Abstract
With the advent of genomic sequences and next generation sequencing technologies (RNA-Seq), multiple repertoires of olfactory proteins in various insect species are being unraveled. However, functional analyses are lagging behind due in part to the lack of simple and reliable methods for heterologous expression of odorant receptors (ORs). While the Xenopus oocyte recording system fulfills some of this lacuna, this system is devoid of other olfactory proteins thus testing only the “naked” ORs. Recently, a moth OR was expressed in the majority of neurons in the antennae of the fruit fly by using Orco-GAL4 to drive expression of the moth OR. Electroantennogram (EAG) was used to de-orphanize the moth OR, but generic application of this approach was brought to question. Here, we describe that this system works with ORs not only from taxonomically distant insect species (moth), but also closely related species (mosquito), even when the fruit fly has highly sensitive innate ORs for the odorant being tested. We demonstrate that Orco-GAL4 flies expressing the silkworm pheromone receptor, BmorOR1, showed significantly higher responses to the sex pheromone bombykol than the control lines used to drive expression. Additionally, we show that flies expressing an OR from the Southern house mosquito, CquiOR2, gave significantly stronger responses to the cognate odorants indole and 2-methylphenol than the “background noise” recorder from control lines. In summary, we validate the use of Orco-GAL4 driven UAS-OR lines along with EAG analysis as a simple alternative for de-orphanization and functional studies of insect ORs in an intact olfactory system.
Keywords: Electroantennogram, EAG, Orco, Bombyx mori, Culex quinquefasciatus, Spodoptera littoralis
Introduction
The advent of genome sequencing coupled with next generation sequencing technologies (RNA-Seq) opened new opportunities for the field of insect olfaction. It is now possible to identify the complete repertoire of olfactory proteins in an insect species and pave the way to exploit these molecular targets for reducing populations of medically important insects and agricultural pests [1]. However, the characterization and/or de-orphanization of these newly identified putative odorant receptors (ORs) remain the rate-determining step in our progress towards a full understanding of how insects perceive the environment. While hundreds of OR genes have been identified from multiple insect species, the largest majority of them are yet to be characterized and remain as putative ORs. This is mainly due to the lack of a simple and reliable method for heterologous expression of ORs and functional analysis.
The Xenopus oocyte recording system was employed in insect olfaction soon after OR genes were identified from Drosophila melanogaster [2,3] to de-orphanize for the first time an insect OR, DmelOR43a [4]. It was later realized that the odorant receptor co-receptor (Orco) [5] is necessary for the proper formation of ion-channels [6]. Thereafter, many ORs have been de-orphanized by co-expression with their respective Orcos in Xenopus oocytes. Albeit simple, this heterologous expression system lacks other olfactory proteins and this may prevent full characterization of ORs and addressing questions of specificity and sensitivity. Additionally, for unknown reason(s) some ORs remain silent when expressed in Xenopus oocytes. The most elegant system for de-orphanization and functional studies of insect ORs is probably the “empty neuron” system [7,8]. However, it relies on single sensillum recordings, which in turn requires skilled hands and sophisticated instrumentation. Furthermore, this heterologous system has limitations when testing ORs from taxonomically distant insect species. For example, it works very well for dipteran insects (same order as Drosophila) like the malaria mosquito [9], but performs poorly with moth ORs [10], which are fully functional only when expressed in trichoid sensilla using a different system [11]. Recently, the fruit fly has been employed to de-orphanize a moth OR by using the Orco-GAL4 line to drive expression of a moth OR in the majority of olfactory receptor neurons (ORNs) [12]. Then, a simple and readily available technique in chemical ecology laboratories, electroantennagram (EAG), was employed to record from flies expressing the moth OR. While the results were encouraging, it raised the question whether this facile technique would be applicable to ORs from closely related species due to Drosophila “background” responses (to common odorants). To address this concern we tested the system with the expression of ORs from insects taxonomically distant as well as a closely related species. Surprisingly, flies expressing bombykol receptor from the silkworm moth, BmorOR1, gave a significant signal to “noise” ratio even considering the “background” detection of bombykol by the fruit fly [11]. More importantly, flies expressing a mosquito OR sensitive to a common odorant, indole, responded with remarkable sensitivity. Our data thus validate the Orco-driven expression of allospecific ORs as a complementary tool for de-orphanization and functional studies of insect ORs.
