Abstract
Heterogeneous nuclear ribonucleoprotein (hnRNP) C plays a key role in RNA processing. More recently hnRNP C has also been shown to function as a DNA binding protein exerting a dominant-negative effect on transcriptional responses to the vitamin D hormone,1,25-dihydroxyvitamin D (1,25(OH)2D), via interaction in cis with vitamin D response elements (VDREs). The physiologically active form of human hnRNPC is a tetramer of hnRNPC1 (huC1) and C2 (huC2) subunits known to be critical for specific RNA binding activity in vivo, yet the requirement for heterodimerization of huC1 and C2 in DNA binding and downstream action is not well understood. While over-expression of either huC1 or huC2 alone in mouse osteoblastic cells did not suppress 1,25(OH)2D-induced transcription, over-expression of huC1 and huC2 in combination using a bone-specific polycistronic vector successfully suppressed 1,25(OH)2D-mediated induction of osteoblast target gene expression. Over-expression of either huC1 or huC2 in human osteoblasts was sufficient to confer suppression of 1,25(OH)2D-mediated transcription, indicating the ability of transfected huC1 and huC2 to successfully engage as heterodimerization partners with endogenously expressed huC1 and huC2. The failure of the chimeric combination of mouse and human hnRNPCs to impair 1,25(OH)2D-driven gene expression in mouse cells was structurally predicted, owing to the absence of the last helix in the leucine zipper (LZ) heterodimerization domain of hnRNPC gene product in lower species, including the mouse. These results confirm that species-specific heterodimerization of hnRNPC1 and hnRNPC2 is a necessary prerequisite for DNA binding and down-regulation of 1,25(OH)2D-VDR-VDRE-directed gene transactivation in osteoblasts.
Introduction
1,25-dihydroxyvitamin D (1,25(OH)2D) is the active form of vitamin D in target tissues such as bone. Upon interaction with its cognate intracellular receptor, vitamin D receptor (VDR), and dimerizing with the unliganded retinoid X receptor (RXR), the 1,25(OH)2D-VDR-RXR complex acts to transregulate the expression of genes by directly binding to specific vitamin D response elements (VDREs). This binding event influences transcription of vitamin D-responsive genes via concerted interaction of the activated VDR with the chromatin remodeling apparatus as well as with receptor co-activators and co-repressors in the transcriptional machinery of the cell that ultimately control the diverse physiological actions of vitamin D [1–4]. In previous studies we have described an additional regulatory component of intracellular 1,25(OH)2D-VDR-RXR signaling involving alternatively-spiced members of the heterogeneous nuclear ribonucleoprotein C family of nucleic acid binding proteins (hnRNP1 and hnRNP C) [5–9].
hnRNPs are abundantly expressed nucleo-cytoplasmic shuttle proteins that share a wide array of nucleic acid and protein targets that impact a multitude of cellular and molecular functions throughout the cell [10–12]. For example, the functional hnRNPC1/C2 tetramer (C13C21) [13,14] is known to play a role in i) chromatin remodeling [15], ii) the early steps of spliceosome assembly and pre-mRNA splicing [16,17], and iii) in modulating the stability, export and level of translation of bound mRNA molecules by interacting with the 5'-UTR of mRNAs and noncoding RNAs (ncRNAs) [18,19]. Although classically characterized as a RNA binding and regulating protein, hnRNPC1/C2 can also interact with double-stranded DNA. In chromatin immunoprecipitation studies we have shown that hnRNPC1/C2 binds to VDREs in the absence of the VDR, with hnRNPC1/C2-VDRE occupancy being displaced by the 1,25(OH)2D-bound VDR-RXR complex [8]. In this capacity hnRNPC1/C2 functions as a dominant-negative-acting VDRE-binding protein (VDRE-BP). This action of hnRNPC1/C2 appears to contribute to normal VDR signaling capacity by occupying VDREs in the basal state and by participating in reciprocal cyclical VDRE binding following exposure to the VDR-activating ligand, 1,25(OH)2D. This reciprocal relationship between VDR and hnRNPC1/C2 is disrupted when the VDRE-BP is over-expressed, leading to 1,25(OH)2D insensitivity in the target cell [8,20–23]. In vivo, over-expression of hnRNPC1/C2 has been associated with vitamin D resistant rachitic bone disease in vitamin D-deprived New World primates [24] and in man [7]. Based on these molecular and phenotypic observations, the aim of the present study was to recapitulate and translate the VDRE-BP-induced vitamin D-resistant state in human bone cells by developing and characterizing murine osteoblastic cell lines which stably over express tetrameric HNRNPC proteins.
Results
Effects of human hnRNPC1/C2 on 1,25(OH)2D-mediated gene transactivation in mouse osteoblasts
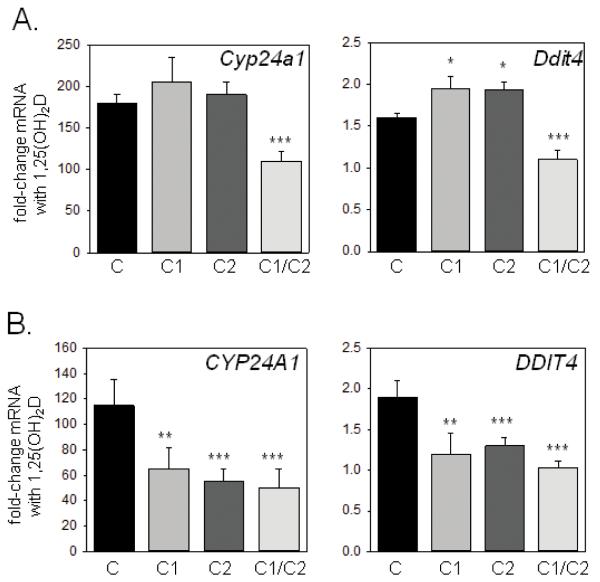
As shown in Figure 1A when transfected into mouse MC3T3 osteoblasts, suppression of 1,25(OH)2D-mediated transcription was only observed when huC1 and huC2 were used in combination. In fact, expression of Ddit4, a gene we have previously shown to be induced by 1,25(OH)2D in osteoblasts [20], was enhanced not suppressed, in mouse MC3T3 cells transfected with either huC1 or huC2 alone in the presence of 1,25(OH)2D. By contrast, over-expression of either huC1 or huC2 was sufficient to suppress 1,25(OH)2D-induced gene expression in human MG-63 osteoblast-like cells, with combined over-expression of huC1 and huC2 having an additive suppressive effect (Figure 1B).
