Abstract
Background
The epilepsies are a clinically heterogeneous group of disorders. Despite strong evidence for heritability, genome-wide association studies in epilepsy have had limited success in identifying risk loci probably due to relatively small sample sizes and lack of power. Here we report three meta-analyses conducted for all epilepsy and its two largest clinical subtypes – genetic generalized epilepsy (GGE) and focal epilepsy.
Methods
We examined 12 case/control cohorts comprising 8,696 cases and 26,157 controls. Cases were predominantly Caucasian from the United Kingdom, Western Europe, Finland, USA and Australia, with some African-American and Han Chinese subjects. Controls were ethnically matched. Subjects were phenotyped into categories of GGE, focal epilepsy and unclassified epilepsy. A fixed-effects meta-analysis was performed after standardized imputation, to account for different genotyping platforms across sites. Standardized association protocols were applied locally, prior to combining summary statistics for meta-analysis. The genome-wide significance threshold was set at p<1·66×10−8.
Findings
Meta-analysis of the ‘all epilepsy’ cohort identified loci at 2q24.3 (p=8·71×10−10) implicating SCN1A, a well-established monogenic epilepsy gene encoding the alpha1 sodium channel subunit and at 4p15.1 (p=5·44×10−9) harboring PCDH7 as the lead candidate, which encodes a protocadherin molecule not previously implicated in epilepsy. For the GGE cohort, a single signal at 2p16.1 (p=9·99×10−9) was observed, implicating VRK2 or FANCL respectively encoding a protein kinase involved in signal transduction and apoptosis and a ubiquitin ligase involved in a DNA repair pathway. No single nucleotide polymorphism (SNP) achieved genome-wide significance for focal epilepsy.
Interpretation
This first meta-genome-wide association study in epilepsy has uncovered new loci for the common forms of epilepsy and is a further step in understanding the genetic architecture of the epilepsies with the ultimate aim of assisting in biological understanding, classification and prognostication. The data suggest thatspecific loci can actpleiotropically raising risk for epilepsy broadly, or have effects limited to a specific epilepsy subtype. This suggests that future genetic analysis may benefit from both “lumping”, where all epilepsies are grouped together, and “splitting”, where specific clinical subtypes are analyzed.
Funding
International League Against Epilepsy and multiple governmental and philanthropic agencies.
INTRODUCTION
Epilepsy is a common disorder, affecting up to 4% of people at some time in life,1 comprising heterogeneous syndromes defined by clinical, electroencephalographic (EEG) and brain imaging criteria.2 Broadly, the epilepsies are divided clinically into generalized and focal forms. Genetic factors contribute to both, as shown by familial aggregation and twin studies.3 Causative mutations in many genes, including some coding for ion channel subunits and others affecting synaptic function or brain development, have been discovered.3,4 The majority of these discoveries are for relatively rare familial epilepsies segregating in a Mendelian fashion, or epilepsies (particularly the severe infantile epilepsies) arising from de novo mutations.5-7
The genetic determinants underlying the common epilepsies, where clinical genetic data suggest complex inheritance, remain largely unknown. There is some evidence to suggest a role for rare sequence and copy number variants,8-10 whereas the contribution of common polymorphisms is still unclear,11,12 partly reflecting the relatively small sample sizes analyzed to date.
The largest genome-wide association study (GWAS) in epilepsy so far, consisting of 3,445 focal epilepsy cases,13 found no variants of genome-wide significance. More recently, a study of 1,018 cases of mesial temporal lobe epilepsy with hippocampal sclerosis, a subtype of focal epilepsy, implicated the 2q24.3 region around the sodium channel SCN1A,14 and an independent study of Han Chinese patients with known or suspected lesional focal epilepsy found evidence for a risk allele at 1q32 based on a discovery sample of 504 cases.15
For generalized epilepsy, a GWAS incorporating 1,527 European cases with genetic generalized epilepsies (GGE) in the discovery analysis and 1,493 GGE cases in the replication cohort found evidence for common risk alleles at 2p16.1 and 17q21.32, and suggestive evidence at the SCN1A locus.16 In addition, associations were reported for the GGE subtype juvenile myoclonic epilepsy at 1q43 and for generalized absence epilepsy at 2q22.3.16
Here we present the first large multicenter collaboration designed to discover variants that may increase risk for common epilepsies, consisting of a meta-analysis of (prior to quality control (QC)) 40,789 subjects comprising 10,064 epilepsy cases from twelve cohorts and 30,725 control subjects.
In view of clinical evidence that there may be genetic factors that raise risk for epilepsy broadly and in a syndrome-specific manner,17-19 we pre-specified three analyses as part of the study. Variants were sought that influence risk for all epilepsies, for genetic generalized epilepsy (previously idiopathic generalized epilepsy)2,20 and for focal epilepsy.
METHODS
Study Design and Participants
A meta-analysis was performed on twelve previously published and unpublished epilepsy cohorts from EPICURE,16 EPIGEN,13 Philadelphia, the Melbourne -Imperial - Liverpool Collaboration,21 Finland13 and Hong Kong15 (see Supplementary Table 1 (pre-QC numbers)). These cohorts were identified from the literature (PubMed - using search terms of epilepsy, seizures, association studies) and through publicity via Chapters of the International League Against Epilepsy (ILAE) and during international conferences. All twelve case cohorts (and their associated controls) broadly aligned with European, Asian or African ancestry (see Table 1 (post-QC numbers) and Supplementary Fig.1).
Table 1.
Post QC case/control numbers, as structured for analysis.
| Index GWAS | Ethnicity1 | Epilepsy cases | GGE | Focal | Population controls3 |
|---|---|---|---|---|---|
| EPIGEN-Dublin | Irish | 638 | - | 520 | 2232 |
| EPIGEN-Brussels | Belgian | 505 | 48 | 406 | 1675 |
| EPIGEN-Duke | AA2& EA | 760 | 102 | 551 | 504 |
| EPIGEN-London | British + other | 1007 | 93 | 773 | 2494 |
| ILM Collaboration | European-descent | 1703 | 212 | 1263 | 2699 |
| GenEpa | Finnish | 422 | - | 422 | 1963 |
| EPICURE | NW- European | 1440 | 1440 | - | 2454 |
| Philadelphia_550_AA2 | African American | 324 | 81 | 222 | 2746 |
| Philadelphia_550_CAU | European American | 819 | 440 | 378 | 5736 |
| Philadelphia_Omni_AA | African American | 106 | - | - | 97 |
| Philadelphia_Omni_CAU | European American | 485 | 190 | 288 | 682 |
| Hong Kong | Asian-Han | 487 | - | 487 | 2875 |
| TOTAL | 8696 | 2606 | 5310 | 26157 |
Broad ethnicity of the cohort. AA: African American. EA: European American. Other: indicates mixed, as would be expected in a cosmopolitan population. European-descent: European-caucasian. NW-European: North-West European.
EPIGEN-Duke individuals of AA ancestry were merged with Philadelphia_550_AAcohort.
See Supplementary Table 2 for further details on control cohorts.
A combination of population-based datasets were employed as controls. These cohorts were both screened and unscreened for neurological conditions. Further details are available in Table 1 and Supplementary Table 2.
All study participants provided written informed consent for DNA analysis. Local institutional review boards reviewed and approved study protocols at each site.
Procedures
Phenotyping
Seizures and epilepsy syndromes were classified according to the ILAE terminology.2,20 For all cases, epilepsy specialists evaluated their phenotype at the source center. Patients with epilepsy were assigned to one of three phenotypic categories: 1) GGE, 2) focal epilepsy or 3) unclassified epilepsy. Cases for each were defined as follows:
– GGE: Criteria were tonic-clonic, absence or myoclonic seizures with generalized spike-wave discharges on EEG and no evidence of an acquired cause. In rare instances the criterion for a diagnostic EEG was waived when there was clear clinical evidence of myoclonic or absence seizures with tonic-clonic seizures, and no evidence for an acquired cause. The ILAE has adopted the term “GGE” for syndromes previously known as “idiopathic” or “primary” generalized epilepsies in view of strong evidence for a genetic basis from genetic epidemiological and twin studies and an absence of identified acquired factors.2,20
– Focal epilepsy: This comprised patients with a confirmed diagnosis of focal epilepsy, including cases with focal structural brain lesions. The samples were predominantly comprised of adults, so cases of benign epilepsy of childhood with centro-temporal spikes were not specifically included.
