Abstract
Rapid and specific quantitation of a variety of proteins over a wide concentration range is highly desirable for bio-sensing at the point-of-care, in clinical laboratories, and in research settings. Our recently developed electrochemical proximity assay (ECPA) is a target-flexible, DNA-directed, direct-readout protein quantitation method with detection limits in the low femtomolar range, making it particularly amenable to point-of-care detection. However, consistent quantitation in more complex matrices is required at the point-of-care, and improvements in measurement speed are needed for clinical and research settings. Here, we address these concerns with a reusable ECPA, where a gentle regeneration of the surface DNA monolayer (used to capture the proximity complex) is achieved enzymatically through a novel combination of molecular biology and electrochemistry. Strategically placed uracils in the DNA sequence trigger selective cleavage of the backbone, releasing the assembled proximity complex. This allows repeated protein quantitation by square-wave voltammetry (SWV)—as quickly as 3 min between runs. The process can be repeated up to 19 times on a single electrode without loss of assay sensitivity, and currents are shown to be highly repeatable with similar calibrations using seven different electrodes. The utility of reusable ECPA is demonstrated through two important applications in complex matrices: 1) direct, quantitative monitoring of hormone secretion in real-time from as few as 5 murine pancreatic islets, and 2) standard addition experiments in unspiked serum for direct quantitation of insulin at clinically relevant levels. Results from both applications distinguish ECPA as an exceptional tool in protein quantitation.
Keywords: antibody-oligonucleotide conjugates, aptamer, biosensor, regenerate, hormone secretion, islet, deoxyuridine
INTRODUCTION
In the emerging era of personalized medicine, it is critically important to improve biomarker detection technologies. Medical diagnoses and treatments would be revolutionized by technology capable of rapid and specific quantitation of an arbitrary protein in real time, over a wide concentration range.1 These qualities are highly desirable for assays used at the point-of-care (POC), as well as in clinical and research laboratories. In general, advantages of POC devices include short turnaround times for acquiring critical data, low cost, and improved patient compliance with diagnosis and therapeutic regimens through in-home POC self-testing.2 For example, Rossi and Khan have shown that point-of-care testing can significantly reduce mortality in patients undergoing congenital heart surgery when used in combination with goal-directed therapy.3 However, current POC devices are limited in their reliance on single-use test cartridges and dried reagents, often leading to reduced performance metrics (LOD and dynamic range) and increased measurement variability compared to analogous clinical determinations.4 Reusable or reversible sensors would facilitate continuous POC device calibration, resulting in minimally invasive, higher accuracy assays. In this way, reusability is a critical area for the development of improved POC devices moving forward.
Historically, many strategies have been employed to regenerate sensor surfaces and make them reusable. In the case of affinity chromatography, for example, lowering the pH of the eluent buffer solution disrupts affinity binding, leading to the release of the target protein. The downside of this approach is that the capture antibody is often denatured, greatly limiting the number of measurement cycles. The introduction of reusability into capture-based surface assays—e.g. aptamer switches5,6 or electrochemical proximity assays (ECPA)7—can be confounded by the analyte’s irreversible binding to the sensor surface during measurement.
Electrochemical detection is employed in a wide range of biosensors because of its inherent signal stability, high sensitivity and ease of calibration compared to optical techniques. In addition, the instrumentation can easily be integrated with miniaturized POC devices8 and microfluidic or lab-on-a-chip (LOC) platforms.9-12 Electrochemical biosensing has seen renewed interest of late, based on the high performance of DNA-directed sensing by square-wave voltammetry (SWV)5,2,7,10,12-15, since analyte presence can be encoded into a DNA “signal” for readout. Micropatterned aptamer-modified electrodes have been used to quantify cellular secretions6,12, and antibodies have been detected in whole blood13 using this approach. In fact, some groups have achieved continuous, realtime monitoring of small-molecule therapeutics in blood serum10 and even in whole animals.15 By combining electrochemical DNA recognition with the proximity immunoassay concept16-18 we have developed a more generalizable protein assay system termed the electrochemical proximity assay (ECPA).7 Using direct-readout methodology, ECPA has been shown capable of detecting hormones at levels as low as 20 fM with high selectivity and wide dynamic range. Compared to other electrochemical protein quantitation methods6,12,13, target flexibility is a major advantage of ECPA, where the protein-to-probe binding is decoupled from the electrochemical signal generation using a DNA-based “circuit.” While the response of other methods such as aptamer folding approaches will vary depending on aptamer structure, ECPA allows accurate positioning of the electrochemical label, regardless of the type of probes (aptamers or antibodies) or the identity of the target protein. For ECPA to be of widespread practical utility, however, the analysis speed needs improvement, and the ability to make measurements in complex matrices must be demonstrated.
In this paper, we describe the development of a reusable ECPA capable of performing multiple iterative measurements in complex matrices. Reusable ECPA is a “signal-on” protein sensor that combines electrochemistry with molecular biological techniques, accomplishing a unique and “soft” surface regeneration chemistry. Two biological test beds are described: hormone quantitation in murine blood serum and secretion quantitation from endocrine tissue. With results that highlight the superior reusability, selectivity, and sensitivity of ECPA, the work presented herein represents a significant improvement in protein quantitation technology.
