Abstract
Gibbons are small arboreal apes that display an accelerated rate of evolutionary chromosomal rearrangement and occupy a key node in the primate phylogeny between Old World monkeys and great apes. Here we present the assembly and analysis of a northern white-cheeked gibbon (Nomascus leucogenys) genome. We describe the propensity for a gibbon-specific retrotransposon (LAVA) to insert into chromosome segregation genes and alter transcription by providing a premature termination site, suggesting a possible molecular mechanism for the genome plasticity of the gibbon lineage. We further show that the gibbon genera (Nomascus, Hylobates, Hoolock and Symphalangus) experienced a near-instantaneous radiation ~5 million years ago, coincident with major geographical changes in Southeast Asia that caused cycles of habitat compression and expansion. Finally, we identify signatures of positive selection in genes important for forelimb development (TBX5) and connective tissues (COL1A1) that may have been involved in the adaptation of gibbons to their arboreal habitat.
Gibbons (Hylobatidae) are critically endangered1 small apes that inhabit the tropical forests of Southeast Asia (Fig. 1) and belong to the superfamily Hominoidea along with great apes and humans. In the primate phylogeny, gibbons diverged between Old World monkeys and great apes, providing a unique perspective from which to study the origins of hominoid characteristics.
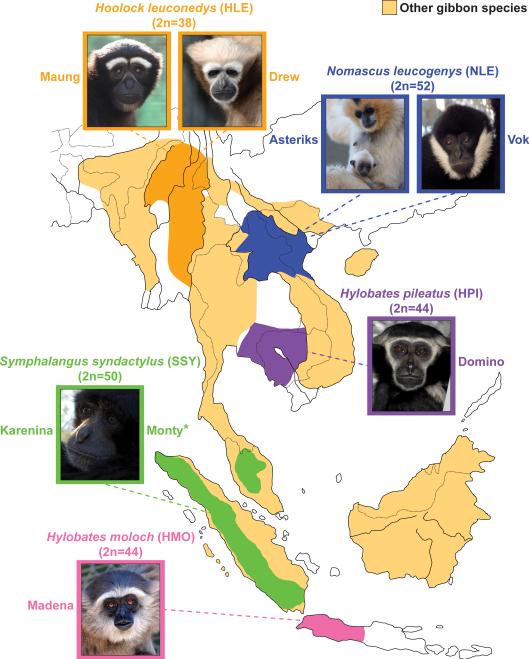
Figure 1. Geographic distribution of gibbon species used in the study.
We sequenced two individuals from each gibbon genus and two different species (H. moloch and H. pileatus) for the genus Hylobates. The extant geographic localization for each genus is illustrated on the map. Individuals in the photos are the ones sequenced in this study. (*deceased animal)
Gibbons have several distinctive traits, the most striking of which is the unusually high number of large-scale chromosomal rearrangements in comparison to the inferred ancestral ape karyotype2. The four gibbon genera (Nomascus, Hylobates, Hoolock, and Symphalangus) occupy different regions of Southeast Asia and bear distinctive karyotypes, with diploid chromosome numbers ranging from 38 to 52 (Fig. 1). Given the relatively recent differentiation of these genera (4-6 million years ago (mya)), this constitutes an extraordinary rate of karyotype change.
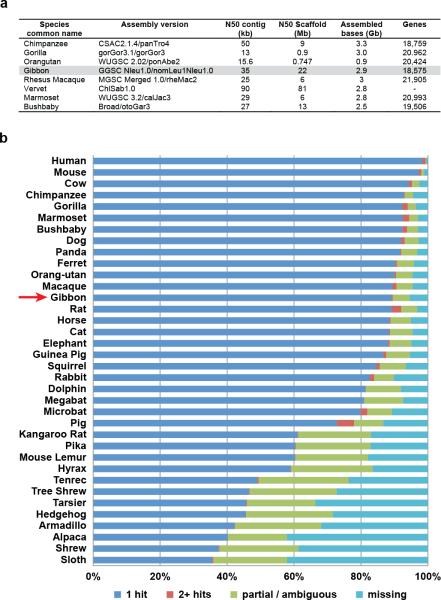
In order to investigate the mechanisms behind the plasticity of the gibbon genome, understand the evolutionary relationships among the four extant gibbon genera, and study the evolution of putatively functional sequences related to gibbon-specific adaptations, we sequenced and assembled the genome of a female northern white-cheeked gibbon (Nomascus leucogenys) named ‘Asia’. The reference assembly (Nleu1.0) provides on average 5.7-fold Sanger read coverage over 2.9 gigabase pairs (Gbp) (Table 1) (Table ST1.1). Our quality assessment (EDF 1) confirmed its equivalence to other Sanger sequence-based non-human primate draft assemblies (e.g., orangutan, rhesus3,4) (Supplementary Information S1, Supplementary Files 1-2). We also obtained ~15x whole-genome shotgun (WGS) short-read data (Illumina) for two individuals of each gibbon genus and high-coverage exome data (>60X) for two of the same individuals in order to derive error models for single nucleotide polymorphism (SNP) calls (Supplementary Information S2; Tables ST2.1-3). the gibbon genome was especially evident when human-gibbon chromosome alignments were compared with those between human and great apes, rhesus macaque (Old World monkey), and marmoset (New World monkey) (Fig. 2a). Interestingly, this higher rate of reshuffling applied only to large-scale chromosomal rearrangements (>10 Mbp), while smaller scale rearrangements (10-100 kbp) were comparable with other species (Fig. 2b) (Supplementary Information S1).
Table 1.
Gibbon assembly statistics
| Assembly (Nleu1.0/nomLeu1) | |
|---|---|
| Total sequence length | 2,936,052,603 bp |
| Ungapped length | 2,756,591,777 bp |
| Total contig length | 2.77 Gbp (92.36%) |
| Number of contigs >1 kbp | 197,908 |
| N50 contig length | 35,148 bp |
| Number of scaffolds >3 kbp | 17,976 |
| N50 scaffold length | 22,692,035 bp |
| Average read depth | 5.6X |
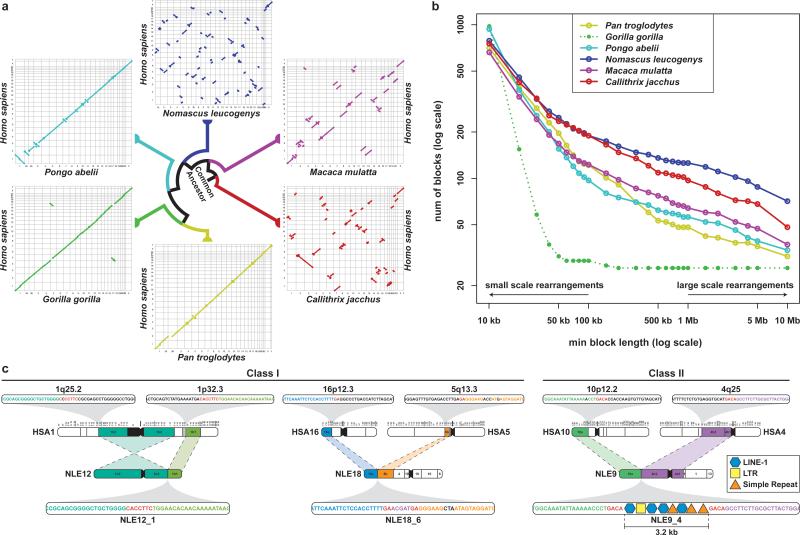
Figure 2. Analysis of gibbon-human synteny and breakpoints.
