Abstract
Geographic patterns of genetic variation are shaped by multiple evolutionary processes, including genetic drift, migration, and natural selection. Switchgrass (Panicum virgatum L.) has strong genetic and adaptive differentiation despite life history characteristics that promote high levels of gene flow and can homogenize intraspecific differences, such as wind-pollination and self-incompatibility. To better understand how historical and contemporary factors shape variation in switchgrass, we use genotyping-by-sequencing to characterize switchgrass from across its range at 98,042 SNPs. Population structuring reflects biogeographic and ploidy differences within and between switchgrass ecotypes and indicates that biogeographic history, ploidy incompatibilities, and differential adaptation each have important roles in shaping ecotypic differentiation in switchgrass. At one extreme, we determine that two Panicum taxa are not separate species but are actually conspecific, ecologically divergent types of switchgrass adapted to the extreme conditions of coastal sand dune habitats. Conversely, we identify natural hybrids among lowland and upland ecotypes and visualize their genome-wide patterns of admixture. Furthermore, we determine that genetic differentiation between primarily tetraploid and octoploid lineages is not caused solely by ploidy differences. Rather, genetic diversity in primarily octoploid lineages is consistent with a history of admixture. This suggests that polyploidy in switchgrass is promoted by admixture of diverged lineages, which may be important for maintaining genetic differentiation between switchgrass ecotypes where they are sympatric. These results provide new insights into the mechanisms shaping variation in widespread species and provide a foundation for dissecting the genetic basis of adaptation in switchgrass.
Keywords: GBS, polyploidy, ecotypes, perennial grass
Introduction
A major goal of evolutionary biology is to understand how genetic variation is generated, shaped, and maintained within a species. Gene flow and recombination act to homogenize a species, while a myriad of processes, including mutation, range shifts, and natural selection, counteract those homogenizing forces to generate genetic differences (Hewitt 1996, Novembre and Di Rienzo 2009, Nosil et al. 2009). Many widespread, outcrossing plant species have strong population structure (Lowry et al. 2008, Eckert et al. 2010, Zalapa et al. 2011) despite the potential for high levels of gene flow to homogenize genomic differences throughout these species (ex: Keller et al. 2010, Prunier et al. 2011), making these species appealing systems for understanding how different mechanisms can oppose gene flow to generate spatial and genome-wide patterns of variation within a species.
Diversity in widespread plants is affected by multiple factors, each of which leaves a signature on patterns of intraspecific variation. For instance, many widespread species underwent major range expansions and contractions during the Pleistocene glacial cycles, and contemporary population structure often reflects this biogeographic history, such as genetic divisions or geographic patterns that originated from these historical range shifts (Hewitt 1996, Soltis et al. 2006). Adaptive differences can also greatly influence patterns of variation, particularly in species with different ecotypes, where differential adaptation can reduce gene flow via immigrant and hybrid inviability and thus promote genetic differentiation between ecotypes (Porter 1966, Lowry et al. 2008). Additionally, in many plants, genomic events such as polyploidy and chromosomal rearrangements are associated with intra- and interspecific reproductive isolation and differentiation (Soltis et al. 2007, Rieseberg et al. 1995, Lowry and Willis 2010).
Polyploidy is of particular interest because of the major impact it can have on gene flow in a species. Multiple ploidy levels exist within many species, and gene flow between ploidy levels is restricted by the low fitness of the progeny produced by crosses between ploidy levels (Martinez-Reyna and Vogel 2002, Henry et al. 2005). This reproductive isolation occurs immediately upon polyploidization, thus polyploidy can generate differentiation independent of geographic allopatry (Coyne and Orr 2004). In addition, physiological and genetic effects of polyploidy may directly result in adaptive differentiation within a species (Otto and Whitton 2000). However, the role of polyploidy in shaping intraspecific diversity is poorly understood because ploidy differences often correlate with adaptive and geographic patterns, and distinguishing the individual effects of each factor on patterns of diversity remains a major challenge (Levin 1983, Ramsey and Shemske 2002). There may also be some aspects of polyploidy that can only be better understood by incorporating information about the other factors that shape diversity. For example, competition between cytotypes and fitness costs from interploidy matings favor the exclusion of new cytotypes (Levin 1975, Petit et al. 1999); therefore other factors may have key roles in the establishment and maintenance of multiple cytotypes that is seen in many species. Thus, by characterizing the association of species-wide population structure with ploidy, geography, phenotype, and habitat, we can assess the effects of the multiple factors that affect gene flow and examine how they combine to generate patterns of variation.
In the perennial North American grass switchgrass (Panicum virgatum L.), ploidy is one of the many factors that shapes variation across its extensive native range. Switchgrass is a foundational species for habitat restoration, emerging bioenergy feedstock, and forage crop (Sanderson 1996), and the resources developed for these purposes, including extensive germplasm collections, make switchgrass an attractive system for examining how diversity is shaped throughout a widespread species. Switchgrass is divided into two clades, the lowland and upland ecotypes, which are genetically distinct despite substantial sympatry in their distributions (Zhang et al. 2011b). The geographic distribution of genetic differences between and within the ecotypes suggests that switchgrass was periodically partitioned into separate refugia during glacial maxima, with some genetic differences resulting from those periods of allopatry persisting until today (McMillan 1959, Zhang et al. 2011b). The lowland and upland ecotypes are adapted to different edaphic conditions and have low viability in the habitat of the other ecotype (Porter 1966), and throughout the species, there is strong local adaptation to growing season (McMillan 1964). In addition, switchgrass has multiple ploidy levels (Costich et al. 2010), and a ploidy-related incompatibility system restricts gene flow between ploidy levels (Martinez-Reyna and Vogel 2002). The lowland ecotype is primarily tetraploid while the upland ecotype has both tetraploid and octoploid lineages (Costich et al. 2010), thus ploidy differences may be important in shaping differences within and between switchgrass ecotypes. Furthermore, switchgrass hybridizes with its sister species, Panicum amarum and P. amarulum, in portions of the range (Palmer 1975, Barkworth et al. 2007). However, the roles and relative importance of these different factors in shaping the overall diversity in switchgrass remains poorly understood. For instance, ploidy may have a major impact on diversification and the patterns of gene flow in switchgrass, but key details about ploidy are not known, such as the timing of polyploidization in different lineages or the frequency of natural ploidy shifts. Thus it is difficult to decipher the effect of ploidy on generating and maintaining diversity in the species.
Recent studies examining genomic diversity in switchgrass have begun to better resolve patterns of population structure (Morris et al. 2011, Lu et al. 2013). However, these studies have focused primarily on the upland ecotype, so patterns of genomic diversity across the species and within the lowland ecotype have been largely unexplored. Therefore, we use genotyping-by-sequencing (GBS; Elshire et al. 2011) to genotype more than 100 switchgrass samples from across the US range, as well as three related Panicum taxa at 98,042 SNPs. With these data, we can better understand how geography, adaptive differences, and ploidy shape diversity throughout this ecologically and economically important grass.
Materials and Methods
Plant materials
To survey species-wide patterns of diversity in switchgrass, we genotyped 123 Panicum virgatum samples from 41 populations (Tables 1 and S1). The switchgrass plants used in this study represent collections from remnant habitats and from cultivated varieties which are seed increases of source-identified collections that have undergone few generations of amplification and are considered good representations of the native genetic variation from original collection locations (Zalapa et al. 2011). These sample included two di-haploids, which have only one copy of each chromosome from each sub-genome (Young et al. 2010) and should have no heterozygosity. We also included one sample each of P. amarum, P. amarulum, and P. anceps. The P. amarum and P. amarulum lineages are alternately referred to as either separate species or subspecies of the same taxon (P. amarum; Barkworth et al. 2007, Triplett et al. 2012), and their taxonomic definition is further complicated by reports of hybridization between P. amarulum and switchgrass (Palmer 1975). We included the P. amarum and P. amarulum samples, as well as a sample from a more distantly related species, P. anceps (Aliscioni et al. 2003), to assess their relationships with switchgrass and as outgroups for the within-switchgrass comparisons.
Table 1.