Figure 1. Effects of over-expressed human hnRNPC1 and C2 on vitamin D receptor (VDR)-mediated gene transcription in human and mouse osteoblast-like cells.
Effect of over-expression of human hnRNPC1 (C1) and/or human hnRNPC2 (C2) on 1,25(OH)2D-induction of the vitamin D-target genes CYP24A1/Cyp24a1 and DDIT4/Ddit4 in A) mouse MC3T3 osteoblastic cells and B)human MG-63 osteoblastic cells. Cells were treated with or without 10 nM 1,25(OH)2D for 6 hrs, 24 hrs post-transfection with either empty vector (C), C1, C2 or C1 and C2 (C1/C2). Data show mean±SD (n=3 separate cultures) fold-change in mRNA expression for CYP24A1/Cyp24a1 and DDIT4/Ddit4 for each transfected cell type following treatment with 1,25(OH)2D relative to vehicle-treated control cells. ***statistically different from empty vector controls (C), p < 0.001, **statistically different from C, p < 0.01.
To determine whether the lack of efficacy of huC1 and huC2 individually to squelch 1,25(OH)2D-directed gene expression was due to relatively low transfection efficiency and subsequent expression of the human subunits in mouse osteoblasts, similar studies were also carried out using a bone-specific vector to drive expression of huC1 in mouse cells harboring the endogenously expressed oligomerization partner, muC2 (Figure 2A,B). Despite 50–3000-fold higher levels of expression relative to the endogenous muC1 and muC2 expression levels, transfection with huC1 continued to have no significant effect on 1,25(OH)2D-induced gene expression (Figure 2C).
Figure 2. Stable expression of only human hnRNPC1 protein in mouse MC3T3-E1 osteoblasts does not confer resistance to 1,25(OH)2D.
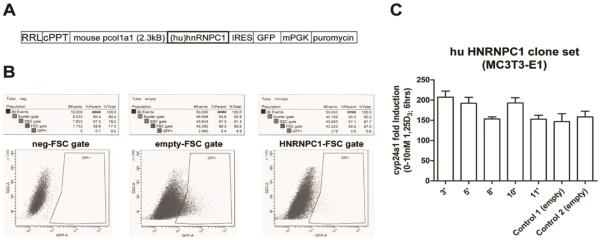
A) Osteoblastic transduction was performed using the lentivirus RRL transfer vector harboring a stop codon-less human (hu) hnRNPC1 cDNA and a green fluorescent protein (GFP) cassette driven by the bone-specific mouse col1a1 2.3kB promoter. The lenti vector backbone also contains the central polypurine tract (cPPT) from the HIV-1 integrase gene to increase the copy number of lentivirus integrating into the host genome to enhance viral titer. B) FACS sorting of GFP-positive MC3T3-E1 osteoblasts transduced with the hu hnRNPC1 and empty lentivirus vectors for the isolation and generation of stable clones. C) 15 hu hnRNPC1-over expressing stable lines were created (data not shown), whereby five (clones 3', 5', 8', 10', 11') lines with variable hnRNPC1 over expression were further evaluated for 1,25(OH)2D (10 nM, 6 hrs) mediated induction of endogenous mouse (mu) Cyp24a1 mRNA.
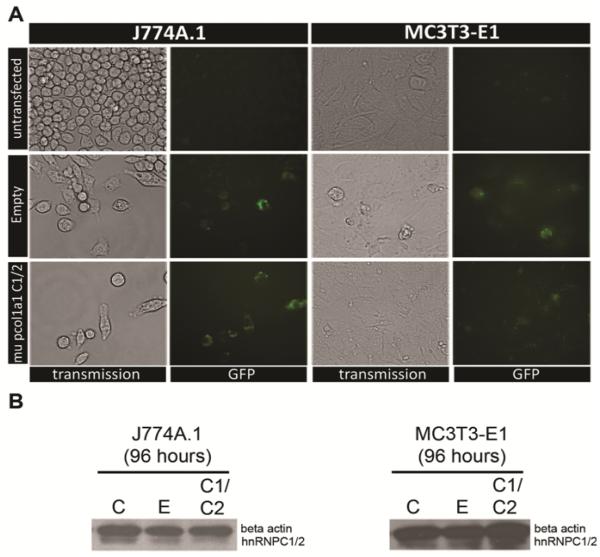
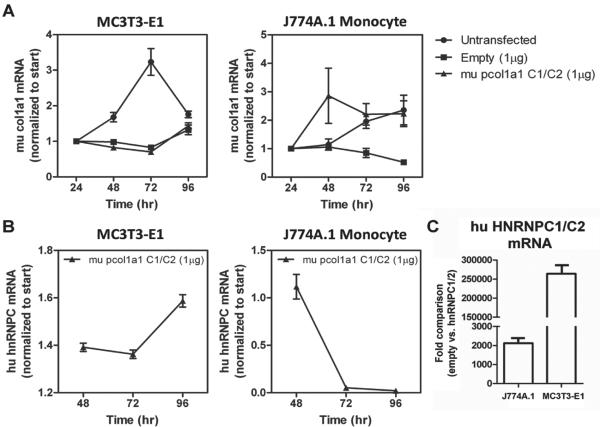
The apparent requirement for both huC1 and huC2 to suppress 1,25(OH)2D-directed transcription in mouse osteoblasts was further illustrated in MC3T3 transfected cell lines where huC1 and huC2 were co-expressed under the control of the osteoblast-specific mouse col1a1 promoter. In contrast to the previous experiments, vectors used in this part of the study encoded the huC1 and huC2 linked to one another by the self-cleaving 23 amino acid picornaviral 2A-like sequence from the porcine teschovirus-1 (P2A) to allow for efficient functional stoichiometric expression of multiple flanking proteins under the influence of the bone-specific col1a1 2.3kB promoter (Figure 3A; [25]). The advantage of combined transgene expression with the P2A over transgenes linked by an internal ribosomal entry site or introduced by individual constructs is its independence of copy number required for integration, its relatively small size and its high cleavage efficiency [26]. Using this system, both the huC1 and huC2 isoforms were equally over-expressed specifically in the nuclear compartment of MC3T3 cells (Figure 3B). The efficiency of transfection of the single open reading frame was assessed by GFP expression within various cell lines (Figure 4A), and the absence of uncleaved product in Western blot analyses indicated efficient cleavage of the P2A protein (Figure 4B). Cell (tissue)-specific expression of huC1/huC2 was confirmed by parallel analysis of the temporal pattern of endogenous mouse Col1a1 mRNA levels over time in MC3T3 bone cells but not J774A mouse monocytes (24–96 hrs) (Figure 5).
