Abstract
LKB1 is commonly thought of as a tumor suppressor gene because its hereditary mutation is responsible for a cancer syndrome, and somatic inactivation of LKB1 is found in non-small cell lung cancer, melanoma, and cervical cancers. However, unlike other tumor suppressors whose main function is to either suppress cell proliferation or promote cell death, one of the functions of LKB1-regulated AMPK signaling is to suppress cell proliferation in order to promote cell survival under energetic stress conditions. This unique, pro-survival function of LKB1 has led to the discovery of reagents, such as phenformin, that specifically exploit the vulnerability of LKB1-null cells in their defect in sensing energetic stress. Such targeted agents represent a novel treatment strategy because they induce cell killing when LKB1 is absent. This review article summarizes various vulnerabilities of LKB1-mutant cells that have been reported in the literature and discusses the potential of using existing or developing novel reagents to target cancer cells with defective LKB1.
Keywords: Tumor Suppressor, tumor vulnerability, targeted therapy, metabolic stress
Introduction
The current cancer treatment paradigm is to inhibit biological pathways that are hyperactive in cancer cells with pharmaceutical reagents. While these approaches have proven successful in the clinic, they share two common limitations. First, the targeted proteins or pathways are likely to play important physiological roles in some normal tissues, and their inhibition thus leads to toxic side effects. Second, cancer cells have defective DNA damage/repair checkpoint(s) which make them genetically unstable 1. Consequently, cancer cells are genetically heterogeneous, and each cell contains numerous pre-existing mutations that are not normally selected. Systemic therapy creates an environment for the selection of cancer cells with mutated target proteins that no longer interact with the drug. Therefore, side effects and secondary mutation-related drug-resistance are two inevitable consequences of current cancer treatment approaches.
Tumor suppressor genes are not normally perceived as viable drug targets, and a common quote from pharmaceutical companies is “how can you target something that is not there?” It is important to note that even though Dr. Frank McCormick pioneered the concept of killing p53-null cells with the ONYX virus, the most successful clinical study of this virus was accompanied by evidence that ONYX-015 kills cancer cells through a mechanism that is not related to p53 inactivation 2,3. Recent studies have indicated that defects in the LKB1-AMPK signaling pathway make tumors vulnerable to varieties of stress, which can be exploited therapeutically.
Liver kinase 1 (LKB1 also known as STK11) is a tumor suppressor gene that is inactivated by bi-allelic mutation in non-small cell lung cancer (NSCLC), malignant melanoma and cervical cancer 4–6. Our knowledge of the biological roles of LKB1 has rapidly expanded over the past decade. Initial research focused on its roles in cell polarity, cell motility, protein translation and energy metabolism, and recent advances indicated that LKB1 is also involved in the regulation of other cellular process, such as DNA damage checkpoint, liposome function, and various signal transduction pathways. Hence, the inactivation of LKB1in human tumors will lead to the de-regulation of multiple cellular processes, but it is still unclear which of them is related to tumorigenesis. From a treatment perspective, the lack of proper regulation should make cancer cells vulnerable to reagents that specifically inhibit these pathways. A therapeutic approach that can specifically eliminate LKB1-deficient cells will have at least two advantages. First, LKB1 is ubiquitously expressed in all tissue types, and therapeutic approaches against LKB1-null cells should have less toxic effects on normal tissues (i.e. fewer side effects). Second, LKB1 is frequently inactivated by bi-allelic genetic inactivation. Because the genetic codes for LKB1 are lost in these cancer cells, they are unlikely to be restored by genetic instability. Even if cancer cells find another way to restore LKB1 function, its tumor suppressor function should inhibit the growth of these cells. This review article will focus on the nature of these vulnerabilities and recent advances in the development of clinical reagents that target these vulnerabilities in LKB1-null cancer cells (Table 1).
Table 1. A summary of agents may be used to target LKB1 mutants and their stages of development.
Only LKB1 independent targets and mechanisms are listed. The information of clinical trials was obtained from www.clinicaltrials.gov. Although some of the listed agents have already been enrolled in clinical trials, only very few of them were specifically designed for LKB1 mutants. Also, some of the listed agents are still in silencing RNA formula, but due to their potentiality and good mechanistic studies, they are included here as well.
| Class | Agents | Targets / Mechanisms that are LKB1 independent |
Stage of development | Comments | Refs |
|---|---|---|---|---|---|
|
AMPK activators / stress inducers |
Metformin | Inhibits mitochondria complex 1 and induces higher AMP/ATP ratio in LKB1 mutants (25–27). |
Multiple phase 1 to 3 trials in malignancy and diabetes. Some of them included AMPK in the outcome measures (e.g. the phase 2 trial NCT01266486) |
The phase 2 NA_00052073 also included LKB1 status in the secondary outcome measures. |
25–27 |
| Phenformin | Targets mitochondria complex 1 and induces more severe energy stress in LKB1 mutants (27); |
Was withdrawn from market in 1978 due to rare but severe lactic acidosis in diabetic pts. Not currently in clinical trial. |
LKB1 farnesylation is required for activation of AMPK by phenformin (30) |
27,30 | |
| AICAR | Induces apoptosis in LKB1-null MEF cells and ovarian cancer cells (34,35) |
Phase 1-2, but none for malignancies at this moment |
34,35 | ||
|
mTOR / HIF-1α / LOX inhibitors |
Rapalogues (Everolimus, sirolimus, temsirolimus) |
mTORC1 | Now in multiple phase 1-3 clinical trials |
NCT01178151 is a phase 2 trial specifically for PJS |
32,42,43 |
| AZD8055 | ATP competitive inhibitor for both mTORC1 and mTORC2 |
Phase 1 but not specifically for LKB1 mutants |
Preferentially reduced both LDH and PDH levels in a STK11-/-/NIC breast cancer model |
81 | |
| BAPN | LOX | Laboratory | 82 | ||
|
FAK/Src inhibitor FAK/Src inhibitor CHK1 inhibitors |
PF573228 | FAK inhibition in LKB1-null background (53) |
Laboratory | 52, 53 | |
| Defactinib (PF- 4554878, VS-6063) |
FAK inhibition | Phase 1 & 2 including a trial for KRAS mutant NSCLC, but not specific for LKB1 |
Needs more studies for LKB1 mutants |
52 | |
| Dasatinib | Src inhibition | Phase 1 & 2 for various malignancies. Not LKB1 specific |
52 | ||
| AZD7762 | CHK1 inhibition | Phase 1 (solid tumor). Not LKB1 specific. |
Cardiac toxicity (83), neutropenia (84) |
54 | |
|
CHK1 inhibitors Nucleotide metabolism inhibitors |
CHIR124 | CHK1 inhibition | Laboratory | 54 | |
| DTYMK shRNA | DTYMK inhibition, reduce dTTP biosynthesis |
Laboratory | 54 | ||
|
COPI / lysosomal maturation inhibitors |
Bafilomycin A1 (bafA) | Inhibits vacuolar ATPase | Laboratory | 66 | |
|
COPI / lysosomal maturation inhibitors YAP inhibitors / Hippo activators |
saliphenylhalamide A (saliPhe) |
Inhibits vacuolar ATPase | Laboratory | 66 | |
| Dox-inducible YAP shRNA (iYAP shRNA) |
Silences YAP | Laboratory | 72,73 | ||
| Verteporfin | Disrupts the TEAD-YAP interaction | Laboratory | Suppresses YAP, but needs to be tested in the setting of LKB1 mutation |
74 | |
| Super-TDU | Compete with YAP for the interaction with TEAD |
Laboratory | Suppressed YAP and gastric cancer growth. Needs to be tested in LKB1 mutants. |
75 |
Abbreviations AICAR 5-aminoimidazole-4-carboxamide-1-beta-4-ribofuranoside; DTYMK deoxythymidylate kinase; LOX lysyl oxidase; BAPN β-aminopropionitrile; PJS Peutz-Jeghers Syndrome; LDH lactate dehydrogenase; PDH pyruvate dehydrogenase; NIC Neu/HER2-MMTV-Cre.
