Abstract
Mucosal surface epithelial cells are equipped with several defense mechanisms that guard against pathogens. Recent studies indicate that microRNAs (miRNAs) mediate post-transcriptional gene suppression and may be a critical component of the complex regulatory networks in epithelial immune responses. Transcription of miRNA genes in epithelial cells can be elaborately controlled through pathogen recognition receptors, such as Toll-like receptors (TLRs), and associated nuclear factor kappaB (NF-κB) and mitogen-activated protein kinase (MAPK) pathways, and ultimately nuclear transcription factor associated-transactivation and transrepression. Activation of these intracellular signaling pathways may also modulate the process of miRNA maturation. Functionally, miRNAs may modulate epithelial immune responses at every step of the innate immune network, including production and release of cytokines/chemokines, expression of adhesion and costimulatory molecules, shuttling of miRNAs through release of exosomes and feedback regulation of immune homeostasis. Therefore, miRNAs act as critical regulators to the fine-tuning of epithelial immune responses.
Keywords: epithelial cells, immune responses, miRNAs, posttranscriptional regulation, TLRs
INTRODUCTION
Epithelial cells along mucosal surfaces form a physical barrier that separates the host’s internal milieu from the external environment.1 These cells are also equipped with several defense mechanisms to guard against infection by pathogens. Recent studies indicate that epithelial cells express a variety of pathogen pattern recognition receptors (PRRs), such as the Toll-like receptors (TLRs) and nucleotide binding and oligomerization domain-like receptors (NLRs), which recognize pathogens or pathogen-associated molecular patterns. TLRs recognize microbes on the cell surface and in endosomes, whereas NLRs sense microbial molecules in the cytosol. Upon specific microbial recognition, these receptors recruit adaptor proteins and activate downstream signaling cascades that regulate the activity of nuclear factor kappaB (NF-κB), mitogen-activated protein kinases (MAPK), or caspase-dependent signaling pathways.2 This activation induces the expression of several adhesion molecules, inflammatory mediators (for example, cytokines/chemokines) and antimicrobial peptides, initiating innate epithelial immune responses against microbial infection.2 However, the immune response is a double-edged sword, as excessive inflammation can exacerbate tissue damage and cause chronic inflammatory diseases.3 Hence, the innate immune system has developed complicated self-regulatory systems so that this ‘sword’ will not damage the host. Various mechanisms have evolved for this purpose, for example, the release of extracellular soluble decoy TLRs and activation of intracellular antagonists to downregulate TLR signaling.3,4
Among numerous regulatory molecules, microRNAs (miRNAs) have received much attention as a newly identified family of regulators in animal and plant cells. miRNAs comprise a large family of about 21-nucleotide-long RNAs that have emerged as key post-transcriptional regulators of gene expression.5,6 In mammals, miRNAs are predicted to control the expression of ~50% of protein-coding genes.7 Accumulating data indicate that miRNAs are an essential part of the complex regulatory networks that control various cellular processes, including differentiation and fate of epithelial and immune cells.8 This review briefly summarizes the current understanding of miRNA regulation of epithelial immunity, with a focus on TLR-associated epithelial immune responses.
REGULATION OF MIRNA BIOGENESIS BY DOWNSTREAM SIGNALING PATHWAYS OF PRRS
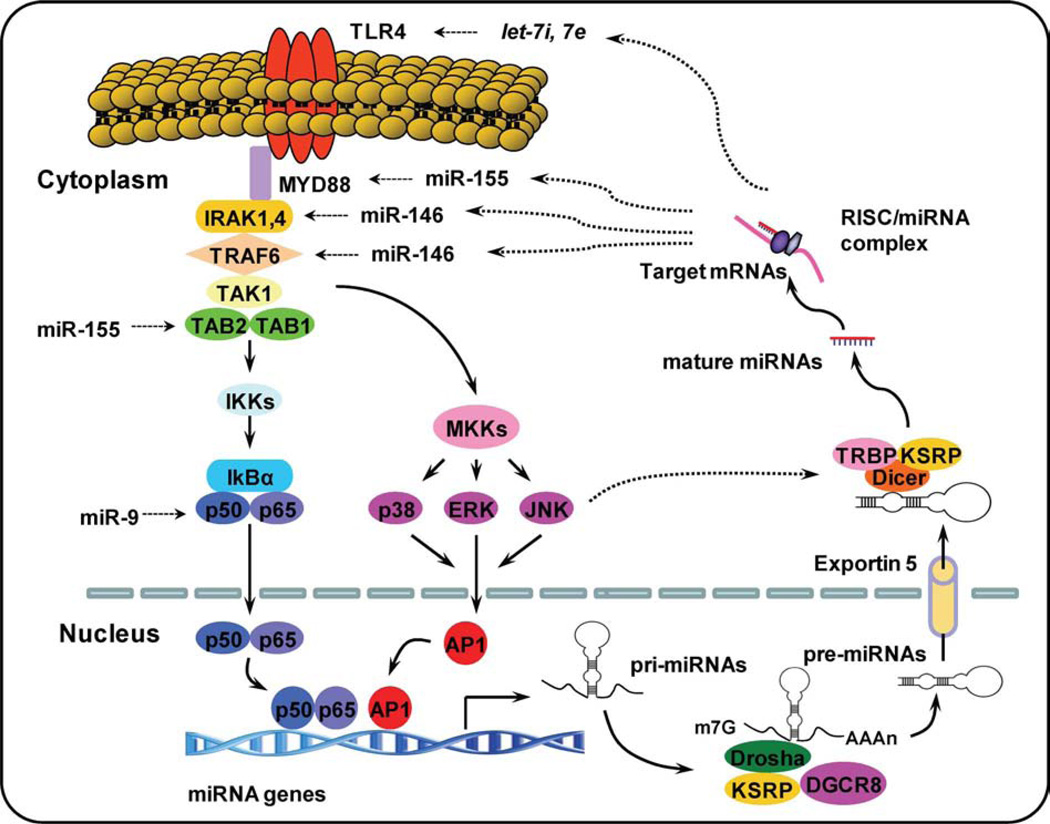
miRNAs are initially transcribed as primary transcripts known as pri-miRNAs by RNA polymerase II (RNA pol II) and cropped into about 70- to 100-nucleotide-long hairpin precursors (termed pre-miRNAs) in the nucleus by the RNAse III, Drosha.9 Pre-miRNAs are actively transported by exportin-5 to the cytoplasm where they are cleaved by the enzyme, Dicer, to form mature miRNAs. This cleavage event gives rise to a double-stranded ~22-nt product comprised of the mature miRNA guide strand and the miRNA* passenger strand. The mature miRNA is then loaded into the RNA-induced silencing complex, while the passenger strand is degraded. The RNA-induced silencing complex identifies target mRNA by base-pair complementarity resulting in mRNA cleavage and/or translational suppression10 (Figure 1). Whereas current research on miRNAs has focused on their physiological and pathological functions, the specific mechanisms that regulate miRNA expression remain largely unknown. So far, a total of 1048 mature miRNAs have been identified in humans.11 miRNA expression may be regulated in a similar manner to that of protein-coding genes through both transcriptional and post-transcriptional mechanisms.7,12 Activation of downstream signaling cascades of PRRs can regulate miRNA expression through both mechanisms.
