SUMMARY
Diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) is one of the mammalian PPIP5K isoforms responsible for synthesis of diphosphoinositol polyphosphates (“inositol pyrophosphates”; PP-InsPs), regulatory molecules that function at the interface of cell signaling and organismic homeostasis. The development of drugs that inhibit PPIP5K2 could have both experimental and therapeutic applications. Here, we describe the first synthetic strategy for producing naturally-occurring 5- PP-InsP4, as well as several new inositol polyphosphate analogues, and we study their interactions with PPIP5K2 using biochemical and structural approaches. These experiments uncover an additional ligand binding site on the surface of PPIP5K2, adjacent to the catalytic pocket. This site facilitates substrate capture from the bulk phase, prior to its transfer into the catalytic pocket. In addition to demonstrating a “catch-and-pass” reaction mechanism in a small molecule kinase, we demonstrate that binding of our analogues to the substrate capture site inhibits PPIP5K2. The work suggests that the substrate binding site offers new opportunities for targeted drug design.
INTRODUCTION
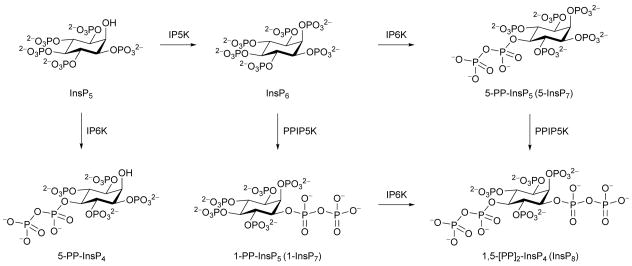
The process of signal transduction that governs many cellular activities frequently relies upon evolutionarily-conserved families of small, regulatory molecules. Among them are the diphosphoinositol polyphosphates (“inositol pyrophosphates”: 5-PP-InsP4, 1-PP-InsP5 (“1-InsP7”), 5-PP-InsP5 (“5-InsP7”), and 1,5-[PP]2-InsP4 (“InsP8”); Figure 1) in which six to eight phosphate groups are crammed around the six-carbon inositol ring. These “high-energy” molecules are synthesized by two distinct classes of kinases, IP6Ks and PPIP5Ks. The IP6Ks add the 5-diphosphate group (Draskovic et al., 2008); mammals express three IP6K isoforms (Thomas and Potter, 2013). The PPIP5Ks synthesize the 1-diphosphate (Wang et al., 2012); there are two isoforms in mammals (Thomas and Potter, 2013). Interest in this field has recently been heightened by demonstrations that diphosphoinositol polyphosphates operate at the interface of cell signaling and organismic homeostasis (Choi et al., 2005; Szijgyarto et al., 2011; Shears, 2009; Illies et al., 2007; Chakraborty et al., 2010; Pulloor et al., 2014). Here, a dynamic balance between the activities of IP6Ks and PPIP5Ks is of particular significance. For example, the synthesis of 5-PP-InsP5 by IP6Ks inhibits the PtdIns(3,4,5)P3 /PDK1/AKT/mTOR cascade (Chakraborty et al., 2010) that controls cell growth and metabolism in response to changes in levels of nutrients, growth factors and bioenergetic status (Benjamin et al., 2011). This inhibitory action of 5-PP-InsP5 is reversed through its further phosphorylation by the PPIP5Ks (Gokhale et al., 2013). There may be therapeutic value in inhibiting PPIP5K activity in order to elevate 5-PP-InsP5 levels and attenuate the mTOR pathway, which is hyperactivated in 70% of human tumors, contributing to the derangement of cell growth and metabolism that accompanies cancer development and progression (Benjamin et al., 2011). We (Gokhale et al., 2013) recently published proof-of-principle of the latter idea by demonstrating that AKT phosphorylation in myoblasts is inhibited when PPIP5K1 expression is “knocked-down”. It is just such therapeutic motives that frequently drive the development of drugs that can specifically target kinases such as PPIP5Ks. Candidate molecules may be rationally designed when information on protein structure is available. To this end, we recently solved the structure of the N-terminal kinase domain of PPIP5K2 (PPIP5K2KD) in complex with natural substrate within the catalytic site (Wang et al., 2012). However, the architecture of the active site exhibits substantial geometric and electrostatic constraints that raise challenges for the design of an effective yet specific inhibitor.
Figure 1. Biosynthesis of diphosphoinositol phosphates.
IP5K, inositol pentakisphosphate 2-kinase; IP6K, inositol hexakisphosphate 5-kinase; PPIP5K, diphosphoinositol pentakisphosphate 1-kinase.
In the current study we set out to prepare new substrate analogues that might modify PPIP5K2 activity. The synthesis of analogues of diphosphoinositol polyphosphates presents particular technical challenges due to the reactive nature of the diphosphate group and the protected diphosphate intermediates (Best et al., 2010). The high negative charge density of these materials also presents purification problems (Capolicchio et al., 2013). Although several of the naturally-occurring diphosphoinositol polyphosphates have been synthesized (Albert et al., 1997; Best et al., 2010; Wu et al., 2013; Capolicchio et al., 2013), the preparation of useful analogues has only recently been accomplished (Riley et al., 2012; Wu et al., 2013). In the latter studies, analogues of 5-PP-InsP4 and 5-PP-InsP5 were synthesized in which the diphosphate groups were replaced with metabolically stabilized phosphonoacetate (PA) or methylenebisphosphonate (PCP) groups. In the current study we describe the synthesis of a new series of diphosphoinositol polyphosphate analogues. We demonstrate how we used these new reagents to gain insight into a previously-described (Weaver et al., 2013) substrate-stimulated ATPase activity of PPIP5K2KD. These experiments also led us to uncover a hitherto unsuspected second ligand binding site in PPIP5K2 KD that performs an important aspect of the catalytic cycle by enhancing capture of substrate from the bulk phase.
