Abstract
Highly virulent bacterial pathogens have evolved rapid means to suppress host inflammatory responses by unknown mechanisms. Here, we use virulent Francisella tularensis, the cause of lethal tularemia in humans, as a model to elucidate these mechanisms. We show that following infection of murine macrophages F. tularensis rapidly and selectively destabilizes mRNA containing AU-rich elements that encode for cytokines and chemokines important in controlling bacterial infection. Degradation of host mRNA encoding IL-1β, IL-6, and CXCL1 did not require viable bacteria or de novo protein synthesis, but did require escape of intracellular organisms from endocytic vesicles into the host cytosol. The specific targeting of host mRNA encoding inflammatory cytokines and chemokines for decay by a bacterial pathogen has not been previously reported. Thus, our findings represent a novel strategy by which a highly virulent pathogen modulates host inflammatory responses critical to the evasion of innate immunity.
Keywords: mRNA stability, intracellular bacteria, inflammation, macrophage
Introduction
Rapid induction of inflammatory responses is required for development of effective innate and adaptive immunity against invading pathogens. The host has a wide array of strategies to detect, respond, and eliminate microorganisms. These include pattern recognition receptors, compartmentalization in toxic vacuoles and production of pro-inflammatory cytokines and chemokines to directly activate anti-microbial pathways and/or recruit host cells capable of aiding in eradication of the pathogen [1]. Virulent organisms must evade and modulate these host factors to ensure their successful infection, replication and dissemination in the host. Identification of the mechanisms by which highly virulent bacteria negatively regulate host responsiveness to infection is central to identification of novel therapeutics and vaccines.
Francisella tularensis ssp. tularensis (Ftt) is a gram negative facultative intracellular bacterium and the causative agent of tularemia. Isolates of Ftt are extremely virulent, causing acute lethal disease in humans and mice following inhalation with as few as 10 bacteria [2]. Ftt infects the host through a variety of routes, including inhalation, ingestion, through skin abrasions, and by arthropod bites [3]. Following inoculation, Ftt primarily targets macrophages and dendritic cells in the lung and peripheral organs, e.g. the spleen, for intracellular replication [3]. After being engulfed by the host cell via phagocytosis, the bacterium quickly escapes the endocytic compartment gaining access to the cytosol, where it replicates to high numbers. The existence of reproducible in vitro and in vivo models that closely replicate features of human disease make Ftt an ideal model organism for study of immunity to highly pathogenic organisms.
There are several subspecies and strains of Francisella that can be categorized by their relative virulence for humans (as reviewed, [2]). As stated above Ftt causes an acute, lethal disease in humans. F. tularensis ssp. holarctica (Fth) is also capable of causing significant disease in humans, but is not typically fatal. Over 60 years ago, a live vaccine strain (LVS) was generated from Fth following serial passage in mice [4]. As a vaccine strain, LVS is largely attenuated in humans. Lastly, F. novicida is an avirulent species of Francisella. The avirulent nature of F. novicida has made this bacterium useful for incorporation into comparative studies performed with Ftt.
An important aspect of virulence embodied by Ftt, like other highly pathogenic organisms, is its ability to evade detection by the host and suppress induction of inflammatory responses in vitro and in vivo [5–7]. Here we uncovered a unique mechanism by which Ftt down-regulates proinflammatory responses. Performing comparative and co-infection studies with avirulent F. novicida, we show that Ftt impacted the host inflammatory responses within the first 2 hours of infection without the requirement for replication or de novo protein synthesis. Our data identified that the rapid impairment of inflammatory responses by Ftt was due to its ability to induce selective decay of host mRNA encoding cytokines and chemokines required for resolution of infection. This strategy may represent a common mechanism used by other intracellular pathogens to effectively target and dismantle the host inflammatory response.
Materials and Methods
Mice and generation of bone marrow derived macrophages (BMM)
Specific pathogen free C57BL/6 NHsd mice were bred at Rocky Mountain Laboratories (RML). C57BL/6J wild type controls (WT) and TLR2 deficient mice (TLR2−/−) were purchased from Jackson Laboratories (Bar Harbor, ME). TTPflox/flox (WT) controls and TTPflox/flox LysM-Cre (TTP−/−) were provided by Dr. Peter Murray (St. Jude Children’s Research Hospital, Memphis, TN). All research involving animals was conducted in accordance with Animal Care and Use guidelines and animal protocols were approved by the Animal Care and Use Committee at RML. BMM were generated as previously described, except that media was supplemented with 20 ng/mL recombinant murine MCSF and heat inactivated fetal bovine serum was used in medium for all BMM cultures (Peprotech, Rocky Hill, NJ) [8]. BMM were primed with either 500 ng/mL Pam3CSK4 (P3C) or 25 ng/mL monophosphoryl lipid A (MPLA) (both from Invivogen, San Diego, CA) as indicated in every experiment 16 h prior to infection.
Bacteria
Stock cultures of F. tularensis ssp. tularensis SchuS4 (Dr. Jeannine Peterson, CDC, Fort Collins, CO), SchuS4 ΔfevR (ΔFTT0383) (Dr. Jean Celli, Washington State University, Pullman, WA), Live Vaccine Strain – ATCC29684 (LVS; Dr. Karen Elkins, CBER/FDA, Rockville, MD), F. tularensis ssp. holarctica strain FSC200 (Dr. Anders Sjostedt, University of Umea, Umea, Sweden) and F. novicida U112 (Dr. Denise Monack, Stanford University, Stanford, CA) were generated and utilized as previously described [6]. As indicated, killed SchuS4 was generated following treatment with 1.5% paraformaldehyde (PFA) for 30 min and washed twice prior to inoculation. Alternatively, SchuS4 was killed by γ-irradiation (250 kilorads, Co60 source) and washed once prior to inoculation. Successful killing of SchuS4 was verified by incubation of bacteria 72 h on modified Mueller Hinton (MMH) agar. Where specified, SchuS4 was pretreated with chloramphenicol (10 μg/mL) 30 min prior to infection to prevent de novo bacterial protein synthesis, as previously described [9]. Chloramphenicol was retained in the media for the duration of the experiment. All experiments were performed under approved BSL-3 safety protocols at RML.
Infection and exposure of BMM to Francisella
Bacteria were resuspended at the indicated MOI (or corresponding MOI for killed bacteria) in 250 μL complete DMEM (cDMEM, DMEM with Glutamine, HEPES, and NEAA added [Life Technologies, Grand Island, NY], plus 10% heat inactivated FBS [Thermo Fisher, Waltham, MA]), added to BMM on ice, and plates were centrifuged for 15 min at 850 × g at 4°C. MOIs were selected to ensure uptake of similar numbers of each strain of bacteria by BMM. Cells were then incubated at 37°C for 45 min, washed twice with DMEM, followed by incubation with gentamicin (50 μg/mL for 45 min at 37°C) to eliminate extracellular bacteria. Cells were washed once with DMEM and cultured in cDMEM. Intracellular bacteria were enumerated as previously described [10]. Briefly, culture medium was removed and cells were lysed with sterile water. Cell lysates were serially diluted and plated on modified Mueller Hinton (MMH) agar plates for enumeration of colonies. For co-infection experiments, SchuS4 and F. novicida were mixed and added to BMM in a total volume of 250 μL and infection proceeded as above. In co-infected cultures, ~95% of cells were infected with both SchuS4 and F. novicida, as determined by confocal microscopy. Where specified, BMM were pretreated with cytochalasin D (10 μg/mL, A.G. Scientific, San Diego, CA) or bafilomycin A1 (100 nM, A.G. Scientific) 1 h prior to infection to inhibit phagocytosis and endosomal acidification, respectively. As indicated, the transcription inhibitor actinomycin D (2.5 μg/mL, Sigma, St. Louis, MO) or the p38 inhibitor SB203580 (1 μg/mL, Cell Signaling Technologies, Danvers, MA) was added at the initiation of infection. All drugs were retained in the media throughout the course of the infection. At the time points assessed none of the chemicals significantly affected viability of francisella.
