Abstract
Lysine-specific demethylase 1 (LSD1) is an epigenetic enzyme that oxidatively cleaves methyl groups from monomethyl and dimethyl Lys4 of histone H3 (H3K4Me1, H3K4Me2) and can contribute to gene silencing. This study describes the design and synthesis of analogues of a monoamine oxidase antidepressant, phenelzine, and their LSD1 inhibitory properties. A novel phenelzine analogue (bizine) containing a phenyl-butyrylamide appendage was shown to be a potent LSD1 inhibitor in vitro and was selective versus monoamine oxidases A/B and the LSD1 homologue, LSD2. Bizine was found to be effective at modulating bulk histone methylation in cancer cells, and ChIP-seq experiments revealed a statistically significant overlap in the H3K4 methylation pattern of genes affected by bizine and those altered in LSD1−/− cells. Treatment of two cancer cell lines, LNCaP and H460, with bizine conferred a reduction in proliferation rate, and bizine showed additive to synergistic effects on cell growth when used in combination with two out of five HDAC inhibitors tested. Moreover, neurons exposed to oxidative stress were protected by the presence of bizine, suggesting potential applications in neurodegenerative disease.
Reversible histone lysine methylation is a major mechanism for regulating chromatin dynamics and gene expression. Lysine-specific demethylase 1 (LSD1), the first histone demethylase identified, is responsible for oxidatively cleaving one or two methyl groups from Lys4 of histone H3 (H3K4).1– 7 In this way, LSD1 is thought to play a role in gene silencing, since methylation of H3K4 in promoter regions is a well-established chromatin mark linked to transcriptional activation.8,9 Since its discovery, LSD1 histone demethylase activity has been investigated as a pharmacologic target for cancer and other diseases. It has been found that LSD1 levels are often elevated in various cancers, including prostate, non-small cell lung, and ER-negative breast cancer.10–12 Moreover, a variety of tumor suppressors that have been shown to be silenced in cancer by epigenetic mechanisms could theoretically be reactivated by LSD1 blockers,13–16 as has been achieved with histone deacetylase and DNA methyltransferase inhibitors.17
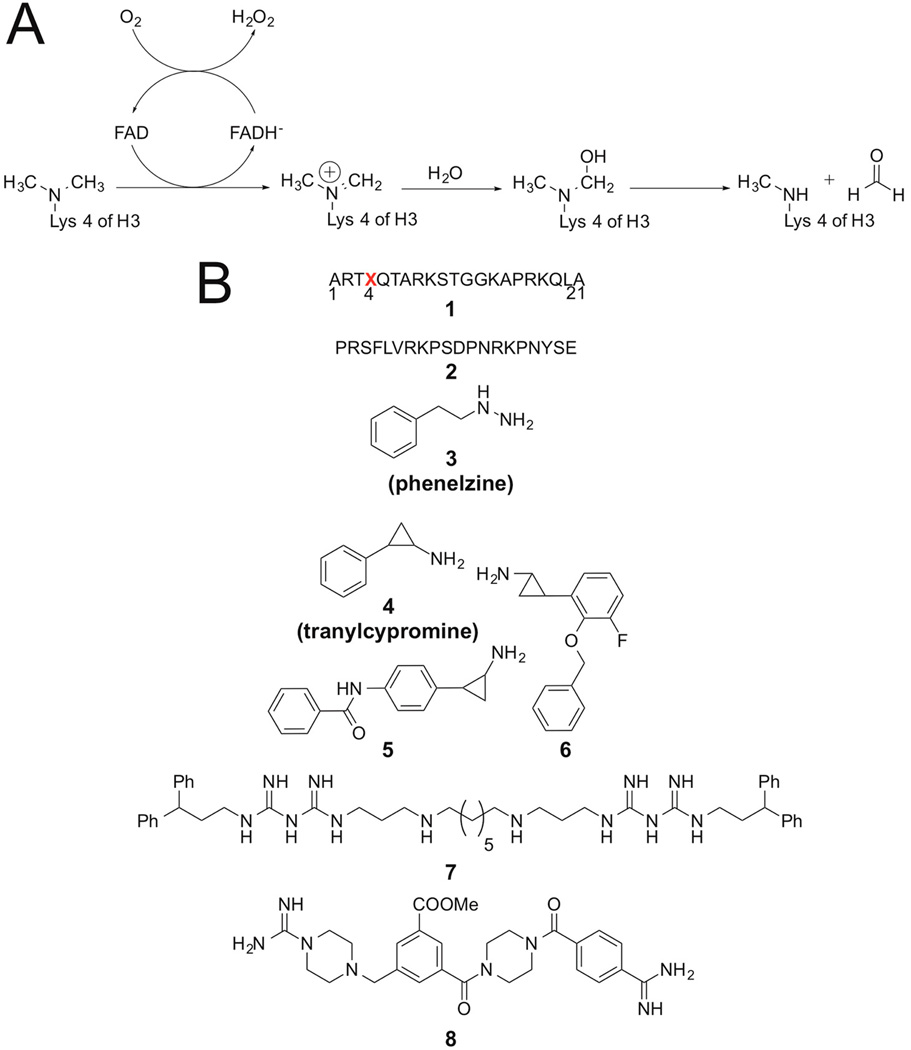
LSD1 is a 90 kDa flavin-bound enzyme that belongs to the amine oxidase protein superfamily, which uses molecular oxygen as a cosubstrate and generates hydrogen peroxide and formaldehyde as byproducts (Figure 1A).1,7,18,19 Based on its enzymatic mechanism, LSD1 cannot demethylate trimethylated H3 Lys4 (H3K4Me3), but members of the iron-dependent Jmj histone demethylases are known to serve this function.1,20 In addition to the C-terminal amine oxidase catalytic domain, LSD1 also contains an N-terminal SWIRM domain and a 105 aa Tower domain insert, which is located in the amine oxidase domain that can bind CoREST. In cells, LSD1 is often found in CoREST complexes that include HDAC1/2.4,21–25 The LSD1 homologue, LSD2, also catalyzes demethylation of H3K4Mel and H3K4Me2 but lacks the CoREST binding Tower domain insert and exhibits significant sequence and local structure differences compared to LSD1.26,27 Mechanistically and structurally, LSD1 is also related to the flavin-dependent monoamine oxidases (MAO A/B), as well as polyamine oxidase enzymes.15,25,28
Figure 1.
(A) LSD1 demethylation mechanism. (B) LSD1 inhibitor structures published previously: (1) Histone H3-21mer peptides with various modified lysine residues, X; (2) N-terminal SNAIL1 20-mer peptide; (3) phenelzine; (4) tranylcypromine; (5, 6) tranylcypromine analogues; (7) polyamine analogue; (8) guanidinium-containing compound.
Several prior LSD1 demethylase inhibitors have been reported including peptides (1, 2), MAOIs and derivatives thereof (3–6), polyamines (7), and guanidine containing compounds (8) (Figure 1B).2,29–40’ One strategy that has shown promise has been the development of tranylcypromine analogues.37,38 Tranylcypromine is a classical MAO inhibitor and mechanism-based inactivator involving an oxidative cyclopropylamine ring-opening reaction, used for the treatment of clinical depression, and is weakly potent as an LSD1 mechanism-based inactivator (Ki(inact) = 0.5 mM, kinact = 0.67 min−1).41–43 However, it has been shown that tranylcypromine can be modified by aryl attachment to produce more selective LSD1 inhibitors with enhanced potency.32,33,44–48 Nevertheless, no LSD1 inhibitor has yet been evaluated in clinical trials.
Prior work from our group showed that the antidepressant MAO inhibitor phenelzine is considerably more potent than tranylcypromine as an LSD1 inhibitor.29 Like tranylcypromine, phenelzine is an LSD1 mechanism-based inactivator, but in this case the key initiating step involves hydrazine rather than cyclopropylamine oxidation. In this manuscript, we generated a series of phenelzine analogues informed by the general concept derived from tranylcypromine studies that linkage of phenelzine to an aromatic functionality might improve its LSD1 potency.32,33 Below we disclose structure–activity relationships with various phenelzine analogues, ultimately developing a compound with considerable potency and selectivity for LSD1 versus MAO and LSD2 enzymes. This phenelzine analogue bizine was further examined in cell-based assays assessing bulk as well as gene-specific histone methylation changes, antiproliferative activity in several cancer cell lines, and neuroprotection in response to oxidative stress. Below we describe these findings and place their significance in the context of prior LSD1 studies.