LKB1 as a serine/threonine kinase
LKB1 protein has a central kinase domain, two N-terminal nuclear leading sequences, and a C-terminal regulatory motif. The protein also contains a CAAX motif at C-terminal end and can be farnesylated. The formation of an LKB1/Strad/MO25 complex is essential for LKB1’s kinase activity 7, and there are at least 13 known substrates of LKB1, which includes AMPKs, BRSKs, and MARKs 8. LKB1 is ubiquitously expressed in all tissues, so the mere presence of this protein does not inhibit cell proliferation 9. Existing data indicate that LKB1 kinase activity can be regulated either by posttranslational modification of LKB1 itself or by the regulation of conformational configuration of its downstream targets. The upstream regulators of LKB1 include ERK, RSK, ATM, and DNA-PK, most of which will be discussed in this review.
Mechanism of LKB1 inactivation in human tumors
Genetic inactivation of LKB1
LKB1 was first found to be associated with Peutz-Jeghers syndrome 10. This is an autosomal dominant disease that is characterized by gastrointestinal hamartomatous polyps, and elevated risk of various neoplasms 11. Many patients carry germline mutations that inactivate the kinase function of LKB1 12. This finding initiated intensive surveys for somatic LKB1 mutation analysis in human tumors, and LKB1 was subsequently found to be frequently mutated in non-small cell lung cancer (NSCLC), melanoma, and cervical cancers 4–6. The mutation frequency of LKB1 in NSCLC is still undetermined. Initial analysis in lung cancer cell lines indicated that LKB1 is inactivated in 30% of samples 4, but sub-sequence analysis in adenocarcinomas only revealed 5–15% point mutations. Part of this confusion is due to the fact that LKB1 is frequently inactivated by homozygous deletion which will not be detected by sequencing analysis alone. A combined analysis of homozygous deletion and LOH with somatic mutation indicated LKB1 is inactivated in 39% of NSCLC samples 13. Similarly, a combined sequencing and LKB1-large deletion analysis revealed 27% of Chinese lung adenocarcinomas contain LKB1 genetic alteration 14. The NextGen sequencing method should provide data on both somatic mutation and copy-number variations, and such analysis in the future should be able to provide a definitive number on LKB1 mutation frequencies in these tumor types.
LKB1 mutation was found to be rare in most other tumor types, which suggests that the deletion of LKB1 can only provide a growth advantage in certain tumor types. In the case of lung cancers, LKB1 mutation is limited to NSCLC. Adenocarcinoma of the lung contains the highest mutation frequency of LKB1, and LKB1 was also found to be mutated at lower frequencies in large cell and squamous cancer of the lung 15,16. Interestingly, a recent study in a mouse model shows that Lkb1-deficient lung adenocarcinoma (ADC) progressively transdifferentiates into squamous cell carcinoma (SCC) through pathologically mixed ADC-SCC intermediates 17. This finding immediately raised the following questions: (i) does human LKB1-mutant SCC also transdifferentiate from ADC, (ii) if so, does this transdifferentiation provides an advantage for tumor survival in the lung tissue microenvironment, and (iii) do human LKB1-mutant ADC and SCC have differential drug responses? Addressing the third question may have significant implications for the treatment approach to LKB1-mutated squamous lung cancer.
Epigenetic inactivation of LKB1
Although LKB1 is bona fide tumor suppressor gene, the lack of somatic LKB1 mutations in other major cancer types was puzzling. This led to the investigation of whether LKB1 can be inactivated in other cancer types through epigenetic modification. Initial analysis in 51 cancer cell lines indicated that three colorectal and one cervical carcinoma cell lines contain LKB1 promoter methylation. Similar analysis in 195 carcinomas of various tissue types revealed alterations in one colorectal carcinoma and three testicular carcinomas 18. The low frequency of LKB1 promoter methylation was also confirmed in colorectal cancer, hepatocellular carcinoma, and astrocytoma 19–21. The only exception reported is a recent study by a Korean group who found LKB1 promoter methylation in 13% of samples, and argued that LKB1 genetic and epigenetic alteration may vary depending on patient ethnicity 22. In general, LKB1 is not frequently inactivated in human cancers by epigenetic modification.
Biological inactivation of LKB1
Even though the epigenetic inactivation of LKB1 is a rare event in human cancer, a recent development is the discovery that the phosphorylation of AMPK by LKB1 is attenuated in melanoma cells with BRAF V600E activating mutation23,24. Mechanistically, the presence of BRAF V600E mutation activates MEK/ERK signaling and its downstream target RSK. The phosphorylation of LKB1 at Ser325 and Ser428 by ERK2 and p90RSK was shown to be sufficient to inactivate LKB1 as an energy sensor through the disruption of LKB1’s interaction with its downstream effect molecule AMPK 23. Interestingly, LKB1’s interaction with Strad and MO25 was not affected in this specific scenario, and it is unknown whether LKB1 is still capable of interacting with its other downstream targets in the presence of BRAF V600E mutation. In any case, this example begs the question whether LKB1 can interact with other cellular components, and whether similar types of protein-protein interactions are also capable of inhibiting LKB1 kinase function. If these types of inhibitory proteins are present in other tumor types, targeted approaches against LKB1-deficient tumors may also be applicable in these tumors.
The unanticipated use of AMPK activators against LKB1-null tumor cells
Metformin, phenformin and AICAR are commonly used as AMPK activators even though their mechanisms of activation are different. Metformin and phenformin treatments inhibit mitochondria complex I 25–27, which can lead to the depletion of intracellular ATP and a rise in AMP. The binding of AMP to γ-regulator domain promotes the phosphorylation of AMPK at threonine 172 by upstream kinases, such as LKB1 28. This binding also protects the enzyme against dephosphorylation by phosphatases and causes allosteric activation of AMPK 29. LKB1 is also required in the process of AMPK activation because the farnesylation of LKB1 was recently found to be required for the activation of AMPK by phenformin 30.
Extensive experimental data indicates that the activation of AMPK by metformin or phenformin has anti-proliferative activity in many cancer types, but the underlying mechanisms are not completely understood 31. mTOR is one of the downstream targets that is inhibited by AMPK, which leads to inhibition of protein synthesis and cell proliferation 32. Another hypothesis is that tumor cells have defects in various aspects of lipid metabolism, and they are dependent on de-regulated lipid metabolism. The activation of AMPK is capable of inhibiting lipid metabolism, thus suppressing cell proliferation33. In summary, current data indicate that AMPK-dependent growth inhibition by metformin and phenformin in human cancers is LKB1-dependent.
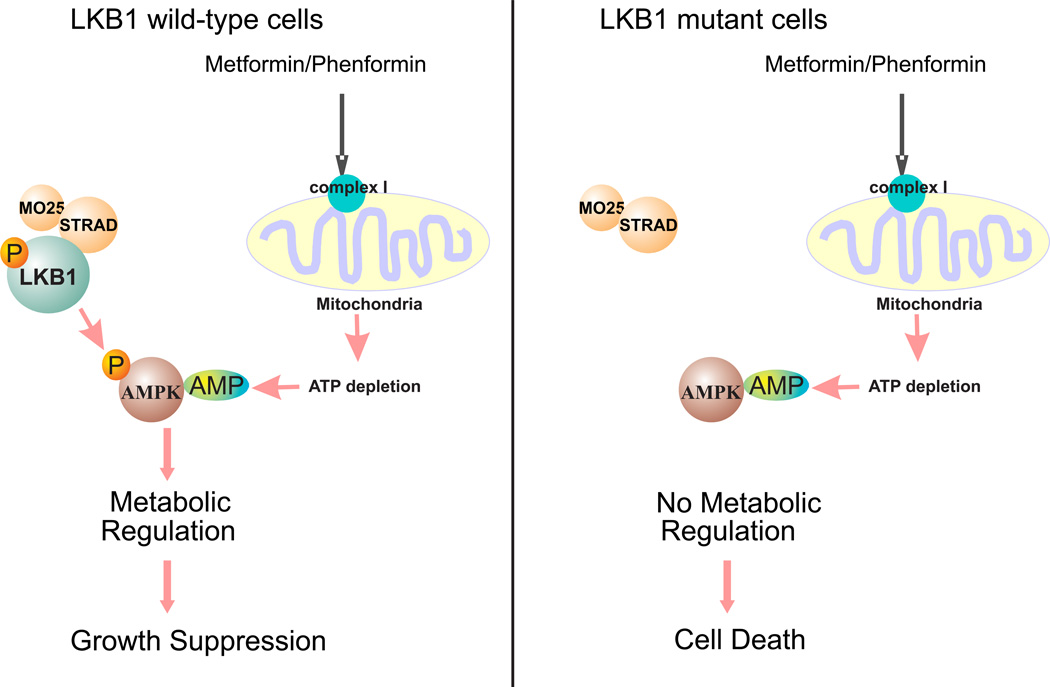
An unexpected finding from studies of metformin and phenformin is that treatment of LKB1-null cells with these reagents leads to cell death 27. AMPK is not a mediator in this process because activated AMPK cannot be detected after such treatment. Therefore, LKB1-dependent AMPK activation is not the cause of this apoptosis. It should be noted that the inhibition of mitochondria complex I and the depletion of intracellular ATP by metformin and phenformin are LKB1-independent, so these reagents generate energetic stress regardless of LKB1 status. The mechanism underlying phenformin-induced cell death in LKB1-null cells was recently proposed 27. It was hypothesized that phenformin-induced ATP depletion is usually detected by the LKB1/AMPK metabolic checkpoint, whose activation re-balances the energy consumption in cancer cells to promote cell survival. However, LKB1-null cells cannot sense this defect, and the lack of proper energy consumption control eventually leads to cell death (Figure 1). This hypothesis suggests that LKB1/AMPK plays a pro-survival role against phenformin-induced cell killing.
