Abstract
Differentiation of B cells into antibody secreting cells induces changes in gene transcription, Igh RNA processing, the unfolded protein response, and cell architecture. The transcription elongation factor ELL2 (eleven nineteen lysine-rich leukemia gene) stimulates the processing of the secreted form of the Igh mRNA from the heavy chain gene. Mice (mus musculus) with the ELL2 gene floxed in either exon 1 or exon 3 were constructed and crossed to CD19 driven cre/ CD19+. The B-cell specific ELL2 conditional knockouts (ell2loxp/loxp CD19cre/+) exhibit curtailed humoral responses both in NP-ficoll and NP-KLH immunized animals; recall responses were also diminished. The number of immature and recirculating B cells in the bone marrow is increased in the conditional knockouts while plasma cells in spleen are reduced relative to control animals. There are fewer IgG1 antibody producing cells in the bone marrow of conditional knockouts. LPS ex vivo stimulated B220loCD138+ cells from ELL2 deficient mouse spleens are 4-fold less abundant than from control splenic B-cells, have a paucity of secreted Igh, and distended, abnormal appearing ER. IRE1alpha is efficiently phosphorylated but the amounts of Ig kappa, ATF6, BiP, Cyclin B2, OcaB (BOB1, Pou2af1), and XBP1 mRNAs, unspliced and spliced, are severely reduced in ELL2 deficient cells. ELL2 enhances the expression of BCMA, important for long term survival. Transcription yields from the cyclin B2 and the canonical UPR promoter elements are up-regulated by ELL2 cDNA. Thus ELL2 is important for many aspects of antibody secretion, XBP1 expression, and the unfolded protein response.
INTRODUCTION
Activation of B-cells by cognate antigen or polyclonal stimulators like lipopolysaccharide (LPS) results in a large shift in RNA processing to the secretory-specific form of Immunoglobulin (Ig) heavy chain mRNA (1) and to a considerable up-regulation of the unfolded protein response (UPR) to accommodate the massive quantity of Igh protein that results (2). This also causes major structural accommodations and altered endoplasmic reticulum (ER) cellular architecture. B cell expansion follows after LPS or antigen stimulation to initially create extrafollicular plasmablasts. B cells can also enter the follicles and form germinal centers where memory and plasma cells (PCs) can form (3). Long lived plasma cells can reside in the bone marrow where they express a number of receptors for survival factors (4). Opposing suites of transcription factors either maintain the B cell program (e.g. Pax5, Bach2, Bcl6) or promote and facilitate differentiation to antibody secretion (e.g. IRF4, Blimp1, XBP1) (5). The mRNA for transcription elongation factor ELL2 was elevated when Blimp-1 and IRF-4 were turned on (2, 6) and (7); we showed that addition to B cells of exogenous cDNA for ELL2 stimulates alternative RNA processing resulting in the use of the Igh secretory-specific poly(A) site and the skipping of weak alternative exons in Igh and test substrates (1). ELL2 drives the association of positive transcription elongation factor b (pTEFb) to RNAP-II, thereby stimulating phosphorylation of ser-2 on the carboxyl-termini of the elongating polymerase near the start of the Igh locus (1, 8, 9). Thus the binding of ELL2 with the highly phosphorylated carboxyl-terminal domain of RNA polymerase II, polyadenylation factors, Dot1L (the histone H3K79 methylase), and with histone H3 K79me3 modifications are all seminal to the changes in RNA processing seen at the Igh locus. However it was not known whether ELL2 is required for secretory Igh production in the animal and at what stage.
It was also not known whether the related members in the ELL family could substitute for ELL2. Three mammalian ELL (eleven-nineteen lysine-rich leukemia) genes encode proteins that are engaged in transcription elongation with pTEFb and RNA polymerase II (10). ELL1 is ubiquitously expressed while ELL3, found in stem cells (11) and in unstimulated B cells (12), is reduced upon LPS stimulation. ELL2, and not the other ELLs, was found in the super elongation complex associated with HIV-1 TAR and Tat with pTEFb (13). ELL2 is elevated in plasma cell differentiation (1) and germinal center cells and is highly expressed in all myeloma cells surveyed for survival along with IRF4 (6), making it unique among its related family members. We therefore reasoned that deleting ELL2 specifically in B cells could alter the production of the Igh secretory specific mRNA after activation of those cells.
Two different ELL2 conditional knockout mice were generated, one in exon 1 and one in exon 3; both models showed impaired plasma cell differentiation in vivo and in vitro. The endogenous levels of serum Ig were decreased in the CD19cre driven conditional ELL2 knockout mice. These mice showed impaired responses to immunizations with NP-ficoll, a T-independent antigen, or NP-Keyhole limpet hemocyanin (KLH), a T-dependent antigen; antigen-specific Ig production was significantly reduced in the knockouts relative to wild type mice. Recall responses were also affected. The splenic Transitional 3 (T3) cells and plasma cells were significantly reduced in the conditional knockouts while bone marrow recirculating B cells were significantly increased in the knockouts. But IgG1 producing bone marrow cells were decreased in the knockout. Studies reported here with spleen cells from the CD19cre conditional knockouts of ELL2 in B cells show that following LPS stimulation of resting B cells the processing of Igh mRNA to the secretory-specific form was severely reduced, even in the small number of sorted CD138+B220lo so called plasma cells produced. The amount of the Ig chaperones and activators in the unfolded protein response was diminished, most especially ATF6, BiP, cyclin B2, and XBP1; Ig kappa light chain and BCMA were also impaired in the knockouts. Thus ELL2 is a key player not only in Igh mRNA processing but also in subsequent antibody secreting cell differentiation, making it exceptional in the ELL family of factors and crucial for plasma cell development.
MATERIALS AND METHODS
Materials and methods
Generation of conditional knockout mice
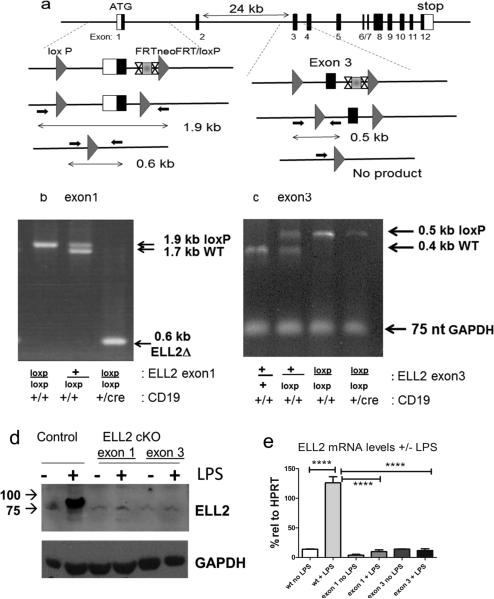
ELL2 is a single gene that resides on mouse chromosome 13. In collaboration with genOway, www.genoway.com, ~1 kb upstream of the transcription start site and aa 1 ~50 of exon 1 were surrounded by loxP sites in a conditional targeting vector (Figure 1a), the vector was inserted into the genome in ES cells (129Sv/pas ) and insertion of the construct selected for the neomycin marker in the targeting vector; the neo cassette was flanked by frt sites, see Supplemental Figure 1 for the southern blots showing proper integration into the ELL2 gene. The insertion/and subsequent deletion was also detected by PCR (Figure 1). After germline transmission of the targeted allele was established, the frt flanked neo cassette was removed by crossing to mice expressing Flpe. The mice were made homozygous for the exon1 loxp/loxp locus, then crossed to CD19 cre/+ mice (Jackson labs Cd19tm1(cre)Cgn Ighb/J # 006785, C57BL/6) to produce progeny that were ELL2 exon1 loxp/loxp CD19cre/+. The CD19cre was chosen because it is B cell-specific, is expressed throughout B cell development, maps to chromosome 7, and was previously used to elucidate the action of many other genes. The loxp to loxp distance after deletion is ~1.5 kb and there is a XmaI site just upstream of the frt cassette. The deletion was detected using primers (CM # 82437293 &-4) just beyond the loxp sites as shown in Figure 1.
Figure 1.
ELL2 loxp/loxp mice have efficient deletion of ELL2 in B cells
a. Strategy for generating alleles of ELL2 with either exon 1 or exon 3 flanked by loxp sites (triangles). The FRTneo gene flanked by sites indicated as X...X is deleted in ES cells in both models. Loxp sites are indicated by the grey triangles. The small arrows indicate the primers used to generate PCR fragments of 1.9 kb (plus 2 loxP sites) and 1.7 kb (no loxP) for exon 1. Deletion between the two loxP sites in the presence of CR19cre/+ generates a 0.6 kb PCR fragment. The small arrows in exon 3 flank one of the loxP sites to generate PCR fragments of 0.5 kb (plus loxP) and 0.4 kb (no loxP). Absence of a 0.5 kb product indicates deletion between the two loxP sites with CD19cre/+ .
b. PCR analysis to detect the 0.6 kb deletion product in the presence of CD19cre/+ in B cells.
c. PCR analysis showing deletion of the 0.5 kb fragment in the presence of CD19cre/ +. The GAPDH DNA PCR product indicates that there is the same amount of DNA in each sample.
d. Immunoblot of nuclear lysates from splenocytes +/− 20 ug/ml LPS for 4 days of ELL2 loxp/loxp mice with CD19 cre/+ or CD19+/+. Proteins were run on a denaturing polyacrylamide gel, blotted and probed with affinity purified rabbit anti-ELL2 peptide (R4502) as previously described.
e. mRNA levels of ELL2 with or without LPS stimulation four days after addition in control and ELL2loxp/loxp CD19 cre/+ mice was quantified relative to HPRT control RNA, set as 100% by real-time, quantitative PCR. N>5 each group. P=0.001 (***). Error bars indicate SEM, here and throughout.