Population Information
| Population Name | n | State | Latitude | Longitude | Inferred Gene Pools*1 | Population Type*2 |
Seed Source*3 |
|---|---|---|---|---|---|---|---|
| Rocky Run 1 | 2 | WI | 43.47 | −89.43 | Upland Northern Great Plains | Wild | SW112 |
| Hwy 59 | 4 | WI | 42.9 | −87.55 | Upland Northern Great Plains | Wild | SW127 |
| Bald Bluff | 3 | WI | 42.85 | −88.63 | Upland Northern Great Plains Lowland Atlantic Coastal Plain;Upland Midwest;Upland |
Wild | SW128 |
| Staten Island | 5 | NY | 40.5859 | −74.1482 | Eastern Savanna | Wild | SW781 |
| Blackwell | 7 | OK | 35.963 | −97.07 | Upland Northern Great Plains Upland Eastern Savanna;Mixed |
SIC | PI 421520;ECS |
| Cave-in-Rock | 6 | IL | 37.47 | −88.1658 | Upland Upland Northern Great Plains;Upland Eastern |
SIC | PI 469228;ECS |
| Rt 72/563 NJ | 4 | NJ | 39.817 | −74.533 | Savanna;Mixed Upland | Wild | ECS-1 |
| Howard | 1 | IN | 40.4508 | −86.1281 | Upland Northern Great Plains | Wild | SW33 |
| Waterford | 2 | WI | 42.78 | −88.3 | NA | Wild | SW123 |
| NRCS 9064224 | 1 | IN | 40.481 | −86.22 | Mixed Upland | Wild | NRCS-PMC |
| Columbiana | 1 | OH | 40.616 | −80.695 | Upland Eastern Savanna | Wild | SW64 |
| Wadena | 1 | MN | 46.4433 | −95.1349 | Upland Northern Great Plains | Wild | SW60 |
| Chiwaukee 1 | 1 | WI | 42.55 | −87.8 | Upland Midwest | Wild | SW124 |
| NRCS 9084291 | 2 | MI | 42.983 | −86.059 | Upland Midwest | Wild | NRCS-PMC |
| Dacotah | 3 | ND | 46.3845 | −100.9398 | Upland Eastern Savanna | SIC | NRCS-PMC |
| Pathfinder | 2 | KS | 39.82 | −98.48 | Upland Northern Great Plains Upland Eastern Savanna;Mixed |
SIC | USDA-ARS |
| Shelter | 4 | WV | 39.396 | −81.199 | Upland | SIC | NRCS-PMC |
| Sunburst | 3 | SD | 42.872 | −97.3957 | Upland Northern Great Plains | Bred | SDCIA;ECS |
| Toledo, OH | 1 | OH | 41.583 | −83.667 | Upland Northern Great Plains | Wild | ECS-2 |
| Allegheny River, PA | 1 | PA | 40.95 | −79.617 | Upland Eastern Savanna | Wild | ECS-10 |
| Hoffman | 1 | NC | 35.0306 | −79.5456 | Lowland/Upland Hybrid | Wild | PI 315723 |
| Sprewell Bluff | 3 | GA | 32.899 | −84.436 | Lowland/Upland Hybrid | Wild | UGA-SPB |
| Kanlow | 7 | OK | 35.3288 | −96.2408 | Lowland Southern Great Plains | SIC | PI 421521 |
| AW-314/MS-155 | 1 | AR | 35.4266 | −91.836 | Lowland Southern Great Plains | Wild | PI 421999 |
| PMT-785 | 4 | TX | 29.443 | −96.94 | Lowland Western Gulf Coast | Wild | PI 422003 |
| T 2086 | 3 | NC | 34.2358 | −77.9412 | Lowland Atlantic Coastal Plain | Wild | PI 476290 |
| Oscar Scherer S.P. | 2 | FL | 27.1859 | −82.4565 | Lowland Atlantic Coastal Plain | Wild | UGA-OSP |
| Pasco County | 1 | FL | 28.33 | −82.42 | Upland Midwest | Wild | UGA-PCF |
| Wabasso | 2 | FL | 27.747 | −80.435 | Lowland Atlantic Coastal Plain | Wild | PI 422000 |
| Chippewa | 3 | MN | 45.52 | −95.307 | Upland Northern Great Plains | Wild | SW48 |
| Jackson | 3 | MI | 42.2537 | −84.3101 | Upland Midwest | Wild | SW43 |
| Ipswich Prairie 2 | 4 | WI | 42.57 | −90.4 | Upland Midwest | Wild | SW115 |
| Albany, NY | 4 | NY | 42.717 | −73.833 | Upland Northern Great Plains | Wild | ECS-12 |
| Pangburn | 5 | AR | 35.4266 | −91.836 | Lowland Southern Great Plains | Wild | PI 414065 |
| BN-12323-69 | 5 | KS | 38.81 | −98.27 | Upland Midwest | Wild | PI 414070 |
| Panicum amarum | 1 | NA | NA | NA | Lowland | Wild | NA |
| Panicum amarulum | 1 | NA | NA | NA | Lowland Atlantic Coastal Plain | Wild | NA |
| Panicum anceps | 1 | NA | NA | NA | NA | Wild | NA |
| Alamo | 1 | TX | 28.3305 | −98.1163 | Lowland Southern Great Plains | SIC | ECS |
| Indiana Dunes S.P. | 16 | IN | 41.6582 | −87.0577 | Upland Midwest | Wild | PPG-IDSP |
| Forestburg | 1 | SD | 44.022 | −98.105 | Upland Northern Great Plains | SIC | ECS |
| Shawnee | 1 | IL | 37.47 | −88.1658 | Upland Eastern Savanna | Bred | ECS |
| Southlow | 1 | MI | NA | NA | Mixed Upland | Ecopool | NRCS-PSMC |
: based on samples above read count cutoff
: Bred, a product of one or more cycles of selection and breeding; SIC, source-identified cultivar derived from a random seed increase without selection and breeding; Wild, seed harvested from remnant population.
: USDA-ARS, switchgrass breeding program (Lincoln, NE); SDCIA, South Dakota Crop Improvement Association (Brookings, SD); NRCS-PMC, NRCS Plant Materials Centers (Bismarck, ND; Rose Lake, MI; Big Flats, NY; Cape May, NJ; Americus, GA; Coffeeville, MS); PI-xxxxxx, NRCS-GRIN; Germplasm Resources Information Network, USDA-ARS (Beltsville, MD); ECS-xx, Ernst Conservation Seeds (Meadville, PA); SWxxx, seeds collected directly from prairie remnant site and processed in Madison, WI; UGA-xxx, seeds collected directly from prairie remnant site and processed in in Athens, GA; PPG-IDSP seeds collected directly from remnant habitat and processed in Chicago, IL.
Ploidy and Phenotypic Data
Switchgrass is allopolyploid with a base chromosome count of 9, and the two most common ploidy levels are tetraploid (2n = 4X = 36) and octoploid (2n = 8X = 72; Costich et al. 2010). Tetraploid switchgrass shows disomic inheritance and can be genotyped like a diploid, with three genotype classes of AA, AB, BB (Okada et al. 2010). Octoploid switchgrass has sets of four homologous chromosomes and therefore has with five potential genotype classes of AAAA, AAAB, AABB, ABBB, BBBB. Ploidy data was previously obtained using flow cytometry for a majority of the samples (Zhang et al. 2011a, Table S1). For others, ploidy information was inferred from previously characterized cultivars with well-known and stable ploidy levels (Costich et al. 2010, Zalapa et al. 2011, Lu et al. 2013). For the IDSP population, ploidy was inferred from flow cytometry of a daughter plant of one sample (Personal communication, D. Lowry). We do not have flow cytometry data for the three other Panicum samples. Panicum amarum and P. amarulum are generally hexaploid and tetraploid, respectively, but there is evidence of multiple ploidy levels in these lineages (Triplett et al. 2012). The phenotypes of the samples were characterized as "Lowland Ecotype," "Upland Ecotype," or "Intermediate" based on traits that are characteristic of each ecotype, including plant architecture and leaf color (Zhang et al. 2011a).
Genotyping-By-Sequencing
Seven pooled genotyping-by-sequencing lanes, each composed of 36 multiplexed samples were sequenced on an Illumina GA-IIx sequencer (Illumina, San Diego, CA, USA) using 100bp paired-end reads. Technical replicates of each sample were sequenced in separate libraries. GBS library preparation was adapted from Elshire et al. (2011), using PstI as the restriction enzyme. Additional information about DNA extraction, library preparation, and sequencing adapters is in the supporting material. One subtle but important improvement in library construction was to amplify and normalize individual libraries before pooling and sequencing. This reduced the range in coverage per sample by nearly an order of magnitude.
Custom R-scripts (R Development Core Team, 2010) were used to process and assign sequencing reads to the appropriate sample based on the sample-specific DNA barcode at the beginning of each first-end sequencing read. Any reads containing sequence from the sequencing adapters or PCR primers were omitted. Candidate SNPs were identified according to Morris et al. (2011). Any loci with SNPs at three or more consecutive positions likely contain indels and were removed. We also excluded any SNPs more than 50 bp from a restriction site to remove potential artifacts due to lower quality at the ends of sequencing reads. To address challenges associated with measuring relatedness in a species with multiple ploidy levels, we generated pseudo-haploid genotypes by randomly sampling one sequencing read for each sample at each locus, and calling the genotype based on the allele in that sequencing read, as previously described (Morris et al. 2011). For computational reasons, a randomized subset of the GBS loci were used for the pseudo-haploid genotypes, and the pseudo-haploid genotypes are used for all analysis using genetic distances.
Tetraploid switchgrass has disomic inheritance (Okada et al. 2010), so we also generated diploid genotypes to estimate levels of heterozygosity and visualize genome-wide patterns of admixture in hybrid samples, with the understanding that heterozygosity estimates will be elevated in octoploids because they contain twice as many alleles of each locus as do the tetraploids. We used the following cutoffs for assigning diploid genotypes: heterozygous A:B genotypes require at least 1 read each of allele A and allele B. Homozygous A:A genotypes require 6 or more reads of allele A without any reads of allele B, otherwise they are assigned a partial, A:NA genotype.