Figure 3. Equimolar expression of human hnRNPC1 and C2 confers resistance to 1,25(OH)2D in mouse MC3T3-E1 osteoblasts.
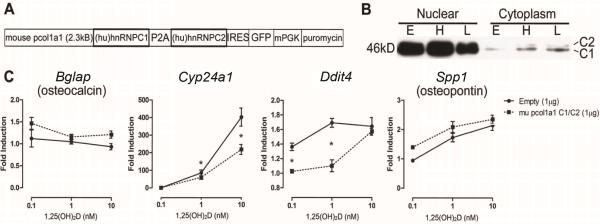
A) Schematic representation of the mouse col1a1 2.3kB promoter-driven piconoviral P2A sequence construct for the co-expression of human (hu) hnRNPC1 and C2 proteins with a GFP reporter. B) Western blot analyses showing over-expression of hnRNPC1 and C2 proteins using the P2A construct enriched in the nuclear fraction of MC3T3-E1 cells at 96 hrs post transfection. Empty vector (E), and high (H, 2μg/well) and low (L, 0.1 μg/well) levels of P2A expression vector cDNA were transfected into MC3T3 cells. C) 1,25(OH)2D ([0.1–10nM]; 6 hrs) induction of mouse osteoblastic genes after transfection of empty vector or hnRNPC1/C2 P2A expression construct in MC3T3-E1 96 hrs post transfection.
Figure 4. Efficient transfection of the P2A hnRNPC1/C2 construct.
A) Transient transfection of P2A hnRNPC1/C2 construct with IRES-GFP reporter shows efficient labeling of both J774A.1 mouse monocytic and MC3T3-E1 osteoblastic cells after 96 hrs post transfection. The empty vector control also depicts efficient and comparable GFP labeling. B) Western analysis of whole cell lysate for beta actin and hnRNPC1/C2 expression 96 hours post transfection.
Figure 5. Cell-type specific action and temporal regulation of the P2A hnRNPC1/C2 construct.
A) Endogenous mouse (mu) type I collagen expression was monitored over a 96 hr period under the different transient transfection conditions. For both transfected constructs into MC3T3-E1 cells, col1a1 mRNA expression was delayed when compared to the untransfected controls. In J774A.1 mouse monocytes the expression of endogenous mu type I collagen was variable between the transfected P2A construct and untransfected control, and unregulated in the empty vector control. B) Human (hu) hnRNPC mRNA levels were monitored in the mouse col1a1-promoter driven P2A construct transfected cells at 96 hrs post transfection. Hu hnRNPC mRNA was temporally regulated in a similar fashion as mu type I collagen in MC3T3-E1 osteoblasts. No temporal regulation of hu hnRNPC mRNA was observed in J774A.1 mu monocytes transfected with the P2A construct, suggesting the mu col1a1 2.3kB promoter was bone-specific. C) After 96 hrs of transfection, MC3T3-E1 cells harboring the P2A hnRNPC1/C2 construct express hu hnRNPC with mRNA levels approximately 125-fold higher relative to the matching empty vector control when compared to the J774A.1 mouse monocytic cell line.
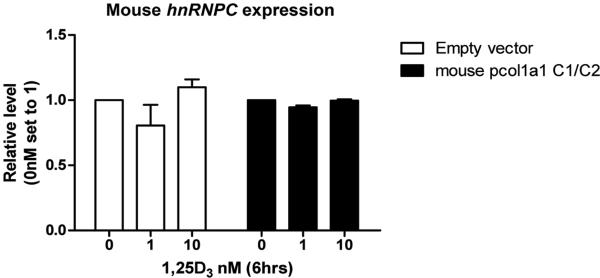
As shown in Figure 3C, co-expression of huC1 and huC2 using the P2A vector acted as a dominant-negative inhibitor of 1,25(OH)2D-induced expression of Cyp24a1 and Ddit4 target genes in mouse osteoblasts. Endogenous muC1/muC2 transcript levels were unchanged under both basal and 1,25(OH)2D-treated conditions (Figure 6), suggesting no interference by endogenous mouse hnRNPC protein subunits.
Figure 6. Endogenous mouse hnRNPC not responsible for functional effects.
Endogenous mouse (mu) hnRNPC expression was monitored in MC3T3-E1 cells after transfection and 6 hour treatment with various concentrations of 1,25(OH)2D.
Species-specific variations in the structure of the hnRNPC oligomerization domains dictate function
The hnRNPC proteins associate with RNA as tetramers with a composition of 3 hnRNP C1 subunits and 1 hnRNP C2 subunit (C13-C21). Studies in vivo have shown that the formation of the tetrameric complex, which is highly stable in solution, is driven by a 28 residue helical region (residues 180–207) referred as hnRNPC leucine zipper-like (CLZ) oligomerization domain [27] (Figure 7A). The CLZ domain promotes the oligomerization by forming a coiled coil tetramer structure; hnRNPC proteins possessing mutations in the CLZ domain bind weakly to nucleic acid substrates [27]. Amino acid sequences of the hnRNPC CLZ oligomerization domain are well conserved within vertebrates, exhibiting 74–98% identity to the human CLZ domain (Figure 7B). Like many oligomerizing proteins, hnRNPC isoforms employ α-helical coiled coils as a oligomerization mechanism with the parts stabilized by continuous interhelical contacts formed between hydrophobic faces of amphipathic helices [28]. For each set of seven residues (heptad) along the helix (denoted a–g; see Figure 7B) the contact geometry is repeated for the ideal coiled coil. Hydrophobic residues occupying the a and d heptad positions form interhelical contacts in the coiled coil core as well as other contact surfaces at interfacial e and g heptad positions. Comparison of the human and mouse hnRNPC sequences in Figure 7B revealed that the Ser at position 200 in the mouse is replaced by Asp in man; as such, the most distal interfacial g position heptad of the hnRNPC CLZ domain used for hnRNPC1-hnRNPC2 oligomerization in the mouse is disrupted. .