Figure 1.
Intricate network of TLR signaling and miRNAs. Upon specific microbial recognition, TLR recruit adaptor proteins and activate downstream signaling cascades that activate NF-κB signaling pathway and MAPK signaling pathway. This activation induces the expression of inflammatory mediators and miRNA genes. After stimulation, pri-miRNAs are transcribed by RNA polymerase II and cropped into pre-miRNAs. Pre-miRNAs are actively transported to the cytoplasm byexportin-5 where they are cleaved by the enzyme, Dicer, to form mature miRNAs. miRNAs have been shown to regulate multistep of TLR signaling indicated by dotted line. AP-1, activator protein-1; IKK, IkappaB kinase; IRAK, IL-1 receptor-associated kinase 1; KSRP, KH-type splicing regulatory protein; miRNAs, microRNAs; MKK, mitogen-activated protein kinase (MAPK) kinase; MyD88, myeloid differentiation primary response gene 88; RISC, RNA-induced silencing complex; TAB1, transforming growth factor activated kinase-1-binding protein 1; TAK1, transforming growth factor activated kinase-1; TLR, Toll-like receptor; TRAF, TNF receptor-associated factor; TRBP, TAR RNA-binding protein.
Transcriptional regulation of miRNA genes by PRR-associated signaling pathways
Approximately 50% of the human miRNA genes are expressed from non-protein-coding transcripts. The majority of miRNA genes are located in intergenic regions or in antisense orientation to annotated genes, indicating that they form independent transcription units.13 Lee et al., by demonstrating promoter regulated expression of the polycistronic miRNA gene, mir-23a-27a-24-2, provided the first direct evidence that miRNAs can exist as independent transcriptional units.9 Other miRNAs are located within introns of annotated genes, which may be transcribed as part of their ‘host genes’.14 For example, human mir-22 gene is located in the second exon of the non-coding transcript MGC14376. Correlated expression of miR-22 and MGC14376 had been demonstrated in various cells and tissues.15 Nevertheless, an intronic miRNA does not necessarily cotranscribe with its host gene. Nucleosome mapping studies indicate that one-third of intronic miRNAs have transcription initiation regions independent from their host gene promoters.16 A significant fraction of primary transcripts of intergenic miRNAs are 3–4 kb in length, with clearly defined 5′ and 3′ boundaries.13,17 Whereas some pri-miRNAs contain only one miRNA, other pri-miRNA transcripts contain several miRNAs. These cotranscribed miRNAs are referred to as ‘cluster’ miRNA genes. Indeed, the human miR-17 cluster contains six miRNAs, while the human miR-302 cluster includes eight miRNAs.18,19 Recent studies indicate that transcription of miRNA genes in epithelial cells can be elaborately controlled through various regulatory mechanisms including transactivation and transrepression by nuclear transcription factors associated with the downstream signaling pathways of TLR/NLRs, in particular, the NF-κB and MAPK pathways.
The NF-κB pathway
Activation of the NF-κB signal pathway through TLRs/NLRs is a common response in many epithelial cells following microbial infection. The NF-κB family of transcription factors consists of five members, p50, p52, p65 (RelA), c-Rel and RelB. In most cells, NF-κB exists in a latent state in the cytoplasm bound to inhibitory κBs that mask its nuclear localization signal. Activation of NF-κB causes it to move into the nucleus and regulate the expression of a number of host genes, including miRNAs.20 The transcription activation domain necessary for the positive regulation of gene expression is present only in p65, c-Rel and RelB.20–22 Thus, promoter binding of p65, c-Rel and RelB is usually associated with gene transactivation. Because they lack transcription activation domains, p50 and p52 may repress transcription unless they are associated with a transcription activation domain-containing NF-κB family member or another protein capable of coactivator recruitment.23,24
It was first described in 2007 in human THP-1 monocytes that transcription of miR-146a gene in response to TLR signaling is activated in an NF-κB-dependent manner.25 Since then, a subset of miRNA genes has been identified as NF-κB-dependent (Table 1). Many of the studies were carried out in non-epithelial cells or in malignant cells. For example, lipopolysaccharide (LPS) induces miR-27b expression in a NF-κB-dependent manner in human macrophages.26 Additionally, LPS directly induces miR-9 expression via the myeloid differentiation primary response gene 88 (MyD88)/NF-κB-dependent pathway in human neutrophils, monocytes and macrophages.27 miR-155, an important miRNA related to inflammation, has been shown to be activated by NF-κB signaling pathway in various cell types in response to many stimuli, including LPS and LMP1 (the viral latent protein of Epstein–Barr virus).28,29 Similar to miR-155, miR-146a expression can be induced through NF-κB-dependent mechanisms in response to various immune-mediators such as LPS, IL-1β, LMP1 and tumor necrosis factor (TNF)-α.25,30,31 Furthermore, miR-16 and miR-21 are directly regulated by NF-κB in gastric cancer cells32 and NF-κB activation promotes miR-301a expression in pancreatic cancer cells.33
Table 1.