Population Structure
Population structure was investigated with Principal Coordinate Analysis (PCoA) using the dist.dna ('ape' package, Paradis et al. 2004) and cmdscale functions in R (R Development Core Team, 2010) using pseudo-haploid genotypes. Pseudo-haploid genotypes were also used to generate neighbor-joining trees using the bionj function ('ape' R package, Paradis et al. 2004). We also used STRUCTURE (Pritchard et al. 2000) to assess population structure and admixture using pseudo-haploid genotypes. For runs using all loci, we used a 500k-cycle burn-in followed by 2 million MCMC cycles. We also generated 5 random subsets of 1000 loci, and used a 100k-cycle burn-in followed by 500k MCMC cycles. We followed Evanno et al. (2005) to determine the number of demes best supported by the results. Isolation-by-distance was evaluated by calculating the full and partial correlations of genetic distance and log-transformed geographic distance in R (R Development Core Team, 2010), including ploidy, ecotype, and/or regional gene pool as covariates.
Genome-wide Patterns of Admixture
To visualize the genome-wide patterns of admixture in identified hybrid samples, we mapped the GBS loci from this analysis to the Setaria italica reference genome (Bennetzen et al. 2012) using 80% homology across 50% of the GBS sequence (clc_assemble_ref_long, CLC Assembly Cell, CLC bio, Cambridge, MA, USA), and retaining only loci with unique hits to the S. italica reference. Grasses have high levels of conserved synteny (Devos 2005), and S. italica is closely related to switchgrass (Bennetzen et al. 2012), so the genomic location of the loci mapped to S. italica should closely resemble their relative location in the switchgrass genome. Because S. italica is diploid while switchgrass is polyploid, homeologous switchgrass GBS loci will map to the same location in the S. italica genome, thus partially masking the true patterns of admixture. Therefore, this method provides a preview of the possibilities when using the assembled switchgrass genome that is in progress. Alleles were considered private in an ecotype if the SNP is genotyped in at least 10 individuals in the focal ecotype and is either completely absent or genotyped in at least 10 individuals in the other ecotype without seeing the allele.
Estimating Genome-Wide Nucleotide Diversity
The pseudo-haploid genotypes are equivalent to the genotype of a single copy of the genome within each sample, so genetic distance represents the amount of differences between two copies of the genome. We used the average pairwise genetic distance within a population as an estimate of the genome-wide level of nucleotide diversity in a population and therefore as a way to compare relative levels of diversity. This raw diversity quantification is another advantage of GBS data over predefined SNP data. We calculated average within-population genetic distance for all populations with more than one sample using dist.dna ('ape' package, Paradis et al. 2004) in R (R Development Core Team, 2010). Populations that contained samples from different gene pools based on population structure results were divided into subgroups based on gene pool. We did not include samples that show a strong signal of admixture between regional gene pools within each ecotype based on PCoA results.
Simulated Octoploids
To test if differences in heterozygosity between tetraploid and octoploid groups are due strictly to their different number of chromosomes, we generated simulated octoploid genotypes by combining diploid genotypes of two samples from the primarily tetraploid gene pools. We used the following rules: the 8X genotype is heterozygous if both alleles are found in either sample. The 8X genotype is homozygous if both samples are homozygous or partial (eg: A:NA) for the same allele. The 8X genotype has a partial genotype if one sample is partial and the other has no genotype. We simulated within-gene pool octoploids using all pairwise combinations of samples within each tetraploid gene pools and admixed octoploids using all pairwise combinations of samples from different tetraploid gene pools.
Results
Sequencing and genotyping results
We generated genotyping-by-sequencing libraries for 126 samples, producing 141.8 million paired-end sequence reads on the Illumina GA-IIx platform. Of these reads, 132.8 million (93.6 %) passed preliminary filtering criteria. The mean per-sample read count was 1.05 million reads, but varied from 53,243 to 2,802,834 reads per sample (Table S1). We removed 14 samples with less than 310,000 sequencing reads from further analysis, as these had an excess of SNPs with missing data (Fig S1a), and because above that threshold, diversity measures are minimally affected by sequencing coverage (Fig S1c). We identified 98,042 SNPs on 27,666 short contigs (called GBS loci in the rest of the text). Of these, 71,020 (72.4%) SNPs are genotyped in at least 80 samples, 19,182 (19.6%) are genotyped in all the switchgrass samples, and 3,549 (3.6%) are also genotyped in the Panicum amarum, P. amarulum, and P. anceps samples. There are 6,896 (7.0%) SNPs where the minor allele is found in a single individual, with the number of singleton SNPs per sample ranging from 2 to 252 in the switchgrass samples and 763, 58, and 399 in the P. amarum, P. amarulum, and P. anceps samples, respectively. Our ability to detect singletons is reduced as we require significant variation in allele counts across samples due to a chi-squared filter we apply to our genotyping results (as in Morris et al. 2011, Myles et al. 2010). For computational reasons, 19,907 SNPs (20.3%) were randomly selected to use for the pseudo-haploid genotypes in all the individuals.
Using the di-haploid samples, we identify 9.2% (9,016) of the putative SNPs representing co-assembled homeologs rather than alleles at single genomic loci. The di-haploid sample are both part of the same gene pool, lowland Southern Great Plains, and therefore cannot identify homeologous SNPs that are variable only in other gene pools. Therefore, we retain the homeologous SNPs in the analysis because removing them would disproportionately remove variation from the lowland Southern Great Plains gene pool and bias diversity measures. However, PCoA results are qualitatively the same whether the detected homeologous SNPs are included or removed.
Population Structure
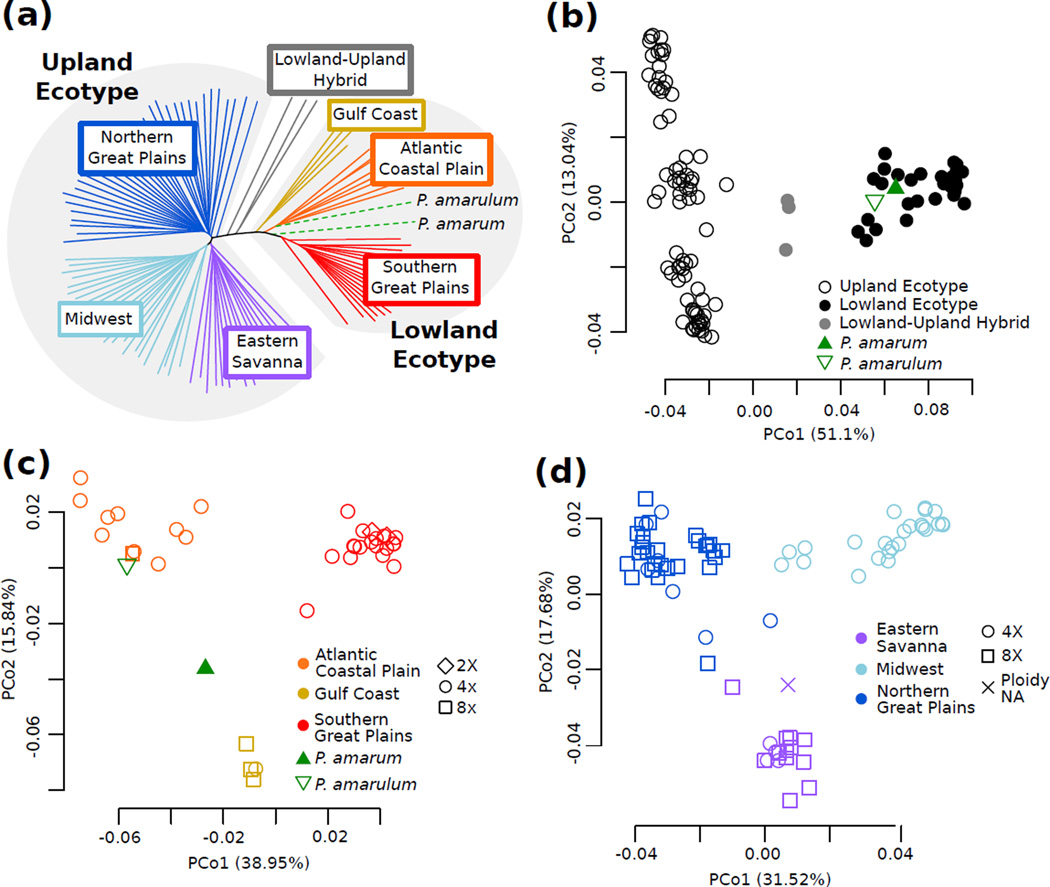
To characterize species-wide patterns of population structure and genome-wide nucleotide diversity, we used pseudo-haploid genotypes to evaluate the relationships of switchgrass from 41 populations across the United States. PCoA and STRUCTURE resolve two main groups corresponding to the upland and lowland switchgrass ecotypes (Figs. 1b, S2a, S3c, S4), as expected (Morris et al. 2011, Zalapa et al. 2011). We further detect additional population stratification within both major switchgrass ecotypes. Within the lowland ecotype, three major gene pools are resolved (Figs. 1a,c, S2b) corresponding to three US geographic areas: the Southern Great Plains, the Western Gulf Coast, and the Atlantic Coastal Plain (Fig 2a). While the Western Gulf Coast gene pool is represented by a single population, the log-likelihood values from STRUCTURE are consistent with three demes, and McMillan (1964) also identified a morphologically distinct group in the same region. Within the upland ecotype, three main gene pools are identified, as well, (Figs 1a,d, S2c) corresponding to the Northern Great Plains, the Midwest, and the Eastern Savanna of the United States (Fig 2a). These gene pools correspond with the three upland clades identified by Lu et al. (2013). Several upland samples cluster between gene pools (Table S1, Fig 1d), and these sample are removed when calculating average genetic distances within populations and between gene pools.