a, Oxford plots for human chromosomes (on the y-axis) vs. chimpanzee, gorilla, orangutan, gibbon, rhesus macaque, and marmoset chromosomes (on the x-axis). Each line represents a collinear block larger than 10 Mbp. The gibbon genome displays a significantly larger number of large-scale rearrangements than all the other species. In the gorilla plot, chromosomes 4 and 19 stand out as the product of a reciprocal translocation between chromosomes syntenic to human chromosomes 5 and 17. b, The graph shows the number of collinear blocks in primate genomes with respect to the human genome. The number of collinear blocks is a proxy for the number of rearrangements and decreases as the size of the blocks becomes larger. The gibbon genome has undergone a greater number of large-scale rearrangements; however, the number of small-scale rearrangements is comparable with the other species. [Note: the extremely low number of large rearrangements in the gorilla genome (dotted green line) is a reflection of the use of the human genome as a template in the assembly process]. c, Examples of gibbon-human synteny breakpoints. The first two are Class I breakpoints (i.e., base-pair resolution) originated through non-homology based mechanisms. NLE12_1 is the result of an inversion in human chromosome 1and NLE18_6 is the result of a translocation between human chromosomes 16 and 5 with an untemplated insertion in the gibbon sequence shown in purple; in both cases, microhomologies in the human sequences are shown in red. The last example (NLE9_4) is a Class II breakpoint (3.2 kbp) containing a mixture of repetitive sequences.
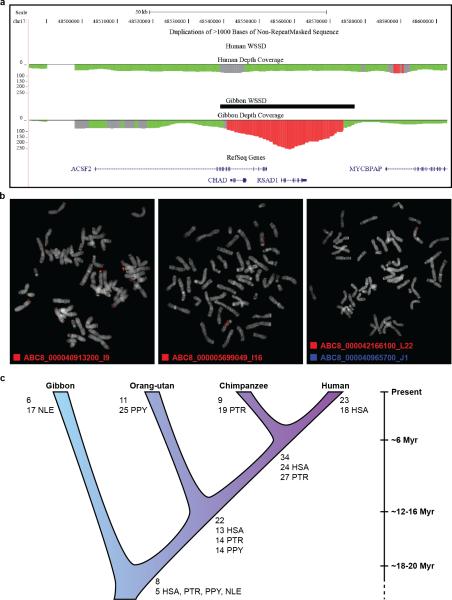
We identified 96 gibbon-human synteny breakpoints in Nleu1.0 and classified them as to whether they could be defined at the base-pair level (Class I, N=42) or only narrowed to an interval due to greater complexity (Class II, N=54). As previously reported5, breakpoints were significantly depleted of genes (Fig. SF5.2 and Supplementary File 3) and breakpoint intervals contained a mixture of repetitive sequences that inserted exclusively into the gibbon genome2,5,6 (Fig. 2c). To assess breakpoint segmental duplication (SD) content, we identified gibbon-specific SDs using in silico methods followed by experimental validation (EDF 2) (Fig. SF3.1, Supplementary Information S3 and File 4). Of note, both gibbon-specific SDs and gene family expansion analyses suggested the gibbon genome has not undergone a greater rate of duplication than other hominoids, further supporting a model in which accelerated evolution has been limited to gross chromosomal rearrangements (Supplementary Information S6; Fig. SF6.1).
SD enrichment was the best predictor of gibbon-human synteny breakpoints, as shown through permutation analyses (p-value <0.0001); however, breakpoints were also enriched for Alu elements (Table ST5.1; Supplementary Information S5; Fig. SF5.2). While non-allelic homologous recombination (NAHR) between highly similar sequences can mediate large-scale rearrangements7, the majority of gibbon chromosomal breakpoints bore signatures of non-homology based mechanisms (Fig. 2c). These included the insertion of non-templated sequences (2-51 nt) and/or the absence of identity, suggesting non-homologous end joining (NHEJ). The presence of micro-homologies (2-26 nt) in a small portion of the breakpoints (13/42) pointed to additional alternative mechanisms such as microhomology-mediated end joining (MMEJ)8 or microhomology-mediated break-induced replication (MMBIR)9. The origin of the complex breakpoint interval structures was less obvious and reinforced the observation that breakpoints tend to be receptacles for repeats.
To explore the possibility that chromatin conformation, rather than sequence, might predispose regions to breakage, we investigated the relationship between gibbon breakpoints and CCCTC-binding factor (CTCF), an evolutionarily conserved protein with multiple functions, including mediating intra-and interchromosomal interactions10. We therefore performed chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) of CTCF-bound DNA using lymphoblast cell lines established from eight gibbon individuals (Supplementary Information S5). We observed an enrichment of gibbon-human breakpoints in CTCF-binding events (p-value = 0.0028), heightened when we considered a ~20 kbp window centered around each breakpoint (p-value of <0.0001). Notably, this enrichment was maintained only for CTCF-binding events shared with other primates (human, orangutan and rhesus macaque)11 but not those specific to gibbon (p-value=0.0019) (Fig. SF5.4).
Thus, gibbon-human breakpoints co-localized with distinct genomic features and epigenetic marks; however, since many of these features were shared with other primates, other factors unique to the gibbon lineage must have been present to trigger the increased frequency of chromosomal rearrangements.
LAVA insertions in the gibbon genome
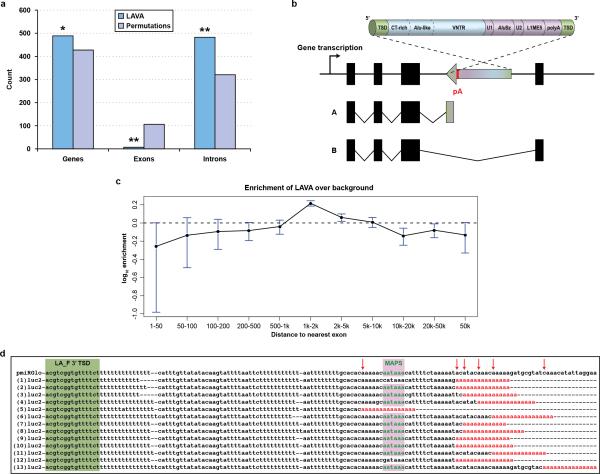
The gibbon genome contains all previously described classes of transposable elements that are mostly shared with the other primates. One exceptional addition is the LAVA element, a novel retrotransposon that emerged exclusively in gibbons12 and has a composite structure comprised of portions of other repeats (3’- L1-AluS-VNTR-Alu-like -5’) (Fig. 3a). Searches of Nleu1.0 retrieved 1,797 LAVA insertions, 1,256 of which were 3’-intact elements, many carrying signs of target-primed reverse transcription (TPRT)13. The distribution of 3’-intact LAVA elements uncovered a significant overlap with genes (Pearson chi-squared, p=0.017) and Gene Ontology (GO) analyses using the Database for Annotation, Visualization, and Integrated Discovery (DAVID)14 showed a significant functional enrichment exclusive to the ‘microtubule cytoskeleton’ category (FDR=0.031, p-value=0.001) (Supplementary Information S7 and File 6) (EDF 3). Additional analyses with meta-pathway database tools15-16 refined this enrichment to pathways related to chromosome segregation, including ‘establishment of sister chromatid cohesion’ and ‘mitotic metaphase and anaphase’ (Table ST7.3). Genes with LAVA insertions include proteins that function as checkpoints for cell division and for spindle integrity/architecture (e.g., MAP4, CEP164, BUB1B)17-19, participate in kinetochore assembly and attachment to the spindle (e.g., MAD1L1, CLASP2)20,21, and play a role in chromosome segregation during cell division (e.g., KIFAP3, KIF27)22 (EDT 1).