Figure 7. hnRNPC1/C2 tetramer stability based on species-specific amino acid substitutions.
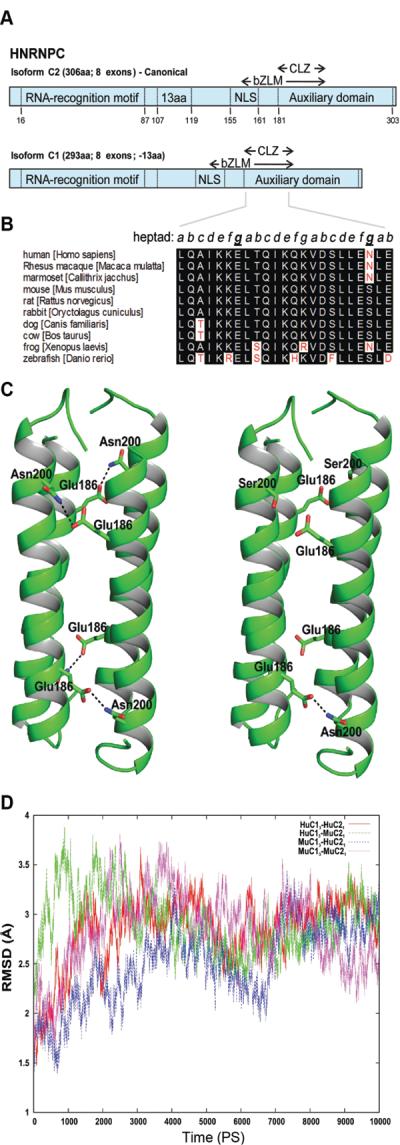
A) Schematic representation of human hnRNPC isoforms. Each protein contains a single RNA recognition motif (RRM), a delineated nuclear localization signal (NLS), a basic leucine zipper-like motif (bZLM), and an encompassing acidic auxiliary domain. hnRNPC2 contains an additional 13 a.a. and is expressed at one-third the level of hnRNPC1. An oligomerization domain (CLZ) is also present in C proteins. Numbers represent a.a. position. B) Alignment of CLZ sequences derived from the Ensembl database (www.ensembl.org). Key asparagine (N) to serine (S) change within the mouse sequences occupying an interhelical contact surface heptad residue within the coiled coil core of hnRNP C. Similar residues are highlighted in black while discrepant residues are highlighted in white. The heptad arrangement is in italics, with the key intermolecular residues forming anti-parallel chain contacts underlined and bolded. Numbering based on the human C2 full-length isoform. C) The hnRNPC protein CLZ tetramers of human (Hu) and mouse (Mu) C1 and C2 proteins. HuC13-HuC21 (left panel) and MuC13-HuC21 (right panel). The Asn200, Ser200 and Glu186 residues are shown in stick representation. In HuC13-HuC21 CLZ domains, pairs from the adjacent helices make four hydrogen bonds in the coiled coil tetramer. In contrast, in the MuC13-HuC21 tetramer, only one hydrogen bond is possible. The hydrogen bonds formed between Asn200 and Glu186 are shown in dashed lines. D) Molecular dynamic simulations for CLZ tetramers. The root mean square fluctuations for the backbone atoms of the tetramer complexes for different combinations of human (Hu) and mouse (Mu) hnRNPC1 and C2. HuC13-HuC21, red; HuC13-MuC21, green; MuC13-HuC21, blue and MuC13-MuC21, magenta. Data are based on experimental structure over 10 nanosec simulations.
Free energy and stability analysis of tetramers formed by combinations of mouse and human hnRNPC1/C2
To determine whether interaction between the Asp-200 in the human and Ser-200 in the mouse hnRNPC protein was sufficient to cause destabilization of the CLZ coiled hnRNPC1/C2 tetramer, the binding free energy of interspecies tetramers harboring three hnRNPC1 and one hnRNPC2 subunit combinations, huC13-muC21 and muC13 -huC21, was calculated using the MMPBSA method implemented in Amber 9.0. When calculating the binding free energy, the holo-tetramer was considered as a complex, the three hnRNPC1 subunits as a receptor and the hnRNPC2 subunit and its CLZ domain as a binding ligand partner. In all cases, the negative binding free energy indicated favorable interactions between the C13-C21 CLZ domains regardless of the species derivation of the subunits. As shown in Table 1, the highest negative binding energy was observed with huC13-huC21, possibly due to the additional favorable interactions that could occur between Asn-200 and Glu-186 in the tetramer (Figure 7C, left panel). By contrast, only one hydrogen bond is possible for the muC13-huC21 complex (Figure 7C, right panel), providing a potential explanation for the relatively low negative binding energy for the mouse hnRNPC1/C2 tetramer. The negative free energy of the interspecies oligomeric combinations, muC13-huC21 (−107.22 Kcal/mol) and huC13-muC21 (−103.25 Kcal/mol), were intermediate to the species-specific tetramers (Table 1). These results indicate that the Asn-200 in the human hnRNPC CLZ domain plays a pivotal role in stabilization of the hnRNPC1/C2 oligomer in man. To assess the potential impact of hnRNPC subunit binding energy variations on tetramer stabilization, molecular dynamic simulations were carried out (Figure 7D). Data are shown as the root mean square deviations (RMSD) for the backbone atoms (N-CA-C-O) from the experimental structure, over 10 nanoseconds for the four tetrameric combinations depicted in Table 1. The huC13-muC21 inter-species tetramer complex showed large fluctuations (instability) during the initial phases of the 10 nanosecond period of simulation but eventually reached a state comparable the other tetramers after 7 nanoseconds.