11856cre-MICI AAGAGGATTGCTGTAGGCACCCTCC
11857cre-MICI TCCCAGAAAACTCTGACCGCTCG
The wild type allele produces a PCR band of 1806 base pairs (bp), insertion of loxp produces a band of 1983 bp, while after cre-mediated excision the band is reduced to ~0.6 kb, see Figure 1. The PCR conditions: were 10 ng genomic DNA with 5% DMSO in reaction, denature 94°C 120 sec, 1 cycle, then 35 cycles of: denature 94°C 30 sec, anneal 65°C 30 sec, extend 68°C 300 sec, 1 cycle of Completion 68°C 480 sec.
The ELL2 conditional knockout mouse with the deletion of exon 3 (encoding amino acids 66-97) was cloned by June Liu from Dr. Zhou Wang's laboratory. We used ES cells (129Sv/pas ) and insertion of the construct was selected for the neomycin marker in the targeting vector; the neo cassette was flanked by frt sites as described (above) for ELL2 exon 1 and shown in Supplemental Figures. The insertion of the targeting vector was detected using Southern blotting and increased size of the EcoRI fragment on the 3’ side and insertion of a new SacI site on the 5’ side, Supplemental Figures. Genotyping the ELL2 mice from the Wang lab was conducted using PCR with:
ELL2ckoC(90446583): 5’-AGG AGT TCA AGG TCT GCA TC-3’
ELL2ckoF(90446584): 5’-GGT GGA AAT CAC TCC TGT TC-3’
The wild type allele produces a PCR band of 400 bp and insertion of loxp produces a band of 500 bp as Figure 1c. When exon 3 is deleted the 500 nt band disappears. GAPDH was used as control for DNA content. The PCR conditions: were 10 ng genomic DNA, denature 94°C 120 sec, 1 cycle, then 35 cycles of: denature 94°C 30 sec, anneal 55°C 30 sec, extend 72°C 40 sec, 1 cycle of Completion 72°C 300 sec.
For genomic GAPDH, the primer sequences were as following.
GAPDH-F(13947763): GAG ACA GCC GCA TCT TCT TGT
GAPDH-R (13947764): CAC ACC GAC CTT CAC CAT TTT
The PCR condition is the same as in the genotyping of ELL2 and the expected PCR band size is 75 bp.
To genotype CD19cre/+ mice we used primers for CD19 flanking the potential cre insertion site:
CD19creF(50312489): 5’-GCG GTC TGG CAG TAA AAA CTA TC-3’
CD19creR(50312490): 5’-GTG AAA CAG CAT TGC TGT CAC TT-3’
CD19wtF(50312491): 5’-CCT CTC CCT GTC TCC TTC CT-3’
CD19wtR(50312492): 5’-TGG TCT GAG ACA TTG ACA ATC A-3’
CD19 wt gene produces a fragment of ~477 bp while the CD19 cre shows a fragment of 100 bp. Heterozygotes show both bands. The PCR condition is the same as in the genotyping of ELL2. Isolation of genomic DNA was done using whole blood samples collected from tail vein bleeds using DNeasy Blood and Tissue Kit (Qiagen 69504) according to manufacturer's instructions.
All mice were maintained at the University of Pittsburgh animal facilities and experiments undertaken and conducted in accordance with institutional policies, as per Animal Welfare Assurance number: A3187-01.
Flow cytometry
Bone marrow and spleen were harvested from mice and processed as previously described (14) and (15). Cell staining was performed using antibodies to murine surface markers obtained from eBioscience or BD Pharmingen. Primary anti-mouse Abs were B220 (clone RA3−6B2), CD19 (clone MB19−1), CD43 PE (clone S7), AA4.1 (clone AA4.1), IgM (clone 331), IgD (clone 11-26), CD138 (clone 281-2), CD21 (clone eBioD9), CD23 (clone B3B4), CD5 (clone 53-7.3). Secondary reagents were streptavidin-Cy7PE or streptavidin-eFluor 450. Dead cells were excluded using DAPI. Flow cytometry was performed on a four laser, twelve detector LSR II or a four laser, thirteen detector LSR Fortessa (BD Biosciences). Data were analyzed using FlowJo software Version. The schemes were derived from (16, 17) and described more fully in Supplemental data Table 1.
To sort cells for antibody secreting plasma cells, LPS induced splenocytes were incubated with APC conjugated Anti-Human/Mouse CD45R (B220) (eBioscience #47-0452) and PE conjugated Rat Anti-Mouse CD138 (BD Biosciences #553714) for 30 minutes on ice in the dark. Dead cells were excluded by DAPI staining. BCMA staining was done using Monoclonal Anti-mouse BCMA-Fluorescein (R&D Systems Inc. #FAB593F).
ELISA
Assays were performed following standard procedures using Clonotyping System-AP (SouthernBiotech, Birmingham AL) on the 96-well plates (Dynex Immulux HB, Chantilly, VA). The kit with IgG2c antisera is necessary for the C57B/L6 mice which lack IgG2a (18). The plate was coated with 100 μl/well of capture antibody (Goat anti-mouse Ig(H+L)-UNLB) to a concentration of 5 μg/ml in 1 × PBS, pH 7.4. After overnight incubation at 4 °C, the wells were treated with 10 % BSA for 3 hours at room temperature. Mouse serum was diluted 5000 times for IgG1 or 1000 times for other Ig isotypes and incubated with the plates overnight at 4 °C. The wells were washed and alkaline phosphatase (AP)-labelled detection antibodies (diluted 5000 times) were added and incubated for 1 hour at room temperature. The wells were washed and developed using 1 mg/ml AP Substrate solution (Thermo Scientific, Waltham, MA) containing PNPP for 15 min or more. The plate was read at A450. Standards were run for the various isotypes. For NP-specific Ig detection, plates were coated with NPBSA (Biosearch Technologies, Petaluma, CA).
Immunizations
Six to 8 week old ELL2 conditional knockout mice and litter mate control mice were immunized i.p. with either 4-hydroxy-3-nitrophenyl acetyl (NP)-Ficoll (F1420, Biosearch technologies) at 25ug in 0.1ml PBS or 100 ug NP-keyhole limpet hemocyanin (KLH) (Biosearch technologies N-5060) precipitated with alum (Pierce, 77161) as previously described for blimp knockouts (19). Serum was collected at 1, 2 and 3 weeks post-injection using NP-BSA coated plates in an ELISA. For the recall response the same dose of NP-KLH was given 6 weeks after the initial dose and serum collected at 7 and 14 days.
Western Blot
Protein samples were obtained from nuclear and cytoplasmic extracts of 0 days and 3 or 4 days post LPS exposure using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific #78833) according to manufacturer's instructions. Protein samples were then measured for concentration by Bradford Assay. Western protocol followed was the Bio-Rad General Protocol for Western Blotting (Bio-Rad Bulletin 6376 Rev A). Molecular weight markers used in the gels were the Precision Plus Protein™ Kaleidoscope™ markers from BioRad (#161-0375). Before running protein samples on 10% Acrylamide Mini-PROTEAN TGX precast Gels (Bio-Rad Laboratories #456-1034), samples were boiled at 95°C for 5 minutes and centrifuged at 16000 × g in a microcentrifuge for 1 min. Samples were then loaded onto gels and run for 5 minutes at 50 V. The voltage was then increased to 165 V for approximately 1 hour. After gels had run to desired length, the gel was placed in a 1X transfer buffer (25 mM Tris, 190 mM glycine 20% methanol, 1% SDS) for 15 minutes. After transfer to polyvinyldifluoride (PVFD) membrane, samples were analyzed by immunoblot and visualized by enhanced chemiluminescence using Pierce ECL Western Blotting Substrate (Thermo Scientific #32209). Blots were imaged on a ProteinSimple FluorChem™ M System.
