Abstract
Emerging and re-emerging mosquito-borne viruses continue to pose a significant threat to human health throughout the world. Over the past decade, West Nile virus (WNV), Dengue virus (DENV), and Chikungunya virus (CHIKV), have caused annual epidemics of virus-induced encephalitis, hemorrhagic fever\shock syndromes, and arthritis, respectively. Currently, no specific antiviral therapies or vaccines exist for use in humans to combat or prevent these viral infections. Thus, there is a pressing need to define the virus-host interactions that govern immunity and infection outcome. Recent technological breakthroughs in ‘omics’ resources and high-throughput based assays are beginning to accelerate antiviral drug discovery and improve on current strategies for vaccine design. In this review, we highlight studies with WNV and discuss how traditional and systems based approaches are being used to rapidly identify novel host targets for therapeutic intervention and develop a deeper conceptual understanding of the host response to virus infection.
Emerging mosquito-borne viruses
Emerging and re-emerging mosquito-borne viruses continue to pose a significant threat to human health throughout the world. Over the past decade, West Nile virus (WNV), Dengue virus (DENV), and Chikungunya virus (CHIKV), have caused annual epidemics of virus-induced encephalitis, hemorrhagic fever\shock syndromes, and arthritis, respectively. Since its introduction to the United States in 1999, WNV has been estimated to cause more than 3 million infections, resulting in over 780,000 illnesses, 38,000 clinically confirmed cases, and 1,500 deaths between 1999-2014 [1,2]. Since the 1960s, DENV has emerged in the Americas, southeast Asia, and the Indian subcontinent and has been estimated to cause 50 to 100 million infections per year and a total of 2.5 billion people worldwide are at risk of infection. In 2005, a major outbreak of CHIKV occurred on the Reunion Island off the western coast of Africa, resulting in annual epidemics of CHIK infection in Africa, southeast Asia, India and Australia [3]. Most recently, CHIKV has spread to the Americas, with over 8000 suspected cases within the Caribbean islands [4]. This is the first documented outbreak of autochthonous CHIKV in the Americas. Currently, no specific antiviral therapies or vaccines exist for use in humans to combat or prevent these mosquito-borne infections. Thus, there is a pressing need to define the virus-host interactions that govern immunity and infection outcome. Recent technological advancements in ‘omics’ resources and high-throughput based assays are beginning to accelerate antiviral drug discovery and improve on current strategies for vaccine design. In this review, we highlight studies with WNV and discuss how traditional and systems based approaches are being used rapidly identify novel host targets for therapeutic intervention and develop a deeper conceptual understanding of the host response to virus infection.
West Nile virus pathogenesis
WNV infection of mice has provided valuable insight into the pathogenesis of virus infection in humans (reviewed in [5]). Three distinct stages of WNV pathogenesis have been defined through studies in mice: initial infection and spread (early phase), peripheral virus amplification (viremic phase), and neuroinvasion (central nervous system (CNS) phase). The early phase is defined by WNV infection and replication at the site of inoculation, in keratinocytes [6], dermal dendritic cells and skin-resident Langerhans cells [7]. The viremic phase is defined by virus spread to the spleen, a primary site for peripheral virus replication, and non-productive infection of other peripheral organs (e.g. liver, kidney, etc.). During these first two stages, dendritic cells, macrophages, and possibly neutrophils are believed to be the key target cells of infection [8-10]. While the specific dendritic cell or macrophage subsets that amplify WNV in vivo have yet to be identified, genetic deletion of CD8+α DCs or antibody-mediated depletion of macrophages lead to dysregulated host control of virus replication, increased mortality, and defects in adaptive immunity [9,11-13]. The final stage involves WNV neuroinvasion into the central nervous system, where the virus targets and replicates in neuronal subsets. These distinct stages of pathogenesis are believed to recapitulate what occurs in humans following WNV transmission by a mosquito bite.