Table 1.
Binding free energy of the hnRNPC CLZ domain according to tetramer combinations of mouse and human hnRNPC1 and C2. The total binding free energy for different combinations of human (hu) and mouse (mu) hnRNPC1 and C2 CLZ domains, estimated from MMPBSA calculations.
| CLZ tetramer | ΔGMM/PBSA (Kcal/mol) |
|---|---|
| HuC13-HuC21 | −112.07 |
| HuC13-MuC21 | −103.25 |
| MuC13-HuC21 | −107.22 |
| MuC13-MuC21 | −94.59 |
Discussion
The goal of the present study was to develop a mouse model that recapitulated the hnRNPC1/C2 (VDRE-BP) over-expression-induced vitamin D-resistant state observed in vivo in the skeletons of growing nonhuman and human primates [7]. Before developing the animal model, we assessed the effects of hnRNPC1/C2 over expression using a mouse osteoblast cell line. The studies here show that over-expression of huC1 and huC2 in combination or transfection with a bone-specific, polycistronic vector suppressed trans-species 1,25(OH)2D-mediated induction of mouse osteoblast target gene expression. This finding suggests that human hnRNPC1/C2 can function as a dominant-negative-acting inhibitor of 1,25(OH)2D-driven gene expression in mouse bone cells and that species-specific tetramerization is a critical determinant of its actions as a regulator of VDR-directed transactivation.
A monomer of the hnRNPC protein has a primary structure that is common to hnRNPs and which can be divided into four distinct domains: 1] the RNA recognition motif (RRM) consisting of beta-sheets; 2] the basic high affinity RNA binding domain or bZLM, residues 180–207; 3] the leucine zipper containing four heptad repeats toward the carboxyl-terminus (CLZ); and 4) the negatively-charged carboxyl-terminal domain (CTD) [29,30]. It is the bZLM domain, rather than the RRM, which plays the major role in binding to pre-mRNA and snRNA with high affinity [31]. Based on detailed sequence-specific labeling and solution structural analyses, the current model suggests that hnRNPC proteins exist in an anti-parallel four-helix tetrameric bundle which is centrally positioned and that these bundles assemble through highly stable associations of their leucine zipper CLZ domains [27,32]. The human and mouse CLZ domains are well conserved with the only difference being an Asp to Ser change at the terminal helix of the coiled coil structure (Figure 7B). The CTD is thought to be involved in protein-protein interactions that serve to increase specificity of nucleotide binding. Although there is increasing awareness of the composition, function, and mechanisms associated with hnRNPC and homologous proteins, the protein-protein interactions that participate in their recruitment to cis elements along genomic DNA have yet to be fully defined. Emerging evidence shows that the CTD of hnRNPs interacts with transcription elongation factors, chromatin-modifying complexes and pre-mRNA maturation enzymes, thus providing a molecular basis for the coupling between gene transcription and RNA processing [15]. While the hnRNPC protein domain responsible for interaction in cis with VDREs regulated expression of 1,25(OH)2D-driven target genes remains uncertain, our modeling indicates that the DNA trans-binding domain may be located within a hinge portion of the acidic CTD. This would place it apart from the core bundle stabilization domain known to bind cis elements which vary among higher and lower mammalian species [15,32].
hnRNPs were first recognized for their ability to bind to single-stranded RNA [10,33,34]. However, it was subsequently determined that hnRNPs have the ability to bind both single- and double-stranded DNA [5,7,15] with specific cis-acting DNA sequences being important determinants in transcriptional regulation of gene expression under the control of steroid hormone receptors such as VDR and the estrogen receptor [9]. In the current study over-expression of either huC1 or huC2 in human osteoblasts was sufficient to suppress 1,25(OH)2D-VDR-mediated signaling. Unexpectedly, repression of 1,25(OH)2D-VDR-VDR-directed transcription was only achieved by co-over-expression of both huC1 and huC2 in murine osteoblasts. These data, together with molecular dynamic simulation analyses presented here Figure 7 suggest that optimal DNA binding and the subsequent dominant-negative action of hnRNPC1/C2 on transcription occurs with species-specific tetramerization patterns involving the C1 and C2 isoforms (Figure 7). Binding free energy modeling further suggests that this differential response is due to subtle variations in the protein-protein interactions required for hnRNPC1/C2 tetramerization.
While the computational molecular dynamic studies presented here showed that interspecies tetramers of C1 and C2 can form (Figure 7), it is also known that tetramer stability is influenced by elements in the negatively charged CTD of hnRNPC that have diverged most between mouse and man [31]. For example, a deletion construct of the human hnRNPC lacking the CTD beyond the leucine zipper can exist as a dimer [32], highlighting the importance of this region of hnRNPC protein for tetramer assembly. Further, velocity sedimentation analysis of deletion constructs of the hnRNPC CTD demonstrate that the residues between S-240 and G-263 play a critical role in the native tetramer formation and stabilization [32]. Interestingly, the only proximal variation between mouse and human sequences within the CTD is at amino acid position 242, where mouse harbors an alanine and humans a glycine (data not shown), suggesting that differences in the hydropathy index of that portion of the CTD may alter CTD-dependent functions. Importantly, site-specific mutagenesis experiments that either removed or reversed the charges at specific residues within this proximal CTD resulted in a decrease in tetramer assembly [32]. Either lysine or alanine (non-charged) substitutions significantly attenuated tetramer assembly; the mouse harbors an alanine at position 242 in the CTD of hnRNPC. These observations suggest that the incompletely conserved CLZ domain and the carboxyl-terminal half of the CTD between mouse and man may disturb the function of hnRNPC by affecting subunit oligomerization and by altering interactions between the non-homologous residues of the CTD and the four-helix bundle of native hnRNPC proteins. With these observations in mind, we postulate that changes in the helix-interaction structures of hnRNPC tetramers can fundamentally alter its binding affinity for cis elements controlling 1,25(OH)2D-VDR-driven target genes.