Antibodies Used
Antibodies used included: Primaries: XBP1 (M-186) (Santa Cruz Biotechnology sc-7160) mw29/40 kDa; pAb anti-IRE1 alpha [p Ser 724] antibody (Novus Biologicals NB100-2323) mw 110 kDa; Rb pAb to IRE1 (Abcam ab37073); ELL2 R4502 affinity purified rabbit antibody (1); ATF-6α (H-280) (Santa Cruz Biotechnology sc-22799) mw75-85 kDa; BIP (C50B12) (Cell Signaling Technology 3177S) mw 75 kDa; Anti-Mouse IgM (μ-chain specific) antibody produced in goat (Sigma M8644-1MG)MW >55kDa; Anti-Hu/Mo Blimp1 purified clone: 6D3 (eBioscience 14-5963-82) Mw 110 kDa and 150 kDa with sumolyation; IgM kappa chain mw 25 kDa; Cyclin B2 (H-105) (Santa Cruz Biotechnology sc-22776) mw 43kDa; YY1 (H-414) (Santa Cruz Biotechnology sc-1703); Monoclonal Mouse Anti-Actin Clone C4 (MP Biologicals 691001)mw 43 kDa. Secondaries: Goat anti-rabbit IgG-HRP (Santa Cruz Biotechnology sc-2004); Donkey anti-goat IgG-HRP (Santa Cruz Biotechnology sc-2020); Goat anti-mouse IgG-HRP (Santa Cruz Biotechnology sc-2005); Goat anti-rat IgG-HRP (Santa Cruz Biotechnology sc-2006)
ELISPOT
Millipore MultiScreen 96-well Filter Plates (Millipore #MSIPS4W10) were coated with 5-6 μg/ml goat anti-mouse heavy and light-chain, purified immunoglobulins (Southern Biotech #5300-04) for 2 hours at RT. Wells were then washed and blocked with cell media + 10% FCS for 1.5 hours at RT. Live cells (sorted with DAPI), after 72 hours post LPS exposure (20 ug/mL) were then added to the wells and allowed to incubate overnight at 37 °C. After incubation with Goat anti-mouse IgM-AP antibodies (Southern Biotech #5300-04) for 1.5 hours at RT, spots were visualized with 1-Step NBT/BCIP solution (Thermo Scientific #34042). Counting and imaging of spots was done on an Immunospot S6 Micro Analyzer using Immunspot 5.0 Professional software. For bone marrow samples anti IgG1-AP antibody was used.
B cell cultures-
Splenocytes were extracted from mice and naïve B cells selected by autoMACS using a B cell Isolation Kit (Miltenyi Biotec # 130-090-862) utilizing a cocktail of biotin-conjugated antibodies against CD43 (Ly-48), CD4 (L3T4), and Ter-119, as well as Anti-Biotin MicroBeads. The splenocytes were counted by hemocytometer and cultured at a density of 1-5 x106 cells per mL. Cells were cultured for 72 or 96 hours with LPS at 20ug/ml (LPS from E. coli 00111:B4 Sigma #L3012-10MG) in RPMI 1640 media with 50 uM 2- mercaptoethanol, 2 mM glutamine, 10% FCS, sodium pyruvate, non-essential amino acids, Pen/Strep and HEPES buffer. A cell density of ~5 ×106 cells/ml was maintained by dilution in medium with LPS.
RNA isolation and RT-QPCR
NucleoSpin RNA II kits (Clontech 740955.50) were used to isolate RNA from cells at 0 and 72/96 hours post LPS exposure. To create cDNA SuperScript First-Strand (Invitrogen 11904-018) kits were used according to manufacturer's instructions and dT primers. cDNA was then used in RT-QPCR reactions using SYBR Green PCR Master Mix (Applied Biosystems 4309155) reagents. Primers used for RT-QPCR are listed in the Supplemental data, Table 2.
Luciferase
The mouse cyclin B2 promoter (-1188) cloned into the firefly luciferase pGL4.10 vector at the Kpn1 and Nco1 sites, was a generous gift of Dr. Kurt Engeland, Universitatt Leipzig, Germany. This is similar to the previous cyclin constructs in which the inhibitory effect of p53 was demonstrated on the cyclin B2 promoter (20, 21). We cloned portions of the human blimp-1 (-2973 to 0) , ELL2 (-3000 to 0), and IRF4 (-2182 to 0) promoters into pGL4.11 (Promega, Madison, WI). The expression cDNA plasmids for blimp-1 (CM#:632), c-Myc (CM#:633), ELL2 [CM#:56 7 (1)], IRF4 (CM#:594), p53 wt (CM#:634), mutant p53R175H (CM#:635) and the p65 subunit of NF-kB (Dr. Gutian Xiao at UPCI) were transfected in 12-well plates (Falcon, Franklin Lakes, NJ) with the indicated reporters in 293T cells using GenJuice(Novagen, San Diego, CA) as the transfection reagent. After 2 days, cells were lysed with 100 μl/well of 1 × Reporter Lysis Buffer (Promega, Madison, WI), and luciferase activity was assayed with 20 μl of each lysate using the dual Luciferase Assay System (Promega) with a Luminometer. The SR proteins were cloned into the mammalian expression plasmids pEF4his (Invitrogen). The original cDNAs were a generous gift of Martha Peterson (22) and Gavin Screaton (23). For each transfection, 200ng of the firefly luciferase reporter plasmid 0.6 ng of Renilla luciferase reporter (pRL-TK, Promega) was used as a control. Firefly luciferase values were normalized to the Renilla plasmid activity values.
Statistics
All experiments reported here were performed with samples from at least three different mice of the indicated genotypes or three different cell transfections. The results were plotted and analyzed using Graphpad PRISM 5; the P values * = 0.05, ** =0.01, *** = 0.001 are indicated on graphs. The probability/ P value was determined by either a two-tailed Student's T test when two samples were analyzed, or in the case of multiple samples using ANOVA with Tukey's or Bonferonni's post tests. Error bars represent S.E.M.
RESULTS
Conditional knock-out of ELL2 in B cells
Mice deficient in ELL1 (aka MEN1) are embryonic lethal (24). To avoid the lethality that might accompany a complete loss of ELL2 in mice, we deleted ELL2 specifically in B cells. The strategy we used to make and detect deletions is drawn in Figure 1a and described more completely in the methods section while the Southern blots showing proper integration are shown in the Supplemental data section. We independently targeted exon 1 in one strain of mice and exon 3 in another, since we were initially concerned that the loxp insertion in the ELL2 promoter region might compromise normal ELL2 expression elsewhere. Mice bearing the ELL2 exon1 loxP/loxP or exon3 loxP/loxP genes, diagramed in Figure 1a, were crossed with mice carrying one copy of the cre recombinase coding sequence, driven by the B cell specific CD19 promoter designated CD19 cre/+ (25). As illustrated in Figure 1b, the deletion that occurs in splenic B-cells of exon1 in the ELL2 conditional knockout, generating the shorter 0.6 kb PCR product, is > 90%. The ELL2 exon3 loxp/loxp is also deleted in splenic B cells of mice carrying that insert, see Figure 1c. In this assay the specific PCR product encompassing the 5’ loxp site is virtually absent in the ELL2 exon 3 floxed splenic B cells, with the PCR product corresponding to the GAPDH gene serving as a positive loading control (see Figure 5d for a more complete deletion). To assess ELL2 protein expression in the conditional knockout mice (ELL2 loxp/loxp CD19 cre/+), B220+ resting splenocytes were purified and stimulated with Lipopolysaccharide for 3 or 4 days, a treatment that normally induces B cell proliferation and plasma cell differentiation. When compared with control (ELL2 +/+ CD19 cre/+ or ELL2 loxp/loxp CD19 +/+) splenocytes treated the same way, there was very little or no ELL2 protein expression in the conditional knockout mice (Figure 1d). The RNA from splenic B cells was isolated with and without 4 days of LPS exposure. The level of HPRT mRNA (set as 100%) was used as an internal RT-PCR control. As shown in Figure 1e, the mRNA for ELL2 rises >6-fold relative to no LPS in the control mouse spleens, achieving levels greater than HPRT mRNA, a housekeeping gene that is consistently expressed throughout B cell differentiation (26). The mRNA for ELL2 does not increase after LPS in either the ELL2 exon 1 or the exon 3 targeted mice, consistent with the gene deletion and protein results. Thus the cre/loxp strategy is effective at eliminating ELL2 expression in these late stages of B cell development.
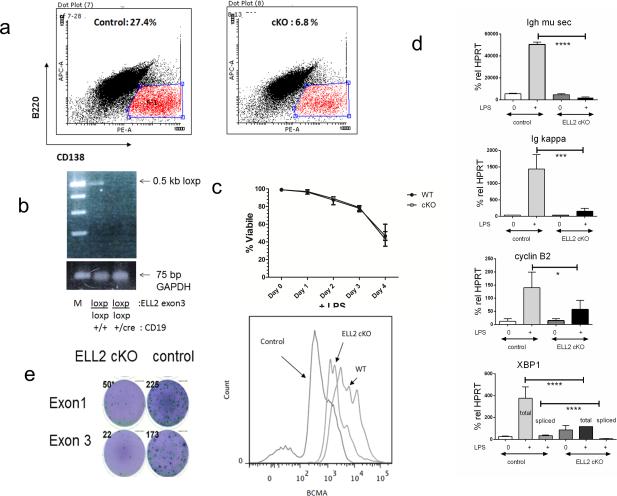
Figure 5.