NF-κB-dependent miRNAs
| miRNA | Stimulus | Alteration | Transcription factors/ other cofactors |
Cell lines | Reference |
|---|---|---|---|---|---|
| miR-146a | LPS; TNF-α; IL-1β; LMP1; H2O2 |
Up | P65 | Human monocytic cell; human neural cells; human alveolar epithelial A cells; human monocyte-derived dendritic cells; human PMN and monocytes |
25, 27, 30, 31, 92 |
| miR-21 | LPS; C. parvum; nicotine |
Up | P65; P50 | Human biliary epithelial cells; gastric cancer cells; human peripheral blood mononuclear cells |
32, 34, 35, 85 |
| miR-23b | LPS; C. parvum | Up | P65 | Human biliary epithelial cells | 34, 35 |
| miR-27b | LPS; C. parvum | Up | P65 | Human biliary epithelial cells; human macrophages | 26, 34, 35 |
| miR-125ba | LPS; C. parvum | Up | P65 | Human biliary epithelial cells; human fibroblast-like synoviocytes; LPS-tolerized THP1 cells |
34, 35, 93, 94 |
| Down | ND | Mouse Raw 264.7 macrophage; mouse primary macrophages; human monocyte-derived dendritic cells |
58, 83, 92 | ||
| miR-30b | LPS; C. parvum | Up | P65 | Human biliary epithelial cells | 34, 35 |
| miR-16 | Nicotine | Up | P65; P50 | Gastric cancer cells | 32 |
| miR-301a | TNF-α | Up | P65; P50 | Pancreatic cancer cells | 33 |
| miR-155 | LMP1; LPS; alcohol | Up | P65; P50 | Human B cells; mouse macrophage cells; mouse Kupffer cells; human PMN and monocytes |
27–29 |
| miR-9 | LPS; TNF-α; IL-1β | Up | ND | Human PMN and monocytes, macrophages | 27 |
| miR-29b | TNF-α | Down | P65; YY1; SP1; HDAC3; HDAC1 |
Human myoblast cell; human acute myeloid leukemia cells | 37, 38 |
| let-7i | LPS; C. parvum | Down | P50; C/EBP-β; HDAC3 | Human biliary epithelial cells | 36 |
Abbreviations: C/EBP-β, CCAAT/enhancer-binding protein beta; C. parvum, Cryptosporidium parvum; HDAC1, histone deacetylase 1;HDAC3, histone deacetylase3;LMP1, Epstein–Barr virus-encoded latent membrane protein-1; LPS, lipopolysaccharide; ND, not determined; PMN, polymorphonuclear neutrophil; SP1, specificity protein 1; TNF-α, tumor necrosis factor-α; YY1, Yin Yang 1.
The expression of miR-125 after TLR activation is still controversial now.
We recently showed that transcription of a subset of miRNA genes is regulated through NF-κB activation in human biliary epithelial cells in response to LPS stimulation.34 Infection of biliary epithelial cells by Cryptosporidium parvum, a protozoan parasite that activates TLR4/ NF-κB signaling pathway in infected cells, also displayed a similar transcription profile of these miRNA genes.35 Specifically, inhibition of NF-κB activation by SC-514, an IkappaB kinase 2 (IKK2) inhibitor, blocked LPS- and C. parvum-induced upregulation of a subset of pri-miRNAs, including pri-miR-125b-1, pri-miR-21, pri-miR-23b-27b-24-1 and pri-miR-30b. Moreover, direct binding of NF-κB p65 subunit to the promoter elements of mir-125b-1, mir-21, mir-23b-27b-24-1 and mir-30b genes was identified by chromatin immunoprecipitation analysis and confirmed by the luciferase reporter assay using the constructs covering the potential promoter elements of these miRNA genes.34,35
In addition to the upregulation of some miRNAs, NF-κB signaling may be also involved in the downregulation of miRNA genes. Transcription of the let-7i gene in biliary epithelial cells in response to LPS stimulation and C. parvum infection has been reported to be suppressed through promoter binding by NF-κB subunit p50 along with CCAAT/enhancer-binding protein beta.36 Furthermore, NF-κB negatively regulates miR-29b transcription through interacting with other transcription factor Yin Yang 1 or specificity protein 1 in various cell lines.37,38
The MAPK pathways
Similar to regulation of protein-coding RNA genes, transcription of miRNA genes appears to be regulated by multiple signaling pathways including the MAPK signaling pathways. In mammals, three major MAPK pathways (MAPK/ERK, MAPK/JNK and MAPK/p38 pathways) are closely linked with inflammation. The activator protein 1 (AP-1) transcription factor is composed of heterodimers of the c-Fos and c-Jun families and can be activated through different MAPK pathways.39 The MAPK pathways regulate miRNA expression not only at the transcription level but also at the post-transcription level. Some AP-1-dependent miRNAs have already been confirmed as listed in Table 2.
Table 2.
AP-1-dependent miRNAs
| miRNA | Stimulus | Alteration | Transcription factors | Cell lines | Reference |
|---|---|---|---|---|---|
| miR-21 | PMA; ectopic expression of HER2/neu |
Up | c-Fos; c-Jun | Side population cells from various cancer cell lines; human breast cancer cell line; human promyelocytic leukemia cell line |
40, 41, 95 |
| miR-155 | B-cell receptor cross-linking | Up | FosB and JunB | Human lymphoma cell | 43 |
| miR-146b | The two PDGF ligands AA and BB | Up | c-fos | Human glioblastoma and ovarian cancer cells | 42 |
| miR-99a | The tyrosine kinase c-Src | Down | ND | Human c-Src-transformed cells | 44 |
Abbreviations: ND, not determined; PDGF, platelet-derived growth factor; PMA, phorbol 12-myristate 13-acetate.