RESULTS AND DISCUSSION
Phenelzine Analogue LSD1 Inhibitors
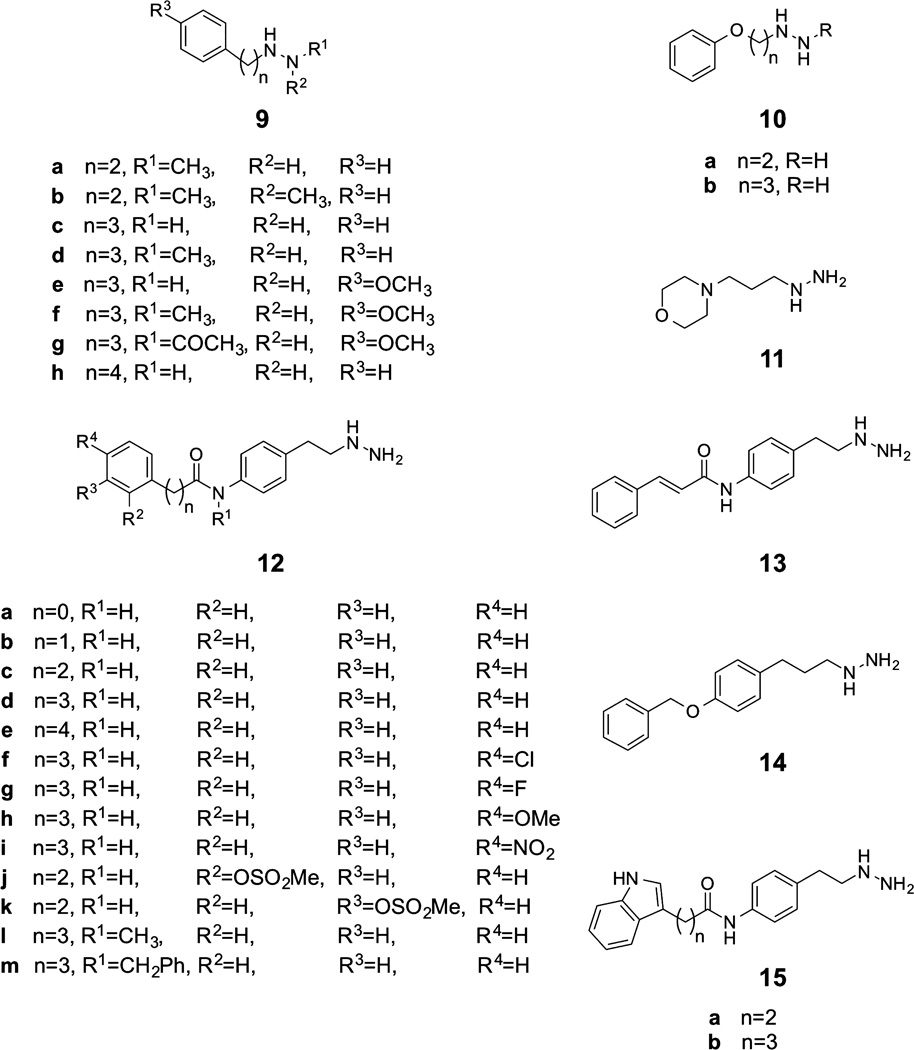
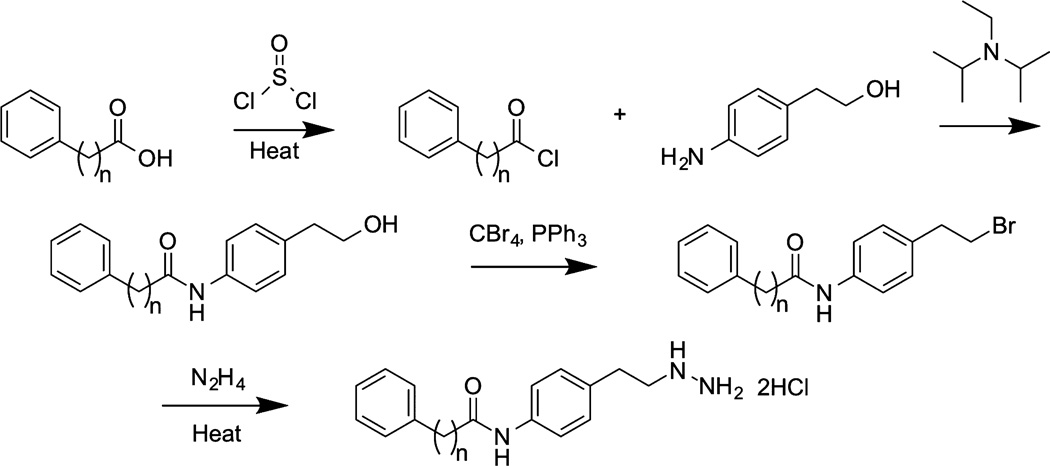
On the basis of prior findings that phenelzine was a moderately potent, mechanism-based inactivator of LSD1, we synthesized a series of phenelzine analogues exploring hydrazine modifications, variations in alkyl chain length and rigidity, phenyl replacement, and phenyl ring substitution (compounds 9–15, Figure 2)29. Synthetic routes generally involved late stage hydrazine introduction by converting a terminal alkyl hydroxy group to either the corresponding bromide or mesylate followed by hydrazine displacement reactions as exemplified in Figure 3 (additional detailed routes are shown in Supplementary Figures 1–4). Compounds were assayed for their ability to inhibit recombinantly purified GST-LSD1 using a dimethyl-Lys4 histone H3-21mer peptide substrate by monitoring peroxide formation via a colorimetric peroxidase assay.49 These results (Table l) showed that adjusting the alkyl chain length (9c, 9h, 10a,b) could lead to modest increases or decreases in LSD1 inhibitory potency compared with that of phenelzine (Ki(inact) = 5.6 µM; kinact = 0.35 min−1) (Supplementary Figure 5), whereas methyl or acetyl substitutions on the hydrazine (9a,b,d,f,g) negated LSD1 inhibitory action. Additionally, morpholine replacement of the phenyl ring (11) was not compatible with LSD1 inhibition. Furthermore, incorporation of a methoxy substituent at the 4-position of the phenyl ring of phenelzine made little difference (9e). Important LSD1 inhibitory potency enhancements were achieved by linking aryl groups through various tethers to the phenelzine core (12–15). This trend was loosely related to the previously reported results with tranylcypromine analogue 5.32
Figure 2.
Phenelzine analogues tested as LSD1 inhibitors.
Figure 3.
General synthesis of novel phenelzine analogues.
Table 1.