Quantification of cell death and cytokines
Cell death was measured using Cytotox 96 Non-radioactive cytotoxicity kit according to the manufacturer’s instructions (Promega, Madison, WI). Concentrations of IL-1β, IL-6, and CXCL1 in culture supernatants were determined by ELISA (R&D Systems, Minneapolis, MN).
Western blotting and qRT-PCR
At the indicated time points medium was removed from BMM. Cells were lysed in 150 μl Nu PAGE LDS Sample Buffer (Life Technologies) by vigorous pipetting and scraping of the cells. Cell lysates were heated at 95°C and immediately placed on ice prior to loading onto 4–12% SDS NuPAGE gradient gels (Life Technologies). Western blots were generated from SDS-NuPAGE gels as previously described [11]. Blots were probed with antibodies to phospho-T180/Y182 p38, p38, phospho-T334 MK2 (27B7), MK2, and β-actin (13E5), (all from Cell Signaling Technologies). RNA was purified using RNeasy kits (Qiagen), cDNA generated using the Superscript VILO kit (Life Technologies) and real-time quantitative PCR was run using primer/probe sets (Table 1) (all from Life Technologies) and an ABI 7900HT (Life Technologies). Input RNA was normalized to GAPDH, and fold change of the indicated genes as compared to untreated, uninfected controls was quantified as ΔΔCT. The mRNA half-life (t1/2) was calculated as −ln2/m, where m is the slope of the line fit on a semi-logarithmic plot of mRNA concentration as a function of time using least-squares regression as previously described [12].
Table 1.
Taqman Gene Expression Assays and Assay ID numbers used in this study
| Gene | ID number |
|---|---|
| IL-1b | Mm00434228_m1 |
| IL-6 | Mm00446190_m1 |
| CXCL1 | Mm04207460_m1 |
| IL-18 | Mm00434225_m1 |
| CCL2 | Mm00441242_m1 |
| CCL3 | Mm00441259_g1 |
| CCL4 | Mm00443111_m1 |
| CCL5 | Mm01302427_m1 |
| CCL22 | Mm00436439_m1 |
| IL-1a | Mm00439620_m1 |
| IL-12b | Mm00434174_m1 |
| TNFa | Mm00443258_m1 |
| GADPH | Mm99999915_g1 |
Phagosomal Integrity Assay
To differentiate between vacuolar and cytosolic bacteria, phagosomal integrity assays were performed, as previously described [8]. Anti-F. tularensis LPS (US Biological, Salem, MA) antibodies were conjugated with either Alexa Fluor 546 or Alexa Fluor 488 using Zenon Alexa Fluor 546 mouse IgG2a or Zenon Alexa Fluor 488 mouse IgG2a labeling kits according to manufacturer’s instructions (Invitrogen). Briefly, BMM were treated with 50 μg/mL digitonin for 1 min to selectively permeabilize the plasma membrane, and then stained with Alexa Fluor 546-conjugated anti-F. tularensis LPS to label cytoplasmic bacteria. Next, BMM were fixed with 3% PFA for 20 min at 37°C and further permeabilized in 0.1% saponin/10% horse serum in PBS for 30 min. All intracellular bacteria were labeled using Alexa Fluor 488-conjugated anti-F. tularensis LPS (US Biological). Cytoplasmic bacteria exhibited 546/488 dual fluorescence whereas vacuole-bound bacteria exhibited only 488 single fluorescence allowing differential enumeration of organisms. Detection and quantitation of cytoplasmic versus vacuole-bound bacteria was accomplished by visualizing infected cells using an Axio Imager epifluoresence microscope (Zeiss).
Statistical Analysis
Statistical differences between two groups were determined using a two-tailed Student’s t test. For comparison between three or more groups, analysis was done by one-way ANOVA followed by Tukey’s multiple comparisons test. Significance was determined at p<0.05.
Results
SchuS4 inhibits production of IL-1β, IL-6, and CXCL1 in previously activated cells by reducing mRNA transcripts
F. tularensis ssp. tularensis SchuS4 both evades provoking an inflammatory response upon infection and suppresses the ability of the host cell to mount an inflammatory response after infection [5,11]. However, the majority of this work was conducted in resting cells. Given the dramatic ability of SchuS4 to dampen inflammation we postulated that SchuS4 may also down modulate inflammatory programs in cells activated prior to infection. We chose to examine IL-1β, IL-6, CXCL1, and IL-18 as representative cytokines/chemokines produced by macrophages that are important for host defense against bacterial infections [13]. Macrophages were primed with P3C prior to infection resulting in increased phosphorylation of MAPK p38, increased expression of genes encoding IL-1β, IL-6, and CXCL1, and secretion of IL-6 and CXCL1 (Supplemental Figure 1). SchuS4 infection of P3C-primed BMM did not induce secretion of IL-1β, IL-6, CXCL1, or IL-18 that was significantly greater than primed, uninfected cells, despite replication of the bacterium (Fig 1A–B). Rather, SchuS4 infection significantly decreased the amount of IL-6 and CXCL1 present in culture supernatants compared to primed, uninfected controls (Fig 1B). This demonstrated that SchuS4 evades provoking an inflammatory response and readily inhibits an established inflammatory program in previously-activated cells.
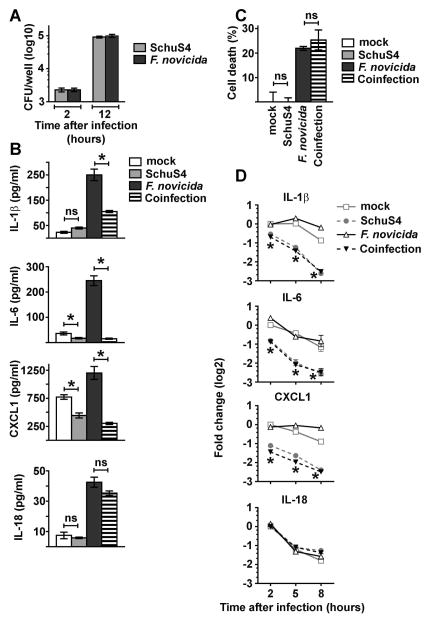
Figure 1. SchuS4 infection reduces IL-1β, IL-6, and CXCL1 RNA transcripts to inhibit cytokine/chemokine production in previously activated BMM.