– Unclassified epilepsy: This group consisted of patients in whom there was neither electro-clinical evidence for generalized epilepsy, nor evidence for a focal seizure onset, or patients with evidence for generalized and focal epilepsy.
The phenotyping committee curated patient phenotypes into a single database. Details relating to individual case cohorts are provided in Supplementary Methods. Analyses were performed on three phenotypic groups – GGE, focal epilepsy and ‘all epilepsy’ consisting of all patients with a confirmed diagnosis of epilepsy, including GGE, focal and unclassified epilepsy.
Statistical analysis
Imputation
As contributing sites had employed different genotyping platforms, we conducted imputation to infer genotypes for common genetic variants that were not directly genotyped. This allowed us to combine results across sites. Each of the five sites imputed their study datasets according to a standardized protocol. This used IMPUTE2 to infer haplotypes and impute, usingthe 1000 Genomes Phase I (interim) June 2011 reference panel (see Supplementary Methods).
Association analysis
Every site performed linear mixed model (LMM) association analysis for each of their datasets, using the software FaSTLMM (version 1.09).22 This performs linear regression, including a polygenic term designed to account for the contributions of population stratification and causal variants aside from the one being tested. Although evaluating a binary trait, it is valid to use linear regression (rather than logistic regression) because effect sizes are expected to be small.
This analysis was performed separately for each of the pre-selected phenotypic categories of epilepsy: 1) all epilepsy; 2) GGE; 3) focal epilepsy. Gender was included as a covariate.
Meta-analysis
Fixed-effects meta-analysis was conducted using the software METAL.23 As the vast majority of the epilepsy cases considered were of European descent (see Table 1), we chose a fixed-effects model to optimize power. SNPs showing significant amounts of heterogeneity (p<0.05) were removed before applying the fixed-effects analysis. Genomic correction was applied to the association analysis results for each dataset before combining for meta-analysis. Again, these steps were performed separately for each of the three phenotypic tests.
Significance threshold for meta-analysis
We set our genome-wide threshold for statistical significance at 1·66×10−8, reflecting an empirical Bonferroni correction of the 5×10−8 genome-wide significance threshold for three tests. We regarded signals between 1·66×10−8 and 5×10−7 as suggestive evidence of association.
Power calculations
We calculated the proportion of heritability a variant must explain for the detection power to be at least 80%; we considered variance explained on the liability scale,24 for which we assumed a point prevalence of 0·5% for all epilepsy, 0·2% for GGE and 0·3% for focal epilepsy25 (Supplementary Fig.2).
Logistic regression
In addition to the main association analysis, we also performed logistic regression for variants in a 1Mb window centered on each variant that showed suggestive evidence of association (p<5×10−7) from any of the three meta-analyses (all epilepsy, GGE and focal). The purpose of this analysis was 1) a technical validation and 2) to estimate odds ratios. For this we analyzed the dosage data, including gender and the first 20 principal components, using PLINK,26 and then combined the results from each site again using a fixed effect meta-analysis.
Conditional analysis
Conditional analysis was performed using FaSTLMM on variants in the same regions as defined for logistic regression. The purpose of the conditional analysis was to determine if any other genetic variants in the region associated with the disease phenotype, independent of the strongest signal from that region. We conditioned on the most significant variants within each of the three regions, i.e. rs6732655 and rs28498976 for all epilepsy, and rs2947349 for GGE. Gender was included as a covariate in the conditional analysis.
Significance threshold for conditional analysis
We applied Bonferroni correction to control for multiple testing in the conditional analysis and set the threshold for significance at 5×10−6 (each 1Mb region contained approximately 10,000 SNPs).
Confirmatory genotyping
To examine the accuracy of the imputation across regions showing signals satisfying genome-wide significance, we conducted genotyping in a subset of patients included in the meta-analysis and compared hard genotypes with imputation dosage files. We selected a subset of individuals to represent each of the three broad ethnicities included in our analysis (i.e. Caucasian, African-American and Han Chinese). Genotyping was conducted using TaqMan®(Life Technologies) for rs28498976, Sanger sequencing for rs6732655 and Kasper KASP™ (LGC Genomics) for rs2947349. Results are shown in Supplementary Table 3.
Enrichment analysis
Enrichment analysis was conducted using the interval-based enrichment analysis tool as integrated in the package INRICH.27 Briefly, INRICH takes a set of independent, nominally associated genomic intervals and tests for enrichment of predefined gene sets using permutation. We considered variants with a p value < 1×10−5 and defined the interval around index SNPs using an r2 threshold of 0·2. Gene sets as defined by GO ontology pathways were tested for enrichment.
Role of the funding source
The sponsors of the study had no role in the study design, data collection, data analysis, data interpretation, or writing of the report. The Strategy and Analysis committee members had full access to all data in the study. The Strategy committee takes final responsibility for the decision to submit for publication.
RESULTS
All epilepsy
After application of our quality control (QC) criteria (see Supplementary Methods), we included a total of 34,853 individuals (8,696 epilepsy cases and 26,157 controls) across twelve cohorts in the ‘all epilepsy’ meta-analysis (see Table 1). We estimated 80% power to detect a variant explaining 0·07%, or greater, of the liability variance (see Supplementary Fig.2).
Principal component analysis indicated that patient cohorts clustered in three broad ethnicities (European, Asian and admixed African-American) as expected (see Supplementary Fig.1). We observed an inflation factor of 1·031, suggesting adequate control for possible cryptic stratification (see Supplementary Fig. 3a).
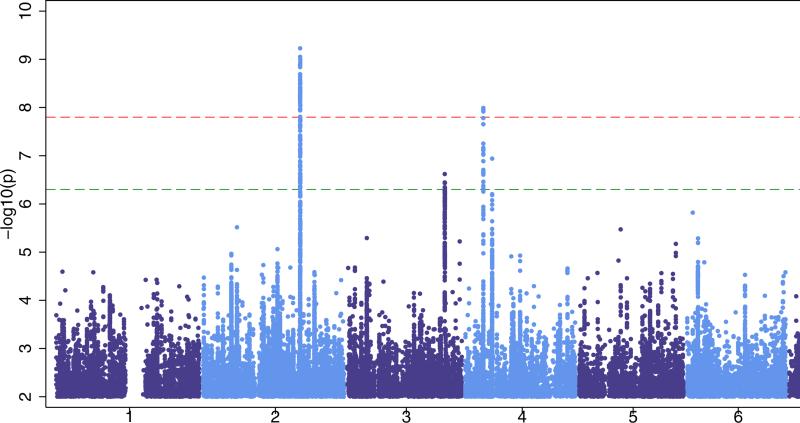
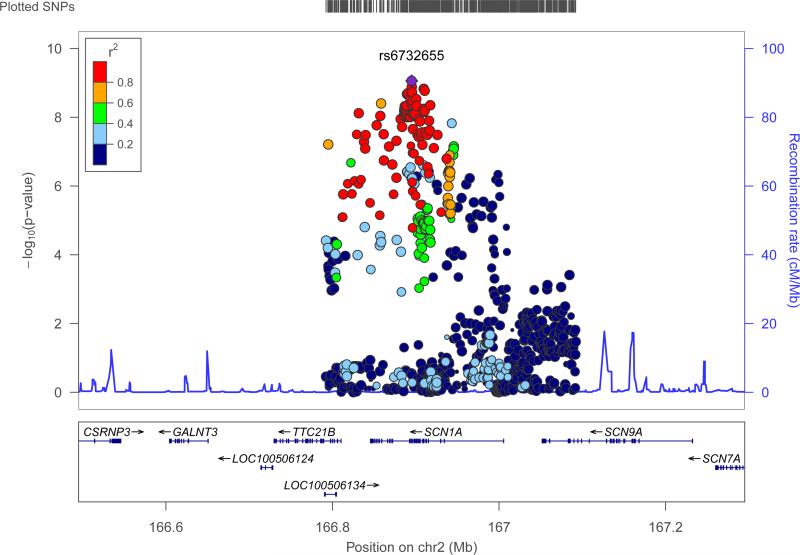
We identified two loci at genome-wide significance (p<1·66×10−8, see Fig. 1a). The first signal was located at 2q24.3 (Fig. 2). This signal was centered on the voltage-gated sodium channel SCN1A gene, which is a knowngene for certain monogenic epilepsies.28,29 The most strongly associated variant in this interval was rs6732655 (p=8·71×10−10, OR 0·89 (CI: 0·86-0·93), see Table 2 and Supplementary Fig.4), located in intron 16 of SCN1A. Seventy other variants in this region satisfied the threshold for genome-wide significance. Logistic regression validated the association with 2q24.3 (Supplementary Fig.5). The direction of effect was consistent across most cohorts, and there was no evidence of significant heterogeneity.