Methods and Materials
Transgenic flies and cross
The transgenic lines UAS-SlitOR6 and Orco-GAL4 were kindly provided by Dr. Nicolas Montagne. The UAS-BmorOR1 line was maintained in the laboratory from previous work [10]. In order to obtain transformants of the odorant receptor CquiOR2 [13] from Culex quinquefasciatus, the complete open read frame (ORF) was cloned into the pUAST vector (obtained from the Drosophila Genomic Research Center). Recombinant plasmids were purified using Plasmid Midi Kit (Qiagen). Transgenic lines (UAS-CquiOR2) were generated and balanced by BestGene Inc. (Chino Hills, CA, USA). Flies were maintained at 25°C in a 12:12-h light:dark cycle on standard corn meal agar medium. UAS-SlitOR6, UAS-BmorOR1, and UAS-CquiOR2 lines were crossed separately with Orco-GAL4 flies to generate F1 progeny, which were used for EAG measurements.
Stimuli
(9Z,12E)-Tetradecadienyl acetate (Z9,E12-14-OAc) and (10E,12Z)-hexadecadien-1-ol (bombykol) were purchased from Plant Research International (Wageningen, The Netherlands). 2-Heptanone, indole, 3-methylindole and 2-methylphenol were purchased from Sigma Aldrich (St. Louis, MO). These odorants were dissolved in hexane to make stock solutions of 10 μg/μl from which decadic dilutions were made. A 10-μl aliquot of a stimulus dissolved in hexane in the desired dose was loaded on a lter paper strip, the solvent was evaporated for 30 s, and the strip was placed in a disposable syringe. Hexane alone served as negative control, whereas 2-heptanone was used as positive control.
Electroantennographic (EAG) analysis
Individual F1 progeny were analyzed 5 days after emergence. Typically male flies were used, except for systematic comparison of male and female responses to bombykol. Flies were collected using an aspirator, inserted into a micropipette tip and immobilized, as previously reported [14]. Glass electrodes were manufactured using micropipette puller P-97 (Sutter Instrument Company, Novato, CA). The electrodes were filled with a solution of 1M potassium chloride and 1% polyvinylpyrrolidone. The reference electrode was inserted in the eye of an immobilized fly and the recording electrode was placed in contact with the tip of the antennae using a micromanipulator MP-12 and a high impedance AC/DC pre-amplifier (Syntech, Germany). The preparation was bathed in a high humidity air stream flowing from a Stimulus Controller CS-55 (Syntech) at 160 ml/min to which compensatory flow or stimulus pulse (125 ml/s, 300 ms) was added. Signal from the antenna induced by stimulus or control puff was recorded for 10 s. Data were processed with the EAG2000 software (Syntech).
Quantification of DmelOrco transcripts
Five days old male and female flies W1118 were collected and antennae were dissected and pooled (30 antennae per tube; triplicate male and female samples). Total RNA was isolated using TRizol Reagent (Invitrogen), according to manufacturer’s instruction, and, subsequently, treated with 1U of Dnase I (Promega). First strand cDNAs were synthesized from 1 μg of total RNA using oligo dT(18) primer (Invitrogen) and MMLV reverse transcriptase (Promega). Genomic DNA contamination was monitored by PCR with a pair of primer designed on the basis of the gene encoding Actin42A protein and spanning an intron.
