitor domain in the dorsal spinal cord, and the expression level decreases further in mature neurons located more laterally as described previously (Gamer et al., 1999) (Fig. 1G). The expression level of Gdf11 continues to decline through e12.5 (Fig. 1H), and by e13.5 practically no Gdf11 expression can be detected in the brachial (forelimb) level spinal cord (supplemental Fig. 2A, available at www.jneurosci.org as supplemental material). These results demonstrate that Gdf11 is expressed transiently in newly born neurons in the developing spinal cord, and its expression level decreases as neurons mature and move laterally away from the progenitor domain.
Figure 1.
Transient expression of Gdf11 in the developing spinal cord. Detection of Gdf11 expression by in situ hybridization on cross-sectioned spinal cord tissues. A–C, At e10 Gdf11 is expressed in cells located lateral to the progenitor domain at brachial (A), thoracic (B) and lumbar (C) levels of the spinal cord. D–F, Downregulation of Gdf11 expression is observed in mature neurons at e10.5 (D, arrow). G, H, The expression level of Gdf11 continues to decline through e11.5 (G) and e12.5 (H ). Asterisk (*) indicates dorsal root ganglia. Scale bars, 100 μm.
Loss of Gdf11 function reduces the rate of neuronal differentiation
To evaluate a possible function of Gdf11 in neuronal differentiation, we first injected BrdU into pregnant female mice at e9.5, e10.5, e11.5, and e12.5 and harvested their embryos 24 h later. Cross-sectioned spinal cord tissue was collected from Gdf11−/− embryos and their WT littermates at various R-C levels and stained with antibodies against Sox2 (labeling progenitor cells) and BrdU, and the numbers and distribution of BrdU+ cells outside the Sox2+ progenitor domain (BrdU+ Sox2− ), which represent neurons derived from progenitors that were in S-phase 24 h previously, were then examined. At brachial levels, fewer Brdu+ Sox2− cells in the mantle zone of Gdf11−/− embryos were present at e10.5 as compared to the controls (Fig. 2A,B). In addition, a slower progression of neuronal differentiation can be observed in the Gdf11−/− embryos as indicated by similar distribution patterns of Brdu+ Sox2− cells in the ventral spinal cord of e11.5 Gdf11−/− compared to e10.5 WT embryos, and in e12.5 Gdf11−/− compared to e11.5 WT embryos (compare Fig. 2, A with D and C with F). Moreover, a higher level of neuronal differentiation in the dorsal spinal cord of Gdf11−/− embryos was observed at e13.5 compared to the controls (Fig. 2G,H). In contrast to the effect of Gdf11 on R-C patterning, which affects predominantly cells located at the thoracic and lumbar level spinal cord (Liu, 2006), its effect on neuronal differentiation can be observed throughout the length of the spinal cord in Gdf11−/− embryos. Therefore, to examine the role of Gdf11 in neuronal differentiation independently of its function in R-C patterning, we focused our analyses on the forelimb level (approximately C5–T1, designated as brachial level) spinal cord, where early function of Gdf11 in R-C patterning has minimal effect.
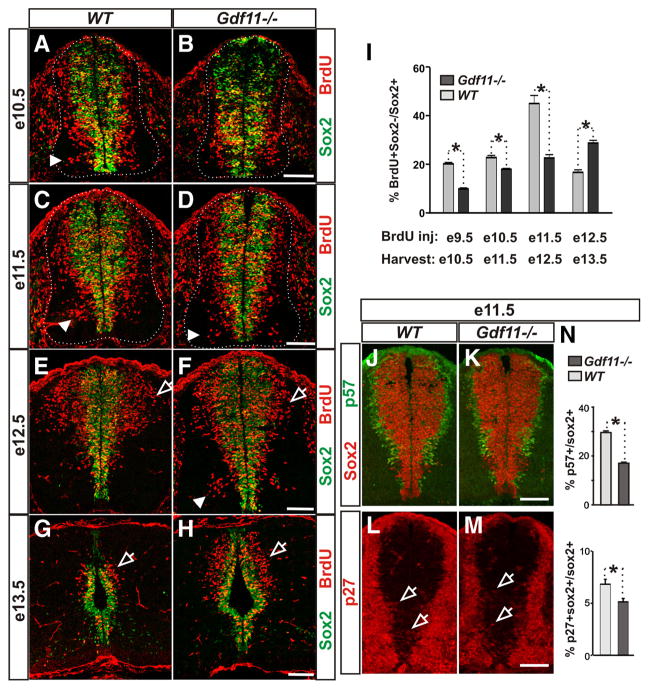
Figure 2.
Gdf11 facilitates cell cycle exit and neuronal differentiation. A–H, Sox2 and BrdU staining of cross-sectioned brachial level spinal cords from embryos labeled with BrdU for 24 h before harvesting at e10.5 (A, B), e11.5 (C, D), e12.5 (E, F ), and e13.5 (G, H ). Brdu+ Sox2− cells represent S-phase progenitors at the time of BrdU labeling that differentiated during the 24 h chase. White dotted lines delineate the margin of the spinal cord. Arrowheads point to neurons generated in the ventral spinal cord (compare A with D and C with F ). Open arrows indicate neurons generated in the dorsal spinal cord. I, The percentage of BrdU+ Sox2−/Sox2+ cells in Gdf11−/− and WT littermates injected with BrdU at e9.5–e12.5 and harvested 24 h later. A slower and prolonged neuronal differentiation is observed in the Gdf11−/− embryos. *p <0.05. J–M, p57 kip2 and Sox2 staining (J, K ) and p27 kip1 and Sox2 staining (L, M ) of cross-sectioned brachial level spinal cord obtained from e11.5 WT and Gdf11−/− embryos. Open arrows point to p27+ Sox2+ cells (Sox2 signal is removed for easier visualization of the faint p27 staining). N, Percentage of progenitors that are exiting the cell cycle (p57+/Sox2+ and p27+ Sox2+/Sox2+) in e11.5 embryos. Higher p57+/Sox2+ and p27+ Sox2+/Sox2+ ratios were observed in WT compared to Gdf11−/− embryos. *p <0.05. Scale bars, 100 μm.
Quantification obtained from 3–5 pairs of Gdf11−/− and WT littermates revealed that the number of BrdU+ Sox2− cells increases between e10.5 and e12.5 but decreases sharply afterward in WT embryos (supplemental Fig. 2C). When the percentage of progenitor cells that differentiated into neurons (estimated by the BrdU+ Sox2−/Sox2+ ratio) is compared between WT embryos at different stages, an increase was observed from e10.5 (~20%) to e12.5 (~45%), followed by a decrease to ~17% at e13.5 (Fig. 2 I), suggesting that the majority of neurons were born by e12.5 in the brachial level spinal cord of WT embryos. In contrast, fewer BrdU+ Sox2− cells are present in the Gdf11−/− embryos than in the controls between e10.5 and e12.5, but significantly more are present at e13.5 (supplemental Fig. 2C, available at www.jneurosci.org as supplemental material). The percentage of differentiated progenitors in the Gdf11−/− embryos is lower than that of the controls between e10.5 and e12.5 but increases steadily from e10.5 (~10%) to e13.5 (~29%) and is higher than that of the controls by e13.5 (Fig. 2 I), suggesting that neuronal differentiation progresses at a slower rate and lasts for a longer period of time in the absence of Gdf11.