Following virus infection of a target host (i.e. humans infected with WNV), the immune system is rapidly engaged and drives antiviral immune responses necessary for controlling virus replication, limiting virus-mediated pathology, and providing immunity to re-infection (Fig. 1A). Accordingly, the innate and adaptive immune systems are essential for providing protection against WNV infection [5]. In particular, type I IFN and related antiviral defenses are triggered following recognition of WNV infection and activation of the RIG-I like receptor (RLR), Toll-like receptor (TLR), and NOD-like receptor (NLR) signaling pathways. Infection analysis of RIG-I−/− or MDA5−/− macrophages, dendritic cells and fibroblasts revealed that RIG-I is activated early during infection whereas MDA5 is required for enhancing and sustaining type I IFN and interferon-stimulated gene (ISG) expression [14-16]. Furthermore, in vivo studies have demonstrated that RLR signaling is required for protection as well as controlling peripheral organ and CNS viral burden, limiting virus-mediated pathology, and programming protective immunity to WNV infection [5,17]. Similarly, the TLRs [18,19] and NLRs [20,21] have been shown to restrict virus replication in a cell and tissue-specific manner and regulate protective CNS immunity during WNV infection. Additionally, components of the innate immune cellular responses, including natural killer cells [22,23], neutrophils [8], and γδ T cells [24], and cell-mediated and humoral adaptive immune responses are critical for protection against WNV infection (as reviewed in greater detail in [5]). Studies in humans infected with WNV have been limited, however, certain risk factors for symptomatic infection outcome include advanced age, immunocompromised status [25], genetic factors [26-28], and reduced expansion of regulatory T cells [29].
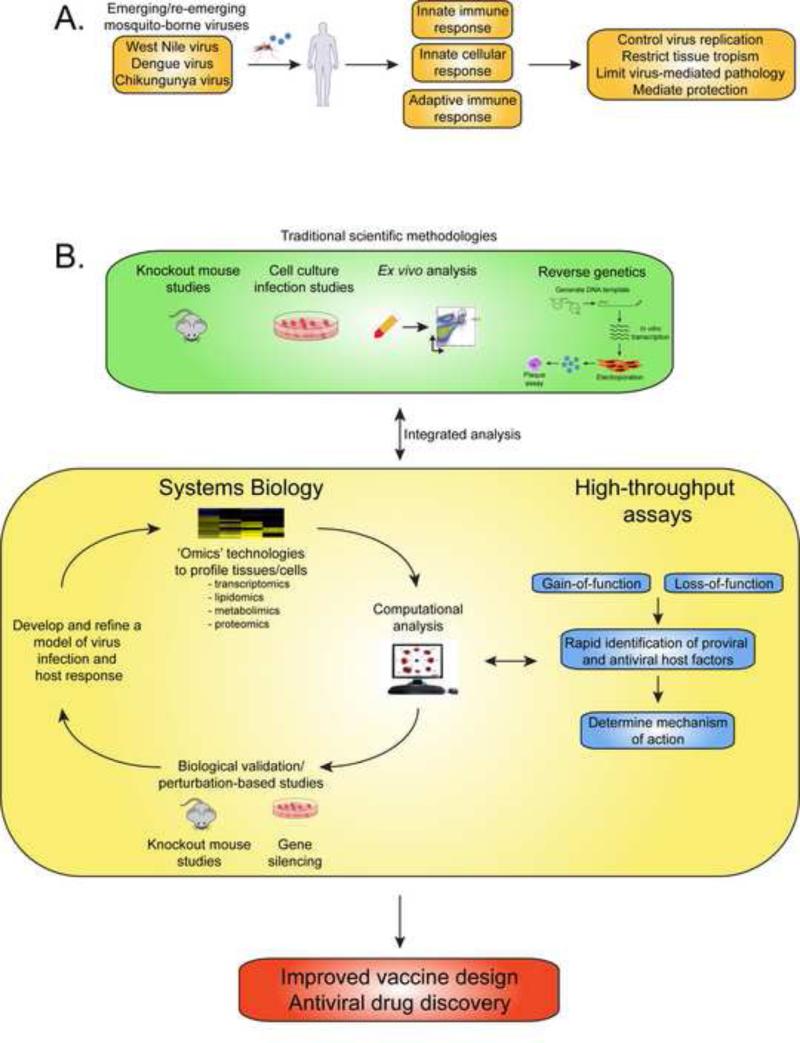
Figure 1. Systems analysis to study the host response to emerging mosquito-borne viruses.
(A) Schematic representing the host immune response to virus infection. (B) Traditional scientific methodologies to study the immune response to virus infection have predominantly involved knockout mouse, cell culture, ex vivo infection analysis and the use of a reverse genetics systems to manipulate viruses. Integrating this approach with systems biology and high-throughput based assays can provide a platform to accelerate identification of host targets of therapeutic intervention and improve on current strategies for vaccine design.