Relative quantification qPCR experiment was performed using an ABI Prism 7300 Sequence Detection System (Applied Biosystems). The reactions were carried out in a mix containing SYBR Green dye (Power SYBR Green PCR Master Mix, Applied Biosystems), 5 μmol of each primers and 2 μL of cDNA. All the sample were run in biological triplicate. The thermal cycling was set for one cycle at 50°C, 2 min, 95°C for 10 min, followed by 40 cycles at 95°C, 15 s, and 59°C for 1 min. Reactions without cDNA template were ran as negative controls. The specificity of the PCR products was verified by melting curve analysis. A fragment of gene encoding the RpL32 ribosomal protein was used as an internal control gene. The expression data analysis was made using the equation 2−ΔΔCT. The following PCR primers were designed using ABI Primer Express 3.0 software (Applied Biosystems): DmelOrco (FlyBase ID: FBgn0037324, For- CAGTGCCAGAAGGCGATGA; Rev- GCGAATGCCAAGAAGCCA), DmelRpL32 (FlyBase ID: FBgn0002626; For- GACCATCCGCCCAGCATAC; Rev- AACAGAGTGCGTCGCCG), and DmelAct42A (FlyBase ID: FBgn0000043; For- GCGTCTGTCATTGTGCTAAGTGT; Rev- ATCCGGCATGTGCAAAGC).
Statistical Analyses
Statistical analyses were performed with the software GraphPad Prism 6 (La Jolla, CA). Differences between means were tested for significance with student’s t-test (comparison with a control), Tukey’s test (comparison by groups of means), and non-parametric Mann-Whitney (qPCR) tests.
Results and Discussion
Spodoptera littoralis pheromone
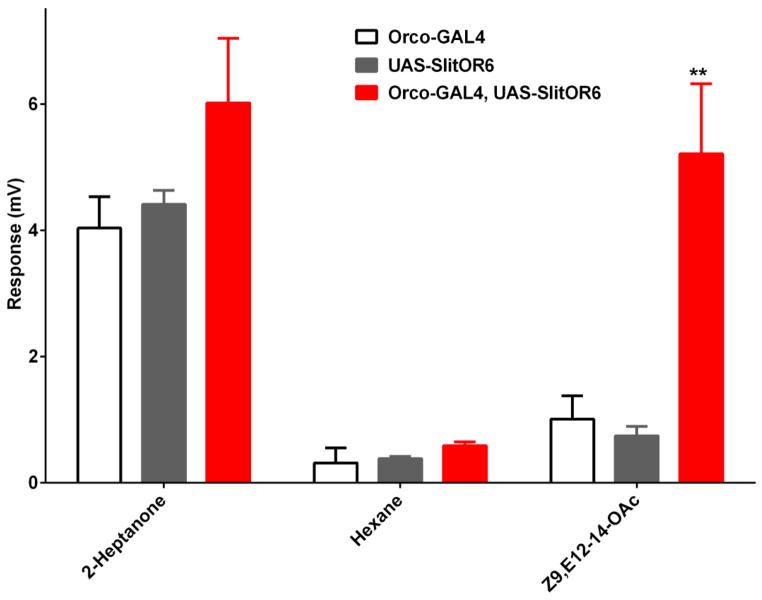
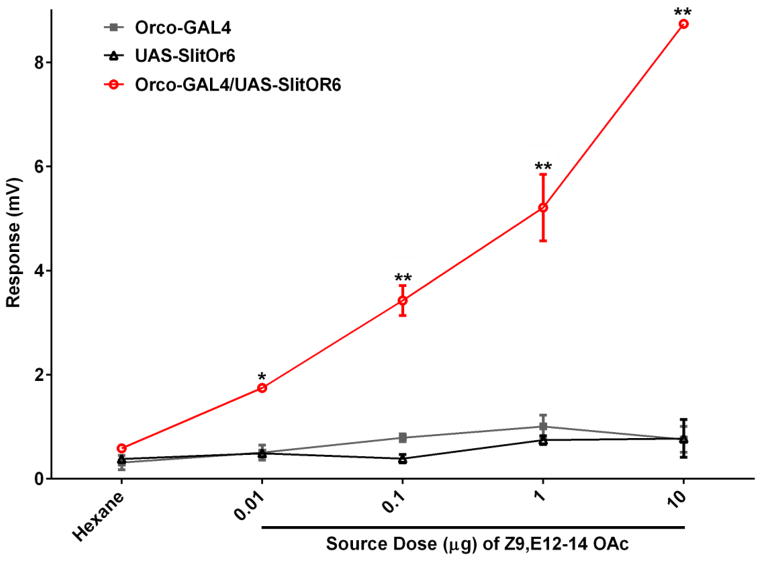
We recorded EAG from antennae of the GAL4 driver line (Orco-GAL4), the UAS responder line (UAS-SlitOR6) and their F1 progeny. Antennal preparations were challenged with two stimuli, i.e., 2-heptanone and Z9,E12-14-OAc. 2-Heptanone is an innate odorant for the flies [8] so it was used here as a “quality