The slower progression of neuronal differentiation in the absence of Gdf11 suggests that Gdf11 might affect cell cycle exit. We therefore examined several proteins involved in cell cycle regulation, including the cyclin-dependent kinase inhibitors p27 kip1 and p57 kip2, which inhibit the G1-S transition and cause cell cycle exit (Mainprize et al., 2001). In the developing spinal cord, p57 kip2 is expressed in almost all newly born neurons located lateral to the progenitor domain, except the V2 interneurons and MNs (Gui et al., 2007). A higher number of p57 Kip2+ cells were observed in the WT compared to the Gdf11−/− spinal cord at e10.5 and e11.5 (Fig. 2 J, K; data not shown). Quantification obtained from three pairs of Gdf11−/− and WT littermates between e10.5 and e12.5 revealed that the percentages of progenitors that are leaving the cell cycle (represented by the p57 Kip2+/Sox2+ ratio) are significantly higher in WT compared to Gdf11−/− embryos at e10.5 and e11.5, but not at e12.5 (Fig. 2 N; data not shown). p27 kip1 is expressed at high levels in neurons located in the mantle zone of the developing spinal cord (supplemental Fig. 3, available at www.jneurosci.org as supplemental material); however, low levels of p27 kip1 expression can be observed in a subset of Sox2+ progenitors that could represent progenitors that are leaving the cell cycle (Fig. 2 L, M; data not shown). The p27 kip1+ Sox2+/ Sox2+ ratio in WT embryos is also significantly higher than that in their Gdf11−/− littermates at e10.5 and e11.5 (Fig. 2 N; data not shown). These results, in conjunction with the Gdf11 expression pattern, indicate that Gdf11 secreted from the newly born neurons acts on neighboring progenitors to promote their cell cycle exit and differentiation.
Loss of Gdf11 function causes an increase in progenitor proliferation
An apparent increase in the number of Sox2+ cells was observed in the Gdf11−/− embryos compared to their control littermates from e12.5 onwards (Fig. 2 E–H ); this could result from the effect of Gdf11 on cell cycle exit or, alternatively, Gdf11 could also have a function in progenitor proliferation. To examine this possibility, we performed short-term BrdU labeling to evaluate the percentage of S-phase progenitors at various developmental stages. BrdU was injected into pregnant female mice at e10.5, e11.5, e12.5, and e13.5, and embryos were harvested 30 min later. Cross-sectioned brachial level spinal cord tissues from 5–7 pairs of WT and Gdf11−/− littermates were labeled with BrdU, Sox2, and Topro 3 (Fig. 3A–F; data not shown) for cell counting.
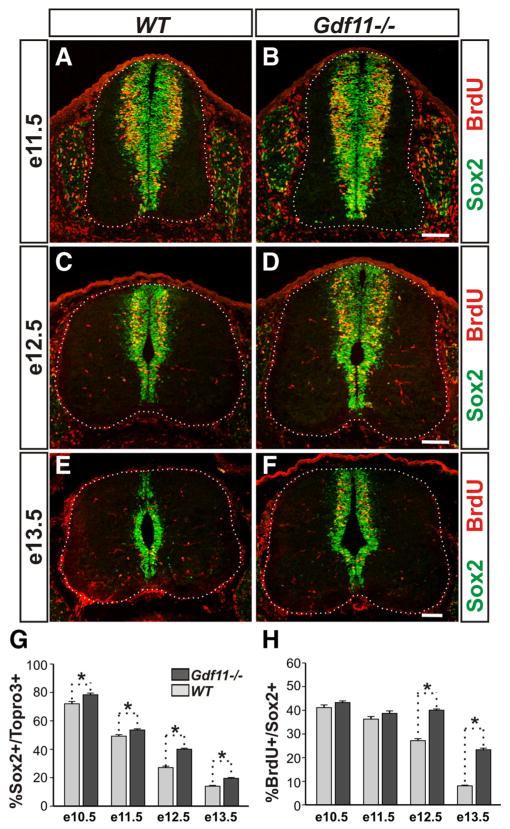
Figure 3.
Delayed declination of progenitor proliferation in the Gdf11−/− spinal cord. A–F, Sox2 and BrdU staining of cross-sectioned brachial level spinal cords from Gdf11−/− and WT littermates labeled with BrdU for 30 min at e11.5 (A, B), e12.5 (C, D), and e13.5 (E, F ). White dotted lines delineate the margin of the spinal cord. Scale bars, 100 μm. G, H, Percentages of progenitors (Sox2+) to total cells (Topro3+) and S-phase progenitors (BrdU+) to Sox2+ progenitors. A declination of progenitor proliferation is observed in the WT embryos between e11.5 and e12.5 and continues through e13.5. This declination is delayed for ~1 d in the Gdf11−/− embryos. *p <0.05.
A continuous increase in total cell (Topro3+) numbers between e10.5 and e13.5 was observed in both Gdf11−/− and WT embryos (supplemental Fig. 2 D). In contrast, the number of Sox2+ progenitors and the number of BrdU+ S-phase progenitors increased from e10.5 to e11.5 but decreased afterward in WT embryos (supplemental Fig. 2 E, F ). As neurogenesis progresses, the percentage of progenitors (represented by the Sox2+/To-pro3+ ratio) declines from ~72% at e10.5 to ~14% at e13.5 in WT embryos (Fig. 3G). Similarly, the percentage of S-phase progenitors (represented by the BrdU+/Sox2+ ratio) is reduced from ~41% at e10.5 to ~8% at e13.5 in WT embryos (Fig. 3H ). These data demonstrate that a decline in progenitor numbers and their proliferative ability occurs normally during neurogenesis, and by e13.5 only a few progenitors with limited proliferative ability remain in the brachial level spinal cord of WT embryos (Fig. 3E).
A similar trend can be observed in the Gdf11−/− embryos but at a slower pace. Excess numbers of Sox2+ progenitors and BrdU+ S-phase progenitors were observed in Gdf11−/− embryos when compared to the controls (supplemental Fig. 2 E, F ), resulting in the higher percentage of progenitors observed at e10.5 (78%) through e13.5 (20%) in the Gdf11−/− mutants (Fig. 3G). The percentage of S-phase progenitors is similar between the mutants and the controls before e11.5 but is significantly higher in the Gdf11−/− mutant compared to the controls at e12.5 (40 vs 27%) and e13.5 (23 vs 8%) (Fig. 3H ). Examination of M-phase progenitors using an anti-phosphorylated histone H3 antibody also revealed a higher percentage of M-phase progenitors in the Gdf11−/− spinal cord at e12.5 and e13.5 (data not shown). The increased progenitor proliferative ability in the Gdf11−/− spinal cord persists into e15.5, when few proliferating progenitors are present in their WT littermates (supplemental Fig. 2 B), while no significant difference in either the percentage of progenitors or the percentage of S-phase progenitors was observed between Gdf11−/− and WT littermates at e9.5 (data not shown).
These data demonstrate that during neurogenesis, a reduction in the number and proliferative ability of spinal cord progenitors occurs following the peak period of Gdf11 expression and neuronal differentiation. In the absence of Gdf11, the progression of neurogenesis is delayed and progenitor cells maintain their proliferative ability for a longer period of time, resulting in an increased number of progenitors.
In addition to decreased neuronal differentiation and increased progenitor proliferation, decreased cell death could also cause an increase in progenitor numbers. We therefore used cleaved caspase3 as a marker for apoptotic cells to examine progenitor cell death in cross-sectioned spinal cord tissue from Gdf11−/− and WT littermates. No caspase3+ cells were detected in the progenitor domains of either Gdf11−/− or WT spinal cord at e11.5 or e12.5 (supplemental Fig. 3C–F). In contrast, a ~3-fold increase in caspase3+ progenitor cells was observed in Gdf11−/− spinal cord compared to the controls at e10.5 (supplemental Fig. 3 A, B). We do not know why increased progenitor apoptosis is only present in Gdf11−/− embryos at e10.5. One possibility is that some of the differentiated progenitors could die if they cannot exit cell cycle in the absence of Gdf11. Alternatively, this could represent a compensatory mechanism that limits the numbers of progenitors present in Gdf11−/− embryos.