BMM were primed with P3C (500 ng/mL) and infected with SchuS4 MOI 50, F. novicida MOI 10, or co-infected with both SchuS4 MOI 50 and F. novicida MOI 10. Cells were infected with the indicated MOIs to ensure equivalent uptake of F. novicida and SchuS4. Mock infected cells served as negative controls. (A) Intracellular bacteria were enumerated at the indicated times after infection. (B) Supernatants were harvested 12 h after infection and the quantity of IL-1β, IL-6, CXCL1 and IL-18 was measured by ELISA. *= p<0.05 and ns = not significant when comparing the indicated groups. (C) Supernatants were harvested 12 h after infection and LDH levels were quantified to calculate the percentage of cell lysis. ns = not significant compared to the indicated samples. (D) RNA was harvested at the indicated times after infection. qRT-PCR was performed to examine the quantity of the indicated transcripts, normalized to GAPDH. *= p<0.05 SchuS4 and F. novicida/SchuS4 coinfected cells compared to mock and F. novicida infected cells. Each data point depicts the mean of triplicate samples. Error bars represent SEM. Data are representative of three independent experiments.
We next sought to test the ability of SchuS4 to modulate cytokine responses in host cells activated with a more complex stimulus rather than only a single defined ligand. We utilized avirulent F. novicida to increase production of inflammatory cytokines among infected cells. As expected, F. novicida infection of primed cells resulted in secretion of significantly greater concentrations of IL-1β, IL-6, CXCL1, and IL-18 compared to cultures of uninfected and SchuS4 infected cells (Fig 1A–B). BMM co-infected with SchuS4 and F. novicida secreted similar amounts of IL-18 compared to cells infected with F. novicida alone (Fig 1B). However, BMM co-infected with SchuS4 and F. novicida did secrete significantly less IL-1β, IL-6, and CXCL1 than cells infected with F. novicida alone (Fig 1B). The dampened release of IL-1β, IL-6 and CXCL1 observed in SchuS4-F. novicida co-infected cells could not be attributed to reduced viability of macrophages since cell death in SchuS4-F. novicida co-infected cultures was similar to that of F. novicida-only infected cells (Fig 1C). We next determined if the reduced secretion of IL-1β, IL-6, and CXCL1 was correlated with a reduction in mRNA transcripts encoding these proteins. All cultures infected with SchuS4 had significantly less IL-1β, IL-6, and CXCL1 mRNA transcripts compared to uninfected cells or macrophages infected with F. novicida alone (Fig 1D). In contrast, and in agreement with similar concentrations of IL-18 detected in culture supernatants of F. novicida and SchuS4-F. novicida co-infected cells, the amount of IL-18 mRNA was unaffected by SchuS4 infection at each time point tested (Fig 1D). Thus, inhibition of IL-1β, IL-6, and CXCL1 among SchuS4 infected cells was correlated with decreased numbers of mRNA transcripts encoding these proteins. These data also indicated that the impact of SchuS4 infection was selective for specific genes rather than a global reduction of mRNA transcripts among infected cultures. We next tested the ability of other closely related subspecies and strains of Francisella to reduce levels of mRNA among BMM. Similar to SchuS4 infected cells, virulent F. tularensis ssp. holarctica strain FSC200 reduced the number of detectable mRNA transcripts and did not induce secretion of IL1β, IL-6 or CXCL1 above that detected in uninfected cells (Fig 2A and B). In contrast, similar to cells infected with F. novicida, BMM infected with attenuated LVS increased mRNA transcripts compared to uninfected and SchuS4 or FSC200 infected cells (Fig 2A). LVS and F. novicida also triggered secretion of significantly more IL-1β, IL-6 and CXCL1 compared to uninfected and SchuS4 or FSC200 infected cells (Fig 2B). Therefore, reduction of mRNA transcripts encoding specific pro-inflammatory cytokines and chemokines is a feature of infection with virulent, but not avirulent or attenuated, strains of Francisella.
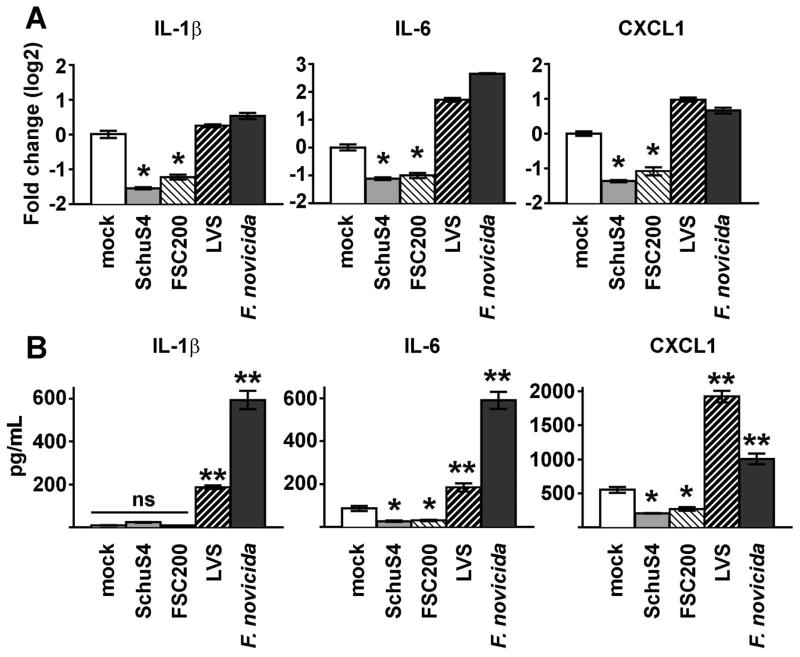
Figure 2. Reduction of IL-1β, IL-6, and CXCL1 mRNA correlates with virulence of Francisella.
BMM were primed with P3C (500 ng/mL) and infected with SchuS4 MOI 50, FSC200 MOI 50, LVS MOI 25, or F. novicida MOI 10. Cells were infected with the indicated MOIs to ensure equivalent uptake. (A) RNA was harvested 5 h after infection. qRT-PCR was performed to examine the quantity of the indicated transcripts, normalized to GAPDH. (B) Supernatants were harvested 12 h after infection and the indicated cytokines were quantified by ELISA. *= significantly reduced (p<0.05) compared to the mock infected cells. ** = significantly greater than uninfected or SchuS4 or FSC200 infected cells. ns = not significant compared to the indicated samples. Each data point depicts the mean of triplicate samples. Error bars represent SEM. Data are representative of two independent experiments.
Reduction of IL-1β, IL-6, and CXCL1 mRNA by SchuS4 requires phagosomal escape, but not bacterial protein synthesis or viability
We next identified the cellular location from which SchuS4 affected numbers of mRNA transcripts. Upon Francisella engagement of receptors at the plasma membrane, the bacterium is phagocytosed into an endosome and then approximately one hour after infection escapes into the cytoplasm, where it replicates exponentially [14,15]. SchuS4 could be initiating the observed decrease in IL-1β, IL-6, and CXCL1 mRNA upon contact with the cell surface, while transiting the endosome, or after escape into the cytosol. Thus, using a combination of chemical inhibitors, BMM with specific mutations in surface receptors, mutagenized SchuS4 and two different techniques to kill the bacterium, we empirically determined the requirements for SchuS4 to inhibit production of IL-1β, IL-6 and CXCL1.