control” for all lines tested. Z9,E12-14-OAc is an essential constituent of the sex pheromone of S. littoralis [15] and the cognate ligand for SlitOR6 [12]. High EAG responses were recorded from all lines when stimulated with 2-heptanone (Figure 1), but only the F1 progeny responded to Z9,E12-14-OAc with robust signal (Figure 1), as previously observed [12]. To provide more detail about the sensitivity of this surrogate system, we performed concentration response analysis for Z9,E12-14-OAc. Clearly, the flies carrying SlitOR6 showed robust dose-dependent responses, with a threshold of 10 ng (Figure 2). These results, confirming previous findings [12], are not surprising, particularly considering that the fruit fly is devoid of innate ORs sensitive to the sex pheromone of S. littoralis. This is not the case with bombykol, the sex pheromone of the silkworm moth, Bombyx mori, which is detected with high sensitivity by DmelOR7a [11] housed in ab4 sensilla [10]. Intriguingly, the olfactory receptor neurons (ORNs) housing DmelOR7a in the fruit fly are more sensitive to bombykol than the ORNs in the antennae of the silkworm moth that house the sex pheromone receptor BmorOR1 [11]. Thus, we asked the question whether the Orco system could be used to analyze an OR under such a high background “noise.”
Figure 1.
EAG recorded from male antennae of transgenic flies expressing a pheromone receptor, SlitOR6, from S. littoralis (Orco-GAL4, UAS-SlitOR6) and from the lines used to obtain the crossings: Orco-GAL4 and UAS-SlitOR6. 2-Heptanone was used as positive control and hexane as negative control. Z9,E12-14-OAc is a constituent of S. littoralis sex pheromone [15]. Source doses: 2-heptanone, 100 μg; Z9,E12-14-OAc, 1 μg. N=6; error bars in all figures represent SEM, statistical significance at 5% and 1% levels are denoted with one (*) and two (**) asterisks, respectively.
Figure 2.