RESULTS AND DISCUSSION
Stimulation of the ATPase activity of PPIP5K2KD by 5-PA-InsP5 and 2-O-Bn-5-PA-InsP4
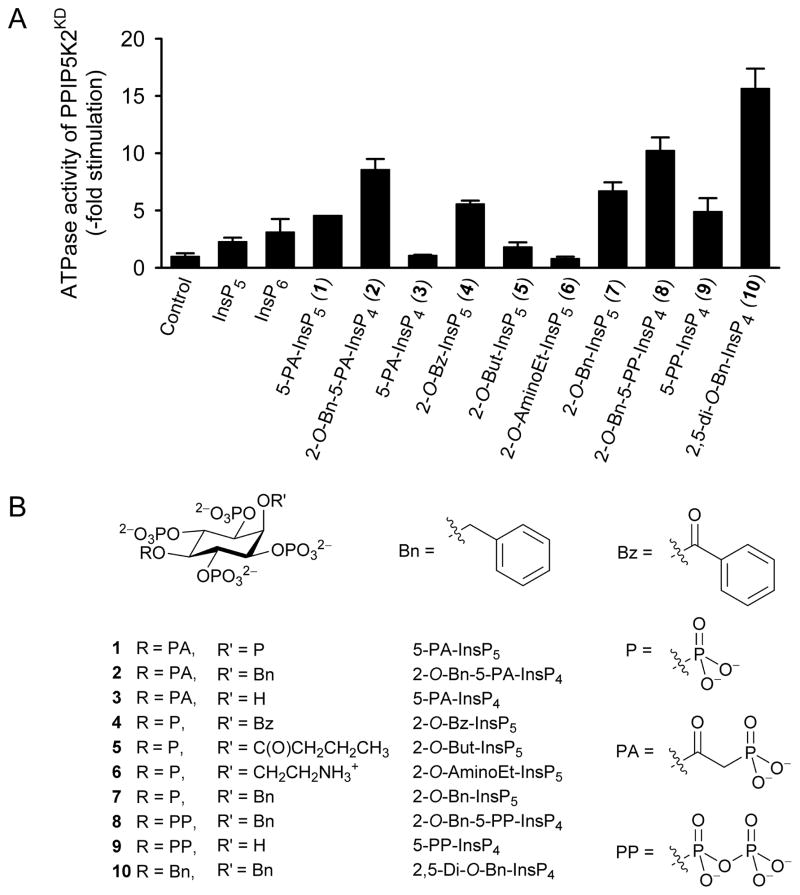
We (Weaver et al., 2013) recently reported that PPIP5K2KD exhibits an unusual, non-productive, substrate-stimulated ATPase activity (e.g. we observed a 2 to 3-fold activation by 25 μM of either Ins(1,3,4,5,6)P5 or InsP6; Figure 2A). We now report that 25 μM of either of two previously described analogues of diphosphoinositol polyphosphates (Riley et al., 2012) also stimulate ATP hydrolysis: 5-fold by 5-O-α-phosphonoacetyl-myo-inositol 1,2,3,4,6-pentakisphosphate [5-PA-InsP5 (1)], and 9-fold by 2-O-benzyl-5-O-α-phosphonoacetyl-myo- inositol 1,3,4,6-tetrakisphosphate [2-O-Bn-5-PA-InsP4 (2)] (Figure 2A,B). In view of the precise geometric and electrostatic specificity constraints within the active site (Wang et al., 2012), we did not anticipate that it could accommodate 2-O-Bn-5-PA-InsP4, which sports a bulky hydrophobic group. It was therefore unexpected that 2-O-Bn-5-PA-InsP4 should be a more effective activator of ATPase activity than the two natural substrates, InsP6 (Figure 2A) and PP- InsP5 (Weaver et al., 2013). We sought further information on this phenomenon.
Figure 2. The effects of inositol phosphates and analogues upon ATPase activity of PPIP5K2KD.
Panel A shows the effects of the indicated inositol phosphates and analogues (25 μM) upon ATPase activity of PPIP5K2KD. Data represent means and standard errors from 3 experiments. Data for Ins(1,2,3,4,5)P5 (InsP5) and InsP6 are taken from (Weaver et al., 2013). Panel B describes the structures of the reagents that were used.
Design and synthesis of inositol phosphate and diphosphoinositol phosphate analogues
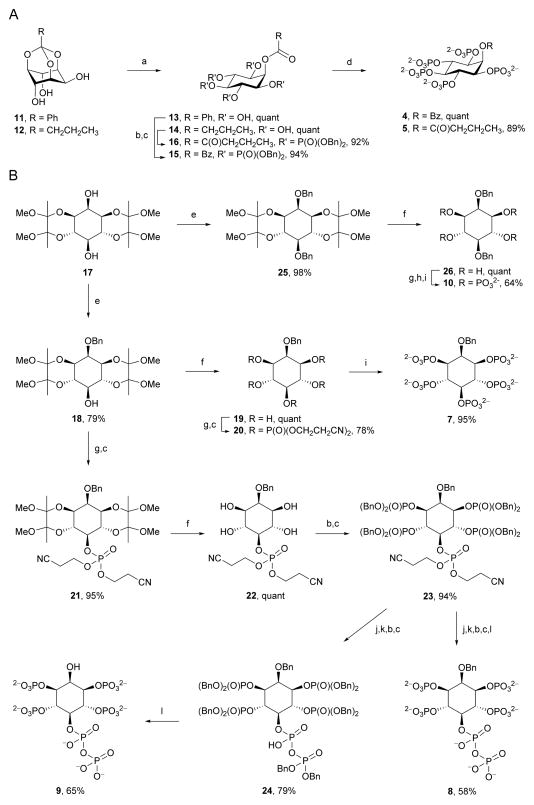
To increase insight into the origin of ligand-stimulated ATPase activity of PPIP5K2KD, we explored the contributions of the benzyl (Bn) and α-phosphonoacetyl (PA) groups in 2-O-Bn-5-PA-InsP4 (2). Our approach was to synthesize a series of analogues (Figure 2B) that exchanged Bn for either OH, monophosphate, or alternative hydrophobes, while the PA group was replaced by either a monophosphate or a diphosphate. For analogues containing acyl groups at O-2, the appropriate synthetic precursors are orthoesters of myo-inositol, because acid hydrolysis of these compounds gives rise to 2-O-acyl esters with very high regioselectivity (Godage et al., 2006; Godage et al., 2013) Thus, 2-O-benzoyl-InsP5 (4) and 2-O-butanoyl-InsP5 (5) (Figure 2B,3A) were synthesized from myo-inositol orthobenzoate (11) and myo-inositol orthobutanoate (12), respectively. Acid hydrolysis of 11 and 12 (Godage et al., 2013) gave pentaols 13 and 14 respectively and subsequent phosphorylation and deprotection provided 2-O-benzoyl-InsP5 (4) and 2-O-butanoyl-InsP5 (5).
For analogues containing alkyl ethers or hydroxyl groups at C-2, synthetic routes proceeding from the symmetrical butane-2,3-diacetal (BDA) protected diol 17 are expedient, as in our recent synthesis of 2-O-(2-aminoethyl)-InsP5 (6) (Riley et al., 2013). The remaining compounds 7–10 were also synthesized from diol 17 using a unified approach (Figure 2B, 3B). Thus, regioselective benzylation of the axial hydroxyl group of diol 17 gave 2-O-benzyl ether 18, and removal of BDA protecting groups gave pentaol 19. Phosphorylation of 19 afforded the symmetrical pentakisphosphate 20 and cleavage of phosphate protection gave 2-O-Bn-InsP5 (7) in 59% overall yield from 17.
Figure 3. Chemical Syntheses.
A, Syntheses of 2-O-Bz-InsP5 (4) and 2-O-But-InsP5 (5) and B, Syntheses of 2-O-Bn-InsP5(7), 2-O-Bn-5-PP-InsP4 (8), 5-PP-InsP4 (9), and 2,5-di-O-Bn-InsP4 (10). Reagents and conditions: (a) TFA:H2O, 10:1; (b) (BnO)2PN(iPr)2, 5-phenyl-1-H-tetrazole, DCM; (c) mCPBA, −40 °C to RT; (d) Pd(OH)2, H2, MeOH, H2O; (e) BnBr, NaH, DMF; (f) aq TFA (90%):DCM, 1:1; (g) (NCCH2CH2O)2PN(iPr)2, 5-phenyl-1-H-tetrazole, DCM; (h) tBuOOH, −40 °C to RT; (i) aq ammonia, 70 °C; (j) DBU, BSTFA; (k) MeOH, TFA; (l) NaHCO3, Pd(OH)2, H2, tBuOH, H2O.