ELL2 loxp/loxp CD19 cre/+ influences B220loCD138+ production, Igh secretory mRNA, and XBP1.
a. The flow cytometric profile of the LPS treated splenic B cells from the conditional knockout mice, left or the controls, right. B220lo CD138+ cells (boxed in) were enumerated as a % of the total live cell pool and used for subsequent analyses in panels b, d, e, and f. Splenic B cells were treated with 20ug/ml LPS for 3 days.
b. PCR of the DNA from the sorted cells, exon 3-/- mice shown, see Figure 1 for details.
c. Total cell viability in the LPS culture prior to sorting was determined by trypan blue staining.
d. RNAs from the sorted B220loCD138+ cells were quantified relative to HPRT using real-time quantitative PCR. Probes specific for the various species are indicated in the Supplemental material. Control or ELL2 cKO ( ELL2 loxp/loxp CD19 cre/+ ). Ig mu heavy chain mRNA secretory specific mRNA (Igh mu sec), top, was quantified using probes for the 3’ region in the secretory specific form. The RTQPCR probes for Ig kappa light chain, cyclin B2 and spliced versus unspliced forms of XBP1 mRNA are indicated in text or supplement. Error bars and P values as described in previous figures.
e. ELISPOT. Control or conditional knockout mice spleen cells stimulated with LPS for 3 days and sorted for B220loCD138+ (100,000/ well) were assayed for the production of IgM with anti-mouse IgM Alkaline Phospahatase antibodies and visualized with NBT/BCIP reagent. Spots were counted in an ELISPOT reader and the data enumerated in Table 1.
f. Cells in the B220loCD138+ selected pools were surface stained for BCMA. Control is with no antibody, WT is the ELL2loxp/loxp CD19+/+, ELL2 cKO is ELL2 loxp/loxp CD19 cre/+ ).
Reduction in Ig secretion in ELL2 loxp/loxp CD19cre/+ mice
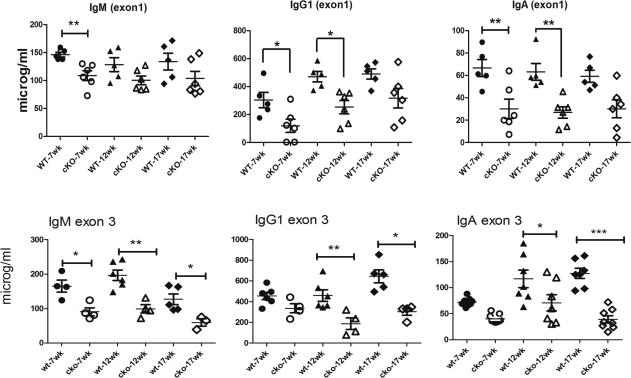
To determine if ELL2 has an effect on Ig secretion in the animal, we then conducted ELISA assays for the amount of secreted Ig in the serum of unimmunized control ELL2 loxp/loxp CD19+/+ or ELL2+/+ CD19 cre/+ mice (combined data labeled wt on graphs) and the ELL2 loxp/loxp CD19 cre/+ mice (cKO). We saw a decrease in the ELL2 exon 1 cKOs in serum levels of secreted IgM, IgG1, IgA isotypes as shown in Figure 2 and for all the other IgG isotypes (data not shown). With the ELL2 exon 3 cKO mice, the Ig levels were decreased 2-fold or greater for IgM, IgG1 and IgA (Figure 2) as well as for the other IgG isotypes (data not shown). ELL2 expression on stimulated B cells is thus required for Ig secretion in the whole animal.
Figure 2.
Serum levels in naïve ELL2loxp/loxp CD19cre/+ mice are reduced with both exon 1 and exon 3 deletions. Control mice with either ELL2loxp/loxp CD19+/+ or ELL2+/+ CD19cre/+ (filled symbols, designated wt) and the ELL2 loxp/loxp CD19 cre/+ mice (open symbols, designated cKO) were bled at 7, 12 and 17 weeks. Serum was analyzed by ELISA for the indicated isotypes. P= 0.05 (*), 0.01 (**), or 0.001 (***) as indicated on graphs.
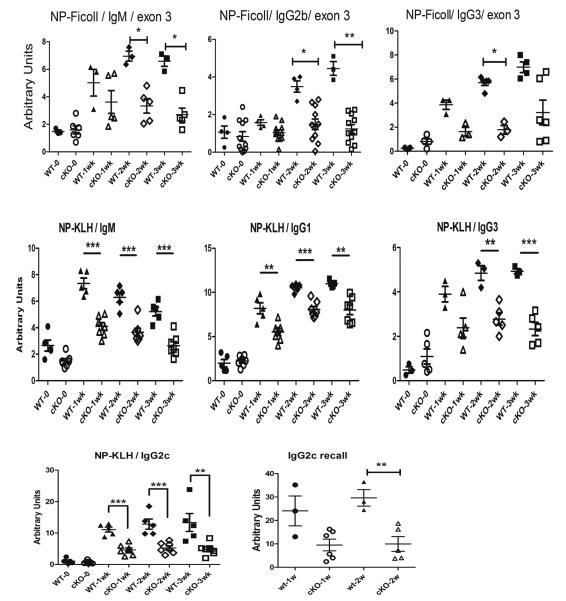
The effect of the conditional knockout of ELL2 is even more evident when the system is challenged by antigen. Mice with a conditional knockout of exon 3 of ELL2 (cKO) between 7-8 weeks of age were immunized with NP-ficoll and serum was drawn at 1, 2 and 3 weeks post immunization. ELISAs for NP specific antibodies were conducted. As summarized in Figure 3, there was significantly less anti-NP antibody of the IgM, IgG2b, and IgG3 isotypes in the exon 3 conditional knockout mice relative to control mice. Next we immunized naive exon 3 ELL2 cKO and control mice with NP-KLH, an antigen which requires T-cell help for the B cell responses. As shown in Figure 3 there was a significantly lower amount of specific anti-NP antibody in the exon 3 NP-KLH immunized conditional knockout mice of the IgM, IgG1, IgG3 and IgG2c isotypes relative to the control mice. Other isotypes in exon 3 targeted mice were also decreased (data not shown). Thus we conclude that deletion of ELL2 in stimulated B cells impairs the specific antibody responses to the T-independent antigen, NP-ficoll and to the T-dependent antigen, NP-KLH. Upon re-stimulation with NP-KLH we observed a recall response in the control mice that was decreased in the conditional knockout mice, data for IgG2c shown in Figure 3. Thus ELL2 is important for antigen specific responses and immunological memory.
Figure 3.
NP-ficoll and NP-KLH specific antibody response diminished in immunized ELL2 conditional exon 3 knockout mice. Control mice (filled symbols) and ELL2 loxp/loxp CD19 cre/+ mice (open symbols) were immunized and serum was drawn at 7, 14 and 21 days post immunization. The amount of NP-specific antibody was determined by ELISA. NP-KLH IgG2c recall response was elicited by immunization 6 weeks after the initial dose, serum was collected after 7 and 14 days.
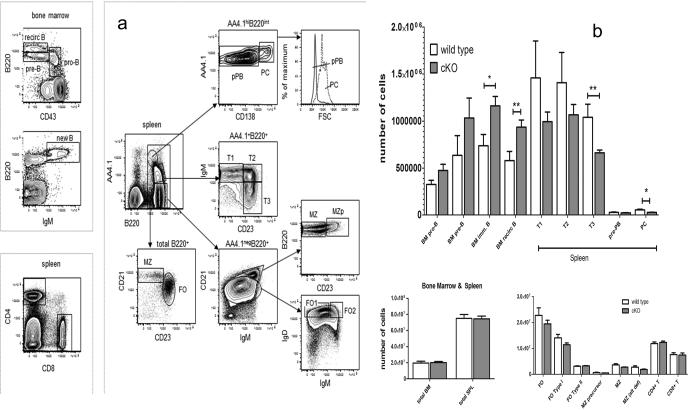
B cell subsets in vivo
The cell surface markers used for this study are summarized in Table 1 Supplemental data. Analysis by flow cytometry for both the exon 1 and exon 3 cKO naïve mice indicates that there is no significant change in B cell numbers, B1 vs B2 ratios, T cells, or their distribution (data not shown). However, when the ELL2 exon 3 targeted mice were challenged with antigen (NP-ficoll or NP-KLH) there was a significantly higher percentage of immature and recirculating B cells in the bone marrow after immunization than in the control (Figure 4). There is a trend to fewer Transitional 1 and 2 type B-cells (T1, T2) in the spleen and a significant drop in T3 cells in the conditional knockouts (Figure 4b). The immgen.org profile for ELL2 shows that it is expressed at an intermediate level in these cells. When we determined the number of CD138+ cells we saw a significant drop in their numbers in the immunized exon 3 ELL2 loxp/loxp CD19 cre/+ mice relative to control mice treated the same way. With the loss of ELL2 in B cells, differentiation can continue to the B220+ IgM+ (new B cell) stage but the increase in recirculating cells in the bone marrow shown in Figure 4a indicates a potential problem with further maturation in the spleen, which is evident in the lower numbers of T3 and CD138+ cells produced. We performed ELISPOTS on bone marrow cells from unimmunized exon1 and exon 3 ELL2 cKO mice using anti-IgG1 antibody. As shown in the results enumerated in Table 1, there were significantly fewer IgG1 producing cells in the bone marrow of the cKO mice. These IgG+ cells may represent long-lived plasma cells or memory B cells (27).
Figure 4.
ELL2 loxp/loxp CD19 cre/+ mice have increased naïve bone marrow cells and decreased T3 and plasma cells in the spleen. a. Bone marrow and spleen were harvested from mice 21 days after NP-ficoll or NP-KLH immunization and subjected to flow cytometry using the indicated antibodies. The surface markers used to designate each population are indicated in Materials and Methods and Supplemental data. b. The number of cells in each of the harvested categories were quantified using the flow data. Error bars are +/− S.E.M. and P values as described in Figure 2.