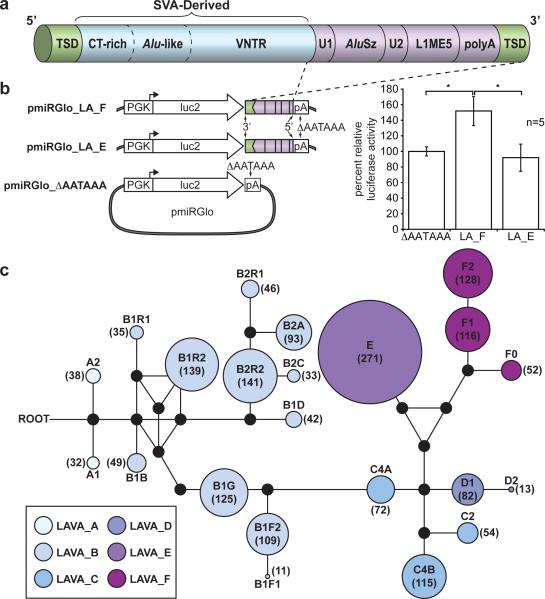
Figure 3. The LAVA element and evidence for LAVA-mediated early transcription termination.
a, Schematic view of the LAVA element highlights the main components that originated from common repeats (L1, Alu, VNTR, and Alu-like). Target site duplications (TSDs) and polyA-tail are also indicated. b, Luciferase reporter constructs used to assay for LAVA-mediated early transcriptional termination (left panel) and results of the luciferase reporter assay (right panel) showing increased luciferase activity by ~50% relative to the background for pmiRGlo_LA_F (*P=0.0013) (see Supplementary Information S7.8). c, A median-joining network showing the relationships among the 22 LAVA subfamilies generated by comparing the 3’-intact LAVA elements. Colored circles represent subfamilies and their size is proportional to the number of elements in the subfamily (numbers inside each circle). Black dots represent hypothetical sequences connecting adjacent subfamilies. All possible relationships are shown. (Branch lengths are not drawn to scale.)
Intragenic LAVA insertions were skewed toward introns (Pearson chi-squared, p=0.0001) and were less frequent than expected when within <1 kbp of the nearest exon junction (EDF 3). The majority (74%) of intronic LAVA elements were found in the antisense orientation. We hypothesized that intronic antisense LAVA insertions may cause early transcription termination (ETT) by providing a polyadenylation site in antisense orientation, as previously described for L1 elements23,24 (EDF 3). Indeed, we found 84.1% of the 3’-intact LAVA elements encoded a perfect polyadenylation signal at their 3’-end in antisense orientation.
To obtain experimental evidence that LAVA elements disrupt transcription, we performed a reporter assay in which the 3’-end of a luciferase gene construct lacking a transcriptional termination site was fused to the 3’-terminal fragments of LAVA_E and LAVA_F elements, mimicking the arrangement observed in gibbon genes (Fig. 3b-left). Luciferase activity exceeding background level by ~50% was observed from the LAVA_F reporter construct (Fig. 3b-right), indicating faithful termination of luciferase transcription. Further, 3’ Rapid Amplification of cDNA Ends (RACE) experiments confirmed that the transcription termination site had been supplied from the LAVA element (EDF 3). Thus antisense intronic LAVA insertions can cause ETT with some variability possibly due to the genomic context of the polyadenylation site, which explained the difference between the two reporter constructs.
We also investigated LAVA induced ETT in vivo by analyzing RNA-seq data generated for Asia (Table ST2.4). Specifically, we looked for paired-end reads only partially aligning to an antisense LAVA element due to untemplated residues and then identified cases for which presence of a poly(A) tail was preventing full-length alignment. This analysis revealed that elements from a variety of sub-families have the potential to cause ETT, including those identified for LAVA elements inserted in the microtubule cytoskeleton genes (e.g. B2R2, C4B, B1R2) (EDT 1). Of note, we observed that ETT occurred at relatively low levels as we identified a significant number of read pairs indicative of normal transcription and splicing for LAVA-terminated genes (Table ST7.5). This is to be expected, as full inactivation of many of these genes would be incompatible with life. On the other hand, as alternative splicing and RNA-pol II transcript termination/ polyadenylation are tightly coupled processes, LAVA-mediated ETT could also act by differently affecting distinct isoforms and/or influence the ratio between isoforms. Finally, LAVA insertions may also impact gene expression by functioning as exon traps, as shown for SVA elements25. One putative example of an exon trapping event was identified for HORMAD2, a gene that monitors the formation of synapsis during crossover26 (Supplementary Information S7, Table ST7. 6, Fig. SF7.1-2).
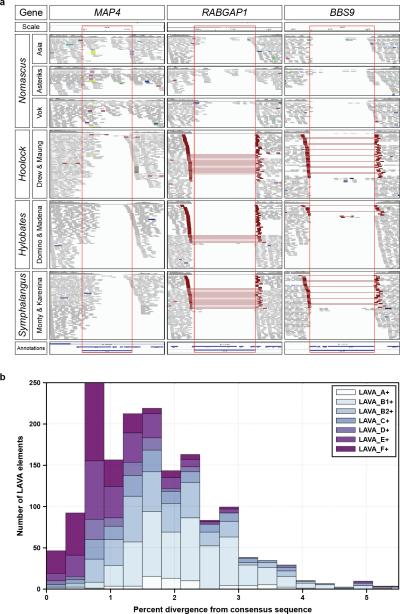
Since genome reshuffling began in the common ancestor of all extant gibbon species, LAVA insertions must have occurred in key genes before the four genera diverged. We experimentally confirmed the mode and tempo of all 23 LAVA insertions in genes from the microtubule cytoskeleton category using both site-specific PCR and in silico methods (EDF 4) and found that most of the insertions (15/23) were shared by the four gibbon genera (Supplementary File 6). Eleven of the genes match the structural requirements for ETT and five of them are also shared. These genes include MAP4, involved in spindle architecture, and CEP164, a G2/M checkpoint whose inactivation results in an aberrant spindle during cell division18,19 (EDT 1).
The complex evolutionary history of gibbons
We explored the relationship between LAVA family expansion and evolution of the gibbon lineage and, through analyses of diagnostic mutations, identified 22 LAVA subfamilies (Fig. 3c). In addition, we tested for presence/absence of 200 LAVA loci from among the evolutionarily youngest elements in each subfamily (EDF 4) across 17 unrelated gibbon individuals and found that 52% of loci were shared among all four genera, whereas 27% were Nomascus-specific. The remaining LAVA insertions showed a variety of confounding phylogenetic relationships consistent with incomplete lineage sorting (ILS) of ancestral polymorphisms, perhaps as a result of a rapid radiation of gibbon genera (Supplementary Information S7; Table ST7.1-2). We used a maximum likelihood method27 to obtain age estimates for the 22 LAVA subfamilies. In the case of the two oldest subfamilies, LAVA_A1 and LAVA_A2, we obtained estimates of ~18 mya and ~17 mya, respectively (Table ST7.3). A coalescent-based methodology implemented in the software G-PhosCS28 using Nleu1.0 estimated a gibbon-great ape population divergence time of ~16.8 mya (95% CI: 15.9-17.6 mya) assuming a split time with macaque of 29 mya (Supplementary Information S4). Hence, the LAVA element likely originated around the time of the divergence of gibbons from the ancestral great ape/human lineage.