To determine if SchuS4 interaction with the cell membrane was sufficient to reduce mRNA transcripts, the actin depolymerizing agent cytochalasin D was used to inhibit bacterial phagocytosis. As expected, SchuS4 infection of cytochalasin D-treated BMM decreased bacterial uptake >97% compared to untreated cells (data not shown). Cytochalasin D-treated cells infected with SchuS4 did not reduce IL-1β, IL-6, or CXCL1 transcripts (Fig 3A). Therefore, phagocytosis of the bacterium was necessary to decrease these mRNA.
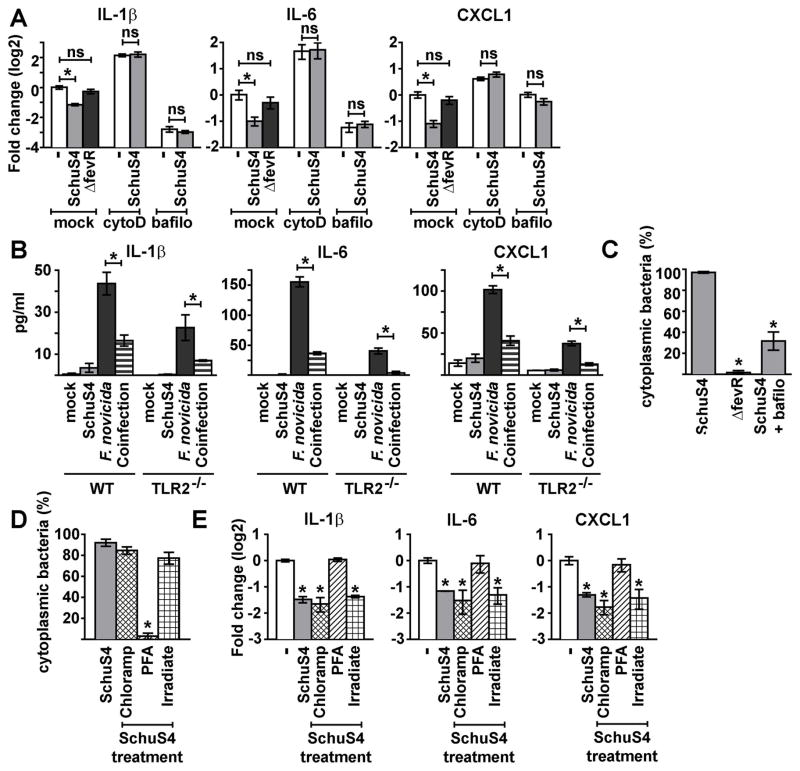
Figure 3. Phagosomal escape, but not bacterial protein synthesis or viability, is required for SchuS4-mediated reduction of IL-1β, IL-6, and CXCL1 mRNA.
(A–C) BMM were primed with P3C (500 ng/ml) and treated with cytochalasin D (CytoD) to inhibit bacterial phagocytosis or bafilomycin (Bafilo) to inhibit bacterial escape from endosomes prior to infection, where indicated. BMM were infected with SchuS4 MOI 50 or SchuS4 ΔfevR MOI 50. Mock infected cells (−) served as negative controls. (A) RNA was harvested 5 h after infection. qRT-PCR was performed to examine the quantity of the indicated transcripts, normalized to GAPDH. *= p<0.05 compared to the indicated samples. ns = not significant compared to the indicated samples. (B) Wild type (WT) or TLR2−/− BMM were primed with MPLA (25 ng/mL) and infected with SchuS4 MOI 50, F. novicida MOI 10, or co-infected with both SchuS4 MOI 50 and F. novicida MOI 10. Secretion of IL-1β, IL-6 and CXCL1 was assessed by ELISA. *=p<0.05 F. novicida only infected compared to F. novicida/SchuS4 co-infected cells. (C) Cytoplasmic bacteria were quantified 5 h after infection using a phagosomal-integrity assay. *= p<0.05 compared to WT SchuS4 infection of untreated cells. (D–E) BMM were primed with P3C (500 ng/mL) and infected with MOI 50 of SchuS4, SchuS4 pretreated with chloramphenicol (Chloramp) to inhibit bacterial protein synthesis, or inoculated with SchuS4 killed by treatment with PFA or irradiation (Irradiate). (D) Cytoplasmic bacteria were quantified 5 h after infection using a phagosomal-integrity assay. *= p<0.05 compared to untreated SchuS4. (E) RNA was harvested 5 h after infection. qRT-PCR was performed to examine the quantity of the indicated transcripts, normalized to GAPDH. *= p<0.05 compared to uninfected cells. Each data point depicts the mean of triplicate samples. Error bars represent SEM. Data are representative of three independent experiments.
Previous studies have shown that interaction with the surface receptor TLR2 was required for immunosuppressive mechanisms of SchuS4 [16,17]. To determine if ligation of TLR2 was required for modulation of secretion of IL-1β, IL-6 and CXCL1 by SchuS4 we compared the ability of SchuS4 to affect production of these cytokines among wild type and TLR2−/− BMM. The absence of TLR2 had no effect on the ability of SchuS4 to either evade induction or dampen secretion of IL-1β, IL-6 and CXCL1 among infected BMM (Fig 3B).
We next determined if reduction of IL-1β, IL-6, and CXCL1 transcripts required escape of SchuS4 from the endosome via two methods. First, cells were treated with bafilomycin A1, which inhibits SchuS4 escape from the endosome (Fig 3C) [8]. SchuS4 infection of bafilomycin-treated cells did not decrease the number of IL-1β, IL-6, or CXCL1 mRNA transcripts beyond that observed in treated, uninfected cells (Fig 3A). However, bafilomycin treatment alone resulted in reduction of mRNA transcripts independent of infection (Fig 3A), suggesting that bafilomycin had an off target effect on the ability of host to increase transcription of host mRNA. Thus, we utilized another method to confirm our finding that phagosomal escape of SchuS4 was required for reduction of host mRNA. Next, we used a targeted deletion mutant of SchuS4 which is defective for phagosomal escape, SchuS4 ΔfevR (Fig 3C) [18]. SchuS4 ΔfevR does not exhibit defects in growth in axenic medium and is phagocytosed by primary cells with similar efficiency as wild type SchuS4. However, these mutants fail to escape the phagolysosome following engulfment by macrophages [18]. Infection of BMM with SchuS4 ΔfevR did not result in the reduction of IL-1β, IL-6, or CXCL1 transcripts (Fig 3A). Although it was possible that fevR is important for expression of bacterial proteins that may contribute to reduction of host mRNA transcripts, taken together with data from cells treated with bafilomycin these results suggest that reduction of IL-1β, IL-6, and CXCL1 mRNA was dependent on SchuS4 escape into the cytosol.
We then assessed whether de novo bacterial protein synthesis was required to decrease IL-1β, IL-6, and CXCL1 mRNA transcripts following SchuS4 infection. Chloramphenicol was used to inhibit SchuS4 protein synthesis. Chloramphenicol-treated SchuS4 escaped the endosome with similar kinetics as untreated bacteria, but were unable replicate, demonstrating the effectiveness of the antibiotic treatment (Fig 3D and data not shown). There were no significant differences in the reduction of transcripts encoding IL-1β, IL-6, and CXCL1 among cells infected with chloramphenicol-treated SchuS4 versus untreated SchuS4 (Fig 3E). Thus, de novo protein synthesis was not required for SchuS4 to decrease these mRNA.