It is clear that the full range of hnRNPC activities has not been elucidated. For example, members of the hnRNPC family can bind single-stranded DNA [35], suggesting that they may have distinct roles in DNA replication and recombination events as well as in transcription and transcript handling. The identification of species-specific cellular functions of hnRNPC proteins and their specific DNA targets will greatly help to elucidate the broader role of this versatile nuclear protein, in particular its role in vitamin D physiology and diseases that are marked by relative degrees of target tissue resistance to active vitamin D metabolites.
Materials and Methods
Reagents and cell culture
Crystalline 1,25(OH)2D (Biomol, Plymouth Meeting, PA USA) was reconstituted in absolute ethanol. Cells were treated with 0-10nM of 1,25(OH)2D incubated in the respective media with 1% FCS. MC3T3-E1 murine and MG63 human osteoblastic cells (both ATCC, Manassas, VA USA) were maintained in α-MEM plus 10% FBS and incubated at 37°C in a 95% air/5% CO2 atmosphere until 80–90% confluent and then passaged. The J774A.1 mouse monocyte/macrophage cell line (ATCC) was cultured in DMEM with 10% FCS. For over-expression studies, the hnRNPC1 and C2 constructs were previously cloned and ligated into the pCDNA 3.1/V5-His-TOPO vector (Invitrogen, USA; [8]); the constructs were transfected using the BioT reagent (Bioland Scientific, Paramount, CA USA) in 12-well tissue culture plates (0.1–1.0 μg/well total recDNA). Cells were analyzed at designated time points post transfection or transduction.
Cell fractionation and Western blot
Treated cells were harvested and washed with ice-cold PBS. Cell lysates were prepared in RIPA buffer with 1 X ProteoBlock protease inhibitor cocktail (Fermentas, Glen Burnie, MD USA) and 1mM PMSF. For separation of nuclear and cytoplasmic fractions for extraction, the NE-PER nuclear and cytoplasmic extraction kit (Thermo Fisher Scientific Inc., Rockford, IL USA) was utilized. Protein samples (20μg) were separated by SDS-PAGE. Primary anti-hnRNP C1/C2 (Santa Cruz Biotechnology sc-32308, Santa Cruz, CA USA) and secondary antibodies were used at 1:1000. I-Block™ blocking reagent (Applied Biosystems, Foster City, CA USA) was used during incubation steps.
Quantitative real-time PCR (qPCR) analysis
RNA was prepared using the RNeasy minikit (Qiagen Inc., Valencia, CA USA). cDNA was synthesized by SuperScript Reverse Transcriptase III (Invitrogen) utilizing random hexamers. qPCR analysis was performed with a Stratagene MX-3005P instrument utilizing TaqMan® system reagents from Applied Biosystems, Foster City, CA USA (Table S1). Target genes were normalized to 18S rRNA expression. All cDNAs were amplified under the following conditions: 50°C for 2 min; 95°C for 10 min followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. All reactions were performed in triplicate.
Bicistronic human hnRNPC1 and C2 vector construction
Construct pRRL-sin-cPPT-mouse (mu) col1a1 2.3kB promoter-IRES-GFP-hPGK-puromycin was produced by PCR of mu pcol1a1 with XbaI at the 5' end (primer 5-col1a1-XbaI (Table S2)) and blunted at the 3' end (primer 3-col1a1-MluI) prior to digestion. The mouse col1a1 2.3kB promoter was kindly provided by Benoit de Crombrugghe (Univ. Texas, M. D. Anderson Cancer Center). Construct pRRL-sin-cPPT-mu col1a1 2.3kB promoter-human (hu) hnRNPC1-IRES-GFP-hPGK-puromycin was produced by PCR of hu hnRNPC1 [8] with a MluI site at the 5' end (primer 5-hu hnRNPC1-MluI) and EcoRI at the 3' end (primer 3-hu hnRNPC1-EcoRI). The products were then digested and ligated into pRRL-sin-cPPT-mu col1a1 2.3kB promoter-IRES-GFP-hPGK-puromycin cut with MluI and EcoRI. Construct pRRL-sin-cPPT-mu col1a1 2.3kB promoter-hu hnRNPC1-P2A-hu hnRNPC2-IRES-GFP-PGK-puromycin was produced by PCR of hu hnRNPC2 blunted at the 5' end (primer 5-hu hnRNPC2) and with ClaI at the 3' end (primer 3-hu hnRNPC2-ClaI). This product was then digested and ligated with pRRL-sin-cPPT-CMV-GFP-P2A-MCS cut with SmaI and EcoRI. This plasmid was then used to PCR P2A-hu hnRNPC2 blunted at the 5' end (primer 5-P2A) and with EcoRI at the 3' end (primer 3-hu hnRNPC2-EcoRI). PCR of hu hnRNPC1 was produced with EcoRI at the 5' end (primer 5-hu hnRNPC1-EcoRI) and blunted without the stop codon at the 3' end (primer 3-hu hnRNPC1-(-TAA)). Both were digested and ligated into plasmid pRRL-sin-cPPT-mu col1a1 2.3kB promoter-IRES-GFP-hPGK-puromycin digested with EcoRI. Corresponding empty vectors were also constructed.
Lentiviral production and transduction
Lentiviral production was performed for construct pRRL-sin-cPPT-mu col1a1 2.3kB promoter-hnRNPC1-IRES-GFP-hPGK-puromycin. The construct pMD.G was used for the production of the VSV-G viral envelope in combination with the packaging constructs pHit60 [36] and the pBabe constructs correspond to the different transfer vectors. Briefly, 100 mm dishes of nonconfluent 293T cells were co-transfected with 6.5μg of pHit60, 3.5μg of pMDG (encoding the VSV-G envelope) and 10μg of p Babe (hu hnRNPC1 construct), by the CaPi-DNA coprecipitation method [37]. Next day, the medium was adjusted to make a final concentration of 10mM sodium butyrate and the cells were incubated for 8 hrs to obtain high-titer virus production as previously described. After the 8 hr incubation, cells were washed and incubated in fresh medium without sodium butyrate. Conditioned medium was harvested 16 hrs later and passed through 0.45 mm filters. For transduction of MC3T3-E1 cells, cells were seeded at 8×104 cells per well in 12-well plates. Transductions were carried out in 0.5 ml DMEM, including serial dilutions of lentiviral vector supernatant. The cells were then washed with phosphate-buffered saline (PBS) after 24 hrs post-transduction and incubated in regular medium for 48 hrs.