The evolutionary history of the gibbon lineage and, in particular, the timing and order of splitting among the four genera, is still a subject of debate29. To address this issue we generated medium coverage (mean ~15X) WGS short read data for two individuals from each of the four genera, including two different Hylobates species (H. moloch and H. pileatus) (Table ST2.1-2). While phylogenetic analysis of assembled whole mitochondrial DNA genomes using BEAST30 strongly supported monophyletic groupings for each gibbon genus, the branching order of the four genera remained unresolved (Fig. SF9.1-2; Supplementary Information S9).
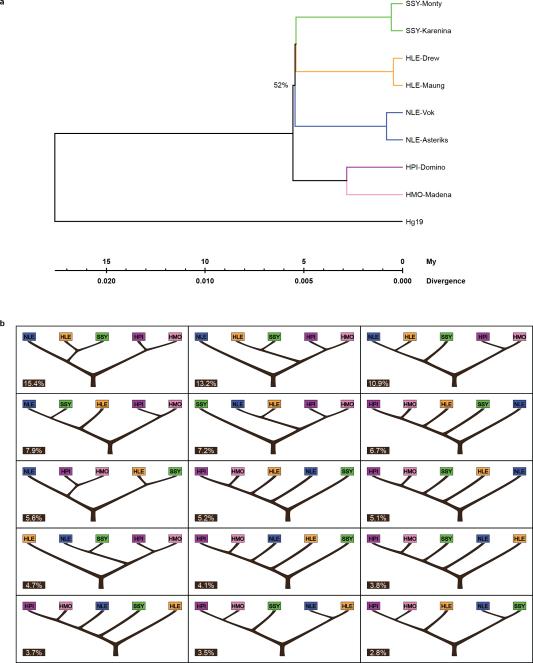
Neighbor Joining trees constructed from pairwise sequence divergence, k, across ~11,000 genic (200 bp) and ~12,000 non-genic (1 kb) autosomal loci supported a supermatrix sequence topology of (((Siamang (SSY), Hoolock (HLE)), Nomascus (NLE)), (H. pileatus (HPL)), H. moloch (HMO)) (Fig. 4a), though bootstrap confidence for the node separating NLE and Hylobates was low (~52%). This topology was also the most frequently observed when constructing k-based Unweighted Pair Group Method with Arithmetic Mean (UPGMA) trees along the genome using non-overlapping 100 kbp sliding windows. However all 15 possible rooted topologies for the four genera were observed at considerable frequencies (EDF 5), consistent with the extensive ILS observed in the LAVA element analysis.
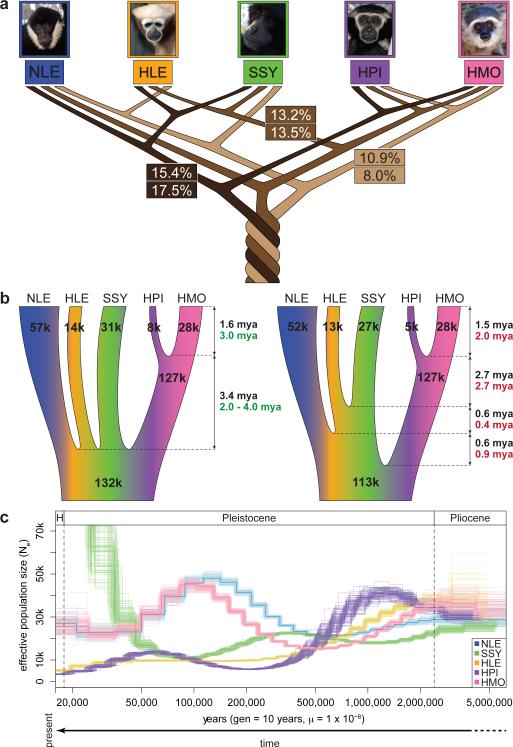
Figure 4. Gibbon phylogeny and demography.
a, The three most frequently observed UPGMA gene trees (numbers at the top) constructed across the genome at 100 kbp sliding windows and posterior probabilities (numbers at the bottom) for the same species topologies from a coalescent-based ABC analysis. The relatively low numbers observed suggest presence of substantial ILS amongst the gibbon genera. b, Parameters estimates describing gibbon population demography assuming an instant radiation for all four genera (left) and the most probable bifurcating species topology (right). Black, green and red numbers indicate divergence times and Ne as calculated by ABC, BEAST and G-PhoCS analysis respectively (Supplementary Information S9). c, PSMC analysis estimating changes in historical Ne. Note: the large increase in Ne observed in our PSMC plot for SSY in recent times is likely exaggerated due to higher sequencing error and mapping biases in non-NLE samples (see details in Supplementary Section S8). A generation time of 10 years45-46 was used to obtain a per generation mutation rate of 1x10-8 per year.
In order to infer the most likely bifurcating species topology amongst the four genera while taking into account ILS, we employed a novel coalescent-based ABC methodology using the autosomal nongenic and genic loci (Veeramah et al. submitted) (Supplementary Information S8). The topology described above had the highest combined posterior probability, though support was relatively low (p(Model)=17%) and other topologies, including one with NLE and Hylobates interchanged as the most external taxa, had comparable probabilities (Fig. 4a).
The estimated internal branch lengths under the best species topology using our ABC framework and G-PhoCS were very short, supporting a rapid speciation process for the four gibbon genera (Fig 4b-right). Given this observation and uncertainty in the best topology, we also estimated parameters under an instantaneous speciation model (Fig. 4b-left). Assuming an overall autosomal mutation rate of 1 x 10−9/site/year, we placed the beginning of the speciation process at ~5 mya under both models, with the two Hylobates species diverging ~1.5 mya.
Consistent with the ABC analysis, SSY and HLE share the largest number of alleles across the whole genome (Table ST8.5). However, NLE and the two Hylobates samples are both significantly closer to SSY than HLE as assessed by the D-statistic31. This result could be explained by two independent gene flow events between SSY and both NLE and Hylobates. However fertile intergenic hybrids have yet to be observed either in the wild or captivity32; an alternative explanation would be long-term population structure in the gibbon ancestral population. Both the ABC and G-PhoCS analyses suggest that the ancestral gibbon effective population size (Ne) was large (80,000-130,000) but neither of these frameworks can distinguish this from a structured ancestral population.
The coalescent-based analysis (Fig 4a), along with estimates of genome-wide heterozygosity (Fig ST8.2), suggests a larger long-term Ne for both N. leucogenys and H. moloch compared to the other species. Analysis using the pairwise sequentially Markovian coalescent (PSMC) model33 indicates that these two species underwent an increase in Ne during the Late Pleistocene era (500-100 thousand years ago (kya) followed by a subsequent decrease in Ne 100-50 kya (Fig. 4c) (Supplementary Information S8). It is important to point out that fluctuation in Ne could result from changes in the actual number of individuals in the population, changes in population structure, and/or variable gene flow.