For the synthesis of 2-O-Bn-5-PP-InsP4 (8) and 5-PP-InsP4 (9), the C-5 hydroxyl group of 18 was first phosphitylated with bis(cyanoethyl)(N,N-diisopropylamino)phosphine, followed by oxidation to provide the protected monophosphate 21. Removal of BDA groups followed by phosphorylation afforded the fully-protected pentakisphosphate 23 in high yield (Figure 3B). We constructed the diphosphate group by modifying a recently described methodology (Capolicchio et al., 2013); compound 23 was treated with DBU (1,8-diazabicyclo[5.4.0]undec-7-ene) and BSTFA (N,O-bis(trimethylsilyl)trifluoroacetamide) to remove both cyanoethyl protecting groups on the 5-phosphate, followed by methanolysis of the temporary trimethylsilyl protection to give the phosphate monoester at the C-5 position. Phosphitylation then afforded a mixed P(III)-P(V) intermediate, which was oxidized to produce 24 (Figure 3B). We next prepared 2-O-Bn-5-PP-InsP4 (8) from crude 24 (which contained DBU), by hydrogenolysis over palladium hydroxide, which removed the benzyl esters from the phosphates; amines (either in DBU or in triethylamine (Riley et al., 2012)) inhibit hydrogenolytic cleavage of the 2-O-benzyl ether. Note that 2-O-Bn-5-PP-InsP4 (8) is the first synthetic analogue of an inositol pyrophosphate that retains the actual diphosphate group.
Hydrogenolysis of purified 24 from which DBU was removed yielded 5-PP-InsP4 (9), in 51% overall yield from 23 (Figure 3B). This is the first description of the chemical synthesis of the only naturally occurring diphosphoinositol polyphosphate to feature an unphosphorylated 2-OH group (Figure 1); 5-PP-InsP4 has been implicated in telomere maintenance (York et al., 2005; Saiardi et al., 2005). Finally, for the synthesis of 2,5-di-O-Bn-InsP4 (10), diol 17 was benzylated followed by cleavage of BDA groups. Subsequent phosphorylation and deprotection of cyanoethyl phosphate esters gave (10) in 63% overall yield from 17 (Figure 3B).
Stimulation of the ATPase activity of PPIP5K2KD by synthetic inositol phosphate analogues
As discussed above (and see Figure 2A), 2-O-Bn-5-PA-InsP4 (2) strongly activated non-productive ATPase activity of PPIP5K2KD. The degree of stimulation was dramatically reduced when the latter’s benzyl group was replaced with a hydroxyl group to give 5-PA-InsP4 (3, the PA analogue of 5-PP-InsP4; (Riley et al., 2012)) (Figure 2). Furthermore, 5-PP-InsP4 (9) stimulated ATPase activity at about half the rate shown by 2-O-Bn-5-PP-InsP4 (8). Taken together, these data (Figure 2) lead to an unexpected conclusion that the ligand’s benzyl group makes an important contribution to its association with PPIP5K2KD.
In 2-O-Bn-5-PP-InsP4 (8) the natural and more negatively charged diphosphate (PP) group replaces the PA group at O-5 in 2-O-Bn-5-PA-InsP4 (2). We found that 2-O-Bn-5-PP-InsP4 (8) was slightly (20%) more effective at stimulating ATPase activity than was 2-O-Bn-5-PA-IP4 (2) (Figure 2). Nevertheless, the PA group still contributes to efficacy more than a monophosphate group because 2-O-Bn-InsP5 (7) was less effective than was 2-O-Bn-5-PA-InsP4 (2) (Figure 2).
Even in 2-O-Bn-InsP5 (7), the 2-O-benzyl ether has an enhancing effect, roughly doubling the rate of ATP hydrolysis that is induced by InsP6, which has a phosphate group at C-2 (Figure 2). To investigate the effect of hydrophobes other than benzyl at O-2, we compared 2-O-Bn-InsP5 (7) with 2-O-Bz-InsP5 (4) and 2-O-But-InsP5 (5). The change from the 2-O-benzyl ether to the structurally related 2-O-benzoyl ester led to only a minimal reduction in ATPase-stimulating activity, while substitution with a 2-O-butanoyl ester gave a sharp reduction in activity (Figure 2). Furthermore, the analogue with a positively charged aminoethyl group at O-2 in (2-O-AminoEt-InsP5 (6)) did not stimulate ATPase activity (Figure 2).
We next tested one further analogue in which the diphosphate group in 2-O-Bn-5-PP-InsP4 (8) was replaced with a second benzyl group (i.e. 2,5-di-O-Bn-InsP4; 10). This compound elicited the highest degree of ATPase activity among all of the analogues that we have synthesized (Figure 2). This was initially a puzzling conclusion, as structural considerations indicate that 2,5-di-O-Bn-InsP4 (10) would at best be a poor ligand for the active site.
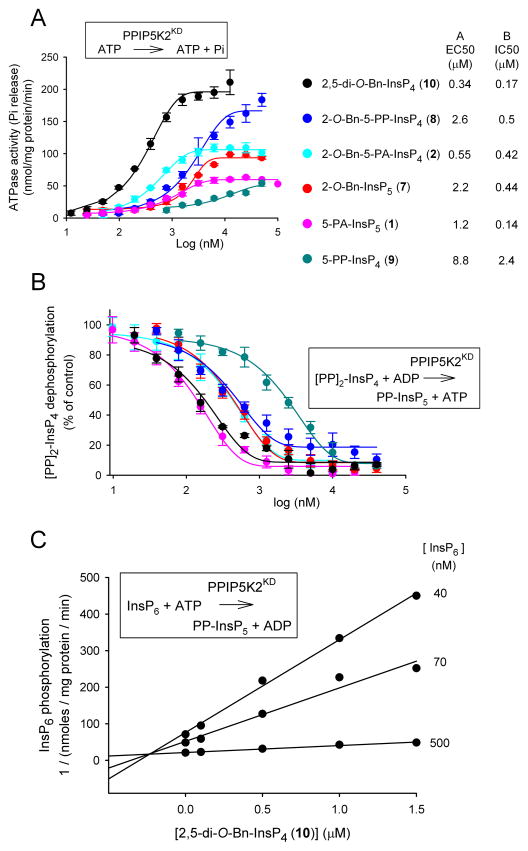
Several of the compounds described above were selected for detailed dose response curves; we used the six compounds that yielded the greatest stimulation of ATPase activity, with the exception of 2-O-Bz-InsP5 (4), which in any case showed similar efficacy to 2-O-Bn-InsP5 (7) (Figure 4A). The most potent of this series of compounds was 2,5-di-O-Bn-InsP4 (10, EC50 = 340 nM; Figure 4A). In general, the rank order of the EC50 values for these compounds approximated the rank order of their maximal effects, with the notable exception of 2-O-Bn-5-PP-InsP4 (8) (Figure 4A).
Figure 4. Effects of inositol phosphate analogues upon PPIP5K2KD activities: ATPase, 1,5-[PP]2-InsP4 dephosphorylation and InsP6 phosphorylation.
Dose-response curves are shown for the indicated inositol phosphate analogues and their stimulation of ATPase activities (Panel A), and their inhibition of ADP-driven dephosphorylation of 100 nM [PP]2-InsP4 (Panel B). Error bars represent standard errors (n=3). The EC50 and IC50 values for the molecules tested are given in the key to the right of panels A and B. Panel C shows a Dixon plot of the inhibition of InsP6 kinase activity of PPIP5K2KD by 2,5-di-O-Bn-InsP4 (10); the data are from a representative experiment (one of three), performed in duplicate. Insets to each panel illustrate the reaction being assayed.