EXPERIMENTAL DESIGN
Reagents and Materials
All solutions were prepared with deionized, ultrafiltered water (Fisher Scientific). The following reagents were used as received: insulin antibodies (clones 3A6 and 8E2; Fitzgerald Industries), 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (99.5%), tris-(2-carboxyethyl) phosphine hydrochloride (TCEP), (Sigma-Aldrich, St. Louis, MO), bovine serum albumin (BSA, 98%; EMD Chemicals Inc.), human thrombin, immunoglobulin E (IgE), and insulin (Sigma Aldrich). HPLC-purified, methylene blue-conjugated DNA (MB-DNA) was purchased from Biosearch Technologies (Novato, CA). Oligonucleotides were obtained from Integrated DNA Technologies (IDT; Coralville, Iowa), with purity and yield confirmed by mass spectrometry and HPLC, respectively. All DNA sequences used in the experimental model and in ECPA measurements are given in Table S1. These sequences were designed and optimized computationally using the nucleic acid package web server (NUPACK).19
Pancreatic Islet Extraction and Sampling
Islets were isolated as described previously20-22 from live male C57BL/6 mice and maintained in RPMI medium (supplemented with 10% fetal bovine serum) with 11 mM glucose, at 37 °C under humidified 5% CO2 for 24 h prior to sampling. For secretion assays, between 5 and 15 islets were loaded into 0.5 mL tubes containing 300 ML imaging medium with basal glucose (3 mM) and maintained at 37 °C for 20 min. Reusable ECPA was carried out for insulin quantitation, using the cyclic methodology described below. During measurements, glucose was increased to 11 mM, and tubes were incubated at 37 °C for 1 h. Four male C57BL/6 mice contributed islets to this study, totaling ~150 islets.
Preparation of Antibody-Oligonucleotide Conjugates
Antibody-oligonucleotide conjugates were prepared as described previously7, by covalent attachment of AbArm1 to insulin antibody 3A6 and AbArm2 to insulin antibody 8E2, respectively. Conjugation and purification were accomplished using the Antibody-Oligonucleotide All-In-One Conjugation Kit (Solulink), according to the manufacturer’s instructions. Oligonucleotides were activated with sulfo-S-4FB, and quantities and qualities were confirmed using absorbance measurements (A260nm and A260nm/A360nm ratios) after oligonucleotide modification. 100 μg quantities of each antibody were activated with S-HyNic, then mixed and incubated with activated oligonucleotides at room temperature for 2 h. Conjugates were purified using the supplied magnetic affinity matrix, and final conjugate concentrations were determined via the BCA protein assay.
Preparation of Sensor Surface
All reusable ECPA sensors were fabricated using a Au working electrode (2 mm diameter, CH instruments, Inc., Austin, TX). Electrodes were polished using a previously reported protocol7, with minor alterations. Au electrodes were cleaned with piranha solution (H2SO4/H2O2; 3:1, V:V) for 20 min, polished with a slurry of 0.05 μm aluminum oxide in water (Buehler, Lake Bluff, IL) for 10 min, then sonicated in ethanol for 10 min. Freshly made piranha solution was then transferred onto the Au surface for 10 min, after which the electrode was rinsed with ddH2O. (Caution: piranha solution is dangerous to human health and should be used with extreme caution and handled only in small quantities.) The Au electrodes were further cleaned by a series of oxidative and reductive scans in 0.5 M H2SO4, as previously described.7 The cleaned Au electrodes were thoroughly washed with ddH2O then ethanol and dried under flowing nitrogen.
Typical experiments herein included preparation of 7 electrodes in parallel, as follows. Thiolated-DNA was reduced with TCEP immediately before immobilization on a clean electrode. 14 ML (2 μL per electrode) of 200 MM thiolated-DNA was mixed with 28 ML (4 μL per electrode) of 10 mM TCEP in a 1.5-mL microcentrifuge tube. This tube was incubated for 120 min at room temperature (~21 °C) in the dark for reduction of disulfide bonds in the thiolated-DNA. This solution was then diluted to a total volume of 1.4 mL in HEPES/NaClO4 buffer (10 mM HEPES and 0.5 M NaClO4, pH 7.0)5,7 to a final concentration of 2 MM thiolated DNA. The MB-DNA was also reduced in a similar way. To generate sufficient reduced MB-DNA for 20-30 measurements, 3 ML of 200 MM MB-DNA was mixed with 6 ML of 10 mM TCEP in a 1.5-mL microcentrifuge tube. This tube was incubated for 120 min at room temperature (~21 °C) in the dark, and then the solution was diluted to a total volume of 600 ML in HEPES/NaClO4 buffer, giving 1 MM MB-DNA. For immobilization, each freshly cleaned electrode was directly transferred to the reduced thiolated-DNA solution and incubated overnight (~12-16 h) at room temperature in the dark. Following incubation for self-assembled monolayer (SAM) formation, excess DNA was washed away through a room temperature ddH2O rinse (~15 s). For further protection against adsorption, the SAM-modified electrode was then immediately transferred to 3 mM 6-mercaptohexanol solution for 40 min at room temperature in the dark.23
Electrochemical Measurements
Electrochemical measurements were performed as described previously7 using a GAMRY Reference 600 electrochemistry workstation with a standard three-electrode configuration, comprised of a Ag|AgCl(s)|KCl(sat) reference electrode (Bioanalytical Systems), an in-house fabricated platinum gauze flag (0.77 cm2) counter electrode, and an Au working electrode (diameter = 2 mm). All potentials are reported relative to Ag|AgCl(s)|KCl(sat). Electrochemical measurements were performed in HEPES/NaClO4 buffer using SWV with a 50 mV amplitude square wave signal at a frequency of 60 Hz, over the range from 0.10 V to −0.45 V. For each measurement, a blank voltammetric trace was recorded upon immersion of the electrode into the sample (t ≈ 0 min), then a voltammetric scan was carried out at a particular point in time (e.g. t = 3 min). The blank scan was subtracted from the scan at time t, and the integrated signal was calculated as the peak area from −330 to −100 mV (centered around MB peak).