Functional sequence evolution
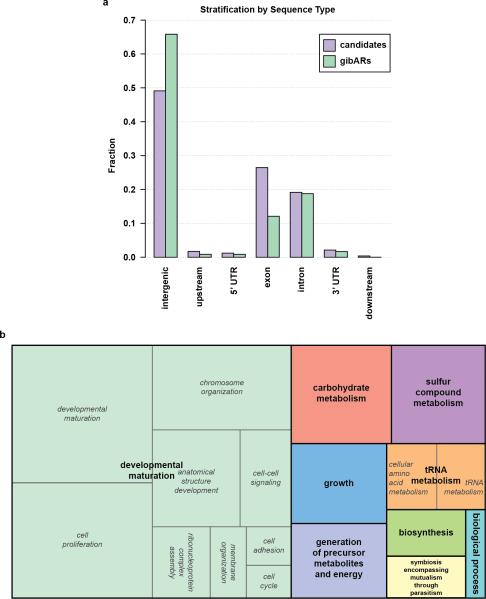
Accelerated substitution rates are a hallmark of adaptive evolution, and genomic regions with excess lineage-specific substitutions have been found to have functional roles34. We identified 240 short (153 bp median length) regions with accelerated substitution rates in the gibbon lineage (gibARs). We observed that gibARs were primarily intergenic (66%) and tended to co-localize near the same genes as LAVA elements (p-value=81E-06; odds ratio of 2.74 (1.79–4.07, 95% CI)). Consistent with this finding, a GO enrichment test for genes within +/−100 kbp of each gibAR (in comparison with background genes) revealed enrichment for the ‘chromosome organization’ category (Benjamini-Hochberg FDR <5%) (EDF 6). Given evidence of functional roles gathered for human accelerated regions35, we speculate that the gibARs may create functional elements (e.g., enhancer, protein-binding domains) to modulate the transcriptional effect of local LAVA insertions (Supplementary Information S12 and File 9).
We assessed the potential presence of positive selection in 13,638 human genes with one-to-one orthologs in gibbon using a branch-site likelihood ratio test36 (Supplementary Information S10). One of the most striking features of gibbons is their use of brachiation (i.e., arboreal locomotion using only the arms). We uncovered evidence related to traits possibly associated with this adaptation such as the gibbon's longer arms, more powerful shoulder flexors, rotator muscles, and elbow flexors37. First, some genes whose functions relate to these anatomical specializations appear to have undergone positive selection in gibbons. They include TBX5 (p-value=0.00015), required for the development of all forelimb elements38; COL1A1 (pro-alpha1 chains of type I collagen) (p-value=3.39E-11), the fibril-forming collagen main protein of bones, tendons, and teeth39; and CHRNA1 (acetylcholine receptor subunit alpha precursor) (p-value=0.00039), involved in skeletal muscle contraction40. These genes have not been identified as positively selected in other primates to date. We also observed that some genes involved in chondrogenesis (SNX19, ID2, and EXT1) were associated with gibARs. Finally, the chondroadherin gene (CHAD)41 coding for a cartilage matrix protein is specifically duplicated in all gibbon genera (EDF 2).
DISCUSSION
Our sequencing, assembling, and analysis of the gibbon genome has provided numerous insights into the accelerated evolution of the gibbon karyotype and identified genetic signatures related to gibbon biology. First, SDs and repetitive sequences were the best predictors of gibbon-human breakpoints, although we excluded a causal role given the predominance of non-homology-based repair signatures. Furthermore, accelerated rearrangement was confined to large-scale chromosomal events, pointing to a mechanism responsible for causing gross chromosomal changes, rather than global genomic instability. This is in line with our hypothesis that the high rate of chromosomal rearrangements may have been due to LAVA-induced premature transcription termination of chromosome segregation genes. This effect may have occurred at a low enough level to be compatible with life but sufficient to increase the frequency of chromosome segregation errors. Of note, the link between erroneous chromosome segregation and increased chromosomal rearrangement has been recently demonstrated by others through in vitro experiments25,26.
The question remains how such a high number of chromosomal rearrangements could become fixed in such a relatively short time. One possibility is that a combination of geographic isolation and post-mating reproductive barriers accelerated the radiation of the four gibbon genera. Our estimates dated the lineage-splitting event to the Miocene-Pliocene transition, when major changes in the distribution of tropical and subtropical forests were caused by the elevation of the Yunnan Plateau and rise in sea levels42,43. Furthermore, fluctuation in sea levels beginning in the Early Pliocene appears to have brought about cycles of forest fragmentation and amalgamation, leading to alternating range compression and expansion for many mammalian groups44.
Together these results advance our knowledge of the unique traits of the small apes and highlight the complex evolutionary history of these species. Moreover, our analyses of the shattered gibbon genome helped gain insight into the mechanisms of chromosome evolution and uncovered a novel source of genome plasticity.
METHODS
Sanger-based whole-genome sequencing was performed as described for other species. The genome assembly was generated using the ARACHNE genome assembler assisted with alignment data from the human genome (Supplementary Information S1). The source DNA for the sequencing was derived from a single female (Asia; studbook no. 0098, ISIS no. NLL605) housed at the Virginia Zoo in Norfolk, VA. Short-read libraries were constructed at the Oregon Health & Science University (OHSU) following standard Illumina protocols and sequenced on an Illumina HiSeq 2000. Analyses were performed with custom analysis pipelines. See Supplementary Information for additional methods.
Extended Data
Extended Data Figure 1. The gibbon assembly: statistics and quality control.
a, The table compares the gibbon assembly statistics to those of other primates sequenced with a similar strategy. b, The plot represents the percentage of the 10,734 single-copy gene HMMs (hidden Markov models) for which just one gene (blue) is found in the different mammalian genomes in Ensembl 70. Other HMMs match more than one gene (red). The missing HMMs (cyan) either do not match any protein or the score is within the range of what can be expected for unrelated proteins. The remaining category (green) represents HMMs for which the best matching gene scores better than unrelated proteins but not as well as expected. (See Supplementary Information section 1.4 for more details.)
Extended Data Figure 2. Analysis of gibbon-human synteny blocks and identification and validation of gibbon segmental duplications.
a, The image shows a representative gibbon-only WSSD (whole-genome shotgun sequence detection) call by Sanger read depth. The duplication identified in this case overlaps with the gene CHAD that codes for a cartilage matrix protein. b, Examples of FISH hybridizations on gibbon metaphases using duplicated human fosmid clones that were identified by the (WGS) detection strategy (red signals). A) Interchromosomal duplication. B) Interspersed intrachromosomal duplication. C) Intrachromosomal tandem duplication confirmed using cohybridization with a single control probe (blue signals). c, Megabases of lineage-specific and shared duplications for primates based on GRChr37 read depth analysis. Copy-number-corrected values by species are shown below.
Extended Data Figure 3. Analysis of LAVA element insertion in genes and early termination of transcription.
a, The histogram shows the results of permutation analyses. We find a significant association between LAVA elements and genes. Moreover, insertions are significantly enriched in introns and depleted in exons, most likely as a result of selection against insertions in exons. b, Schematic representation of the mechanism through which LAVA intronic insertions in anti-sense orientation might cause early termination of transcription: A) truncated transcript; B) normal transcript (pA=polyadenylation site). c, We calculated the distance to the nearest exon for each intronic LAVA and compared this to what would be expected for random insertions (i.e., background). We found fewer insertions than expected by chance within 1 kbp of the nearest exon. d, Identification of pmiRGlo_LA_F polyadenylation sites by 3’RACE. Alignment of thirteen 3’RACE PCR clone sequences and the pmiRGlo_LA_F sequence. LAVA_F 3’ TSD is highlighted by dark background; the major antisense LAVA_F polyadenylation signal (MAPS) is in red. The termination sites are marked with arrows on the LAVA_F sequence. Poly(A)tails of the identified transcripts are colored in red.