Systems biology approach to study the host response to WNV infection
Traditional scientific methodologies to study the host response to WNV infection have been paramount for identifying the viral and host genetic factors that control virus replication and infection outcome (Fig. 1B). While extremely important, this approach often involves studying individual components of the immune system (i.e. knockout mice) and results in providing a narrow and simplified representation of the host response to viral infection. Systems biology is a scientific approach that integrates multiple disciplines, including biology, immunology, virology, computer science, and mathematics, to develop a quantitative and a comprehensive understanding of a biological phenomenon (e.g. host response to virus infection). This approach consists of an iterative cycle that begins with collecting experimental data through various ‘omics’-based technologies (e.g. transcriptomics, proteomics, lipidomics, metabolomics, etc.). Next, these data sets are carefully integrated and analyzed using mathematics and computers to generate complex biological networks that define relationships between gene sets from different experimental conditions (wild type versus mutant virus, multiplicity of infection, time, etc...). More sophisticated computational analysis can identify regulatory nodes, hubs, or bottlenecks that would suggest a regulatory mechanism for a given biological phenomenon. The next step of this process, and probably one of the most important, is to validate these computational models through experimentation. This typically involves perturbation-based analysis using knockout mice or various techniques to silence gene expression (short-hairpin RNAs (shRNA), small interfering RNAs (siRNA), CRISPR/CAS systems, etc...). Finally, the newly generated biological model can be further refined through an additional round of hypothesis-driven research, perturbation-based experiments, and high-throughput based assays. The ultimate goal of this iterative process is to identify novel host targets of therapeutic intervention or pathways that can be modulated for enhancing immunogenicity during vaccination. Such systems biology approaches are now being harnessed to model pathogen-host interactions and immune response networks during hepatitis C virus [30-34], influenza virus [35-38] and severe acute respiratory syndrome-associated coronavirus infections [35,36,39,40].
These types of studies with WNV have been limited, however, our group recently used a systems biology approach to define the innate immune molecular signatures that control tissue tropism to WNV infection. Normally in wild-type mice, WNV replication is limited to the skin, draining lymph node, spleen, and central nervous system [5]. Genetic deletion of innate immune signaling components, such as MAVS [17], IRF-3 [41] or the type I IFN receptors [42], leads to productive virus replication in normally non-permissive organs, such as the liver. Thus, in this study, we compared the molecular signatures between the spleen (permissive) and liver (nonpermissive) compartments following WNV infection [22]. Transcriptional profiling revealed distinct gene expression patterns between these two organ compartments during WNV infection. Furthermore, functional genomics analysis and pathway modeling not only confirmed the importance type I IFN signaling networks for controlling tissue tropism, but revealed a previously unappreciated role for natural killer cell-mediated restriction of WNV replication within the liver. Biological validation and perturbation based studies revealed that natural killer cells indeed expand in the liver following WNV infection and both the RLR and type I IFN signaling pathways are important for mediating natural killer cell expansion and activity. Through these studies, we developed a model, whereby gene networks regulated by the RLR and type I IFN signaling axis impart restriction of virus replication and facilitate natural killer cell recruitment and expansion to prevent productive WNV replication within the liver. Further studies are now focused on identifying the cell types within the liver that support WNV replication and using computational tools to better understand the immune defense programs within these cells.
In a similar line of investigation, Cho et al. discovered that neuronal subtypes from distinct regions of the brain differentially trigger an innate immune response to WNV infection [43]. Specifically, granule cell neurons, which are located within the cerebellum, were found to be less susceptible to WNV infection and more responsive to type I IFN as compared to cortical neurons, which are found within the cerebral cortex. Transcriptional profiling and computational analysis revealed that granule cell neurons have a higher basal expression of a number of genes related to antiviral immunity, autophagy, inflammation, and leukocyte chemotaxis. Molecular analysis linked this differential expression to epigenetic modification and regulation by microRNAs. Combined, these studies reveal a previously unappreciated role for how cell- and tissue-specific innate defense programs are essential for controlling viral replication and tropism. Future studies should continue using traditional scientific approaches and ‘omics’-based technologies to comprehensively define the molecular signatures and immune networks to better predict WNV infection outcome. Particular emphasis should be placed on modeling the immunological signature of humans infected with WNV to better understand the underlying risk factors that contribute to symptomatic versus asymptomatic infection outcome.