Model system
For modeling signal, the SAM-coated sensor was immersed in 10 nM DNA Loop and 15 nM MB-DNA in 500 μL of HEPES/NaClO4 buffer. For modeling background, the sensor was immersed in 15 nM MB-DNA in 500 μL of HEPES/NaClO4 buffer. Both signal and background currents were measured at the 3-min time point. After initial measurements, the electrode surface was regenerated using the enzymatic deoxyuridine-excision process.
Aptamer-Based Reusable ECPA System
Seven different Au working electrodes were used for measurement of each thrombin concentration. Thrombin of seven different concentrations (from 0 to 10 nM) was incubated for 15 min with 10 nM THRaptA and 15 nM THRaptB in HEPES/NaClO4 buffer prior to measurements. The thrombin/aptamer incubations were then added into the glass electrochemical cell. After measurement at the 3 min time point, each sensor was regenerated by immersing in 2 units of Uracil DNA Excision Mix (Epicentre, UEM04100) in 100 μl Uracil Excision Enzyme Buffer (50 mM Tris-HCl at pH 9.0, 20 mM (NH4)2SO4 and 10 mM EDTA) for 7 min at 37 °C. Enzyme solution and cleaved DNA were then removed via a room temperature ddH2O rinse (~15 s), and the electrodes were then dried under a stream of nitrogen gas. At this stage, the sensor with regenerated SAM was ready for the next round of probe loading and measurement. Regeneration and selectivity tests with other proteins (IgE, insulin, or BSA) were carried out under the same conditions.
Antibody-Based Reusable ECPA System
Antibody-oligonucleotide conjugates were prepared as described above; the conjugates are referred to as Ab1 (DNA-conjugated antibody 1: AbArm1–3A6) and Ab2 (DNA-conjugated antibody 2: AbArm2–8E2) in the discussion below. For sensor calibration against insulin and for insulin secretion quantitation from islets, seven gold working electrodes were used in a cyclic methodology, as described below.
Cyclic methodology for insulin quantitation in serum and secretory samples
The sensor was incubated with 50 nM Ab1 for 3 min by dropping 10 μL of the solution on the sensor surface, followed by a wash step of HEPES/NaClO4 buffer for ~15 s. The sensor was then transferred to the sample solution (300 μL secretory sample or serum). The secretory samples were maintained at 37 °C by a heating block, while serum samples were kept near 0 °C by placing on ice. After a 3 min incubation, the sensor was again washed with HEPES/NaClO4 buffer for ~15 s. Next, the electrode was incubated with 50 nM Ab2 for 3 min by dropping 10 μL of the solution onto the Au surface. For SWV scanning, the sensor was then transferred to 500 μL of HEPES/NaClO4 buffer consisting of 50 nM MB-DNA. Integrated signal was measured at the 3 min time point. For consistency, this protocol was not only used for measuring insulin responses in biological samples, but also for calibration standards. Figure 5 and the accompanying discussion provide further details on this cyclic methodology.
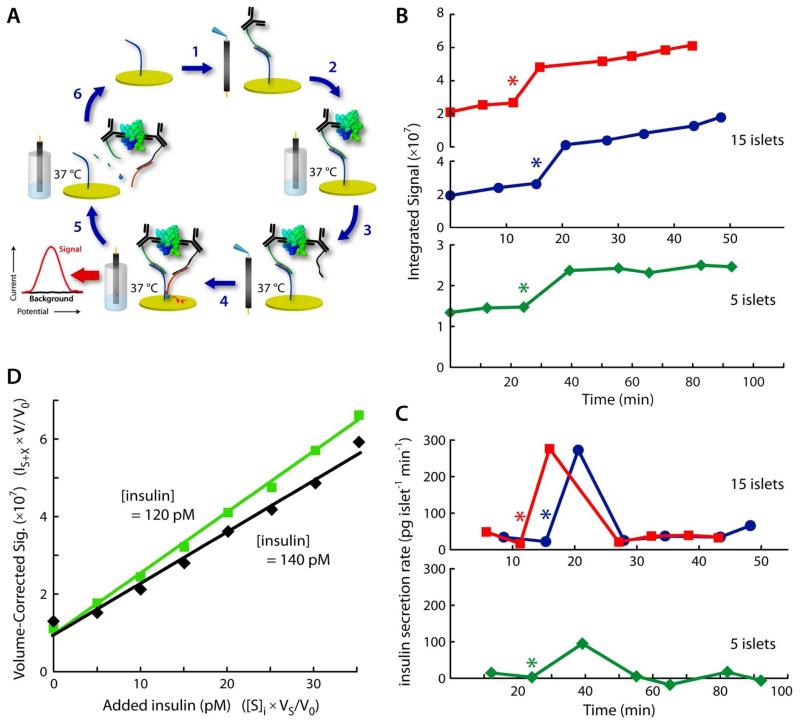
Figure 5.
Real-time measurements using the reusable ECPA cycle for quantitative insulin detection in biological samples. (A) Cyclic methodology for reusable ECPA (discussed in text). (B) Integrated signal traces during real-time monitoring of insulin secretion from as few as 5 living pancreatic islets. Islets were transitioned from basal glucose (3 mM) to stimulatory glucose (11 mM) at the points labeled with asterisks (*). (C) Insulin secretion rates were quantified via calibration. Islets responded to glucose increases with the expected spikes of insulin secretion; increases from tens to hundreds of pg islet−1 min−1 matched well with previous reports. (D) Insulin quantitation in unspiked serum from two different C57BL/6 mice was accomplished through standard addition methods made possible by reusable ECPA.