Inhibition of PPIP5K2KD catalytic activity by inositol phosphate analogues
The six compounds that were selected for the dose-response curves in the assays of ATP hydrolysis (Figure 4A) were now investigated for their effects upon diphosphoinositol polyphosphate metabolism by PPIP5K2KD (Figure 4B). We used a high-throughput “reverse-kinase” assay that records ATP generated from ADP during the dephosphorylation of 100 nM 1,5-[PP]2-InsP4 (Riley et al., 2012; Weaver et al., 2013). Each of the tested analogues inhibited [PP]2-InsP4 metabolism by PPIP5K2KD (Figure 4B). The two most potent of these analogues were 5-PA-InsP5 (1) and 2,5-di-O-Bn-InsP4 (10) (Figure 4B). The inhibitory effect of 5-PA-InsP5 (1) was not in itself surprising, as we have already demonstrated it to be a PPIP5K substrate (Riley et al., 2012). However, the inhibition by 2,5-di-O-Bn-InsP4 (10) was more unexpected, since due to its bulk, and its less negative charge compared to physiological substrates, we predict it to be a poor ligand for the active site of the highly specific PPIP5K2KD (Wang et al., 2012). Nevertheless, its ability to inhibit “reverse-kinase” activity was directly confirmed by HPLC analysis of [PP]2-[3H]InsP4 dephosphorylation (Figure S1A,B,C). We confirmed that 2,5-di-O-Bn-InsP4 (10) also inhibited PPIP5K2KD in the “forward”, kinase direction using InsP6, a physiologically-relevant substrate (Figure S1,D,E). A Dixon plot demonstrated that inhibition by 2,5-di-O-Bn-InsP4 (10) was competitive in nature (Ki = 240 nM; Figure 4C), arguing that its actions are not allosteric in nature.
The rank order of potencies (as IC50 values) with which this group of molecules inhibited [PP]2- InsP4 dephosphorylation by PPIP5K2KD (Figure 4B) is different in two key respects from the rank-order of efficacy (EC50 values) for their separate stimulation of ATPase activity (Figure 4A,B). First, 2-O-Bn-InsP5 (7) is 4-fold less potent than 2-O-Bn-5-PA-InsP4 (2) at stimulating ATPase activity (Figure 4A), whereas the two analogues are equally efficient at inhibiting [PP]2-InsP4 metabolism (Figure 4B). Second, 5-PA-InsP5 (1), the weakest activator of ATPase (Figure 4A), is the most potent inhibitor of [PP]2-InsP4 dephosphorylation (Figure 4B). These are observations that suggest there is some uncoupling of inositol phosphate turnover from the ATPase activity.
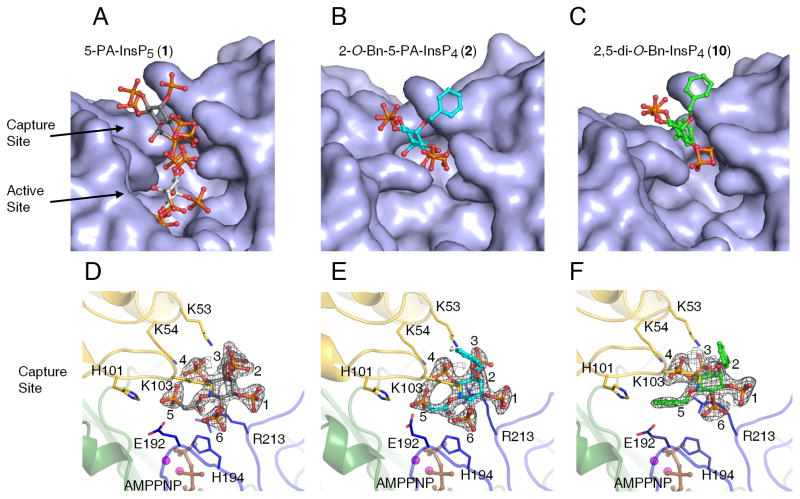
Structural analysis reveals that PPIP5K2KD has two adjacent ligand binding sites
We have previously published an atomic-level description of both 5-PP-InsP5 and InsP6 substrates bound into the catalytic pocket of PPIP5K2KD (Wang et al., 2012). In a separate study, we (Riley et al., 2012) soaked 2 mM 5-PA-InsP5 (1) into crystals of PPIP5K2KD, and reported that this ligand occupies the active site in the same orientation as the natural substrate. At the time of that earlier study, we contoured the simulated annealing omit map at 2σ, and observed some additional but uninterpretable electron density at the entrance of the active site (data not shown). For the current study, we increased the soaking concentration of 5-PA-InsP5 (1) to 10 mM, and have now unequivocally detected an additional ligand-binding site, at 1.7 Å resolution, that is located near the surface of PPIP5K2KD, at the entrance to the catalytic center (Figure 5A,D; Table S1). This new site is too remote from ATP to permit ligand phosphorylation. It should be noted that both the new site and the catalytic pocket cannot be occupied simultaneously, due to steric clashing (Figure 5A, S2A). That is, these particular structural data arise from a mixture of crystal complexes in which 5-PA-InsP5 (1) is separately bound to either of the two sites
Figure 5. X-ray analysis of inositol phosphate analogues bound to PPIP5K2KD uncovers two ligand-binding sites.
X-ray analysis was performed after we soaked crystals of PPIP5K2KD with either 10 mM of 5- PA-InsP5 (1), 5 mM of 2-O-Bn-5-PA-InsP4 (2), or 10 mM of 2,5-di-O-Bn-InsP4 (10). Panels AC give surface representations of ligand binding, with the analogues shown as stick-and-ball models. Atoms are colored blue for nitrogen, red for oxygen, orange for phosphorus, and gray, cyan or green for carbon. Panels D–F provide the corresponding depictions of the protein as a ribbon diagram, with some key residues as stick models and numbered. For increased clarity, panel D only shows the 5-PA-InsP5 (1) that binds to the capture site (see the text for the functional rationalization of this site). The carbons around the inositol ring are numbered. AMPPNP is colored brown and magnesium atoms are colored magenta. Refined 2Fo-Fc maps are contoured at 1.0 σ and are shown in gray mesh.
The stimulation of ATPase activity of PPIP5K2KD by either 2-O-Bn-5-PA-InsP4 (2) or 2,5-di-O-Bn-InsP4 (10) indicate that these compounds also interact with PPIP5K2KD, so we soaked 5–10 mM of either 2-O-Bn-5-PA-InsP4 (2) or 2,5-di-O-Bn-InsP4 (10) into the PPIP5K2KD crystals. X-ray analysis revealed that both of these analogues were exclusively associated with the second ligand binding site (Figure 5B,C,E,F; Table S1). Neither of these analogues were found to occupy the catalytic site. These data suggest the stimulation of ATPase activity by either natural substrates or their analogues is associated with their occupation of the second binding site, not the catalytic site. That explanation is in turn consistent with other data (Figure 4A,B, and see above) that suggest ligand-stimulated ATPase activity is uncoupled from the inositol phosphate kinase activity.