Kinetics of Phenelzine Analogue LSD1 Inhibitors
| inhibitor | Ki(inact) (µM) | kinact (min−1) | kinact/Ki(inact) (µM–1 min−1) | IC50 (µM) |
|---|---|---|---|---|
| phenelzine | 5.6 ± 1.3 | 0.35 ± 0.056 | 0.063 ± 0.018 | N/A |
| 9a | N/A | N/A | N/A | 85.00 |
| 9b | N/A | N/A | N/A | >100.0 |
| 9c | 5.0 ± 1.1 | 0.32 ± 0.010 | 0.064 ± 0.014 | N/A |
| 9d | N/A | N/A | N/A | 46.74 |
| 9e | 8.0 ± 3.5 | 0.15 ± 0.023 | 0.019 ± 0.0087 | N/A |
| 9f | N/A | N/A | N/A | >100.0 |
| 9g | N/A | N/A | N/A | N/A |
| 9h | 22 ± 3.0 | 0.12 ± 0.01 | 0.0055 ± 0.00087 | N/A |
| 10a | 44 ± 9.7 | 0.15 ± 0.010 | 0.0034 ± 0.00079 | N/A |
| 10b | 12 ± 2.1 | 0.22 ± 0.020 | 0.018 ± 0.0036 | N/A |
| 11 | N/A | N/A | N/A | >100.0 |
| 12a | 0.28 ±0.11 | 0.19 ± 0.036 | 0.70 ± 0.31 | N/A |
| 12b | 0.37 ± 0.033 | 0.20 ± 0.0087 | 0.54 ± 0.054 | N/A |
| 12c | 0.26 ± 0.058 | 0.24 ± 0.022 | 0.92 ± 0.22 | N/A |
| 12d | 0.059 ± 0.021 | 0.15 ± 0.017 | 2.5 ± 0.96 | N/A |
| 12e | 0.26 ±0.11 | 0.22 ± 0.038 | 0.86 ± 0.39 | N/A |
| 12f | 0.156 ± 0.047 | 0.17 ± 0.018 | 1.1 ± 0.35 | N/A |
| 12g | 0.138 ± 0.0.48 | 0.17 ± 0.020 | 1.2 ± 0.44 | N/A |
| 12h | 0.207 ± 0.089 | 0.26 ± 0.042 | 1.2 ± 0.57 | N/A |
| 12i | 0.282 ± 0.076 | 0.21 ± 0.024 | 0.74 ± 0.22 | N/A |
| 12j | 0.204 ± 0.098 | 0.18 ± 0.034 | 0.88 ± 0.46 | N/A |
| 12k | 0.223 ± 0.064 | 0.17 ± 0.020 | 0.76 ± 0.24 | N/A |
| 121 | 2.0 ± 0.73 | 0.24 ± 0.033 | 0.12 ± 0.045 | N/A |
| 12m | 1.6 ± 0.49 | 0.22 ± 0.025 | 0.14 ± 0.044 | N/A |
| 13 | 0.10 ± 0.039 | 0.17 ± 0.21 | 1.7 ± 0.68 | N/A |
| 14 | 0.90 ± 0.45 | 0.18 ± 0.038 | 0.20 ± 0.11 | N/A |
| 15a | 0.21 ± 0.076 | 0.21 ± 0.030 | 1.0 ± 0.41 | N/A |
| 15b | 0.10 ± 0.035 | 0.17 ± 0.019 | 1.7 ± 0.60 | N/A |
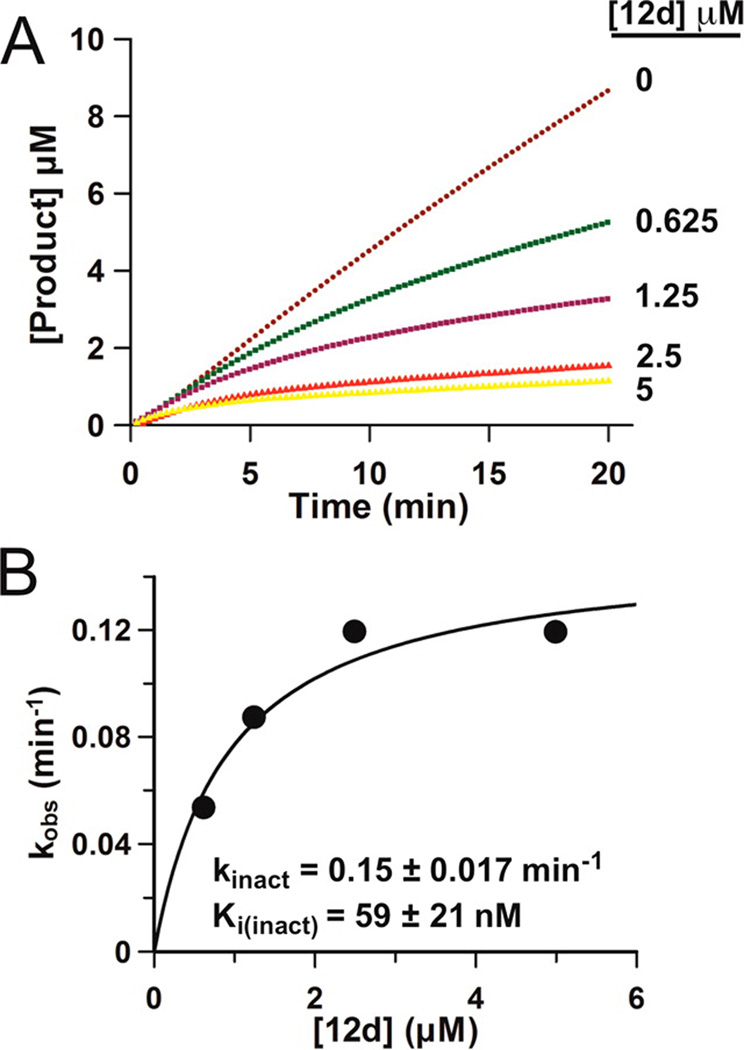
Compounds 12a–e showed that amino-phenelzine fused to phenyl-alkanoic acids via an amide spacer were improved LSD1 inhibitors compared to phenelzine itself. Of this set, compound 12d containing the propanyl spacer was the most potent LSD1 inhibitor with a Ki(inact) of 59 nM and a kinact of 0.15 min−1 (Figure 4A,B). This Ki(inact) for 12d compares favorably to one of the most potent tranylcypromine analogues (Ki(inact) = 0.61 µM) reported in the literature.33 Alternatives to the alkanoic spacers in 12 including an alkenoic acid spacer (13) and an alkyl ether spacer (14) led to reduced LSD1 inhibitory potency. However, replacing the ethanyl tether with a trans-ethenyl group resulted in improved inhibitor potency as can be seen by comparing 12c with 13. Terminal aryl substitutions in the context of the ethanyl and the propanyl spacers represented in 12f–k generally had similar LSD1 potency as that of 12d, suggesting that substitutions at this position are well tolerated. Of note, N-substitution of the amide linker attachment present in 12l,m greatly attenuated LSD1 inhibition relative to 12d, potentially highlighting the importance of the amide NH group in hydrogen bonding to the LSD1 active site. Interestingly, replacement of the terminal phenyl group in 12c,d with an indole group to generate 15a,b largely preserved LSD1 inhibitory potency.
Figure 4.
Inhibition of LSD1 by compound 12d (bizine). (A) Steady-state progress curve of LSD1 inactivation by compound 12d (bizine) ranging from 0 to S µM. (B) kobs values obtained from steady-state data plotted against inhibitor concentration to determine kinact and Ki(inact) values.
To confirm the LSD1 inhibition peroxidase assay results obtained with 12d, we turned to a recently developed isotope-based mass spectrometric assay, MassSQUIRM, to directly and quantitatively assess 12d effects on Lys4-methylation50. This assay is conducted for an extended time period utilizing a high LSD1 concentration, with conditions where LSD1-catalyzed demethylation of the H3-21-K4Me2 substrate nears completion, resulting in extensive conversion of the substrate to mono-and unmethylated H3-21. As reported previously, greater than 10 mM phenelzine is needed to extinguish LSD1 activity under MassSQUIRM conditions50. Thus, we compared 50 µM each of phenelzine and analogue 12d in an identical LSD1 inhibition MassSQUIRM assay. Results showed that 50 µM phenelzine had a negligible impact on LSD1 inhibition, whereas the same concentration of 12d led to very substantial LSD1 inhibition, with the unreacted dimethyl-peptide remaining as the major species at the conclusion of the experiment (Supplementary Figure 6). These experiments corroborate the findings with the spectrophotometric peroxidase assay that showed that 12d was a far more potent LSD1 inhibitor than phenelzine.
To assess the relative selectivity of our potent LSD1 phenelzine analogue 12d, we carried out counter screen enzyme assays versus MAO A, MAO B, and LSD2. As shown in Supplementary Table 1, based on kinact/Ki(inact) measure of inactivation efficiency, 12d is 23-fold selective for inhibiting LSD1 versus MAO A, 63-fold selective versus MAO B, and >100-fold versus LSD2. In contrast, phenelzine preferentially inhibits MAO A and is equipotent in blocking MAO B compared with LSD1. These results support the potential utility of 12d as a selective pharmacologic probe for cellular LSD1 histone demethylase activity.