Dose-dependent EAG responses recorded from the male antennae of transgenic flies (Orco-GAL4,UAS-SlitOR6) stimulated with a component of S. littoralis sex pheromone, Z9,E12-14-OAc. EAG responses of the F1 crosses were compared with those elicited by the Orco-GAL4 driver line and UAS-responder line. N=6
Bombyx mori pheromone
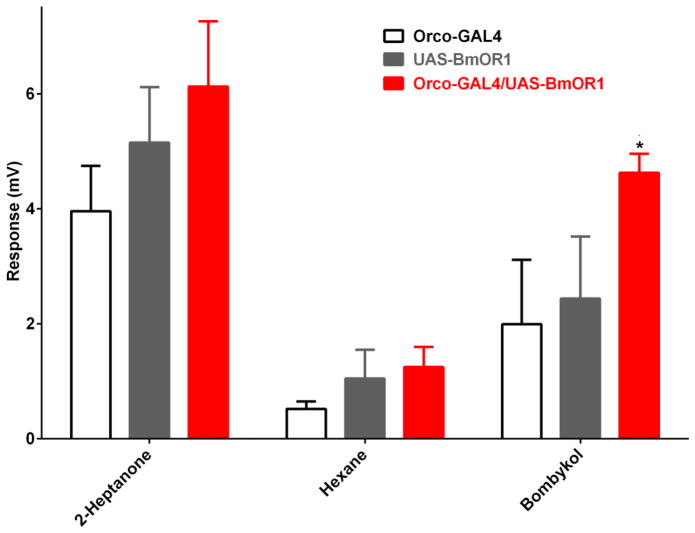
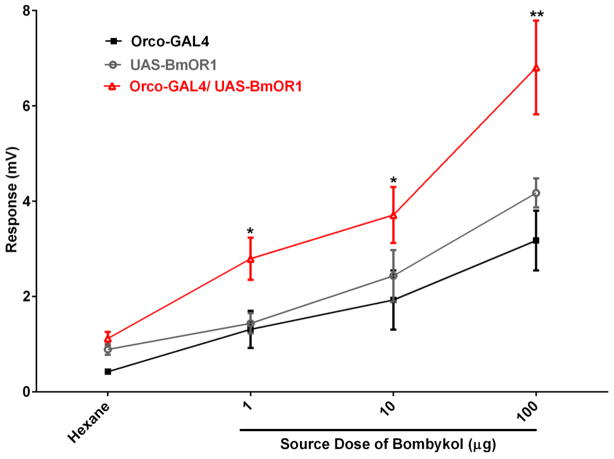
The Orco-GAL4 line was crossed with the UAS-BmorOR1 responder line, and EAG was recorded from the antennae of F1 progeny. Surprisingly, the responses recorded from the antennae of transgenic Orco-GAL4, UAS-BmorOR1 were significantly higher than those recorded with controls lines (Figure 3). Dose-dependent relationships (Figure 4) showed higher responses recorded from Orco-Gal4 flies carrying BmorOR1 than control flies at all doses tested. These findings suggest that even at such a high background level, the Orco system has potential application for heterologous expression and functional analysis of insect ORs.
Figure 3.
EAG responses recorded from the driver, responder, and F1 crossing expressing the pheromone receptor from the silkworm moth, BmorOR1. Responses elicited by bombykol in the Orco-GAL4,UAS-BmorOR1 flies were significantly higher than those recorded from control flies. 2-Heptanone elicited robust responses in all lines. Source doses: 2-heptanone, 100 μg; bombykol, 50 μg. N=6
Figure 4.
Dose-dependent relationships elicited by bombykol. The three lines responded in a dose-dependent fashion considering that an innate OR from the fruit fly is very sensitive to bombykol [11], but responses elicited by flies carrying BmorOR1 were higher than those obtained with control flies. N=6
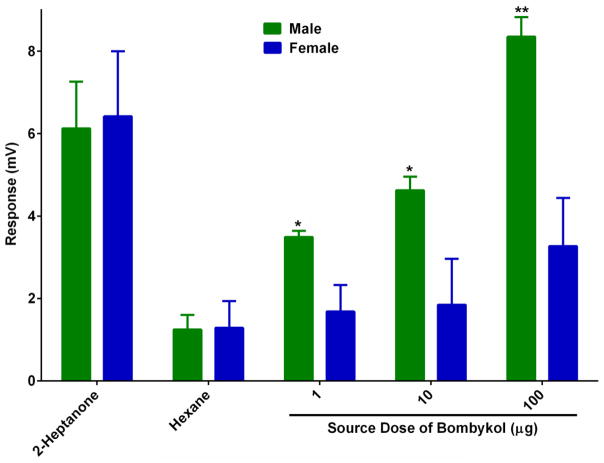
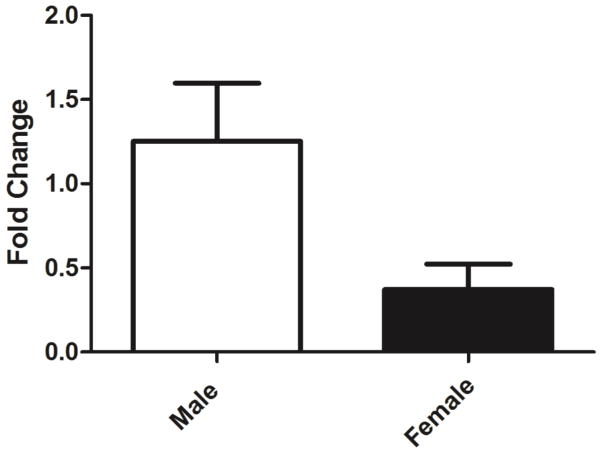
Both male and female antennae of the fruit fly house ORNs sensitive to bombykol. We then compared the EAG responses of male and female transgenic flies (Orco-GAL4,UAS-BmorOR1) to bombykol. EAG recorded from male antennae were significantly higher than those obtained with female antennae at all doses tested (Figure 5). The higher levels of Orco transcripts in male antennae (compared to female antennae) [16] might be manifested in higher levels of expression of a heterologous OR thus the higher EAG responses. To test this hypothesis we quantified Orco transcript by qPCR. Indeed, Orco transcript were detected in significantly higher levels in male than female antennae (Figure 6)