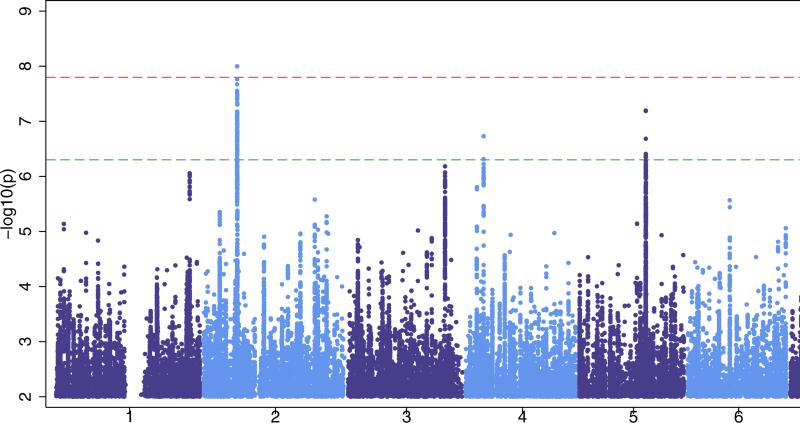
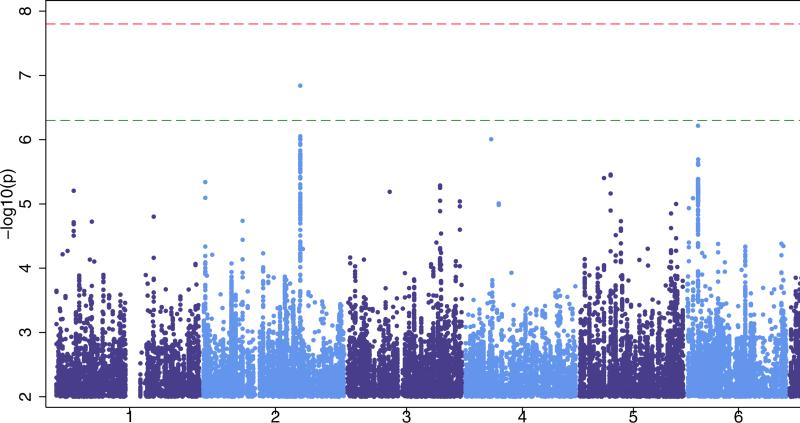
Figure 1. Manhattan plots for our three primary analyses.
‘all epilepsy’ (Fig 1a), GGE (Genetic Generalized Epilepsy, Fig 1b) and focal epilepsy (Fig 1c). The red line shows our threshold of ‘significance’ set at 1·66×10−8 and the greenline indicates the ‘suggestive’ threshold of 5×10−7.
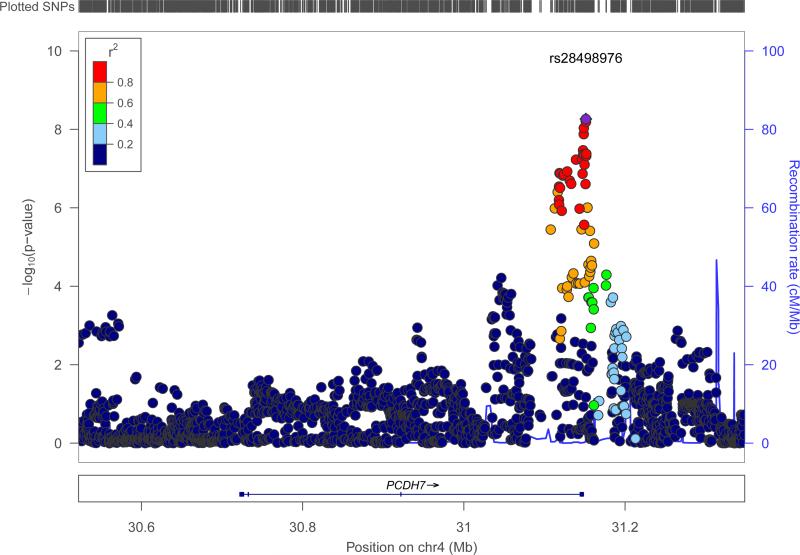
Figure 2. Genomic context of 2q24.3 signal from ‘all epilepsy’ analysis.
Plot created using LocusZoom.60 Linkage disequilibrium data are taken from 1000 Genomes Project, HG19, March 2012.
Table 2.
Summary of loci at p<5·0×10−7.
| rs number | chr | pos (bp) | A1/A2 | MAF | cand. gene | annotation | pheno | OR (CI) | LMM p | condp |
|---|---|---|---|---|---|---|---|---|---|---|
| rs6732655 | 2q24.3 | 166895066 | T*/A | 0.22(A) | SCN1A | intronic | allepi | 0·89 (0·86-0·93) | 8·71×10−10 | 4·95×10−7 |
| rs28498976 | 4p15.1 | 31151357 | A/G* | 0.46(A) | PCDH7 | intergenic | allepi | 0·90 (0·87-0·94) | 5·44×10−9 | 2·29×10−4 |
| rs111577701 | 3q26.2 | 167861408 | T/C* | 0.09(T) | GOLIM4 | intergenic | allepi | 1.16 (1.09-1.24) | 4·42×10−7 | - |
| rs535066 | 4p12 | 46240287 | T/G* | 0.40(G) | GABRA2 | intergenic | allepi | 1.10 (1.05-1.16) | 1·71×10−7 | - |
| rs2947349 | 2p16.1 | 58059803 | A*/C | 0.26(C) | VRK2/FANCL | intergenic | GGE | 1·23 (1·16-1·31 | 9·99×10−9 | 1×10−4 |
| rs1939012 | 11q22.2 | 102595135 | C/T* | 0.40(T) | MMP8 | intronic | GGE | 1.12 (1.07-1.17) | 2·37×10−8 | - |
| rs1044352 | 4p15.1 | 31147874 | T*/G | 0.50(T) | PCDH7 | synonymous | GGE | 0.88 (0.82-0.93) | 1·87×10−7 | - |
| rs55670112 | 5q22.3 | 114268470 | A/C* | 0.47(C) | none | intergenic | GGE | 1.18 (1.1-1.26) | 6·34×10−8 | - |
| rs12987787 | 2q24.3 | 166858391 | C/T* | 0.21(C) | SCN1A | intronic | focal | 1.12 (1.01-1.14) | 1·45×10−7 | - |
Chr: cytogenetic band; pos (bp): base pair position, build 37 (hg19); A1/A2: allele 1, allele 2
indicates ancestral/chimp allele
MAF: minor allele frequency, from all 1000Genomes populations; cand. gene: most plausible candidate gene attributable to the signal; annotation: type of SNP; pheno: phenotype; OR (CI): odds ratio, corresponds to allele 2, computed from logistic regression; 95% confidence interval in brackets; LMM p: p value from linear mixed model meta analysis; cond: p value when conditioning on this specific SNP., to determine independent signals from same locus.