Gdf11 facilitates the temporal progression of neurogenesis
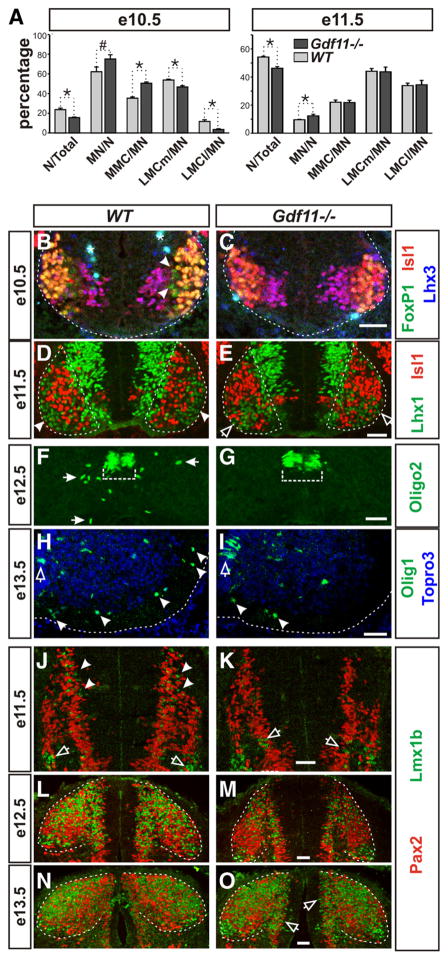
Our data to date demonstrate that Gdf11 affects the pace of neuronal differentiation and the maintenance of progenitor proliferation ability and, thus, suggest that Gdf11 might have a function in controlling the temporal progression of neurogenesis. To evaluate this possibility, we first examined progenitors located in the pMN domain that give rise to subtypes of MNs and oligodendrocytes in a sequential manner. At e9.5, similar numbers of HB9+ MNs are detected in somite number-matched Gdf11−/− and WT littermates (supplemental Fig. 4 A, B, available at www.jneurosci.org as supplemental material), indicating that the onset of neuronal differentiation is not affected by the absence of Gdf11. Examination of cross-sectioned e10.5 brachial spinal cord labeled with Isl1, Lhx3, and FoxP1 revealed the presence of early born medial motor column (MMC) MNs (Isl1+ Lhx3+) (Tsuchida et al., 1994) and LMCm MNs (Isl1+ FoxP1+) (Dasen et al., 2008; Rousso et al., 2008) in both WT and Gdf11−/− embryos, but the later-born LMCl MNs (Isl1− FoxP1+) are only present in WT embryos (Fig. 4 B, C). Quantification of seven pairs of Gdf11−/− and WT littermates revealed that fewer neurons (deduced from the numbers of Topro3+ cells/section minus the numbers of Sox2+ cells/section) were generated in Gdf11−/− embryos compared to the controls at e10.5 (15 vs 24% of Topro3+ cells). However, being the first types of neurons generated in the spinal cord, the percentage of MNs (deduced from the total numbers of Isl1+ Lhx3+, Isl1+ FoxP1+, and Isl1− FoxP1+ cells) to all neurons is higher in the Gdf11−/− embryos compared to the controls (75 vs 62%,) (Fig. 4 A). Examination of MN subtype composition in the Gdf11−/− embryos and the controls revealed a bias toward the earlier born MMC and LMCm MNs in the Gdf11−/− embryos and indicated a delay in the progression of MN subtype generation (Fig. 4 A).
Figure 4.
Slower progression of neurogenesis in the Gdf11−/− embryonic spinal cord. A, Percentage of neurons to total cells (N/Total), motor neurons to neurons (MN/N), and different subtypes of MNs to MNs (MMC/MN, LMCm/MN, and LMCl/MN) in Gdf11−/− and WT littermates at e10.5 and e11.5. *p <0.05, #p =0.05. B–G, Images of cross-sectioned ventral spinal cords stained with MN and OLP markers. B, C, White dotted lines delineate the margin of the spinal cord. Arrowheads indicate the presence of late born LMCl MNs (FoxP1+ Isl1− Lhx3− ) in the WT embryos. Asterisk (*) indicates nonspecific staining. D, E, White dotted lines delineate the ventral horns. Late born LMCl MNs (Lhx1+ Isl1− ) have migrated through LMCm MNs (Lhx1− Isl1+) to form the LMCl motor column (arrowheads) at the lateral border of the ventral horn in the WT embryo. In the Gdf11−/− spinal cords, the LMCl MNs are still migrating and just starting to reach their final positions (open arrows). F, G, Olig2+ OLPs (white arrows) have started to leave the progenitor domain (white dotted bracket) in WT embryos but not in their Gdf11−/− littermates at e12.5. H, I, Images of right ventral spinal cords, midline is at the left edge of the images and white dotted lines delineate the margin of the spinal cord. Many more Olig1+ OLPs (arrowheads) have migrated into the white matter in the WT compared to the Gdf11−/− embryos. Open arrows indicate the Olig1+ OLPs located in the progenitor domain. J–O, Images of cross-sectioned dorsal spinal cords stained with Pax2 and Lmx1b. J, K, Many Lmx1b+ late-born dILB interneurons (arrowheads) are present in the WT, but few are in Gdf11−/− embryos where Lmx1b+ early born dI5 interneurons (open arrows) are still being generated. L, M, A change in the shape of the dorsal horn (white dotted lines) in WT embryos caused by the influx of Lmx1b+ dILB and Pax2+ dILA interneurons is apparent compared to their Gdf11−/− littermates. N, O, Most of the Lmx1b+ dILB and Pax2+ dILA interneurons have finished their migration into the dorsal horn (white dotted lines) in WT embryos, but many are still being generated and migrating (open arrows) in their Gdf11−/− littermates. Scale bars, 50 μm in all images.
Quantification performed on e11.5 embryos revealed a similar MN subtype composition in Gdf11−/− embryos and the controls (Fig. 4 A). However, a qualitative difference can be observed at this stage as the late-born Lhx1+ Isl1− LMCl MNs (Tsuchida et al., 1994) have already migrated to the lateral margin of the ventral horn and formed LMCl columns in WT embryos (Fig. 4 D), but in contrast the Lhx1+ Isl1− LMCl MNs are still migrating and are intermingled with the Isl1+ Lhx1− MNs in Gdf11−/− spinal cords (Fig. 4 E). The LMCl columns become apparent in Gdf11−/− embryos a day later at e12.5 (data not shown), indicating a slower progression of MN differentiation. This prolonged period of MN differentiation also caused a delay in the onset of OLP generation as indicated by the lack of Olig2+, Olig1+, and Sox10+ OLPs (Zhou et al., 2000) outside the progenitor domains in e12.5 Gdf11−/− spinal cords when compared to the controls (Fig. 4 F, G; supplemental Fig. 4 E–H). By e13.5, more Olig1+ OLPs have migrated into the white matter in the WT embryos compared to their Gdf11−/− littermates (Fig. 4 H, I ).
A slower progression of neurogenesis was also observed in the dorsal spinal cord of Gdf11−/− embryos. Progenitors in the dorsal spinal cord first give rise to six types of early born interneurons (dI1–dI6) followed by two types of late born interneurons (dILA and dILB). Using antibodies against Lmx1b, which labels early born dI5 and late born dILB neurons, and Pax2, which labels dI4 and dI6 as well as dILA neurons, we examined the generation of these interneurons in Gdf11−/− embryos. At e10.5, similar patterns of Lmx1b+ dI5 and Pax2+ dI4 and dI6 neurons were observed in WT and Gdf11−/− embryos (supplemental Fig. 4C,D). At e11.5, Lmx1b+ dILB neurons started to migrate out of the progenitor zone in WT (Fig. 4 J), but not in Gdf11−/− embryos where Lmx1b+ dI5 neurons are still being generated (Fig. 4 K). By e12.5, the migration of Lmx1b+ dILB and Pax2+ dILA neurons into the dorsal horns in the control embryos caused a shape change of the dorsal horn that was not observed in their Gdf11−/− littermates (Fig. 4 L, M ). The generation of Lmx1b+ dILB neurons and their migration are nearly complete in the WT embryos by e13.5 (Fig. 4 N), but their generation and migration are still in progress in Gdf11−/− embryos (Fig. 4O). This delay in the progression of neuronal differentiation also caused a delay in the onset of oligodendrocyte generation in the dorsal spinal cord, as few Oligo2+ OLPs were present in Gdf11−/− embryos at e15.5 compared to the controls (supplemental Fig. 4 I, J ). The delayed OLP generation and migration does not seem to affect their later differentiation, as MBP+ cells can be detected in both Gdf11−/− and control spinal cord at e18.5 (supplemental Fig. 4 K, L).