In addition to NF-κB signal pathway, it was reported that miR-21, in human promyelocytic leukemia cells, is upregulated in response to phorbol 12-myristate 13-acetate stimulation through c-Fos and c-Jun binding to the miR-21 promoter.40 Furthermore, miR-21 expression has been shown to be AP-1-dependent in the stem-like side cell populations from various cancer cell lines.41 miR-146b is another identified AP-1-dependent miRNA. It was reported that platelet-derived growth factor regulates the expression of miR-146b via MAPK-dependent induction of c-Fos binding to the AP-1 element in glioblastoma and ovarian cancer cells.42 Activation of the MAPK signaling pathway also induces transcription of mir-155 gene in different cell types in response to various stimuli.28 Indeed, Yin et al. reported that induction of miR-155 expression by B-cell receptor signaling occurs through the extracellular MAPK/ERK and MAPK/JNK but not MAPK/p38 pathway. The binding of transcription factors Jun-B and Fos-B (and possibly also c-Fos) to miR-155 promoter element has been demonstrated upon B-cell receptor cross-linking.43 In addition to upregulation of miRNAs, MAPK pathway is also involved in the downregulation of miRNAs, for example, miR-99a.44
Post-transcriptional regulation of miRNA genes through intracellular signaling
Following transcription, the primary miRNAs undergo two cleavage steps to generate the mature miRNAs. The first cleavage is catalyzed in the nucleus by the RNase III enzyme, Drosha. This enzyme is part of a large multiprotein complex, which also includes DGCR8 (a double-stranded RNA-binding protein) and several associated proteins such as the DEAD-box helicases p68 (DDX5), p72 (DDX17) and numerous heterogenous nuclear RNA complex (hnRNP) protein. After cropping of the pri-miRNAs by Drosha, there is an additional processing by the type III ribonuclease Dicer in the cytoplasm, resulting in the production of mature miRNAs.12,45 Recent studies provide evidence that intracellular signaling pathways may modulate the process of miRNA maturation.
p68, p72 and hnRNPs
Because p68 and p72 are important components of the large Drosha processing complex, regulation of their expression or function by signaling pathways will modulate pri-miRNA processing.46 Activation of the transforming growth factor-β pathway promotes interactions between p68 and the SMAD proteins, signal transducers of the transforming growth factor-β family signaling cascade, facilitating processing of pre-miR-21. Ligand-specific SMAD proteins bind to the Drosha processing complex subunit p68 to facilitate pre-miR-21 accumulation.47 These results indicate that the association of p68/Drosha with accessory factors, such as SMADs, may be important for the maturation of miRNAs in response to extracellular stimuli. Although there is no direct experimental evidence yet, we speculate that activation of downstream signaling pathways of PRRs may modulate miRNA maturation through similar mechanisms.
The family of hnRNPs may serve as accessory factors in the regulated processing of a variety of miRNAs. Studies have shown that hnRNP A1 specifically binds to a miRNA cluster containing miR-18a and facilitates Drosha-mediated processing of miR-18a, but not the other members of the cluster.48 More recent studies showing that hnRNP A1 binds specifically to the conserved terminal loop of the let-7a precursor and blocks its Drosha-mediated processing in somatic cells.49 Moreover, it has been reported that MAPK/p38 pathway can phosphorylate hnRNP A1 and thus, promotes the cytoplasmic translocation of hnRNP A1 and associated miRNA maturation.50 Taken together, these results imply that MAPK signal pathway may be involved in the miRNA processing controlled by hnRNP A1.
The KH-type splicing regulatory protein (KSRP) and human immunodeficiency virus (HIV) TAR RNA-binding protein (TRBP)
Besides these accessory factors of the Drosha complex, other proteins may also be involved in pri-/pre-miRNA maturation process. KSRP is a multifunctional single-stranded RNA-binding protein which was recently demonstrated to be involved in the maturation of a set of miRNA precursors.51 KSRP directly interacts with G-rich regions present within the loop of a subset of miRNAs, promoting both Drosha-and Dicer-mediated miRNA processing. LPS stimulation increases the level of mature miR-155 in macrophages without significantly altering the expression of its primary transcript. Further experimentation indicated that KSRP interacts with pri-miR-155, and knockdown of KSRP prevents LPS-mediated increase of miR-155.52 It is well established that MAPK/p38 signal pathway phosphorylates KSRP.53 Therefore, downstream signaling pathways of PRRs may modulate the miRNA processing through KSRP association with Drosha or Dicer.
TRBP is an integral component of the Dicer-containing complex. The presence of TARBP2 frameshift mutations causes diminished TRBP protein expression and a defect in the processing of miRNAs, resulting in a global downregulation of mature miRNAs.54 Activation of the MAPK/Erk pathway promotes phosphorylation of TRBP. Expression of phospho-mimic TRBP and TRBP phosphorylation enhanced miRNA maturation by increasing stability of the miRNA-generating complex.55 This study provided the first evidence showing a direct connection between a cell signaling pathway and the core miRNA machinery. Results of this study also suggest that other cellular networks may target the miRNA pathway through interaction with TRBP to carry out functional cellular responses. Indeed, a recent report by Melo et al. indicated that the small molecule, enoxacin (a fluoroquinolone used as an antibacterial compound), enhances the production of miRNAs by binding to TRBP.56
REGULATION OF EPITHELIAL IMMUNE RESPONSES BY MIRNAS
Targeting of innate immune effector molecules by miRNAs
miRNAs are predicted to regulate the translation of 50% all human gene transcripts.7 The usual consequence of miRNA and mRNA interaction is the downregulation of protein expression by translational repression and/or mRNA cleavage.10 miRNA-regulated genes may include those innate immune response genes and those that have been experimentally validated are listed in Table 3. Some miRNAs relevant to TLR/NF-κB/MAPK-mediated immune responses are also illustrated in Figure 1.
Table 3.