Table 1.
ELISPOT data, splenic B cells and bone marrow
| Bone marrow IgG1+ | Splenic IgM+, B220loCD138+ sorted 3 days LPS | |||
|---|---|---|---|---|
| # spots | statistics | # spots | statistics | |
| control | 66 | +/− 35 std dev | 318 | +/− 65 std dev |
| cKO ELL2 | 9 | +/− 7 std dev | 65 | +/− 16 std dev |
| P value | 2.6×10−3 | 2×10−5 | ||
Values normalized to 100,000 cells. P value determined by 2 tailed T tests. N= >6 per group.
ELL2 deletion influences Igh processing and plasma cell differentiation
After ex vivo LPS stimulation of the naïve splenic B cell population in both exon 1 and exon 3 floxed, ELL2 conditional knockouts as compared with CD19+/+ controls we noted a decrease in the production of B220loCD138+ cells. As shown in a representative experiment in Figure 5a with an exon 3 deficient mouse there were approximately 4-fold fewer cells in the conditional knock out. The cells that had become B220loCD138+ were sorted and analyzed by PCR to determine if they were still deleted for exon 3 of ELL2. They were still deficient in ELL2 and do not represent the progeny of a population of un-deleted cells (Figure 5b). The conditional knockout and control cells were equally viable during the course of all the LPS stimulation experiments with both types of cKOs. We isolated RNA from the B220loCD138+ cells of the two different ELL2 exon knockouts and the control and subjected the samples to reverse transcriptase and quantitative PCR as described in Materials and Methods. We compared RNA from naïve B cells and normalized all the values to HPRT, which had previously been shown to remain constant throughout the B to plasma cell transition (28). Production of the secretory-specific Igh mu mRNA is induced dramatically in the control ELL2+ cells as expected. In the knockout mice there was no induction of the secretory specific heavy chain mRNA. The overall level of Igh mRNA is reduced in the ELL2 cKO as well compare the total amount of Igh mu mRNA sec plus Igh mu mb (Figure 5 d and Table 2 , 3) cKO versus the control. So ELL2 drives up the amount of mature Igh mRNA as well as influencing processing. We also saw a decrease in Ig kappa mRNA relative to the control (Figure 5d). Cyclin B2 mRNA was also not as robustly induced in the conditional knockout mice relative to the control B220loCD138+ cells. Examination of XBP1 mRNA revealed a dramatic decrease in both total mRNA and the specifically cytoplasmic spliced form that results from induction of the IRE1 phosphorylation. An ELISPOT to measure IgM secretion, conducted with the B220loCD138+ cells after 4 days of LPS, shows a 4.4-fold decreased number of spots, many of which are less intense than those in the control, indicating decreased secretion of IgM in the ELL2 conditional knockout (Figure 5e and Table 1). Thus, as we had predicted based on our previous studies (1), ELL2 is essential for efficient processing to the secretory form of Igh in plasma cells; in the absence of ELL2 the change-over in RNA processing does not occur and secreted Igh mu is not induced. In addition, other mRNAs were also reduced in the cKO mice (Table 2); these include several in the unfolded response pathway like ATF6, BiP and OcaB, the transcription of which thought to be driven by XBP1 (29). Expression of IRF4 and blimp-1 were at or above control levels in the ELL2 cKO B220loCD138+ cells (Table 2) so those early steps in B cell activation are largely intact.
Table 2.
mRNA expression in B220loCD138+ sorted cells relative to HPRT as 100%*
| control | cKO | control/ cKO | |
|---|---|---|---|
| ATF6 | 313 | 65 | 4.8 |
| BiP | 1,790 | 536 | 3.3 |
| Blimp-1 | 22 | 25 | 0.9 |
| ELL1 | 52 | 5 | 10.4 |
| ELL2 | 2,032 | <50 | >37 |
| ELL3 | 5 | 4 | 1.3 |
| Ire1 | 272 | 140 | 1.9 |
| IRF4 | 25 | 32 | 0.8 |
| Ig Mu mb | 2,138 | 3,093 | 0.7 |
| OcaB | 2,680 | 548 | 4.9 |
S.E.M. less than 5% in all cases, not indicated for clarity; n> 3 mice each class, exon 1 and exon 3.
Table 3.
The mRNA expression in wt vs cKO ELL2 mice relative to HPRT set as 100%* total in the LPS stimulated cultures at 3 -4 days
| gene | Control no LPS | Control 4d LPS | cKO no LPS | cKO 4d. LPS | control/cKO 4d. LPS |
|---|---|---|---|---|---|
| AFF1 | 3 | 9 | 0.6 | 5 | 1.8 |
| AICDA | 0.6 | 33 | 1.8 | 21.3 | 1.6 |
| ATF6 | 60 | 320 | 64 | 155 | 2.1 |
| Bcl6 | 1.3 | 5.2 | 1.3 | 2 | 2.6 |
| BiP | 400 | 1400 | 432 | 308 | 4.6 |
| Blimp-1 | 0.5 | 48.7 | 1.0 | 40.1 | 1.2 |
| BRD4 | 100 | 190 | 75 | 80 | 2.4 |
| Cebp-β | 87 | 43 | 90 | 27 | 1.6 |
| CstF64 | 1.5 | 7 | 2 | 3 | 2.3 |
| Cyclin B2 | 15 | 65 | 15 | 8 | 8.1 |
| Eaf1 | 1.8 | 6.6 | 2 | 4 | 1.7 |
| Eaf2 | 7 | 55 | 2 | 35 | 1.6 |
| ELL3 | 37 | 4 | 50 | 3 | 1.3 |
| Hif1a | 33 | 22 | 25 | 22 | 1.0 |
| Hsp40 | 40 | 40 | 38 | 50 | 0.8 |
| Igh mb | 3000 | 2900 | 2200 | 6800 | 0.4 |
| Igh sec | 5400 | 45,000 | 2600 | 1200 | 37.5 |
| IRE1 | 20 | 270 | 20 | 174 | 1.6 |
| IRF4 | 1.8 | 21.2 | 1.7 | 21.3 | 1 |
| NF-kb p50 | 19 | 22 | 22 | 18 | 1.2 |
| NF-kb p65 | 25 | 18 | 6 | 13 | 1.4 |
| Pax5 | 36 | 57 | 35 | 45 | 1.3 |
| PCNA | 70 | 250 | 150 | 300 | 0.8 |
| Sup5H | 4 | 8.5 | 4 | 6.5 | 1.3 |
| XBP1 Total | 70 | 405 | 71 | 100 | 4.0 |
| XBP1 spliced | 4 | 70 | 3 | 3 | 23.3 |
S.E.M. less than 5% in all cases, not indicated for clarity; n> 6 mice each class, exon 1 and exon 3.
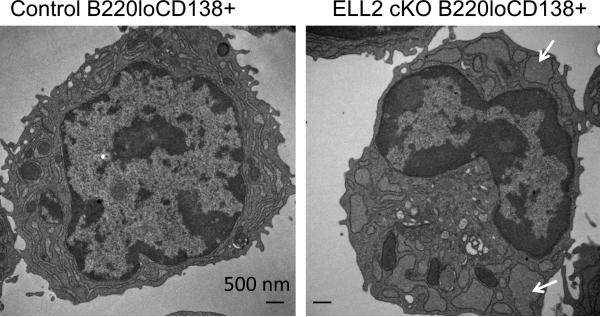
The sorted B220loCD138+ cells were fixed and analyzed in the transmission electron microscope (Figure 6). The conditional knockout cells display an endoplasmic reticulum which is dilated and fragmented. Similar, distended ER is seen in XBP1 knockout plasma cells (30). Based on the accumulated data, we conclude that ELL2 deletion has a significant impact on Igh secretory mRNA and protein, XBP1 production, and the activated cell architecture.
Figure 6.
Transmission EM of sorted B220loCD138+ cells (see previous figure) from controls (a) and the ELL2 conditional knockouts (b). Arrows indicate areas of distended ER in the ELL2 cKO cells.
We then examined the expression of a number of other mRNAs and proteins in the bulk population of control and conditional ELL2 knockout LPS stimulated cells. We observed normal decreases in the conditional knockout mouse cells in some mRNAs that accompany early steps in plasma cell differentiation, compare plus versus minus LPS values, like ELL3 and C/ebp-beta (Table 3). Some mRNAs are induced after LPS, both in control and the conditional knockouts like AICDA, PCNA, and Eaf2, a factor associated with the ELL2:pTEFb complex, although their overall levels at the induced state are somewhat less than control. We observed no significant change in the expression of some “housekeeping” genes like Hif1 alpha and Hsp40 in either the controls or the knockouts. A majority of the probes used in the RT-QPCR spanned exons. When we examined the splicing patterns of several of these genes (Bip, Eaf2, IRF4) we saw no difference between control and the ELL2 deficient cells before or after LPS stimulation, data not shown. Thus some aspects of the plasma cell program are operative. The induction of cyclin B2 mRNA seen in control LPS stimulated cells was lacking in the conditional knockouts. Cyclin B2 protein resides in the Golgi (31) where carbohydrates are added to secreted proteins; this suggests that the Golgi may also be affected by the lack of a normal unfolded protein response.