These results demonstrate that Gdf11 has a function in facilitating temporal progression of neurogenesis in the developing spinal cord, but not in initiating neuronal differentiation. Without Gdf11, neurogenesis proceeds at a slower rate and continues for a longer period of time, resulting in a delay in the onset of gliogenesis.
Gdf11 induced changes in progenitor properties can be maintained in vitro
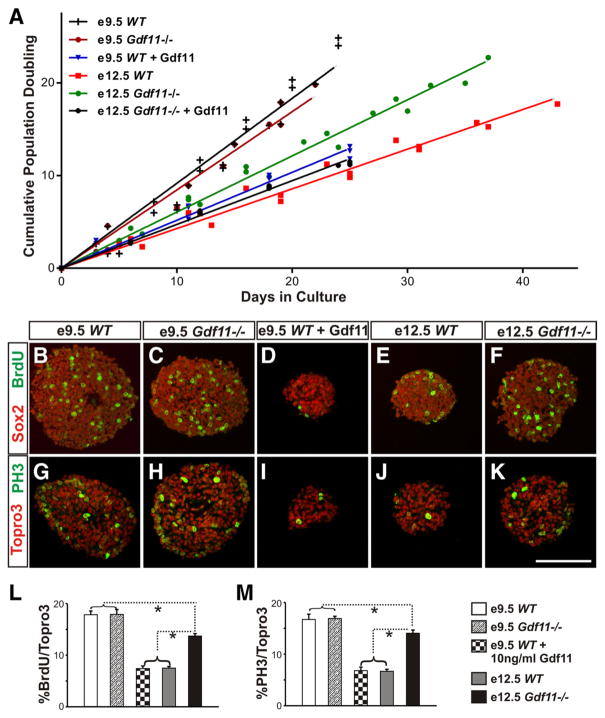
Our data to date indicate that Gdf11 can cause lasting changes in progenitor properties— differences in progenitor differentiation and proliferation abilities between WT and Gdf11−/− embryos remain long after the cessation of Gdf11 expression. We reason that if the transient expression of Gdf11 can indeed cause a lasting change in progenitor cell properties, these differences should be maintained in culture. We therefore isolated neural progenitors from brachial level spinal cord of WT and Gdf11−/− littermates at e9.5 (when little Gdf11 signal has been received) and e12.5 (after the peak of Gdf11 expression) and cultured them in vitro. Under our culture conditions (see Materials and Methods), the majority of neurosphere cells are Sox2+ progenitor cells (supplemental Fig. 5A–E, available at www.jneurosci.org as supplemental material) and do not express Gdf11 (supplemental Fig. 5 K, L). The population doubling time (PDT) is very similar between e9.5 Gdf11−/− and WT neurospheres (~24 h). However, a significant difference in growth rate ( p < 0.05) was observed between neurospheres derived from e12.5 WT (PDT, ~58 h) and Gdf11−/− (PDT, ~41 h) embryos (Fig. 5A). Addition of Gdf11 at 10 ng/ml to e9.5 WT and e12.5 Gdf11−/− neurospheres dramatically reduced their growth rate (PDTs, ~51 h and ~52 h, respectively) (Fig. 5A), but has little effect on the growth rate of e12.5 WT neurospheres (PDT, ~60 h). The slower growth observed in e12.5 neurospheres and in neurospheres cultured with Gdf11 is not caused by increased cell death or cell differentiation, as the percentage of cleaved caspase3+/Topro3+ cells and Sox2+/ Topro3+ cells are similar in all neurospheres (supplemental Fig. 5A–J). In contrast, a lower percentage of S-phase (BrdU+/Topro3+) and M-phase (PH3+/ Topro3+) cells were observed in neurospheres derived from e12.5 WT embryos and e9.5 WT embryos cultured with Gdf11 (Fig. 5B–K ). Quantification obtained from three sets of cultures revealed similar percentages of S-phase and M-phase cells present in neurospheres derived from e9.5 WT and Gdf11−/− embryos (~17%), followed by neurospheres derived from e12.5 Gdf11−/− embryos (~14%) (Fig. 5 L, M ). A significant reduction in the percentage of S-phase and M-phase cells ( p < 0.001) was observed in neurospheres derived from e12.5 WT and from e9.5 WT embryos cultured with Gdf11 (~7%, Fig. 5 L, M ). Because no increase in cell death or cell differentiation was observed, we reason that the reduction of S-phase and M-phase cells and the increased PDT in e12.5 WT neurospheres, as well as in e9.5 WT and e12.5 Gdf11−/− neurospheres cultured with Gdf11, most likely reflects a decrease in progenitor proliferative ability. These data demonstrate that the in vivo action of Gdf11 causes a reduction in the proliferative ability of spinal progenitors that can be maintained in culture and be mimicked by the addition of Gdf11 in vitro.
Figure 5.
Difference in proliferation ability between neurospheres derived from Gdf11−/− and WT embryos. A, Growth curves of neurospheres derived from brachial level spinal cord of e9.5 and e12.5 WT and Gdf11−/− embryos. No difference in growth rate was observed between WT and Gdf11−/− neurospheres derived from e9.5 embryos. Significant differences ( p <0.05) in growth rate are observed between WT and Gdf11−/− neurospheres derived from e12.5 embryos and between neurospheres derived from e9.5 and e12.5 embryos. Addition of Gdf11 at 10 ng/ml to e9.5 WT and e12.5 Gdf11−/− neurosphere cultures effectively reduced their growth rate to that comparable to the e12.5 WT neurospheres. B–F, Sox2 and BrdU staining in cultured neurospheres labeled with BrdU for 30 min. G–K, Topro3 and PH3 labeling in cultured neurospheres. Scale bar, 100 μm. L, M, Percentage of S-phase (L) or M-phase (M) cells in neurospheres. Neurospheres derived from e9.5 embryos have the highest percentage of S-phase or M-phase cells, followed by neurospheres derived from e12.5 Gdf11−/− embryos. Neurospheres derived from either e12.5 WT embryos or from e9.5 WT embryos cultured with 10 ng/ml Gdf11 have the lowest percentage of S-phase or M-phase cells.*p <0.001.
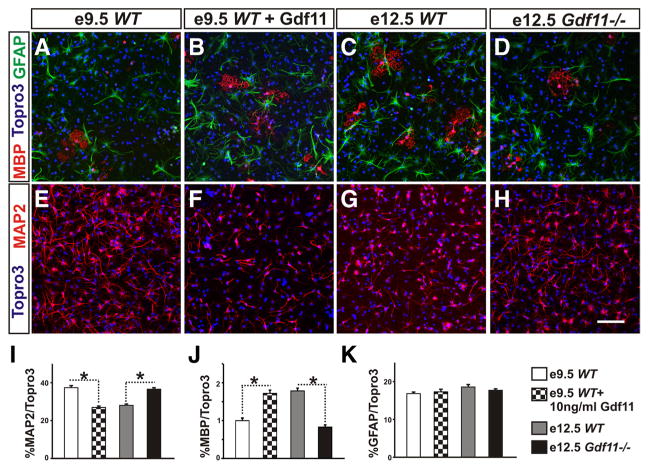
To evaluate the effect of Gdf11 on progenitor differentiation potential, passage 3 neurospheres were dissociated and cultured in differentiation media (see Materials and Methods) for 3 d. Examination of a neuronal marker (MAP2), an oligodendrocyte marker (MBP), and an astrocyte marker (GFAP) revealed a difference in differentiation potential between neurospheres derived from embryos with different genotypes and from different stages (Fig. 6 A–H ). Under our culture conditions, neurospheres derived from e9.5 WT and e12.5 Gdf11−/− embryos differentiated mostly into neurons (~37%) and a small percentage of oligodendrocytes (~0.7%) (Fig. 6 I, J ). In contrast, the percentage of neurons differentiated from e12.5 WT neurospheres and e9.5 WT or e12.5 Gdf11−/− neurospheres cultured with Gdf11 is reduced significantly (~26 –28%), while their ability to produce oligodendrocytes increased >2-fold (~1.8 –2.0%) (Fig. 6 I, J; supplemental Fig. 6, available at www.jneurosci.org as supplemental material). Addition of Gdf11 to e12.5 WT neurosphere culture did not cause significant change (supplemental Fig. 6), and no significant difference in astrocyte differentiation was found among these neurospheres (Fig. 6K; supplemental Fig. 6G).