To determine if SchuS4 viability was required to reduce IL-1β, IL-6, and CXCL1 transcripts, we compared mRNA levels among cells infected with live SchuS4 versus cells exposed to SchuS4 killed with either PFA or irradiation. Unlike live SchuS4, PFA-killed SchuS4 were not able to significantly decrease mRNA transcripts of IL-1β, IL-6 or CXCL1 compared to uninfected controls (Fig 3E). However, PFA-killed SchuS4 were retained in the endosome (Fig 3D). As described above, escape from the phagosome appeared to be essential for reduction of mRNA transcripts by SchuS4 (Fig 3A). Thus, it was not clear if the effect on mRNA transcripts observed among cells treated with PFA killed SchuS4 was due to the absence of viable SchuS4, or their retention in the endosome. To resolve this issue, we utilized irradiated SchuS4. Despite their inability to replicate, irradiated SchuS4 escape the endosome, which allowed us to assess the requirement for bacterial viability independent of escape into the cytosol (Fig 3D). Irradiated SchuS4 reduced IL-1β, IL-6, and CXCL1 mRNA equivalently to live SchuS4 (Fig 3E). Thus, the ability of SchuS4 to decrease IL-1β, IL-6, and CXCL1 transcripts was not dependent on viability of the organism.
SchuS4 infection decreases IL-1β, IL-6, and CXCL1 mRNA concomitant with p38 dephosphorylation
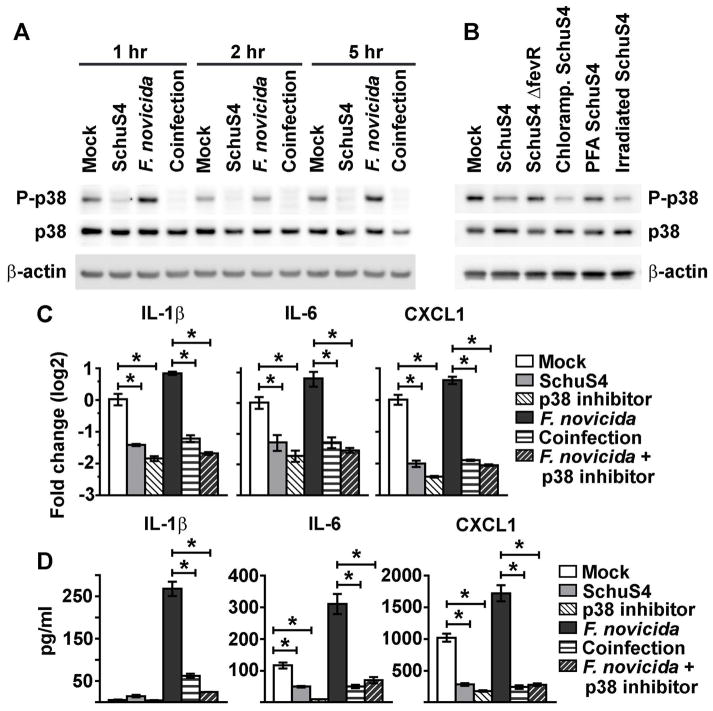
The mitogen activated protein kinases, and specifically p38, play a central role in the production of many proinflammatory cytokines/chemokines following bacterial infection, including IL-1β, IL-6, and CXCL1. Therefore, we determined if SchuS4 infection impacted activation of p38. We observed notable dephosphorylation of p38 as early as 1 h after infection in SchuS4 and SchuS4-F. novicida co-infected cells, compared to both uninfected and F. novicida infected cells (Fig 4A). We next confirmed that dephosphorylation of p38 was a consistent phenotype among BMM infected or exposed to preparations of SchuS4 that were associated with reduction of mRNA. Consistent with their ability to reduce IL-1β, IL-6, and CXCL1 mRNA, infection or exposure of BMM with SchuS4 treated with chloramphenicol or inactivated by irradiation both resulted in dephosphorylation of p38 in BMM (Fig 4B). Further, neither SchuS4ΔfevR nor PFA inactivated SchuS4 reduced phosphorylation of p38 compared to mock controls (Fig 4B). The lack of impact on phosphorylation of p38 by these bacteria is consistent with their inability to reduce host mRNA. Thus, dephosphorylation of p38 in BMM correlates with conditions in which SchuS4 was able to reduce host mRNA transcripts. To determine whether inactivation of the p38 pathway was sufficient to reduce IL-1β, IL-6, and CXCL1 transcript levels and secretion in the absence of infection, we inhibited p38 activity with SB203580. BMM treated with SB203580 exhibited decreased levels of mRNA transcripts and secretion of these proteins compared to uninfected cells (Fig 4 C–D). Similarly, F. novicida infected, SB203580-treated BMM had significantly fewer IL-1β, IL-6 and CXCL1 mRNA transcripts and reduced protein detected in the culture supernatant compared to mock-treated F. novicida infected controls (Fig 4C–D). Together these data demonstrate that SchuS4 inactivates p38 upon infection, and that decreased activation of p38 is sufficient to reduce IL-1β, IL-6, and CXCL1 mRNA and secretion.
Figure 4. Dephosphorylation of p38 by SchuS4 infection.
BMM were primed with P3C (500 ng/mL) and infected with SchuS4 MOI 50, chloramphenicol (Chloramp) treated SchuS4 MOI 50, SchuS4 ΔfevR MOI 50, F. novicida MOI 10, or co-infected with both SchuS4 MOI 50 and F. novicida MOI 10. As indicated, BMM were exposed to SchuS4 inactivated with either PFA or irradiation. Where specified, cells were treated with the p38 inhibitor SB203580 at the onset of infection. Mock infected cells served as negative controls. (A–B) Cell lysates were generated at the indicated times after infection (A) or 5 hours after infection or exposure to SchuS4 (B). Western blots were probed with antibodies to P-p38, and p38 and actin as loading controls. (C) RNA was harvested from cells 5 h after infection. qRT-PCR was performed to examine the indicated transcripts. (D) Supernatants were harvested 12 h after infection and the quantity of IL-1β, IL-6, and CXCL1 was measured by ELISA. *= p<0.05 comparing the indicated samples. Each data point depicts the mean of triplicate samples. Error bars represent SEM. Data are representative of three independent experiments.
SchuS4 infection destabilizes ARE-containing mRNA
Both transcription and mRNA stability are known to be regulated by p38 [19,20]. Thus, the reduction in mRNA encoding IL-1β, IL-6, and CXCL1 following SchuS4 infection could result either from a decreased rate of new transcription, or an increased rate of mRNA decay. To differentiate between these two possibilities, actinomycin D was used to inhibit transcription. Actinomycin D is routinely used to determine if different stimuli are affecting mRNA levels during transcription or as a post-transcriptional event [21]. If the ability of SchuS4 to reduce mRNA transcripts in the presence of actinomycin D was lost it would suggest that SchuS4 was acting to reduce mRNA by inhibiting new transcription. Conversely, if treatment with actinomycin D had no effect on the ability of SchuS4 to reduce the number of mRNA transcripts this would demonstrate that SchuS4 was reducing copies of mRNA via acceleration of mRNA decay. SchuS4 infection of actinomycin D-treated BMM resulted in a similar magnitude of reduction of IL-1β, IL-6, and CXCL1 transcripts compared to untreated, SchuS4 infected cells (Fig 5A). Accordingly, the half-life of these transcripts in SchuS4 infected, actinomycin D-treated cells was significantly decreased compared to uninfected actinomycin D cells (Fig 5A). These data support that SchuS4-mediated reduction of IL-1β, IL-6, and CXCL1 mRNA occurs via decreased mRNA stability. To further confirm that SchuS4 is inducing mRNA destabilization, we examined the activation of MK2, which is directly phosphorylated by p38 and phosphorylates mRNA binding proteins to indirectly regulate mRNA stability [22]. Similar to the kinetics of p38 inactivation following SchuS4 infection, dephosphorylation of MK2 was observed as early as 1 h after infection (Fig 5B). Thus, SchuS4-mediated inactivation of the p38-MK2 pathway upon infection results in the destabilization of IL-1β, IL-6, and CXCL1 mRNA.