Figure 1.
The effects of AMPK activators in LKB1-wild type and mutant cells. Metformin and phenformin inhibit mitochondria complex I, which results in the depletion of intracellular ATP and increases in AMP. The binding of AMP to the AMPK activates AMPK kinase activity and its metabolic regulation function. The lack of LKB1 prevents the activation of AMPK by AMP. As a result, metformin and phenformin induced energetic stress cannot be properly detected in LKB1-mutant cells, which eventually lead to cell death.
5-aminoimidazole-4-carboxamide riboside (AICAR) is another commonly used AMPK activator. Inside a cell, AICAR is converted to AICAr monophosphate (ZMP) by adenosine kinase, and ZMP can act as an AMP analog to activate AMPK in an LKB1-dependent manner. Interestingly, the introduction of AICAR also induces caspase-3 cleavage in LKB1-null MEF cells and ovarian cancer cells 34,35, although the underlying mechanism(s) have not been explored. Even though AICAR-induced activation of AMPK has been shown to promote apoptosis in many cell lines, this type of apoptosis is usually dependent on the activation of AMPK 36,37. This is unlikely to be the mechanism in LKB1-null cells because genetic evidence shows that AMPK activation in response to treatment with AICAR is compromised in LKB1−/− MEFs, and can be restored following reconstitution of LKB1 38,39. Depending on the cell type, the introduction of AICAR may or may not lead to the depletion of ATP 40,41. Hence, it will be interesting to see in the future whether AICAR induces apoptosis in LKB1-null cells through a different mechanism, and whether LKB1/AMPK also provides a pro-survival role for cancer cells in this treatment setting.
Hyper-activated mTOR and ribosomal RNA synthesis in LKB1-null cells
mTOR is one of the earliest signaling pathways that was found to be negatively regulated by LKB1/AMPK signaling. Extensive experimental evidence indicates that mTOR-mediated protein translation is hyper-activated in cancer cells without LKB1, and various rapamycin analogs (rapalogs) have been developed to suppress the activity of mTORC1 (figure 2). This topic has been extensively reviewed elsewhere 32,42,43. Currently, these rapalogs (everolimus, sirolimus, temsirolimus) are in multiple phase 1–3 clinical trials, and NCT00178151 is a phase 2 trial that specifically evaluate the effects of everolimus in the treatment of advanced malignancies in patients with Peutz-Jeghers syndrome. Even though mTOR inhibitors have shown activities in some lung cancer patients, the underlying mechanism is still unknown. In a genetically engineered mouse model, invasive endometrial tumors derived from concurrent loss of Pten and LKB1 rely strongly on dysregulated mTOR signaling, and are hypersensitive to mTOR inhibition 44. Interestingly, the concurrent loss of Lkb1 and Pten also leads to mouse lung SCC that recapitulates the human disease 45, and it will be interesting to determine in the future whether mTOR inhibitors are specifically useful in patients with both LKB1 and PTEN mutations.
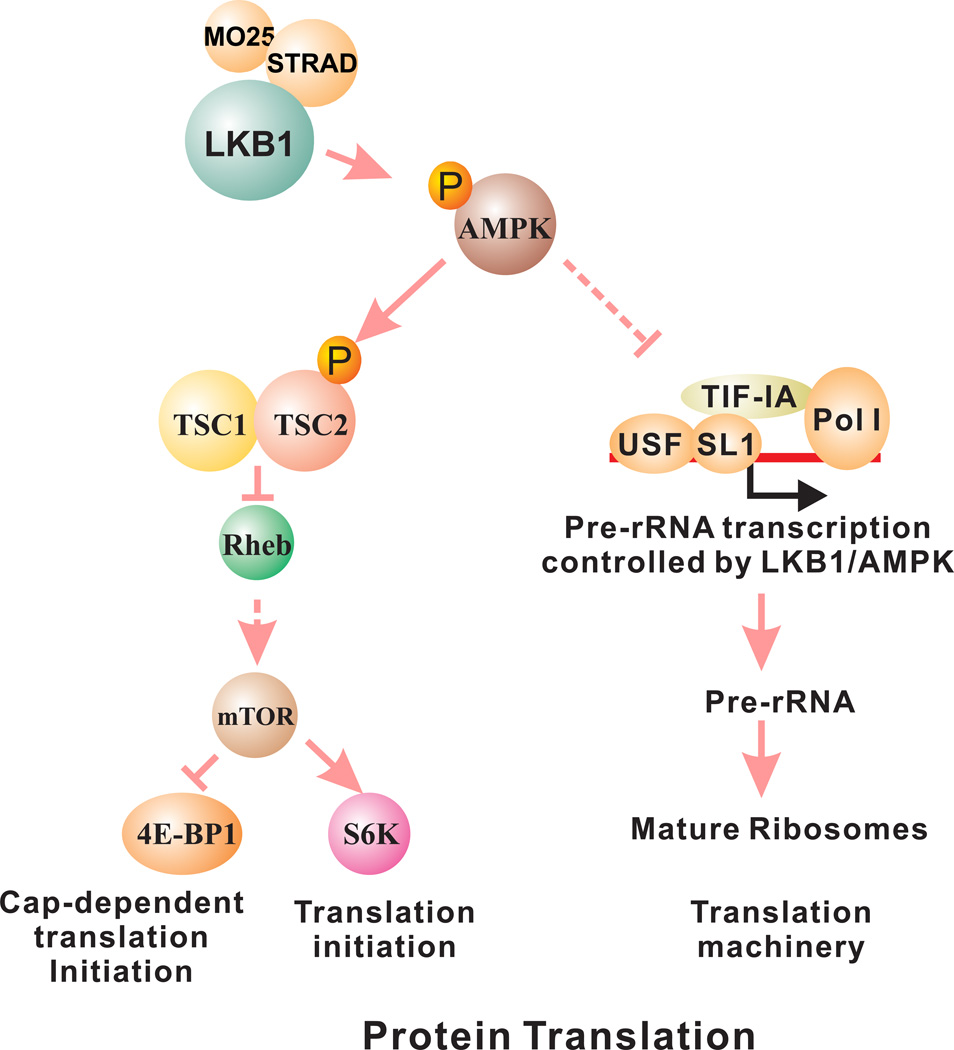
Figure 2.
AMPK regulates protein translation through its effects on mTOR and pre-rRNA synthesis. The activation of AMPK suppresses mTOR activity, thus interfering with translation initiation. AMPK also phosphorylates TIF-IA to prevent the assembly of pre-rRNA transcription initiation complex, thus prevent the synthesis of ribosome which is required for protein translation.