Figure 6.
Difference in differentiation potential between neurospheres derived from Gdf11−/− and WT embryos A–D, MBP (for oligodendrocytes), GFAP (for astrocytes), and Topro3 (for all nuclei) staining of dissociated neurospheres grown in differentiation media for 3 d. E–H, MAP2 (for neurons) and Topro3 staining of dissociated neurospheres grown in differentiation media for 3 d. Scale bar, 100 μm. I–K, Percentage of neurons, oligodendrocytes, or astrocytes differentiated from dissociated neurosphere cells after 3 d in culture. Significantly more neurons and less oligodendrocytes (*p < 0.001) are produced by neurospheres derived from e9.5 WT and e12.5 Gdf11−/− embryos when compared to neurospheres that received Gdf11 signals either in vitro (e9.5 WT neurospheres cultured with 10 ng/ml Gdf11) or in vivo (neurospheres derived from e12.5 WT embryos).
These results demonstrate that neurospheres derived from e9.5 WT embryos that received little Gdf11 signal in vivo, as well as those derived from e12.5 Gdf11−/− embryos, have a greater ability to differentiate into neurons in vitro. In contrast, neurospheres derived from e12.5 WT embryos that have received significant amounts of Gdf11 signals in vivo exhibit an increased ability to differentiate into oligodendrocytes and a reduced ability to differentiate into neurons. This effect of Gdf11 on progenitor differentiation can be simulated in vitro by culturing e9.5 WT or e12.5 Gdf11−/− neurospheres in the presence of Gdf11.
Gdf11 downregulates Pax6 expression in neural progenitors
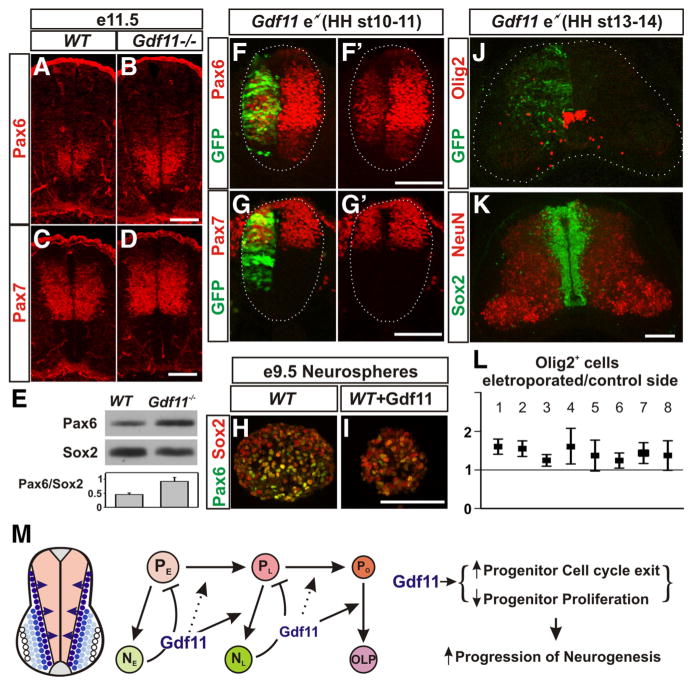
In searching for downstream effectors that could mediate Gdf11 function in causing lasting changes in spinal progenitors, we examined the expression patterns of various transcription factors that are known to be expressed in the progenitor domains of the developing spinal cord. Among the factors examined, Pax6 is expressed initially in a broader region along the D-V axis, but its expression is later restricted to the intermediate zone as neurogenesis progresses (supplemental Fig. 7, available at www.jneurosci.org as supplemental material). A higher level of Pax6 expression was detected in Gdf11−/− embryos when compared to the controls from e10.5 and persisted through e12.5, the last stage examined (Fig. 7 A, B; supplemental Fig. 7). In contrast, no apparent difference in the expression of Pax7, Olig2, and Nkx6.1, three transcription factors expressed in progenitor domains overlapping with the Pax6 domain, was observed between Gdf11−/− embryos and WT littermates (Fig. 7C,D; supplemental Fig. 7G; data not shown). Western analyses performed with e11.5 brachial level spinal cord tissue revealed that the Pax6 level in Sox2+ progenitors is ~2-fold higher in Gdf11−/− embryos compared to the controls (Fig. 7E).
Figure 7.
Gdf11 downregulates Pax6 expression. A–D, Pax6 (A, B) and Pax7 (C, D) expression in cross-sectioned brachial level spinal cord from e11.5 WT and Gdf11−/− embryos. Fewer Pax6+ cells with lower levels of Pax6 expression were observed in WT embryos (A) when compared to Gdf11−/− littermates (B). Pax7 expression is similar between WT (C) and Gdf11−/− embryos (D). E, Western analyses of brachial level spinal cord from e11.5 embryos revealed an ~2-fold difference in the Pax6 expression level in Sox2+ progenitors between Gdf11−/− and WT embryos. F–G′, Pax6 (F, F′) and Pax7 (G, G′) expression in cross-sectioned spinal cord from HH stage 17 chick embryos electroporated with a Gdf11 expression construct at HH stage 10 –11. White dotted lines delineate the margin of the spinal cord. The Pax6 level in the electroporated side (marked by GFP expression) is reduced significantly. H, I, Lower levels of Pax6 expression in e9.5 WT neurospheres cultured with 10 ng/ml Gdf11. J, K, Olig2 and GFP (J) and Sox2 and NeuN (K) expression in cross-sectioned brachial level spinal cord from e7 chick embryos harvested 5 d after Gdf11 electroporation at HH stage 13–14. The white dotted lines delineate the margin of the spinal cord in J. Scale bar, 100 μm in all images. L, Quantification of Olig2+ cells in the electroporated side compared to the control side of eight individual embryos (plots represent mean ±95% confidence to exhibit the degree of variability). Values above one indicate more Olig2+ cells are present in the electroporated side. M, A model for Gdf11 function in controlling the temporal progression of neurogenesis. Gdf11 is expressed transiently in the newly born neurons located lateral to the progenitor domains in the spinal cord. It acts as a feedback signal on neighboring progenitors to promote the transition from early progenitors (PE) that produce early born neurons (NE) to late progenitors (PL) that produce late-born neurons (NL), and to oligodendrocyte progenitors (PO) that produce oligodendrocyte precursors (OLP) by enhancing cell cycle exit and inhibiting progenitor proliferation. This results in facilitating the progression of neurogenesis in the developing spinal cord.
The ability of Gdf11 to downregulate Pax6 expression is further demonstrated by misexpressing Gdf11 in chick embryonic spinal cord using in ovo electroporation. Misexpression of Gdf11 affects the proliferation of neural progenitors and reduces the size of the neural tube at the electroporated side (Fig. 7F–G′ (Liu, 2006). Therefore, a high level of Gdf11 expression could cause deformation of the neural tube, and such embryos were excluded from further analyses. The majority of embryos harvested between HH stage 15–17 (10/16) and HH stage 21–22 (7/9) had relatively normal looking spinal cords, albeit smaller at the electroporated side, and exhibited decreased levels of Pax6 expression (Fig. 7 F, F′;data not shown). In contrast, Pax7 expression is not affected significantly by Gdf11 misexpression (Fig. 7G,G′; data not shown). Moreover, addition of 10 ng/ml Gdf11 to cultured neurospheres isolated from e9.5 WT brachial level spinal cord also downregulated Pax6 expression in vitro (Fig. 7 H, I ). These results demonstrate that Gdf11 facilitates the downregulation of Pax6 expression in the spinal progenitors.