RESULTS AND DISCUSSION
Reusable ECPA
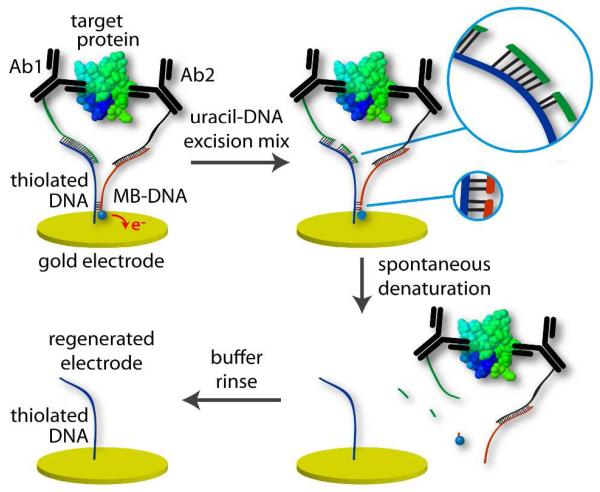
A disadvantage of the original ECPA design7 was the single-use nature of the sensors. Each electrode could only be used for a single measurement without having to clean and re-establish the SAM. To circumvent constant sensor cleaning, a reusable methodology was employed in this work. The principle of the reusable electrochemical proximity assay (reusable ECPA) is depicted in Figure 1, which highlights the enzymatic surface regeneration step borrowed from molecular biological techniques. The scheme begins with the fully assembled, five-part ECPA complex (upper left), comprised of thiolated DNA/Ab1/target protein/Ab2/MB-DNA. The complex forms in proportion to protein amount in solution, permitting electron transfer reactions between the electrode and the methylene blue in MB-DNA—the endpoint of the original ECPA assay.7 However, in this work, selected deoxythymidines (dT) were replaced by deoxyuridines (dU), which are susceptible to enzymatic cleavage by the Uracil DNA Excision Mix. dT moieties were replaced by dUs in sections of Ab1 (green strand) and MB-DNA (orange strand) that were complementary to dAs in the surface-bound thiolated DNA (blue strand), i.e. the portions that served to anchor the probes and signaling strands to the electrode surface. In this way, Ab1 and MB-DNA strands could be enzymatically broken into 3 and 2 shorter segments of DNA, respectively (Figure 1A, upper right). These strands, being no longer energetically stable as double-stranded DNA (dsDNA) at the electrode, spontaneously denatured (Figure 1A, lower right) to release probe/target complex from the electrode surface. Following a short ddH2O rinse, 4 parts of the 5-part complex (Ab1/target protein/Ab2/MB-DNA) were washed away from the electrode, leaving only the monolayer floor composed of thiolated-DNA and mercaptohexanol (not shown) at the sensor surface (Figure 1A, lower left). Through this gentle, enzymatic mechanism, ECPA electrodes were regenerated in a matter of minutes and were ready for probe loading and another measurement.
Figure 1.
Cycle for gentle, enzymatic regeneration of the SAM-coated electrode surface. After measurement (upper left), electrode was incubated in uracil-DNA excision mixture (upper right). Cleavage of the DNA backbone occurred at strategically placed uracils. Resulting shorter DNA sections spontaneously denatured (lower right) and were washed away in buffer (lower left), leaving the SAM of thiolated DNA that was ready for probe loading and another measurement.
DNA-Based Experimental model of reusable ECPA
We have previously shown DNA-based experimental models to offer a pragmatic solution to experimental optimization of proximity ligation assays17, proximity fluorescence assays24, and ECPA.7 The central theme of these models is the use of a ssDNA loop (“DNA Loop”) in lieu of the probe/target complex, eliminating the need for antibodies and target protein in initial optimization studies. For reusable ECPA, we used DNA model strands from our previous work7, except the DNA Loop and MB-DNA strands contained deoxyuridines (sequences in Table S-1). dUs facilitated enzymatic cleavage and removal of the Loop—which represented the assay probes.
In contrast to previous work7, a major goal of reusable ECPA was to reduce measurement time and allow rapid cycling through electrochemical measurements and surface regeneration steps. As such, a higher concentration of MB-DNA was adopted (50 nM) with a 3-min measurement time. It was hypothesized that a shorter number of base pairs between thiolated DNA and MB-DNA would prevent background formation at higher MB-DNA concentrations, while also allowing more rapid equilibration of the probe/target complex. This hypothesis was confirmed using a 5-base connection to thiolated DNA (G5 system, Figure S-1), which showed baseline response for background yet significant signal formation in the presence of the DNA Loop after only 3 min. As such, the G5 system was used throughout this work to allow rapid measurement and sensor regeneration.
Comparison of Sensor Regeneration Methods
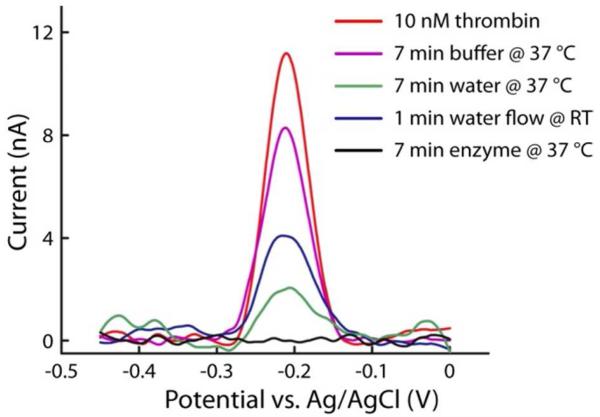
In similar electrochemical DNA and protein assays, others have employed regeneration methods such as water or buffer rinses5, denaturing rinses6,25,26, or even microfluidic systems for serial rinsing12, achieving between 5-10 consecutive measurements. Figure 2 shows the comparison of different regeneration methods for reusable ECPA. As rapid regeneration is more desirable, the methods were compared after no more than 7 min of treatment. The red trace in Figure 2 shows the signal current (~11 nA) after 3 min of equilibration with 10 nM thrombin, thrombin aptamer probes (10 nM THRaptA, 15 nM THRaptB), and 50 nM MB-DNA in buffer. The purple trace shows the measurement after a subsequent 7-min immersion in buffer at 37 °C, where current reduced to only 80% of the original value. A 7-min immersion in water showed improved removal of the probe/target complex, although ~20% of the original current remained (green trace). Others have shown sensors to be regenerated for a few cycles after ~30 s washes with room temperature deionized water25, yet this flowing water method only reduced the ECPA signal to ~35% of the original value (dark blue trace). As shown in Figure 2, the only method that provided a fully cleaned electrode—showing current at baseline levels after treatment—was the uracil-triggered enzymatic cleavage presented in this work (black trace). With this gentle enzymatic sensor cleaning approach, ECPA current was reduced to baseline levels in only 7 min.