Extended Data Figure 4. Evolution of the LAVA element.
a, Screenshots from the Integrative Genomics Viewer (IGV) browser for loci MAP4, RABGAP1, and BBS9. Each column shows portions of the IGV visualization of a LAVA insertion locus identified in Nleu1.0 and its flanking sequence. Red rectangles indicate the margins of each LAVA insertion. Read pairs are colored in red when their insert size is larger-than-expected, indicating the presence of a LAVA insertion. MAP4 is a shared LAVA insertion, while RABGAP1 and BBS9 are Nomascus-specific. b, LAVA elements containing at least 300 bp of the LA section of LAVA elements were selected and reanalyzed using RepeatMasker to determine subfamily affiliation and divergence from the consensus sequence. LAVA elements are grouped based upon their subfamily affiliations (see legend top right). The x-axis shows the percent divergence from the respective consensus sequence, and the y-axis shows the number of elements with a certain percent divergence from the consensus sequence.
Extended Data Figure 5. Analysis of the phylogenetic relationships between gibbon genera.
a, Neighbor-joining trees for gibbons using non-genic loci. b, UPGMA trees for 100 kbp nonoverlapping sliding windows moving along the gibbon genome reporting the top 15 topologies (see also Supplementary Table ST8.3). The percentage of total support for each topology is given within each subpanel.
Extended Data Figure 6. Analysis of the relationship between gibbon accelerated regions (gibARs) and genes.
a, Intergenic regions are enriched in gibARs. Different sequence types are shown on the x-axis, and the y-axis displays the fraction of gibARs and candidate regions annotated to the respective class. gibARs are significantly enriched in intergenic regions (p = 4.7E-6) and significantly depleted in exons (p = 7.3E-6). p-values for each class were calculated with the Fisher's exact test. Introns are comparably prevalent in candidates and gibARs, while in UTR and flanking region counts are too low to draw meaningful conclusions (data not shown). b, TreeMap from REVIGO for GOslim Biological Process terms with a Benjamini-Hochberg FDR of 5%. Each rectangle is a cluster representative; larger rectangles represent ‘superclusters’ including loosely related terms. The size of the rectangles reflects the p-value.
Extended Data Table 1.
Genes from the ‘microtubule cytoskeleton’ GO category with LAVA insertions
| Gene | Function | LAVA strand | Polyadenylation signal | Orthology | Subfamily |
|---|---|---|---|---|---|
| CEP164 | G2/M checkpoint and nuclear divisions | antisense | TTTATT | Shared | LAVA_B2R2 |
| MAP4 | Spindle architecture | antisense | TTTATT | Shared | LAVA_B1R2 |
| STAU2 | RNA-decay | antisense | TTTATT | Shared | LAVA_C4A |
| KIFAP3 | Kinesin, motor protein moving on microtubules | antisense | TTTATT | Nomascus | LAVA_B1B |
| SNTB2 | Syntrophin | antisense | TTTATT | Nomascus | LAVA_B2R2 |
| BBS9 | Localizes to non-membranous centriolar satellites | antisense | TGTTTA | Nomascus | LAVA_E |
| DNHD1 | Dynein, motor protein moving on microtubules during mitosis | antisense | TTTGTT | Shared | LAVA_B2R2 |
| SHROOM3 | Regulator of the microtubule cytoskeleton | antisense | TTTGTT | Shared | LAVA_C2 |
| EVI5 | Centrosome stability and dynamics/completion of cytokinesis | antisense | TTTGTG | Shared | LAVA_B1R2 |
| SMC3 | Cohesin | antisense | TTTAGT | Nomascus | LAVA_B1F2 |
| MAD1L1 | Kinetochore-bound checkpoint protein | antisense | TT--TA | Shared | LAVA_D1 |
| BUB1B | Spindle checkpoint | antisense | TGTTTA | Shared | LAVA_F1 |
| HOOK3 | Centrosomal assembly | antisense | TGTTTA | Nomascus | LAVA_E |
| TRAF5 | TNF receptor-associated factor 5 | antisense | TGTTTA | Nomascus | LAVA_F2 |
| DYNC1LI1 | Intracellular trafficking and mitosis | antisense | TTTATT | Shared | LAVA_C4B |
| C2CD3 | Distal centriole formation | sense | TTTATT | Shared | LAVA_B1G |
| CLASP2 | Regulation of spindle and kinetochore function | sense | CTTACT | Shared | LAVA_B1R2 |
| DNAH3 | Dynein, motor protein moving on microtubules during mitosis | sense | TTTATT | Shared | LAVA_B2R1 |
| INVS | Cell rounding and spindle positioning during mitosis | sense | TTTATT | Shared | LAVA_C4B |
| KIF27 | Kinesin, motor protein moving on microtubules | sense | TTTATT | Shared | LAVA_B1D |
| MFN2 | Mitochondrial fusion | sense | TTTATT | Nomascus | LAVA_E |
| NINL | Centrosome, microtubule organization in interphase cells | sense | TTTATT | Shared | LAVA_B1F2 |
| RABGAP1 | Interaction with Mad2-spindle checkpoint | sense | TGTTTA | Nomascus | LAVA_E |
Genes highlighted in gray carry LAVA insertions that are shared, antisense, and carry a perfect antisense polyadenylation site.
