Abstract
Post-translational covalent modification by ubiquitin and ubiquitin-like proteins (UBLs) is a major eukaryotic mechanism for regulating protein function. In general, each UBL has its own E1 that serves as the entry point for a cascade. The E1 first binds the UBL and catalyzes adenylation of the UBL’s C-terminus, prior to promoting UBL transfer to a downstream E2. Ubiquitin’s Arg 72, which corresponds to Ala72 in the UBL NEDD8, is a key E1 selectivity determinant: swapping ubiquitin and NEDD8 residue 72 identity was shown previously to swap their E1 specificity. Correspondingly, Arg190 in the UBA3 subunit of NEDD8’s heterodimeric E1 (the APPBP1-UBA3 complex), which corresponds to a Gln in ubiquitin’s E1 UBA1, is a key UBL selectivity determinant. Here we dissect this specificity with biochemical and X-ray crystallographic analysis of APPBP1-UBA3-NEDD8 complexes in which NEDD8’s residue 72 and UBA3’s residue 190 are substituted with different combinations of Ala, Arg, or Gln. APPBP1-UBA3’s preference for NEDD8’s Ala72 appears to be indirect, due to proper positioning of UBA3’s Arg190. By contrast, our data are consistent with direct positive interactions between ubiquitin’s Arg72 and an E1’s Gln. However, APPBP1-UBA3’s failure to interact with a UBL having Arg72 is not due to a lack of this favorable interaction, but rather arises from UBA3’s Arg190 acting as a negative gate. Thus, parallel residues from different UBL pathways can utilize distinct mechanisms to dictate interaction selectivity, and specificity can be amplified by barriers that prevent binding to components of different conjugation cascades.
Keywords: ubiquitin, NEDD8, E1, protein interaction, specificity, gating
Post-translational modification by ubiquitin and ubiquitin-like proteins (UBLs) is a predominant eukaryotic regulatory mechanism with roles in cell division, the immune response, development, and many other processes (1). A widely recognized functional consequence of a UBL modification is ubiquitin-mediated proteolysis, although ubiquitin can be attached to targets via a variety of linkages that signal different effects on targets (2). In addition to ubiquitin, there are over 10 UBLs in higher eukaryotes that when conjugated to macromolecules, alter the function of their important targets (3). These include ubiquitin’s closest relative NEDD8, which has its own functions despite 58% sequence identity with ubiquitin. The NEDD8 pathway is essential for the regulation of the cell cycle and plays roles in signaling pathways and embryogenesis (4-7). Among NEDD8’s targets are cullin subunits of cullin-RING ligases, and several ribosomal subunits (8-14).
Prior to conjugation, ubiquitin and NEDD8 precursors are processed by deubiquitinating enzymes (DUBs). Then, ubiquitin and NEDD8 are covalently attached to targets by parallel but distinct hierarchical cascades of enzymes in classes known as E1, E2 and E3 (15-18). First, E1 initiates a cascade by catalyzing C-terminal adenylation of the UBL, and subsequently forming a covalent thioester-linked intermediate between its catalytic cysteine and the UBL’s C terminus. E1 further catalyzes a transthiolation reaction, whereby the UBL is transferred from the E1’s catalytic cysteine to the E2’s catalytic cysteine. Often, an E3 facilitates UBL transfer from an E2 to a target. The cycle can be reset by UBL removal from targets by DUBs.
Despite the close similarity between ubiquitin and NEDD8 sequences, these UBLs have their own conjugation cascades, targets, and downstream functions (9-12). In order to ultimately understand the unique functions of different UBLs, it is important to understand the molecular “code” by which UBLs are distinguished by their enzymatic and effector machineries. To date, some of the best progress in this area has been in deciphering sequences dictating E1 discrimination between ubiquitin and NEDD8. Several previous studies indicate that E1s provide critical UBL selection during initial noncovalent binding, and that this selection hinges on the identity of a UBL’s residue 72, the only C-terminal tail residue differing between ubiquitin (Arg) and NEDD8 (Ala) (Fig. 1A). First, ubiquitin’s Arg 72 was identified as a critical determinant for E1 binding during adenylation (19). Second, “ubiquitinizing” NEDD8 with an Ala72Arg mutation allows binding to ubiquitin’s E1 (20), and “NEDD8ylizing” ubiquitin with Arg72Leu or Arg72Ala mutations allows ubiquitin to be activated by NEDD8’s E1 (21, 22). Third, a “NEDD8ylized” ubiquitin can be transferred to NEDD8’s E2, albeit with ∼80-fold diminished kcat/Km, suggesting either that the same UBL residue dictates both E1 and E2 selectivity, or E1 is fully responsible for dictating an E2’s UBL selection (22). This latter notion was supported by the finding that an E2’s UBL selectivity is dictated by side-chains involved in E1-, but not UBL-binding (23). Interestingly, ubiquitin and NEDD8’s residue 72 also plays a fundamental role in DUB specificity (24-26).
Figure 1. Sequence conservation at a UBL’s residue 72 and E1 residues corresponding to UBA3’s 190.
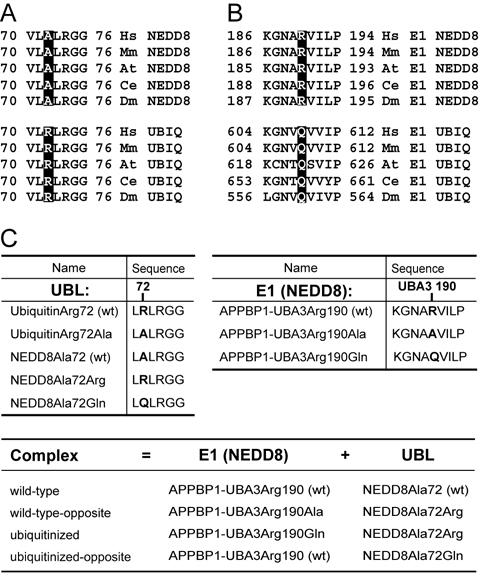
(A) Sequence alignment of the C-terminal tail region of NEDD8 and ubiquitin (UBIQ) from the following organisms: Hs, human; Mm, M. musculus; AT, A. thaliana; Ce, C. elegans; Dm, D. melanogaster. NEDD8 and ubiquitin residue 72 is highlighted. (B) Sequence alignment of the E1 region containing Arg190 (highlighted) from the UBA3 subunit of NEDD8’s E1 (E1 NEDD8) and the corresponding Gln from the UBA1 ubiquitin E1 (E1 UBIQ). (C) Nomenclature and sequences for NEDD8, ubiquitin and E1 NEDD8 (APPBP1-UBA3) mutants.