Given the extent of linkage disequilibrium (LD) between the variants associated with ‘all epilepsy’ in the 2q24.3 region (Fig. 2), we conducted logistic regression conditioned on the most significant variant identified from the univariate analysis (rs6732655). Results indicated a tentative independent signal, coming from rs13406236, an intronic variant in SCN9A (p=1·39×10−4 on conditioning, see Supplementary Fig.6). No further significant signals were identified.
A second signal for the ‘all epilepsy’ phenotype was located at 4p15.1 and included the 3’ end of the protocadherin gene, PCDH7 (Fig. 3). The most strongly associated variant in this region was rs28498976 (p=5·44×10−9, OR=0·90 (CI: 0·87-0·94), see Table 2), located 2·5kb from the 3’-end of PCDH7. Logistic regression across PCDH7 supported the association with this locus (see Supplementary Fig.5). There were no additional significant signals from 4p15.1 on conditioning for rs28498976 (see Supplementary Fig.6). The direction of effect was consistent across all cohorts and there was no evidence of heterogeneity. Although only achieving genome-wide significance for the ‘all epilepsy’ phenotype, the PCDH7 signal appeared stronger in GGE compared to focal epilepsy (see Supplementary Fig.7).
Figure 3. Genomic context of 4p15.1 signal from ‘all epilepsy’ analysis.
Plot created using LocusZoom.60 Linkage disequilibrium data are taken from 1000 Genomes Project, HG19, March 2012.
PCDH7 encodes a calcium-dependent adhesion protein, a member of the cadherin gene family, not previously associated with epilepsy. The gene is expressed in the central nervous system, specifically in thalamocortical circuits and the hippocampus,30,31 and expression of PCDH7 is controlled by MECP2,32mutations in which cause Rett Syndrome. The cytoplasmic domain of the PCDH7 protein binds to protein phosphatase 1α (PPP1CA), which is enriched in dendritic spines and is important in learning and memory,33 and template activation factor 1 (TAF1), which along with PCDH7 plays a role in neurite extension.34,35
Suggestive signals of note (p<5×10−7) for the ‘all epilepsy’ phenotype were detected at 3q26.2 (p=4·42×10−7) and 4p12 (p=1·71×10−7) (see Table 2). The 3q26.2 region contained the 5’ end of GOLIM4 (see Supplementary Fig. 8), encoding Golgi internal membrane protein 4, which is degraded when manganese increases above normal levels, suggesting a role for this protein in manganese regulation.36 The vast majority of brain manganese is in glutamine synthase, an enzyme playing a key role in producing or degrading the neurotransmitters glutamate, glutamine, and gamma-aminobutyric acid (GABA). Decreased brain levels of glutamine synthase and of manganese have been reported in epilepsy.37,38 The 4p12 region contained the 3’ end of the gamma-aminobutyric acid receptor, α2-subunit gene (GABRA2, see Supplementary Fig.9). Mutations in other GABA receptors have been found to cause epilepsy.39
Genetic generalized epilepsy (GGE)
After QC, we considered a total of 21,596 individuals (2,606 cases and 18,990 controls) across six cohorts in the GGE meta-analysis (see Table 1). Individuals considered were a subset of those included in the ‘all epilepsy’ analysis. For the GGE analysis we estimated 80% power to detect a variant explaining 0·17% or greater of the liability variance (see Supplementary Fig.2). Results from the GGE meta-analysis indicated an inflation factor of 1·05 (see Supplementary Fig. 3b).
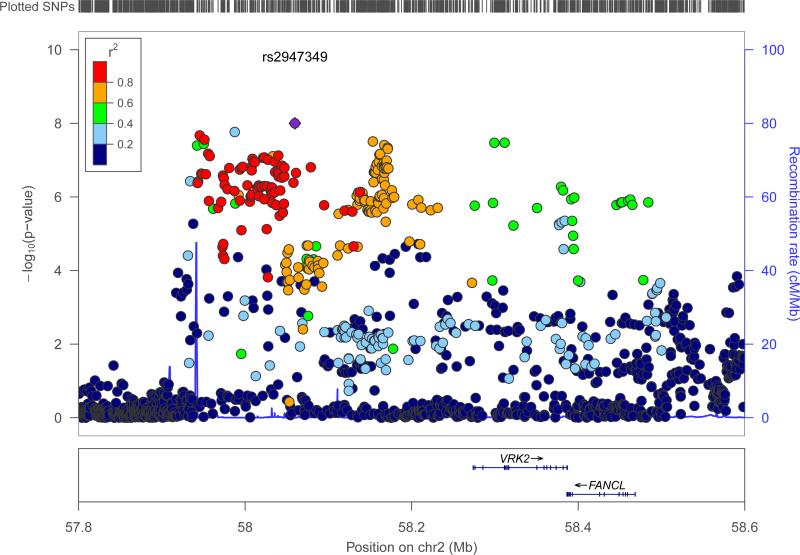
A single signal achieved the threshold of genome-wide significance (see Fig. 1b). Located at 2p16.1, the interval contained genes encoding vaccinia-related kinase 2 (VRK2) and Fanconi Anemia, Complementation Group L (FANCL) (see Fig. 4). The most strongly associated variant in this region was the intergenic variant rs2947349 (p=9·99×10−9, OR=1·23 (CI: 1·16-1·31), see Table 2). Logistic regression analysis supported the association with 2p16.1 (see Supplementary Fig.5). There were no additional significant signals from 2p16.1 on conditioning for rs2947349 (see Supplementary Fig.6). The direction of effect was consistent across all cohorts, and the association appeared to be specific to GGE (see Supplementary Fig.10).
Figure 4. Genomic context of 2p16.1 signal from GGE analysis.
Plot created using LocusZoom60. Linkage disequilibrium data are taken from 1000 Genomes Project, HG19, March 2012.
VRK2 is a serine/threonine protein kinase involved in signal transduction and apoptosis.40,41 Variation in VRK2 has previously been suggested as a risk factor for epilepsy16 and schizophrenia.42-44 Indeed, the schizophrenia risk variant (rs2312147)43 shows a strong signal for GGE (p=2·3×10−6, OR=1·22 (CI:1·14-1·30)) and is in high LD with the strongest variant for GGE (r2=0·82) – although the direction of the effect is opposite. The EPICURE cohort in which 2p16.1 was originally proposed as a risk factor for GGE was included in our meta-analysis. Excluding the EPICURE cohort, the top SNP (rs13026414)16 remains nominally significant at p=7×10−3. These results provide further support to the suggestion that VRK2 is a risk locus for both epilepsy and schizophrenia. The other gene in the region, FANCL, is a RING-type E3 ubiquitin ligase of the Fanconi anemia pathway. FANCL mono-ubiquitinates FANCD2 and FANCI, proteins involved in DNA repair and homologous recombination.45 FANCL has not been previously implicated in epilepsy or any seizure-related phenotype.