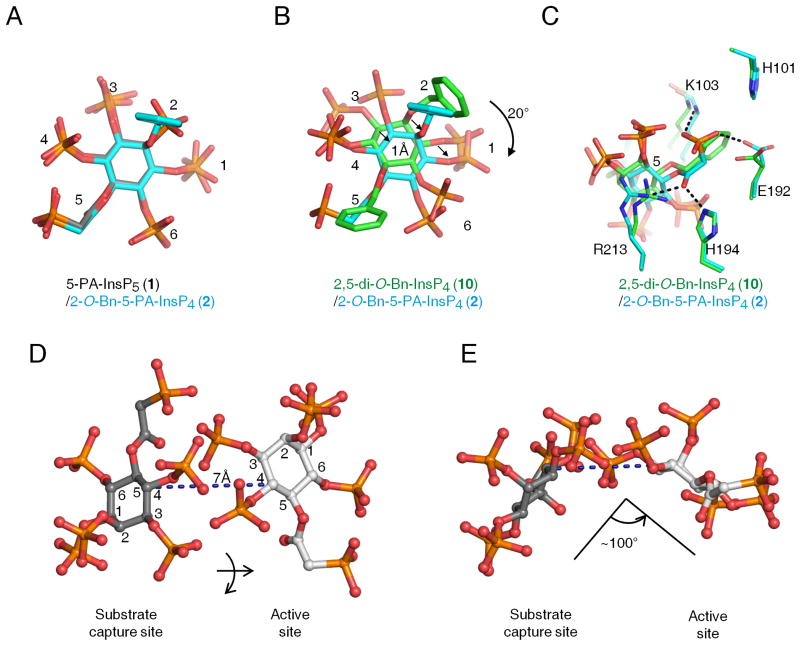
The architecture of this second ligand-binding site is represented by a deep cleft, which is walled on one side by K53, K54 and K103. The opposite face is formed from R213, and a loop created from residues E192 to H194 (Figure 5). In this binding site, the shared groups in 5-PA-InsP5 (1) and 2-O-Bn-5-PA-InsP4 (2) exhibit almost identical conformations (Figure 6A). As for 2,5-di-O- Bn-InsP4 (10) this molecule is rotated about 20º clockwise (Figure 6B). For both 2-O-Bn-5-PA-InsP4 (2) and 2,5-di-O-Bn-InsP4 (10), their phosphate groups at C-3 and C-4 would clash with the positions that the C-4 and C-3 phosphates of InsP6/5-PP-InsP5 normally occupy in the active site (Figure S2,A,B,C). This phenomenon presumably contributes to the potency of both 2-O-Bn- 5-PA-InsP4 (2) and 2,5-di-O-Bn-InsP4 (10) as inhibitors of inositol phosphate turnover by PPIP5K2KD (Figure 4B,C). These structural considerations explain how both compounds can inhibit inositol phosphate kinase activity without occupying the catalytic site.
Figure 6. Relative orientations of inositol phosphate analogues in the second ligand binding site of PPIP5K2KD.
Data for ligand orientation in the second binding site were obtained as described in the Figure 5 legend. Analogs are depicted as stick models. Atoms are colored blue for nitrogen, red for oxygen and orange for phosphorus. Panel A superimposes 5-PA-InsP5 (1) (carbons colored gray) and 2-O-Bn-5-PA-InsP4 (2) (carbons in cyan). Panel B and C superimpose 2,5-di-O-Bn-InsP4 (10) (carbons colored green) and 2-O-Bn-5-PA-InsP4 (2) (carbons in cyan). Arrows in panel B emphasize spatial separation, and panel C includes details of ligand interactions with neighboring amino-acid residues. Panels D and E illustrate that transfer of 5-PA-InsP5 (1) between the two sites involves a lateral migration of 7 A, a ring flip, and a rotation of about 100. Atoms are colored blue for nitrogen, red for oxygen, orange for phosphorus, and gray for carbon. The inositol ring is numbered.
The association of 2-O-Bn-5-PA-InsP4 (2) and 2,5-di-O-Bn-InsP4 (10) with the second binding site is facilitated by their phosphate groups at C-3 and C-4 making multiple electrostatic interactions with K53, K54 and R213 (Figure 5, S2 E,F). The terminal phosphonate of 2-O-Bn-5-PA-InsP4 (2) also forms polar contacts with the backbone of K103 and the H194 side chains (Figure 5, S2E). Additionally, the 5-O-benzyl group in 2,5-di-O-Bn-InsP4 (10) has van der Waals interactions with the side chains of H101 and E192 (Figure 5, S2F). The 2-O-benzyl group in this analogue is disordered, indicative of its mobility. Nevertheless, our catalytic data (Figure 4) indicate that this benzyl group makes a significant contribution to ligand potency.
The data that we have obtained allow important conclusions to be drawn concerning the likely significance of the second binding site. For example, we can exclude the possibility that, as is the case with certain enzymes (Reed et al., 2010), the role of the second, non-catalytic substrate-binding site is to regulate enzyme activity by substrate inhibition; PPIP5K2KD does not exhibit that property (Weaver et al., 2013). Other enzymes use non-catalytic ligand-binding sites to promote enzyme activation allosterically (Grant, 2012). That also seems unlikely to be the case for PPIP5K2KD, since our inositol phosphate analogues do not enhance kinase activity, but inhibit it instead (Figure 4B; (Riley et al., 2012)). The nature of the inhibition was competitive (Figure 4C), further arguing against an allosteric effect. Instead, the proximity to the active site of the second ligand binding site (Figure 4) suggests the latter plays a role in catalysis of natural substrates, by facilitating their capture from the bulk phase. A precedent for such a phenomenon has been reported for microbial anthranilate phosphoribosyltransferases (Castell et al., 2013; Marino et al., 2006); anthranilate substrate is first captured at a surface-mounted binding-site, before delivery to a proximal catalytic site (Castell et al., 2013). A similar substrate transfer is feasible in PPIP5K2KD, and our structural data provide some atomic-level insight into such a phenomenon. For example, we presume that delivery of natural substrate is efficient, so that substrate only occupies the capture site transiently; this may explain why we could not detect substrate in the capture site in our crystal structures. We found that such substrate transfer within PPIP5K2KD requires that the inositol ring be flipped, and rotated by approximately 100º (Figure 6D,E). This motion is presumably facilitated by the flexible amino-acid side chains that comprise the active site (Wang et al., 2012). Such conformational dynamics can reinforce catalytic specificity (Herschlag, 1988), which is indeed a notable feature of PPIP5K2KD (Wang et al., 2012).
Site directed mutagenesis of the substrate capture site
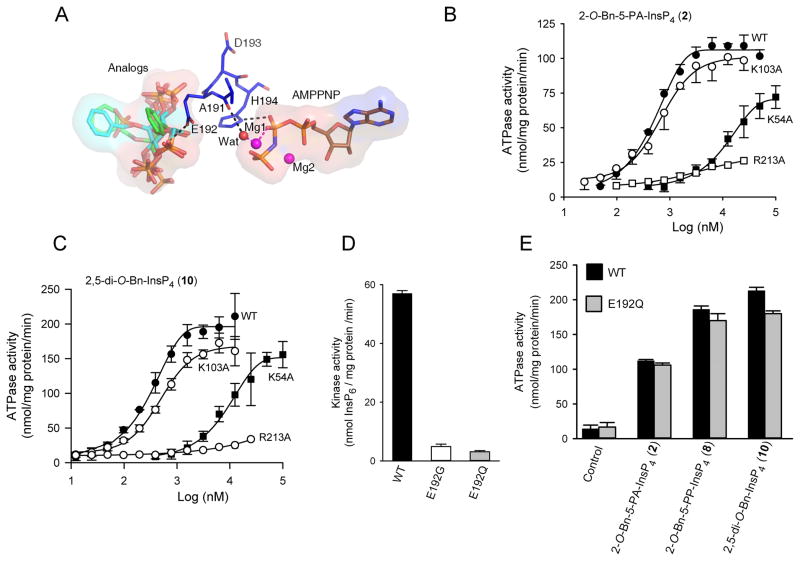
Mutagenesis offers a valuable means of pursuing conclusions drawn from structural analysis. The loop that is formed from residues A191 to H194 screens the catalytic site from ligands that are associated with the second binding site (Figure 5, 7A, S3). The influence of the carbonyl oxygen of A191 may be indirect, through a hydrogen bond with a water molecule that in turn coordinates with a Mg2+ ion that interacts with the nucleotide’s β-phosphate moiety (Figure 7A). The role of H194 seems more direct; it can form a hydrogen bond with the oxygen atom of the nucleotide’s β-phosphate (Figure 7A, S3). This could stabilize the transition state, or have some other catalytic role. These interactions could be relevant to both kinase and ATPase activities of PPIP5K2KD. Consistent with these ideas, an H194A mutant did not exhibit any detectable ATPase activity, either in the presence or absence of 2-O-Bn-5-PA-InsP4 (2) or 2,5-di-O-Bn-InsP4 (10), and its inositol phosphate kinase activity was reduced 80-fold compared to that of the wild-type enzyme (data not shown).