The basis for UBL selectivity has been partially characterized for NEDD8’s E1. The crystal structure of NEDD8’s heterodimeric E1 (APPBP1-UBA3) in complex with NEDD8 and ATP allowed modeling ubiquitin in place of NEDD8 in the structure (21). In the model, a conserved Arg in UBA3 (Arg190) would clash with ubiquitin’s specificity-conferring Arg72. By contrast, the corresponding residue in E1s for ubiquitin is absolutely conserved throughout evolution as a glutamine (Fig. 1B), and a UBA3 Arg190Gln substitution allows adenylation of ubiquitin to some extent (21). Interestingly, in the wild-type E1-UBL complex, although UBA3’s Arg190 faces NEDD8’s Ala72, these two residues do not contact each other. There are two simplistic explanations for how the UBA3 Arg190Gln mutation allows NEDD8’s E1 to catalyze ubiquitin adenylation. First, it is possible that the polar Gln attracts ubiquitin through positive interactions with Arg72, and because NEDD8’s E1 lacks this Gln it fails to bind ubiquitin. Second, it is possible that NEDD8’s E1 actively prevents ubiquitin binding via electrostatic repulsion, with UBA3’s Arg190 repelling ubiquitin’s Arg72. Here we structurally and biochemically dissect roles of NEDD8’s residue 72 and UBA3’s residue 190, in order to gain further insights into mechanisms underlying E1-UBL selectivity.
Experimental procedures
Protein Preparation
Mutations were generated by PCR, and the entire coding sequence for each construct was verified by sequencing. Expression and purification of human APPBP1-UBA3, Ubc12, NEDD8, ubiquitin, and variants has been described (21, 23, 27, 28). Biochemical experiments were performed with full-length versions of APPBP1-UBA3. Surface plasmon resonance experiments examining noncovalent UBL interaction with APPBP1-UBA3 utilize UBA3 mutants harboring the catalytic Cys216Ala mutation, whereas those examining transthiolation retain the catalytic Cys. For BIACORE experiments, GST-APPBP1-UBA3 (Cys216Ala) and mutants were purified by glutathione affinity and gel filtration chromatography in 50 mM Tris-HCl, 150 mM NaCl, 0.5 mM DTT, pH 7.6. NEDD8 and variants were expressed as His-MBP-fusions from pRSF-1b with a GGS linker inserted upstream of NEDD8 to facilitate thrombin cleavage. His-MBP-NEDD8 was purified by nickel affinity, treated with thrombin, and dialyzed at 4°C in 50 mM Tris-HCl and 150 mM NaCl, pH 7.6. NEDD8 was purified further by gel filtration and concentrated to 300 μM. His-ubiquitin was expressed in BL21 (DE3) RIL from pET15b, purified by nickel affinity chromatography and concentrated to 550 μM. Crystallographic experiments utilized truncated APPBP1-UBA3 lacking APPBP1 residues 254-258 and UBA3 residues 1-12 to facilitate crystallization as described previously (21). APPBP1-UBA3-NEDD8 mutant complexes for crystallography were expressed and purified as for the wild-type complex (21), except that NEDD8 and variants were expressed from pGEX-2TK (27) and thus retain additional vector sequences upstream of NEDD8, which we find improve crystal contacts. Proteins/complexes were aliquotted, flash-frozen in liquid nitrogen, and stored at -80 °C until use.
Surface Plasmon Resonance
Kinetic studies were performed at 25 °C using a BIACORE 3000 (Biacore, Inc.) surface plasmon resonance (SPR) instrument. Anti-GST antibodies (Biacore, Inc.) were covalently attached to a carboxymethyl dextran-coated gold surface (CM-4 Chip; Biacore, Inc.). The carboxymethyl groups of dextran were activated with N-ethyl-N’-(3-dimethylaminopropyl) carbodiimide (EDC) and N-hydroxysuccinimide (NHS), and anti-GST antibodies were attached at pH 5.0 in 10 mM sodium acetate. Any remaining reactive sites were blocked by reaction with ethanolamine. Anti-GST antibodies were immobilized at levels of ∼4000 RU for each flow cell.
The kinetics of association and dissociation were monitored at a flow rate of 100 μL/min. GST-APPBP1-UBA3 constructs were captured to a level of ∼ 1250-1450 RU for each experiment. GST was captured on the reference surface to account for any non-specific binding to the GST tag. The UBL analytes were prepared in 10 mM Tris (pH 7.6), 150 mM NaCl, 0.1 mg/mL bovine serum albumin, and 0.005% P20 surfactant. Binding was measured for concentration ranges of 41 nM to 10 μM for all UBLs. To account for injection artifacts, a series of sensorgrams was recorded throughout the experiment after injecting only buffer (blank injections). The analytes dissociated completely from the chip surfaces, eliminating the need for a regeneration step. Data reported are the difference in SPR signal between the flow cell containing the APPBP1-UBA3 construct and the reference cell without APPBP1-UBA3. Additional instrumental contributions to the signal were removed by subtraction of the average signal of the blank injections from the reference-subtracted signal (29). Triplicate injections were made, and the data were analyzed globally by equilibrium affinity analysis using the software package Scrubber 2 (Biologic Software).
Ubc12∼UBL thioester assays
APPBP1-UBA3-catalyzed formation of the thioester-linked Ubc12∼NEDD8 covalent intermediate was assayed in 20 μL volumes containing 100 nM of APPBP1-UBA3 or mutants, 3 μM Ubc12, 4 μM wild-type NEDD8 or mutants, and 50 mM Tris-HCl, 50 mM NaCl, 10 mM MgCl2, 5 mM ATP, 1 mM DTT, pH 7.6. The reactions were started by adding the E1 enzyme to the assay mixture, incubated at room temperature for 5 min, and quenched by the addition of 10 μL 2× non-reducing SDS-PAGE sample buffer. Reaction products were resolved by 15% SDS-polyacrylamide gels, and transferred to nitrocellulose membranes. The membranes were saturated with 3% bovine serum albumin (BSA) in TBS-T (50 mM Tris-HCl, 150 mM NaCl, 0.1% Tween 20, pH 7.4) prior incubation with anti-NEDD8 antibody. Anti-NEDD8 polyclonal antibodies (RCA) were raised in rabbits against the peptide corresponding to residues 12-26 of the human NEDD8 protein, and affinity purified as described (30). After extensive washing, detection of bound antibodies was carried out using horseradish peroxidase-labeled secondary antibody and enhanced chemiluminescence (Amersham). Experiments comparing reactivity across UBLs were performed similarly, except with His-NEDD8 and His-ubiquitin, allowing comparable detection with anti-His antibodies (Qiagen).