Supplementary Material
Acknowledgments
The gibbon genome project was funded by the National Human Genome Research Institute (NHGRI) including grants U54 HG003273 (R.A.G) and U54 HG003079 (R.K.W.) with further support from National Institutes of Health NIH/NIAAA P30 AA019355 and NIH/NCRR P51 RR000163 (L.C.), R01_HG005226 (J.D.W., M.F.H.), NIH P30CA006973 (S.J.W.), fellowship from the National Library of Medicine Biomedical Informatics Research Training Program (N.L.), R01 GM59290 (M.A.B.) and U41 HG007497-01 (M.A.B, M.K.K.), R01 MH081203 (J.M.S.), HG002385 (E.E.E.), National Science Foundation (NSF) CNS-1126739 (B.U., M.A.B., M.K.K.) and DBI-0845494 (M.W.H.), PRIN 2012 (M.R.), Futuro in ricerca 2010 RBFR103CE3 (M.V.), ERC Starting Grant (260372) and MICINN (Spain) BFU2011-28549 (T.M.-B.), grant of the Ministry of National Education, CNCS – UEFISCDI, project number PN-II-ID-PCE-2012-4-0090 (A.D.), grant of the Deutsche Forschungsgemeinschaft SCHU1014/8-1 (G.G.S.), ERC Starting and Advanced Grant and EMBO Young Investigator Award (Z.I., N.V.F.), ERC Starting Grant and EMBO Young Investigator Award (D.T.O.) , Commonwealth Scholarship Commission (M.C.W.). E.E.E. is an investigator of the Howard Hughes Medical Institute. We acknowledge the contributions of the staff of the HGSC, including the operations team – H. Dinh, S. Jhangiani V. Korchina, C. Kovar; the library team – K. Blankenburg, L. Pu, S. Vattathil; the assembly team – D. Rio-Deiros, H. Jiang; the submissions team – M. Batterton, D. Kalra, K. Wilczek-Boney, W. Hale, G. Fowler, J. Zhang; the quality control team – P. Aqrawi, S. Gross, V. Joshi, J. Santibanez; and the sequence production team – U. Anosike, C. Babu, D. Bandaranaike, B. Beltran, D. Berhane-Mersha, C. Bickham, T. Bolden, M. Dao, M. Davila, L. Davy-Carroll, S. Denson, P. Fernando, C. Francis, R. Garcia III, B. Hollins, B. Johnson, J. Jones, J. Kalu, N. Khan, B. Leal, F. Legall III, Y. Liu, J. Lopez, R. Mata, M. Obregon, C. Onwere, A. Parra, Y. Perez, A. Perez, C. Pham, J. Quiroz, S. Ruiz, M. Scheel, D. Simmons, I. Sisson, J. Tisius, G. Toledanes, R. Varghese, V. Vee, D. Walker, C. White, A. Williams, R. Wright, T. Attaway, T. Garrett, C. Mercado, N. Ngyen, H. Paul, Z. Trejos. We thank Dr. Zoltan Ivics for providing some of the reagents. We additionally acknowledge the Production Sequencing Group at The Genome Institute. Wellcome Trust (grant numbers WT095908 and WT098051), NHGRI (U41HG007234) and European Molecular Biology Laboratory. For the production of next-generation sequences, we acknowledge the Massively Parallel Sequencing Shared Resources (MPSSR) at OHSU, the National Center of Genomic Analyses (CNAG) (Barcelona, Spain), the University of Arizona Genetics Core (UAGC), and the UCSF sequencing core. We also acknowledge the Louisiana Optical Network Institute (LONI). We thank the Gibbon Conservation Center and the Fort Wayne Children's Zoo for providing the gibbon samples. Resources for exploring the gibbon genome are available at UCSC (http://genome.ucsc.edu), Ensembl (http://ensembl.org), NCBI (http://ncbi.nlm.nih.gov), and the Baylor College of Medicine Human Genome Sequencing Center (https://www.hgsc.bcm.edu/non-human-primates/gibbon-genome-project). The MAKER annotation pipeline is supported by NSF IOS-1126998.We finally thank Ms. Tonia Brown for proofreading and editing the manuscript.
Footnotes
Author contributions. L.C. led the project and the manuscript preparation. L.C., W.C.W., K.C.W., J.R., E.E.E, M.T.-B., R.A.H., K.R.V., M.F.H. supervised the project and contributed to overall organization of the manuscript. L.C., T.J.M. prepared the figures. Sanger data production, assembly construction and testing: L.F., C.F., D.M.M., L.N., A.C. S.L.L, L.R.L., D.P.L., W.C.W., K.C.W., J.R., S.G., L.W.H., D.R., S.M. Mitochondrial genome assembly: Y.L. Illumina sequencing production and submission: L.C., T.M.-B., J.D.W., M.H., E.T., L.J.W., M.G., I.G., A.B., J.H-R. Provided samples: G.S. Gene set and validation of gene models: D.B., S.W., S.S., B.A., M.M., J.He., P.F., M.S.C., M.Y. Assembly validation: B.L.-G., J.He., T.M.-B. BAC library generation: P.J.dJ., B.tH., B.Z. Cytogenetic analyses: M.R., N.A., O.C. Segmental duplications and structural variations: J.H., C.B., B.L.-G., J.Q., M.F.-C., G.C., F.A., M.V., T.M.-B., E.E.E. cDNA Array CGH: L.D., M.O., A.K., J.M.S. Comparative analysis of gibbon chromosomal rearrangements: J.He. Breakpoint analysis: L.C., C.W., L.J.W. LAVA analysis: L.C., R.A.H., T.J.M., N.H.L., L.J.W., K.N., K.S., A.D., M.A.B., M.K.K., J.A.W., B.U., A.S., R.H. Luciferase assay and 3’ RACE: A.D., B.I., C.O., G.G.S., N.V.F., Z.I. RNA-seq analysis for early transcription termination: S.J.W., C.L.B. Short-read alignments, SNP calling and population genetics analysis (autosomal DNA): L.J., T.K.O., F.L.M., A.E.W., L.J.W., K.R.V., M.F.H., J.D.W. Population genetics analyses (mtDNA): C.R., L.W., M.B., T.M.-B. Positive selection analyses: G.W.C.T., M.W.H. Gene family evolution analyses: M.W.H., C.C. Gibbon accelerated region analyses: K.S.P., D.K. CTCF-binding analyses: M.C.W., D.T.O., P.F., E.T., C.W., L.J.W., J.He., K.B. Biogeography analysis: N.G.J., C.R. Principal Investigators: R.K.W., R.A.G.
Author information. The N. leucogenys WGS project has been deposited in GenBank under the project accession ADFV00000000.1. All short-read data have been deposited into the Short Read Archive (http://www.ncbi.nlm.nih.gov/sra) under the accession SRP043117. E.E.E. is on the scientific advisory board (SAB) of DNAnexus, Inc. and was an SAB member of Pacific Biosciences, Inc. (2009-2013) and SynapDx Corp. (2011-2013).