Figure 2.
Comparison of electrode regeneration methodology. Washing electrodes with buffer or water after measurements show significant signal retention. Enzymatic regeneration was the only method which reduces the signal to baseline levels.
Aptamer-Based Reusable ECPA
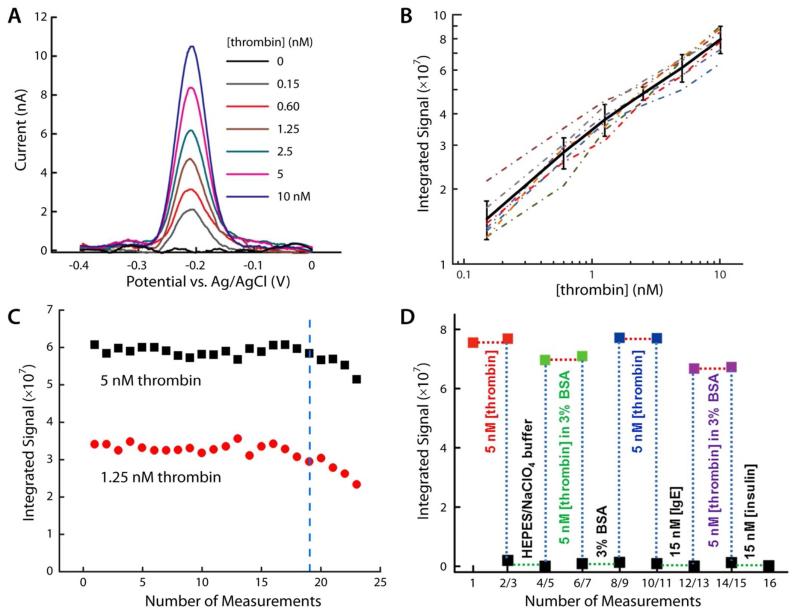
Initial testing of the re-usable ECPA concept was carried out with aptamer-based probes. This approach not only helps confirm the flexibility of ECPA, but it is also a more economical way to begin fine-tuning the methodology. As the aptamers are made from DNA, there is no requirement for probe-oligonucleotide conjugate synthesis and purification; aptamers are also less expensive probes. As in prior work7, the antibody-oligonucleotide conjugates are simply replaced by aptamers with equal arm sequences. Results from aptamer-based reusable ECPA are shown in Figure 3. Two thrombin-binding DNA aptamers (THRaptA, THRaptB) with distinct binding sites were applied as affinity probes. Using conditions optimized by the DNA-based experimental model, background levels were measured in the absence of human thrombin and remained at baseline current at the 3-min time point. Figure 3A shows the background with no thrombin (black trace) and typical MB oxidation peaks appearing at −210 mV in the presence of varying amounts of thrombin, all measured at the 3-min time point. Seven current signals at varying thrombin levels (0 to 10 nM) were measured by seven different sensors, prepared under the same conditions. With current increasing in proportion to thrombin concentration for all electrodes, the results confirmed the robustness and precision of the methodology.
Figure 3.
Aptamer based reusable ECPA for thrombin quantitation. (A) SWV scans for thrombin quantitation in the pM to nM range. Seven different sensors responded proportionally to thrombin, suggesting high method precision. (B) Thrombin calibration curves with seven different sensors (dotted lines), with the average trace (black line) shown; error bars represent standard deviations. The thrombin LOD was found to be 10 pM. (C) Two independent sensors were cycled through the reusable ECPA methodology during measurement of differing thrombin concentrations (red and black points). The gentle enzymatic surface regeneration permitted up to 19 uses without significant loss of signal. (D) High target specificity was confirmed through various challenges to the sensor, where only thrombin generated measurable responses.
Each sensor was then enzymatically regenerated seven times, such that signal currents of seven different concentrations of thrombin were measured by every sensor. In Figure 3B, integrated signal is reported versus thrombin concentration for each of the seven sensors; calibration curves are traced by dash-dotted lines, with each sensor having a different colored line. The solid black curve with error bars shows the averages and standard deviations from the seven sensors, and the linearly extrapolated limit of detection (LOD) for thrombin was calculated to be 10 pM (5 fmol in 500 μL). Remarkably, this LOD using averaged signals from seven different electrodes was slightly lower than previously achieved with a single electrode.7 This result demonstrates high confidence in the sensor-to-sensor reproducibility of reusable ECPA, since seven completely different electrodes—with separately prepared DNA monolayers—were able to deliver a LOD in the low picomolar range.
To further interrogate reusability, two sensors with two different concentrations of thrombin (1.25 and 5.00 nM) were iteratively taken through probe loading, measurement, and enzymatic regeneration; the integrated signal from these experiments is reported in Figure 3C. Significant deviation from the average value occurred at the 21st repeated measurement for the 5.00 nM thrombin sample and at the 20th repeated measurement for the 1.25 nM thrombin sample. From this data, it was concluded that at least 19 consecutive measurements could be made without loss of signal (blue dotted line) via reusable ECPA.