Crystallization, data collection and structure determination
All crystals were grown at 18°C using the hanging-drop vapor diffusion method after mixing 1 μL complexes with an equal volume of the corresponding reservoir solution. Crystals of APPBP1(Δ254-258)-UBA3(ΔN12 C216A)Arg190Gln-NEDD8Ala72Arg (hereafter referred to as APPBP1-UBA3Arg190Gln-NEDD8 Ala72Arg or “ubiquitinized”, Fig. 1C) were grown with reservoir solution containing 0.1 M Tris, 0.2 M NaCl, 10% PEG 10K, 8% PEG400, 5 mM DTT, pH 8.0, and were soaked for 5 min in 50 mM Tris, 0.2 M NaCl, 14% PEG 10K, 8% PEG400, 5 mM DTT, pH 8.0 supplemented with 10% glycerol, and then for 5 min in the same solution supplemented with 20% glycerol, prior to freezing in a solution containing 30% glycerol. Data for these crystals were collected at the X25 beamline at the National Synchrotron Light Source. Crystals of APPBP1(Δ254-258)-UBA3(ΔN12 C216A)Arg190Ala-NEDD8Ala72Arg (hereafter referred to as APPBP1-UBA3Arg190Ala-NEDD8Ala72Arg or “wild-type-opposite”, Fig. 1C) and APPBP1(Δ254-258)-UBA3(ΔN12 C216A)-NEDD8Ala72Gln (hereafter referred to as APPBP1-UBA3Arg190 (wt)-NEDD8Ala72Gln or “ubiquitinized-opposite”, Fig. 1C) were grown with reservoir solution containing 0.1 M Tris, 0.4 M ammonium acetate, 9-10% PEG 10K, 5 mM DTT, pH 7.0-8.0. These crystals were soaked in 0.1 M Tris, 0.4 M ammonium acetate, 12-13% PEG 10K, 5 mM DTT, pH 7.0-8.0 supplemented with increasing concentrations of glycerol (5-30%, in 5% increments) prior to flash-freezing in the presence of 30% glycerol and data collection at the 8.2.2 beamline at the Advanced Light Source. All crystals formed with four complexes in the asymmetric unit in the space group P212121, with unit cells and packing similar to the wild-type complex reported previously (21). Reflection data were indexed, integrated and scaled using HKL2000 (31). The structure of APPBP1-UBA3Arg190Ala-NEDD8Ala72Arg was determined first, by rigid body refinement using a model derived from the structure of wild-type APPBP1-UBA3-NEDD8 (21), in which UBA3’s Arg190 side-chain was truncated after the β-carbon. Rigid body refinement, simulated annealing, energy minimization and restrained B-factor refinement were carried out using the program CNS (32), iteratively with manual rebuilding using O (33). The structure of APPBP1-UBA3Arg190Ala-NEDD8Ala72Arg, but with NEDD8’s residue 72 side-chain truncated after the β-carbon, was then used as a model for the other two structures, which were refined using a similar protocol. Simulated annealing (2000 K) omit maps confirm the presence and the correct placement of mutated residues in all three structures. In one complex from the asymmetric unit of all these structures, additional vector-derived residues N-terminal of NEDD8 participate in crystal packing. The four complexes in the asymmetric unit for the final model of APPBP1-UBA3Arg190Ala-NEDD8Ala72Arg contain: APPBP1 residues 6-204, 207-534; 6-200, 202, 204-205, 208-534; 6-203, 208-253, 261-534; 6-203, 205-534; UBA3 residues -1 and 12-442; -1 and 12-442; -1 and 12-441; -1 and 12-441; and NEDD8 residues -10 to -1 and 1-76; 1-76; 1-76; 1-76, respectively. The four complexes in the asymmetric unit for the final model of APPBP1-UBA3Arg190Gln-NEDD8Ala72Arg contain: APPBP1 residues 6-200, 208-534; 6-201, 211-534; 6-199, 212-252, 262-534; 6-200, 209-534; UBA3 residues -1 and 12-442; -1 and 12-441; -1 and 12-441; -1 and 12-441; and NEDD8 residues -10 to -1 and 1-76; 1-76; -2 to -1 and 1-76; -1 and 1-76, respectively. The four complexes in the asymmetric unit for the final model of APPBP1-UBA3Arg190 (wt)-NEDD8Ala72Gln contain: APPBP1 residues 6-204, 207-534; 6-203, 205-534; 6-200, 202, 204-205, 208-253, 260-534; 6-203, 208-534; UBA3 residues -1 and 12-442; -1 and 12-441; -1 and 12-442; -1 and 12-441; and NEDD8 residues -10 to -1 and 1-76; 1-76; 1-76; 1-76, respectively. Other residues are not modeled, presumably due to disorder. Ramachandran statistics were analyzed with Procheck (34). Structures are shown using Pymol (35).
Results
UBA3’s Arg190 gates against binding to a UBL with a basic side-chain at residue 72
In order to quantify effects of mutations on APPBP1-UBA3-NEDD8 interactions, we developed a binding assay using BIACORE (Table 1, Fig. 2). In our assay, GST-APPBP1-UBA3 binds NEDD8 with a Kd value of 333 ± 1 nM. Furthermore, a NEDD8 mutant harboring the “ubiquitinizing” Ala72Arg mutant does not display any binding in our assay. Thus, we examined the role of UBA3’s Arg190 in this discrimination. Consistent with previous studies, “ubiquitinizing” GST-APPBP1-UBA3 with an Arg190Gln mutation allows binding to NEDD8Ala72Arg, with a Kd of 1.02 ±0.1 μM.
Table 1.
SPR-measured Kd and RU values for interactions between APPBP1-UBA3 position190 variants and NEDD8 position 72 variants
| Interaction | Kd (nM) | Rmax (RU) |
|---|---|---|
| UBA3Arg190 (wt) + NEDD8Ala72 (wt) | 333 (±1) | 50.7 (±0.1) |
| UBA3Arg190 (wt) + NEDD8Ala72Arg | >10000 | n/a |
| UBA3Arg190 (wt) + NEDD8Ala72Gln | 8750 (±70) | 49.5 (±0.2) |
| UBA3Arg190 (wt) + NEDD8Ala72Lys | >10000 | n/a |
| UBA3Arg190Gln + NEDD8Ala72 (wt) | 3460 (±10) | 48.3 (±0.1) |
| UBA3Arg190Gln + NEDD8Ala72Arg | 1020 (±10) | 47.2 (±0.1) |
| UBA3Arg190Gln + NEDD8Ala72Gln | >10000 | n/a |
| UBA3Arg190Gln + NEDD8Ala72Lys | 886 (±4) | 47.3 (±0.1) |
| UBA3Arg190Ala + NEDD8Ala72 (wt) | 5500 (±30) | 43.9 (±0.1) |
| UBA3Arg190Ala + NEDD8Ala72Arg | 1660 (±10) | 43.1 (±0.1) |
| UBA3Arg190Ala + NEDD8Ala72Gln | >10000 | n/a |
| UBA3Arg190Ala + NEDD8Ala72Lys | 2970 (±20) | 54.3 (±0.1) |
Figure 2. Surface Plasmon Resonance Analysis of APPBP1-UBA3 binding to UBLs.