Figure 5. Destabilization of ARE-containing mRNA following SchuS4 infection.
(A) BMM were primed with P3C (500 ng/mL) and infected with SchuS4 MOI 50 and treated with actinomycin D (ActD) at the onset of infection to inhibit transcription. RNA was harvested from cells at the indicated times after infection and qRT-PCR was performed to quantify the indicated mRNA transcripts. mRNA half-life (t1/2) in uninfected and SchuS4-infected actinomcyin D-treated samples is indicated. *= p<0.05 comparing the indicated sample to both uninfected and actinomycin D-only treated cells. (B–C) BMM were primed with P3C (500 ng/ml) and infected with SchuS4 MOI 50, F. novicida MOI 10, or co-infected with both SchuS4 MOI 50 and F. novicida MOI 10. (B) Cell lysates were generated at the indicated times after infection. Western blots were probed with antibodies to P-MK2, and MK2 and actin as loading controls. Mock infected cells served as negative controls. (C) RNA was harvested from cells 5 h after infection and qRT-PCR was performed to quantify the indicated mRNA transcripts. Mock infected cells served as negative controls. * = p<0.05 comparing the indicated sample to both uninfected and F. novicida-only infected cells. ns = not significantly different. (D) WT controls and TTP−/− BMM were primed with P3C (500 ng/mL) and infected with SchuS4 MOI, F. novicida MOI 10, or co-infected with both SchuS4 MOI 50 and F. novicida MOI 10. RNA was harvested 5 h after infection. qRT-PCR was performed to examine the quantity of the indicated transcripts, normalized to GAPDH. *= p<0.05 comparing the indicated groups. Each data point depicts the mean of triplicate samples. Error bars represent SEM. Data are representative of three independent experiments.
The stability of mRNA that encode tightly regulated proteins, such as many cytokines and chemokines, is controlled by the interaction of a variety of mRNA binding proteins with mRNA sequence elements. The most well characterized of these are AU-rich elements (ARE), found in the 3′ untranslated region of many inherently unstable transcripts. ARE consist of one or more AUUUA pentamers which interact with a variety of host proteins which regulate stability of host mRNA. [23]. IL-1β, IL-6, and CXCL1 mRNA contain ARE, whereas IL-18 transcripts, which were not reduced after SchuS4 infection, do not. This suggested that mRNA containing ARE were targeted for degradation during SchuS4 infection [24]. To confirm that SchuS4 infection was inducing selective destabilization of ARE-containing transcripts, we assessed a variety of mRNA transcripts encoding chemokines and cytokines produced by macrophages following encounter with microorganisms. CCL2, CCL3, CCL4, IL-1α, IL-12b, and TNFα mRNA, which contain ARE, were all reduced 5 hours after SchuS4 infection (Fig 5C). In contrast, CCL5 and CCL22 transcripts, which do not have ARE, remain unchanged upon SchuS4 infection (Fig 5C). Thus, SchuS4 infection resulted in selective destabilization of ARE-containing mRNA to limit production of key proinflammatory cytokines and chemokines.
There are many proteins that control the stabilization or decay of host mRNA via interaction with the 3′ ARE. The most well studied of these is tristetraprolin (TTP). Phosphorylation of TTP is dependent on p38. Phosphorylated TTP does not interact with host mRNA. Upon dephosphorylation, TTP interacts with AU-rich sequences in the 3′ region of host mRNA facilitating degradation of the sequence [25]. Given the observed dephosphorylation of p38 among SchuS4 infected cells and the selective targeting of ARE containing mRNA for decay, we hypothesized that SchuS4 may be facilitating TTP activity to degrade host mRNA. However, we did not observe reconstitution of host mRNA stabilization among SchuS4 infected TTP deficient macrophages (Fig 5D). This suggests that alternative, p38 dependent, mechanisms controlling host mRNA stabilization or degradation are targeted by SchuS4 in primary macrophages.
Discussion
It has been well established that SchuS4 both evades and suppresses inflammatory responses among resting resident host cells [5,6,11]. Here, we found that SchuS4 similarly inhibited proinflammatory responses among cells activated prior to challenge, as measured by secretion of key cytokines and chemokines. Further, we identified that a major mechanism by which SchuS4 rapidly inhibited production of these host products was to induce decay of ARE-containing mRNA transcripts.
We first established that ligation of surface receptors and/or phagocytosis of the bacterium was not sufficient for SchuS4 to execute suppression of host responses. Rather, we determined that mRNA decay relied on escape of SchuS4 into the cytosol. Our finding that interaction with surface receptors and uptake of the organisms alone were not adequate for SchuS4 mediated degradation of host mRNA was initially surprising. Previous studies showed that engagement of surface receptors, e.g. TLR2 [16] and CR3 [17], was required for some of the immunosuppressive mechanisms of SchuS4. The involvement of CR3 in mediating inhibition of inflammatory responses was dependent on opsonization of bacteria with serum prior to infection. We did not utilize opsonized bacteria in our study. Thus, it is unlikely that CR3 had a role in the degradation of mRNA. To confirm TLR2 was not required for modulation of IL-1β, IL-6 or CXCL1, we verified that SchuS4 was still capable of inhibiting secretion of these cytokines following co-infection with avirulent F. novicida among TLR2−/− cells (Fig 3C). Thus, to our knowledge, this data represents the first immunosuppressive mechanism induced by SchuS4 that dissociates Francisella-mediated inhibition of inflammatory responses from TLR signaling pathways.
We also determined that neither de novo bacterial protein synthesis nor viability was required for mRNA destabilization following infection, but that escape into the host cytosol was necessary to induce decay of host mRNA. Francisella escape from the endosome requires the Francisella Pathogenicity Island (FPI), therefore it appears likely that the FPI and putative Type VI secretion system of Francisella remain functional in the absence of new protein synthesis and in the majority of irradiated bacteria [26]. Together these data suggest that molecules required by the bacteria for both endosomal escape and suppression of proinflammatory responses via mRNA decay are pre-formed in SchuS4. Interestingly, bacteria killed using PFA did not escape the endosome and as a result also failed to accelerate host mRNA decay. Inactivation of bacteria with PFA or irradiation does not induce significant lysis of bacteria [27,28]. However, PFA renders the bacteria completely inactive whereas gamma irradiation can prevent bacteria replication while leaving bacterial metabolism intact [29]. Thus, it is possible that although neither PFA killed or irradiated bacteria were able to replicate, irradiated bacteria may have retained an active metabolism allowing for a functional Type IV secretion system and escape from the endosome. Regardless of the mechanism of endosomal escape, our data show that the immediate availability of bacterial products capable of inhibiting inflammatory responses is consistent with the rapid ability of SchuS4 to modulate host responses. Identification of the bacterial component(s) required for mRNA decay is currently under investigation by our laboratory.