High-throughput screens to identify WNV restriction factors
High-throughput based screening assays provide a rapid approach to identify host factors that either support or restrict virus replication. Specifically, these studies are designed to identify host factors can either directly antagonize a specific aspect of the viral life cycle (e.g. virus binding, entry, RNA synthesis, budding, etc...) or indirectly by modulating the immune response. Gain-of-function based approaches typically involve ectopic expression of an individual host gene and evaluating the impact on virus replication. In these assays, a reduction in virus replication suggests an antiviral property associated with the gene of interest. These genes are then used in follow-up studies to determine the mechanism of action. An initial small-scale screen by Jiang and colleagues identified RSAD2 (also known as viperin) and ISG20 as cellular enzymes that efficiently suppress WNV infection [44]. In this analysis, over-expression of viperin and ISG20 suppressed WNV-replicon colony formation, suggesting that these antiviral proteins likely target viral RNA or protein biosynthesis. In a similar manner, Schoggins and colleagues screened more than 380 human genes and identified several additional interferon-stimulated genes, including pattern recognition receptors (RIG-I, MDA5, CGAS), transcription factors (IRF1, ATF3, IRF7), and uncharacterized antiviral genes (HPSE, NAMPT, PBEF1, SAA1, and PHF15) that were observed to restrict WNV infection [45].
Loss-of-function based approaches typically involve gene silencing through siRNA or shRNA based technologies. In this assay, an increase in virus replication indicates that the gene of interest possesses antiviral activity. Krishnan and colleagues used siRNAs targeting over 21,000 human genes to screen and identify cellular proteins associated with the early stages of WNV infection which include entry, viral RNA synthesis and translation. This study identified over 300 host proteins that impact WNV infection, of which 283 host genes were found to facilitate WNV infection and 22 host genes reduced WNV infection. In a similar analysis, Li and colleagues used shRNAs to screen 245 human ISGs and identified 47 host genes that negatively impacted WNV replication [46]. This list of ISGs includes previously identified genes (e.g., MAVS, STAT2, IRF1, IFITM2, and PKR) as well as novel ISGs such as DDX24, IFI44L, IFI6, TRIM21, and TRIM6. More recently, Yasunaga et al. used Drosophila to identify cell-intrinsic antiviral genes that restrict WNV infection [47]. This group performed a genome-wide high-content RNA interference screen in Drosophila cells that identified 50 host genes, that when silenced, enhanced WNV replication. Remarkably, many of these genes have defined human orthologs. Follow-up mechanistic analysis on a subset of the candidate genes, found that members of the Tip60 acetylase complex and dXPO1, which controls nuclear export of specific host mRNAs, possess antiviral activity against WNV infection. These high-throughput based screening assays have provided yet another avenue for identifying host genes involved in controlling WNV replication. However, for many of these host genes, little is known about how they mechanistically control viral replication. Future studies should place a greater emphasis on mechanistic analysis to better define the mode of action of these anti-WNV ISGs.
Conclusions
Emerging mosquito-borne flavivirus infections continue to be a significant human health problem worldwide. It is becoming increasingly evident that development of effective vaccines that provide life-long immunity requires a comprehensive understanding of the innate and adaptive immune response to virus infection [48]. In support, systems biology approaches have been used to identify molecular networks that regulate the immune response to vaccination in humans [48,49]. Specifically, transcriptional analysis of blood from individuals vaccinated against yellow fever virus (YF-17D) [50] or influenza virus [51] identified molecular signatures that can predict the magnitude of the immune responses to a vaccine. These studies are beginning to pave the way towards the development of a 'vaccine chip' that could be used to predict vaccine-induced immunity [48,49]. In summary, the use of these technologies will continue to provide valuable insight to overcoming current challenges that have hindered effective vaccine development and prophylactic treatment strategies to prevent or combat emerging and re-emerging virus infection.
Highlights.