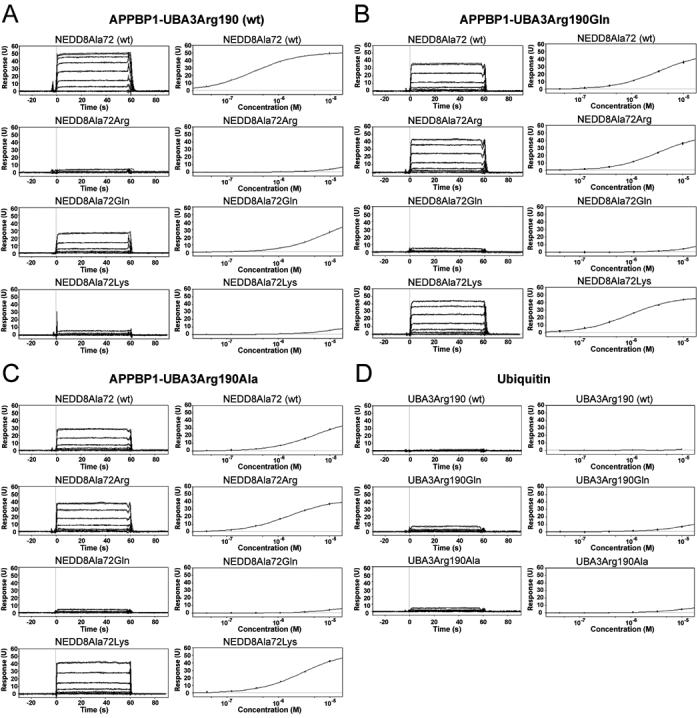
Representative sensorgrams (left) and binding curves (right) from Surface Plasmon Resonance interaction assays, performed as described in Experimental Procedures, for (A) GST-APPBP1-UBA3Arg190 (wt), (B) GST-APPBP1-UBA3Arg190Gln, and (C) - GST-APPBP1-UBA3Arg190Ala with NEDD8Ala72 (wt), NEDD8Ala72Arg, NEDD8Ala72Gln, and NEDD8Ala72Lys, as indicated. (D) Representative sensorgrams (left) and binding curves (right) for ubiquitin binding to GST-APPBP1-UBA3Arg190 (wt), GST-APPBP1-UBA3Arg190Gln, and GST-APPBP1-UBA3Arg190Ala, as indicated.
We next asked whether relief from negative repulsion from the E1’s Arg is sufficient to allow binding of NEDD8Ala72Arg, or whether attractive electrostatic interaction with the E1’s Gln are required. A UBA3 Arg190Ala substitution would address this question by removing barriers from the Arg side-chain, without providing opportunities for favorable polar interactions with the UBL Arg. GST-APPBP1-UBA3Arg190Ala binds NEDD8Ala72Arg with a Kd value of 1.66 ±0.01 μM. The similar Kds (1.02 ±0.1 μM v 1.66 ±0.01 μM) for the Gln and Ala substitutions suggest that UBA3’s Arg190 primarily acts as a gate that negatively selects against a UBL’s Arg72. The UBA3 Arg190Ala substitution also allows binding to a NEDD8 with a Lys substitution at residue 72, although in this case a Gln does appear to impart more positive interactions (3-fold better binding).
UBA3’s residue 190 and NEDD8’s residue 72 do not play equivalent roles in E1-Ubl selectivity
The UBA3Arg190Ala-NEDD8Ala72Arg pair essentially swaps Arg and Ala selectivity determinants between E1 an NEDD8. Thus, we asked whether these E1 and UBL positions play equivalent roles in determining binding with a complete set of Ala, Arg, and Gln substitutions at both positions (Table 1, Fig. 2). The results reveal consistent, non-equivalent, directional effects. First, the wild-type E1-NEDD8 pair binds best, ∼5-fold better than the “opposite” pair in which the identities of UBA3’s residue 190 and NEDD8’s 72 have been swapped [UBA3Arg190Ala and NEDD8Ala72Arg]. Second, the “ubiquitinized” pair (with the UBA3Arg190Gln and NEDD8Ala72Arg substitutions to residues found in ubiquitin’s E1 and ubiquitin) associate with a ∼9-fold lower Kd than the “opposite ubiquitinized” pair, wild-type UBA3 (Arg190) and NEDD8Ala72Gln.
Interestingly, both the APPBP1-UBA3 Arg190Gln and Arg190Ala variants preferentially bind to NEDD8Ala72Arg (or NEDD8Ala72Lys), rather than to wild-type NEDD8 (Ala72), or the NEDD8Ala72Gln mutant. This raises the possibility that an Arg serves a function beyond dictating E1-UBL selectivity.
NEDD8’s E1 contains additional determinants selecting against ubiquitin
Having established UBA3’s residue 190 as gating against a UBL’s Arg72, we asked the extent to which this single interaction establishes APPBP1-UBA3’s selectivity against initial noncovalent binding to ubiquitin. Although binding is too weak for us to measure a Kd within the limits of our assay, some signal is detected for interactions between wild-type GST-APPBP1-UBA3 and ubiquitinArg72Ala, but not for wild-type ubiquitin (Fig. 2D). In a related vein, GST-APPBP1-UBA3 Arg190Gln and Arg190Ala do show evidence of interaction wild-type ubiquitin. Therefore, although the same trends are observed for mutations allowing APPBP1-UBA3 interaction with “ubiquitinized” NEDD8Ala72Arg, additional residues also contribute to NEDD8 E1’s discrimination against noncovalent binding to ubiquitin.
NEDD8’s E1-E2 transthiolation with UBLs harboring Arg at position 72
Although wild-type ubiquitin is excluded from NEDD8’s E1, a previous study found that NEDD8’s E1 could transfer the “NEDD8ylized” ubiquitinArg72Leu mutant to NEDD8’s E2 (Ubc12), albeit with less efficiency overall than the NEDD8 transthiolation cycle (22). This result raised the question as to whether specificity was established entirely by noncovalent binding by the E1, or whether the ubiquitin mutation relieves discrimination against an Arg at the UBL’s position 72 by APPBP1-UBA3 and Ubc12 during transthiolation. To address this question, we assayed the different APPBP1-UBA3 mutants for NEDD8Ala72Arg transfer to Ubc12 (Fig. 3). We observe no detectable activity with wild-type APPBP1-UBA3, consistent with gating against this “ubiquitinizing” mutation. However, both the APPBP1-UBA3 Arg190Gln and Arg190Ala mutants can transfer NEDD8Ala72Arg to Ubc12. Similar results are also observed for ubiquitin, although reactions are less efficient, consistent with the weaker noncovalent binding of ubiquitin to these mutants.
Figure 3. Altered E1 NEDD8 (APPBP1-UBA3) - E2 (Ubc12) transthiolation specificity for UBA3 Arg190 mutants.