There are a number of means by which stability of host mRNA is controlled in the cell. The most commonly studied of these is the interaction of stabilizing/destabilizing proteins with AREs present in the 3′UTR of host mRNA transcripts. There are several excellent reviews discussing the biology of AREs and their role in stabilization of host mRNA [23,30,31]. Briefly, AREs are pentameters of AUUUA. They are highly variable in their abundance and sequence and are divided into specific classes based on the number of ARE pentameters present in the 3′UTR. AREs are not present in every mRNA and thus, represent a mechanism by which the host can selectively control post-transcriptional production of potent cytokines and chemokines. Stabilization of host mRNA that contain AREs occurs through direct and/or indirect interaction of stabilizing or destabilizing proteins present in the host cell. Phosphorylation of these host proteins by kinases, e.g. by p38, dictates their activity. Modulation of ARE interacting proteins has been noted as a mechanism by which viruses influence inflammatory responses in host cells [32,33].
However, destabilization of cytokine mRNA during infection with intact bacteria has not been described. Rather, there is precedent in the literature for increasing mRNA stability by bacteria and/or bacterial components as a mechanism to sustain inflammatory responses. For example, avirulent F. novicida elicits a strong pro-inflammatory response following infection of host cells and it has been proposed that F. novicida inactivates TTP as a mechanism to enhance pro-inflammatory responses [34–36]. Specifically, a F. novicida ΔlpcC mutant provokes an even greater inflammatory response compared to wild type F. novicida. This elevated inflammatory response corresponded with increased deactivation of TTP compared to cells infected with wild type F. novicida. In contrast to infections mediated by F. novicida, we show that SchuS4 suppressed inflammatory responses by enhanced decay of host ARE-containing mRNA. Thus, it was possible that, as opposed to F. novicida, SchuS4 may further activate TTP to destabilize host mRNA. However, we did not observe a difference in the ability of SchuS4 to reduce ARE-containing mRNA among TTP-deficient BMM compared to littermate control BMM (Fig 5D). This suggested that SchuS4 infection may alter the activity of other ARE binding proteins known to interact with MK2, e.g. poly-A binding protein (PABP), HuR and BAF1, which regulate stability of mRNA [37–39]. We are currently examining the ability of SchuS4 to modulate function of multiple ARE binding proteins following infection.
In this report, we describe a novel mechanism used by a highly virulent pathogen to rapidly and effectively dampen components of the host inflammatory response via destabilization of cellular ARE-containing mRNA. Importantly, this mechanism did not require synthesis of bacterial components following infection. Rather, effective induction of mRNA decay was initiated with only the proteins, lipids, and carbohydrates present in the bacterium at the outset of infection. Our data represent an important advance in understanding the pathogenesis of disease following infection with Ftt. Destabilization of host cell mRNA may also represent a common mechanism of virulence employed by other similarly pathogenic bacteria. Continued characterization of virulence mechanisms restricted to highly virulent pathogens such as Ftt will likely be a fruitful approach to understanding the failure of the immune system to properly mount a protective response to these and other intracellular pathogens.
Supplementary Material
Acknowledgments
The authors also thank Dr. Jeffery Wilusz and Dr. Harlan Caldwell for helpful discussion and suggestions pertaining to this manuscript. This work was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Allergy and Infectious Diseases.
References
- 1.Baxt LA, Garza-Mayers AC, Goldberg MB. Bacterial subversion of host innate immune pathways. Science. 2013;340:697–701. doi: 10.1126/science.1235771. [DOI] [PubMed] [Google Scholar]
- 2.Sjostedt A. Tularemia: History, epidemiology, pathogen physiology, and clinical manifestations. Ann N Y Acad Sci. 2007;1105:1–29. doi: 10.1196/annals.1409.009. [DOI] [PubMed] [Google Scholar]
- 3.Hall JD, Woolard MD, Gunn BM, Craven RR, Taft-Benz S, Frelinger JA, Kawula TH. Infected-host-cell repertoire and cellular response in the lung following inhalation of Francisella tularensis schu s4, lvs, or u112. Infect Immun. 2008;76:5843–5852. doi: 10.1128/IAI.01176-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eigelsbach HT, Downs CM. Prophylactic effectiveness of live and killed tularemia vaccines. I. Production of vaccine and evaluation in the white mouse and guinea pig. J Immunol. 1961;87:415–425. [PubMed] [Google Scholar]
- 5.Bosio CM, Bielefeldt-Ohmann H, Belisle JT. Active suppression of the pulmonary immune response by Francisella tularensis Schu4. J Immunol. 2007;178:4538–4547. doi: 10.4049/jimmunol.178.7.4538. [DOI] [PubMed] [Google Scholar]
- 6.Chase JC, Celli J, Bosio CM. Direct and indirect impairment of human dendritic cell function by virulent Francisella tularensis Schu s4. Infect Immun. 2009;77:180–195. doi: 10.1128/IAI.00879-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Telepnev M, Golovliov I, Grundstrom T, Tarnvik A, Sjostedt A. Francisella tularensis inhibits toll-like receptor-mediated activation of intracellular signalling and secretion of tnf-alpha and il-1 from murine macrophages. Cell Microbiol. 2003;5:41–51. doi: 10.1046/j.1462-5822.2003.00251.x. [DOI] [PubMed] [Google Scholar]
- 8.Chong A, Wehrly TD, Nair V, Fischer ER, Barker JR, Klose KE, Celli J. The early phagosomal stage of Francisella tularensis determines optimal phagosomal escape and Francisella pathogenicity island protein expression. Infect Immun. 2008;76:5488–5499. doi: 10.1128/IAI.00682-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chong A, Wehrly TD, Child R, Hansen B, Hwang S, Virgin HW, Celli J. Cytosolic clearance of replication-deficient mutants reveals Francisella tularensis interactions with the autophagic pathway. Autophagy. 2012;8:1342–1356. doi: 10.4161/auto.20808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bosio CM, Elkins KL. Susceptibility to secondary Francisella tularensis live vaccine strain infection in B-cell-deficient mice is associated with neutrophilia but not with defects in specific t-cell-mediated immunity. Infect Immun. 2001;69:194–203. doi: 10.1128/IAI.69.1.194-203.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bauler TJ, Chase JC, Bosio CM. IFN-beta mediates suppression of Il-12p40 in human dendritic cells following infection with virulent Francisella tularensis. J Immunol. 2011;187:1845–1855. doi: 10.4049/jimmunol.1100377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen CY, Ezzeddine N, Shyu AB. Messenger RNA half-life measurements in mammalian cells. Methods in enzymology. 2008;448:335–357. doi: 10.1016/S0076-6879(08)02617-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamamoto Y, Klein TW, Friedman H. Induction of cytokine granulocyte-macrophage colony-stimulating factor and chemokine macrophage inflammatory protein 2 mrnas in macrophages by Legionella pneumophila or Salmonella typhimurium attachment requires different ligand-receptor systems. Infect Immun. 