Misexpression of Gdf11 in early chick embryos caused a reduction in neural progenitors and precluded the examination of the effect of Gdf11 on OLP generation. To examine the effect of Gdf11 on OLP differentiation, we modified our approach by performing experiments in older embryos (HH stage 13–14) with a lower concentration of plasmids (0.5 mg/ml) and only analyzed the brachial level spinal cord where neurogenesis is more advanced. Under these conditions, only a limited difference in tissue size and cell number is present between the electroporated and the control side of the spinal cord (Fig. 7K ), and a low level (~1.5-fold) but consistent increase in Oligo2+ OLPs after Gdf11 electroporation can be observed (Fig. 7J ). Quantification (plotted as mean ± 95% confidence) on serially sectioned spinal cord tissue obtained from eight electroporated embryos revealed a correlation between the level of Gdf11 expression (represented by GFP marker expression) and the increase in Olig2+ OLPs. In those embryos that have higher and more uniform GFP expression throughout the spinal cord (Fig. 7 J, L, embryos 1 and 2), more than a 1.5-fold increase in the number of Olig2+ and Sox10+ OLPs was observed in the electroporated side compared to the control side (Fig. 7J; supplemental Fig. 8E, available at www.jneurosci.org as supplemental material). Higher variability in Olig2+ OLPs numbers is apparent in embryos with splotchy high GFP expression interrupted by areas with no GFP expression (Fig. 7L, embryos 4, 5, 7, 8), while a ~1.25-fold increase in OLPs were observed in embryos with a low level of GFP expression (Fig. 7L, embryos 3, 6). These results demonstrate that Gdf11 has the ability to influence progenitors to differentiate into OLPs in vivo.
Discussion
Gdf11 affects multiple aspects of spinal cord development
Previously, we demonstrated that the early expression of Gdf11 in the node/tail bud region synergizes with Fgf8 signals to induce caudal Hox gene expression for proper R-C patterning of the spinal cord (Liu, 2006). In this study, we examined a later function of Gdf11, which is mediated by its transient expression in newly generated neurons in the developing spinal cord.
Our current data suggest that Gdf11 secreted by newly born neurons has multiple effects on neighboring progenitors during neurogenesis. First, Gdf11 has a more direct function in promoting cell cycle exit. Significantly lower percentages of progenitors are leaving the cell cycle in Gdf11−/− embryos compared to their WT littermates at e10.5–e11.5, but not at e12.5, a time correlating with the peak period of Gdf11 expression, and this effect is most likely mediated by the ability of Gdf11 to up-regulate p27 kip1 and p57 kip2. As a result, a decreased percentage of neurons along with a delay in the generation of late-born LMCl MNs was observed in Gdf11−/− embryos at this stage.
In addition, Gdf11 also has indirect effects on progenitors that result in changes in their differentiation and proliferation properties that remain in the progenitors after the cessation of Gdf11 expression. A change in progenitor differentiation properties, in conjunction with the effect of Gdf11 in promoting cell cycle exit, could facilitate the temporal progression of neurogenesis. Thus, a slower and prolonged period of neuronal differentiation associated with a delay in gliogenesis was observed in the Gdf11−/− embryos. The delayed onset of OLP generation did not seem to reduce the total numbers of OLPs present in the spinal cord toward the end of embryonic development or their differentiation (supplemental Fig. 4 K, L). The excess numbers of progenitor cells in the mutants could give rise to more OLPs over time to compensate for the delay. Alternatively, the OLPs in Gdf11−/− embryos might divide more often to make up for the difference.
The excess numbers of progenitors observed in Gdf11−/− embryos could potentially be the result of reduced cell cycle exit in the absence of Gdf11. However, our BrdU labeling data, along with our observation that many progenitors still maintain their proliferative ability in the Gdf11−/− embryos at e15.5, argue for an independent function of Gdf11 in inhibiting progenitor proliferation. Moreover, an increased population doubling time along with reduced percentages of S-phase and M-phase progenitor cells were observed without any induction of cellular differentiation in cultured neurospheres that have received Gdf11 signals either in vivo or in vitro, further demonstrating that the ability of Gdf11 to inhibit cell proliferation is independent from its function in promoting cell differentiation.
These effects of Gdf11 in the developing spinal cord are similar to those observed in the olfactory epithelium (OE) and the developing retina. In the OE, Gdf11 is expressed in the olfactory receptor neurons (ORNs) and their progenitors. Gdf11 determines the numbers of ORNs by upregulating p27 kip1 expression and blocking cell division in a transit-amplifying progenitor population (INPs). In Gdf11−/− embryos, an increase in the numbers of BrdU+ proliferating INPs, as well as increased numbers of ORNs was observed in the OE (Wu et al., 2003). In the developing retina, Gdf11 controls the numbers of retinal ganglion cells (RGCs), amacrine cells, and rod cells by affecting the temporal competence of their progenitor cells, but not progenitor proliferation. In Gdf11−/− embryos, the temporal window for generating RGCs is extended and, therefore, excess numbers of RGCs are generated at the expense of amacrine and rod cells (Kim et al., 2005). Our results demonstrate that in the developing spinal cord both progenitor proliferation and the temporal progression of neuronal differentiation are affected by Gdf11.
Effect of Gdf11 on progenitor properties could be partially mediated by its ability to downregulate Pax6 expression
Gdf11-induced changes in progenitor property can be preserved in culture, suggesting an involvement of transcriptional changes of downstream effectors. We do not know the exact identifiers of these downstream effectors at present; however, our data indicate that Pax6 is a potential candidate.
Higher levels of Pax6 expression are observed in Gdf11−/− embryos compared to their WT littermates. Pax6 is expressed widely in neural progenitors, including the developing cerebral cortex, retina, and spinal cord, and the involvement of Pax6 in both progenitor proliferation and neuronal differentiation has been demonstrated (Osumi et al., 2008). In the developing spinal cord the initially broad expression of Pax6 is later confined by the inhibitory action of bone morphogenetic protein signals from the roof plate (Timmer et al., 2002) and Shh signals from the floor plate to the intermediate region, where it is maintained by the retinoic acid signal from the somites (Diez del Corral et al., 2003). In addition, both positive and negative auto-regulation further controls the level of Pax6 expression (Aota et al., 2003; Kleinjan et al., 2004; Manuel et al., 2007). The ability of Gdf11 to suppress Pax6 expression is most likely mediated through its binding to the activin receptors and the subsequent phosphorylation of Smad2/3, as activin A has been shown to inhibit Pax6 expression in cultured spinal cord explants (Pituello et al., 1995), and the MH1 domain of Smad3 can interact with Pax6 to repress its expression in a ligand-dependent manner (Grocott et al., 2007).
The function of Pax6 in determining the spatial identities of neurons located in the ventral spinal cord has been well studied (Ericson et al., 1997; Briscoe et al., 2000). More recently, Pax6 was shown to promote neuronal commitment in the developing spinal cord by upregulating Neurogenin2 (Ngn2) expression and thus, facilitating the transition of progenitor cells to neuronal precursors. However, in order for the neuronal precursors to differentiate into neurons it is essential to turn off Pax6 expression, and this is achieved, at least in part, through the feedback inhibition of Pax6 by Ngn2 (Bel-Vialar et al., 2007). Indeed, precocious and excessive neuronal differentiation has been observed in Pax6−/− mouse embryos (Bel-Vialar et al., 2007), supporting the idea that prolonged expression of Pax6 in the Gdf11−/− spinal cord could reduce the rate and increase the period of neuronal differentiation.
In addition, it is proposed that the MN-producing period of the pMN domain is governed by the coexpression of Olig2 with Pax6 and Ngn1/Ngn2 (Kessaris et al., 2001). A downregulation of Pax6, Ngn1, and Ngn2 expression, along with an expansion of Nkx2.2 expression from the neighboring p3 progenitor domain, coincides with the switch from MN to OLP production. The observation of ectopic and precocious generation of OLPs in the Pax6−/− mice (Sun et al., 1998; Sugimori et al., 2007) suggests that Gdf11 could also promote OLP production by downregulating Pax6 expression.