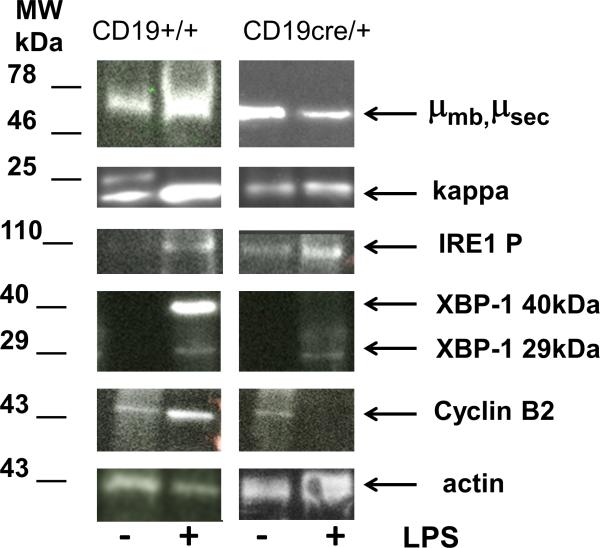
Changes in mRNA expression were confirmed by analysis of the protein in both exon 1 and exon 3 conditional knockouts. In Figure 7 protein results from the exon 3 mice are presented. The unconventionally spliced mRNA of XBP1 (XBPs) encodes a protein (32) of approximately 40 kDa that acts as a transcription factor for up-regulation of the unfolded protein response (UPR) genes (33). Since the unconventional XBP1 splicing is initiated by the IRE1 alpha protein after its aggregation and phosphorylation by endoplasmic reticular stress (34) and (35), we investigated the amount and state of IRE1 in our cultures. The level of IRE1alpha mRNA is only slightly reduced in the conditional ELL2 knockouts relative to the control (Table 3). The level of phosphorylated IRE1 protein was determined by western blotting using anti-pSer724 IRE1 antibody (Figure 7); increases relative to naïve B cells were seen in control and conditional knockouts. These bands were confirmed as IRE1 by reaction with a pan IRE antibody (data not shown). Activation of the novel RNase activity of IRE1 is initiated by dimerization-induced trans-autophosphorylation and requires a homodimer of catalytically functional RNase domains (34). When large amounts of protein like secreted Ig or other secretory-associated proteins that may be unfolded are produced, this causes stress to the ER (36). Here, with reduced levels of BiP, as in the ELL2 conditional knockouts, IRE1 phosphorylation is activated, perhaps in a situation analogous to that of a mutant of IRE1 that cannot bind BiP and remains phosphorylated (37).
Figure 7.
Protein expression. Proteins extracted from control (CD19+/+ ELL2 loxp/loxp) and conditional knockout (CD19 cre/+ ELL2 loxp/loxp ) splenic B cells stimulated for 3 days with LPS (bulk populations) were run on 10% acrylamide SDS PAGE. Data from an exon 3 deleted mouse shown. The indicated primary and secondary antibodies (see supplemental materials and methods) were used in western blots with ECL substrate. Molecular weights are extrapolated from Kaleidoscope colored markers run on each gel (not shown).
Transcription studies
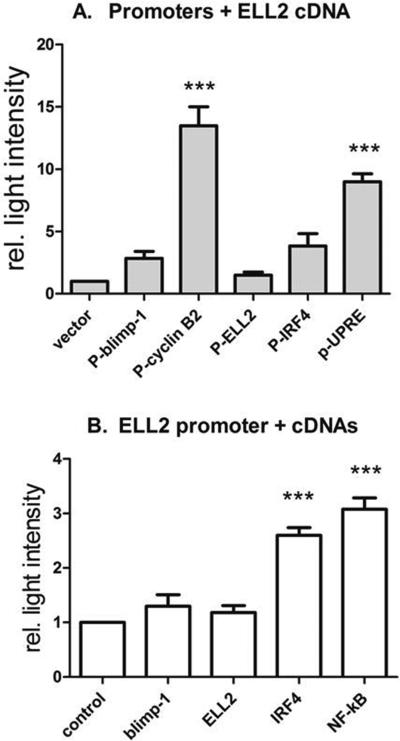
We had previously shown that the alternative processing of the Ig mu heavy chain is directly regulated by the binding and the action of ELL2 of RNAPII (1). Elongation factors not only change processing patterns but they also increase the processivity of RNAPII (38) so mRNA yields increase in their presence. We wanted to determine if ELL2 influences the apparent transcription of some of the other genes we see altered in plasma cell differentiation. We combined ELL2 cDNA with various luciferase reporter constructs bearing promoters in 293 HEK cells and measured luciferase activity (Figure 8) versus transcription without ELL2. The different promoters showed different levels of luciferase transcriptional induction with ELL2. The cyclin B2 promoter had been cloned into a luciferase reporter by Dr. Engeland who generous supplied it to us (20). Addition of a cDNA for ELL2 increased luciferase activity from the cyclin B2 promoter almost 12-fold over vector alone, indicating that ELL2 can directly enhance the yield of cyclin B2 promoter driven mRNA. The positive effect of ELL2 is partially diminished by the addition of p53 which had previously been shown to decrease cyclin B2 transcription (see supplemental data Figure 2). ELL2 cDNA had no effect on its own promoter while it stimulated the blimp-1 and IRF4 promoters about 2 to 4 fold. The luciferase construct bearing the 5X UPRE or unfolded protein response element promoter (39), common to many UPR genes (40), was induced > 8-fold by ELL2 cDNA, the same induction seen with a cDNA for NF-kB p65 subunit protein (data not shown). Transfections conducted in a B cell line and Jurkat T cells demonstrated a similar pattern of induction with all promoters (data not shown). Thus ELL2 may participate in the UPR not only by virtue of directing Igh mRNA processing and activating XBP1, but it may also help to raise the levels of the UPR controlled mRNAs by enhancing their mRNA yields. The ELL2 promoter is stimulated by IRF4 and NF-kB but not by itself or blimp-1 (Figure 8). IRF4 binding to the ELL2 promoter by ChIP had been shown previously (6). Meanwhile IRF4 and blimp-1 have transcriptional stimulatory effects on each other. SRp55, implicated in alternative splicing of CD44 and with DNA damage responses (41), significantly enhances blimp transcription (Supplemental data). SRp55 showed little effect on Igh alternative RNA processing (22). Thus considering all the evidence presented, we conclude that ELL2 is an essential part of the transcription network leading to plasma cell differentiation; it appears to act between the IRF4/ blimp-1 and XBP1 genes.
Figure 8.
ELL2 influences cyclin B2 and UPR expression by stimulating transcription/ RNA yields from their promoters.
a. The mouse cyclin B2 promoter ( -1188 to 0) was cloned into pGL4.10, the luciferase reporter plasmid. 293T cells were co-transfected with the CycB2 reporter and the indicated cDNA plasmids and luciferase activity measures with a luminometer.
b. The ELL2 promoter (-3000 to 0) was cloned into pGL4.11 luciferase reporter and transfected with the indicated cDNA plasmids.
DISCUSSION
ELL2 floxed conditional deletions, caused by the CD19 promoter driven cre, decrease basal Ig levels in the serum. In the exon 3 floxed ELL2 conditional knockouts, the responses to immunization with NP-KLH and NP-ficoll, T dependent and independent antigens respectively, were significantly reduced relative to the controls. In the CD19cre driven deletion of exon 3 floxed ELL2, no gross abnormalities are seen in the number of cells present in the early B cell stages and in total bone marrow and splenic B cells. ELL2 is not maximally expressed until after LPS induction of splenic B cells and then its expression increases as much as 30-fold. This pattern of expression may explain the lack of influence on the early B cell populations since ELL2 is not maximally expressed until after LPS or antigen induction of splenic B cells. In the spleen, deletion of ELL2 in the total B cell population is >90% in the floxed ELL2 alleles and this percentage persists after LPS stimulation. The large increase in transcription of ELL2 at a point when the CD19 promoter is very active may explain why the gene is so successfully targeted by CD19cre.
After ex vivo LPS stimulation of resting B cells, production of secretory specific Igh mu is significantly reduced in both exon 1 and exon 3 knockouts as judged by mRNA, protein, and ELISPOT analyses. Production of ex vivo LPS stimulated, splenic B cells that are B220loCD138+ is reduced by at least 4-fold in the conditional knockouts. Those sorted B220loCD138+ cells lack ELL2 by mRNA and genomic DNA analyses and have a paucity of secreted Igh and XBP1, with distended, abnormal appearing ER. In the whole animal the effect of ELL2 deletion on total Ig production is less profound than that which we see in vitro. Our in vitro experiments were done with splenic B2 cells, not B1 cells, and following LPS stimulation; engagement of TLR4 is not the only way to trigger Ig production. Thus other factors may influence secretory Ig production in B1 cells or in other activation pathways in the whole animal.