Figure 5.
Differential EAG responses recorded from male and female antennae of transgenic line expressing a silkworm moth pheromone receptor (Orco-GAL4,UAS-BmorOR1). Responses elicited in male antennae were significantly higher than those generated by female antennae at all doses tested. Responses to 2-heptanone were not significantly different between male and female antennae. N=6
Figure 6.
Transcript levels of DmelOrco in male and female antennae from 5-days-old adult fruit flies. This odorant receptor co-receptor gene was significantly (5% level, Mann Whitney test) more expressed in male than female antennae.
Mosquito oviposition attractant
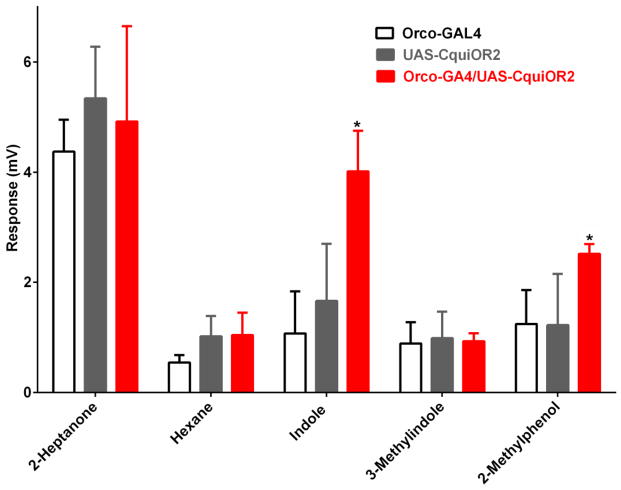
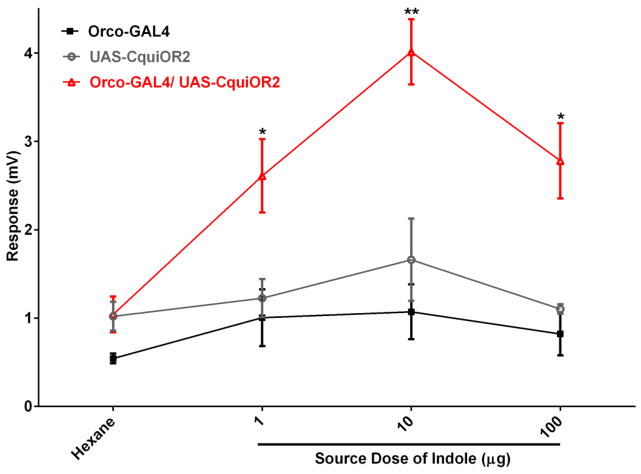
Lastly, we tested the Orco system with the heterologous expression of an OR from a closely related species, the Southern house mosquito, Culex quinquefasciatus. CquiOR2 has been previously de-orphanized using the Xenopus oocyte recording system and demonstrated to be sensitive to indole [13]. Transgenic flies expressing CquiOR2 responded to indole and 2-methylphenol, whereas the EAG responses recorded from the transgenic flies for the other compounds were not significantly different from the controls (Figure 7). Interestingly, the responses obtained with the Orco system differed slightly from the profile obtained with the Xenopus oocyte recording system [13] in that 3-methylindole (=skatole) did not elicit EAG response in the flies, but generated significant currents in the Xenopus oocyte system, close to the response observed for 2-methylphenol. These subtle differences between the two systems may be derived from the role of other olfactory proteins (e.g. odorant-binding proteins, odorant-degrading enzymes), which are absent in Xenopus but present in an intact insect olfactory system. The EAG responses recorded with the major ligand, indole, showed a dose-dependent relationship, but saturated responses at high (100 μg) doses (Figure 8)
Figure 7.
EAG responses recorded from transgenic flies expressing a mosquito receptor, CquiOR2, and the corresponding driver and responder lines. Source dose for all odorants, 100 μg. Responses to 2-heptanone and 3-methylindole (=skatole) were not significantly different, but males of F1 progeny carrying CquiOR2 gave stronger EAG responses to indole and 2-methylphenol. N=6