Suggestive evidence for association with GGE was detected at 4p15.1 (p=1·87×10−7), 5q22.3 (p=6·34×10−8) and 11q22.2 (p=2·37×10−8)(see Table 2). The 4p15.1 PCDH7 signal is the same as that appearing genome-wide significant for the ‘all-epilepsy’ phenotype (see Fig. 3 and Supplementary Fig.11). The 5q22.3 signal is intergenic (see Supplementary Fig.12). The 11q22.2 signal contained the 5’ end of the matrix metallopeptidase gene MMP8 (see Supplementary Fig. 13). The direction of effect was consistent across all cohorts and appeared specific to GGE (see Supplementary Fig. 14). With a p value of 2·37×10−8, it reached the conventional threshold for genome-wide significance (p<5×10−8), but not our more stringent one (p<1·66×10−8). Matrix metaloproteases are zinc-dependent endopeptidases involved in the breakdown of the extracellular matrix in normal physiological processes and of the blood-brain barrier in inflammation.46 Increased expression of MMPs have been recorded in various neurological disease states,47 and epileptogenesis is decreased in MMP9 knockout mice but increased in transgenic rats overexpressing MMP9.48
Focal
Post-QC, 28,916 individuals (5,310 cases, 23,606 controls), across eightcohorts, were included in the ‘focal’ epilepsy analysis. No signal achieved genome-wide significance. For the focal analysis we estimated 80% power to detect a variant explaining 0·10% or greater of the liability variance (see Supplementary Fig.2). Results from the focal meta-analysis indicated an inflation factor of 1·014 (see Supplementary Fig. 3c). We observed one sub-threshold signal of note (rs12987787, p=1·45×10−7) from 2q24.3, the region containing SCN1A (see Table 2 and Supplementary Fig. 15).
Targeted genotyping of the three GWAS-significant signals confirmed that imputation was accurate with a minimum correlation of 0·98 observed between experimentally determined and imputed genotypes (see Supplementary Table 3).
An assessment of enrichment of gene ontology (GO) terms for regions containing variants with nominally significant p values (p<1×10−5) for each of the three phenotypes found enrichment in several pathways (see Supplementary Table 4). Although none of these survived correction for multiple testing, the results highlight pathways of biological plausibility.
Finally, we investigated whether any of the four susceptibility loci at nominal genome-wide significance (p<5×10−8) were associated with outcome of newly treated epilepsy using data from Speed et al., 2014.21 We considered both the index SNP (Table 2) and SNPs within a 20Kb window around each of the 5 genes (SCN1A, PCDH7, VRK2/FANCL, MMP8) (see Supplementary Table 5). The minimum p value of association with outcome of newly treated epilepsy for any susceptibility locus was 8·14×10−4 (MMP8). We found no evidence for an association between SCN1A (the target for sodium channel-blocking class anti-epileptic drugs) and epilepsy outcome.
DISCUSSION
This first GWAS meta-analysis in the common epilepsies identified three loci with genome-wide significance and suggests some loci may show specificity for epilepsy type.
In the whole cohort consisting of all epilepsy, the region of the sodium channel subunit SCN1A was clearly implicated. This gene is a well established cause of genetic epilepsy with febrile seizures plus (GEFS+),28,29 a generally mild, familial form of epilepsy, and Dravet syndrome, a severe epileptic encephalopathy usually arising from de-novo mutation.7 SCN1A was implicated in a recent GWAS of mesial temporal lobe epilepsy and hippocampal sclerosis with febrile seizures (mTLEHS+FS)14 and a meta-analysis of SCN1A rs3812718 in all epilepsy.49 SCN1A mutations are also observed in a spectrum of paroxysmal neurological disorders including familial hemiplegic migraine50 and, more rarely, in certain focal epilepsies.51 It is therefore unclear whether this robust association with all epilepsy is a true common variant association or a synthetic association due to tagged rare variants in cases with GEFS+. Whilst it is possible the cohorts could have included individuals from monogenic GEFS+ families with SCN1A mutations of large effect, review of the phenotyping data suggested inclusion of more than a few such cases is unlikely; moreover SCN1A variants are only found in about 10% of large GEFS+ families.52
Our ‘all epilepsy’ analysis identified a second locus (4p15.1), which also satisfied our threshold for genome-wide significance. This locus is novel for epilepsy and implicates the gene PCDH7. This protocadherin gene is a plausible candidate for common forms of epilepsy with mutations in another protocadherin gene, PCDH19, causing epilepsy and mental retardation limited to females (EFMR).53
For the specific category of GGE, we again observed the association at 2p16.1, previously reported in the EPICURE cohort,16 which comprised approximately half of our GGE cohort (see Table 1). The association maintained nominal significance after removal of EPICURE casesfor this locus, where the genes VRK2 and FANCL are within close proximity. With our additional samples, we did not observe significance for the 17q21 locus reported by EPICURE for GGE (see Supplementary Fig. 16).
For the larger subcategory of focal epilepsy we did not find any locus at genome-wide significance, consistent with the EPIGEN study of focal epilepsy that was negative (samples included here).13 However, a signal at 2q24.3 (containing SCN1A) in focal epilepsy approached, but did not achieve significance (see Supplementary Fig.15). The focal signal was in high LD with that observed for ‘all epilepsy’ (r2 = 0·85). Importantly, the 2q24.3 signal for focal epilepsy observed here is different to that reported in a recent study of the narrow focal epilepsy phenotype of mTLEHS+FS.14 rs7587026 (the previously reported mTLEHS+FS variant) is not significant in our analysis of a broader focal epilepsy phenotype consisting of all focal epilepsies (p=0·01) (see Supplementary Fig. 17). We also failed to observe the previously reported association at 1q32.1, implicating CAMSAP1L1, in the Hong Kong cohort15 (included here, see Supplementary Fig.18), where the majority of cases had focal epilepsy due to known lesions.
Consistent with experience of GWAS analyses in other neuropsychiatric disorders, and common disorders in general, this study reinforces the value of large sample sizes. In the epilepsies, electro-clinical and imaging data permit the identification of clinical syndromes that share common clinical features. Our study suggests that an experimental design that includes fractionation of samples into clinical subtypes may reveal syndrome-specific risk alleles, but the identification of these alleles will be facilitated by the collection and genotyping of larger sample sizes. While this “lumping” versus “splitting” debate in genetic analyses is not unique to the epilepsies, there has been long-standing controversy about this in clinical epileptology54 that genetics will help to inform.
Limitations of our study include sample size; although ours is large, even larger samples have yielded increasing discoveries in other disorders.55-57 Larger samples would enable further analysis of epilepsy subtypes and the ILAE Consortium now provides a useful vehicle for this. Second, our meta-analysis relied on separately generated genotypes on a variety of platforms, an issue common to most meta-analyses. Third, it would be ideal to extend the phenotyping data to include treatment outcome but, in a cross-sectional cohort, this has methodological problems. Finally, we did not have an independent replication sample. However, stringent criteria for statistical significance were set a priori and, for loci achieving our threshold of genome-wide significance, the direction of effects were uniform across the cohorts, and extended over multiple variants in high LD.
Taken together, these data show that, given sufficient sample size, susceptibility loci for common epilepsies can be identified through the analysis of common variation. The role of rare variants of large effect is also well established, particularly in rarer Mendelian epilepsies.3-7 The role of rare variants in the common epilepsies is currently being explored by deep sequencing approaches.11,58,59 A dual approach of identifying both rare and common variation will result in a greater understanding of the genetic architecture for the overall epilepsy population, necessary for precision medicine. While these findings will not be of immediate clinical utility, they are an important first step to understanding the genetic architecture of epilepsies which may lead to clinically relevant markers of prognosis and outcome.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to the patients and volunteers who participated in this research. We thank the following clinicians and research scientists for their contribution through sample collection (cases and controls), data analysis and project support: Wim Van Paesschen, Benjamin Legros, Patrick Tugendhaft, Kevin Shianna, Edouard Louis, Michel Georges, William Gallentine, Aatif Husain, Mohamad Mikati, Saurabh Sinha, Raju Yerra, Chris French, Zelko Matkovic, Steven J Howell, Paul Cooper, Mark Kellett, Richard Roberts, Muna Alwaidh, Oliver C Cockerell, Brendan McLean, Adrian Hughes, Marcus Reuber, Peter Cleland, Kathleen White, Celia Cramp, Peter Goulding, R E Appleton, Mark Lawden, B Sharack, Guiliano Avanzini, Ditte B Kjelgaard, Oebele Brouwer, Floor Jansen, Kees Braun, Hans Carpay, Willem Frans Arts, Paul Boon, Leanne Lehwald, Jorge Vidaurre, Pedro Weisleder, Chang-Yong Tsao, Annie WC Kung, Monica Islam, Emily de los Reyes, Jennifer McKinney, Laurel Slaughter, Bethanie Morgan-Followell, Lori Hamiwka, Deborah Terry, Jacqueline Yinger, Sally Steward, Mary Karn, Jo Ellen Lee, Donna Kring, Sarah Borror, Karen Carter, Cathy Schumer, Guy Rouleau, Micheline Gravel, Virginia Wong, Colin HT Lui, NC Sin, TH Tsoi, Rhian Gwilliamand all contributing clinicians from the Department of Clinical and Experimental Epilepsy at the National Hospital for Neurology and Neurosurgery and UCL Institute of Neurology. The KORA-Study Group consists of A. Peters (speaker), J. Heinrich, R. Holle, R. Leidl, C. Meisinger, K. Strauch, and their coworkers C. Gieger and H. Grallert, who are responsible for the design and conduct of the KORA studies.