Another important component of protein synthesis is the ribosome. The transcription of ribosomal RNA (rRNA) genes by RNA polymerase I and the subsequent maturation processing of rRNA are essential steps in the generation of functional ribosomes. Several hereditary cancer syndromes, such as Blooms syndrome, Werner syndrome, and Shwachman-Diamond syndrome, have defects either in rDNA transcription or rRNA processing 46. In addition, hyperactive Pol I transcription was recently shown to be required for maintaining the malignant phenotype of B-lymphoma 47. The recruitment of TIF-IA/Pol-I complex to the rRNA promoter is essential for the formation of the rRNA transcription initiation complex 48. Emerging evidence indicates that AMPK is also involved in the regulation of pre-ribosomal RNA synthesis (Figure 2). First, mTOR phosphorylation of TIF-IA at Ser-44 activates and at Ser-199 inactivates rRNA transcription 49. Because AMPK negatively regulates mTOR, this is the first indirect evidence that AMPK is involved in the regulation of rRNA transcription. More recently, AMPK was found to directly phosphorylate TIF-IA at Ser-635, which disrupts the interaction of TIF-IA with transcription factor SL1 and prevents the assembly of the transcriptional initiation complex 50. This evidence indicated that AMPK is also a negative regulator of pre-rRNA synthesis. Hence, AMPK may negatively regulate protein synthesis through the inhibition of both mTOR and ribosome synthesis. Currently, it is unknown whether the negative regulation of pre-rRNA synthesis by AMPK is LKB1-dependent. If this is the case, reagents that inhibit rRNA transcription or rRNA maturation may also be used against LKB1-null cancers.
Sensitivity to Fak/Src inhibitors
The role of LKB1 in regulating lung cancer invasion and metastasis is an emerging research field. Early studies in Drosophila indicated that LKB1 is involved in regulation of cell polarity, and the loss of epithelial polarity may be related to epithelial to mesenchymal transition (EMT) 51. Direct evidence of LKB1’s involvement in cancer metastasis came from a study in a mutant Kras mouse model, in which the additional loss of Lkb1 accelerated tumor progression and promoted metastasis 16. This was a groundbreaking study because it provided in vivo evidence that the inactivation of LKB1 not only disrupts the regulation of mTOR signaling but also promotes cancer metastasis. A follow-up analysis in this mouse model indicated that SRC and FAK are activated in Lkb1-deficient primary and metastatic lung tumors 52. The negative correlation between LKB1 and FAK activation was also reported in human cancer cell lines where LKB1 was found to suppress FAK activity 53. These findings suggest that LKB1-null cancer cells may have hyper-activated SRC/FAK function, thus rendering them susceptible to SRC/FAK inhibition. Dasatinib is a Src inhibitor that is already in phase 1 and 2 clinical trials for various malignancies. Defactinib is a FAK inhibitor that is also in phase 1 and 2 clinical trials, one of which is a trial for Kras-mutant NSCLC patients. It will be interesting to see in the future whether these reagents can prevent the metastasis of LKB1-mutant tumors by targeting FAK or Src.
LKB1 as a sensor for DNA damage
A recent synthetic lethality screen revealed that the downregulation of DTYMK and Chk1 expression by RNAi preferentially induced cell killing in LKB1-null cells, and that LKB1-null cells accumulated more DNA damage than their isogenic LKB1-wild type counterparts 54. This finding suggested that LKB1 may play a role as a sensor for DNA damage, and that LKB1-mutant cells are defective in this DNA damage checkpoint. Consequently, these cells rely on Chk1’s function as the last defense against DNA damage, making Chk1 depletion a synthetic lethal combination with LKB1.
The puzzle here is how LKB1 acts as a sensor for DNA damage (Figure 3). The most obvious question is whether nuclear LKB1 plays a role in this process. LKB1 has two nuclear leading sequences and LKB1 expression is present in the nucleus. Furthermore, decreased nuclear LKB1 levels correlate with HNSCC metastasis, suggesting that nuclear LKB1 is capable of suppressing HNSCC 55. A similar phenomenon was found in breast cancer, where the presence of nuclear LKB1 is associated with increased overall survival and disease-free survival 56. Despite these interesting correlations, the nuclear function of LKB1 is still unknown.
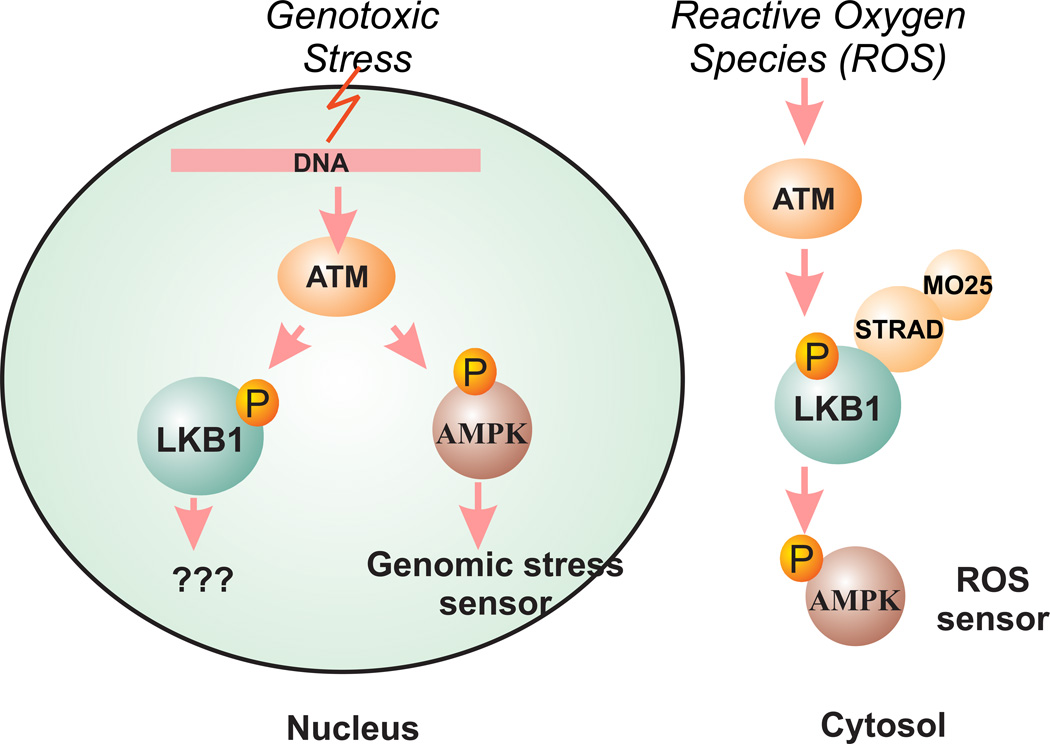
Figure 3.
The role of LKB1 as a ROS sensor but not a genomic stress sensor. Ionization radiation activates ATM and AMPK in a LKB1-independent manner to sense genomic stress. LKB1/AMPK is required for reactive oxygen species (ROS) to activate mTOR through cytosolic ATM.
First, there is no direct evidence indicating that LKB1 can function as a kinase in the nucleus. The kinase function of LKB1 is mainly limited to the cytosol as the LKB1/STRAD/MO25 complex is mostly found in the cytosol. Even though STRADα is capable of passive diffusion into the nucleus, its main function appears to be to relocalize LKB1 into the cytosol 57. Second, even though AMPK was recently proposed as a sensor for genomic stress 58, the phosphorylation of AMPK after ionizing radiation (IR) in the nucleus is mostly mediated by ATM and ATR 59,60 and is observed in both LKB1-wild type and mutant cell lines 59. Therefore, ionization-mediated AMPK response in the nucleus appears to be LKB1-independent. These findings, however, do not necessarily rule out LKB1 as a sensor for genetic damage because the N-terminus of ATM does interact with LKB1 61, and ATM is capable of phosphorylating LKB1 at Thr-366 62. Interestingly, recent data indicated that ATM negatively regulates mTORC1 and induces autophagy via the LKB1/AMPK pathway in the cytoplasm 63, and cytoplasmic ATM is capable of activating LKB1/AMPK in response to reactive oxygen species (ROS) 64. These data provided evidence that ATM and LKB1 may function as a sensor for oxidative stress in the cytosol. Because ROS is an important source of oxidative DNA damage 65, LKB1 may indirectly act as a sensor to control oxidative DNA damage. Consequently, reagents against DNA damage checkpoint proteins may be valuable for the treatment of LKB1-mutant cancer in the future.