Figure 1.
Population structure in switchgrass inferred from neighbor joining tree (a) and PCoA (b,c,d). Color corresponds to inferred ecotype (b) or regional gene pool (a,c,d) and shape corresponds to ploidy (c,d). The P. amarum and P. amarulum samples are labeled individually (a,b,c), and their ploidy in unknown. (a) Neighbor joining tree of all switchgrass samples, P. amarum, and P. amarulum. (b) PCoA with samples from (a). Note that P. amarum and P. amarulum cluster with lowland ecotype samples. (c) PCoA with lowland ecotype samples, P. amarum, and P. amarulum shows three lowland regional gene pools. (d) PCoA with upland ecotype samples shows three upland regional gene pools.
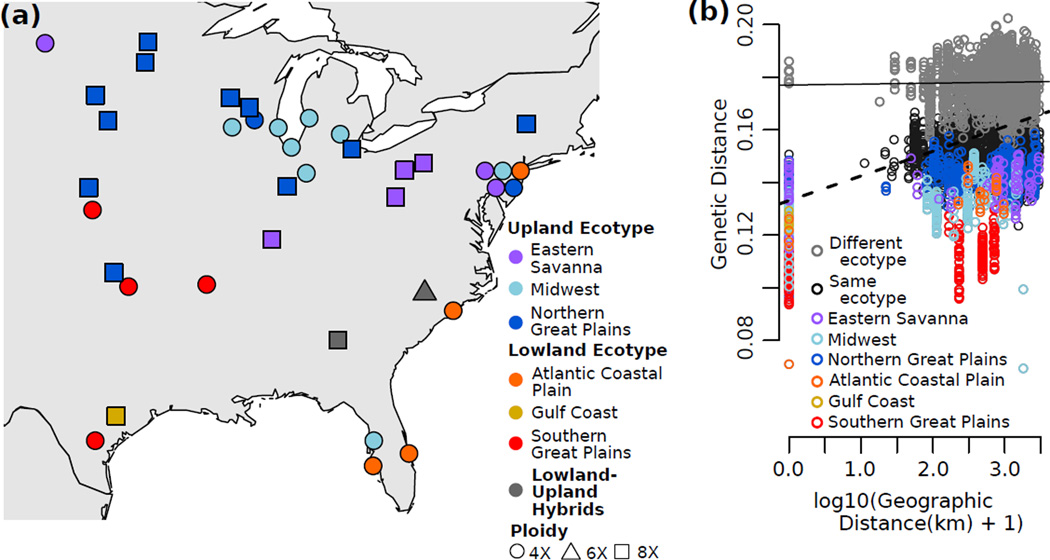
Figure 2.
Geographic patterns of genetic diversity. (a) Map of populations. Color corresponds to inferred regional gene pool and shape corresponds to predominant ploidy. (b) Patterns of isolation-by-distance shown by all pairwise comparisons of genetic and geographic distance. Gray = between ecotypes. Black = within ecotype. Colors = within each regional gene pool. Lines representing the linear regression of all between-ecotype comparisons (solid line, slope = 3.5×10−4) and all within-ecotype comparisons (dashed line, slope = 9.4×10−3) indicate greater IBD within ecotypes than between ecotypes.
The lowland and upland ecotypes remain genetically distinct across their large and partially overlapping ranges (Fig. 2a), as the geographic distance to the most-distant sample of the same ecotype ranges from 1500 to 3000 kms, while the distance to the closest sample of the other ecotype ranges from 0 to 500 kms. Similarly, there is only a minimal relationship between genetic and geographic distances of samples from different ecotypes (Fig. 2b). A pattern of isolation-by-distance (IBD, r = 0.356, Fig 2b, Table 3) in the data set is driven largely by IBD within the gene pools, as the signal of IBD decreases dramatically when accounting for regional gene pools (r = 0.169, Table 3).
Table 3.
Correlations of Genetic Distance and Geographic Distance
| r-value | Lower (2.5%) confidence limit | Higher (97.5%) confidence limit | Ecotype | Regional Gene Pool | Ploidy |
|---|---|---|---|---|---|
| 0.356 | 0.326 | 0.386 | |||
| 0.371 | 0.340 | 0.397 | X | ||
| 0.169 | 0.145 | 0.192 | X | ||
| 0.342 | 0.311 | 0.374 | X | ||
| 0.250 | 0.230 | 0.275 | X | X | |
| 0.360 | 0.329 | 0.392 | X | X | |
| 0.172 | 0.150 | 0.197 | X | X | |
| 0.251 | 0.228 | 0.275 | X | X | X |
Correlation and partial correlation coefficients of genetic distance and log-transformed geographic distance. Xs indicate the covariate(s) used when calculating partial correlations. Confidence limits based on 500 bootstrap replicates.
Each switchgrass gene pool is primarily one ploidy level, either tetraploid (4X) or octoploid (8X). However ploidy is not fixed within the gene pools, as the three primarily 8X gene pools contain at least one 4X sample (Fig 1c,d). In addition, two 8X samples that did not pass the read-count cutoff are part of populations in the primarily 4X gene pools and cluster with those gene pools when included in the analysis (Fig S3d,e). Octoploid samples are more similar to tetraploids from the same gene pool than to octoploids from other gene pools (Fig 1), indicating that ploidy has changed multiple times in switchgrass. Lu et al. (2013) concluded that the upland ecotype was ancestrally octoploid and a haploidization event generated the tetraploid upland Midwest gene pool. However, the presence of tetraploids in all three upland gene pools (Fig 1) suggest that, instead, the upland ecotype was ancestrally tetraploid and that octoploidy arose independently in the upland Eastern Savanna and Northern Great Plains gene pools. Furthermore, since no gene pool is exclusively octoploid, then polyploidy could not have initiated the differentiation between gene pools, nor are any gene pools completely reproductively isolated due to ploidy differences.
Panicum amarum and P. amarulum are ecotypes of P. virgatum
We included P. amarum and P. amarulum samples as outgroups for our switchgrass analysis and to evaluate their relationship to switchgrass, as they are generally considered a sister lineage to P. virgatum (Barkworth et al. 2007). However, the P. amarum and P. amarulum samples in this study cluster within the lowland ecotype (Fig. 1b, S4), as also seen using gene trees (Triplett et al. 2012). Furthermore, P. amarum and P. amarulum are genetically more similar to the lowland ecotype samples than to the upland ecotype samples (Table 2). These results are in contrast to the P. anceps sample that clusters separately from all the other samples (Figs. S3b, S4). These results indicate that P. amarum and P. amarulum are, in fact, switchgrass ecotypes adapted to the harsh conditions of coastal habitats (Barkworth et al. 2007). Interestingly, while P. amarum and P. amarulum are often considered part of the same taxon, the P. amarulum in this study is more similar to the lowland Atlantic Coastal Plain gene pool than it is to the P. amarum sample (Fig 1c, Table 2), supporting the hypothesis that P. amarum and P. amarulum are not part of a single lineage that is distinct from the lowland and upland switchgrass lineages (Triplett et al. 2012). While these patterns need to be confirmed with more samples, they indicate that P. amarulum is a morphologically diverged type of the switchgrass lowland Atlantic Coastal Plain gene pool, or potentially the result of hybridization between the Atlantic Coastal Plain gene pool and the P. amarum ecotype (Palmer 1975, Barkworth et al. 2007).
Table 2.
Average Pairwise Genetic Distance of Gene Pools and Panicum samples.
| Upland Midwest (24) |
Upland Northern Great Plains (32) |
Upland Eastern Savanna (15) |
Lowland Atalantic Coastal Plain (9) |
Lowland Southern Great Plains (18) |
Lowland Western Gulf Coast (4) |
Hybrid-SPB (2) | Hybrid-HOF (1) | P. amarulum (1) | P. amarum (1) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Upland Midwest | ||||||||||
| Upland Northern Great Plains | 0.1518 | |||||||||
| Upland Eastern Savanna | 0.1482 | 0.1529 | ||||||||
| Lowland Atalantic Coastal Plain | 0.1825 | 0.1794 | 0.1788 | |||||||
| Lowland Southern Great Plains | 0.1821 | 0.1773 | 0.1791 | 0.15 | ||||||
| Lowland Western Gulf Coast | 0.1742 | 0.1676 | 0.1699 | 0.1558 | 0.1425 | |||||
| Hybrid-SPB | 0.1605 | 0.1592 | 0.158 | 0.1572 | 0.1596 | 0.1545 | ||||
| Hybrid-HOF | 0.1621 | 0.1559 | 0.1615 | 0.1623 | 0.1581 | 0.1591 | 0.1603 | |||
| P.amarulum | 0.1748 | 0.1707 | 0.1715 | 0.1293 | 0.1479 | 0.1472 | 0.1489 | 0.1559 | ||
| P.amarum | 0.1856 | 0.182 | 0.1824 | 0.153 | 0.1503 | 0.1485 | 0.1629 | 0.1713 | 0.1434 | |
| P.anceps (1) | 0.3498 | 0.345 | 0.3551 | 0.3595 | 0.3314 | 0.346 | 0.3492 | 0.3448 | 0.356 | 0.3421 |
Parentheses indicate the number of samples included in each group. The lowland-upland hybrids are separated into population of origin. Note lowland Western Gulf Coast is more similar to upland ecotype gene pools than are are either other lowland ecotype gene pool.