We have attempted to manipulate Pax6 expression by electroporating Pax6 RNA interference plasmids (pRFPRNAi-Pax6A and pRFPRNAi-Pax6B) (Das et al., 2006) into chick embryos at HH stage 13–14. Unfortunately, these experiments provided limited information, as pRFPRNAi-Pax6A expression had no significant effect on either Pax6 expression or Olig2+ OLP numbers (data not shown), whereas pRFPRNAi-Pax6B expression caused a significant reduction in Pax6 expression level and resulted in the loss of Sox2+ neural progenitors (supplemental Fig. 8AD), therefore prohibiting further analyses. Although we cannot provide direct proof of the involvement of Pax6 in mediating Gdf11 function at present, the fact that Gdf11 is able to reduce the Pax6 expression level in conjunction with the known function of Pax6 in progenitor proliferation/differentiation suggests that Pax6 could at least mediate certain aspects of the effect of Gdf11 on neural progenitors.
A model for Gdf11 function in controlling the temporal progression of neurogenesis and progenitor proliferation in the developing spinal cord
Based on the results presented, we propose a model in which Gdf11 secreted by newly born neurons in the developing spinal cord acts as a positive feedback signal on the neighboring progenitors to enhance their cell cycle exit, limit their proliferation, and facilitate the transition from producing early born neurons to late born neurons and, finally, to OLPs (Fig. 7M ). These functions of Gdf11 are mediated, at least in part, by its ability to upregulate p27 kip1 and p57 kip2 and downregulate Pax6 expression, and by additional, as yet unknown targets of Gdf11.
In our model, Gdf11 has no effect on the onset of neurogenesis, and this is in accordance with the following observations: (1) Gdf11 is expressed in newly differentiated neurons but not in progenitor cells; (2) the onset of neuronal differentiation is not delayed in Gdf11−/− embryos; and (3) misexpression of Gdf11 in chick embryos by electroporation did not induce precocious neuronal differentiation (our unpublished observation).
In addition, our data also suggest that Gdf11 is not the only factor that affects the progenitor proliferation and temporal progression of neurogenesis, as the progenitor proliferation rate still decreases, neuronal differentiation still progresses, and OLPs are eventually generated in the Gdf11−/− embryos, albeit with a delay. Although we do not yet know the identities of these additional factors, our data demonstrate that they cannot completely compensate for Gdf11 function in the Gdf11−/− embryos and suggest an essential role for Gdf11 in spinal cord neurogenesis.
Supplementary Material
Acknowledgments
This study was supported by National Institutes of Health Grant R01 NS045933 to J.-P. L. We thank S.-J. Lee for providing the Gdf11 knock-out mice and in situ probes; J. Dasen, M. Goulding, and T. Jessell, for providing various antibodies; S. Medrano for technical advice in neurosphere culture; and M. Barnett for assistance in cell counting. We also thank S. Zeitlin, H. Scrable, B. Winckler, and B. Novitch for critical reading this manuscript.
References
- Andersson O, Reissmann E, Ibáñez CF. Growth differentiation factor 11 signals through the transforming growth factor-beta receptor ALK5 to regionalize the anterior-posterior axis. EMBO Rep. 2006;7:831–837. doi: 10.1038/sj.embor.7400752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aota S, Nakajima N, Sakamoto R, Watanabe S, Ibaraki N, Okazaki K. Pax6 autoregulation mediated by direct interaction of Pax6 protein with the head surface ectoderm-specific enhancer of the mouse Pax6 gene. Dev Biol. 2003;257:1–13. doi: 10.1016/s0012-1606(03)00058-7. [DOI] [PubMed] [Google Scholar]
- Baxter MA, Wynn RF, Jowitt SN, Wraith JE, Fairbairn LJ, Bellantuono I. Study of telomere length reveals rapid aging of human marrow stromal cells following in vitro expansion. Stem Cells. 2004;22:675–682. doi: 10.1634/stemcells.22-5-675. [DOI] [PubMed] [Google Scholar]
- Bel-Vialar S, Medevielle F, Pituello F. The on/off of Pax6 controls the tempo of neuronal differentiation in the developing spinal cord. Dev Biol. 2007;305:659– 673. doi: 10.1016/j.ydbio.2007.02.012. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Pierani A, Jessell TM, Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–445. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- Cai J, Qi Y, Hu X, Tan M, Liu Z, Zhang J, Li Q, Sander M, Qiu M. Generation of oligodendrocyte precursor cells from mouse dorsal spinal cord independent of Nkx6 regulation and Shh signaling [see comment] Neuron. 2005;45:41–53. doi: 10.1016/j.neuron.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Das RM, Van Hateren NJ, Howell GR, Farrell ER, Bangs FK, Porteous VC, Manning EM, McGrew MJ, Ohyama K, Sacco MA, Halley PA, Sang HM, Storey KG, Placzek M, Tickle C, Nair VK, Wilson SA. A robust system for RNA interference in the chicken using a modified microRNA operon. Dev Biol. 2006;294:554–563. doi: 10.1016/j.ydbio.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Dasen JS, De Camilli A, Wang B, Tucker PW, Jessell TM. Hox repertoires for motor neuron diversity and connectivity gated by a single accessory factor, FoxP1. Cell. 2008;134:304–316. doi: 10.1016/j.cell.2008.06.019. [DOI] [PubMed] [Google Scholar]
- Desai AR, McConnell SK. Progressive restriction in fate potential by neural progenitors during cerebral cortical development. Development. 2000;127:2863–2872. doi: 10.1242/dev.127.13.2863. [DOI] [PubMed] [Google Scholar]
- Diez del Corral R, Storey KG. Opposing FGF and retinoid pathways: a signalling switch that controls differentiation and patterning onset in the extending vertebrate body axis. Bioessays. 2004;26:857–869. doi: 10.1002/bies.20080. [DOI] [PubMed] [Google Scholar]
- Diez del Corral R, Olivera-Martinez I, Goriely A, Gale E, Maden M, Storey K. Opposing FGF and retinoid pathways control ventral neural pattern, neuronal differentiation, and segmentation during body axis extension. Neuron. 2003;40:65–79. doi: 10.1016/s0896-6273(03)00565-8. [DOI] [PubMed] [Google Scholar]
- Ericson J, Rashbass P, Schedl A, Brenner-Morton S, Kawakami A, van Heyningen V, Jessell TM, Briscoe J. Pax6 controls progenitor cell identity and neuronal fate in response to graded Shh signaling. Cell. 1997;90:169–180. doi: 10.1016/s0092-8674(00)80323-2. [DOI] [PubMed] [Google Scholar]
- Frantz GD, McConnell SK. Restriction of late cerebral cortical progenitors to an upper-layer fate. Neuron. 1996;17:55–61. doi: 10.1016/s0896-6273(00)80280-9. [DOI] [PubMed] [Google Scholar]
- Gamer LW, Wolfman NM, Celeste AJ, Hattersley G, Hewick R, Rosen V. A novel BMP expressed in developing mouse limb, spinal cord, and tail bud is a potent mesoderm inducer in Xenopus embryos. Dev Biol. 1999;208:222–232. doi: 10.1006/dbio.1998.9191. [DOI] [PubMed] [Google Scholar]
- Grocott T, Frost V, Maillard M, Johansen T, Wheeler GN, Dawes LJ, Worm-stone IM, Chantry A. The MH1 domain of Smad3 interacts with Pax6 and represses autoregulation of the Pax6 P1 promoter. Nucleic Acids Res. 2007;35:890–901. doi: 10.1093/nar/gkl1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui H, Li S, Matise MP. A cell-autonomous requirement for Cip/Kip cyclin-kinase inhibitors in regulating neuronal cell cycle exit but not differentiation in the developing spinal cord. Dev Biol. 2007;301:14–26. doi: 10.1016/j.ydbio.2006.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. J Morphol. 1951;88:49–92. [PubMed] [Google Scholar]