Validated targets of miRNAs relevant to the innate immunity
| miRNAs | Targets | Innate immune function | Reference |
|---|---|---|---|
| miR-155 | SOCS1 | Positive regulation of host antiviral innate immune response by promoting type I IFN signaling | 77 |
| Regulation of endotoxin sensitivity and tolerance | 92 | ||
| TAB2 | Negative regulation of inflammatory cytokine production in response to microbial stimuli | 83 | |
| FADD, IKKε, Ripk1 | Increase translation of TNF-α | 58 | |
| IL-13Rα1 | Determining M2 phenotype in Macrophage | 96 | |
| BACH1, ZIC3 | Modulation of transcriptional regulatory factors | 97 | |
| C/EBP-β | Regulation of granulocyte CSF expression | 98 | |
| MyD88 | Negative regulation of Helicobacter pylori-induced inflammation | 84 | |
| miR-146b | TRAF6, IRAK1 | Negative regulation of Toll-like receptor and cytokine signaling | 25 |
| miR-146a | TRAF6, IRAK1 | Negative regulation of Toll-like receptor and cytokine signaling | 25 |
| IL-8, RANTES | Negative regulation of severe inflammation | 30 | |
| miR-218 | MAFG | Regulation of the overall epithelial inflammatory response to tobacco smoke exposure | 99 |
| miR-203 | SOCS3 | Regulation of inflammatory responses and keratinocyte functions | 100 |
| miR-192 | MIP-2α | Regulation of inflammatory responses in chronic inflammatory bowel diseases | 101 |
| miR-132 | p300 | A negative effect on the expression of interferon-stimulated genes | 81 |
| miR-26a/miR-145/ miR-34a/let-7b |
IFN-β | Negative regulation of innate immune response to viral infections | 60 |
| miR-16 | TNF-α; IL-8 | Enhancing cytokines/chemokines mRNA degradation | 58 |
| miR-17-3p | ICAM-1 | Negative regulation of neutrophil adhesion to endothelial cells | 64 |
| miR-31 | E-selectin | Negative regulation of neutrophil adhesion to endothelial cells | 64 |
| let-7i | TLR4 | Regulation of epithelial defense responses against C. parvum | 78 |
| let-7e | TLR4 | Regulation of endotoxin sensitivity and tolerance | 92 |
| miR-125b | TNF-α | Regulation of TNF-α translation | 58 |
| miR-98 | CIS | Modulation NF-κB activity | 86 |
| SOCS4 | An inhibitory effect on phosphorylation of signal transducers and activators of transcription proteins |
102 | |
| miR-101 | MKP-1 | Positive regulation of the activation of MAPKs signal pathway | 87 |
| miR-9 | NF-κB1 | A feedback control of the NF-κB-dependent responses | 27 |
| miR-21 | PDCD4 | Negative regulation of TLR4 signaling | 85 |
| miR-126 | TOM1 | Positive regulation of IL-1β and TNF-α-induced signaling pathways | 103 |
| VCAM-1 | Regulation of adhesion molecule expression | 65 | |
| miR-222/miR-339 | ICAM-1 | Regulation of adhesion molecule expression | 61 |
| miR-221 | ICAM-1 | Regulation of the interaction between epithelia cells and T cells | 63 |
| miR-513 | B7-H1 | Regulation of cholangiocyte inflammatory response | 66, 67 |
Abbreviations: BACH1, basic leucine zipper transcription factor 1; C/EBP-β, CCAAT/enhancer binding protein beta; CIS, cytokine-inducible Src homology 2; E-selectin, endothelial-leukocyte adhesion molecule-1; FADD, Fas-associated death domain protein; H. pylori, Helicobacter pylori; ICAM-1, intercellular cell adhesion molecule 1; IKKε, IkappaB kinase epsilon; IL-13Rα1, IL-13 receptor α1; IRAK1, IL-1 receptor-associated kinase 1 genes; MAFG, v-maf musculoaponeurotic fibrosarcoma oncogene homolog G; MIP-1α, macrophage inflammatory protein 1alpha; MKP-1, MAPK phosphatase-1; MyD88, myeloid differentiation primary response gene 88; NF-κB1, nuclear factor-κB1; PDCD4, programmed cell death 4; Ripk1, the receptor (TNFR superfamily)-interacting serine-threonine kinase 1; SOCS1, suppressor of cytokine signaling 1; SOCS3, suppressor of cytokine signaling 3; SOCS4, suppressors of cytokine signaling 4; TAB2, P3K7-binding protein 2; TLR4, Toll-like receptor 4; TRAF6, TNF receptor-associated factor 6; TOM1, target of myb1; VCAM-1, vascular cell adhesion molecule 1; ZIC3, Zic family member 3.
miRNAs as key regulators to epithelial immune responses
miRNAs may modulate epithelial immune responses at every step of the innate immune network, including production and release of cytokines/chemokines, expression of adhesion and costimulatory molecules, shuttling of miRNAs through release of exosomes and feedback regulation of immune homeostasis. Recent studies have also revealed a few principles relevant to miRNA-mediated regulation in epithelial immune responses, which will be integrated into the detailed discussions below. Briefly, if a miRNA strongly inhibits translation of a target at physiological conditions, downregulation of this miRNA may be required for upregulation of this target at the protein level in epithelial cells following immune stimuli. Some TLR/NF-κB-responsive miRNAs are abundantly expressed in epithelial cells; and downregulation of these miRNAs is required for an efficient translation of their targets upon activation of the TLR/NF-κB pathway. Moreover, each miRNA may have multiple targets and several miRNAs may target the same mRNA molecule. Therefore, miRNAs can modulate the coordinated expression of immune response genes in epithelial cells in response to immune stimuli. Finally, miRNAs may provide feedback regulation to NF-κB signaling to maintain epithelial homeostasis. Therefore, miRNAs act as critical regulators to the fine tuning of epithelial immune responses.
Regulation of epithelial expression of cytokines and chemokines
Based on bioinformatics analysis, approximately 29% of cytokine/chemokine mRNAs have potential target sites for miRNAs.57 Tili and colleagues first reported that miR-125b targets the 3’-untranslated region (3′ UTR) of the TNF-α transcript and suppresses TNF-α transcription in mouse Raw 264.7 macrophages.58 Recent studies have also demonstrated that miR-16 is a critical regulator of inflammatory responses. Expression profiling has shown that miR-16 is abundantly expressed in most cell types including epithelial cells. Intriguingly, miR-16 induces rapid degradation of RNAs which contain AU-rich elements (AREs) in their 3′ UTRs. The majority of cytokine and chemokine mRNAs contain AREs within their 3′ UTRs, such as TNF-α, IL-8 and IL-6. miR-16-mediated degradation of mRNAs for cytokines/chemokines requires the miRNA processing components, Dicer, Ago/eiF2C family members and the ARE-binding protein, tristetraprolin, and involves the interactions between miR-16 UAAAUAUU sequence and AREs within 3′ UTRs of targeted RNAs. It has been speculated that the high level of miR-16 in inflammatory cells might restrict the production of inflammatory mediators under non-stimulated conditions.59 Expression of interferon (IFN)-β, the main type I IFN cytokine important to the initiation of innate responses in response to virus infection, is also finely controlled by various miRNAs. It has been experimentally confirmed that miR-26a, −34a, −145 and let-7b directly regulate IFN-β production by targeting IFN-β 3′ UTR. Importantly, HIV and simian immunodeficiency virus infection decreases the expression of these miRNAs in macrophages.60 Because the above miRNAs are transcribed from different genes, it is plausible that multiple miRNAs from different gene loci may target the same mRNA, providing coordinated regulation in host cells in response to microbial challenge.