Figure 7. Impact upon the catalytic properties of PPIP5K2KD following mutation of K54, K103, E192 and K213.
A Shows the spatial separation of AMP-PNP from the PPIP5K2KD substrate capture site. Protein residues are shown as stick models. AMPPNP, and inositol phosphate analogs 5-PA-InsP5 (1), 2- O-Bn-5-PA-InsP4 (2), and 2,5-di-O-Bn-InsP4 (10) and all are shown as stick models. Atoms are colored blue for nitrogen, red for oxygen, orange for phosphorus, and carbon for grey, cyan or green. Hydrogen bonds are shown as black dashed lines. B, C, Dose-response curves for the effects of 2-O-Bn-5-PA-InsP4 (2) and 2,5-di-O-Bn-InsP4 (10) upon the ATPase activities of wild-type PPIP5K2KD(closed circles; data are taken from Figure 2A) and the following mutants: K54A mutant (closed squares), K103A (open circles) and R213A (open squares). Panel D shows the InsP6 kinase activities of wild-type, E192G and E192Q mutants of PPIP5K2KD, determined with 5 μM substrate. Panel E shows the ATPase activities of wild-type and E192Q mutants of PPIP5K2KD obtained in the presence of the indicated analogues, at concentrations of 25 μM. Error bars represent standard errors from three experiments (error bars are not shown when they are smaller than the symbol).
As discussed above, our structural and biochemical data led us to hypothesize that the stimulation of ATPase activity by either natural substrates or their analogues is associated with their occupation of the substrate capture site, not the catalytic site. We attempted to consolidate this idea by selecting for mutation residues that might separate ATPase activity from inositol phosphate kinase activity. For example, our X-ray data (Figure 5) indicate that K54 and R213 interact with ligand at the substrate capture site. However, previous mutagenic work has shown that both K54 and R213 also contribute directly to inositol phosphate kinase activity (Wang et al., 2012). Nevertheless, we did observe that K54A and R213A mutants exhibited a substantially impaired degree of stimulation of ATPase activity by either 2-O-Bn-5-PA-InsP4 (2) or 2,5-di-O-Bn-InsP4 (10) (Figure 7B,C). These mutagenic data are consistent with the idea that K54 and R213 participate in substrate capture.
The side-chain of K103 is also suggested by our structural data to interact with ligand that is bound to the capture site (Figure 5, S2). Nevertheless, a K103A mutant showed only a slight reduction in 2-O-Bn-5-PA-InsP4 (2)-activated and 2,5-di-O-Bn-InsP4 (10)-activated ATPase activity, compared to wild-type enzyme (Figure 7B,C). Likewise, the InsP6 kinase activity of the K103A mutant (71 ± 4 nmol/mg protein/min, n=3) was similar to that of wild-type enzyme (61 ± 4 nmol/mg protein/min).
We also mutated E192. This residue is present in the loop between the two ligand binding sites (Figure 7A). It is too distant from ATP and the catalytic site to influence either directly, and electrostatic repulsion would prevent it from interacting with negatively charged groups on substrates located in the second binding site. Nevertheless, E192 is evolutionarily conserved in PPIP5Ks from mammals to yeast (not shown), suggestive of functional significance. We investigated the significance of E192 by preparing E192G and E192Q mutations that we posited would eliminate electrostatic repulsion between the amino acid side chain and phosphorylated ligands, enhancing ligand binding to the substrate capture site. We further hypothesized that such an effect might reduce the efficiency of transfer of substrate to the catalytic site, which in turn would lead to a reduction in inositol phosphate kinase activity. Indeed, we found that these E192G and E192Q mutations reduced the rate of InsP6 phosphorylation by 12- and 18-fold respectively, compared to wild-type PIP5K2KD (Figure 7D). It was of further note that neither of these particular mutations altered the rate of non-productive, ligand-stimulated ATPase activity elicited by either 2-O-Bn-5-PA-InsP4 (2), 2-O-Bn-5-PP-InsP4 (8), or 2,5-di-O-Bn-InsP4 (10) (Figure 7E), confirming the uncoupling of this aspect of PPIP5K2KD activity from its kinase activity.
We have shown that 2,5-di-O-Bn-InsP4 (10) binds to a substrate-capture site on the surface of PPIP5K2KD (Figure 5), thereby inhibiting the enzyme’s catalytic activity (Figure 4). This new ligand binding site may be exploitable as a pharmacological target. We therefore examined the specificity of 2,5-di-O-Bn-InsP4 (10), by investigating if it interacted with IP6K2, a member of a different class of kinases that also phosphorylate InsP6 and synthesize diphosphoinositol polyphosphates (see Figure 1). Since the affinity of IP6K2 for InsP6 (430 nM; (Saiardi et al., 2000)) is very similar to that for InsP6 phosphorylation by PPIP5K2KD (390 nM; (Weaver et al., 2013)), an identical substrate concentration ([InsP6] = 500 nM) was used to assay both enzymes. As shown in Figure 4C, 2,5-di-O-Bn-InsP4 (10) inhibits PPIP5K2KD with an IC50 value of 1 μM (Figure 4C). This analogue was found to be a much weaker inhibitor of IP6K2 activity (IC50 = 63 μM; data not shown). Thus, 2,5-di-O-Bn-InsP4 (10) may be a useful lead molecule for future development of a drug that can specifically inhibit PPIP5Ks and not IP6Ks, even though both groups of enzymes phosphorylate InsP6.
In conclusion, our studies have uncovered a new, “catch-and-pass” aspect to the catalytic cycle of PPIP5K2KD. Substrate (either InsP6 or 5-PP-InsP5) first associates with a ligand binding site on the surface of the kinase. We propose that this phenomenon enhances kinase activity by improving substrate capture from the bulk phase; substrate is then delivered into the catalytic pocket. As far as we are aware, this is the first indication of a substrate-capture mechanism in a small-molecule kinase. Indeed, the only precedent that we have found in the literature for a dedicated substrate capture site on any enzyme is that observed for certain microbial anthranilate phosphoribosyl-transferases (Castell et al., 2013; Marino et al., 2006). Our structural, biochemical and mutagenic data have also led us to conclude that the stimulation of ATPase activity of PPIP5K by inositol phosphate analogues is associated with their occupation of the substrate capture site, not the catalytic site. Moreover, the previously puzzling observation (Weaver et al., 2013) that a degree of non-productive ATP hydrolysis is also stimulated by natural substrate, can now be rationalized as a consequence of its interaction with the capture site. Furthermore, the fact that InsP6-stimulated ATPase activity is abolished by an R213A mutation (Weaver et al., 2013) can now be viewed as resulting from an impairment to substrate occupation of the capture site.