1996;64:3062–3068. doi: 10.1128/iai.64.8.3062-3068.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clemens DL, Lee BY, Horwitz MA. Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect Immun. 2004;72:3204–3217. doi: 10.1128/IAI.72.6.3204-3217.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Golovliov I, Baranov V, Krocova Z, Kovarova H, Sjostedt A. An attenuated strain of the facultative intracellular bacterium Francisella tularensis can escape the phagosome of monocytic cells. Infect Immun. 2003;71:5940–5950. doi: 10.1128/IAI.71.10.5940-5950.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crane DD, Ireland R, Alinger JB, Small P, Bosio CM. Lipids derived from virulent Francisella tularensis broadly inhibit pulmonary inflammation via Toll-like receptor 2 and Peroxisome Proliferator-Activated Receptor alpha. Clin Vaccine Immunol. 2013;20:1531–1540. doi: 10.1128/CVI.00319-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dai S, Rajaram MV, Curry HM, Leander R, Schlesinger LS. Fine tuning inflammation at the front door: Macrophage complement receptor 3-mediates phagocytosis and immune suppression for Francisella tularensis. PLoS Pathog. 2013;9:e1003114. doi: 10.1371/journal.ppat.1003114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wehrly TD, Chong A, Virtaneva K, Sturdevant DE, Child R, Edwards JA, Brouwer D, Nair V, Fischer ER, Wicke L, Curda AJ, Kupko JJ, 3rd, Martens C, Crane DD, Bosio CM, Porcella SF, Celli J. Intracellular biology and virulence determinants of Francisella tularensis revealed by transcriptional profiling inside macrophages. Cell Microbiol. 2009;11:1128–1150. doi: 10.1111/j.1462-5822.2009.01316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khabar KS. Rapid transit in the immune cells: The role of mRNA turnover regulation. J Leukoc Biol. 2007;81:1335–1344. doi: 10.1189/jlb.0207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arthur JS, Ley SC. Mitogen-activated protein kinases in innate immunity. Nat Rev Immunol. 2013;13:679–692. doi: 10.1038/nri3495. [DOI] [PubMed] [Google Scholar]
- 21.Hao S, Baltimore D. The stability of mrna influences the temporal order of the induction of genes encoding inflammatory molecules. Nat Immunol. 2009;10:281–288. doi: 10.1038/ni.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kotlyarov A, Neininger A, Schubert C, Eckert R, Birchmeier C, Volk HD, Gaestel M. Mapkap kinase 2 is essential for LPS-induced TNF-alpha biosynthesis. Nat Cell Biol. 1999;1:94–97. doi: 10.1038/10061. [DOI] [PubMed] [Google Scholar]
- 23.Anderson P. Post-transcriptional regulons coordinate the initiation and resolution of inflammation. Nat Rev Immunol. 2010;10:24–35. doi: 10.1038/nri2685. [DOI] [PubMed] [Google Scholar]
- 24.Gruber AR, Fallmann J, Kratochvill F, Kovarik P, Hofacker IL. Aresite: A database for the comprehensive investigation of au-rich elements. Nucleic Acids Res. 2011;39:D66–69. doi: 10.1093/nar/gkq990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perez-Ortin JE, Alepuz P, Chavez S, Choder M. Eukaryotic mrna decay: Methodologies, pathways, and links to other stages of gene expression. J Mol Biol. 2013;425:3750–3775. doi: 10.1016/j.jmb.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 26.Barker JR, Chong A, Wehrly TD, Yu JJ, Rodriguez SA, Liu J, Celli J, Arulanandam BP, Klose KE. The Francisella tularensis pathogenicity island encodes a secretion system that is required for phagosome escape and virulence. Mol Microbiol. 2009;74:1459–1470. doi: 10.1111/j.1365-2958.2009.06947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chao Y, Zhang T. Optimization of fixation methods for observation of bacterial cell morphology and surface ultrastructures by atomic force microscopy. Appl Microbiol Biotechnol. 2011;92:381–392. doi: 10.1007/s00253-011-3551-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGrath RA, Williams RW, Swartzendruber DC. Breakdown of DNA in x-irradiated escherichia coli. Biophys J. 1966;6:113–122. doi: 10.1016/S0006-3495(66)86643-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magnani DM, Harms JS, Durward MA, Splitter GA. Nondividing but metabolically active gamma-irradiated Brucella melitensis is protective against virulent b. Melitensis challenge in mice. Infect Immun. 2009;77:5181–5189. doi: 10.1128/IAI.00231-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bakheet T, Williams BR, Khabar KS. Ared 3.0: The large and diverse AU-rich transcriptome. Nucleic Acids Res. 2006;34:D111–114. doi: 10.1093/nar/gkj052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 32.Dickson AM, Anderson JR, Barnhart MD, Sokoloski KJ, Oko L, Opyrchal M, Galanis E, Wilusz CJ, Morrison TE, Wilusz J. Dephosphorylation of hur protein during alphavirus infection is associated with HuR relocalization to the cytoplasm. J Biol Chem. 2012;287:36229–36238. doi: 10.1074/jbc.M112.371203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kummer M, Prechtel AT, Muhl-Zurbes P, Turza NM, Steinkasserer A. Hsv-1 upregulates the are-binding protein tristetraprolin in a stat1- and p38-dependent manner in mature dendritic cells. Immunobiology. 2009;214:852–860. doi: 10.1016/j.imbio.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 34.Parsa KV, Ganesan LP, Rajaram MV, Gavrilin MA, Balagopal A, Mohapatra NP, Wewers MD, Schlesinger LS, Gunn JS, Tridandapani S. Macrophage pro-inflammatory response to Francisella novicida infection is regulated by ship. PLoS Pathog. 2006;2:e71. doi: 10.1371/journal.ppat.0020071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma J, Li Q, Mishra BB, Pena C, Teale JM. Lethal pulmonary infection with Francisella novicida is associated with severe sepsis. J Leukoc Biol. 2009;86:491–504. doi: 10.1189/jlb.1208728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakayasu ES, Tempel R, Cambronne XA, Petyuk VA, Jones MB, Gritsenko MA, Monroe ME, Yang F, Smith RD, Adkins JN, Heffron F. Comparative phosphoproteomics reveals components of host cell invasion and post-transcriptional regulation during Francisella infection. Mol Cell Proteomics. 2013;12:3297–3309. doi: 10.1074/mcp.M113.029850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bollig F, Winzen R, Gaestel M, Kostka S, Resch K, Holtmann H. Affinity purification of ARE-binding proteins identifies polyA-binding protein 1 as a potential substrate in MK2-induced mRNA stabilization. Biochem Biophys Res Commun. 2003;301:665–670. doi: 10.1016/s0006-291x(03)00015-9. [DOI] [PubMed] [Google Scholar]
- 38.Maitra S, Chou CF, Luber CA, Lee KY, Mann M, Chen CY. The AU-rich element mRNA decay-promoting activity of Brf1 is regulated by mitogen-activated protein kinase-activated protein kinase 2. Rna. 2008;14:950–959. doi: 10.1261/rna.983708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tiedje C, Ronkina N, Tehrani M, Dhamija S, Laass K, Holtmann H, Kotlyarov A, Gaestel M. The p38/MK2-driven exchange between tristetraprolin and HuR regulates AU-rich element-dependent translation. PLoS genetics. 2012;8:e1002977. doi: 10.1371/journal.pgen.1002977. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.