IRF4, blimp-1 and XBP1 have been identified as important for plasma cell production (5). When a B cell line (L29mu+) was stimulated to convert to the plasma cell phenotype, it did so in a multistep process; proteomic analyses showed that the metabolic capacity and secretory machinery were put into place prior to the mass production of Ig that normally follows (42). Ig secretion in a blimp-1 knockout is low, indirectly through the downstream effects of pax5 suppressing XBP1 (2). When B cells deficient in secretory-specific mu protein (AID-/- mus-/- or mus-/-) were stimulated with mitogens they showed reduced ability to differentiate into B220loCD138+ cells and reduced survival (43). But the absence of secretory Ig protein alone did not prevent XBP1 accumulation, or XBP1 splicing, which may normally precede up-regulation of secreted Ig (43), (33). The hypothesis that the machinery for secretion is put in place prior to detectable Igh processing to the secreted form was generated. But, what we see in the ELL2 conditional knockouts is that the two processes (establishment of secretory machinery and Igh mRNA processing) appear linked; overall XBP1 production is reduced along with Igh. It is clear that lack of ELL2 has broader effects than lack of secretory Ig alone and our luciferase studies show ELL2 can act on cyclin B2 and genes bearing the Unfolded Protein Response Element (UPRE) to enhance mRNA levels.
Another group used an siRNA mediated knockdown of ELL2 mRNA in cultured plasma cells (12). They showed, using deep mRNA sequencing, that the knockdown of ELL2 influenced Igh processing, and expression of several splicing factors, cyclin B2 (ccnb2), and the B cell maturation antigen (tnfrsf17) aka BCMA. We saw a decrease in Igh secretory mRNA processing, and cyclin B2 and BCMA levels in LPS stimulated splenic B cells in our knockout mice. Loss of BCMA can contribute to long term survival (4) and this may be reflected in the reduced number of IgG1 antibody secreting cells by ELISPOT we see in bone marrow cells in the knockouts. We see no major effects on RNA splicing but rather a significant impairment in the development of the unfolded protein responses, perhaps because ELL2 and secreted Ig are important for the establishment of the UPR but not necessarily for its maintenance which could be missed by looking only at established plasma cells.
In the ELL2 knockouts IRE1alpha is phosphorylated but the expression of the downstream proteins for the unfolded protein response including spliced XBP1 are reduced in the LPS stimulated spleens cells. In mature plasma cells the ER response is unique from that seen in other cells (44). The UPR in many cells typically has three arms, the IRE-1/ XBP pathway, an ATF6 pathway, and the PERK pathway (40). But PERK and ATF6 knockout mice secrete normal amounts of Ig (33, 45). XBP1 conditional deletion mice show defects in PC development (46) and low levels of secretory Ig (47). Thus when B cells are stimulated the primary pathway for ER remodeling appears to reside in the IRE1-1 to XBP1 pathway (48). Aggregation and then auto phosphorylation of IRE1 causes it to acquire the ability to specifically cleave and then splice XBP1 mRNA; the newly spliced XBP1 RNA species encodes XBP1 protein with transcriptional activity on its own promoter and other UPR promoters containing the UPRE (33, 35). However, the low levels of Igh mRNA in XBP1-/- result from the 8-fold increased levels of IRE1P over control; the highly abundant IRE1P cleaves the mu secretory mRNA (49). But a double deletion of XBP1 and IRE1 restores IgM secretion by inhibiting mRNA degradation (49). Taken together, this leads us to a conclusion that Ig secretion can occur without XBP1 cleavage/ splicing and there may be other proteins that allow for the up-regulation of the UPR besides the spliced mRNA encoded XBP1. We believe that ELL2 is one of those proteins; it appears to have a role in enhancing the transcription of other UPR proteins through the UPR element. ELL2 influences more than Igh processing; its effect at enhancing transcription from a variety of promoters, to different extents, implies important specific interactions that remain to be explored.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. O.J. Finn and E.M. for encouragement and the Immunology Department for seed funding for animal model studies.
This work was supported by NSF # MCB-0842725 to C.M.; T32 CA082084 training support to K.S.P. UPCI CCSG shared resources, supported in part by award P30CA047904; R01 AI079047 and R21 AI105846 to L.B.; and 9R01CA186780 and R37DK051193 to Z.W. and J.L.
REFERENCES
- 1.Martincic K, Alkan SA, Cheatle A, Borghesi L, Milcarek C. Transcription elongation factor ELL2 directs immunoglobulin secretion in plasma cells by stimulating altered RNA processing. Nat Immunol. 2009;10:1102–1109. doi: 10.1038/ni.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaffer A, Shapiro-Shelef M, Iwakoshi NN, Lee A, Qian S, Zhao H, Yu X, Yang L, Tan BK, Rosenwald A, Hurt EM, Petroulakis E, Sonenberg N, Yewdell JW, Calame K, Glimcher LH, Staudt LM. XBP1, Downstream of Blimp-1, Expands the Secretory Apparatus and Other Organelles, and Increases Protein Synthesis in Plasma Cell Differentiation. Immunity. 2004;21:81–93. doi: 10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 3.Ochiai K, Maienschein-Cline M, Simonetti G, Chen J, Rosenthal R, Brink R, Chong Anita S., Klein U, Dinner Aaron R., Singh H, Sciammas R. Transcriptional Regulation of Germinal Center B and Plasma Cell Fates by Dynamical Control of IRF4. Immunity. 2013;38:918–929. doi: 10.1016/j.immuni.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Connor BP, Raman VS, Erickson LD, Cook WJ, Weaver LK, Ahonen C, Lin L-L, Mantchev GT, Bram RJ, Noelle RJ. BCMA Is Essential for the Survival of Long-lived Bone Marrow Plasma Cells. J. Exp. Med. 2004;199:91–98. doi: 10.1084/jem.20031330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nutt SL, Taubenheim N, Hasbold J, Corcoran LM, Hodgkin PD. The genetic network controlling plasma cell differentiation. Seminars in Immunology. 2011;23:341–349. doi: 10.1016/j.smim.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 6.Shaffer AL, Emre NCT, Lamy L, Ngo VN, Wright G, Xiao W, Powell J, Dave S, Yu X, Zhao H, Zeng Y, Chen B, Epstein J, Staudt LM. IRF4 addiction in multiple myeloma. Nature. 2008;454:226–231. doi: 10.1038/nature07064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sciammas R. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 2006;25:225–236. doi: 10.1016/j.immuni.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 8.Shell SA, Martincic K, Tran J, Milcarek C. Increased phosphorylation of the carboxyl terminal domain of RNA polymerase II and loading of polyadenylation and co-transcriptional factors contribute to regulation of the Ig heavy chain mRNA in plasma cells. J. Immunol. 2007;179:7663–7673. doi: 10.4049/jimmunol.179.11.7663. [DOI] [PubMed] [Google Scholar]
- 9.Milcarek C, Albring M, Langer C, Park KS. The Eleven-Nineteen Lysine-rich Leukemia gene (ELL2) influences the histone H3 modifications accompanying the shift to secretory Immunoglobulin heavy chain mRNA production. J. Biol. Chem. 2011;286:33795–33803. doi: 10.1074/jbc.M111.272096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith E, Lin C, Shilatifard A. The super elongation complex (SEC) and MLL in development and disease. Genes Dev. 2011;25:661–672. doi: 10.1101/gad.2015411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo Z, Lin C, Shilatifard A. The super elongation complex (SEC) family in transcriptional control. Nat Rev Mol Cell Biol. 2012;13:543–547. doi: 10.1038/nrm3417. [DOI] [PubMed] [Google Scholar]
- 12.Benson MJ, Äijö T, Chang X, Gagnon J, Pape UJ, Anantharaman V, Aravind L, Pursiheimo J-P, Oberdoerffer S, Liu XS, Lahesmaa R, Lähdesmäki H, Rao A. Heterogeneous nuclear ribonucleoprotein L-like (hnRNPLL) and elongation factor, RNA polymerase II, 2 (ELL2) are regulators of mRNA processing in plasma cells. Proc. Nat. Acad.Sci., U.S.A. 2012;109:16252–16257. doi: 10.1073/pnas.1214414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He N, Liu M, Hsu J, Xue Y, Chou S, Burlingame A, Krogan NJ, Alber T, Zhou Q. HIV-1 Tat and Host AFF4 Recruit Two Transcription Elongation Factors into a Bifunctional Complex for Coordinated Activation of HIV-1 Transcription. Molecular Cell. 2010;38:428–438. doi: 10.1016/j.molcel.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Q, Esplin B, B. L. E47 regulates hematopoietic stem cell proliferation and energetics but not myeloid lineage restriction. Blood. 2011;117:3529–3538. doi: 10.1182/blood-2010-07-297689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borghesi L, Aites J, Nelson S, Lefterov P, James P, Gerstein R. E47 is required for V(D)J recombinase activity in common lymphoid progenitors. J. Exp. Med. 2005;202:1669–1677. doi: 10.1084/jem.20051190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santos PM, Ding Y, Borghesi L. Cell-Intrinsic In Vivo Requirement for the E47–p21 Pathway in Long-Term Hematopoietic Stem Cells. J. Immunol. 2014;192:160–168. doi: 10.4049/jimmunol.1302502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winkelmann R, Sandrock L, Porstner M, Roth E, Mathews M, Hobeika E, Reth M, Kahn ML, Schuh W, Jäck H-M. B cell homeostasis and plasma cell homing controlled by Krüppel-like factor 2. Proc.Nat. Acad. Sci., USA. 2011;108:710–715. doi: 10.1073/pnas.1012858108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin RM, Brady JL, Lew A, M the need for IgG2c specific antiserum when isotyping antibodies from C57BL/6 and NOD mice. J Immunol Methods. 1998;212:187–192. doi: 10.1016/s0022-1759(98)00015-5. [DOI] [PubMed] [Google Scholar]