Kras-LKB1 mutation-driven COPI addition
A recent study discovered that cancer cell lines with combined mutations in Kras and LKB1 are particuliarly sensitive to the depletion of coatomer complex I (COPI) subunits, such as ARCN1, COPB1 and COPA 66. These data suggest that Kras/LKB1 mutant cell lines are addicted to some functions mediated by COPI. COPI is involved in various membrane-trafficking events, and is best known for its role in coating vescles to be returned to the ER after they reach the cis or ER-Golgi intermeidate compartment, a.k.a. retrograde transport 67. However, the authors presented evidence that Kras/LKB1 mutant cells are addicted to COPI’s role in lysosome acidification. Lysosomes are cellular vesicles containing many different acid hydrolases, which are capable of degrading various biomolecules, including proteins, carbohydrates, lipids, and nucleic acid. The acidification of lysosomes is an important step in their maturation process because many lysosomal enzyme precursors can only be properly cleaved into their mature form at acidic pH, and most lysosomal hydrolases are active only at acidic pH. In addition, the acidity also denatures proteins to make them easier to digest. Kras/LKB1-null cells were found to have more acidic lysosomes in their steady-state organelle accumulation. Therefore, these cells may be selectively dependent upon lysosomal maturation. Consistent with this, the vacuolar ATPase inhibitor bafilomycin A1 and saliphenylhalamide A were found to have selective toxicity against Kras/LKB1-mutant cell lines. Even though these reagents have only been tested in the laboratory setting, future development of clinical grade vacuolar ATPase inhibitors may have important implications in the treatment of Kras/LKB1 mutant lung cancers.
Hyper-activated YAP1 as a therapeutic target in LKB1-mutant cancers
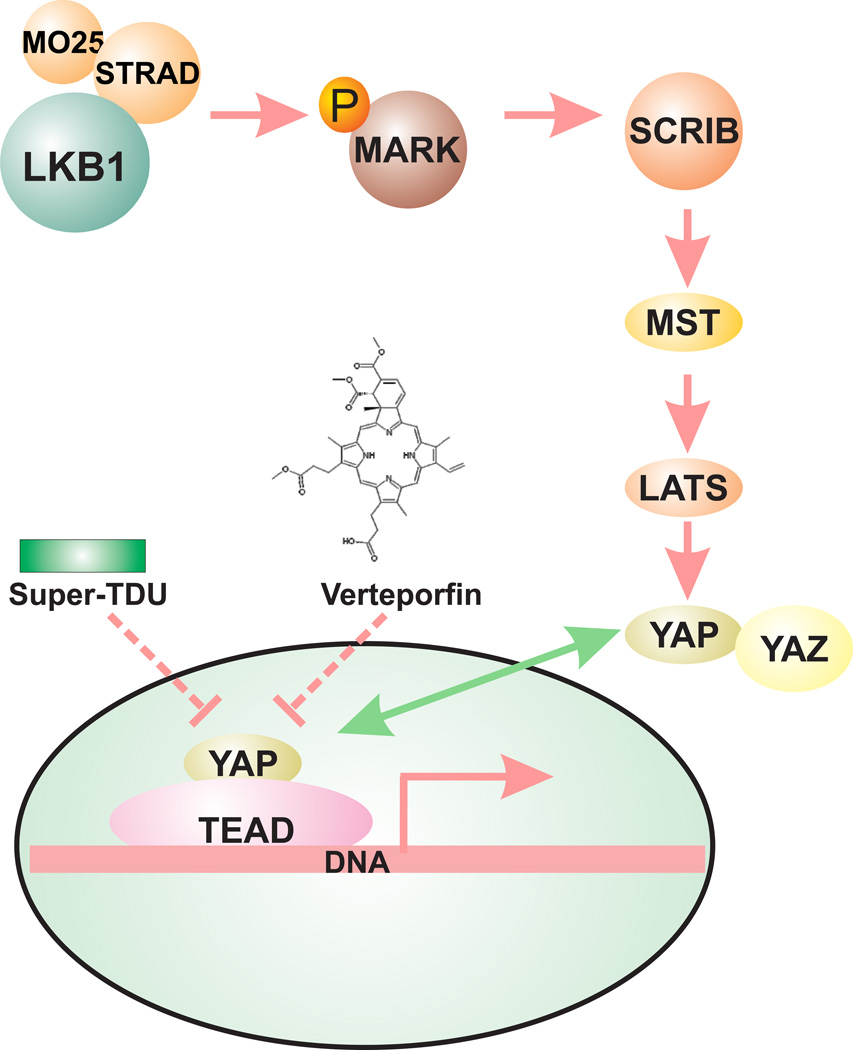
The Hippo pathway is an ancient developmental pathway that regulates organ size and proliferation. It inhibits cell proliferation upon high cell density or stress, and can act as a tumor suppressor pathway in many tissues. One of the main downstream targets that is suppressed by the Hippo pathway is YAP1 (Yes-associated protein 1), a transcription co-activator that associates with the TEAD family of transcription factors to promote cell proliferation 68. In some cancers, such as melanoma, YAP1 is amplified to act as an oncogene 69. YAP1 has not been reported to be amplified in lung cancer, but elevated nuclear YAP1 expression has been observed and it negatively correlates with patient survival and LKB1 expression 70–73. In addition, RNAi knockdown of LKB1 significantly enhanced the transcription of YAP1-activated genes, and the over-expression of LKB1 inhibits YAP1-dependent transcription. Interestingly, YAP1 has also been shown functionally essential since the depletion of YAP1 reduced the growth of LKB1 mutant tumors, and its mutant form (YAP-S217A) was able to significantly overcome all of LKB1’s tumor suppressive effects (73). LKB1 is a serine/threonine kinase. Even though LKB1 does not directly interact with YAP1, the over-expression of LKB1 correlates with an increase in the phosphorylation of YAP1 at Serine 127 and the degradation of YAP1 by the proteasome 72. RNAi analysis indicated that three LKB1 substrates (MARK1, 3 and 4) are capable of modulating YAP1-dependent transcription, and protein-protein interaction studies suggested that LKB1, MARK, MST1, LATS1 and SCRIB may form a Hippo regulator protein complex 73. Therefore, LKB1 is a suppressor of YAP1 function, and aberrant activation of YAP1 function may be another biological basis for the mutational inactivation of LKB1 in lung cancer. Because YAP1 is hyper-activated and is functionally essential in LKB1-mutant cells, therapeutic strategies against YAP1 activation may be beneficial in the treatment of LKB1-mutant tumors (Figure 4). Existing strategies include (i) the delivery of shRNA against YAP1, (ii) reagents that disrupt the YAP1/TEAD interaction (verteporfin) 74, and (iii) reagents that compete with YAP for its interaction with STEAD (Super-TDU) 75. These reagents are currently in preclinical development but they certainly represent an appealing strategy to eliminate LKB1-mutant cancers.
Figure 4.
LKB1 negatively regulates YAP1 function. LKB1 regulates SCRIB cellular localization through its phosphorylation of MARK. The proper cellular localization of SCRIB is required for the regulation of YAP phosphorylation by its upstream kinases, such as MST and LATS.
Vulnerability of LKB1-mutant cells to Hsp90 inhibitor, 17-AAG
The Genomics of Drug Sensitivity in Cancer (GDSC) recently published drug sensitivity data on 138 anticancer drugs in approximately 700 cancer cell lines 76, and LKB1-mutant cancer cell lines were found to be most sensitive to 17-AAG (http://www.cancerrxgene.org/translation/Gene/1367). 17-AAG (17-N-allylamino-17-demethoxygeldanamycin) is a derivative of geldanamyin, which was initially used to reveal a protein-protein interaction between LKB1 and Hsp90 77,78. Initially, Hsp90 and Cdc37 were found to bind specifically to the kinase domain of LKB1, and prevent ubiquitin-mediated LKB1 degradation by the proteasome 77,78. More recent data indicated that LKB1 in complex with Hsp90/Cdc37 has a longer half-life but lacks its autophosphorylation 79. Its kinase activity, however, can be transiently stimulated when it is released from Hsp90/Cdc37 79,80. The released LKB1 interacts with Hsp/Hsc70, which recruits the ubiquitin ligase CHIP to mark LKB1 for proteasome-mediated degradation 79. A G163D point mutation in LKB1 was previously found to weaken the interaction between Hsp90/Cdc37 77, but it is unclear whether the weakened LKB1/Hsp90/Cdc37 interaction is responsible for the increased cancer cell sensitivity to 17-AAG. Furthermore, most LKB1-mutant cancer cell lines contain nonsense or frameshift mutations that usually abolish the production of LKB1 protein. Once again, LKB1 appears to play an unknown protective role in cancer cells against the toxicity of 17-AAG, and additional research is required to delineate the molecular basis for 17-AAG sensitivity of LKB1 mutant tumors.