Figure 8.
Dose-dependent relationships recorded from male antennae of F1 progeny, driver and responder lines. EAG responses recorded from flies carrying the Southern house mosquito OR, CquiOR2, were significantly higher than the “background” response from control lines. Although EAG saturation was observed with 100 μg dose, the response was significantly higher than those from control flies. N=6
In conclusion, the Orco system has a potential application in insect olfaction. EAG recordings are simple and the possible “background noise” generated by the regular reception of odorants by the fruit fly is not an impediment. For example, the innate bombykol ORN from the fruit fly is very sensitive, yet bombykol elicited significantly higher responses from flies carrying BmorOR1 than control flies. One added value to the Orco driver system is that test ORs are expressed in both basiconic and trichoid sensilla. While the biochemical machinery of basiconic sensilla suffice for the reception of general odorants by allospecific ORs [9], moth ORs expressed in basiconic sensilla are much less sensitive to cognate pheromone [10] than when the same OR is expressed in trichoid sensilla [11]. Thus, with the Orco system one does not have to select the most suitable site for expression of a test OR as EAG records responses from both types of sensilla. Although preparations of Xenopus oocytes for recordings and transgenic lines can be completed in weeks and months, respectively, the Orco driver system provides a complete repertoire of olfactory proteins, including OBPs and ODEs.
Acknowledgments
We thank Dr. Nicolas Montagne (UPMC- Université Paris) for sharing flies, Dr. Julien Pelletier for initial molecular work for the preparation of pUAST-CquiOR2, Madhulika Battu for help maintaining fly colonies, and laboratory members for insightful discussions. Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award R01AI095514. The content is solely the responsibility of the authors and does not necessarily represent the official views of N.I.H. C. U.-V. and W.J.deC (Universidade Federal de Uberlandia, Brazil) were supported by the National Council of Scientific and Technological Development (CNPq, Brazil), Science Without Borders Program (237054/2012-4).