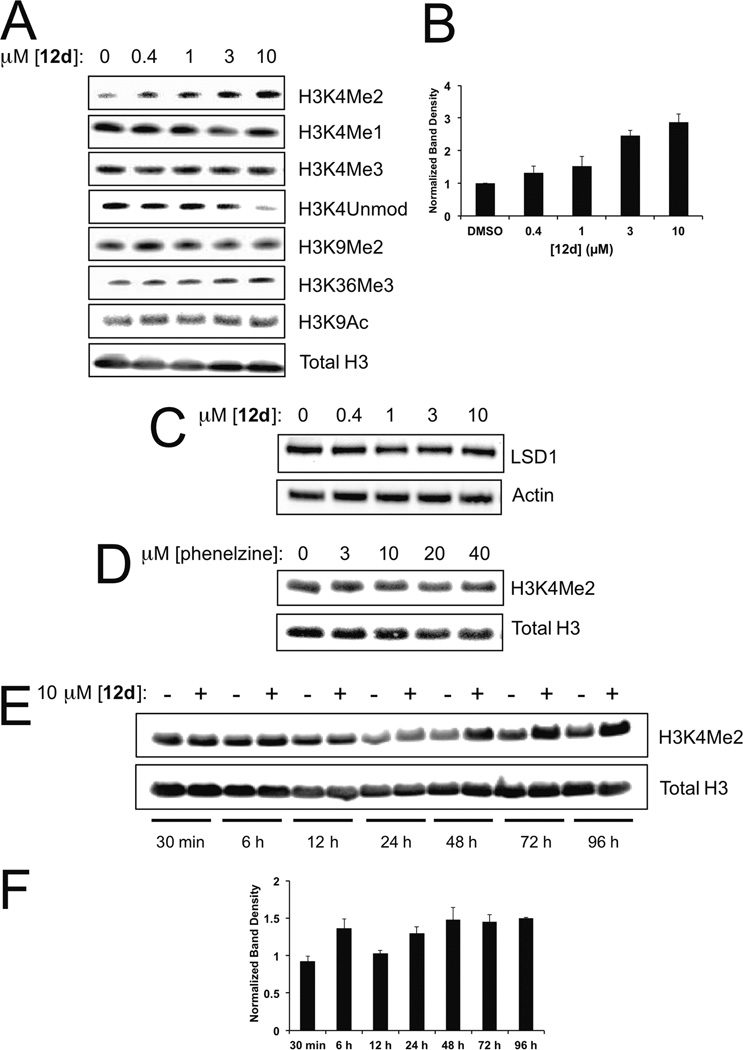
Compound 12d Effects on Cellular H3K4 Methylation
The ability of compound 12d, that we hereafter call bizine, to induce bulk histone H3-Lys4 methylation was assessed using Western blots in the prostate cancer LNCaP cell line, which has been used successfully in previous LSD1 inhibitor studies,51,52 with histone H3 methylation-state-specific antibodies. As can be seen, after 48 h treatment with bizine, there was a dose-dependent increase in H3K4Me2 signal (Figure 5A,B). The EC50 of this bizine effect was ~2 µM. There were no significant reproducible changes in H3K4Mel, H3K4Me3, unmethylated H3K4, or other histone H3 marks examined including H3K9Me2, H3K9Ac, and H3K36Me3 (Figure 5A). Furthermore, there was no discernible effect of bizine on LSD1 protein levels (Figure 5C). The increase in cellular global H3K4Me2 levels after treatment with bizine is a primary effect that is consistent with prior studies with less selective LSD1 inhibitors and genetic LSD1 alterations.14,32,37 However, in our hands with LNCaP cells, the MAO inhibitor phenelzine, which is an ~ 100-fold weaker LSD1 inhibitor than bizine, did not induce H3K4Me2 changes at concentrations up to 40 µM (Figure 5D). Additionally, the ~30-fold weaker N-methyl bizine analogue 12l did not show changes in H3K4Me2 at concentrations up to 10 µM (Supplementary Figure 7A,B). Taken together, these results are consistent with the hypothesis that the Western blot effects related to bizine are mediated through LSD1 inhibition.
Figure 5.
LSD1 inhibition by compound 12d (bizine) in LNCaP cells. (A) Cells were treated with compound 12d (bizine) (0.4–10 µM) for 48 h and blotted against indicated proteins. (B) H3K4Me2 band density quantification plot. Statistically significant increases were observed at 3 µM and 10 µM 12d (bizine) treatment as determined by three biological replicates. (C) Cells were treated with compound 12d (bizine) (0.4–10 µM) for 48 h and blotted against LSD1 and actin. (D) Cells were treated with phenelzine (3–40 µM) for 48 h and blotted against H3K4Me2 and total H3. (E) Cells were treated with 10 µM compound 12d (bizine) and collected at various indicated time points and blotted against H3K4Me2 and total H3. (F) H3K4Me2 band density quantification plot normalized to vehicle at each indicated time point after 10 µM 12d (bizine) treatment. Statistically significant increases were observed at 6, 24, 48, 72, and 96 h but not at 30 min and 12 h based on 3 biological replicates.
We further examined the effects of bizine on histone H3K4 methylation by assaying additional cancer cell lines (Supplementary Figure 8), H460, A549, and MDA-MB-231, each of which has been used in previous LSD1 inhibitor studies.10,53 With the lung cancer line H460, there were comparable dose-response effects of bizine on H3K4Me2 levels. The lung cancer line A549 and the breast cancer line MDA-MB-231 also showed increases in H3K4Me2 in response to bizine, but a higher concentration (20 µM) was required for reproducible effects.
We also measured the kinetics of bizine’s effect on H3 methylation in the LNCaP cell line. This time course experiment revealed that changes in H3K4Me2 could be detected within 6 h of compound exposure and effects can be observed up to 96 h (Figure 5E,F). However, there was a reproducible drop in H3K4Me2 at 12 h, which suggests a somewhat complex dynamic process involving competing waves of lysine methyltransferase and demethylase action (Supplementary Figure 9). However, it seems that cellular turnover of H3K4-methylation can be a relatively rapid process, on a time scale that is commensurate with many dynamic protein acetylation and phosphorylation events.54,55
To examine the effect of LSD1 inhibition on chromatin H3 Lys-methylation with individual gene resolution, we carried out a ChIP-seq experiment in LNCaP cells treated for 48 h with bizine. Differential peaks between samples with two biological replicates were identified by diffReps.56 In total, we obtained 17,542 differential H3K4Me2 peaks between cells treated with 10 µM bizine versus vehicle (Supplementary Table 2). Among those, 10,874 peaks were found to be upregulated (cut off p-value: p < 0.0001) with LSD1 inhibition. Out of those peaks, there were 2,432 genes identified that showed an increase in H3K4Me2 with LSD1 inhibition near the genes’ promoter regions (Supplementary Table 3 and Supplementary Figure 10). Furthermore, gene ontology (GO) analysis of these 2,432 genes revealed many processes related to LSD1 function (Supplementary Table 4).
After culling the list to exclude microRNA and nonstandard gene names from the 2,432 gene list, we compared the remaining 1,767 genes to the 1,587 genes identified in a ChIP-seq experiment that used a LSD1−/− hematopoietic cell line, which also analyzed H3K4Me2 increases at gene promoters57. There were 146 genes (p-value = 0.0028) that overlapped in the chemical inhibition and LSD1 knockout experiments (Supplementary Table 5). This indicated the presence of a statistically significant overlap in genes affected despite the different LSD1 inhibition methods and cell lines used. GO analysis performed on the 146 genes showed that gene regulation was one of the top five statistically significant processes affected (Supplementary Table 6). Of note, many (26) (p-value = 5.80 × 10 −9) of the 146 overlapped genes (Supplementary Table 7) are established or proposed to be tumor suppressors, including CDH1 and CDKN2A, which have been validated to be affected by LSD1 inhibitors in prior studies.13,58 This is consistent with the proposal that LSD1 inhibitors might have anticancer applications.