On a side note, GDSC screen also included many therapeutic reagents currently used for the treatment of human cancers, such as paclitaxel, docetaxel, cisplatin, etc. None of these reagents have a growth inhibitory bias against LKB1-mutant cancer cell lines at statistical significant level. Hence, there is no direct evidence at this time for a selective or preferential reduction of LKB1-mutant cells by conventional cancer therapy, further supporting the needs in developing novel therapeutic reagents against LKB1-mutant tumors.
Conclusions
Therapeutic strategies against LKB1-mutant cancer are emerging, and their discovery can be classified into three categories. The first group is based on the biological function of LKB1 in regulating other signaling pathways. LKB1 was found to negatively regulate the activities of mTOR, FAK/Src, and YAP1, and LKB1-mutant cells may become dependent on the hyper-activation of these signaling pathways. Hence, inhibitory reagents against the function of these proteins should be useful for the treatment of LKB1-mutant cells. However, these reagents are not specifically designed against LKB1-mutant cells because these pathways may be aberrantly activated by mutations in other genes. The second group is based on unbiased synthetic lethality screening, and targeted inhibitions of proteins, such as Dtymk and Chk1, preferentially induces the killing of LKB1-null cells. The third group is associated with the function of LKB1 as a sensor for a varieties of enviornment stresses, and cells with defective LKB1 appear unfit to detect such enviornmental insults. An interesting challenge in the future is to identify reagents, whose incorporation into the DNA, RNA or proteins will be detrimental for cell survival but can not be properly detected in LKB1-null cells. Such approach may lead to the development of unorthodox agents against LKB1-mutant cells which is not based on the inhibition of any proteins. It should be noted that these strategies may not be limited to cancer cells with LKB1 mutations because LKB1 can also be inactivated at the protein level by other proteins.
Acknowledgments
We would like to thank Dr. Anthea Hammond for editing this manuscript. WZ is an Anise McDaniel Brock Scholar, a Georgia Cancer Coalition Distinguished Cancer, and an American Cancer Society Research Scholar.
This work was supported in part by R01-CA140571, P01 CA116676, Anise McDaniel Brock Scholar fund to WZ, 1RO1CA142858 to A.M., and P30CA138292 to Winship Cancer Institute.
References
- 1.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386(6625):623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 2.Khuri FR, Nemunaitis J, Ganly I, et al. a controlled trial of intratumoral ONYX-015, a selectively-replicating adenovirus, in combination with cisplatin and 5-fluorouracil in patients with recurrent head and neck cancer. Nat. Med. 2000 Aug;6(8):879–885. doi: 10.1038/78638. [DOI] [PubMed] [Google Scholar]
- 3.O'Shea CC, Johnson L, Bagus B, et al. Late viral RNA export, rather than p53 inactivation, determines ONYX-015 tumor selectivity. Cancer Cell. 2004 Dec;6(6):611–623. doi: 10.1016/j.ccr.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 4.Sanchez-Cespedes M, Parrella P, Esteller M, et al. Inactivation of LKB1/STK11 is a common event in adenocarcinomas of the lung. Cancer Res. 2002 Jul 1;62(13):3659–3662. [PubMed] [Google Scholar]
- 5.Guldberg P, thor Straten P, Ahrenkiel V, Seremet T, Kirkin AF, Zeuthen J. Somatic mutation of the Peutz-Jeghers syndrome gene, LKB1/STK11, in malignant melanoma. Oncogene. 1999 Mar 4;18(9):1777–1780. doi: 10.1038/sj.onc.1202486. [DOI] [PubMed] [Google Scholar]
- 6.McCabe MT, Powell DR, Zhou W, Vertino PM. Homozygous deletion of the STK11/LKB1 locus and the generation of novel fusion transcripts in cervical cancer cells. Cancer Genet. Cytogenet. 2010 Mar;197(2):130–141. doi: 10.1016/j.cancergencyto.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baas AF, Boudeau J, Sapkota GP, et al. Activation of the tumour suppressor kinase LKB1 by the STE20-like pseudokinase STRAD. EMBO J. 2003 Jun 16;22(12):3062–3072. doi: 10.1093/emboj/cdg292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lizcano JM, Goransson O, Toth R, et al. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 2004 Feb 25;23(4):833–843. doi: 10.1038/sj.emboj.7600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rowan A, Churchman M, Jefferey R, Hanby A, Poulsom R, Tomlinson I. In situ analysis of LKB1/STK11 mRNA expression in human normal tissues and tumours. J. Pathol. 2000 Oct;192(2):203–206. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH686>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 10.Hemminki A, Markie D, Tomlinson I, et al. A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature. 1998 Jan 8;391(6663):184–187. doi: 10.1038/34432. [DOI] [PubMed] [Google Scholar]
- 11.Amos CI, Frazier ML, Wei C, McGarrity TJ. Peutz-Jeghers Syndrome. 1993 [Google Scholar]
- 12.Alessi DR, Sakamoto K, Bayascas JR. LKB1-dependent signaling pathways. Annu. Rev. Biochem. 2006;75:137–163. doi: 10.1146/annurev.biochem.75.103004.142702. [DOI] [PubMed] [Google Scholar]
- 13.Gill RK, Yang SH, Meerzaman D, et al. Frequent homozygous deletion of the LKB1/STK11 gene in non-small cell lung cancer. Oncogene. 2011 Sep 1;30(35):3784–3791. doi: 10.1038/onc.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang R, Zheng C, Sun Y, et al. Integrative genomic analysis reveals a high frequency of LKB1 genetic alteration in Chinese lung adenocarcinomas. J Thorac Oncol. 2014 Feb;9(2):254–258. doi: 10.1097/JTO.0000000000000056. [DOI] [PubMed] [Google Scholar]
- 15.Zhong D, Guo L, de Aguirre I, et al. LKB1 mutation in large cell carcinoma of the lung. Lung Cancer. 2006 Sep;53(3):285–294. doi: 10.1016/j.lungcan.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 16.Ji H, Ramsey MR, Hayes DN, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007 Aug 16;448(7155):807–810. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- 17.Han X, Li F, Fang Z, et al. Transdifferentiation of lung adenocarcinoma in mice with Lkb1 deficiency to squamous cell carcinoma. Nature communications. 2014;5:3261. doi: 10.1038/ncomms4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esteller M, Avizienyte E, Corn PG, et al. Epigenetic inactivation of LKB1 in primary tumors associated with the Peutz-Jeghers syndrome. Oncogene. 2000 Jan 6;19(1):164–168. doi: 10.1038/sj.onc.1203227. [DOI] [PubMed] [Google Scholar]
- 19.Trojan J, Brieger A, Raedle J, Esteller M, Zeuzem S. 5'-CpG island methylation of the LKB1/STK11 promoter and allelic loss at chromosome 19p13.3 in sporadic colorectal cancer. Gut. 2000 Aug;47(2):272–276. doi: 10.1136/gut.47.2.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu J, Zhang HY, Ma ZZ, Lu W, Wang YF, Zhu JD. Methylation profiling of twenty four genes and the concordant methylation behaviours of nineteen genes that may contribute to hepatocellular carcinogenesis. Cell Res. 2003 Oct;13(5):319–333. doi: 10.1038/sj.cr.7290177. [DOI] [PubMed] [Google Scholar]
- 21.Yu J, Zhang H, Gu J, et al. Methylation profiles of thirty four promoter-CpG islands and concordant methylation behaviours of sixteen genes that may contribute to carcinogenesis of astrocytoma. BMC Cancer. 2004 Sep 14;4:65. doi: 10.1186/1471-2407-4-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee SM, Choi JE, Na YK, et al. Genetic and epigenetic alterations of the LKB1 gene and their associations with mutations in TP53 and EGFR pathway genes in Korean non-small cell lung cancers. Lung Cancer. 2013 Aug;81(2):194–199. doi: 10.1016/j.lungcan.2013.04.013. [DOI] [PubMed] [Google Scholar]