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
REFERENCES CITED
- 1.Mittermeier RA, Rylands AB, Wilson DE. Handbook of the mammals of the world. Lynx Edicions; Barcelona: 2013. [Google Scholar]
- 2.Carbone L, et al. A high-resolution map of synteny disruptions in gibbon and human genomes. PLoS Genet. 2006;2:e223. doi: 10.1371/journal.pgen.0020223. doi:06-PLGE-RA-0357R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Locke DP, et al. Comparative and demographic analysis of orang-utan genomes. Nature. 2011;469:529–533. doi: 10.1038/nature09687. doi:10.1038/nature09687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gibbs RA, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. doi:10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 5.Girirajan S, et al. Sequencing human-gibbon breakpoints of synteny reveals mosaic new insertions at rearrangement sites. Genome Res. 2009;19:178–190. doi: 10.1101/gr.086041.108. doi:10.1101/gr.086041.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carbone L, et al. Evolutionary breakpoints in the gibbon suggest association between cytosine methylation and karyotype evolution. PLoS Genet. 2009;5:e1000538. doi: 10.1371/journal.pgen.1000538. doi:10.1371/journal.pgen.1000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bailey JA, Eichler EE. Primate segmental duplications: crucibles of evolution, diversity and disease. Nat Rev Genet. 2006;7:552–564. doi: 10.1038/nrg1895. doi:10.1038/nrg1895. [DOI] [PubMed] [Google Scholar]
- 8.Yan CT, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. doi:nature06020. [DOI] [PubMed] [Google Scholar]
- 9.Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. doi:10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merkenschlager M, Odom DT. CTCF and cohesin: linking gene regulatory elements with their targets. Cell. 2013;152:1285–1297. doi: 10.1016/j.cell.2013.02.029. doi:10.1016/j.cell.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 11.Schwalie PC, et al. Co-binding by YY1 identifies the transcriptionally active, highly conserved set of CTCF-bound regions in primate genomes. Genome Biology. 2013;14:R148. doi: 10.1186/gb-2013-14-12-r148. doi:10.1186/gb-2013-14-12-r148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carbone L, et al. Centromere remodeling in Hoolock leuconedys (Hylobatidae) by a new transposable element unique to the gibbons. Genome Biol Evol. 2012;4:648–658. doi: 10.1093/gbe/evs048. doi:10.1093/gbe/evs048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 14.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. doi:10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 15.Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008;9(Suppl 1):S4. doi: 10.1186/gb-2008-9-s1-s4. doi:10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamburov A, Wierling C, Lehrach H, Herwig R. ConsensusPathDB--a database for integrating human functional interaction networks. Nucleic Acids Res. 2009;37:D623–628. doi: 10.1093/nar/gkn698. doi:10.1093/nar/gkn698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baker DJ, Jin F, Jeganathan KB, van Deursen JM. Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell. 2009;16:475–486. doi: 10.1016/j.ccr.2009.10.023. doi:10.1016/j.ccr.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samora CP, et al. MAP4 and CLASP1 operate as a safety mechanism to maintain a stable spindle position in mitosis. Nat Cell Biol. 2011;13:1040–1050. doi: 10.1038/ncb2297. doi:10.1038/ncb2297. [DOI] [PubMed] [Google Scholar]
- 19.Leber B, et al. Proteins required for centrosome clustering in cancer cells. Sci Transl Med. 2010;2:33ra38. doi: 10.1126/scitranslmed.3000915. doi:10.1126/scitranslmed.3000915. [DOI] [PubMed] [Google Scholar]
- 20.Schuyler SC, Wu YF, Kuan VJ. The Mad1-Mad2 balancing act - a damaged spindle checkpoint in chromosome instability and cancer. J Cell Sci. 2012;125:4197–4206. doi: 10.1242/jcs.107037. doi:10.1242/jcs.107037. [DOI] [PubMed] [Google Scholar]
- 21.Maia AR, et al. Cdk1 and Plk1 mediate a CLASP2 phospho-switch that stabilizes kinetochore microtubule attachments. J Cell Biol. 2012;199:285–301. doi: 10.1083/jcb.201203091. doi:10.1083/jcb.201203091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haraguchi K, Hayashi T, Jimbo T, Yamamoto T, Akiyama T. Role of the kinesin-2 family protein, KIF3, during mitosis. J Biol Chem. 2006;281:4094–4099. doi: 10.1074/jbc.M507028200. doi:M507028200. [DOI] [PubMed] [Google Scholar]
- 23.Han JS, Szak ST, Boeke JD. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429:268–274. doi: 10.1038/nature02536. doi:10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- 24.Wheelan SJ, Aizawa Y, Han JS, Boeke JD. Gene-breaking: a new paradigm for human retrotransposon-mediated gene evolution. Genome Res. 2005;15:1073–1078. doi: 10.1101/gr.3688905. doi:gr.3688905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Damert A, et al. 5'-Transducing SVA retrotransposon groups spread efficiently throughout the human genome. Genome Res. 2009;19:1992–2008. doi: 10.1101/gr.093435.109. doi:10.1101/gr.093435.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wojtasz L, et al. Meiotic DNA double-strand breaks and chromosome asynapsis in mice are monitored by distinct HORMAD2-independent and -dependent mechanisms. Genes Dev. 2012;26:958–973. doi: 10.1101/gad.187559.112. doi:10.1101/gad.187559.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchani EE, Xing J, Witherspoon DJ, Jorde LB, Rogers AR. Estimating the age of retrotransposon subfamilies using maximum likelihood. Genomics. 2009;94:78–82. doi: 10.1016/j.ygeno.2009.04.002. doi:10.1016/j.ygeno.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gronau I, Hubisz MJ, Gulko B, Danko CG, Siepel A. Bayesian inference of ancient human demography from individual genome sequences. Nat Genet. 2011;43:1031–1034. doi: 10.1038/ng.937. doi:10.1038/ng.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wall JD, et al. Incomplete lineage sorting is common in extant gibbon genera. PLoS One. 2013;8:e53682. doi: 10.1371/journal.pone.0053682. doi:10.1371/journal.pone.0053682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC evolutionary biology. 2007;7:214. doi: 10.1186/1471-2148-7-214. doi:10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Durand EY, Patterson N, Reich D, Slatkin M. Testing for Ancient Admixture between Closely Related Populations. Molecular Biology and Evolution. 2011;28:2239–2252. doi: 10.1093/molbev/msr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirai H, Hirai Y, Domae H, Kirihara Y. A most distant intergeneric hybrid offspring (Larcon) of lesser apes, Nomascus leucogenys and Hylobates lar. Hum Genet. 2007;122:477–483. doi: 10.1007/s00439-007-0425-0. [DOI] [PubMed] [Google Scholar]
- 33.Li H, Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011;475:493–496. doi: 10.1038/nature10231. doi:10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prabhakar S, et al. Human-specific gain of function in a developmental enhancer. Science. 2008;321:1346–1350. doi: 10.1126/science.1159974. doi:10.1126/science.1159974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pollard KS, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. doi: 10.1038/nature05113. doi:10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- 36.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Molecular biology and evolution. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. doi:10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 37.Michilsens F, Vereecke EE, D'Aout K, Aerts P. Functional anatomy of the gibbon forelimb: adaptations to a brachiating lifestyle. J Anat. 2009;215:335–354. doi: 10.1111/j.1469-7580.2009.01109.x. doi:10.1111/j.1469-7580.2009.01109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Browne ML, et al. Evaluation of genes involved in limb development, angiogenesis, and coagulation as risk factors for congenital limb deficiencies. Am J Med Genet A. 2012;158A:2463–2472. doi: 10.1002/ajmg.a.35565. doi:10.1002/ajmg.a.35565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marini JC, et al. Consortium for osteogenesis imperfecta mutations in the helical domain of type I collagen: regions rich in lethal mutations align with collagen binding sites for integrins and proteoglycans. Hum Mutat. 2007;28:209–221. doi: 10.1002/humu.20429. doi:10.1002/humu.20429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Masuda A, et al. hnRNP H enhances skipping of a nonfunctional exon P3A in CHRNA1 and a mutation disrupting its binding causes congenital myasthenic syndrome. Hum Mol Genet. 2008;17:4022–4035. doi: 10.1093/hmg/ddn305. doi:10.1093/hmg/ddn305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hessle L, et al. The skeletal phenotype of chondroadherin deficient mice. PLoS One. 2013;8:e63080. doi: 10.1371/journal.pone.0063080. doi:10.1371/journal.pone.0063080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cane MA, Molnar P. Closing of the Indonesian seaway as a precursor to east African aridification around 3-4 million years ago. Nature. 2001;411:157–162. doi: 10.1038/35075500. doi:10.1038/35075500. [DOI] [PubMed] [Google Scholar]
- 43.Jing-Xian Xu DKF, Li Cheng-Sen, Wang Yu-Fei. Late Miocene vegetation and climate of the Lühe region in Yunnan, southwestern China. Review of Palaeobotany & Palynology. 2008:36–59. [Google Scholar]
- 44.David S, Woodruff LMT. The Indochinese–Sundaic zoogeographic transition: a description and analysis of terrestrial mammal species distributions. Journal of Biogeography. 2009:803–821. [Google Scholar]
- 45.Harvey PH, Martin RD, Clutton-Brock TH. Life histories in comparative perspective. Chicago. 1987 [Google Scholar]
- 46.Kim SK, et al. Patterns of genetic variation within and between Gibbon species. Mol Biol Evol. 2011;28:2211–2218. doi: 10.1093/molbev/msr033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.