Regulation of epithelial expression of adhesion and costimulatory molecules
miRNAs have recently been implicated in regulating the expression of adhesion and costimulatory molecules critical to epithelial cell-immune cell interactions. Vascular cell adhesion molecule 1, endothelial-leukocyte adhesion molecule (E-selectin) and intercellular adhesion molecule 1 (ICAM-1) are common inducible adhesion molecules in epithelial cells which play important roles in regulating leukocyte trafficking during inflammation. miRNA targeting of ICAM-1 was first reported in Dicer-disrupted cells by Ueda et al. They reported that ICAM-1 expression was unregulated in the isogenic Dicer (ex5−/−) cells (cells that lack mature miRNAs). miR-222 and miR-339 target ICAM-1 and regulate its expression at post-transcriptional level.61 Recently, we confirmed that miR-221 can target ICAM-1 3′ UTR in human biliary epithelial cells. It appears that downregulation of miR-221 is required for the induction of ICAM-1 in biliary epithelial cells in response to IFN-γ stimulation or C. parvum infection. Given that miR-221 is the second most abundantly expressed miRNA in human biliary epithelial cells,62 miR-221 may assure the low basal expression of ICAM-1 protein in cells under normal physiological conditions. The relief of miR-221-mediated translational suppression may be necessary for the stimulated expression of ICAM-1 during inflammation.63 It was recently also reported that miR-31 and miR-17-3p target endothelial adhesion molecules, E-selectin and ICAM-1, respectively. Interestingly, TNF-α induces expression of miR-31 and miR-17-3p in endothelial cells and therefore, it has been speculated that miR-31 and miR-17-3p comprise a negative feedback loop controlling TNF-α-associated inflammatory responses in endothelial cells.64 miR-126 has recently been demonstrated to suppress vascular cell adhesion molecule 1 expression and induction of miR-126 decreases leukocyte interactions with endothelial cells.65
B7-H1 is a key member of the B7 family of costimulatory molecules important in regulation of immune response, in particular, T-cell homeostasis. Expression of B7-H1 by epithelial cells in response to microbial infection is tightly regulated to ensure an appropriate antimicrobial immune response. Although expression of B7-H1 mRNA is common in many cells, B7-H1 protein is usually undetectable, suggesting post-transcriptional suppression. We recently showed that B7-H1 is a target of miR-513 and miR-513 targeting may account for the absence of B7-H1 protein in cells under non-stimulation conditions. Moreover, downregulation of miR-513 is required for upregulation of B7-H1 protein level in human biliary epithelial cells following IFN-γ stimulation or microbial challenge, suggesting a role of miR-513 in regulating biliary inflammatory responses through targeting of B7-H1.66,67
Shuttling of miRNAs through epithelial cell-derived exosomes
Exosomes are small (30–90 nm), extracellular vesicles derived from the multivesicular body sorting pathway and are produced by a variety of cells including epithelial cells. Cellular gene products, including proteins, mRNAs and miRNAs, are packaged in exosomes and these molecules can be transferred, by exosome secretion, to recipient cells.68–71 Although the function of exosomes remains largely unknown, some studies suggest that exosomes may be involved in a broad range of biological processes, including stimulation of the immune system, modulation of selected cellular activities, intercellular communication, virus egression and immune evasion, and bacterial and viral sequestration.68–71 Valadi and colleagues recently reported that vesicles released from mast cell lines contain approximately 121 miRNA molecules. Intriguingly, exosome-shuttled miRNA molecules can be delivered to another cell type through uptake of these exosomes.68 Therefore, exosome-mediated transport of miRNAs may provide a novel mechanism of gene regulation between cells. Ohshima et al. also showed that there is the enrichment of let-7 miRNA family in the exosomes from AZ-P7a cells.71 Given the importance of miRNAs in epithelial innate immune responses, it would be interesting to determine if exosomes from epithelial cells also carry miRNAs and thus modulate epithelial–immune cell interactions and epithelial antimicrobial defense, via exosomal delivery of miRNAs.
Regulation of epithelial antimicrobial defense
Cellular miRNA expression is profoundly influenced by microbial infection, which can be attributed to both host antimicrobial defenses and altering the cellular environment to favor microbial replication. It was reported that Dicer knockout mice are more susceptible to vesicular stomatitis virus infection.72 miRNAs can directly target the microbial genome to attenuate microbial replication. Host miR-24 and miR-93 have been reported to target viral large protein of vesicular stomatitis virus.72 In hepatocytes (a type of epithelial cell in the liver), miR-196, miR-296, miR-351, miR-431 and miR-448 directly influence hepatitis C virus genomes to downregulate viral accumulation.73 Ahluwalia and colleagues identified that miR-29a can specifically target the 3′ UTR region of HIV-1 RNAs.74 Several other miRNAs, including miR-28, miR-125b, miR-150, miR-223 and miR-382, are also capable of inhibiting HIV-1 replication via binding to sequences located within the viral genome.75 Host miR-32 was reported to limit the retrovirus primate foamy virus type 1 replication in 293T cells.76 Besides influencing the replication of viruses, miRNAs also can alter cellular proteins to increase host antimicrobial innate immune response. Recently, Wang et al. identified that RNA virus infection induces miR-155 expression in macrophages. Upregulation of miR-155 provides positive feedback regulation to host antiviral innate immune response by promoting type I IFN signaling via targeting suppressor of cytokine signaling 1 (SOCS1).77
Our recent studies indicate that activation of NF-κB signaling in epithelial cells regulates transcription of miRNA genes to orchestrate host anti-C. parvum immune responses through modulation of miRNA-mediated post-transcriptional suppression. Distinct alterations in the miRNA expression profile were detected in epithelial cells following C. parvum infection.35 Activation of NF-κB signaling regulates transcription of a subset of miRNA genes in infected cells. Functional manipulation of several NF-κB-dependent miRNAs (e.g., miR-27b and let-7i) in epithelial cells influences C. parvum infection burden in vitro, raising the possibility that these miRNAs may directly regulate production of antimicrobial molecules important to epithelial anti-microbial defense.34,78 However, the molecular mechanisms by which C. parvum-responsive miRNAs modulate epithelial anti-C. parvum defense are largely unclear. Various immune-related genes are identified as potential targets for these C. parvum-responsive miRNAs using computational analyses. Nevertheless, no complementarity to IFN-γ or anti-microbial peptide mRNA has been identified for these miRNAs.