Bizine Antiproliferation Effects
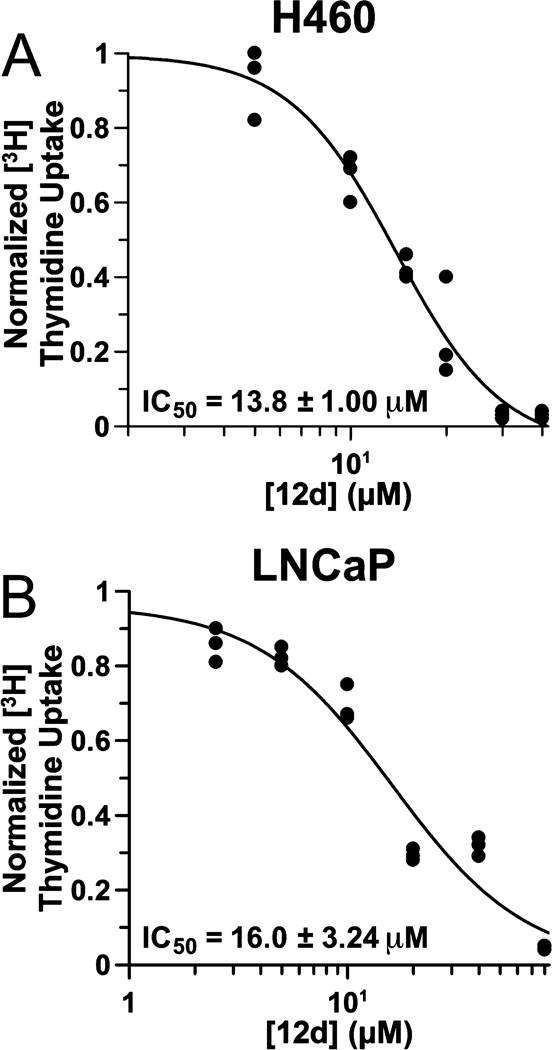
We next pursued the effects of bizine on cell proliferation using a 3H-thymidine incorporation assay as a measure of the rate of DNA synthesis. These studies revealed that bizine can slow the rate of cellular proliferation with an IC50 of 14 and 16 µM in treated H460 and LNCaP cancer cell lines, respectively (Figure 6A,B). These IC50’s are considerably higher than the EC50’s for Western blot changes in H3K4Me2. These findings raised concerns about the mechanistic basis of the cancer cell antiproliferative effects by bizine. To further address this issue, we tested the impact of phenelzine on H460 cell proliferation. There was less than a 50% reduction in 3H-thymidine incorporation in H460 cells after 48 h with 80 µM phenelzine (Supplementary Figure 11), indicating that MAO inhibition by bizine does not primarily contribute to its cell growth inhibitory effects. These experiments suggest that LSD1 inhibition by bizine likely contributes to cancer cell growth inhibition and perhaps that nearly complete LSD1 inhibition at concentrations well above the bizine Western blot EC50 are necessary for reducing cell growth.
Figure 6.
DNA replication dose response curves using a [3H] thymidine assay in (A) H460 cells and (B) LNCaP cells after 48 h treatment with compound 12d (bizine).
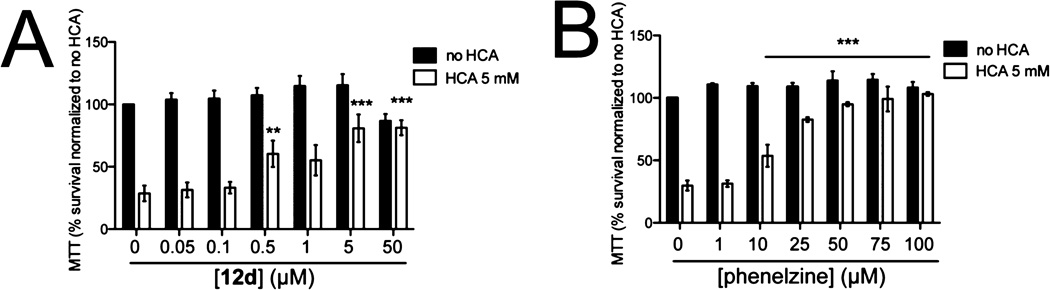
As LSD1 is an enzyme implicated in gene silencing, it is plausible that LSD1 inhibitors combined with histone deacetylase (HDAC) or DNA methyltransferase (DNMT) inhibitors might result in additive or synergistic effects. This concept has been evaluated previously with LSD1 inhibitors that have low selectivity and potency but nevertheless show additivity and synergy with various HDAC and DNMT inhibitors.53,59 Here we examine bizine in binary combinations with one DNMT inhibitor, azacytidine, as well as five HDAC inhibitors, SAHA, TSA, MGCD0103, MS-275, and LBH-589, using 3H-thymidine incorporation in H460 cells after 48 h treatment. The combination index (CI) was calculated for each inhibitor pair.60 Unexpectedly, four of the agents, azacytidine, SAHA, TSA, and MGCD0103, when combined with bizine, exhibited moderate antagonism, CI > 1, on inhibition of H460 cell proliferation at all ratios of the two agents examined (Supplementary Figure 12A – F). The basis of this antagonism is uncertain but may be related to various factors including changes in bizine uptake by cells or metabolism as well as complex pathway effects. MS-275 and LBH-589, in combination with bizine, showed additive to synergistic effects on inhibition of H460 cell proliferation, with the most synergy observed at the highest concentrations of compounds employed. These results reveal that in H460 cells, dual LSD1/HDAC inhibition may be promising, provided a suitable combination of inhibitors is identified that may reflect the precise specificities of the compounds involved.
LSD1 Inhibition and Neuroprotection
HDAC inhibition has previously been reported to protect against oxidative stress in neurons subjected to homocysteic acid (HCA) treatment, which induces glutathione depletion.61,62 We thus explored whether bizine might confer neuroprotection against HCA-induced oxidative stress. Indeed, 0.5 µM bizine led to significantly enhanced survival of neurons after HCA treatment in a dose-dependent fashion (Figure 7A,B). This level of neuroprotection was comparable to the effect of 10 µM phenelzine, consistent with the greater potency of bizine versus phenelzine as an LSD1 inhibitor. These results suggest that LSD1 might serve as an attractive target to treat or protect against neurologic disease, such as stroke, which can be placed in the context of prior work that investigated LSD1 functions in the brain.48,63
Figure 7.
LSD1 inhibition protects neurons against oxidative stress-mediated cell death. (A) Compound 12d (bizine) and (B) phenelzine halt neuronal cell death. (Two-way ANOVA, Bonferroni post hoc test; **p < 0.01; ***p < 0.0001 compared to no HCA).
Conclusion
This study describes a potent and selective LSD1 inhibitor, bizine, derived from the MAO inhibitor phenelzine. Structure-activity relationships demonstrate the key roles of the hydrazine functionality, the secondary amide linker, and the second aryl group in achieving potent LSD1 inhibition. Bizine shows potent action in cancer cells as demonstrated by modulating histone H3K4 methylation and exhibiting moderate antiproliferative properties. Interestingly, some HDAC inhibitors show additive to synergistic effects in combination with bizine in reducing H460 cell growth, whereas other HDAC inhibitors and azacytidine did not. A potentially promising direction is the application of LSD1 inhibition in neuroprotection against oxidative stress. In conclusion, we believe that bizine should be a useful probe in the continuing functional evaluation of LSD1’s demethylase activity in physiologic and pathophysiologic conditions.