Our atomic-level description of this new ligand-binding site on PPIP5K2KD indicates its structural determinants of specificity differ substantially from those of the catalytic site. We have further shown that synthesis of diphosphoinositol polyphosphates by PPIP5K2KD is inhibited by ligands that bind to the capture site but not the catalytic site. These findings in turn raise the possibility that there may be cellular constituents that might inhibit catalytic activity by binding to this site. Finally, the substrate capture site offers a new and selective target for the purposes of rational drug design, including screening in silico (Kitchen et al., 2004), that is free from many of the usual specificity constraints within the catalytic site that typically complicate pharmacological targeting of small-molecule kinases. It may also be possible to design a ligand that occupies both sites.
SIGNIFICANCE
PPIP5K1/2 are small-molecule kinases that synthesize diphosphoinositol polyphosphates, which function at the interface of cell signaling and organismic homeostasis. Synthetic chemical modulators of cell-signaling enzymes such as the PPIP5Ks can provide valuable insight into catalytic mechanisms, they decipher the biological roles of the enzymes in situ, and they generate leads for therapeutic drug development. Substrate analogues offer one approach for the preparation of such chemical reagents. However, the architecture of the active site of PPIP5Ks, as revealed by recent X-ray analysis, has identified substantial geometric and electrostatic constraints that limit the options for designing a substrate analogue that might be an effective modulator. In the current study, we describe the first chemical synthesis of both the naturally-occurring 5-PP-InsP4, and a new family of inositol polyphosphate analogues that include molecules with hydrophobes at the 2- and/or 5-positions. Importantly, these molecules have led us to discover the existence of a second, less constrained substrate-binding site on the surface of PPIP5K2, adjacent to the catalytic pocket. We provide an atomic-level description of this new site, and we describe its role in the catalytic cycle in capturing substrate from the bulk phase. With the assistance of site-directed mutagenesis of this site, we show that its occupation is associated with an unusual, ligand-activated ATPase activity. This considerable amount of new information had not previously been accessible from our experiments with natural substrates, and so represents a considerable success of our chemical biology approach. In addition to adding a new facet to the repertoire of catalytic specializations of the PPIP5Ks, our structural and functional characterization of this ligand-binding site offers a promising new target for drug development; to this end, 2,5-di-O-Bn-InsP4 is a significant lead compound.
EXPERIMENTAL PROCEDURES
Protein Expression, Purification, Crystallization and Structure Determination
The kinase domain of human PPIP5K2 (PPIP5K2KD; residues 1–366) and the N-terminally truncated domain used for the crystallography studies (residues 41–366) were sub-cloned, expressed and purified as before. The latter was crystallized by hanging drop vapor diffusion against a well buffer of 12% (w/v) PEG 3350, 20 mM MgCl2, 0.1 M HEPES, pH 7.0, 1 mM AMP-PNP and 2 mM CdCl2 at 4 °C. The crystals were then soaked with 5–10 mM compounds in a stabilizing buffer containing 22% (w/v) PEG 3350, 10 mM MgCl2, 0.1 M sodium acetate, pH 5.2 or 7.0 at 4 °C for 3 days. Cryosolvent was prepared by adding 33% ethylene glycol into the soaking buffer. Diffraction data were collected using APS beamlines 22-BM and 22-ID. All data were processed with the program HKL2000. The structure was determined using rigid body and direct Fourier synthesis, and refined with the equivalent and expanded test sets. The structure was further manually rebuilt with COOT and refined with PHENIX and REFMAC from the CCP4 package. Ligand topology and parameter files were prepared using the PRODRG server. The molecular graphics representations were prepared with the program PyMol (Schrödinger, LLC). The 2D ligand-protein interaction diagrams were generated by LigPlot+. Atomic coordinates and structure factors have been deposited with the Protein Data Bank with accession codes: 4NZM, 4NZN, 4NZO.
Enzyme assays
HPLC was used to assay [3H]InsP6 phosphorylation by human PPIP5K2KD or human IP6K2 in 100 μl buffer containing 50 mM KCl, 20 mM HEPES pH 7.0, 7 mM MgSO4, 5 mM ATP, 1 mM EDTA (for IP6K2, [MgSO4] was 12 mM and [ATP] was 10 mM). The initial InsP6 concentration was either 500 nM, 70 nM or 40 nM. Various concentrations of 2,5-di-O-Bn-InsP4 were present as indicated. Radioactivity was assessed using an in-line Flo1 detector. The ADP-driven dephosphorylation of 100 nM 1,5-[PP]2-InsP4 by PPIP5K2KD was usually determined by a luminescence-based assay of ATP accumulation (Riley et al., 2012; Weaver et al., 2013). The IC50 values were determined using GraphPad Prism v6.02 (n ≥ 3). In some experiments, 1,5-[PP]2-[3H]InsP4 dephosphorylation by PPIP5K2KD was assayed in 50 μl buffer containing 50 mM KCl, 20 mM HEPES pH 7.0, 7 mM MgSO4, 5 mM ADP, 1 mM EDTA. Reactions were then quenched, neutralized, and analyzed by HPLC using a Partisphere SAX column (Weaver et al., 2013); 1 ml fractions were collected for liquid scintillation spectrometry. The ATPase activity of PPIP5K2KD (10–270 μg/ml) was assayed from Pi release following incubation at 37°C for 120–180 min in 20 μl reaction mixtures containing 20 mM Tris/HCl, pH 7.5, 10 mM ATP, 100 mM KCl, 0.1 mM EDTA and 13 mM MgCl2.
Statistical Analysis
In the figures, error bars represent standard errors from three experiments.
Supplementary Material
Highlights.
First chemical synthesis of 5-PP-InsP4 and a diphosphorylated analogue
Chemical synthesis of inositol polyphosphate analogues with hydrophobic groups
An inositol pyrophosphate kinase has a surface-mounted, substrate capture site
Structural and biochemical characterization of a catch-and-pass catalytic cycle
Acknowledgments
We thank the Wellcome Trust (Programme Grant 082837 to AMR and BVLP) for support. BVLP is a Wellcome Trust Senior Investigator (Grant 101010). This research was also supported by the Intramural Research Program of the NIH / National Institute of Environmental Health Sciences. X-ray data were collected at Southeast Regional Collaborative Access Team (SER-CAT) 22- beamline at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions may be found at www.ser-cat.org/members.html. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. We also thank the NIEHS Collaborative crystallography group for assistance with data collection.