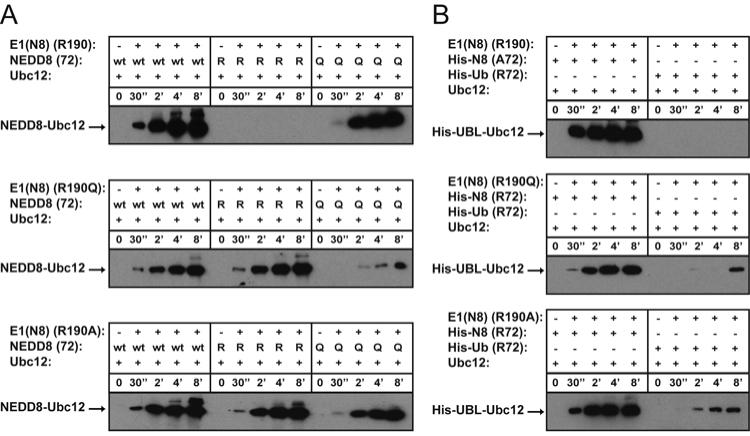
(A) Time-course of forming the Ubc12∼NEDD8 thioester complexes with 100 nM wild-type and indicated mutants of APPBP1-UBA3, 4 μM wild-type and indicated mutants of NEDD8, and 3 μM Ubc12. Reactions were stopped at the indicated times, products were separated by SDS-PAGE and detected by western blotting with anti-NEDD8 antibodies. (B) Western blots of reactions performed as in panel A, except using the indicated His-NEDD8 variants and His-ubiquitin, and probed with anti-His-tag antibodies.
Crystallographic dissection of Arg, Ala, and Gln at UBA3’s residue 190 and NEDD8’s residue 72
Our biochemical data pointed toward a directional gating mechanism by which NEDD8’s E1’s Arg190 influences interaction with a UBL’s residue 72. In order to understand the structural basis underlying this specificity, we determined crystal structures of three mutant APPBP1-UBA3-NEDD8 complexes: “wild-type-opposite” [UBA3Arg190Ala-NEDD8Ala72Arg, 2.85 Å resolution]; “ubiquitinized” [UBA3Arg190Gln-NEDD8Ala72Arg, 3.05 Å resolution]; and “ubiquitinized-opposite” [UBA3Arg190 (wt)-NEDD8Ala72Gln, 2.90 Å resolution]. Details of data and refinement statistics are given in Table 2. Overall, for all three complexes, the electron density 2Fo-Fc maps were continuous and well-defined over all four copies in the asymmetric units (data not shown). Moreover, the side chains at both positions NEDD8’s E1 residue 190 and NEDD8 residue 72 were clearly present in simulated annealing omit maps (Fig. 5), generated from a model lacking the residues UBA3’s 190 and NEDD8’s 72. For each mutant complex, the four complexes in the asymmetric unit are similar: “wild-type-opposite”, 0.5Å rmsd, “ubiquitinized”, 0.6 Å rmsd, and “ubiquitinized-opposite”, 0.5 Å rmsd. Therefore, the figures display only one copy for each mutant complex. The overall structures all superimpose well with the prior wild-type APPBP1-UBA3-NEDD8 structure, with rmsds between 0.4 and 0.5 Å.
Table 2.
Data and refinement statistics
| APPBP1-UBA3Arg190Gln-NEDD8Ala72Arg | APPBP1-UBA3Arg190Ala-NEDD8Ala72Arg | APPBP1-UBA3Arg190 (wt)-NEDD8Ala72Gln | |
|---|---|---|---|
| Data Collection | |||
| Beamline | NSLS X25 | ALS 8.2.2 | ALS 8.2.2 |
| Space group | P212121 | P212121 | P212121 |
| Cell dimensions | |||
| a, b, c (Å) | 134.3, 198.5, 208.8 | 135.6, 198.1, 210.9 | 136.1, 198.9, 210.0 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Resolution (Å) | 3.05 | 2.85 | 2.90 |
| Rmerge (%) | 11.4 (56.6) | 11.0 (39.3) | 16.3 (46.8) |
| I/σI | 12.3 (2.7) | 14.4 (1.5) | 13.2 (1.8) |
| Completeness (%) | 95.9 (97.2) | 96.0 (81.9) | 93.7 (78.6) |
| Refinement | |||
| Resolution (Å) | 50.0-3.05 | 50.0-2.85 | 50.0-2.90 |
| No. reflections | 100137 | 127325 | 117851 |
| Rwork/ Rfree (%) | 22.9/28.0 | 22.4/27.4 | 22.5/27.4 |
| R.m.s deviations | |||
| Bond lengths (Å) | 0.009 | 0.008 | 0.008 |
| Bond angles (°) | 1.59 | 1.51 | 1.44 |
| Ramachandran plot statistics | |||
| Most favoured regions (%) | 81.2 | 83.4 | 83.3 |
| Additional allowed regions (%) | 18.6 | 16.4 | 16.5 |
| Generously allowed regions (%) | 0.2 | 0.2 | 0.3 |
| Disallowed regions (%) | 0.0 | 0.0 | 0.0 |
Rwork = Σ| Fobs - Fcalc | / Σ| Fobs|, where Fobs and Fcalc are the observed and calculated structure factors, respectively. Rfree = Rwork calculated with 5% of the reflection data chosen randomly and omitted from the start of refinement. Parentheses denote the last resolution shell.
Figure 5. Differential APPBP1-UBA3 interactions with NEDD8 for UBA3 residue 190 and NEDD8 residue 72 mutants.
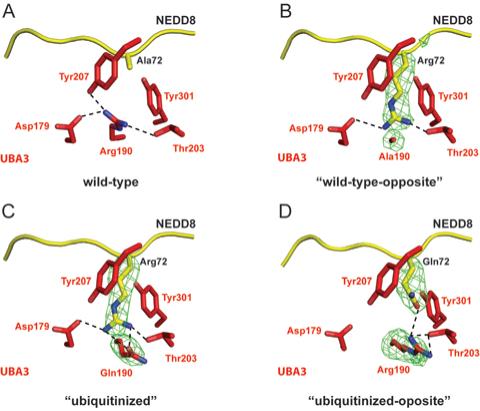
Stick-representation close-up views, with UBA3 colored red and NEDD8 colored yellow, nitrogen blue, oxygen light-red, and hydrogen bonds shown as dashed lines for (A) APPBP1-UBA3-NEDD8 (wild-type), (B) APPBP1-UBA3Arg190Ala-NEDD8Ala72Arg (“wild-type-opposite”), (C) APPBP1-UBA3Arg190Gln-NEDD8Ala72Arg (“ubiquitinized”), and (D) APPBP1-UBA3Arg190 (wt)-NEDD8Ala72Gln (“ubiquitinized-opposite”) complexes. Simulated annealing omit Fo-Fc electron density maps are shown in green mesh, contoured at 3σ over UBA3’s residue 190 and NEDD8’s residue 72 in panels B-D. The maps were calculated using the program CNS (32), after simulated annealing at 2000K omitting both UBA3’s residue 190 and NEDD8’s residue 72.