- Helms AW, Johnson JE. Specification of dorsal spinal cord interneurons. Curr Opin Neurobiol. 2003;13:42–49. doi: 10.1016/s0959-4388(03)00010-2. [DOI] [PubMed] [Google Scholar]
- Hollyday M, Hamburger V. An autoradiographic study of the formation of the lateral motor column in the chick embryo. Brain Res. 1977;132:197–208. doi: 10.1016/0006-8993(77)90416-4. [DOI] [PubMed] [Google Scholar]
- Kessaris N, Pringle N, Richardson WD. Ventral neurogenesis and the neuron-glial switch. Neuron. 2001;31:677–680. doi: 10.1016/s0896-6273(01)00430-5. [DOI] [PubMed] [Google Scholar]
- Kim J, Wu HH, Lander AD, Lyons KM, Matzuk MM, Calof AL. GDF11 controls the timing of progenitor cell competence in developing retina. Science. 2005;308:1927–1930. doi: 10.1126/science.1110175. [DOI] [PubMed] [Google Scholar]
- Kleinjan DA, Seawright A, Childs AJ, van Heyningen V. Conserved elements in Pax6 intron 7 involved in (auto)regulation and alternative transcription. Dev Biol. 2004;265:462–477. doi: 10.1016/j.ydbio.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Liu JP. The function of growth/differentiation factor 11 (Gdf11) in rostrocaudal patterning of the developing spinal cord. Development. 2006;133:2865–2874. doi: 10.1242/dev.02478. [DOI] [PubMed] [Google Scholar]
- Lumsden A, Krumlauf R. Patterning the vertebrate neuraxis. Science. 1996;274:1109–1115. doi: 10.1126/science.274.5290.1109. [DOI] [PubMed] [Google Scholar]
- Mainprize TG, Taylor MD, Rutka JT, Dirks PB. Cip/Kip cell-cycle inhibitors: a neuro-oncological perspective. J Neurooncol. 2001;51:205–218. doi: 10.1023/a:1010671908204. [DOI] [PubMed] [Google Scholar]
- Manuel M, Georgala PA, Carr CB, Chanas S, Kleinjan DA, Martynoga B, Mason JO, Molinek M, Pinson J, Pratt T, Quinn JC, Simpson TI, Tyas DA, van Heyningen V, West JD, Price DJ. Controlled overexpression of Pax6 in vivo negatively autoregulates the Pax6 locus, causing cell-autonomous defects of late cortical progenitor proliferation with little effect on cortical arealization. Development. 2007;134:545–555. doi: 10.1242/dev.02764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matise M. A dorsal elaboration in the spinal cord. Neuron. 2002;34:491–493. doi: 10.1016/s0896-6273(02)00710-9. [DOI] [PubMed] [Google Scholar]
- McConnell SK, Kaznowski CE. Cell cycle dependence of laminar determination in developing neocortex. Science. 1991;254:282–285. doi: 10.1126/science.254.5029.282. [DOI] [PubMed] [Google Scholar]
- McPherron AC, Lawler AM, Lee SJ. Regulation of anterior/posterior patterning of the axial skeleton by growth/differentiation factor 11. Nat Genet. 1999;22:260–264. doi: 10.1038/10320. [DOI] [PubMed] [Google Scholar]
- Medrano S, Burns-Cusato M, Atienza MB, Rahimi D, Scrable H. Regenerative capacity of neural precursors in the adult mammalian brain is under the control of p53. Neurobiol Aging. 2009;30:483–497. doi: 10.1016/j.neurobiolaging.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima M, Toyono T, Akamine A, Joyner A. Expression of growth/differentiation factor 11, a new member of the BMP/TGFbeta superfamily during mouse embryogenesis. Mech Dev. 1999;80:185–189. doi: 10.1016/s0925-4773(98)00205-6. [DOI] [PubMed] [Google Scholar]
- Novitch BG, Chen AI, Jessell TM. Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron. 2001;31:773–789. doi: 10.1016/s0896-6273(01)00407-x. [DOI] [PubMed] [Google Scholar]
- Oh SP, Yeo CY, Lee Y, Schrewe H, Whitman M, Li E. Activin type IIA and IIB receptors mediate Gdf11 signaling in axial vertebral patterning. Genes Dev. 2002;16:2749–2754. doi: 10.1101/gad.1021802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osumi N, Shinohara H, Numayama-Tsuruta K, Maekawa M. Concise review: Pax6 transcription factor contributes to both embryonic and adult neurogenesis as a multifunctional regulator. Stem Cells. 2008;26:1663–1672. doi: 10.1634/stemcells.2007-0884. [DOI] [PubMed] [Google Scholar]
- Pituello F, Yamada G, Gruss P. Activin A inhibits Pax-6 expression and perturbs cell differentiation in the developing spinal cord in vitro. Proc Natl Acad Sci U S A. 1995;92:6952–6956. doi: 10.1073/pnas.92.15.6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Shen Q, Goderie SK, He W, Capela A, Davis AA, Temple S. Timing of CNS cell generation: a programmed sequence of neuron and glial cell production from isolated murine cortical stem cells. Neuron. 2000;28:69– 80. doi: 10.1016/s0896-6273(00)00086-6. [DOI] [PubMed] [Google Scholar]
- Rousso DL, Gaber ZB, Wellik D, Morrisey EE, Novitch BG. Coordinated actions of the forkhead protein Foxp1 and Hox proteins in the columnar organization of spinal motor neurons. Neuron. 2008;59:226–240. doi: 10.1016/j.neuron.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roztocil T, Matter-Sadzinski L, Alliod C, Ballivet M, Matter JM. NeuroM, a neural helix-loop-helix transcription factor, defines a new transition stage in neurogenesis. Development. 1997;124:3263–3272. doi: 10.1242/dev.124.17.3263. [DOI] [PubMed] [Google Scholar]
- Shen Q, Wang Y, Dimos JT, Fasano CA, Phoenix TN, Lemischka IR, Ivanova NB, Stifani S, Morrisey EE, Temple S. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nat Neurosci. 2006;9:743–751. doi: 10.1038/nn1694. [DOI] [PubMed] [Google Scholar]
- Sugimori M, Nagao M, Bertrand N, Parras CM, Guillemot F, Nakafuku M. Combinatorial actions of patterning and HLH transcription factors in the spatiotemporal control of neurogenesis and gliogenesis in the developing spinal cord. Development. 2007;134:1617–1629. doi: 10.1242/dev.001255. [DOI] [PubMed] [Google Scholar]
- Sun T, Pringle NP, Hardy AP, Richardson WD, Smith HK. Pax6 influences the time and site of origin of glial precursors in the ventral neural tube. Mol Cell Neurosci. 1998;12:228–239. doi: 10.1006/mcne.1998.0711. [DOI] [PubMed] [Google Scholar]
- Timmer JR, Wang C, Niswander L. BMP signaling patterns the dorsal and intermediate neural tube via regulation of homeobox and helix-loop-helix transcription factors. Development. 2002;129:2459–2472. doi: 10.1242/dev.129.10.2459. [DOI] [PubMed] [Google Scholar]
- Tsuchida T, Ensini M, Morton SB, Baldassare M, Edlund T, Jessell TM, Pfaff SL. Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell. 1994;79:957–970. doi: 10.1016/0092-8674(94)90027-2. [DOI] [PubMed] [Google Scholar]
- Wilson L, Maden M. The mechanisms of dorsoventral patterning in the vertebrate neural tube. Dev Biol. 2005;282:1–13. doi: 10.1016/j.ydbio.2005.02.027. [DOI] [PubMed] [Google Scholar]
- Wu HH, Ivkovic S, Murray RC, Jaramillo S, Lyons KM, Johnson JE, Calof AL. Autoregulation of neurogenesis by GDF11. Neuron. 2003;37:197–207. doi: 10.1016/s0896-6273(02)01172-8. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Wang S, Anderson DJ. Identification of a novel family of oligodendrocyte lineage-specific basic helix-loop-helix transcription factors. Neuron. 2000;25:331–343. doi: 10.1016/s0896-6273(00)80898-3. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Choi G, Anderson DJ. The bHLH transcription factor Olig2 promotes oligodendrocyte differentiation in collaboration with Nkx2.2. Neuron. 2001;31:791–807. doi: 10.1016/s0896-6273(01)00414-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.