- 19.Shapiro-Shelef M, Lin K-I, McHeyzer-Williams LJ, Liao J, McHeyzer-Williams MG, Calame K. Blimp-1 Is Required for the Formation of Immunoglobulin Secreting Plasma Cells and Pre-Plasma Memory B Cells. Immunity. 2003;19:607–620. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- 20.Krause K, Wasner M, Reinhard W, Haugwitz U, Lange-zu Dohna C, Mössner J, Engeland K. The tumour suppressor protein p53 can repress transcription of cyclin B. Nuc. Acids Res. 2000;28:4410–4418. doi: 10.1093/nar/28.22.4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salsi V, Caretti G, Wasner M, Reinhard W, Haugwitz U, Engeland K, Mantovani R. Interactions between p300 and Multiple NF-Y Trimers Govern Cyclin B2 Promoter Function. J. Biol. Chem. 2003;278:6642–6650. doi: 10.1074/jbc.M210065200. [DOI] [PubMed] [Google Scholar]
- 22.Bruce SR, Dingle RWC, Peterson ML. B-cell and plasma-cell splicing differences: A potential role in regulated immunoglobulin RNA processing. RNA. 2003;9:1264–1273. doi: 10.1261/rna.5820103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Screaton GR, Caceres JF, Mayeda A, Bell MV, Plebanski M, Jackson DG, John I Bell JI, Krainer AR. Identification and characterization of three members of the human SR family of pre-mRNA splicing factors. EMBO Journal. 1995;14:4336–4349. doi: 10.1002/j.1460-2075.1995.tb00108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitani K, Yamagata T, Iida C, Oda H, Maki K, Ichikawa M, Asai T, Honda H, Kurokawa M, Hirai H. Nonredundant Roles of the Elongation Factor MEN in Postimplantation Development. Biochem. Biophys. Res. Comm. 2000;279:563–567. doi: 10.1006/bbrc.2000.3970. [DOI] [PubMed] [Google Scholar]
- 25.Rickert R, Roes J, Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucl. Acids Res. 1997;25:1317–1318. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Genovese C, Harrold S, Milcarek C. Differential mRNA stabilities affect mRNA levels in mutant mouse myeloma cells. Som Cell & Mol Genet. 1991;17:69–81. doi: 10.1007/BF01233206. [DOI] [PubMed] [Google Scholar]
- 27.Zabel F, Mohanan D, Bessa J, Link A, Fettelschoss A, Saudan P, Kündig TM, Bachmann MF. Viral Particles Drive Rapid Differentiation of Memory B Cells into Secondary Plasma Cells Producing Increased Levels of Antibodies. J. Immunol. 2014;192:5499–5508. doi: 10.4049/jimmunol.1400065. [DOI] [PubMed] [Google Scholar]
- 28.Genovese C, Milcarek C. Increased half-life of mu Ig mRNA during mouse B-cell development increases abundancy. Mol Immunol. 1990;17:69–81. doi: 10.1016/0161-5890(90)90082-b. [DOI] [PubMed] [Google Scholar]
- 29.Shen Y, Hendershot LM. Identification of ERdj3 and OBF-1/BOB-1/OCA-B as Direct Targets of XBP-1 during Plasma Cell Differentiation. J. Immunol. 2007;179:2969–2978. doi: 10.4049/jimmunol.179.5.2969. [DOI] [PubMed] [Google Scholar]
- 30.Taubenheim N, Tarlinton DM, Crawford S, Corcoran LM, Hodgkin PD, Nutt SL. High Rate of Antibody Secretion Is not Integral to Plasma Cell Differentiation as Revealed by XBP-1 Deficiency. J. Immunol. 2012;189:3328–3338. doi: 10.4049/jimmunol.1201042. [DOI] [PubMed] [Google Scholar]
- 31.Jackman M, Firth M, Pines J. Human cyclins B1 and B2 are localized to strikingly different structures: B1 to microtubules, B2 primarily to the Golgi apparatus. EMBO J. 1995;14:1646–1654. doi: 10.1002/j.1460-2075.1995.tb07153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 33.Gass JN, Gifford NM, Brewer JW. Activation of an Unfolded Protein Response during Differentiation of Antibody-secreting B Cells. J. Biol. Chem. 2002;277:49047–49054. doi: 10.1074/jbc.M205011200. [DOI] [PubMed] [Google Scholar]
- 34.Tirasophon W, Lee K, Callaghan B, Welihinda A, Kaufman RJ. The endoribonuclease activity of mammalian IRE1 autoregulates its mRNA and is required for the unfolded protein response. Genes Dev. 2000;14:2725–2736. doi: 10.1101/gad.839400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee K, Tirasophon W, Shen X, Michalak M, Prywes R, Okada T, Yoshida H, Mori K, Kaufman RJ. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002;16:452–466. doi: 10.1101/gad.964702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davenport EL, Moore HE, Dunlop AS, Sharp SY, Workman P, Morgan GJ, Davies FE. Heat shock protein inhibition is associated with activation of the unfolded protein response pathway in myeloma plasma cells. Blood. 2007;110:2641–2649. doi: 10.1182/blood-2006-11-053728. [DOI] [PubMed] [Google Scholar]
- 37.Pincus D, Chevalier MW, Aragon T, van Anken E, Vidal SE, El-Samad H, Walter P. BiP binding to the ER-stress sensor Ire1 tunes the homeostatic behavior of the unfolded protein response. PLoS Biol. 2010;8:e1000415. doi: 10.1371/journal.pbio.1000415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shilatifard A. Factors regulating the transcriptional elongation activity of RNA polymerase II. FASEB J. 1998;12:1437–1446. doi: 10.1096/fasebj.12.14.1437. [DOI] [PubMed] [Google Scholar]
- 39.Yoshida H, Okada T, Haze K, Yanagi H, Yura T, Negishi M, Mori K. ATF6 Activated by Proteolysis Binds in the Presence of NF-Y (CBF) Directly to the cis-Acting Element Responsible for the Mammalian Unfolded Protein Response. Mol. Cell. Biol. 2000;20:6755–6767. doi: 10.1128/mcb.20.18.6755-6767.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takayanagi S, Fukuda R, Takeuchi Y, Tsukada S, Yoshida K. Gene regulatory network of unfolded protein response genes in endoplasmic reticulum stress. Cell Stress and Chaperones. 2013;18:11–23. doi: 10.1007/s12192-012-0351-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Filippov V, Schmidt EL, Filippova M, Duerksen-Hughes PJ. Splicing and splice factor SRp55 participate in the response to DNA damage by changing isoform ratios of target genes. Gene. 2008;420:34–41. doi: 10.1016/j.gene.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Anken E, Romijn EP, Maggioni C, Mezghrani A, Sitia R, Braakman I, Heck AJR. Sequential Waves of Functionally Related Proteins Are Expressed When B Cells Prepare for Antibody Secretion. Immunity. 2003;18:243–253. doi: 10.1016/s1074-7613(03)00024-4. [DOI] [PubMed] [Google Scholar]
- 43.Kumazaki K, Tirosh B, Maehr R, Boes M, Honjo T, Ploegh HL. AID−/−μs−/− Mice Are Agammaglobulinemic and Fail to Maintain B220−CD138+ Plasma Cells. J. Immunol. 2007;178:2192–2203. doi: 10.4049/jimmunol.178.4.2192. [DOI] [PubMed] [Google Scholar]
- 44.Ma Y, Shimizu Y, Mann MJ, Jin Y, Hendershot LM. Plasma cell differentiation initiates a limited ER stress response by specifically suppressing the PERK-dependent branch of the unfolded protein response. Cell Stress and Chaperones. 2010;15:281–293. doi: 10.1007/s12192-009-0142-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gass JN, Jiang H-Y, Wek RC, Brewer JW. The unfolded protein response of B-lymphocytes: PERK-independent development of antibody-secreting cells. Mol. Immunol. 2008;45:1035–1043. doi: 10.1016/j.molimm.2007.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reimold A, Iwakoshi N, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese E, Friend D, Grusby M, Alt F, Glimcher L. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- 47.Tirosh B, Iwakoshi NN, Glimcher LH, Ploegh HL. XBP-1 specifically promotes IgM synthesis and secretion, but is dispensable for degradation of glycoproteins in primary B cells. J. Exp. Med. 2005;202:505–516. doi: 10.1084/jem.20050575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aragon IV, Barrington RA, Jackowski S, Mori K, Brewer JW. The specialized unfolded protein response of B lymphocytes: ATF6α-independent development of antibody-secreting B cells. Mol. Immunol. 2012;51:347–355. doi: 10.1016/j.molimm.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Benhamron S, Hadar R, Iwawaky T, So J-S, Lee A-H, Tirosh B. Regulated IRE-1 dependent decay participates in curtailing immunoglobulin secretion from plasma cells. Eur. J. Immunol. 2014;44:867–876. doi: 10.1002/eji.201343953. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.