Interestingly, miRNA-mediated posttranscriptional suppression appears to be hijacked by some virus to create a more favorable intracellular environment for microbial replication. It was reported that miR-122, a liver-specific miRNA, binds to hepatitis C virus genomes and positively regulates hepatitis C virus RNA accumulation in hepatocytes.79 Additionally, Epstein–Barr virus infection induces miR-146a expression, resulting in suppression of the IFN-mediated antiviral function,33 while it is proposed that miR-155 contributes to Epstein–Barr virus immortalization through modulation of NF-κB signaling.80 Furthermore, it was recently reported that CREB-induced miR-132 is highly upregulated after herpes simplex virus and human cytomegalovirus infection, resulting in a negative effect on the expression of IFN-stimulated genes and facilitating viral replication.81 Decreased miRNA expression has also been implicated in efficient HIV-1 replication. Indeed, HIV-1 infection suppressed miR-17/92 expression. Decreased expression of miR-17/92 cluster resulted in increased histone acetyltransferase Tat cofactor (p300/CBP-associated factor) expression and hence viral replication.82
Feedback regulation of epithelial immune responses
Inflammation, while an essential physiological response to insult or injury, is potentially injurious to host tissues and is therefore a highly regulated process. A variety of extracellular and intracellular feedback pathways have evolved to prevent an inappropriate inflammatory response.3,4 Accumulating data suggest that miRNAs are also essential components in the feedback regulation of epithelial immune responses. Some miRNAs may act as negative regulators, while other miRNAs may provide positive feedback regulation.4,8
The miR-146 family is composed of two members, miR-146a and miR-146b. Evidence showing that miR-146a and miR-146b might be involved in the feedback regulation of innate immune response was first provided by Taganov et al. Targets of miR-146 include IL-1 receptor-associated kinase 1 (IRAK1) and TNF receptor-associated factor 6 (TRAF6).25 IRAK1 and TRAF6 are known to be part of TLR/NF-κB signaling pathway. Therefore, upregulation of miR-146 following LPS stimulation could provide a negative feedback regulation to inhibit TLR/NF-κB signaling in macrophages and monocytes.25 Increased miR-146a was also confirmed in alveolar epithelial cells and shown to negatively regulate IL-1β-induced IL-8 and RANTES.30 Significantly, this negative feedback was only seen at high IL-1β concentrations, which indicated that this might be an important mechanism during severe inflammation.30
miR-155 is another miRNA which plays an important role in the feedback regulation of innate immune response. Studies showed that miR-155 exerts both positive and negative feedback regulations of the immune response depending on different cell types. Tili et al. showed that miR-155 enhances TNF-α translation by targeting a series of proteins of the NF-κB signaling components, such as Fas-associated death domain protein, IKKε, the receptor (TNFR superfamily)-interacting serine-threonine kinase 1.58 Moreover, overexpression miR-155 in B cells increases the level of serum TNF-α and enhances cellular susceptibility to septic shock.58 On the other hand, a recent report by Ceppi et al. demonstrated that miR-155 is a part of a negative feedback loop that downmodulates inflammatory cytokine production in mature human Dendritic cells in response to microbial stimuli.83 Their data showed that miR-155 directly controls the level of MAP3K7 binding protein 2 (TAB2), an important signal transduction molecule, and thus provides negative feedback regulation to inhibit TAB2-associated gene transcription. More recently, Tang et al. identified that MyD88 is a novel target of miR-155 and suppression of MyD88 through induced expression of miR-155 attenuates Helicobacter pylori-induced inflammation.84
miR-21 may also act as a negative regulator of TLR4 signaling through targeting of PDCD4. It was reported that LPS decreases expression of PDCD4 through induction of miR-21, resulting in subsequent inhibition of NF-κB signaling activity and promotion of IL-10 production in human peripheral blood mononuclear cells.85 Similarly, targeting of PDCD4 by miR-21 was shown to influence tumor necrosis factor-induced activation of NF-κB. Similarly, miR-9 targets NFKB1, a transcriptional regulator with a key role in the TLR/NF-κB signaling pathway, and consequently, forms an inhibitory regulatory circuitry controlling cell inflammatory responses.27
Other miRNAs may exert positive feedback regulation to innate immune response. We recently demonstrated that miR-98 and let-7 target the cytokine-inducible Src homology 2-containing protein (CIS), one member of the suppressors of cytokine signaling family of proteins that acts as an important negative regulator of inflammatory cytokine signaling. LPS stimulation and C. parvum infection induces CIS expression in human biliary epithelial cells through TLR/NF-κB-suppressed expression of miR-98 and let-7. Induction of CIS expression enhances IκBα degradation promoting NF-κB activation.86 In addition, TLR-dependent induction of miR-101 appears to provide a positive feedback loop to facilitate TLR-mediated immune responses through miR-101-mediated suppression of MAPK phosphatase-1, an inhibitory regulator to TLR signaling.87
CONCLUSION AND PERSPECTIVES
The miRNA–target mRNA interactions are very complex. It has been proposed that a single miRNA can repress hundreds of target transcripts and multiple miRNAs may target the same transcript. Such redundant functions of miRNAs add additional complexity to the regulatory networks with multiple pathways and feedback control of epithelial immune responses. New technologies will help to identify miRNA targeting globally, such as cross-linking argonaute/RNA immunoprecipitation, proteomic approaches and high-throughput sequencing assays.88, 89 The development of miRNA knockouts has greatly advanced our understanding of miRNA-mediated immune responses in vivo. Meanwhile, new in vivo delivery methods are being introduced to assess miRNA targeting and miRNA function, such as AAV8-mediated miRNA delivery.90,91 In addition, identification of miRNAs of major pathogenic significance in persistent inflammatory reactions of the skin and at mucosal sites could provide rationale for the design and implementation of new immunotherapeutic strategies for treatment of these diseases. Unraveling the regulatory functions of miRNAs in epithelial biology is still in its infancy, but will likely yield new insights into our understanding of epithelial immunobiology and immunopathology.