Hybridization and gene flow
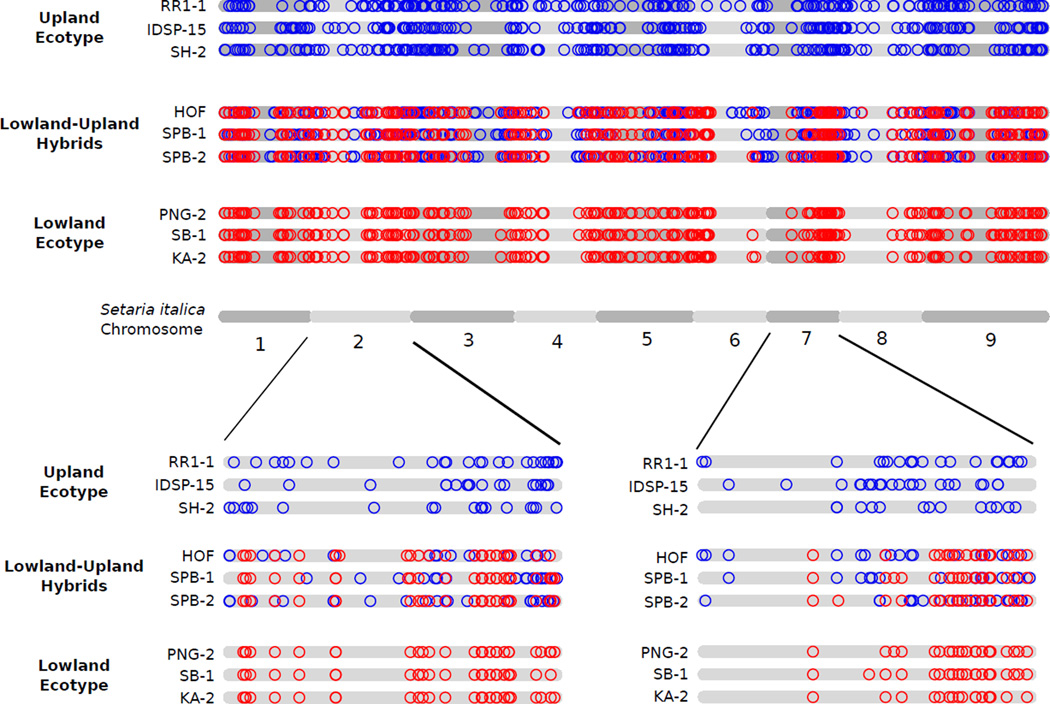
There is a large overlap in the ranges of the lowland and upland switchgrass ecotypes, but reports of natural hybrids between the ecotypes are rare (Zhang et al. 2011a), despite the close proximity of many upland and lowland switchgrass populations (Fig. 2). However, we identify three samples that group between the lowland- and upland ecotype clusters in the PCoA (Fig. 1b), show strong patterns of lowland-upland admixture (Fig S2a), and are equally related to the upland and lowland ecotypes (Table 2), indicating that they are the result of recent hybridization between the ecotypes. The hybrid nature of these sample is further supported by their genome-wide distribution of ecotype-specific alleles (Fig 3) and previous results using EST-SSR markers (Zhang et al. 2011a). The hybrid populations are both in the Atlantic Coastal Plain (Fig. 2a), and further sampling is needed to examine if natural hybridization between the ecotypes is common in that region.
Figure 3.
Genome-wide distribution of ecotype-specific alleles in lowland-upland hybrids. Alleles private to either ecotype are mapped to the reference genome of Setaria italica. Patterns are shown in the three identified lowland-upland hybrid samples as well as three upland ecotype and three lowland ecotype samples. Red = lowland ecotype allele. Blue= upland ecotype allele. To normalize for the overall higher number of upland private alleles in the data set (due to the higher number of upland versus lowland samples), a random subset of 25% of upland alleles is plotted. Genomic regions of predominantly upland or lowland ancestry can be identified. Magnifications of chromosomes 2 and 7 show chromosome-wide patterns at higher resolution.
In addition to the hybrids, we also detect gene flow from the upland ecotype into the lowland Western Gulf Coast gene pool, as that gene pool has a signal of admixture with the upland ecotype (Fig S2a) and is more similar to the upland ecotype than is either other lowland gene pool (Table 2). These findings are supported by EST-SSR results that see lowland-upland ecotype admixture in the same population (Zhang et al. 2011a)
Diversity measures indicate admixture promotes polyploidy
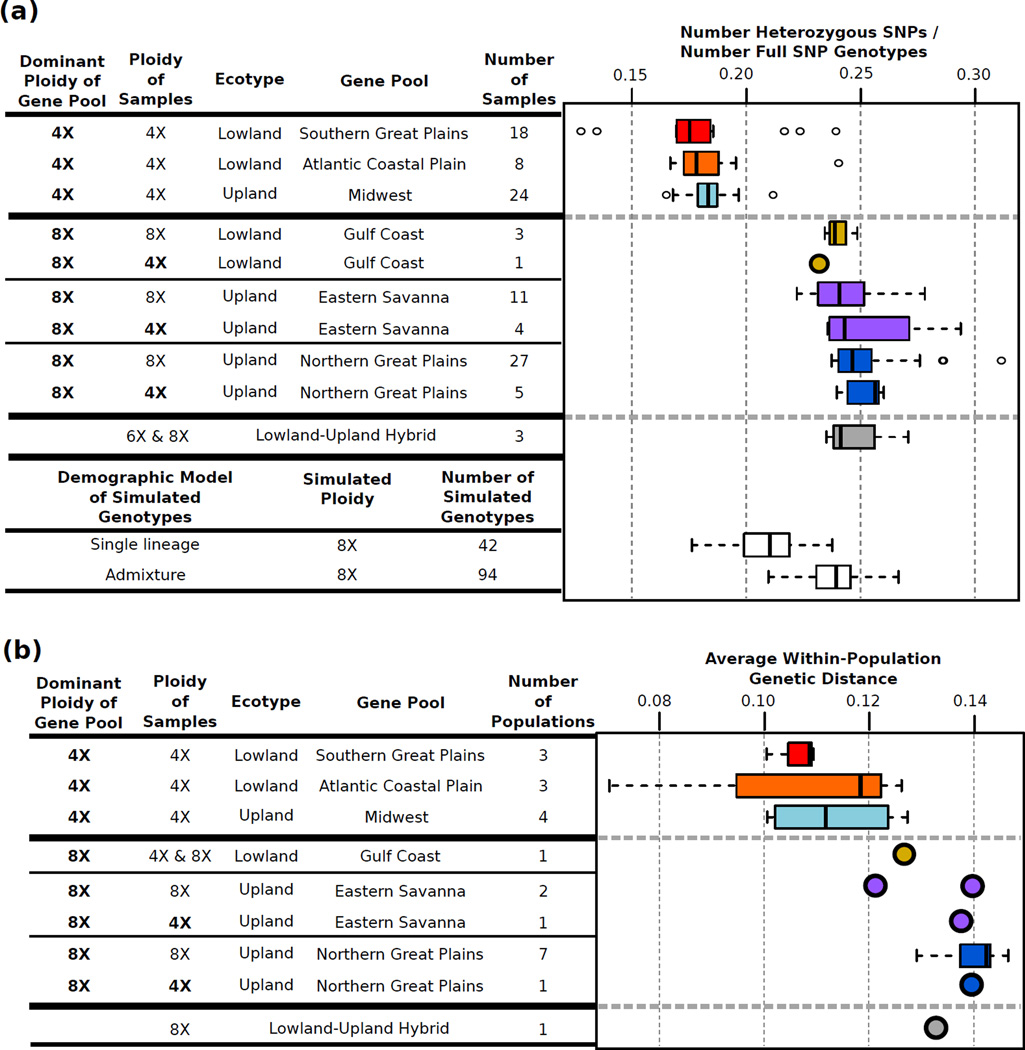
The gene pools have different levels of estimated heterozygosity and nucleotide diversity corresponding to their primary ploidy levels, with the primarily-8X gene pools having higher levels of both measures than the 4X gene pools (Fig 4). However, the tetraploid members of the primarily-8X gene pools also have elevated levels of both heterozygosity and nucleotide diversity (Fig 4), indicating that the elevated diversity measures in the primarily-8X gene pools is not exclusively due to higher ploidy. To estimate how diversity measures would be affected by a shift from tetraploid to octoploid, we simulated octoploid genotypes by combining data from tetraploid samples in the same gene pool. The simulated single lineage octoploids have lower heterozygosity than the true octoploid gene pools (Fig 4), further indicating that ploidy alone does not explain the elevated diversity in the primarily-8X gene pools.