Emerging/re-emerging mosquito-borne viruses are a significant public health threat
There is a need define the virus-host interactions that govern immunity to infection
Systems biology provides a comprehensive analysis of the host response
High-throughput based assays provide rapid identification of host targets
Acknowledgements
The Suthar laboratory is supported by start-up funds provided by the Children's Healthcare of Atlanta, Emory Vaccine Center, the Georgia Research Alliance, and grants from the Emory University Research Council and NIH grants R03AI109194 and 5U19AI057266. The Pulendran laboratory is supported by funding from the NIH grants 5U19AI057266 and U19AI090023.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Petersen LR, Carson PJ, Biggerstaff BJ, Custer B, Borchardt SM, Busch MP. Estimated cumulative incidence of West Nile virus infection in US adults, 1999-2010. Epidemiol Infect. 2012:1–5. doi: 10.1017/S0950268812001070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease C, Prevention West Nile virus and other arboviral diseases--United States, 2012. MMWR Morb Mortal Wkly Rep. 2013;62:513–517. [PMC free article] [PubMed] [Google Scholar]
- 3.Thiberville SD, Moyen N, Dupuis-Maguiraga L, Nougairede A, Gould EA, Roques P, de Lamballerie X. Chikungunya fever: epidemiology, clinical syndrome, pathogenesis and therapy. Antiviral Res. 2013;99:345–370. doi: 10.1016/j.antiviral.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.PAHO/WHO Number of Reported Cases of Chikungunya Fever in the Americas, by Country or Territory 2013-2014. 2014 [Google Scholar]
- 5.Suthar MS, Diamond MS, Gale M. Jr.: West Nile virus infection and immunity. Nat Rev Microbiol. 2013;11:115–128. doi: 10.1038/nrmicro2950. [DOI] [PubMed] [Google Scholar]
- 6*.Lim PY, Behr MJ, Chadwick CM, Shi PY, Bernard KA. Keratinocytes are Cell Targets of West Nile Virus In Vivo. J Virol. 2011;85:5197–5201. doi: 10.1128/JVI.02692-10. [WNV is transmitted by a mosquito-bite, however, the initial targets cells at the site of infection have not been well characterized. In this study, the authors identify that keratinocytes support WNV replication and should be considered as playing an important role in pathogenesis.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnston LJ, Halliday GM, King NJ. Langerhans cells migrate to local lymph nodes following cutaneous infection with an arbovirus. J Invest Dermatol. 2000;114:560–568. doi: 10.1046/j.1523-1747.2000.00904.x. [DOI] [PubMed] [Google Scholar]
- 8.Bai F, Kong KF, Dai J, Qian F, Zhang L, Brown CR, Fikrig E, Montgomery RR. A paradoxical role for neutrophils in the pathogenesis of West Nile virus. J Infect Dis. 2010;202:1804–1812. doi: 10.1086/657416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ben-Nathan D, Huitinga I, Lustig S, van Rooijen N, Kobiler D. West Nile virus neuroinvasion and encephalitis induced by macrophage depletion in mice. Arch Virol. 1996;141:459–469. doi: 10.1007/BF01718310. [DOI] [PubMed] [Google Scholar]
- 10.Samuel MA, Whitby K, Keller BC, Marri A, Barchet W, Williams BR, Silverman RH, Gale M., Jr. Diamond MS: PKR and RNase L contribute to protection against lethal West Nile Virus infection by controlling early viral spread in the periphery and replication in neurons. J Virol. 2006;80:7009–7019. doi: 10.1128/JVI.00489-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11**.Hildner K, Edelson BT, Purtha WE, Diamond M, Matsushita H, Kohyama M, Calderon B, Schraml BU, Unanue ER, Diamond MS, et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322:1097–1100. doi: 10.1126/science.1164206. [In this study, the authors generate a mouse which lacks CD8alpha+ DCs by genetically ablating the transcription factor Batf3. These mice were used in a study to demonstrate that CD8alpha+ DCs are required for promoting CD8+ T cell responses during WNV infection.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pinto AK, Daffis S, Brien JD, Gainey MD, Yokoyama WM, Sheehan KC, Murphy KM, Schreiber RD, Diamond MS. A temporal role of type I interferon signaling in CD8+ T cell maturation during acute West Nile virus infection. PLoS Pathog. 2011;7:e1002407. doi: 10.1371/journal.ppat.1002407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Purtha WE, Chachu KA, Virgin HW. Diamond MS: Early B-cell activation after West Nile virus infection requires alpha/beta interferon but not antigen receptor signaling. J Virol. 