Molecular dynamic simulations and free energy calculations for hnRNPC
The coordinates of the human hnRNP C leucine zipper-like oligomerization domain (CLZ) tetramer (PDB code 1TXP) [27] were used for the simulations. PDB structure contains Ile at position 180 [27]; to get the correct human or mouse sequence, the Ile was mutated to Leu using the program PyMol (www.pymol.org). The mouse hnRNPC tetramer was built by mutating the residue Asn200 to Ser. Molecular dynamics simulations were performed in explicit water using NAMD [38] with CHARMM27/CMAP force field [39]. The tetramers were solvated in a cubic TIP3P solvent box of 9.0 Å. Periodic boundary conditions, Particle Mesh Ewald (PME) summation and SHAKE-enabled 2-femto seconds time steps were used in all molecular dynamics simulations. Initial configurations were subjected to a 500-step minimization with the harmonic constraints of 10 kcal/mol/Å2 on the protein heavy atoms. The systems were gradually heated from 0–300 Kelvin over a period of 30 picosec with harmonic constraints. The simulation at 300 Kelvin was then continued for 200 picosec during which the harmonic constraints were gradually lifted. The systems were then equilibrated for a period of 300 picosec before the 10 nanosec production run. All simulations were carried out in the NPT ensemble. The binding free energy of the hnRNPC2 to hnRNPC1 for the different (interspecies) combinations were estimated using the MMPBSA module in AMBER 9.0 [40] by taking snapshots (2000) at every 5 picosec from the 10 nanosec production run.
Data normalization and statistical analysis
The real-time qPCR methodology is based on determining the delta cycle threshold (ΔCt) values. The average ΔCt value was taken for triplicate sets. The fold induction was calculated by the formula: 2(avg ΔCt 0nm - avg ΔCt i), where i = (0-10nM values). An analysis of variance (ANOVA) statistical test was conducted with a Bonferroni multiple comparison post hoc analysis where p≤0.05 was considered statistically significant.
Acknowledgements
Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Number 5R01AR037399. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank the UCLA Vector Core (Dr. Emmanuelle Faure and Kip Hermann) for vector and viral preparations supported by JCCC/P30 CA016042 and CURE/P30 DK41301. We thank Dr. Duilio Casio for the computational facility at UCLA-DOE Institute of Genomics and Proteomics.
References
- 1.Carlberg C, Campbell MJ. Vitamin D receptor signaling mechanisms: Integrated actions of a well-defined transcription factor. Steroids. 2013;78:127–136. doi: 10.1016/j.steroids.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pike JW, Meyer MB. The vitamin D receptor: new paradigms for the regulation of gene expression by 1,25-dihydroxyvitamin D3. Rheum Dis Clin North Am. 2012;38:13–27. doi: 10.1016/j.rdc.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haussler MR, Whitfield GK, Kaneko I, Haussler CA, Hsieh D, et al. Molecular mechanisms of vitamin d action. Calcif Tissue Int. 2013;92:77–98. doi: 10.1007/s00223-012-9619-0. [DOI] [PubMed] [Google Scholar]
- 4.Haussler MR, Haussler CA, Whitfield GK, Hsieh JC, Thompson PD, et al. The nuclear vitamin D receptor controls the expression of genes encoding factors which feed the “Fountain of Youth” to mediate healthful aging. J Steroid Biochem Mol Biol. 2010;121:88–97. doi: 10.1016/j.jsbmb.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen H, Hu B, Allegretto EA, Adams JS. The vitamin D response element-binding protein. A novel dominant-negative regulator of vitamin D-directed transactivation. J Biol Chem. 2000;275:35557–35564. doi: 10.1074/jbc.M007117200. [DOI] [PubMed] [Google Scholar]
- 6.Adams JS, Chen H, Chun RF, Nguyen L, Wu S, et al. Novel regulators of vitamin D action and metabolism: Lessons learned at the Los Angeles zoo. J Cell Biochem. 2003;88:308–314. doi: 10.1002/jcb.10333. [DOI] [PubMed] [Google Scholar]
- 7.Chen H, Hewison M, Hu B, Adams JS. Heterogeneous nuclear ribonucleoprotein (hnRNP) binding to hormone response elements: a cause of vitamin D resistance. Proc Natl Acad Sci U S A. 2003;100:6109–6114. doi: 10.1073/pnas.1031395100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H, Hewison M, Adams JS. Functional characterization of heterogeneous nuclear ribonuclear protein C1/C2 in vitamin D resistance: a novel response element-binding protein. J Biol Chem. 2006;281:39114–39120. doi: 10.1074/jbc.M608006200. [DOI] [PubMed] [Google Scholar]
- 9.Lisse TS, Hewison M, Adams JS. Hormone response element binding proteins: novel regulators of vitamin D and estrogen signaling. Steroids. 2011;76:331–339. doi: 10.1016/j.steroids.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krecic AM, Swanson MS. hnRNP complexes: composition, structure, and function. Curr Opin Cell Biol. 1999;11:363–371. doi: 10.1016/S0955-0674(99)80051-9. [DOI] [PubMed] [Google Scholar]
- 11.Carpenter B, MacKay C, Alnabulsi A, MacKay M, Telfer C, et al. The roles of heterogeneous nuclear ribonucleoproteins in tumour development and progression. Biochim Biophys Acta. 2006;1765:85–100. doi: 10.1016/j.bbcan.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 12.Dreyfuss G. Structure and function of nuclear and cytoplasmic ribonucleoprotein particles. Annu Rev Cell Biol. 1986;2:459–498. doi: 10.1146/annurev.cb.02.110186.002331. [DOI] [PubMed] [Google Scholar]