This work was in part supported by an award (2009/001) from Brainwave–the Irish Epilepsy Association; by the Medical Research Charities Group of Ireland, the Health Research Board and by a Translational Research Scholars award from the Health Research Board of Ireland (C.W.).Further funding sources include: Wellcome Trust (grant 084730), NIHR (08-08-SCC), GIHE: NIH R01-NS-49306-01 (R.J.B.), GSCFE: NIH R01NS064154-01 (R.J.B. and H.H.), NIH: UL1TR001070, Development Fund from The Children's Hospital of Philadelphia (H.H.), NHMRC Program Grant (ID: 628952; S.F.B., I.E.S., K.L.O.); The Royal Melbourne Hospital Foundation Lottery Grant (S.P.), The RMH Neuroscience Foundation (T.J.O'B.), European Union's Seventh Framework Programme (FP7/2007-2013) under grant agreement n° 279062 and 602102, Department of Health's NIHR Biomedical Research Centers funding scheme, European Community (EC: FP6 project EPICURE: LSHM-CT-2006-037315); German Research Foundation (DFG: SA434/4-1/4-2), EuroEPINOMICS Consortium (European Science Foundation/DFG: SA434/5-1, NU50/8-1, LE1030/11-1, HE5415/3-1); the German Federal Ministry of Education and Research, National Genome Research Network (NGFNplus/EMINet: 01GS08120, and 01GS08123; IntenC, TUR 09/I10); The Netherlands National Epilepsy Fund (grant 04-08); EC (FP7 project EpiPGX 279062). Research Grants Council of the Hong Kong Special Administrative Region, China project numbers HKU7623/08M (S.S.C, P.K., L.W.B., P.C.S), HKU7747/07M (S.S.C., P.C.S.) and CUHK4466/06M (P.K., L.W.B). Collection of Belgian cases was supported by the Fonds National de la Recherche Scientifique, Fondation Erasme, Université Libre de Bruxelles. Glaxo Smith Kline funded the recruitment and data collection for the GenEpA Consortium samples. We acknowledge the support of Nationwide Children's hospital in Columbus, Ohio, USA. The Wellcome Trust (WT066056) and The NIHR Biomedical Research Centres Scheme (P31753) supported UK contributions. Further support was received through the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development (Contract: N01HD33348). The project was also supported by the popgen 2.0 network through a grant from the German Ministry for Education and Research (01EY1103). The KORA research platform (KORA, Cooperative Research in the Region of Augsburg) was initiated and financed by the Helmholtz ZentrumMünchen - German Research Center for Environmental Health, which is funded by the German Federal Ministry of Education and Research and by the State of Bavaria. Furthermore, KORA research was supported within the Munich Center of Health Sciences (MC Health), Ludwig-Maximilians-Universität, as part of LMUinnovativ.
The International League Against Epilepsy (ILAE) facilitated the Consortium through the Commission on Genetics and by financial support; however, the opinions expressed in the manuscript are not necessarily those of the ILAE.
Appendix
AUTHOR CONTRIBUTIONS
Analysis committee:Association analysis: J.P.B., S.S.C., C.G.F.dK., H.G., C.L., D.S., Z.W., C.W.Coordination:G.L.C.Imputation:J.P.B.,S.S.C., C.G.F.dK.,H.G., D.S.,C.W.Protocol development:J.P.B., G.L.C., C.L., D.S., Z.W.
Phenotyping committee: C.D., D.J.D., W.S.K., P.K., D.H.L., A.G.M., M.R.S., P.S.
Strategy committee: L.B., S.F.B., R.J.B., H.Hak, E.L.H, M.R.J., B.P.C.K., P.K., H.L., T.J.O'B., K.L.O. (ex-officio), S.Sis.
Governance committee: S.F.B., A.C., A-E.L., D.H.L.
Writing committee: S.F.B., G.L.C., M.R.J.
Patient recruitment and phenotyping: A.A., F.B., R.J.B., C.B.C., K.C., P.C., G-J.dH., P.DJ., N.D., C.D., D.J.D., C.P.D., C.E.E., T.N.F., M.F., J.F., V.G., H.Hja, I.H., M.J., J.J-K., D.J., M.R.J., R.K., A-M.K., D.K-N.T., W.S.K., P.K., H.L., D.L., W.Lo., A.G.M., M.Moe, R.S.M., H.M., M.N., P-W.N., T.J.O'B., M.P., R.R., P.S.R., F.R., T.San, T.Sca, St.Sc, Su.Sc,S.Sis, C.S., I.E.S., B.S., D.F.S., P.E.S., M.R.S., U.S., P.S., H.S., R.S., K.M.T, M.T., A.T., R.T., F.V., S.V., N.M.W., Y.G.W., C.W., F.Zim.
Genotyping and bioinformatics: R.J.L.A., D.B., L.B., J.P.B., G.L.C., S.S.C., A.J.C, C.G.F.dK, D.B.G., H.G., Y.G., H.Hak, E.L.H., I.H., D.K., C.L., A.M., M.McC, P.N., S.P., AK.R., T.San, P.C.S., D.S., M.S., H.T., Z.W., C.W., F.Zar.
Control cohorts: L.C.B., A.F., H.Hak, Y-L.L., W.Li., J.L.M., A.M.M., M.M.N, A.P., F.P., H.S., G.N.T., The KORA study group, W.Y.