2008;82:10964–10974. doi: 10.1128/JVI.01646-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fredericksen BL, Keller BC, Fornek J, Katze MG, Gale M. Establishment and maintenance of the innate antiviral response to west nile virus involves both RIG-I and MDA5 signaling through IPS-1. Journal of Virology. 2008;82:609–616. doi: 10.1128/JVI.01305-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Errett JS, Suthar MS, McMillan A, Diamond MS, Gale M., Jr. The Essential, Nonredundant Roles of RIG-I and MDA5 in Detecting and Controlling West Nile Virus Infection. J Virol. 2013;87:11416–11425. doi: 10.1128/JVI.01488-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lazear HM, Pinto AK, Ramos HJ, Vick SC, Shrestha B, Suthar MS, Gale M, Jr., Diamond MS. Pattern Recognition Receptor MDA5 Modulates CD8+ T Cell-Dependent Clearance of West Nile Virus from the Central Nervous System. J Virol. 2013;87:11401–11415. doi: 10.1128/JVI.01403-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suthar MS, Ma DY, Thomas S, Lund JM, Zhang N, Daffis S, Rudensky AY, Bevan MJ, Clark EA, Kaja MK, et al. IPS-1 is essential for the control of West Nile virus infection and immunity. PLoS Pathog. 2010;6:e1000757. doi: 10.1371/journal.ppat.1000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daffis S, Samuel MA, Suthar MS, Gale M, Jr., Diamond MS. Toll-like receptor 3 has a protective role against West Nile virus infection. 82. J Virol. 2008:10349–10358. doi: 10.1128/JVI.00935-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Town T, Bai F, Wang T, Kaplan AT, Qian F, Montgomery RR, Anderson JF, Flavell RA, Fikrig E. Toll-like receptor 7 mitigates lethal West Nile encephalitis via interleukin 23-dependent immune cell infiltration and homing. 30. Immunity. 2009:242–253. doi: 10.1016/j.immuni.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramos HJ, Lanteri MC, Blahnik G, Negash A, Suthar MS, Brassil MM, Sodhi K, Treuting PM, Busch MP, Norris PJ, et al. IL-1beta signaling promotes CNS-intrinsic immune control of West Nile virus infection. 8. PLoS Pathog. 2012:e1003039. doi: 10.1371/journal.ppat.1003039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar M, Roe K, Orillo B, Muruve DA, Nerurkar VR, Gale M, Jr., Verma S. Inflammasome adaptor protein Apoptosis-associated speck-like protein containing CARD (ASC) is critical for the immune response and survival in west Nile virus encephalitis. J Virol. 2013;87:3655–3667. doi: 10.1128/JVI.02667-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suthar MS, Brassil MM, Blahnik G, McMillan A, Ramos HJ, Proll SC, Belisle SE, Katze MG, Gale M., Jr. A systems biology approach reveals that tissue tropism to West Nile virus is regulated by antiviral genes and innate immune cellular processes. PLoS Pathog. 2013;9:e1003168. doi: 10.1371/journal.ppat.1003168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang M, Daniel S, Huang Y, Chancey C, Huang Q, Lei YF, Grinev A, Mostowski H, Rios M, Dayton A. Anti-West Nile virus activity of in vitro expanded human primary natural killer cells. BMC Immunol. 2010;11:3. doi: 10.1186/1471-2172-11-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang T, Scully E, Yin Z, Kim JH, Wang S, Yan J, Mamula M, Anderson JF, Craft J, Fikrig E. IFN-γ-producing γδ T cells help control murine West Nile virus infection. J Immunol. 2003;171:2524–2531. doi: 10.4049/jimmunol.171.5.2524. [DOI] [PubMed] [Google Scholar]
- 25.Fischer SA. Emerging viruses in transplantation: there is more to infection after transplant than CMV and EBV. Transplantation. 2008;86:1327–1339. doi: 10.1097/TP.0b013e31818b6548. [DOI] [PubMed] [Google Scholar]
- 26.Lim JK, Lisco A, McDermott DH, Huynh L, Ward JM, Johnson B, Johnson H, Pape J, Foster GA, Krysztof D, et al. Genetic variation in OAS1 is a risk factor for initial infection with West Nile virus in man. PLoS Pathog. 2009;5:e1000321. doi: 10.1371/journal.ppat.1000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27**.Lim JK, McDermott DH, Lisco A, Foster GA, Krysztof D, Follmann D, Stramer SL, Murphy PM. CCR5 deficiency is a risk factor for early clinical manifestations of West Nile virus infection but not for viral transmission. 