METHODS
GST-LSD1 Enzymatic Assays
GST-LSD1 production from an E. coli expression system followed by purification using glutathione affinity chromatography were performed as previously described.64 Rate measurements were performed using a peroxidase-coupled assay as previously described.6 To determine LSD1 activity, 100 µL reactions were initiated by the addition of 2 µL of GST-LSD1 (to obtain 96–154 nM GST-LSD1 final concentration) to reaction mixtures consisting of 50 mM HEPES buffer (pH 7.5), 0.1 mM 4-aminoantipyrine, 1 mM 3,5-dichloro-2-hydroxybenzenesulfonic acid, 0.04 mg mL−1 horseradish peroxidase (Worthington Biochemical Corporation), and appropriate concentration of buffered substrate (dimethyl-Lys-4 H3-21, ARTKme2QTARKSTGGKAPRKQLA, synthesized and purified as described previously36). Absorbance changes were measured at 515 nm using a Beckman Instruments DU series 600 spectrophotometer equipped with a thermostatted cell holder (T = 25 °C), and product formation was calculated using the extinction coefficient of 26,000 M−1. Under these conditions, GST-LSD1 displayed a kcat of ~3 min−1 and a Km for dimethyl-Lys-4 H3-21 of ~40 µM, but the specific parameters were measured for each batch and used for inhibitor parameter calculations. For inhibition studies, phenelzine analogue compounds were dissolved in dimethylsulfoxide (DMSO) to make 5 mM stock solutions that were diluted into reactions at appropriate concentrations. Reactions were run at similar conditions as previously stated with 60–300 µM dimethyl-Lys-4 H3-21 substrate. Progress curves conducted for 20 min were then fit to the following eq 1:
| (1) |
The Kitz and Wilson method was then used to analyze the kobs values to obtain kinact and Ki(inact) values with the following eq 2:
| (2) |
The following Cheng-Prusoff equation, eq 3, was then used to extrapolate the Ki(inact) value to zero substrate:
| (3) |
Each experiment was repeated at least two independent times and repeat measured values were typically within 20% of each other.
MAO A/B Activity and Inhibition Assays
MAO A was purchased from Sigma (product number M 7316). MAO B was purchased from Sigma (product number M 7441). MAO A/B activity was measured spectrophotometrically using a peroxidase-coupled assay as previously described.6 First, 100 µL reactions were initiated by the addition of 2 µL of MAO A/B (to obtain 100–200 nM final concentration for MAO A and 0.837 µM final concentration for MAO B) to reaction mixtures consisting of 50 mM HEPES buffer (pH 7.5), 0.1 mM 4-aminoantipyrine, 1 mM 3,5-dichloro-2-hydroxybenzenesul-fonic acid, 0.04 mg mL−1 horseradish peroxidase (Worthington Biochemical Corporation), and appropriate concentration of buffered substrate (tyramine). Absorbance changes were measured at 515 nm using a Beckman Instruments DU series 600 spectrophotometer equipped with a thermostatted cell holder (T = 25 °C), and product formation was calculated using the extinction coefficient of 26,000 M−1. Under these conditions, MAO A displayed a kcat of 3 ± 0.1 min−1 and a Km for tyramine of 26 ± 3 µM. MAO B displayed a kcat of 0.2 ± 0.02 min−1 and a Km for tyramine of 94 ± 26.0 µM. For inhibitor studies, phenelzine analogue compounds were dissolved in dimethylsulfoxide (DMSO) to make 5 mM stock solutions that were diluted into reactions at appropriate concentrations. Reactions were run at similar conditions as previously stated with 125 µM tyramine substrate for MAO A and with 125–1,000 µM tyramine substrate for MAO B. Progress curves were then fit accordingly to eqs 1–3 as previously stated. Each experiment was repeated at least two independent times, and repeat measured values were typically within 20% of each other.
Cell Culture
LNCaP, H460, and A549 cells were maintained in RPMI 1640 + GlutaMAX (Invitrogen 61870-036) supplemented with 10% fetal bovine serum (FBS, Gibco 10437-028) and 1 unit mL−1 penicillin, 1 µg mL−1 streptomycin (Gibco 15140-122). MDA-MB-231 cells were maintained in DMEM (Gibco 11965) supplemented with 10% FBS and 1 unit mL−1 penicillin, 1 µg mL−1 streptomycin, and 292 µg mL−1 L-glutamine (Corning 30-009-Cl). All cells were grown at 37 °C in 5%/95% CO2/air.
Western Blot
Cells were seeded in 150 × 25 mm plastic tissue culture dishes (Corning 430599). Cells were treated at ~70% confluency with phenelzine analogues (>97% purity as determined by NMR) or vehicle in serum-free media for 48 h. Whole-cell extracts were isolated using RIPA buffer (Sigma R0278) and 1× protease inhibitor cocktail (Roche 11836170001). Histone extracts were isolated as described previously.65 Concentration of whole cell lysates and histone extracts were determined using a Micro BCA Protein Assay Kit (Thermo Scientific 23235). Proteins were resolved by 10– 12% NuPAGE Novex Bis-Tris gels (Invitrogen) and transferred to nitrocellulose membranes (Invitrogen) by iBlot. Data are presented from one representative experiment. Each experiment was repeated at least three independent times with nearly identical results.
Oxidative Toxicity and Neuron Viability Assays
Immature primary cortical neurons were obtained from fetal Sprague–Dawley rats at embryonic day 17 (E17) as previously described66 and plated at a density of 106 cells mL−1 in 96-well plates for the viability experiments. The next day cells were rinsed and then placed in medium containing 5 mM homocysteic acid (HCA). In the dose-curve experiments, increasing concentrations of 12d (bizine) or phenelzine were added at the time of HCA treatment. The next day, cell viability was assessed by the MTT assay (Promega).67 The 2-way ANOVA followed by the Bonferroni post-test was used to measure statistical significance. p < 0.05 was considered to be statistically significant. Each bar represents four different rat cultures. The use of animals and procedures were approved by the Institutional Animal Care and Use Committees of Weill Medical College of Cornell University.
Supplementary Material
ACKNOWLEDGMENTS
We thank R. Chivukula and J. Mendell for providing the mouse specimens and help in cDNA cloning. We thank the NIH and Samuel Waxman Research Foundation for financial support.
Footnotes
ASSOCIATED CONTENT
Supporting Information
This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Culhane JC, Cole PA. LSD1 and the chemistry of histone demethylation. Curr. Opin. Chem. Biol. 2007;11:561–568. doi: 10.1016/j.cbpa.2007.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang M, Culhane JC, Szewczuk LM, Gocke CB, Brautigam CA, Tomchick DR, Machius M, Cole PA, Yu H. Structural basis of histone demethylation by LSD1 revealed by suicide inactivation. Nat. Struct. Mol. Biol. 2007;14:535–539. doi: 10.1038/nsmb1255. [DOI] [PubMed] [Google Scholar]
- 3.Forneris F, Binda C, Battaglioli E, Mattevi A. LSD1: oxidative chemistry for multifaceted functions in chromatin regulation. Trends Biochem. Sci. 2008;33:181–189. doi: 10.1016/j.tibs.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Forneris F, Binda C, Adamo A, Battaglioli E, Mattevi A. Structural basis of LSD1-CoREST selectivity in histone H3 recognition. J. Biol. Chem. 2007;282:20070–20074. doi: 10.1074/jbc.C700100200. [DOI] [PubMed] [Google Scholar]
- 5.Forneris F, Binda C, Dall’Aglio A, Fraaije MW, Battaglioli E, Mattevi A. A highly specific mechanism of histone H3-K4 recognition by histone demethylase LSD1. J. Biol. Chem. 2006;281:35289–35295. doi: 10.1074/jbc.M607411200. [DOI] [PubMed] [Google Scholar]
- 6.Forneris F, Binda C, Vanoni MA, Battaglioli E, Mattevi A. Human histone demethylase LSD1 reads the histone code. J. Biol. Chem. 2005;280:41360–41365. doi: 10.1074/jbc.M509549200. [DOI] [PubMed] [Google Scholar]
- 7.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 8.Liang G, Lin JCY, Wei V, Yoo C, Cheng JC, Nguyen CT, Weisenberger DJ, Egger G, Takai D, Gonzales FA, Jones PA. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc. Natl. Acad. Sci. U.S.A. 2004;101:7357–7362. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 10.Lv T, Yuan D, Miao X, Lv Y, Zhan P, Shen X, Song Y. Over-expression of LSD1 promotes proliferation, migration and invasion in non-small cell lung cancer. PLoS One. 2012 doi: 10.1371/journal.pone.0035065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim S, Janzer A, Becker A, Zimmer A, Schüle R, Buettner R, Kirfel J. Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis. 2010;31:512–520. doi: 10.1093/carcin/bgp324. [DOI] [PubMed] [Google Scholar]