To demonstrate specificity, a single sensor was challenged with nonspecific proteins including human IgE, insulin, and BSA. The sensor was taken through a cascade of different treatments in duplicate, alternating between thrombin and a non-specific treatment such as buffer, 3% BSA, 15 nM IgE, or 15 nM insulin (Figure 3D). Essentially no response was observed in the presence of any treatments other than the thrombin-containing solutions; even with 3-fold lower thrombin (5 nM), the signal was 110- to 160-fold larger than that of insulin or IgE (15 nM). Recovery of signal from 5 nM thrombin in 3% BSA was 90%. These data demonstrated high selectivity toward thrombin and high recoveries in complex matrices through 16 consecutive measurements. Such a result is encouraging for future application of reusable ECPA to continuous measurement of biological samples and for quantitation in clinical laboratories or in point-of-care settings.
Antibody-Based Reusable ECPA
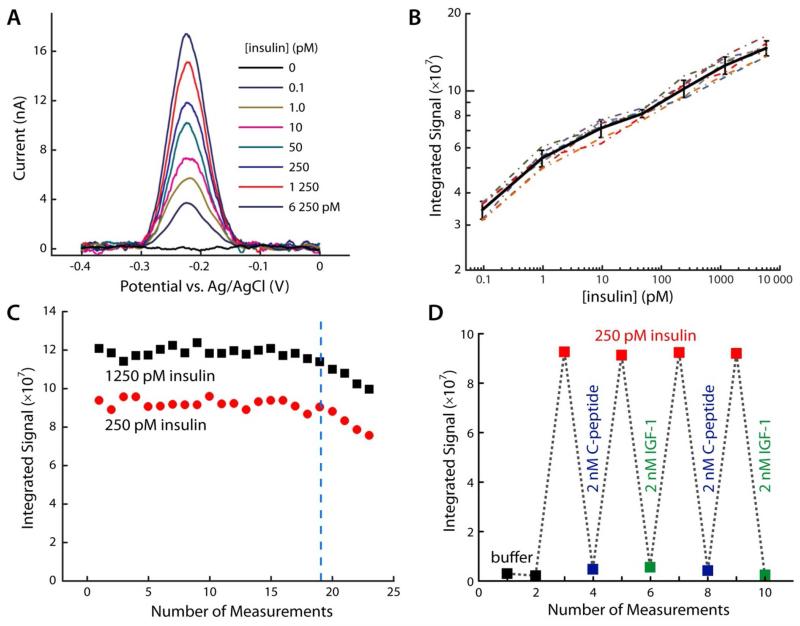
Because reusable ECPA decouples target/probe binding from electrochemical readout, the technique is flexible enough to utilize antibody-based probes as well. This flexibility is useful, considering that aptamer pairs exist for only a few select proteins. Since standard ECPA with antibody-based probes was already proven functional7, conditions for this work were modified toward interrogation of biological systems. A cyclic protocol was adopted to prevent exposure of cells to antibody-based probes (discussed below), and the temperatures of assay solutions were held constant at 37 °C. Figure 4A shows the background with no insulin (black trace) and typical MB oxidation peaks in the presence of varying amounts of insulin measured at the 3-min time point, using seven different electrodes. The system was then calibrated versus insulin concentration, with responses of seven insulin concentrations recorded from seven sensors (Figure 4B). Using direct electrochemical readout at 37 °C, reusable ECPA was capable of easily detecting insulin levels as low as 100 fM, with a dynamic range extending to 6.25 nM. The solid black curve with error bars shows the averages and standard deviations from the seven sensors, and the linearly extrapolated LOD for insulin was found to be 10 fM (5 amol in 500 μL). This LOD using averaged signals from seven different electrodes—incubated with sample at 37 °C—was lower than that previously achieved with a single electrode.7 The result again bodes well for the repeatability of the technique. Furthermore, the dynamic range achieved here (~6 × 105) is 6-fold wider than shown in the previous work.
Figure 4.
Antibody based ECPA for insulin quantitation. (A) SWV scans for insulin quantitation in the fM to nM range. Seven different sensors responded proportionally, suggesting high method precision. (B) Insulin calibration curves with seven different sensors (dotted lines), with the average trace (black line) shown; error bars represent standard deviations. The insulin LOD was found to be 10 fM. (C) Two independent sensors were cycled through the reusable methodology during measurement of varying insulin levels (red and black points). Again, up to 19 uses were permitted without significant loss of signal. (D) High target specificity was confirmed by challenging the sensor with a protein of similar structure (IGF-1) and a co-secreted hormone (C-peptide). Only insulin generated measurable responses.
Reusability of the antibody version was assayed with two different concentrations of insulin (250 and 1250 pM), and integrated signal is reported in Figure 4C. Similar to the aptamer system, the method was capable of at least 19 repeated measurements without significant signal loss. It is interesting that in both aptamer and antibody versions of the assay, the surface degrades measurably after 19 repeated uses in a seemingly probe-independent manner. A systematic investigation of the degradation mechanism was not undertaken in this study, but the effect after 19 uses is likely an aggregate of well-known SAM degradation factors. Plausible explanations include the use of multiple voltammetric scans, repeated enzymatic treatments, repeated buffer washes, and the persistence of reaction side products on the sensor surface.
Finally, selectivity was confirmed with insulin-like growth factor 1 (IGF-1), which has similar molecular structure to insulin, and with C-peptide, which is co-secreted with insulin into the bloodstream. As expected, the dual antibody sensor showed minimal response to 8-fold higher concentrations of either IGF-1 or C-peptide (2 nM) compared to 250 pM insulin (Figure 4D).