Figure 4.
Genetic diversity of switchgrass gene pools. Samples (a) and populations (b) from gene pools with both 4X and 8X samples are divided into subgroups by ploidy. (a) Number of heterozygous SNPs divided by the number of SNPs with full diploid genotype calls as an estimate of heterozygosity for each sample. Simulated octoploid genotypes were generated by combining genotypes from tetraploids either from the same gene pool ("Single lineage") or different gene pools ("Admixture"). Heterozygosity levels in predominantly-8X gene pools is more similar to the admixed rather than the single lineage simulated octoploids. (b) Average within-population genetic distance as an estimate of nucleotide diversity. Note that heterozygosity (a) and nucleotide diversity (b) is the same for both 4X and 8X samples from primarily-8X gene pools. Also, primarily-8X gene pools have similar diversity levels as natural (Lowland-Upland Hybrids) and simulated admixed samples.
Interestingly, the upland-lowland hybrid samples have similar levels of nucleotide diversity and heterozygosity as the primarily-8X gene pools (Fig 4), and one primarily-8X gene pool, lowland Western Gulf Coast, shows evidence of admixture between the lowland and upland ecotypes (Table 2, Fig S2a). This suggest that elevated diversity in the primarily-8X gene pools may be due to admixture. Therefore, to estimate how a combination of admixture and octoploidy would affect diversity, we simulated admixed octoploid genotypes by combining tetraploid samples from different gene pools. The simulated admixed octoploids have heterozygosity similar to the levels seen in the primarily-8X gene pools (Fig 4a), supporting a hypothesis that the primarily-8X gene pools have a history of admixture. Results using gene trees are also consistent with octoploid switchgrass being derived from the combination of two distinct tetraploid lineages (Triplett et al. 2012).
Discussion
Geospatial and genome-wide patterns of nucleotide diversity provide a window into the historical and contemporary processes that shape variation in a species (Novembre and Di Rienzo 2009). Thus, to characterize how different processes have contributed to differentiation in switchgrass, we genotyped 123 switchgrass individuals from 41 populations across the US range at 98,042 SNPs. The major genetic division detected within switchgrass is between the lowland and upland ecotypes, consistent with previous results (Fig 1b, Morris et al. 2011, Zalapa et al. 2011, Lu et al. 2013), and we also identify three regional gene pools within both ecotypes (Figs. 1a, 1c, 1d). Patterns of ploidy within and between gene pools (Figs. 1c, 1d) indicate that the upland and lowland ecotypes were ancestrally tetraploid (4X) and that octoploidy (8X) has independently arisen multiple times. In addition, we detect signatures of historical and contemporary hybridization between the upland and lowland ecotypes (Figs. 1b, S2a; Table 2) and determine that Panicum amarum and P. amarulum are actually switchgrass ecotypes and not separate taxa (Figs 1a, 1b, 1c; Table 2). Furthermore, elevated diversity in the primarily-octoploid gene pools is not the result of polyploidy, but rather is consistent with a history of admixture in these gene pools.
Biogeographic history, adaptation, and ploidy shape switchgrass variation
We detect three regional gene pools within both the upland and lowland ecotypes (Fig. 1). Each gene pool has a dominant ploidy, either tetraploid (4X) or octoploid (8X); however, samples of the alternate ploidy are seen in several gene pools, including all three primarily-8X gene pools (Fig 1, and Fig S3). The three upland gene pools correspond to the three main upland clades identified by Lu et al. (2013). Lu et al. (2013) propose that the upland ecotype was ancestrally octoploid, and that the upland tetraploid gene pool underwent haploidization to become tetraploid and therefore reproductively isolated from the other upland lineages. However, the presence of tetraploids in the upland Eastern Savanna and Northern Great Plains gene pools instead indicates that the upland ecotype was ancestrally tetraploid, and octoploids arose independently in the two primarily-8X gene pools.
Because tetraploids are present in all the observed switchgrass gene pools, it is unlikely that polyploidy itself generated the genetic differences seen in switchgrass. Instead, geography was likely the main driver of differentiation in switchgrass. The geographic patterns of population structure are consistent with the hypothesis that switchgrass was restricted to multiple refugia during glacial maxima (McMillan 1959, Zalapa et al. 2011, Lu et al. 2013). For example, the geographic distribution of the lowland ecotype gene pools (Fig 2a) correspond with predicted distinct glacial refugia in Florida and along the Gulf Coast (McMillan 1959, Zalapa et al. 2011b), and correspond with phylogeographic patterns seen in other species (Soltis et al. 2006). Similarly, the genetic differentiation and geographic distribution of upland gene pools suggest that the upland ecotype was restricted to multiple distinct glacial refugia, as well (Fig 2a). Ploidy differences between gene pools likely arose subsequent to genetic divergence due to vicariance.
Despite this historical vicariance within both ecotypes, populations of the same ecotype that were in different refugia are more closely related than are populations of different ecotypes just meters away (Fig 2). Similarly, when accounting for the spatial structure of the gene pools within each ecotype, there is little isolation-by-distance (Table 3), indicating that one or more mechanisms are acting to restrict gene flow between the ecotypes, resulting in the observed range-wide differentiation between ecotypes.
In many species, differential adaptation restricts gene flow between ecotypes via immigrant and hybrid inviability (ex: Lowry et al. 2008). The lowland and upland ecotypes are adapted to different edaphic conditions and have lower fitness in the habitat of the other ecotype (Porter 1966), suggesting that differential adaptation may restrict gene flow in switchgrass, as well. Most populations show little signature of gene flow from the other ecotype (Fig. S2a), even when in close proximity to populations of the other ecotype. However, we do detect gene flow from the upland ecotype into the lowland Western Gulf Coast gene pool (Table 2) as well as contemporary hybridization between the lowland and upland ecotypes (Fig 3; Fig S2), showing that adaptive differences are not sufficient to restrict all gene flow between the ecotypes and the low levels of inter-ecotype gene flow in most samples are unlikely due solely to differential adaptation.
While intra-ploidy gene flow is at least possible between any gene pool of either ecotype (Fig 1; Fig S3), in many areas inhabited by both ecotypes, particularly the Great Plains, the primary ploidy level of the lowland and upland ecotypes differ (Figs 1, 2, Zhang et al. 2011b). As noted by Lowry et al. (2014), these ploidy differences may help to restrict contemporary gene flow between the ecotypes in these areas. In fact, ploidy differences acting to prevent maladaptive hybridization and maintain differentiation between ecotypes may be a mechanism common to many species (ex: Martin and Husband 2013).
Admixture promotes polyploidy in switchgrass
Within both the lowland and upland switchgrass ecotypes, the frequency of the major ploidy levels differs substantially between genetically distinct regional gene pools (Fig 1c,d). In many plants, hybridization and admixture are associated with subsequent higher rates of polyploidy (Ramsey and Schemske 1998, Chapman and Burke 2007; Paun et al. 2009; Perisod et al. 2010). Interestingly, we see a similar pattern in switchgrass. Elevated ploidy is seen in lineages with signals of admixture between the lowland and upland ecotypes (Fig 1, Fig S2a, Table 2). In the upland gene pools with prevalent octoploidy, we do not detect genetic signals of admixture between ecotypes (Fig. S2a), but there is indication that these lineages may have a history of admixture, as well. First, tetraploid samples within the upland primarily-8X gene pools have the same levels of diversity as the octoploid samples (Fig 4), so increased diversity in these lineages is not due strictly to higher ploidy. Second, the Eastern Savanna and Northern Great Plains gene pools have similar levels of heterozygosity and nucleotide diversity as the lowland-upland hybrids and admixed lowland Western Gulf Coast gene pool (Fig 4). Third, the levels of heterozygosity seen in the Eastern Savanna and Northern Great Plains gene pools are only replicated in synthetic admixed octoploids and not in synthetic octoploids made using samples from the same gene pool (Fig 4a). Furthermore, gene tree analysis indicates that upland octoploids are derived from the combination of two separate upland lineages (Triplett et al. 2012). Combined, these results strongly suggest that the upland primarily-8X gene pools have a history of admixture, most likely of diverged upland populations. The overall association of octoploidy with admixture suggests that polyploidization (ie: increases in ploidy from baseline tetraploid) in switchgrass is promoted by the admixture of genetically diverged lineages, both between and within ecotypes.
Several studies show a positive link between the genetic divergence between species and the formation of polyploids upon hybridization (Chapman and Burke 2007, Paun et al. 2009). However, the universality of this pattern has been doubted because these studies omit polyploids that contain genome copies from only a single progenitor species, generally termed autopolyploids (Buggs et al. 2009, Soltis et al. 2010). Predating this debate, Stebbins' proposed that even within species, polyploidy is often due to the interaction of historically isolated and diverged lineages (Stebbins 1984), forming what can be considered "intraspecific allopolyploids" because they contain genome copies from diverged lineages but the same species. In fact, in several species, polyploidy is seen in suture zones where lineages that were previously isolated in different glacial refugia have come into secondary contact and interbred (Parisod et al. 2010). Our data indicate that switchgrass lineages in which octoploidy is common have a history of admixture (Fig 4). These results further support Stebbins' secondary contact hypothesis for intraspecific polyploids and suggest that hybridization between diverged lineages increases the likelihood of polyploidy even within species.