Footnotes
Supplementary information includes three figures, one table, some methods, and details of the synthetic chemistry and NMR spectra for all new compounds.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Albert C, Safrany ST, Bembenek ME, Reddy KM, Reddy KK, Falck JR, Bröker M, Shears SB, Mayr GW. Biological variability in the structures of diphosphoinositol polyphosphates in Dictyostelium discoideum and mammalian cells. Biochem J. 1997;327:553–560. doi: 10.1042/bj3270553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin D, Colombi M, Moroni C, Hall MN. Rapamycin passes the torch: a new generation of mTOR inhibitors. Nat Rev Drug Discov. 2011;10:868–880. doi: 10.1038/nrd3531. [DOI] [PubMed] [Google Scholar]
- Best MD, Zhang H, Prestwich GD. Inositol polyphosphates, diphosphoinositol polyphosphates and phosphatidylinositol polyphosphate lipids: Structure, synthesis, and development of probes for studying biological activity. Nat Prod Rep. 2010;27:1403–1430. doi: 10.1039/b923844c. [DOI] [PubMed] [Google Scholar]
- Capolicchio S, Thakor DT, Linden A, Jessen HJ. Synthesis of Unsymmetric Diphospho-Inositol Polyphosphates. Angew Chem Int Ed Engl. 2013;52:6912–5916. doi: 10.1002/anie.201301092. [DOI] [PubMed] [Google Scholar]
- Castell A, Short FL, Evans GL, Cookson TV, Bulloch EM, Joseph DD, Lee CE, Parker EJ, Baker EN, Lott JS. The Substrate Capture Mechanism of Mycobacterium tuberculosis Anthranilate Phosphoribosyltransferase Provides a Mode for Inhibition. Biochemistry. 2013;52:1776–1787. doi: 10.1021/bi301387m. [DOI] [PubMed] [Google Scholar]
- Chakraborty A, Koldobskiy MA, Bello NT, Maxwell M, Potter JJ, Juluri KR, Maag D, Kim S, Huang AS, Dailey MJ, Saleh M, Snowman AM, Moran TH, Mezey E, Snyder SH. Inositol pyrophosphates inhibit akt signaling, thereby regulating insulin sensitivity and weight gain. Cell. 2010;143:897–910. doi: 10.1016/j.cell.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Mollapour E, Shears SB. Signal transduction during environmental stress: InsP8 operates within highly restricted contexts. Cell Signal. 2005;17:1533–1541. doi: 10.1016/j.cellsig.2005.03.021. [DOI] [PubMed] [Google Scholar]
- Draskovic P, Saiardi A, Bhandari R, Burton A, Ilc G, Kovacevic M, Snyder SH, Podobnik M. Inositol hexakisphosphate kinase products contain diphosphate and triphosphate groups. Chem Biol. 2008;15:274–286. doi: 10.1016/j.chembiol.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Godage HY, Riley AM, Woodman TJ, Potter BVL. Regioselective hydrolysis of myo-inositol 1,3,5-orthobenzoate via a 1,2-bridged 2′-phenyl-1′,3′-dioxolan-2′-ylium ion provides a rapid route to the anticancer agent Ins(1,3,4,5,6)P5. Chem Commun. 2006:2989–2991. doi: 10.1039/b605392k. [DOI] [PubMed] [Google Scholar]
- Godage HY, Riley AM, Woodman TJ, Thomas MP, Mahon MF, Potter BVL. Regioselective opening of myo-inositol orthoesters: mechanism and synthetic utility. J Org Chem. 2013;78:2275–2288. doi: 10.1021/jo3027774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale NA, Zaremba A, Janoshazi AK, Weaver JD, Shears SB. PPIP5K1 Modulates Ligand Competition Between Diphosphoinositol Polyphosphates and PtdIns(3,4,5)P3 for Polyphosphoinositide-Binding Domains. Biochem J. 2013;453:413–426. doi: 10.1042/BJ20121528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant GA. Kinetic evidence of a noncatalytic substrate binding site that regulates activity in Legionella pneumophila L-serine dehydratase. Biochemistry. 2012;51:6961–6967. doi: 10.1021/bi3008774. [DOI] [PubMed] [Google Scholar]
- Herschlag D. The role of induced fit and conformational changes in specificity and catalysis. Bioorganic Chemistry. 1988;16:62–96. [Google Scholar]
- Illies C, Gromada J, Fiume R, Leibiger B, Yu J, Juhl K, Yang SN, Barma DK, Falck JR, Saiardi A, Barker CJ, Berggren PO. Inositol pyrophosphates determine exocytic capacity. Science. 2007;318:1299–1302. doi: 10.1126/science.1146824. [DOI] [PubMed] [Google Scholar]
- Kitchen DB, Decornez H, Furr JR, Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discov. 2004;3:935–949. doi: 10.1038/nrd1549. [DOI] [PubMed] [Google Scholar]
- Marino M, Deuss M, Svergun DI, Konarev PV, Sterner R, Mayans O. Structural and mutational analysis of substrate complexation by anthranilate phosphoribosyltransferase from Sulfolobus solfataricus. J Biol Chem. 2006;281:21410–21421. doi: 10.1074/jbc.M601403200. [DOI] [PubMed] [Google Scholar]
- Pulloor NK, Nair S, Kostic AD, Bist P, Weaver JD, Tyagi R, Uchil PD, York JD, Snyder SH, Garcia-Sastre A, Lin R, Shears SB, Xavier RJ, Krishnan MN. Human Genome-Wide RNAi Screen Identifies an Essential Role for Inositol Pyrophosphates in Type-I Interferon Response. PLoS Pathog. 2014;10:e1003981. doi: 10.1371/journal.ppat.1003981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed MC, Lieb A, Nijhout HF. The biological significance of substrate inhibition: a mechanism with diverse functions. Bioessays. 2010;32:422–429. doi: 10.1002/bies.200900167. [DOI] [PubMed] [Google Scholar]
- Riley AM, Wang H, Weaver JD, Shears SB, Potter BVL. First synthetic analogues of diphosphoinositol polyphosphates: interaction with PPIP5 kinase. Chem Commun. 2012;48:11292–11294. doi: 10.1039/c2cc36044f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley AM, Windhorst S, Lin H-Y, Potter BVL. Cellular internalisation of an inositol phosphate visualised using fluorescent InsP5. Chembiochem. 2013;15:57–67. doi: 10.1002/cbic.201300583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiardi A, Caffrey JJ, Snyder SH, Shears SB. The Inositol Hexakisphosphate Kinase Family: Catalytic Flexibility, and Function in Yeast Vacuole Biogenesis. J Biol Chem. 2000;275:24686–24692. doi: 10.1074/jbc.M002750200. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Resnick AC, Snowman AM, Wendland B, Snyder SH. Inositol pyrophosphates regulate cell death and telomere length via PI3K-related protein kinases. Proc Nat Acad Sci USA. 2005;102:1911–1914. doi: 10.1073/pnas.0409322102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shears SB. Diphosphoinositol polyphosphates: metabolic messengers? Mol Pharmacol. 2009;76:236–252. doi: 10.1124/mol.109.055897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szijgyarto Z, Garedew A, Azevedo C, Saiardi A. Influence of inositol pyrophosphates on cellular energy dynamics. Science. 2011;334:802–805. doi: 10.1126/science.1211908. [DOI] [PubMed] [Google Scholar]
- Thomas MP, Potter BVL. The enzymes of human diphosphoinositol metabolism. FEBS Journal. 2013;281:14–33. doi: 10.1111/febs.12575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Falck JR, Hall TM, Shears SB. Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding. Nat Chem Biol. 2012;8:111–116. doi: 10.1038/nchembio.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver JD, Wang H, Shears SB. The kinetic properties of a human PPIP5K reveal that its kinase activities are protected against the consequences of a deteriorating cellular bioenergetic environment. Biosci Rep. 2013;33:228–241. doi: 10.1042/BSR20120115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Dul BE, Trevisan AJ, Fiedler D. Synthesis and characterization of non-hydrolysable diphosphoinositol polyphosphate second messengers. Chem Sci. 2013;4:405–410. doi: 10.1039/C2SC21553E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York SJ, Armbruster BN, Greenwell P, Petes TD, York JD. Inositol diphosphate signaling regulates telomere length. J Biol Chem. 2005;280:4264–4269. doi: 10.1074/jbc.M412070200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.