- 13.Nakagawa TY, Swanson MS, Wold BJ, Dreyfuss G. Molecular cloning of cDNA for the nuclear ribonucleoprotein particle C proteins: a conserved gene family. Proc Natl Acad Sci U S A. 1986;83:2007–2011. doi: 10.1073/pnas.83.7.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merrill BM, Barnett SF, LeStourgeon WM, Williams KR. Primary structure differences between proteins C1 and C2 of HeLa 40S nuclear ribonucleoprotein particles. Nucleic Acids Res. 1989;17:8441–8449. doi: 10.1093/nar/17.21.8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahajan MC, Narlikar GJ, Boyapaty G, Kingston RE, Weissman SM. Heterogeneous nuclear ribonucleoprotein C1/C2, MeCP1, and SWI/SNF form a chromatin remodeling complex at the beta-globin locus control region. Proc Natl Acad Sci U S A. 2005;102:15012–15017. doi: 10.1073/pnas.0507596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Irimura S, Kitamura K, Kato N, Saiki K, Takeuchi A, et al. HnRNP C1/C2 may regulate exon 7 splicing in the spinal muscular atrophy gene SMN1. Kobe J Med Sci. 2009;54:E227–236. [PubMed] [Google Scholar]
- 17.Izquierdo JM. Heterogeneous ribonucleoprotein C displays a repressor activity mediated by T-cell intracellular antigen-1-related/like protein to modulate Fas exon 6 splicing through a mechanism involving Hu antigen R. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shetty S. Regulation of urokinase receptor mRNA stability by hnRNP C in lung epithelial cells. Mol Cell Biochem. 2005;272:107–118. doi: 10.1007/s11010-005-7644-2. [DOI] [PubMed] [Google Scholar]
- 19.McCloskey A, Taniguchi I, Shinmyozu K, Ohno M. hnRNP C tetramer measures RNA length to classify RNA polymerase II transcripts for export. Science. 2012;335:1643–1646. doi: 10.1126/science.1218469. [DOI] [PubMed] [Google Scholar]
- 20.Lisse TS, Liu T, Irmler M, Beckers J, Chen H, et al. Gene targeting by the vitamin D response element binding protein reveals a role for vitamin D in osteoblast mTOR signaling. Faseb J. 2011;25:937–947. doi: 10.1096/fj.10-172577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lisse TS, Hewison M. Vitamin D: A new player in the world of mTOR signaling. Cell Cycle. 2011;10 doi: 10.4161/cc.10.12.15620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lisse TS, Hewison M, Adams JS. Hormone response element binding proteins: Novel regulators of vitamin D and estrogen signaling. Steroids. 2011 doi: 10.1016/j.steroids.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adams JS, Chen H, Chun R, Gacad MA, Encinas C, et al. Response element binding proteins and intracellular vitamin D binding proteins: novel regulators of vitamin D trafficking, action and metabolism. J Steroid Biochem Mol Biol. 2004;89–90:461–465. doi: 10.1016/j.jsbmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 24.Chen H, Arbelle JE, Gacad MA, Allegretto EA, Adams JS. Vitamin D and gonadal steroid-resistant New World primate cells express an intracellular protein which competes with the estrogen receptor for binding to the estrogen response element. J Clin Invest. 1997;99:669–675. doi: 10.1172/JCI119210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Felipe P, Martin V, Cortes ML, Ryan M, Izquierdo M. Use of the 2A sequence from foot-and-mouth disease virus in the generation of retroviral vectors for gene therapy. Gene Ther. 1999;6:198–208. doi: 10.1038/sj.gt.3300811. [DOI] [PubMed] [Google Scholar]
- 26.Kim JH, Lee SR, Li LH, Park HJ, Park JH, et al. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One. 2011;6:e18556. doi: 10.1371/journal.pone.0018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whitson SR, LeStourgeon WM, Krezel AM. Solution structure of the symmetric coiled coil tetramer formed by the oligomerization domain of hnRNP C: implications for biological function. J Mol Biol. 2005;350:319–337. doi: 10.1016/j.jmb.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 28.Liu J, Zheng Q, Deng Y, Cheng CS, Kallenbach NR, et al. A seven-helix coiled coil. Proc Natl Acad Sci U S A. 2006;103:15457–15462. doi: 10.1073/pnas.0604871103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burd CG, Dreyfuss G. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 1994;13:1197–1204. doi: 10.1002/j.1460-2075.1994.tb06369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swanson MS, Nakagawa TY, LeVan K, Dreyfuss G. Primary structure of human nuclear ribonucleoprotein particle C proteins: conservation of sequence and domain structures in heterogeneous nuclear RNA, mRNA, and pre-rRNA-binding proteins. Mol Cell Biol. 1987;7:1731–1739. doi: 10.1128/mcb.7.5.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McAfee JG, Shahied-Milam L, Soltaninassab SR, LeStourgeon WM. A major determinant of hnRNP C protein binding to RNA is a novel bZIP-like RNA binding domain. RNA. 1996;2:1139–1152. [PMC free article] [PubMed] [Google Scholar]
- 32.Shahied L, Braswell EH, LeStourgeon WM, Krezel AM. An antiparallel four-helix bundle orients the high-affinity RNA binding sites in hnRNP C: a mechanism for RNA chaperonin activity. J Mol Biol. 2001;305:817–828. doi: 10.1006/jmbi.2000.4331. [DOI] [PubMed] [Google Scholar]
- 33.Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 34.Dreyfuss G, Matunis MJ, Pinol-Roma S, Burd CG. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 35.Takimoto M, Tomonaga T, Matunis M, Avigan M, Krutzsch H, et al. Specific binding of heterogeneous ribonucleoprotein particle protein K to the human c-myc promoter, in vitro. J Biol Chem. 1993;268:18249–18258. [PubMed] [Google Scholar]
- 36.Soneoka Y, Cannon PM, Ramsdale EE, Griffiths JC, Romano G, et al. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 1995;23:628–633. doi: 10.1093/nar/23.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brooks BR, Brooks CL, 3rd, Mackerell AD, Jr., Nilsson L, Petrella RJ, et al. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Case DA, Cheatham TE, 3rd, Darden T, Gohlke H, Luo R, et al. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]