201. J Infect Dis. 2010:178–185. doi: 10.1086/649426. [Previous reports suggested a link between CCR5Delta32, a loss-of-function mutant in the chemokine receptor 5, and a risk factor for WNV infection. In this study, the authors clarify that CCR5 likely promotes immunity to WNV infection and the loss of function mutant is a risk factor for symptomatic outcome of WNV infection.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bigham AW, Buckingham KJ, Husain S, Emond MJ, Bofferding KM, Gildersleeve H, Rutherford A, Astakhova NM, Perelygin AA, Busch MP, et al. Host genetic risk factors for West Nile virus infection and disease progression. PLoS One. 2011;6:e24745. doi: 10.1371/journal.pone.0024745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29*.Lanteri MC, O'Brien KM, Purtha WE, Cameron MJ, Lund JM, Owen RE, Heitman JW, Custer B, Hirschkorn DF, Tobler LH, et al. Tregs control the development of symptomatic West Nile virus infection in humans and mice. J Clin Invest. 2009;119:3266–3277. doi: 10.1172/JCI39387. [Regulatory T cells are important for controlling the immune response during a pathogen infection. In this study, the authors demonstrate that symptomatic human patients infected with WNV exhibited a lower regulatory T cell response as compared to asymptomatic patients. Similarly, WNV-infected mice in which regulatory T cells were ablated exhibited more severe disease outcomes, implicating these immune cells in regulating protective immunity to WNV infection.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith MW, Walters KA, Korth MJ, Fitzgibbon M, Proll S, Thompson JC, Yeh MM, Shuhart MC, Furlong JC, Cox PP, et al. Gene expression patterns that correlate with hepatitis C and early progression to fibrosis in liver transplant recipients. Gastroenterology. 2006;130:179–187. doi: 10.1053/j.gastro.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 31.Walters KA, Smith MW, Pal S, Thompson JC, Thomas MJ, Yeh MM, Thomas DL, Fitzgibbon M, Proll S, Fausto N, et al. Identification of a specific gene expression pattern associated with HCV-induced pathogenesis in HCV- and HCV/HIV-infected individuals. Virology. 2006 doi: 10.1016/j.virol.2006.02.030. [DOI] [PubMed] [Google Scholar]
- 32.Rasmussen AL, Diamond DL, McDermott JE, Gao X, Metz TO, Matzke MM, Carter VS, Belisle SE, Korth MJ, Waters KM, et al. Systems virology identifies a mitochondrial fatty acid oxidation enzyme, dodecenoyl coenzyme A delta isomerase, required for hepatitis C virus replication and likely pathogenesis. J Virol. 2011;85:11646–11654. doi: 10.1128/JVI.05605-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDermott JE, Diamond DL, Corley C, Rasmussen AL, Katze MG, Waters KM. Topological analysis of protein co-abundance networks identifies novel host targets important for HCV infection and pathogenesis. BMC Syst Biol. 2012;6:28. doi: 10.1186/1752-0509-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rasmussen AL, Wang IM, Shuhart MC, Proll SC, He Y, Cristescu R, Roberts C, Carter VS, Williams CM, Diamond DL, et al. Chronic immune activation is a distinguishing feature of liver and PBMC gene signatures from HCV/HIV coinfected patients and may contribute to hepatic fibrogenesis. Virology. 2012;430:43–52. doi: 10.1016/j.virol.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng X, Gralinski L, Armour CD, Ferris MT, Thomas MJ, Proll S, Bradel-Tretheway BG, Korth MJ, Castle JC, Biery MC, et al. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. MBio. 2010;1 doi: 10.1128/mBio.00206-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng X, Gralinski L, Ferris MT, Frieman MB, Thomas MJ, Proll S, Korth MJ, Tisoncik JR, Heise M, Luo S, et al. Integrative deep sequencing of the mouse lung transcriptome reveals differential expression of diverse classes of small RNAs in response to respiratory virus infection. MBio. 2011;2 doi: 10.1128/mBio.00198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bottomly D, Ferris MT, Aicher LD, Rosenzweig E, Whitmore A, Aylor DL, Haagmans BL, Gralinski LE, Bradel-Tretheway BG, Bryan JT, et al. Expression quantitative trait Loci for extreme host response to influenza a in pre-collaborative cross mice. G3 (Bethesda) 2012;2:213–221. doi: 10.1534/g3.111.001800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38**.Ferris MT, Aylor DL, Bottomly D, Whitmore AC, Aicher LD, Bell TA, Bradel-Tretheway B, Bryan JT, Buus RJ, Gralinski LE, et al. Modeling host genetic regulation of influenza pathogenesis in the collaborative cross. PLoS Pathog. 