References
- 1.Leal WS. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annual review of entomology. 2013;58:373–391. doi: 10.1146/annurev-ento-120811-153635. [DOI] [PubMed] [Google Scholar]
- 2.Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, Carlson JR. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron. 1999;22 (2):327–338. doi: 10.1016/s0896-6273(00)81093-4. [DOI] [PubMed] [Google Scholar]
- 3.Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999;96 (5):725–736. doi: 10.1016/s0092-8674(00)80582-6. [DOI] [PubMed] [Google Scholar]
- 4.Wetzel CH, Behrendt HJ, Gisselmann G, Stortkuhl KF, Hovemann B, Hatt H. Functional expression and characterization of a Drosophila odorant receptor in a heterologous cell system. Proceedings of the National Academy of Sciences of the United States of America. 2001;98 (16):9377–9380. doi: 10.1073/pnas.151103998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, Vosshall LB. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43 (5):703–714. doi: 10.1016/j.neuron.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 6.Nakagawa T, Sakurai T, Nishioka T, Touhara K. Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science. 2005;307 (5715):1638–1642. doi: 10.1126/science.1106267. [DOI] [PubMed] [Google Scholar]
- 7.Dobritsa AA, van der Goes van Naters W, Warr CG, Steinbrecht RA, Carlson JR. Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron. 2003;37 (5):827–841. doi: 10.1016/s0896-6273(03)00094-1. [DOI] [PubMed] [Google Scholar]
- 8.Hallem EA, Ho MG, Carlson JR. The molecular basis of odor coding in the Drosophila antenna. Cell. 2004;117 (7):965–979. doi: 10.1016/j.cell.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 9.Carey AF, Wang G, Su CY, Zwiebel LJ, Carlson JR. Odorant reception in the malaria mosquito Anopheles gambiae. Nature. 2010;464 (7285):66–71. doi: 10.1038/nature08834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Syed Z, Ishida Y, Taylor K, Kimbrell DA, Leal WS. Pheromone reception in fruit flies expressing a moth’s odorant receptor. Proceedings of the National Academy of Sciences of the United States of America. 2006;103 (44):16538–16543. doi: 10.1073/pnas.0607874103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Syed Z, Kopp A, Kimbrell DA, Leal WS. Bombykol receptors in the silkworm moth and the fruit fly. Proceedings of the National Academy of Sciences of the United States of America. 2010;107 (20):9436–9439. doi: 10.1073/pnas.1003881107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Montagne N, Chertemps T, Brigaud I, Francois A, Francois MC, de Fouchier A, Lucas P, Larsson MC, Jacquin-Joly E. Functional characterization of a sex pheromone receptor in the pest moth Spodoptera littoralis by heterologous expression in Drosophila. The European journal of neuroscience. 2012;36 (5):2588–2596. doi: 10.1111/j.1460-9568.2012.08183.x. [DOI] [PubMed] [Google Scholar]
- 13.Pelletier J, Hughes DT, Luetje CW, Leal WS. An odorant receptor from the southern house mosquito Culex pipiens quinquefasciatus sensitive to oviposition attractants. PloS one. 2010;5 (4):e10090. doi: 10.1371/journal.pone.0010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Syed Z, Leal WS. Electrophysiological measurements from a moth olfactory system. Journal of visualized experiments. JoVE. 2011;(49) doi: 10.3791/2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamaki Y, Yushima T. Sex pheromone of the cotton leaf worm, Spodoptera littoralis. J Insect Physiol. 1974;20:1005–1014. doi: 10.1016/0022-1910(74)90142-5. [DOI] [PubMed] [Google Scholar]
- 16.Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, Brown JB, Cherbas L, Davis CA, Dobin A, Li R, Lin W, Malone JH, Mattiuzzo NR, Miller D, Sturgill D, Tuch BB, Zaleski C, Zhang D, Blanchette M, Dudoit S, Eads B, Green RE, Hammonds A, Jiang L, Kapranov P, Langton L, Perrimon N, Sandler JE, Wan KH, Willingham A, Zhang Y, Zou Y, Andrews J, Bickel PJ, Brenner SE, Brent MR, Cherbas P, Gingeras TR, Hoskins RA, Kaufman TC, Oliver B, Celniker SE. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471 (7339):473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]