The International League Against Epilepsy Consortium on Complex Epilepsies
Richard J L Anney, Andreja Avbersek, David Balding, Larry Baum, Felicitas Becker, Samuel F Berkovic, Jonathan P Bradfield, Lawrence C Brody, Russell J Buono, Claudia B Catarino, Gianpiero L Cavalleri, Stacey S Cherny, Krishna Chinthapalli, Alison J Coffey, Alastair Compston, Patrick Cossette, Gerrit-Jan de Haan, Peter De Jonghe, Carolien G F de Kovel, Norman Delanty, Chantal Depondt, Dennis J Dlugos, Colin P Doherty, Christian E Elger, Thomas N Ferraro, Martha Feucht, Andre Franke, Jacqueline French, Verena Gaus, David B Goldstein, Hongsheng Gui, Youling Guo, Hakon Hakonarson, Kerstin Hallmann, Erin L Heinzen, Ingo Helbig, Helle Hjalgrim, Margaret Jackson, Jennifer Jamnadas-Khoda, Dieter Janz, Michael R Johnson, Reetta Kälviäinen, Anne-Mari Kantanen, Dalia Kasperavičiūte, Dorothee Kasteleijn-Nolst Trenite, Bobby P C Koeleman, Wolfram S Kunz, Patrick Kwan, Yu Lung Lau, Anna-Elina Lehesjoki, Holger Lerche, Costin Leu, Wolfgang Lieb, Dick Lindhout, Warren Lo, Daniel H Lowenstein, Alberto Malovini, Anthony G Marson, Mark McCormack, James L Mills, Martina Moerzinger, Rikke S Møller, Anne M Molloy, Hiltrud Muhle, Mark Newton, Ping-Wing Ng, Markus M Nöthen, Peter Nürnberg, Terence J O'Brien, Karen L Oliver, Aarno Palotie, Faith Pangilinan, Katharina Pernhorst, Slave Petrovski, Michael Privitera, Rodney Radtke, Philipp S Reif, Felix Rosenow, Ann-Kathrin Ruppert, Thomas Sander, Theresa Scattergood, Steven Schachter, Christoph Schankin, Ingrid E Scheffer, Bettina Schmitz, Susanne Schoch, Pak C Sham, Sanjay Sisodiya, David F Smith, Philip E Smith, Doug Speed, Michael R Sperling, Michael Steffens, Ulrich Stephani, Pasquale Striano, Hans Stroink, Rainer Surges, K Meng Tan, The KORA study group, G Neil Thomas, Marian Todaro, Anna Tostevin, Rossana Tozzi, Holger Trucks, Frank Visscher, Sarah von Spiczak, Nicole M Walley, Yvonne G Weber, Zhi Wei, Christopher Whelan, Wanling Yang, Federico Zara, Fritz Zimprich
Please see Supplementary material for author affiliations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
COMPETING FINANCIAL INTERESTS
The authors have no competing financial interests to declare.
DATA ACCESS
A complete list of p values for all SNPs analyzed can be found at the Epilepsy Genetic Association Database (epiGAD)http://www.epigad.org
REFERENCES
- 1.Hesdorffer DC, Logroscino G, Benn EK, Katri N, Cascino G, Hauser WA. Estimating risk for developing epilepsy: a population-based study in Rochester, Minnesota. Neurology. 2011;76:23–7. doi: 10.1212/WNL.0b013e318204a36a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg AT, Berkovic SF, Brodie MJ, et al. Revised terminology and concepts for organization of seizures and epilepsies: report of the ILAE Commission on Classification and Terminology, 2005-2009. Epilepsia. 2010;51:676–85. doi: 10.1111/j.1528-1167.2010.02522.x. [DOI] [PubMed] [Google Scholar]
- 3.Helbig I, Scheffer IE, Mulley JC, Berkovic SF. Navigating the channels and beyond: unravelling the genetics of the epilepsies. Lancet Neurol. 2008;7:231–45. doi: 10.1016/S1474-4422(08)70039-5. [DOI] [PubMed] [Google Scholar]
- 4.Poduri A, Lowenstein D. Epilepsy genetics--past, present, and future. Curr Opin Genet Dev. 2011;21:325–32. doi: 10.1016/j.gde.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Epi4K Consortium, Epilepsy Phenome/Genome Project. Allen AS, et al. De novo mutations in epileptic encephalopathies. Nature. 2013;501:217–21. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carvill GL, Heavin SB, Yendle SC, et al. Targeted resequencing in epileptic encephalopathies identifies de novo mutations in CHD2 and SYNGAP1. Nat Genet. 2013;45:825–30. doi: 10.1038/ng.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Claes L, Del-Favero J, Ceulemans B, Lagae L, Van Broeckhoven C, De Jonghe P. De novo mutations in the sodium-channel gene SCN1A cause severe myoclonic epilepsy of infancy. Am J Hum Genet. 2001;68(6):1327–32. doi: 10.1086/320609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y, Lu J, Pan H, et al. Association between genetic variation of CACNA1H and childhood absence epilepsy. Ann Neurol. 2003;54:239–43. doi: 10.1002/ana.10607. [DOI] [PubMed] [Google Scholar]
- 9.Heinzen EL, Radtke RA, Urban TJ, et al. Rare deletions at 16p13.11 predispose to a diverse spectrum of sporadic epilepsy syndromes. Am J Hum Genet. 2010;86:707–18. doi: 10.1016/j.ajhg.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Helbig I, Mefford HC, Sharp AJ, et al. 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat Genet. 2009;41:160–2. doi: 10.1038/ng.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Epi4K Consortium Epi4K: gene discovery in 4,000 genomes. Epilepsia. 2012;53:1457–67. doi: 10.1111/j.1528-1167.2012.03511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dibbens LM, Heron SE, Mulley JC. A polygenic heterogeneity model for common epilepsies with complex genetics. Genes Brain Behav. 2007;6:593–7. doi: 10.1111/j.1601-183X.2007.00333.x. [DOI] [PubMed] [Google Scholar]
- 13.Kasperaviciute D, Catarino CB, Heinzen EL, et al. Common genetic variation and susceptibility to partial epilepsies: a genome-wide association study. Brain. 2010;133:2136–47. doi: 10.1093/brain/awq130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kasperaviciute D, Catarino CB, Matarin M, et al. Epilepsy, hippocampal sclerosis and febrile seizures linked by common genetic variation around SCN1A. Brain. 2013;136:3140–50. doi: 10.1093/brain/awt233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo Y, Baum LW, Sham PC, et al. Two-stage genome-wide association study identifies variants in CAMSAP1L1 as susceptibility loci for epilepsy in Chinese. Hum Mol Genet. 2012;21:1184–9. doi: 10.1093/hmg/ddr550. [DOI] [PubMed] [Google Scholar]
- 16.EPICURE Consortium. EMINet Consortium. Steffens M, et al. Genome-wide association analysis of genetic generalized epilepsies implicates susceptibility loci at 1q43, 2p16.1, 2q22.3 and 17q21.32. Hum Mol Genet. 2012;21:5359–72. doi: 10.1093/hmg/dds373. [DOI] [PubMed] [Google Scholar]
- 17.Berkovic SF, Howell RA, Hay DA, Hopper JL. Epilepsies in twins: genetics of the major epilepsy syndromes. Ann Neurol. 1998;43:435–45. doi: 10.1002/ana.410430405. [DOI] [PubMed] [Google Scholar]
- 18.Ottman R, Lee JH, Hauser WA, Risch N. Are generalized and localization-related epilepsies genetically distinct? Archives of neurology. 1998;55:339–44. doi: 10.1001/archneur.55.3.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peljto AL, Barker-Cummings C, Vasoli VM, et al. Familial risk of epilepsy: a population-based study. Brain. 2014;137:795–805. doi: 10.1093/brain/awt368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Commission on Classification and Terminology of the International League Against Epilepsy Proposal for revised classification of epilepsies and epileptic syndromes. Epilepsia. 1989;30:389–99. doi: 10.1111/j.1528-1157.1989.tb05316.x. [DOI] [PubMed] [Google Scholar]
- 21.Speed D, Hoggart C, Petrovski S, et al. A genome-wide association study and biological pathway analysis of epilepsy prognosis in a prospective cohort of newly treated epilepsy. Hum Mol Genet. 2014;23:247–258. doi: 10.1093/hmg/ddt403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lippert C, Listgarten J, Liu Y, Kadie CM, Davidson RI, Heckerman D. FaST linear mixed models for genome-wide association studies. Nat Methods. 2011;8:833–5. doi: 10.1038/nmeth.1681. [DOI] [PubMed] [Google Scholar]