- 12.Metzger E, Wissmann M, Yin N, Müller JM, Schneider R, Peters AHFM, Günther T, Buettner R, Schüle R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 13.Murray-Stewart T, Woster PM, Casero RA. The re-expression of the epigenetically silenced e-cadherin gene by a polyamine analogue lysine-specific demethylase-1 (LSD1) inhibitor in human acute myeloid leukemia cell lines. Amino Acids. 2013 doi: 10.1007/s00726-013-1485-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang Y, Greene E, Murray Stewart T, Goodwin AC, Baylin SB, Woster PM, Casero RA., Jr Inhibition of lysine-specific demethylase 1 by polyamine analogues results in reexpression of aberrantly silenced genes. Proc. Natl. Acad. Sci. U.S.A. 2007;104:8023–8028. doi: 10.1073/pnas.0700720104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Y, Stewart TM, Wu Y, Baylin SB, Marton LJ, Perkins B, Jones RJ, Woster PM, Casero RA., Jr Novel oligoamine analogues inhibit lysine-specific demethylase 1 and induce reexpression of epigenetically silenced genes. Clin. Cancer Res. 2009;15:7217–7228. doi: 10.1158/1078-0432.CCR-09-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin L, Hanigan CL, Wu Y, Wang W, Park BH, Woster PM, Casero RA. Loss of LSD1 (lysine-specific demethylase 1) suppresses growth and alters gene expression of human colon cancer cells in a p53- and DNMT1(DNA methyltransferase 1)-independent manner. Biochem. J. 2013;449:459–468. doi: 10.1042/BJ20121360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takai N, Narahara H. Array-based approaches for the identification of epigenetic silenced tumor suppressor genes. Curr. Genomics. 2008;9:22–24. doi: 10.2174/138920208783884892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaweska H, Henderson Pozzi M, Schmidt DMZ, McCafferty DG, Fitzpatrick PF. Use of pH and kinetic isotope effects to establish chemistry as rate-limiting in oxidation of a peptide substrate by LSD1. Biochemistry. 2009;48:5440–5445. doi: 10.1021/bi900499w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forneris F, Binda C, Vanoni MA, Mattevi A, Battaglioli E. Histone demethylation catalysed by LSD1 is a flavin-dependent oxidative process. FEBS Lett. 2005;579:2203–2207. doi: 10.1016/j.febslet.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 20.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 21.Hakimi M-A, Dong Y, Lane WS, Speicher DW, Shiekhattar R. A candidate X-linked mental retardation gene is a component of a new family of histone deacetylase-containing complexes. J. Biol. Chem. 2003;278:7234–7239. doi: 10.1074/jbc.M208992200. [DOI] [PubMed] [Google Scholar]
- 22.Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat. Rev. Mol. Cell Biol. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 23.Hwang S, Schmitt AA, Luteran AE, Toone EJ, McCafferty DG. Thermodynamic characterization of the binding interaction between the histone demethylase LSD1/KDM1 and CoREST. Biochemistry. 2011;50:546–557. doi: 10.1021/bi101776t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baron R, Binda C, Tortorici M, McCammon JA, Mattevi A. Molecular mimicry and ligand recognition in binding and catalysis by the histone demethylase LSD1-CoREST complex. Structure. 2011;19:212–220. doi: 10.1016/j.str.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forneris F, Battaglioli E, Mattevi A, Binda C. New roles of flavoproteins in molecular cell biology: histone demethylase LSD1 and chromatin. FEBS J. 2009;276:4304–4312. doi: 10.1111/j.1742-4658.2009.07142.x. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Q, Qi S, Xu M, Yu L, Tao Y, Deng Z, Wu W, Li J, Chen Z, Wong J. Structure-function analysis reveals a novel mechanism for regulation of histone demethylase LSD2/AOF1/KDM1b. Cell Res. 2013;23:225–241. doi: 10.1038/cr.2012.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karytinos A, Forneris F, Profumo A, Ciossani G, Battaglioli E, Binda C, Mattevi A. A novel mammalian flavin-dependent histone demethylase. J. Biol. Chem. 2009;284:17775–17782. doi: 10.1074/jbc.M109.003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Murray-Stewart T, Devereux W, Hacker A, Frydman B, Woster PM, Casero RA., Jr Properties of purified recombinant human polyamine oxidase, PAOh1/SMO. Biochem. Biophys. Res. Commun. 2003;304:605–611. doi: 10.1016/s0006-291x(03)00636-3. [DOI] [PubMed] [Google Scholar]
- 29.Culhane JC, Wang D, Yen PM, Cole PA. Comparative analysis of small molecules and histone substrate analogues as LSD1 lysine demethylase inhibitors. J. Am. Chem. Soc. 2010;132:3164–3176. doi: 10.1021/ja909996p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dancy BCR, Ming SA, Papazyan R, Jelinek CA, Majumdar A, Sun Y, Dancy BM, Drury WJ, 3rd, Cotter RJ, Taverna SD, Cole PA. Azalysine analogues as probes for protein lysine deacetylation and demethylation. J. Am. Chem. Soc. 2012;134:5138–5148. doi: 10.1021/ja209574z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tortorici M, Borrello MT, Tardugno M, Chiarelli LR, Pilotto S, Ciossani G, Vellore NA, Bailey SG, Cowan J, O’Connell M, Crabb SJ, Packham G, Mai A, Baron R, Ganesan A, Mattevi A. Protein recognition by short peptide reversible inhibitors of the chromatin-modifying LSD1/CoREST lysine demethylase. ACS Chem. Biol. 2013;8:1677–1682. doi: 10.1021/cb4001926. [DOI] [PubMed] [Google Scholar]
- 32.Binda C, Valente S, Romanenghi M, Pilotto S, Cirilli R, Karytinos A, Ciossani G, Botrugno OA, Forneris F, Tardugno M, Edmondson DE, Minucci S, Mattevi A, Mai A. Biochemical, structural, and biological evaluation of tranylcypromine derivatives as inhibitors of histone demethylases LSD1 and LSD2. J. Am. Chem. Soc. 2010;132:6827–6833. doi: 10.1021/ja101557k. [DOI] [PubMed] [Google Scholar]
- 33.Mimasu S, Umezawa N, Sato S, Higuchi T, Umehara T, Yokoyama S. Structurally designed trans-2-phenyl-cyclopropylamine derivatives potently inhibit histone demethylase LSD1/KDM1. Biochemistry. 2010;49:6494–6503. doi: 10.1021/bi100299r. [DOI] [PubMed] [Google Scholar]
- 34.Zhu Q, Huang Y, Marton LJ, Woster PM, Davidson NE, Casero RA., Jr Polyamine analogs modulate gene expression by inhibiting lysine-specific demethylase 1 (LSD1) and altering chromatin structure in human breast cancer cells. Amino Acids. 2012;42:887–898. doi: 10.1007/s00726-011-1004-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J, Lu F, Ren Q, Sun H, Xu Z, Lan R, Liu Y, Ward D, Quan J, Ye T, Zhang H. Novel histone demethylase LSD1 inhibitors selectively target cancer cells with pluripotent stem cell properties. Cancer Res. 2011;71:7238–7249. doi: 10.1158/0008-5472.CAN-11-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Culhane JC, Szewczuk LM, Liu X, Da G, Marmorstein R, Cole PA. A mechanism-based inactivator for histone demethylase LSD1. J. Am. Chem. Soc. 2006;128:4536–4537. doi: 10.1021/ja0602748. [DOI] [PubMed] [Google Scholar]
- 37.Pollock JA, Larrea MD, Jasper JS, McDonnell DP, McCafferty DG. Lysine-specific histone demethylase 1 inhibitors control breast cancer proliferation in ERα-dependent and -independent manners. ACS Chem. Biol. 2012;7:1221–1231. doi: 10.1021/cb300108c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gooden DM, Schmidt DMZ, Pollock JA, Kabadi AM, McCafferty DG. Facile synthesis of substituted trans-2-arylcyclopropylamine inhibitors of the human histone demethylase LSD1 and monoamine oxidases A and B. Bioorg. Med. Chem. Lett. 2008;18:3047–3051. doi: 10.1016/j.bmcl.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dulla B, Kirla KT, Rathore V, Deora GS, Kavela S, Maddika S, Chatti K, Reiser O, Iqbal J, Pal M. Synthesis and evaluation of 3-amino/guanidine substituted phenyl oxazoles as a novel class of LSD1 inhibitors with anti-proliferative properties. Org. Biomol. Chem. 2013;11:3103–3107. doi: 10.1039/c3ob40217g. [DOI] [PubMed] [Google Scholar]
- 40.Hazeldine S, Pachaiyappan B, Steinbergs N, Nowotarski S, Hanson AS, Casero RA, Jr, Woster PM. Low molecular weight amidoximes that act as potent inhibitors of lysine-specific demethylase 1. J. Med. Chem. 2012;55:7378–7391. doi: 10.1021/jm3002845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang M, Culhane JC, Szewczuk LM, Jalili P, Ball HL, Machius M, Cole PA, Yu H. Structural basis for the inhibition of the LSD1 histone demethylase by the antidepressant trans-2-phenylcyclopropylamine. Biochemistry. 2007;46:8058–8065. doi: 10.1021/bi700664y. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt DMZ, McCafferty DG. trans-2-Phenylcyclopropylamine is a mechanism-based inactivator of the histone demethylase LSD1. Biochemistry. 2007;46:4408–4416. doi: 10.1021/bi0618621. [DOI] [PubMed] [Google Scholar]
- 43.Lee MG, Wynder C, Schmidt DM, McCafferty DG, Shiekhattar R. Histone H3 lysine 4 demethylation is a target of nonselective antidepressive medications. Chem. Biol. 2006;13:563–567. doi: 10.1016/j.chembiol.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 44.Ueda R, Suzuki T, Mino K, Tsumoto H, Nakagawa H, Hasegawa M, Sasaki R, Mizukami T, Miyata N. Identification of cell-active lysine specific demethylase 1-selective inhibitors. J. Am. Chem. Soc. 2009;131:17536–17537. doi: 10.1021/ja907055q. [DOI] [PubMed] [Google Scholar]
- 45.Guibourt N, Ortega MA, Castro-Palomino LJ. WO2010043721 A1 2010 Oxidase inhibitors and their use. Patent application.