Measurement of Hormone Secretion from Endocrine Tissue
One key test of the robustness of reusable ECPA is to apply the technique to quantitation of hormones secreted by living cells over time, as these measurements take place in complex milieu amenable to cellular culture and imaging. Here, the technique is applied to monitoring of insulin secretion from living murine pancreatic islets during glucose-stimulated insulin secretion. Since islets cannot be exposed to freely diffusing antibody-oligo conjugates, the cyclic methodology was designed specifically for this application, with a schematic shown as steps 1-6 in Figure 5A. In this cycle, Ab1 solution was first dropped onto the gold surface of the sensor (step 1) then incubated for 3 min. After washing with buffer, the sensor was lowered into cellular imaging media containing the living islets at varying glucose levels at 37 °C (step 2). Secreted insulin was bound by Ab1, keeping it at the sensor surface. Following a second buffer wash, Ab2 solution was pipetted onto the sensor surface and incubated for 3 min (step 3), then the sensor was immersed into MB-DNA solution. Here, signal current was measured at the 3 min time point (step 4). Following each measurement, the sensor was regenerated enzymatically using the Uracil-DNA Excision Mix (step 6), readying the system for another measurement.
The cyclic methodology (Figure 5A) was employed for hormone secretion quantitation and was applied during calibration to permit quantitative monitoring. To simulate a fasting-to-feeding transition, islets were conditioned in 3 mM glucose then exposed to 11 mM glucose.22,27 Figure 5B shows insulin secretion quantitation by reusable ECPA, where islets from two different mice were transitioned from 3 to 11 mM glucose prior to the fourth measurement (* at 10-15 min time point). Groups of only 5 to 15 islets registered a sharp increase in secreted insulin following the increased glucose. Using the average calibration curve shown in Figure 4B, insulin secretion rates were quantified. Islets at low and high glucose secreted tens to hundreds of pg islet−1 min−1, respectively (Figure 5C). These readings matched well with previous reports22,27,28, providing validation for reusable ECPA as a novel tool in quantitative biology. To our knowledge, the technique is unique in its capability for direct yet selective electrochemical hormone secretion quantitation from small amounts of living tissue in real time.
Measurement of Hormone Levels in Murine Blood Serum
Accurate analytical calibrations can be confounded in complex matrices such as serum, and one of the most effective countermeasures is the use of standard addition methods.29 A unique advantage provided by direct and rapid quantitation with reusable ECPA is the opportunity to implement standard addition, precluding the need for separate calibration curves in complex backgrounds. With a slight modification to the cyclic methodology prescribed above (sample loading on ice instead of at 37 °C), insulin standards were added to 20-fold diluted mouse serum in increments of 5 pM. This approach allowed accurate quantitation of insulin in the native, unspiked serum of two different C57BL/6J mice within a matter of minutes (Figure 5D). Results of these assays (120 and 140 pM) fell precisely within the expected clinical range of insulin in wild-type murine blood (114 ± 31 pM)30, providing another solid validation of our methodology for quantitative biological studies. In particular, these results suggest that reusable ECPA could be useful in clinical or point-of-care settings, to assay a wide variety of protein targets with high sensitivity and selectivity. Although eight separate measurements per sample were conducted here, standard addition curves with fewer data points could be readily implemented to provide high measurement confidence in complex matrices such as serum.
CONCLUSIONS
As noted in a recent review by Plaxco and Soh,1 a universal approach for quantitation of arbitrary molecular analytes could revolutionize healthcare; the authors further suggest that electrochemical, switch-based biosensors could be the answer. While this has now been demonstrated for small molecule monitoring,15 continuous and sensitive protein detection with a generalizable assay has yet to be developed. Our reusable ECPA incorporates many of these aspects for protein quantitation. High selectivity and sensitivity are given by dual probe recognition (aptamers or antibodies), arbitrary protein detection is afforded by decoupling probe recognition from DNA-based readout, and reusability allows real-time detection in biological matrices. As such, the presented work represents a major step toward the desired system of continuous protein quantitation. For example, our physiologically validated electrochemical insulin monitoring provides an attractive framework for assembly of a miniaturized “closed-loop” artificial pancreas for diabetic patients31,32; insulin ECPA could be partnered with the well-established commercial methods for electrochemical glucose monitoring.8,31
Of course, barriers to successful in vivo ECPA still exist. Improvements are needed, such as built-in assay reversibility and methods for preloading of confined surface probes. None-theless, the work herein represents a significant improvement in protein detection technology. When considering the generalizability of the approach to target any protein with antibody pairs, reusable ECPA is likely to be a valuable technique for biologists, chemists, clinicians, and others.
Supplementary Material
ACKNOWLEDGMENT
Funding for this work was provided by the National Science Foundation, CBET-1067779 (C. J. E.); by the National Institutes of Health, R01 DK093810 (C. J. E.); by the Auburn University Intramural Grants Program (C. J. E.and C. S.); and by the Department of Chemistry and Biochemistry at Auburn University.
Footnotes
Notes
The authors declare no competing financial interest. The ECPA methodology presented in this article was included in a U.S. Patent Application (No. 13/650,303) filed on October 12, 2012.
ASSOCIATED CONTENT
Supporting Information Available. Additional information is included in the Supporting Information document, including DNA sequences and results from ECPA experimental model system. This information is available free of charge via the internet at http://pubs.acs.org.