- 23.Zheng B, Jeong JH, Asara JM, et al. Oncogenic B-RAF negatively regulates the tumor suppressor LKB1 to promote melanoma cell proliferation. Mol. Cell. 2009 Jan 30;33(2):237–247. doi: 10.1016/j.molcel.2008.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Esteve-Puig R, Canals F, Colome N, Merlino G, Recio JA. Uncoupling of the LKB1-AMPKalpha energy sensor pathway by growth factors and oncogenic BRAF. PLoS ONE. 2009;4(3):e4771. doi: 10.1371/journal.pone.0004771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El-Mir MY, Nogueira V, Fontaine E, Averet N, Rigoulet M, Leverve X. Dimethylbiguanide inhibits cell respiration via an indirect effect targeted on the respiratory chain complex I. J. Biol. Chem. 2000 Jan 7;275(1):223–228. doi: 10.1074/jbc.275.1.223. [DOI] [PubMed] [Google Scholar]
- 26.Owen MR, Doran E, Halestrap AP. Evidence that metformin exerts its antidiabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain. Biochem. J. 2000 Jun 15;348 Pt 3:607–614. [PMC free article] [PubMed] [Google Scholar]
- 27.Shackelford DB, Abt E, Gerken L, et al. LKB1 inactivation dictates therapeutic response of non-small cell lung cancer to the metabolism drug phenformin. Cancer Cell. 2013 Feb 11;23(2):143–158. doi: 10.1016/j.ccr.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oakhill JS, Chen ZP, Scott JW, et al. beta-Subunit myristoylation is the gatekeeper for initiating metabolic stress sensing by AMP-activated protein kinase (AMPK) Proc. Natl. Acad. Sci. U.S.A. 2010 Nov 9;107(45):19237–19241. doi: 10.1073/pnas.1009705107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanders MJ, Grondin PO, Hegarty BD, Snowden MA, Carling D. Investigating the mechanism for AMP activation of the AMP-activated protein kinase cascade. Biochem. J. 2007 Apr 1;403(1):139–148. doi: 10.1042/BJ20061520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Houde VP, Ritorto MS, Gourlay R, et al. Investigation of LKB1 Ser431 phosphorylation and Cys433 farnesylation using mouse knockin analysis reveals an unexpected role of prenylation in regulating AMPK activity. Biochem. J. 2014 Feb 15;458(1):41–56. doi: 10.1042/BJ20131324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Viollet B, Guigas B, Sanz Garcia N, Leclerc J, Foretz M, Andreelli F. Cellular and molecular mechanisms of metformin: an overview. Clin Sci (Lond) 2012 Mar;122(6):253–270. doi: 10.1042/CS20110386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou W, Marcus AI, Vertino PM. Dysregulation of mTOR activity through LKB1 inactivation. Chin J Cancer. 2013 Aug;32(8):427–433. doi: 10.5732/cjc.013.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lettieri Barbato D, Vegliante R, Desideri E, Ciriolo MR. Managing lipid metabolism in proliferating cells: New perspective for metformin usage in cancer therapy. Biochim. Biophys. Acta. 2014 Apr;1845(2):317–324. doi: 10.1016/j.bbcan.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 34.Shaw RJ, Kosmatka M, Bardeesy N, et al. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc. Natl. Acad. Sci. U.S.A. 2004 Mar 9;101(10):3329–3335. doi: 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nafz J, De-Castro Arce J, Fleig V, Patzelt A, Mazurek S, Rosl F. Interference with energy metabolism by 5-aminoimidazole-4-carboxamide-1-beta-Dribofuranoside induces HPV suppression in cervical carcinoma cells and apoptosis in the absence of LKB1. Biochem. J. 2007 May 1;403(3):501–510. doi: 10.1042/BJ20061053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim WH, Lee JW, Suh YH, et al. AICAR potentiates ROS production induced by chronic high glucose: roles of AMPK in pancreatic beta-cell apoptosis. Cell. Signal. 2007 Apr;19(4):791–805. doi: 10.1016/j.cellsig.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 37.Kim YM, Hwang JT, Kwak DW, Lee YK, Park OJ. Involvement of AMPK signaling cascade in capsaicin-induced apoptosis of HT-29 colon cancer cells. Ann. N.Y. Acad. Sci. 2007 Jan;1095:496–503. doi: 10.1196/annals.1397.053. [DOI] [PubMed] [Google Scholar]
- 38.Hawley SA, Boudeau J, Reid JL, et al. Complexes between the LKB1 tumor suppressor, STRADalpha/beta and MO25alpha/beta are upstream kinases in the AMP-activated protein kinase cascade. J Biol. 2003;2(4):28. doi: 10.1186/1475-4924-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moller DE. New drug targets for type 2 diabetes and the metabolic syndrome. Nature. 2001 Dec 13;414(6865):821–827. doi: 10.1038/414821a. [DOI] [PubMed] [Google Scholar]
- 40.Corton JM, Gillespie JG, Hawley SA, Hardie DG. 5-aminoimidazole-4-carboxamide ribonucleoside. A specific method for activating AMP-activated protein kinase in intact cells? Eur. J. Biochem. 1995 Apr 15;229(2):558–565. doi: 10.1111/j.1432-1033.1995.tb20498.x. [DOI] [PubMed] [Google Scholar]
- 41.Woollhead AM, Sivagnanasundaram J, Kalsi KK, et al. Pharmacological activators of AMP-activated protein kinase have different effects on Na+ transport processes across human lung epithelial cells. Br. J. Pharmacol. 2007 Aug;151(8):1204–1215. doi: 10.1038/sj.bjp.0707343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang S, Houghton PJ. Targeting mTOR signaling for cancer therapy. Curr Opin Pharmacol. 2003 Aug;3(4):371–377. doi: 10.1016/s1471-4892(03)00071-7. [DOI] [PubMed] [Google Scholar]
- 43.Wang X, Sun SY. Enhancing mTOR-targeted cancer therapy. Expert Opin Ther Targets. 2009 Oct;13(10):1193–1203. doi: 10.1517/14728220903225008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng H, Liu P, Zhang F, et al. A genetic mouse model of invasive endometrial cancer driven by concurrent loss of Pten and Lkb1 Is highly responsive to mTOR inhibition. Cancer Res. 2014 Jan 1;74(1):15–23. doi: 10.1158/0008-5472.CAN-13-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu C, Fillmore CM, Koyama S, et al. Loss of Lkb1 and Pten Leads to Lung Squamous Cell Carcinoma with Elevated PD-L1 Expression. Cancer Cell. 2014 Apr 30; doi: 10.1016/j.ccr.2014.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hannan KM, Sanij E, Rothblum LI, Hannan RD, Pearson RB. Dysregulation of RNA polymerase I transcription during disease. Biochim. Biophys. Acta. 2013 Mar-Apr;1829(3–4):342–360. doi: 10.1016/j.bbagrm.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bywater MJ, Poortinga G, Sanij E, et al. Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53. Cancer Cell. 2012 Jul 10;22(1):51–65. doi: 10.1016/j.ccr.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao J, Yuan X, Frodin M, Grummt I. ERK-dependent phosphorylation of the transcription initiation factor TIF-IA is required for RNA polymerase I transcription and cell growth. Mol. Cell. 2003 Feb;11(2):405–413. doi: 10.1016/s1097-2765(03)00036-4. [DOI] [PubMed] [Google Scholar]
- 49.Mayer C, Zhao J, Yuan X, Grummt I. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev. 2004 Feb 15;18(4):423–434. doi: 10.1101/gad.285504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoppe S, Bierhoff H, Cado I, et al. AMP-activated protein kinase adapts rRNA synthesis to cellular energy supply. Proc. Natl. Acad. Sci. U.S.A. 2009 Oct 20;106(42):17781–17786. doi: 10.1073/pnas.0909873106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marcus AI, Zhou W. LKB1 regulated pathways in lung cancer invasion and metastasis. J Thorac Oncol. 2010 Dec;5(12):1883–1886. doi: 10.1097/JTO.0b013e3181fbc28a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carretero J, Shimamura T, Rikova K, et al. Integrative genomic and proteomic analyses identify targets for Lkb1-deficient metastatic lung tumors. Cancer Cell. 2010 Jun 15;17(6):547–559. doi: 10.1016/j.ccr.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kline ER, Shupe J, Gilbert MM, Zhou W, Marcus AI. LKB1 represses focal adhesion kinase (FAK) signaling via a FAK-LKB1 complex to regulate FAK site maturation and directional persistence. J. Biol. Chem. 2013 May 1; doi: 10.1074/jbc.M112.444620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Y, Marks K, Cowley GS, et al. Metabolic and functional genomic studies identify deoxythymidylate kinase as a target in LKB1-mutant lung cancer. Cancer Discov. 2013 Aug;3(8):870–879. doi: 10.1158/2159-8290.CD-13-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kline ER, Muller S, Pan L, Tighiouart M, Chen ZG, Marcus AI. Localization-specific LKB1 loss in head and neck squamous cell carcinoma metastasis. Head Neck. 2011 Oct;33(10):1501–1512. doi: 10.1002/hed.21638. [DOI] [PubMed] [Google Scholar]