Interdependence of ploidy, adaptation, and biogeography
In many plant species, ploidy differences are associated with adaptive differences (Manzaneda et al. 2012, Martin and Husband 2013, see Parison et al. 2010 Table 2 for list of examples). Polyploidy has immediate phenotypic effects (Otto 2007) and could directly result in adaptive differences, potentially because of elevated genetic diversity or because genetic redundancy allows for neo- and subfunctionalization of genes for adaptive traits (Lynch et al. 2001). Alternatively, adaptive differences may develop unrelated to ploidy, but ploidy-related reproductive isolation could prevent the flow of maladapted alleles between differentially adapted cytotypes. Our results indicate that polyploidy was not directly associated with the adaptive differentiation of the major lowland and upland switchgrass ecotypes, though it is possible that polyploidy generates adaptive differences within the ecotypes. However, in many areas where the lowland and upland ecotypes are sympatric, the ecotypes have different ploidy (Zhang et al. 2011b, Lowry et al. 2014). Similarly, ploidy difference exist between the primarily 4X lowland Atlantic Coastal Plain switchgrass gene pool (including P. amarulum) and the P. amarum coastal ecotype, which is often hexaploid (Barkworth et al. 2007). Thus, in regions of sympatry, ploidy often seems to have a role in restricting gene flow between differentially adapted ecotypes.
An intriguing possibility is that the interaction of sympatric ecotypes is important for promoting the establishment and maintenance of alternate ploidy levels. The establishment of polyploids is repressed by minority cytotype exclusion and competition between ploidy levels (Levin 1975, Petit et al. 1999). In switchgrass, octoploidy itself has negative fitness consequences because octoploids produce high numbers of aneuploid offspring (Costich et al. 2010), and no adaptive advantage associated with octoploidy has been demonstrated. The negative effect from polyploidization could, however, be offset by the fitness advantage from preventing maladaptive hybridization, thereby increasing the prevalence of polyploids in these areas.
Furthermore, our results indicate that admixture promotes polyploidy in switchgrass (Fig 4), and that previously isolated switchgrass lineages have come into secondary contact possibly as a result of post-glacial range expansion. (Fig 2a). Thus, the biogeographic history of switchgrass may have been crucial for generating the admixed upland lineages prone to becoming octoploid and therefore able to coexist and avoid maladaptive hybridization with sympatric tetraploid lowland populations.
Germplasm for dissecting the genetic basis of phenotypes
Adaptive differences have been studied in switchgrass for more than 50 years (McMillan 1959, Porter 1966), and recent advances in genomic methods have opened up new avenues for dissecting the genetic basis of these classic cases of ecotypic adaptation (Li et al. 2010, Parchman et al. 2012). Switchgrass is also an important foundational species for grassland restoration efforts and an emerging bioenergy feedstock, and understanding the genetic basis of adaptive traits is key for accelerating improvement efforts, including developing genetic markers for marker-assisted selection (Varshney et al. 2005). Genome-wide association studies, which can use natural populations to identify common genetic variation associated with traits, are a powerful approach for dissecting the genetic basis of phenotypes, particularly for species like switchgrass where phenological differences and long generation times complicate the development of mapping populations. And with the release of a reference switchgrass genome on the horizon, genome-wide association studies will be an important part of improvement efforts in the near-future.
Our study provides a foundation for future genome-wide association studies in switchgrass, such as the regional gene pools (Fig. 1,2) which need to be accounted for when designing species-wide association panels (Myles et al. 2009, Brachi et al. 2011) and are candidates for regional mapping approaches (ex: Brachi et al. 2013). In addition, P. amarum and P. amarulum, which we identify as a switchgrass ecotypes, are underused resources for studying adaptation to marginal conditions in switchgrass, as they are adapted to the sand dunes along marine coasts. However, many adaptive traits are characteristic differences between the lowland, upland, and coastal switchgrass ecotypes (Porter 1966, Palmer 1975), and the correlation of these traits with ecotypic genetic differentiation will confound most association studies (Myles et al. 2009). As such, natural hybrid populations where the intermating of the ecotypes breaks up the associations of phenotypes with population structure, such as the Hoffman and Sprewell Bluff populations identified here (Table 1, Figure 3), are a valuable resource for dissecting the genetic basis of important ecotype-specific traits.
Supplementary Material
Acknowledgements
We thank Indiana Dunes State Park and Indiana Department of Natural Resources for help with sampling, David Lowry for flow cytometry data, Robert Elshire for discussions about GBS library preparation, Jonathan Pritchard for analysis advice, Nina Noah for laboratory assistance, and three anonymous reviewers for helpful suggestions. P.P.G. was partially supported by National Institutes of Health Training Grant T32 GM007197.
Footnotes
Data Accessibility
Raw sequencing reads are available at NCBI SRA under BioProject Accession PRJNA252891 (BioProject ID: 252891). Diploid genotypes, pseudo-haploid genotypes, and additional data used in the analysis are available from the DRYAD repository, doi:10.5061/dryad.k77nh. Table 1 contains collection locations of the samples.
Author Contributions
P.P.G. designed research, performed library preparation, analyzed genetic data, and wrote the paper. G.P.M. designed research, processed sequencing data to generate genotypes, and edited the paper. M.D.C. provided genetic material and information about samples and edited the paper. J.O.B. designed research and edited the paper.
References
- Aliscioni S, Bell HL, Besnard G, et al. New grass phylogeny resolves deep evolutionary relationships and discovers C4 origins. New Phytologist. 2012;193:304–312. doi: 10.1111/j.1469-8137.2011.03972.x. [DOI] [PubMed] [Google Scholar]
- Barkworth ME, Anderton LK, Capels KM, Long S, Piep MB, editors. Manual of Grasses for North America. Logan, Utah: Utah State University Press; 2007. [Google Scholar]
- Bennetzen JL, Schmutz J, Wang H, Percifield R, et al. Reference genome sequence of the model plant Setaria. Nature Biotechnology. 2012;30:555–561. doi: 10.1038/nbt.2196. [DOI] [PubMed] [Google Scholar]
- Brachi B, Morris GP, Borevitz JO. Genome-wide association studies in plants: the missing heritability is in the field. Genome Biology. 2011;12:232. doi: 10.1186/gb-2011-12-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachi B, Villoutreix R, Faure N, Hautekeete N, et al. Investigating the geographical scale of adaptive phenological variation and its underlying genetics in Arabidopsis thaliana. Molecular Ecology. 2013;22:4222–4240. doi: 10.1111/mec.12396. [DOI] [PubMed] [Google Scholar]
- Buggs RJA, Soltis PS, Soltis DE. Does hybridization between divergent progenitors drive whole-genome duplication? Molecular Ecology. 2009;18:3334–3339. doi: 10.1111/j.1365-294X.2009.04285.x. [DOI] [PubMed] [Google Scholar]
- Chapman MA, Burke JM. Genetic divergence and hybrid speciation. Evolution. 2007;61:1773–1780. doi: 10.1111/j.1558-5646.2007.00134.x. [DOI] [PubMed] [Google Scholar]
- Costich DE, Friebe B, Sheehan M, Casler MD, Buckler ES. Genome-size variation in switchgrass (Panicum virgatum): flow cytometry and cytology reveal rampant aneuploidy. The Plant Genome. 2010;3:130–141. [Google Scholar]
- Coyne JA, Orr HA. Speciation. Sunderland MA: Sinauer Associates; 2004. [Google Scholar]
- Devos KM. Updating the 'Crop circle.'. Current Opinion in Plant Biology. 2005;8:155–162. doi: 10.1016/j.pbi.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Eckert AJ, Van Heerwaarden J, Wegrzyn JL, Nelson CD, Ross-Ibarra J, Gonzalez-Martinez SC, Neale DB. Patterns of population structure and environmental associations to aridity across the range of loblolly pine (Pinus taeda L., Pinaceae) Genetics. 2010;185:969–982. doi: 10.1534/genetics.110.115543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE. 2011;6:e19379. doi: 10.1371/journal.pone.0019379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software structure: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Henry IM, Dilkes BP, Young K, Watson B, Wu H, L Comai L. Aneuploidy and genetic variation in the Arabidopsis thaliana triploid response. Genetics. 2005;170:1979–1988. doi: 10.1534/genetics.104.037788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt GM. Some genetic consequences of ice ages, and their role in divergence and speciation. Biological Journal of the Linnean Society. 1996;58:247–276. [Google Scholar]