2013;9:e1003196. doi: 10.1371/journal.ppat.1003196. [Host genetic variation can influence the outcome of a pathogen infection. In this study, the authors used the novel Collaborative Cross model system to understand how variations in host genetics influences the immune response to influenza infection. The authors identified several unique quantitative trait loci that correlated to various aspects of the host response to infection, including immune cell recruitment to the lungs, weight loss, and viral replication.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gralinski LE, Bankhead A, 3rd, Jeng S, Menachery VD, Proll S, Belisle SE, Matzke M, Webb-Robertson BJ, Luna ML, Shukla AK, et al. Mechanisms of severe acute respiratory syndrome coronavirus-induced acute lung injury. MBio. 2013;4 doi: 10.1128/mBio.00271-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitchell HD, Eisfeld AJ, Sims AC, McDermott JE, Matzke MM, Webb-Robertson BJ, Tilton SC, Tchitchek N, Josset L, Li C, et al. A network integration approach to predict conserved regulators related to pathogenicity of influenza and SARS-CoV respiratory viruses. PLoS One. 2013;8:e69374. doi: 10.1371/journal.pone.0069374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daffis S, Samuel MA, Keller BC, Gale M, Jr., Diamond MS. Cell-specific IRF-3 responses protect against West Nile virus infection by interferon-dependent and -independent mechanisms. PLoS Pathog. 2007;3:e106. doi: 10.1371/journal.ppat.0030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Samuel MA, Diamond MS. Alpha/beta interferon protects against lethal West Nile virus infection by restricting cellular tropism and enhancing neuronal survival. J.Virol. 2005;79:13350–13361. doi: 10.1128/JVI.79.21.13350-13361.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43**.Cho H, Proll SC, Szretter KJ, Katze MG, Gale M, Jr., Diamond MS. Differential innate immune response programs in neuronal subtypes determine susceptibility to infection in the brain by positive-stranded RNA viruses. Nat Med. 2013;19:458–464. doi: 10.1038/nm.3108. [One of the main target cells of WNV infection are neurons, and thus, the virus is found to infect and replicate to high titers within the central nervous system. However, little is known about the individual neuronal subsets and how they may differentially respond to virus infection. The authors in the study demonstrate that not all neurons respond to virus infection in the same manner. In fact, cortical neurons are less effective at controlling WNV replication than granule cell neurons.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang D, Weidner JM, Qing M, Pan XB, Guo H, Xu C, Zhang X, Birk A, Chang J, Shi PY, et al. Identification of five interferon-induced cellular proteins that inhibit west nile virus and dengue virus infections. J Virol. 2010;84:8332–8341. doi: 10.1128/JVI.02199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J, Ding SC, Cho H, Chung BC, Gale M, Jr., Chanda SK, Diamond MS. A short hairpin RNA screen of interferon-stimulated genes identifies a novel negative regulator of the cellular antiviral response. MBio. 2013;4:e00385–00313. doi: 10.1128/mBio.00385-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yasunaga A, Hanna SL, Li J, Cho H, Rose PP, Spiridigliozzi A, Gold B, Diamond MS, Cherry S. Genome-Wide RNAi Screen Identifies Broadly-Acting Host Factors That Inhibit Arbovirus Infection. PLoS Pathog. 2014;10:e1003914. doi: 10.1371/journal.ppat.1003914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pulendran B. Learning immunology from the yellow fever vaccine: innate immunity to systems vaccinology. Nat Rev Immunol. 2009;9:741–747. doi: 10.1038/nri2629. [DOI] [PubMed] [Google Scholar]
- 49.Pulendran B, Ahmed R. Immunological mechanisms of vaccination. Nat Immunol. 2011;12:509–517. doi: 10.1038/ni.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Querec TD, Akondy RS, Lee EK, Cao W, Nakaya HI, Teuwen D, Pirani A, Gernert K, Deng J, Marzolf B, et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol. 2009;10:116–125. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakaya HI, Wrammert J, Lee EK, Racioppi L, Marie-Kunze S, Haining WN, Means AR, Kasturi SP, Khan N, Li GM, et al. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol. 2011;12:786–795. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]