- 56.Bouchekioua-Bouzaghou K, Poulard C, Rambaud J, et al. LKB1 when associated with methylatedERa is a marker of bad prognosis in breast cancer. Int. J. Cancer. 2014 Feb 12; doi: 10.1002/ijc.28781. [DOI] [PubMed] [Google Scholar]
- 57.Dorfman J, Macara IG. STRADalpha regulates LKB1 localization by blocking access to importin-alpha, and by association with Crm1 and exportin-7. Mol. Biol. Cell. 2008 Apr;19(4):1614–1626. doi: 10.1091/mbc.E07-05-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanli T, Steinberg GR, Singh G, Tsakiridis T. AMP-activated protein kinase (AMPK) beyond metabolism: a novel genomic stress sensor participating in the DNA damage response pathway. Cancer Biol Ther. 2014 Feb;15(2):156–169. doi: 10.4161/cbt.26726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanli T, Rashid A, Liu C, et al. Ionizing radiation activates AMP-activated kinase (AMPK): a target for radiosensitization of human cancer cells. Int. J. Radiat. Oncol. Biol. Phys. 2010 Sep 1;78(1):221–229. doi: 10.1016/j.ijrobp.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 60.Amatya PN, Kim HB, Park SJ, et al. A role of DNA-dependent protein kinase for the activation of AMP-activated protein kinase in response to glucose deprivation. Biochim. Biophys. Acta. 2012 Dec;1823(12):2099–2108. doi: 10.1016/j.bbamcr.2012.08.022. [DOI] [PubMed] [Google Scholar]
- 61.Fernandes N, Sun Y, Chen S, et al. DNA damage-induced association of ATM with its target proteins requires a protein interaction domain in the N terminus of ATM. J. Biol. Chem. 2005 Apr 15;280(15):15158–15164. doi: 10.1074/jbc.M412065200. [DOI] [PubMed] [Google Scholar]
- 62.Sapkota GP, Deak M, Kieloch A, et al. Ionizing radiation induces ataxia telangiectasia mutated kinase (ATM)-mediated phosphorylation of LKB1/STK11 at Thr-366. Biochem. J. 2002 Dec 1;368(Pt 2):507–516. doi: 10.1042/BJ20021284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alexander A, Cai SL, Kim J, et al. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc. Natl. Acad. Sci. U.S.A. 2010 Mar 2;107(9):4153–4158. doi: 10.1073/pnas.0913860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alexander A, Walker CL. Differential localization of ATM is correlated with activation of distinct downstream signaling pathways. Cell Cycle. 2010 Sep 15;9(18):3685–3686. doi: 10.4161/cc.9.18.13253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scott TL, Rangaswamy S, Wicker CA, Izumi T. Repair of oxidative DNA damage and cancer: recent progress in DNA base excision repair. Antioxidants & redox signaling. 2014 Feb 1;20(4):708–726. doi: 10.1089/ars.2013.5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim HS, Mendiratta S, Kim J, et al. Systematic identification of molecular subtype-selective vulnerabilities in non-small-cell lung cancer. Cell. 2013 Oct 24;155(3):552–566. doi: 10.1016/j.cell.2013.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dice JF. lysosomal pathways for protein degradation. Eurekah.com Landes Bioscience. 2000 [Google Scholar]
- 68.Johnson R, Halder G. The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nature reviews. Drug discovery. 2014 Jan;13(1):63–79. doi: 10.1038/nrd4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Menzel M, Meckbach D, Weide B, et al. In melanoma, Hippo signaling is affected by copy number alterations and YAP1 overexpression impairs patient survival. Pigment cell & melanoma research. 2014 Apr 4; doi: 10.1111/pcmr.12249. [DOI] [PubMed] [Google Scholar]
- 70.Steinhardt AA, Gayyed MF, Klein AP, et al. Expression of Yes-associated protein in common solid tumors. Hum. Pathol. 2008 Nov;39(11):1582–1589. doi: 10.1016/j.humpath.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Y, Dong Q, Zhang Q, Li Z, Wang E, Qiu X. Overexpression of yesassociated protein contributes to progression and poor prognosis of non-smallcell lung cancer. Cancer science. 2010 May;101(5):1279–1285. doi: 10.1111/j.1349-7006.2010.01511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nguyen HB, Babcock JT, Wells CD, Quilliam LA. LKB1 tumor suppressor regulates AMP kinase/mTOR-independent cell growth and proliferation via the phosphorylation of Yap. Oncogene. 2013 Aug 29;32(35):4100–4109. doi: 10.1038/onc.2012.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mohseni M, Sun J, Lau A, et al. A genetic screen identifies an LKB1-MARK signalling axis controlling the Hippo-YAP pathway. Nat Cell Biol. 2014 Jan;16(1):108–117. doi: 10.1038/ncb2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu-Chittenden Y, Huang B, Shim JS, et al. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012 Jun 15;26(12):1300–1305. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jiao S, Wang H, Shi Z, et al. A peptide mimicking VGLL4 function acts as a YAP antagonist therapy against gastric cancer. Cancer Cell. 2014 Feb 10;25(2):166–180. doi: 10.1016/j.ccr.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 76.Yang W, Soares J, Greninger P, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013 Jan;41(Database issue):D955–D961. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nony P, Gaude H, Rossel M, Fournier L, Rouault JP, Billaud M. Stability of the Peutz-Jeghers syndrome kinase LKB1 requires its binding to the molecular chaperones Hsp90/Cdc37. Oncogene. 2003 Dec 11;22(57):9165–9175. doi: 10.1038/sj.onc.1207179. [DOI] [PubMed] [Google Scholar]
- 78.Boudeau J, Deak M, Lawlor MA, Morrice NA, Alessi DR. Heat-shock protein 90 and Cdc37 interact with LKB1 and regulate its stability. Biochem. J. 2003 Mar 15;370(Pt 3):849–857. doi: 10.1042/BJ20021813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xu W, Neckers L. The double edge of the HSP90-CDC37 chaperone machinery: opposing determinants of kinase stability and activity. Future Oncol. 2012 Aug;8(8):939–942. doi: 10.2217/fon.12.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gaude H, Aznar N, Delay A, et al. Molecular chaperone complexes with antagonizing activities regulate stability and activity of the tumor suppressor LKB1. Oncogene. 2012 Mar 22;31(12):1582–1591. doi: 10.1038/onc.2011.342. [DOI] [PubMed] [Google Scholar]
- 81.Andrade-Vieira R, Xu Z, Colp P, Marignani PA. Loss of LKB1 expression reduces the latency of ErbB2-mediated mammary gland tumorigenesis, promoting changes in metabolic pathways. PLoS One. 2013;8(2):e56567. doi: 10.1371/journal.pone.0056567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gao Y, Xiao Q, Ma H, et al. LKB1 inhibits lung cancer progression through lysyl oxidase and extracellular matrix remodeling. Proc. Natl. Acad. Sci. U.S.A. 2010 Nov 2;107(44):18892–18897. doi: 10.1073/pnas.1004952107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sausville E, Lorusso P, Carducci M, et al. Phase I dose-escalation study of AZD7762, a checkpoint kinase inhibitor, in combination with gemcitabine in US patients with advanced solid tumors. Cancer Chemother. Pharmacol. 2014 Mar;73(3):539–549. doi: 10.1007/s00280-014-2380-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Seto T, Esaki T, Hirai F, et al. Phase I, dose-escalation study of AZD7762 alone and in combination with gemcitabine in Japanese patients with advanced solid tumours. Cancer Chemother. Pharmacol. 2013 Sep;72(3):619–627. doi: 10.1007/s00280-013-2234-6. [DOI] [PubMed] [Google Scholar]