- Keller SR, Olson MS, Silim S, Schroeder W, Tiffin P. Genomic diversity, population structure, and migration following rapid range expansion in the Balsam Poplar, Populus balsamifera. Molecular Ecology. 2010;19:1212–1226. doi: 10.1111/j.1365-294X.2010.04546.x. [DOI] [PubMed] [Google Scholar]
- Levin DA. Minority cytotype exclusion in local plant populations. Taxon. 1975;24:35–43. [Google Scholar]
- Levin DA. Polyploidy and novelty in flowering plants. American Naturalist. 1983;122:1–25. [Google Scholar]
- Li Y, Huang Y, Bergelson J, Nordborg M, Borevitz JO. Association mapping of local climate-sensitive quantitative trait loci in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21199–21204. doi: 10.1073/pnas.1007431107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry DB, Behrman KD, Grabowski P, Morris GP, Kiniry JR, Juenger TE. Adaptation between ecotypes and along environmental gradients in Panicum virgatum. The American Naturalist. 2014;183:682–692. doi: 10.1086/675760. [DOI] [PubMed] [Google Scholar]
- Lowry DB, Rockwood CR, Willis JH. Ecological reproductive isolation of coast and inland races of Mimulus guttatus. Evolution. 2008;62:2169–2214. doi: 10.1111/j.1558-5646.2008.00457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry DB, Willis JH. A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biology. 2010;8:e1000500. doi: 10.1371/journal.pbio.1000500. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Lipka AE, Glaubitz J, Elshire R, Cherney JH, Casler MD, Buckler ES, Costich DE. Switchgrass genomic diversity, ploidy, and evolution: novel insights from a network-based SNP discovery protocol. PLoS Genetics. 2013;9:e1003215. doi: 10.1371/journal.pgen.1003215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, O’Hely M, Walsh B, Force A. The probability of preservation of a newly arisen gene duplicate. Genetics. 2001;159:1789–1804. doi: 10.1093/genetics/159.4.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzaneda AJ, Rey PJ, Bastida JM, Weiss-Lehman C, Raskin E, Mitchell-Olds T. Environmental aridity is associated with cytotype segregation and polyploidy occurrence in Brachypodium distachyon (Poaceae) New Phytologist. 2011;193:798–805. doi: 10.1111/j.1469-8137.2011.03988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SL, Husband BC. Adaptation of diploid and tetraploid Chamerion angustifolium to elevation but not local environment. Evolution. 2013;67:1780–1791. doi: 10.1111/evo.12065. [DOI] [PubMed] [Google Scholar]
- Martinez-Reyna JM, Vogel KP. Incompatibility systems in switchgrass. Crop Science. 2002;42:1800–1805. [Google Scholar]
- McMillan C. The role of ecotypic variation in the distribution of the central grassland of North America. Ecological Monographs. 1959;29:285–308. [Google Scholar]
- McMillan C. Ecotypic differentiation within four North American prairie grasses. I. Morphological variation within transplanted community fractions. American Journal of Botany. 1964;51:1119–1128. [Google Scholar]
- Morris GP, Grabowski PP, Borevitz JO. Genomic diversity in switchgrass (Panicum virgatum): from the continental scale to a dune landscape. Molecular Ecology. 2011;20:4938–4952. doi: 10.1111/j.1365-294X.2011.05335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myles S, Chia J-M, Hurwitz B, Simon C, Zhong GY, et al. Rapid Genomic Characterization of the Genus Vitis. PLoS ONE. 2010;5:e8219. doi: 10.1371/journal.pone.0008219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myles S, Peiffer J, Brown PJ, Ersoz ES, Zhang Z, Costich DE, Buckler ES. Association mapping: critical considerations shift from genotyping to experimental design. The Plant Cell. 2009;21:2194–2202. doi: 10.1105/tpc.109.068437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P, Funk DJ, Ortiz-Barrientos D. Divergent selection and heterogeneous genomic divergence. Molecular Ecology. 2009;18:375–402. doi: 10.1111/j.1365-294X.2008.03946.x. [DOI] [PubMed] [Google Scholar]
- Novembre J, Di Rienzo A. Spatial patterns of variation due to natural selection in humans. Nature Reviews Genetics. 2009;10:745–755. doi: 10.1038/nrg2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M, Lanzatella C, Saha MC, Bouton J, Wu R, Tobias CM. Complete switchgrass genetic maps reveal subgenome collinearity, preferential pairing and multilocus interactions. Genetics. 2010;185:745–760. doi: 10.1534/genetics.110.113910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto SP. The evolutionary consequences of polyploidy. Cell. 2007;131:452–462. doi: 10.1016/j.cell.2007.10.022. [DOI] [PubMed] [Google Scholar]
- Otto SP, Whitton J. Polyploid incidence and evolution. Annual Review of Genetics. 2000;34:401–437. doi: 10.1146/annurev.genet.34.1.401. [DOI] [PubMed] [Google Scholar]
- Palmer PG. A biosystematic study of the Panicum amarum-P. amarulum complex (Gramineae) Brittonia. 1975;27:142–150. [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: analyses of phyogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Parchman TL, Gompert Z, Mudge J, Schilkey FD, Benkman CW, Buerkle CA. Genome-wide association genetics of an adaptive trait in lodgepole pine. Molecular Ecology. 2012;21:2991–3005. doi: 10.1111/j.1365-294X.2012.05513.x. [DOI] [PubMed] [Google Scholar]
- Parisod C, Holderegger R, Brockmann C. Evolutionary consequence of autopolyploidy. New Phytologist. 2010;186:5–17. doi: 10.1111/j.1469-8137.2009.03142.x. [DOI] [PubMed] [Google Scholar]
- Paun O, Forest F, Fay MF, Chase MW. Hybrid speciation in angiosperms: parental divergence drives ploidy. New Phytologist. 2009;182:507–518. doi: 10.1111/j.1469-8137.2009.02767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit C, Bretagnolle F, Felber F. Evolutionary consequences of diploid–polyploid hybrid zones in wild species. Trend in Ecology and Evolution. 1999;14:306–311. doi: 10.1016/s0169-5347(99)01608-0. [DOI] [PubMed] [Google Scholar]
- Porter CL. An analysis of variation between upland and lowland switchgrass, Panicum virgatum L., in central Oklahoma. Ecology. 1966;47:980–992. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunier J, Laroche J, Beaulieu J, Bousquet J. Scanning the genome for gene SNPs related to climate adaptation and estimating selection at the molecular level in boreal black spruce. Molecular Ecology. 2011;20:1702–1716. doi: 10.1111/j.1365-294X.2011.05045.x. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2010. [Google Scholar]
- Ramsey J, Schemske DW. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annual Review of Ecology and Systematics. 1998;29:467–501. [Google Scholar]
- Ramsey J, Schemske DW. Neopolyploidy in flowering plants. Annual Review of Ecology and Systematics. 2002;33:589–639. [Google Scholar]
- Rieseberg LH, Linder RC, Seiler GJ. Chromosomal and genic barriers to introgression in Helianthus. Genetics. 1995;141:1163–1171. doi: 10.1093/genetics/141.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson MA, Reed RL, McLaughlin SB, et al. Switchgrass as a sustainable bioenergy crop. Bioresource Technology. 1996;56:83–93. [Google Scholar]
- Soltis DE, Buggs RJA, Doyle JJ, Soltis PS. What we still don't know about polyploid. Taxon. 2010;59:1387–1403. [Google Scholar]
- Soltis DE, Morris AB, McLachlan JS, Manos PS, Soltis PS. Comparing phylogeography of unglaciated eastern North America. Molecular Ecology. 2006;15:4261–4293. doi: 10.1111/j.1365-294X.2006.03061.x. [DOI] [PubMed] [Google Scholar]
- Soltis D, Soltis P, Schemske D, Hancock J, Thompson J, Husband B, Judd W. Autopolyploidy in angiosperms: have we grossly underestimated the number of species? Taxon. 2007;56:13–30. [Google Scholar]
- Stebbins GL. Polyploidy and the distribution of the arctic-alpine flora: new evidence and a new approach. Botanica Helvetica. 1984;94:1–13. [Google Scholar]
- Triplett JK, Wang Y, Zhong J, Kellogg EA. Five nuclear loci resolve the polyploid history of switchgrass (Panicum virgatum L.) and relatives. PLoS ONE. 2012;7:e38702. doi: 10.1371/journal.pone.0038702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney RK, Graner A, Sorrells ME. Genomics-assisted breeding for crop improvement. Trends in Plant Science. 2005;10:621–630. doi: 10.1016/j.tplants.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Young HA, Hernlem BJ, Anderton AL, Lanzatella CL, Tobias CM. Dihaploid stocks of switchgrass isolated by a screening approach. Bioenergy Research. 2010;3:305–313. [Google Scholar]
- Zalapa JE, Price DL, Kaeppler SM, Tobias CM, Okada M, Casler MD. Hierarchical classification of switchgrass genotypes using SSR and chloroplast sequences: ecotypes, ploidies, gene pools, and cultivars. Theoretical and Applied Genetics. 2011;122:805–817. doi: 10.1007/s00122-010-1488-1. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zalapa JE, Jakubowski AR, Price DL, Acharya A, Wei Y, Brummer EC, Kaeppler SM, Casler MD. Natural hybrids and gene flow between upland and lowland switchgrass. Crop Science. 2011a;51:2626–2641. [Google Scholar]
- Zhang Y, Zalapa JE, Jakubowski AR, Price DL, Acharya A, Wei Y, Brummer EC, Kaeppler SM, Casler MD. Post-glacial evolution of Panicum virgatum: centers of diversity and gene pools revealed by SSR markers and cpDNA sequences. Genetica. 2011b;139:933–948. doi: 10.1007/s10709-011-9597-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.