Structures of “wild-type-opposite” and “ubiquitinized” complexes: unlocking UBA3’s gate against a UBL’s Arg72
In order to understand how UBA3’s Arg190 gates against a UBL’s Arg72, we compared the structures of the previously-determined wild-type APPBP1-UBA3-NEDD8 complex, in which UBA3 harbors an Arg, and the “wild-type-opposite” and “ubiquitinized” complexes, in which NEDD8 has an Arg. Strikingly, in all three complexes, the Arg guanidium group is in the same relative location (Fig. 4). This explains how UBA3’s Arg would gate against a UBL’s Arg72 by clashing and repulsion. The results also show how simply removing UBA3’s Arg190 side-chain with an Ala mutation allows a UBL’s Arg.
Figure 4. Structural basis for UBA3’s Arg190’s negative selectivity against a UBL’s Arg72.
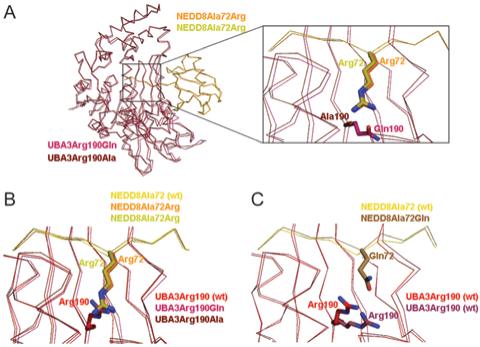
Superimposition of wild-type (21) and mutant APPBP1-UBA3-NEDD8 structures was performed using least-squares fitting over all atoms in O (33). UBA3’s residue 190 is shown in various shades of red; NEDD8’s residue 72 in yellow; nitrogen, blue; and oxygen, light red. (A) Overall superimposition of APPBP1-UBA3Arg190Gln (rose) - NEDD8Ala72Arg (melon, “ubiquitinized”) and APPBP1-UBA3Arg190Ala (maroon) - NEDD8Ala72Arg (chartreuse, “wild-type-opposite”) complexes, with close-up view around the NEDD8 mutant’s Arg72 and UBA3’s residue 190. (B) Close-up view showing NEDD8 mutant Arg72 from APPBP1-UBA3Arg190Gln-NEDD8Ala72Arg (melon) and from APPBP1-UBA3Arg190Ala-NEDD8Ala72Arg (chartreuse), and Arg190 (red) from wild-type APPBP1-UBA3-NEDD8. (C) Close-up view showing Arg190 (violet) and NEDD8 mutant Gln72 from APPBP1-UBA3Arg190 (wt)-NEDD8Ala72Gln, and Arg190 (red) from wild-type APPBP1-UBA3-NEDD8.
In all three structures, the Arg makes many favorable contacts (Fig. 5). In the wild-type complex, Arg190 forms hydrogen bonds with UBA3’s Tyr207 and Thr203, and a salt-bridge with UBA3’s Asp179. These latter two electrostatic interactions are preserved when the Arg comes from the mutant NEDD8. The aliphatic portion of the Arg side-chain also preserves hydrophobic interactions with Tyr207 and Tyr321. In the “ubiquitinized” complex, a hydrogen bond between NEDD8’s mutant Arg72 and UBA3’s mutant Gln190 explains the moderate preference for the “ubiquitinizing” Gln over an Ala at position 190 in UBA3.
Insights into directionality of the UBA3 190 and UBL 72 gate from “wild-type-opposite” structure
We wished to obtain a structural understanding of the directional preference toward residue identity at UBA3’s position 190 and NEDD8’s position 72, revealed from our binding assay. Comparison of the wild-type (UBA3Arg190, NEDD8Ala72) with the “wild-type-opposite” (UBA3 mutant Ala190, NEDD8 mutant Arg72) reveals that swapping the Arg side-chain between UBA3 and NEDD8 removes van der Waals contacts, between UBA3 Arg190’s gamma and delta carbons and Asp179’s beta carbon (Fig. 5A-B). Asp179 is responsible for orienting UBA3’s Arg151, which in turn aligns the backbone of UBA3’s Mg2+-binding Asp146 and NEDD8’s C-terminal Gly-Gly motif. Although at the resolution of our structures subtle structural differences are not detectable, we speculate that slight variations in this region influences affinity for NEDD8.
Although Arg-Gln and Gln-Arg electrostatic interactions are observed in both the “ubiquitinized” (UBA3 mutant Gln190, NEDD8 mutant Arg72) and “ubiquitinized-opposite” (UBA3 Arg190, NEDD8 mutant Gln72) complexes, the “ubiquitinized-opposite” (UBA3 Arg190, NEDD8 mutant Gln72) interaction is lower affinity. The Arg190 hydrophobic contact to UBA3’s Asp179 is lost in both of these complexes, as in the “wild-type-opposite” complex (Fig. 5C-D). Moreover, the UBA3 Asp179 - Arg salt-bridge is lost in the “ubiquitinized-opposite”, in which the UBA3 Arg guanidium group has moved away, in order to accommodate and form a hydrogen bond with the NEDD8 mutant Gln side-chain (Fig. 4C).
The structure of the “ubiquitinized-opposite” complex suggests that the relatively higher affinity interaction between wild-type UBA3, with an Arg190, and wild-type NEDD8, with an Ala72 is an indirect effect. Despite the lack of positive electrostatic 190-72 interactions, an Ala at NEDD8’s position 72 allows proper positioning of UBA3’s Arg. By contrast, in order to accommodate a Gln at NEDD8’s position 72, other interactions are disrupted for relocation of UBA3’s Arg190 (Fig. 4C, 5D).
Discussion
UBL modifications regulate a vast array of eukaryotic pathways. Thus, it is of great interest to understand the detailed mechanisms by which different UBLs are directed to their targets, and thereby modify function. Ubiquitin’s and NEDD8’s residue 72 has emerged as a key determinant distinguishing protein-protein interactions between the two cascades. Here we dissect the structural mechanisms by which the identity of a UBL’s residue 72, and the corresponding E1’s residue 190, dictates interaction selectivity. We find that ubiquitin’s Arg72 actively prevents misactivation by NEDD8’s E1 due to direct negative exclusion by UBA3’s Arg190. This contrasts with a model where NEDD8’s E1 simply lacks residues dictating positive interactions with ubiquitin’s Arg72.
Negative selectivity has emerged as a general mechanism separating the ubiquitin and NEDD8 pathways. In addition to barriers preventing NEDD8’s E1 from interacting with ubiquitin, NEDD8’s E2 (Ubc12) also displays negative selectivity determinants that oppose mismatching with ubiquitin (23). Ubc12’s UBL specificity, from NEDD8 to ubiquitin, was switched 1010-fold simply by removing surface elements, without substituting sequences of ubiquitin E2s (23). NEDD8’s E1 also displays barriers against structures conserved among ubiquitin E2s (36). It seems likely that negative interactions will be found to play major roles in distinguishing members of other UBL cascades. Indeed, negative interactions may hinder binding between the HECT E3 E6AP and the ubiquitin E2 UbcH5 (37), and the altered specificity of a charge-swapped RING E3 CNOT4-UbcH5B pair may involve altered negative selectivity as well as designed positive interactions (38).