- 23.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dempster ER, Lerner IM. Heritability of Threshold Characters. Genetics. 1950;35:212–36. doi: 10.1093/genetics/35.2.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Banerjee PN, Filippi D, Allen Hauser W. The descriptive epidemiology of epilepsy-a review. Epilepsy Res. 2009;85:31–45. doi: 10.1016/j.eplepsyres.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee PH, O'Dushlaine C, Thomas B, Purcell SM. INRICH: interval-based enrichment analysis for genome-wide association studies. Bioinformatics. 2012;28:1797–9. doi: 10.1093/bioinformatics/bts191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Escayg A, MacDonald BT, Meisler MH, et al. Mutations of SCN1A, encoding a neuronal sodium channel, in two families with GEFS+2. Nat Genet. 2000;24:343–5. doi: 10.1038/74159. [DOI] [PubMed] [Google Scholar]
- 29.Wallace RH, Scheffer IE, Barnett S, et al. Neuronal sodium-channel alpha1-subunit mutations in generalized epilepsy with febrile seizures plus. Am J Hum Genet. 2001;68:859–65. doi: 10.1086/319516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim SY, Chung HS, Sun W, Kim H. Spatiotemporal expression pattern of non-clustered protocadherin family members in the developing rat brain. Neuroscience. 2007;147:996–1021. doi: 10.1016/j.neuroscience.2007.03.052. [DOI] [PubMed] [Google Scholar]
- 31.Kim SY, Mo JW, Han S, et al. The expression of non-clustered protocadherins in adult rat hippocampal formation and the connecting brain regions. Neuroscience. 2010;170:189–99. doi: 10.1016/j.neuroscience.2010.05.027. [DOI] [PubMed] [Google Scholar]
- 32.Miyake K, Hirasawa T, Soutome M, et al. The protocadherins, PCDHB1 and PCDH7, are regulated by MeCP2 in neuronal cells and brain tissues: implication for pathogenesis of Rett syndrome. BMC Neurosci. 2011;12:81. doi: 10.1186/1471-2202-12-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Braithwaite SP, Stock JB, Lombroso PJ, Nairn AC. Protein phosphatases and Alzheimer's disease. Prog Mol Biol Transl Sci. 2012;106:343–79. doi: 10.1016/B978-0-12-396456-4.00012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim SY, Yasuda S, Tanaka H, Yamagata K, Kim H. Non-clustered protocadherin. Cell Adh Migr. 2011;5:97–105. doi: 10.4161/cam.5.2.14374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piper M, Dwivedy A, Leung L, Bradley RS, Holt CE. NF-protocadherin and TAF1 regulate retinal axon initiation and elongation in vivo. J Neurosci. 2008;28:100–5. doi: 10.1523/JNEUROSCI.4490-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masuda M, Braun-Sommargren M, Crooks D, Smith DR. Golgi phosphoprotein 4 (GPP130) is a sensitive and selective cellular target of manganese exposure. Synapse. 2013;67:205–15. doi: 10.1002/syn.21632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eid T, Tu N, Lee TS, Lai JC. Regulation of astrocyte glutamine synthetase in epilepsy. Neurochem Int. 2013;63:670–81. doi: 10.1016/j.neuint.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gonzalez-Reyes RE, Gutierrez-Alvarez AM, Moreno CB. Manganese and epilepsy: a systematic review of the literature. Brain Res Rev. 2007;53:332–6. doi: 10.1016/j.brainresrev.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 39.Lerche H, Shah M, Beck H, Noebels J, Johnston D, Vincent A. Ion channels in genetic and acquired forms of epilepsy. J Physiol. 2013;591:753–64. doi: 10.1113/jphysiol.2012.240606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fernandez IF, Perez-Rivas LG, Blanco S, Castillo-Dominguez AA, Lozano J, Lazo PA. VRK2 anchors KSR1-MEK1 to endoplasmic reticulum forming a macromolecular complex that compartmentalizes MAPK signaling. Cell Mol Life Sci. 2012;69:3881–93. doi: 10.1007/s00018-012-1056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Monsalve DM, Merced T, Fernandez IF, Blanco S, Vazquez-Cedeira M, Lazo PA. Human VRK2 modulates apoptosis by interaction with Bcl-xL and regulation of BAX gene expression. Cell Death Dis. 2013;4:e513. doi: 10.1038/cddis.2013.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Genome-wide association study implicates HLA-C*01:02 as a risk factor at the major histocompatibility complex locus in schizophrenia. Biol Psychiatry. 2012;72:620–8. doi: 10.1016/j.biopsych.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li M, Wang Y, Zheng XB, et al. Meta-analysis and brain imaging data support the involvement of VRK2 (rs2312147) in schizophrenia susceptibility. Schizophr Res. 2012;142:200–5. doi: 10.1016/j.schres.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 44.Steinberg S, de Jong S, Andreassen OA, et al. Common variants at VRK2 and TCF4 conferring risk of schizophrenia. Hum Mol Genet. 2011;20:4076–81. doi: 10.1093/hmg/ddr325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dao KH, Rotelli MD, Brown BR, et al. The PI3K/Akt1 pathway enhances steady-state levels of FANCL. Mol Biol Cell. 2013;24:2582–92. doi: 10.1091/mbc.E13-03-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vandenbroucke RE, Dejonckheere E, Van Lint P, et al. Matrix metalloprotease 8-dependent extracellular matrix cleavage at the blood-CSF barrier contributes to lethality during systemic inflammatory diseases. J Neurosci. 2012;32:9805–16. doi: 10.1523/JNEUROSCI.0967-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yong VW. Metalloproteinases: mediators of pathology and regeneration in the CNS. Nat Rev Neurosci. 2005;6:931–44. doi: 10.1038/nrn1807. [DOI] [PubMed] [Google Scholar]
- 48.Wilczynski GM, Konopacki FA, Wilczek E, et al. Important role of matrix metalloproteinase 9 in epileptogenesis. J Cell Biol. 2008;180:1021–35. doi: 10.1083/jcb.200708213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baum L, Haerian BS, Ng HK, et al. Case-control association study of polymorphisms in the voltage-gated sodium channel genes SCN1A, SCN2A, SCN3A, SCN1B, and SCN2B and epilepsy. Hum Genet. 2014;133:651–9. doi: 10.1007/s00439-013-1405-1. [DOI] [PubMed] [Google Scholar]
- 50.Dichgans M, Freilinger T, Eckstein G, et al. Mutation in the neuronal voltage-gated sodium channel SCN1A in familial hemiplegic migraine. Lancet. 2005;366:371–7. doi: 10.1016/S0140-6736(05)66786-4. [DOI] [PubMed] [Google Scholar]
- 51.Abou-Khalil B, Ge Q, Desai R, et al. Partial and generalized epilepsy with febrile seizures plus and a novel SCN1A mutation. Neurology. 2001;57:2265–72. doi: 10.1212/wnl.57.12.2265. [DOI] [PubMed] [Google Scholar]
- 52.Hirose S, Scheffer IE, Marini C, et al. SCN1A testing for epilepsy: application in clinical practice. Epilepsia. 2013;54:946–52. doi: 10.1111/epi.12168. [DOI] [PubMed] [Google Scholar]
- 53.Dibbens LM, Tarpey PS, Hynes K, et al. X-linked protocadherin 19 mutations cause female-limited epilepsy and cognitive impairment. Nat Genet. 2008;40:776–81. doi: 10.1038/ng.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Berg AT, Blackstone NW. Of cabbages and kings: Perspectives on classification from the field of systematics. Epilepsia. 2003;44:8–12. [Google Scholar]
- 55.International Multiple Sclerosis Genetics Consortium. Beecham AH, Patsopoulos NA, et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet. 2013;45:1353–60. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.International Multiple Sclerosis Genetics Consortium. Wellcome Trust Case Control Consortium 2. Sawcer S, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–9. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ripke S, O'Dushlaine C, Chambert K, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45:1150–9. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heinzen EL, Depondt C, Cavalleri GL, et al. Exome sequencing followed by large-scale genotyping fails to identify single rare variants of large effect in idiopathic generalized epilepsy. Am J Hum Genet. 2012;91:293–302. doi: 10.1016/j.ajhg.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Klassen T, Davis C, Goldman A, et al. Exome sequencing of ion channel genes reveals complex profiles confounding personal risk assessment in epilepsy. Cell. 2011;145:1036–48. doi: 10.1016/j.cell.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.