- 46.Guibourt N, Ortega MA, Castro-Palomino LJ. WO2010084160 A1. Phenylcyclopropylamine derivatives and their medical use. Patent application. 2010
- 47.Ortega MA, Castro-Palomino LJ, Fyfe MCT. WO2011131697 A1. Lysine specific demethylase-1 inhibitors and their use. Patent application. 2011
- 48.Neelamegam R, Ricq EL, Malvaez M, Patnaik D, Norton S, Carlin SM, Hill IT, Wood MA, Haggarty SJ, Hooker JM. Brain-penetrant LSD1 inhibitors can block memory consolidation. ACS Chem. Neurosci. 2012;3:120–128. doi: 10.1021/cn200104y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holt A, Sharman DF, Baker GB, Palcic MM. A continuous spectrophotometric assay for monoamine oxidase and related enzymes in tissue homogenates. Anal. Biochem. 1997;244:384–392. doi: 10.1006/abio.1996.9911. [DOI] [PubMed] [Google Scholar]
- 50.Blair LP, Avaritt NL, Huang R, Cole PA, Taverna SD, Tackett AJ. MassSQUIRM: An assay for quantitative measurement of lysine demethylase activity. Epigenetics. 2011;6:490–499. doi: 10.4161/epi.6.4.14531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Willmann D, Lim S, Wetzel S, Metzger E, Jandausch A, Wilk W, Jung M, Forne I, Imhof A, Janzer A, Kirfel J, Waldmann H, Schüle R, Buettner R. Impairment of prostate cancer cell growth by a selective and reversible lysine-specific demethylase 1 inhibitor. Int. J. Cancer. 2012;131:2704–2709. doi: 10.1002/ijc.27555. [DOI] [PubMed] [Google Scholar]
- 52.Benelkebir H, Hodgkinson C, Duriez PJ, Hayden AL, Bulleid RA, Crabb SJ, Packham G, Ganesan A. Enantioselective synthesis of tranylcypromine analogues as lysine demethylase (LSD1) inhibitors. Bioorg. Med. Chem. 2011;19:3709–3716. doi: 10.1016/j.bmc.2011.02.017. [DOI] [PubMed] [Google Scholar]
- 53.Huang Y, Vasilatos SN, Boric L, Shaw PG, Davidson NE. Inhibitors of histone demethylation and histone deacetylation cooperate in regulating gene expression and inhibiting growth in human breast cancer cells. Breast Cancer Res. Treat. 2012;131:777–789. doi: 10.1007/s10549-011-1480-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qiao Y, Molina H, Pandey A, Zhang J, Cole PA. Chemical rescue of a mutant enzyme in living cells. Science. 2006;311:1293–1297. doi: 10.1126/science.1122224. [DOI] [PubMed] [Google Scholar]
- 55.Crump NT, Hazzalin CA, Bowers EM, Alani RM, Cole PA, Mahadevan LC. Dynamic acetylation of all lysine-4 trimethylated histone H3 is evolutionarily conserved and mediated by p300/CBP. Proc. Natl. Acad. Sci. U.S.A. 2011;108:7814–7819. doi: 10.1073/pnas.1100099108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen L, Shao N-Y, Liu X, Maze I, Feng J, Nestler EJ. diffReps: detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS One. 2013 doi: 10.1371/journal.pone.0065598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kerenyi MA, Shao Z, Hsu Y-J, Guo G, Luc S, O’Brien K, Fujiwara Y, Peng C, Nguyen M, Orkin SH. Histone demethylase Lsd1 represses hematopoietic stem and progenitor cell signatures during blood cell maturation. eLife. 2013 doi: 10.7554/eLife.00633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu Y, Steinbergs N, Murray-Stewart T, Marton LJ, Casero RA. Oligoamine analogues in combination with 2-difluoromethylornithine synergistically induce re-expression of aberrantly silenced tumour-suppressor genes. Biochem. J. 2012;442:693–701. doi: 10.1042/BJ20111271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Han H, Yang X, Pandiyan K, Liang G. Synergistic re-activation of epigenetically silenced genes by combinatorial inhibition of DNMTs and LSD1 in cancer cells. PLoS One. 2013 doi: 10.1371/journal.pone.0075136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv. Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 61.Langley B, D’Annibale MA, Suh K, Ayoub I, Tolhurst A, Bastan B, Yang L, Ko B, Fisher M, Cho S, Beal MF, Ratan RR. Pulse inhibition of histone deacetylases induces complete resistance to oxidative death in cortical neurons without toxicity and reveals a role for cytoplasmic p21(waf1/cip1) in cell cycle-independent neuroprotection. J. Neurosci. 2008;28:163–176. doi: 10.1523/JNEUROSCI.3200-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kozikowski AP, Chen Y, Subhasish T, Lewin NE, Blumberg PM, Zhong Z, D’Annibale MA, Wang W-L, Shen Y, Langley B. Searching for disease modifiers-PKC activation and HDAC inhibition - a dual drug approach to Alzheimer’s disease that decreases Abeta production while blocking oxidative stress. ChemMedChem. 2009;4:1095–1105. doi: 10.1002/cmdc.200900045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Y-Z, Zhang Q-H, Ye H, Zhang Y, Luo Y-M, Ji X-M, Su Y-Y. Distribution of lysine-specific demethylase 1 in the brain of rat and its response in transient global cerebral ischemia. Neurosci. Res. 2010;68:66–72. doi: 10.1016/j.neures.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 64.Szewczuk LM, Culhane JC, Yang M, Majumdar A, Yu H, Cole PA. Mechanistic analysis of a suicide inactivator of histone demethylase LSD1. Biochemistry. 2007;46:6892–6902. doi: 10.1021/bi700414b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shechter D, Dormann HL, Allis CD, Hake SB. Extraction, purification and analysis of histones. Nat. Protoc. 2007;2:1445–1457. doi: 10.1038/nprot.2007.202. [DOI] [PubMed] [Google Scholar]
- 66.Ratan RR, Murphy TH, Baraban JM. Oxidative stress induces apoptosis in embryonic cortical neurons. J. Neurochem. 1994;62:376–379. doi: 10.1046/j.1471-4159.1994.62010376.x. [DOI] [PubMed] [Google Scholar]
- 67.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.