Subtle structural differences, which may be distributed over ubiquitin’s and NEDD8’s entire surfaces, also contribute substantial selectivity. Such differences appear to add E1-UBL specificity, on top of the predominant role of a UBL’s residue 72 (19, 22) (also Fig. 2D, 3B). Not surprisingly, the native UBA3Arg190 - NEDD8Ala72 interaction is most favored. Our structural data suggest this may be an indirect effect of proper positioning of UBA3’s Arg190, rather than from direct positive interactions with NEDD8’s Ala72. Fine variation also appears to contribute at least a factor of ten to the differential binding of NEDD8 and ubiquitinArg72Ala to the NEDD8-specific ubiquitin-like protease DEN1/NEDP1/SENP8 (25, 26). Insights into how slight differences in presentation of ubiquitin surface residues can lead to large differences in affinities come from the finding that hydrophobic core mutations selectively diminish binding to ubiquitin-interacting motifs, but not to ubiquitin-associated domains (39).
In contrast to apparently indirect functions of NEDD8’s residue 72, ubiquitin’s Arg 72 mediates important positive interactions with many cognate partners. The most striking effect is observed for ubiquitin interactions with the ubiquitin C-terminal hydrolase Yuh1, where the NEDD8Ala72Arg mutant binds Yuh1 as well as ubiquitin (24). The structure of a Yuh1-ubiquitin aldehyde complex revealed essential positive interactions between Yuh1’s Asp35 and a UBL’s Arg72 (24). In a related vein, a ubiquitin Arg72Ala mutation decreases cleavage by the ubiquitin-specific protease HAUSP (26). It seems that ubiquitin’s Arg72 likely makes positive interactions with ubiquitin’s E1 (19, 20). Consistent with this notion, we observe hydrogen bonding between a UBL’s Arg72 and a “ubiquitinized” UBA3 Gln190 mutant. Ubiquitin’s E1 also contains an Asp in place of UBA3’s Leu206 and Tyr207, which further contact NEDD8’s residue 72. Thus, residue 72 in ubiquitin and NEDD8 appear to mediate specificity in distinct ways.
It seems likely that structures mediating key positive interactions in some other UBL pathways may also serve as negative determinants that prevent interaction with non-cognate conjugation cascades. Indeed, it is possible that conceptually related, but distinct mechanisms confer specificity even among E1-UBL antecedents found in prokaryotes. The bacterial enzymes MoeB and ThiF are prokaryotic ancestors of E1s, with roles in molybdopterin and thiamin biosynthesis, respectively (3, 40). MoeB and ThiF resemble the E1 adenylation domain both structurally and functionally, catalyzing adenylation of the bacterial ubiquitin-fold proteins MoaD and ThiS. Here, specificity may be dictated by UBL insertions, between the βA and βB strands in MoaD and the βD and βE strands in ThiS, which make unique contacts to MoeB and ThiF, respectively (41, 42). Interestingly, the insertions in both MoaD and ThiS contain arginines, which may also gate against non-cognate interactions.
Knowledge of the “code” for selective interactions will lay a foundation for manipulating UBL ligation machineries to transform protein activities as desired.
Acknowledgement
We are indebted to D.J. Miller for assistance with X-ray crystallography, to R. Mathew for assistance in generating the anti-NEDD8 antibody, to D. Duda, L. Borg, D. Huang, D. Scott, D.W. Miller, H. Kamadurai, O. Ayrault, and A. Forget for advice, to C. Ross for computational support, and to S. Bozeman for administrative support. This work was supported in part by ALSAC (American Syrian Lebanese Associated Charities), grants from the NIH (R01GM069530 and R01GM077053 to B.A.S., 5P01CA0719075 to M.F.R., P30CA021765 to St. Jude Cancer Center) and the Howard Hughes Medical Institute. Use of the National Synchrotron Light Source - Brookhaven National Laboratory and the Advanced Light Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contracts No. DE-AC02-98CH10886 and DE-AC02-05CH11231, respectively. B.A.S. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
RCSB Access Codes: 3DBH, 3DBL, 3DBR
References
- 1.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 2.Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 3.Hochstrasser M. Evolution and function of ubiquitin-like protein-conjugation systems. Nature cell biology. 2000;2:E153–157. doi: 10.1038/35019643. [DOI] [PubMed] [Google Scholar]
- 4.Pozo JC, Timpte C, Tan S, Callis J, Estelle M. The ubiquitin-related protein RUB1 and auxin response in Arabidopsis. Science (New York, N.Y. 1998;280:1760–1763. doi: 10.1126/science.280.5370.1760. [DOI] [PubMed] [Google Scholar]
- 5.Osaka F, Saeki M, Katayama S, Aida N, Toh EA, Kominami K, Toda T, Suzuki T, Chiba T, Tanaka K, Kato S. Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast. The EMBO journal. 2000;19:3475–3484. doi: 10.1093/emboj/19.13.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tateishi K, Omata M, Tanaka K, Chiba T. The NEDD8 system is essential for cell cycle progression and morphogenetic pathway in mice. J Cell Biol. 2001;155:571–579. doi: 10.1083/jcb.200104035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kurz T, Pintard L, Willis JH, Hamill DR, Gonczy P, Peter M, Bowerman B. Cytoskeletal regulation by the Nedd8 ubiquitin-like protein modification pathway. Science (New York, N.Y. 2002;295:1294–1298. doi: 10.1126/science.1067765. [DOI] [PubMed] [Google Scholar]
- 8.Lammer D, Mathias N, Laplaza JM, Jiang W, Liu Y, Callis J, Goebl M, Estelle M. Modification of yeast Cdc53p by the ubiquitin-related protein rub1p affects function of the SCFCdc4 complex. Genes & development. 1998;12:914–926. doi: 10.1101/gad.12.7.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liakopoulos D, Doenges G, Matuschewski K, Jentsch S. A novel protein modification pathway related to the ubiquitin system. The EMBO journal. 1998;17:2208–2214. doi: 10.1093/emboj/17.8.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osaka F, Kawasaki H, Aida N, Saeki M, Chiba T, Kawashima S, Tanaka K, Kato S. A new NEDD8-ligating system for cullin-4A. Genes & development. 1998;12:2263–2268. doi: 10.1101/gad.12.15.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gong L, Yeh ET. Identification of the activating and conjugating enzymes of the NEDD8 conjugation pathway. The Journal of biological chemistry. 1999;274:12036–12042. doi: 10.1074/jbc.274.17.12036. [DOI] [PubMed] [Google Scholar]