REFERENCES
- (1).Plaxco KW, Soh HT. Trends Biotechnol. 2011;29:1–5. doi: 10.1016/j.tibtech.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).von Lode P. Clin. Biochem. 2005;38:591–606. doi: 10.1016/j.clinbiochem.2005.03.008. [DOI] [PubMed] [Google Scholar]
- (3).Rossi AF, Khan D. Clin. Biochem. 2004;37:456–461. doi: 10.1016/j.clinbiochem.2004.04.004. [DOI] [PubMed] [Google Scholar]
- (4).Gubala V, Harris LF, Ricco AJ, Tan MX, Williams DE. Anal. Chem. 2012;84:487–515. doi: 10.1021/ac2030199. [DOI] [PubMed] [Google Scholar]
- (5).Kang D, Zuo X, Yang R, Xia F, Plaxco KW, White RJ. Anal. Chem. 2009;81:9109–9113. doi: 10.1021/ac901811n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Liu Y, Tuleouva N, Ramanculov E, Revzin A. Anal. Chem. 2010;82:8131–8136. doi: 10.1021/ac101409t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Hu J, Wang T, Kim J, Shannon C, Easley CJ. J. Am. Chem. Soc. 2012;134:7066–7072. doi: 10.1021/ja3000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Heller A, Feldman B. Chem. Rev. 2008;108:2482–2505. doi: 10.1021/cr068069y. [DOI] [PubMed] [Google Scholar]
- (9).Fanguy JC, Henry CS. Electrophoresis. 2002;23:767–773. doi: 10.1002/1522-2683(200203)23:5<767::AID-ELPS767>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- (10).Swensen JS, Xiao Y, Ferguson BS, Lubin AA, Lai RY, Heeger AJ, Plaxco KW, Soh T. J. Am. Chem. Soc. 2009;131:4262–4266. doi: 10.1021/ja806531z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Selimovic A, Martin RS. Electrophoresis. 2013;34:2092–2100. doi: 10.1002/elps.201300163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Zhou Q, Kwa T, Gao YD, Liu Y, Rahimian A, Revzin A. Lab Chip. 2014;14:276–279. doi: 10.1039/c3lc50953b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Vallée-Bélisle A, Ricci F, Uzawa T, Xia F, Plaxco KW. J. Am. Chem. Soc. 2012;134:15197–15200. doi: 10.1021/ja305720w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Huang KC, White RJ. J. Am. Chem. Soc. 2013;135:12808–12817. doi: 10.1021/ja4060788. [DOI] [PubMed] [Google Scholar]
- (15).Ferguson BS, Hoggarth DA, Maliniak D, Ploense K, White RJ, Woodward N, Hsieh K, Bonham AJ, Eisenstein M, Kippin TE, Plaxco KW, Soh HT. Sci. Transl. Med. 2013;5:213ra165. doi: 10.1126/scitranslmed.3007095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Gullberg M, Gústafsdóttir SM, Schallmeiner E, Jarvius J, Bjarnegård M, Betsholtz C, Landegren U, Fredriksson S. Proc. Natl. Acad. Sci. USA. 2004;101:8420–8424. doi: 10.1073/pnas.0400552101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Kim J, Hu J, Sollie RS, Easley CJ. Anal. Chem. 2010;82:6976–6982. doi: 10.1021/ac101762m. [DOI] [PubMed] [Google Scholar]
- (18).Zhang H, Li F, Li XF, Le XC. Methods. 2013;64:322–330. doi: 10.1016/j.ymeth.2013.10.005. [DOI] [PubMed] [Google Scholar]
- (19).Zadeh JN, Steenberg CD, Bois JS, Wolfe BR, Pierce MB, Khan AR, Dirks RM, Pierce NA. J. Comput. Chem. 2011;32:170–173. doi: 10.1002/jcc.21596. [DOI] [PubMed] [Google Scholar]
- (20).Scharp DW, Kemp CB, Knight MJ, Ballinger WF, Lacy PE. Transplantation. 1973;16:686–689. doi: 10.1097/00007890-197312000-00028. [DOI] [PubMed] [Google Scholar]
- (21).Stefan Y, Meda P, Neufeld M, Orci L. J. Clin. Invest. 1987;80:175–183. doi: 10.1172/JCI113045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Godwin LA, Pilkerton ME, Deal KS, Wanders D, Judd RL, Easley CJ. Anal. Chem. 2011;83:7166–7172. doi: 10.1021/ac201598b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Xiao Y, Lai RY, Plaxco KW. Nat. Protocols. 2007;2:2875–2880. doi: 10.1038/nprot.2007.413. [DOI] [PubMed] [Google Scholar]
- (24).DeJournette C, Kim J, Medlen H, Li X, Vincent L, Easley CJ. Anal. Chem. 2013;85:10556–10564. doi: 10.1021/ac4026048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Zuo X, Xiao Y, Plaxco KW. J. Am. Chem. Soc. 2009;131:6944–6945. doi: 10.1021/ja901315w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Liu Y, Kwa T, Revzin A. Biomaterials. 2012;33:7347–7355. doi: 10.1016/j.biomaterials.2012.06.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Dishinger J, Reid K, Kennedy R. Anal. Chem. 2009;81:3119–3127. doi: 10.1021/ac900109t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Lomasney A, Yi L, Roper M. Anal. Chem. 2013;85:7919–7925. doi: 10.1021/ac401625g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Harris DC. Quantitative Chemical Analysis. 8th ed. W. H. Freeman and Co.; New York: 2010. [Google Scholar]
- (30).Surwit RS, Kuhn CM, Cochrane C, McCubbin JA, Feinglos MN. Diabetes. 1988;37:1163–1167. doi: 10.2337/diab.37.9.1163. [DOI] [PubMed] [Google Scholar]
- (31).Wang J. Chem. Rev. 2008;108:814–825. doi: 10.1021/cr068123a. [DOI] [PubMed] [Google Scholar]
- (32).Brown L, Edelman ER. Sci. Transl. Med. 2010;2:27ps18. doi: 10.1126/scitranslmed.3001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.