- 12.Pan ZQ, Kentsis A, Dias DC, Yamoah K, Wu K. Nedd8 on cullin: building an expressway to protein destruction. Oncogene. 2004;23:1985–1997. doi: 10.1038/sj.onc.1207414. [DOI] [PubMed] [Google Scholar]
- 13.Jones J, Wu K, Yang Y, Guerrero C, Nillegoda N, Pan ZQ, Huang L. A targeted proteomic analysis of the ubiquitin-like modifier nedd8 and associated proteins. Journal of proteome research. 2008;7:1274–1287. doi: 10.1021/pr700749v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xirodimas DP, Sundqvist A, Nakamura A, Shen L, Botting C, Hay RT. Ribosomal proteins are targets for the NEDD8 pathway. EMBO reports. 2008;9:280–286. doi: 10.1038/embor.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 16.Dye BT, Schulman BA. Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu Rev Biophys Biomol Struct. 2007;36:131–150. doi: 10.1146/annurev.biophys.36.040306.132820. [DOI] [PubMed] [Google Scholar]
- 17.Capili AD, Lima CD. Taking it step by step: mechanistic insights from structural studies of ubiquitin/ubiquitin-like protein modification pathways. Current opinion in structural biology. 2007;17:726–735. doi: 10.1016/j.sbi.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knipscheer P, Sixma TK. Protein-protein interactions regulate Ubl conjugation. Current opinion in structural biology. 2007;17:665–673. doi: 10.1016/j.sbi.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 19.Burch TJ, Haas AL. Site-directed mutagenesis of ubiquitin. Differential roles for arginine in the interaction with ubiquitin-activating enzyme. Biochemistry. 1994;33:7300–7308. doi: 10.1021/bi00189a035. [DOI] [PubMed] [Google Scholar]
- 20.Whitby FG, Xia G, Pickart CM, Hill CP. Crystal structure of the human ubiquitin-like protein NEDD8 and interactions with ubiquitin pathway enzymes. The Journal of biological chemistry. 1998;273:34983–34991. doi: 10.1074/jbc.273.52.34983. [DOI] [PubMed] [Google Scholar]
- 21.Walden H, Podgorski MS, Huang DT, Miller DW, Howard RJ, Minor DL, Jr., Holton JM, Schulman BA. The structure of the APPBP1-UBA3-NEDD8-ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1. Mol Cell. 2003;12:1427–1437. doi: 10.1016/s1097-2765(03)00452-0. [DOI] [PubMed] [Google Scholar]
- 22.Bohnsack RN, Haas AL. Conservation in the mechanism of Nedd8 activation by the human AppBp1-Uba3 heterodimer. The Journal of biological chemistry. 2003;278:26823–26830. doi: 10.1074/jbc.M303177200. [DOI] [PubMed] [Google Scholar]
- 23.Huang DT, Zhuang M, Ayrault O, Schulman BA. Identification of conjugation specificity determinants unmasks vestigial preference for ubiquitin within the NEDD8 E2. Nature structural & molecular biology. 2008;15:280–287. doi: 10.1038/nsmb.1387. [DOI] [PubMed] [Google Scholar]
- 24.Johnston SC, Riddle SM, Cohen RE, Hill CP. Structural basis for the specificity of ubiquitin C-terminal hydrolases. The EMBO journal. 1999;18:3877–3887. doi: 10.1093/emboj/18.14.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reverter D, Wu K, Erdene TG, Pan ZQ, Wilkinson KD, Lima CD. Structure of a complex between Nedd8 and the Ulp/Senp protease family member Den1. Journal of molecular biology. 2005;345:141–151. doi: 10.1016/j.jmb.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 26.Shen LN, Liu H, Dong C, Xirodimas D, Naismith JH, Hay RT. Structural basis of NEDD8 ubiquitin discrimination by the deNEDDylating enzyme NEDP1. The EMBO journal. 2005;24:1341–1351. doi: 10.1038/sj.emboj.7600628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walden H, Podgorski MS, Schulman BA. Insights into the ubiquitin transfer cascade from the structure of the activating enzyme for NEDD8. Nature. 2003;422:330–334. doi: 10.1038/nature01456. [DOI] [PubMed] [Google Scholar]
- 28.Huang DT, Schulman BA. Expression, purification, and characterization of the E1 for human NEDD8, the heterodimeric APPBP1-UBA3 complex. Methods Enzymol. 2005;398:9–20. doi: 10.1016/S0076-6879(05)98002-6. [DOI] [PubMed] [Google Scholar]
- 29.Myszka DG. Improving biosensor analysis. J Mol Recognit. 1999;12:279–284. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<279::AID-JMR473>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 30.Huang DT, Miller DW, Mathew R, Cassell R, Holton JM, Roussel MF, Schulman BA. A unique E1-E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8. Nature structural & molecular biology. 2004;11:927–935. doi: 10.1038/nsmb826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. In: Carter CW Jr., Sweet RM, editors. Methods in Enzymology. 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 32.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 33.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 34.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1995;26:283–291. [Google Scholar]
- 35.DeLano WL. The PyMOL Molecular Graphics System. 2002 [Google Scholar]
- 36.Huang DT, Paydar A, Zhuang M, Waddell MB, Holton JM, Schulman BA. Structural basis for recruitment of Ubc12 by an E2 binding domain in NEDD8′s E1. Mol Cell. 2005;17:341–350. doi: 10.1016/j.molcel.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 37.Eletr ZM, Kuhlman B. Sequence determinants of E2-E6AP binding affinity and specificity. Journal of molecular biology. 2007;369:419–428. doi: 10.1016/j.jmb.2007.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winkler GS, Albert TK, Dominguez C, Legtenberg YI, Boelens R, Timmers HT. An altered-specificity ubiquitin-conjugating enzyme/ubiquitin-protein ligase pair. Journal of molecular biology. 2004;337:157–165. doi: 10.1016/j.jmb.2004.01.031. [DOI] [PubMed] [Google Scholar]
- 39.Haririnia A, Verma R, Purohit N, Twarog MZ, Deshaies RJ, Bolon D, Fushman D. Mutations in the hydrophobic core of ubiquitin differentially affect its recognition by receptor proteins. Journal of molecular biology. 2008;375:979–996. doi: 10.1016/j.jmb.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang DT, Walden H, Duda D, Schulman BA. Ubiquitin-like protein activation. Oncogene. 2004;23:1958–1971. doi: 10.1038/sj.onc.1207393. [DOI] [PubMed] [Google Scholar]
- 41.Lake MW, Wuebbens MM, Rajagopalan KV, Schindelin H. Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex. Nature. 2001;414:325–329. doi: 10.1038/35104586. [DOI] [PubMed] [Google Scholar]
- 42.Lehmann C, Begley TP, Ealick SE. Structure of the Escherichia coli ThiS-ThiF complex, a key component of the sulfur transfer system in thiamin biosynthesis. Biochemistry. 2006;45:11–19. doi: 10.1021/bi051502y. [DOI] [PMC free article] [PubMed] [Google Scholar]