ACKNOWLEDGEMENTS
This work was supported by National Institute of health (NIH) grant AI071321, AI095532 and by the Nebraska Tobacco Settlement Biomedical Research Program (LB692 and LB595) (to XM Chen) and NIH grant AI089713 (to SP O’Hara).
References
- 1.Viswanathan VK, Hecht G. Innate immunity and the gut. Curr Opin Gastroenterol. 2000;16:546–551. doi: 10.1097/00001574-200011000-00015. [DOI] [PubMed] [Google Scholar]
- 2.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 3.Kobayashi KS, Flavell RA. Shielding the double-edged sword: negative regulation of the innate immune system. J Leukoc Biol. 2004;75:428–433. doi: 10.1189/jlb.0703321. [DOI] [PubMed] [Google Scholar]
- 4.Chen XM, O’Hara SP, LaRusso NF. The immunobiology of cholangiocytes. Immunol Cell Biol. 2008;86:497–505. doi: 10.1038/icb.2008.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 7.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 8.Liu J, Drescher KM, Chen XM. MicroRNAs and epithelial Immunity. Int Rev Immunol. 2009;28:139–154. doi: 10.1080/08830180902943058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 11.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis BN, Hata A. Regulation of MicroRNA biogenesis: a miRiad of mechanisms. Cell Commun Signal. 2009;7:18. doi: 10.1186/1478-811X-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saini HK, Griffiths-Jones S, Enright AJ. Genomic analysis of human microRNA transcripts. Proc Natl Acad Sci USA. 2007;104:17719–17724. doi: 10.1073/pnas.0703890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X, Liu J, Zhou R, Huang S, Huang S, Chen XM. Gene silencing of MIR22 in acute lymphoblastic leukaemia involves histone modifications independent of promoter DNA methylation. Br J Haematol. 2010;148:69–79. doi: 10.1111/j.1365-2141.2009.07920.x. [DOI] [PubMed] [Google Scholar]
- 16.Corcoran DL, Pandit KV, Gordon B, Bhattacharjee A, Kaminski N, Benos PV. Features of mammalian microRNA promoters emerge from polymerase II chromtatin immunoprecipitation data. PLoS One. 2009;4:e5279. doi: 10.1371/journal.pone.0005279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, et al. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005;33:2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu J, Wang F, Yang GH, Wang FL, Ma YN, Du ZW, et al. Human microRNA clusters: genomic organization and expression profile in leukemia cell lines. Biochem Biophys Res Commun. 2006;349:59–68. doi: 10.1016/j.bbrc.2006.07.207. [DOI] [PubMed] [Google Scholar]
- 20.Hayden MS, Ghosh S. Shared principles in NF-κB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 21.Han J, Ulevitch RJ. Limiting inflammatory responses during activation of innate immunity. Nat Immunol. 2005;6:1198–1205. doi: 10.1038/ni1274. [DOI] [PubMed] [Google Scholar]
- 22.Harada K, Ohira S, Isse K, Ozaki S, Zen Y, Sato Y, et al. Lipopolysaccharide activates nuclear factor-kappaB through Toll-like receptors and related molecules in cultured biliary epithelial cells. Lab Invest. 2003;83:1657–1667. doi: 10.1097/01.lab.0000097190.56734.fe. [DOI] [PubMed] [Google Scholar]
- 23.Poppelmann B, Klimmek K, Strozyk E, Voss R, Schwarz T, Kulms D. NFkappaB-dependent down-regulation of tumor necrosis factor receptor-associated proteins contributes to interleukin-1-mediated enhancement of ultraviolet B-induced apoptosis. J Biol Chem. 2005;280:15635–15643. doi: 10.1074/jbc.M413006200. [DOI] [PubMed] [Google Scholar]
- 24.Kim S, Domon-Dell C, Kang J, Chung DH, Freund JN, Evers BM. Down-regulation of the tumor suppressor PTEN by the tumor necrosis factor-alpha/nuclear factor-kappaB (NF-kappaB)-inducing kinase/NF-kappaB pathway is linked to a default IkappaB-alpha autoregulatory loop. J Biol Chem. 2004;279:4285–4291. doi: 10.1074/jbc.M308383200. [DOI] [PubMed] [Google Scholar]
- 25.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jennewein C, von Knethen A, Schmid T, Brüne B. MicroRNA-27b contributes to lipopolysaccharide-mediated peroxisome proliferator-activated receptor gamma (PPARgamma) mRNA destabilization. J Biol Chem. 2010;285:11846–11853. doi: 10.1074/jbc.M109.066399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bazzoni F, Rossato M, Fabbri M, Gaudiosi D, Mirolo M, Mori L, et al. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc Natl Acad Sci USA. 2009;106:5282–5287. doi: 10.1073/pnas.0810909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gatto G, Rossi A, Rossi D, Kroening S, Bonatti S, Mallardo M. Epstein-Barr virus latent membrane protein 1 trans-activates miR-155 transcription through the NF-kappaB pathway. Nucleic Acids Res. 2008;36:6608–6619. doi: 10.1093/nar/gkn666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perry MM, Moschos SA, Williams AE, Shepherd NJ, Larner-Svensson HM, Lindsay MA. Rapid changes in microRNA-146a expression negatively regulate the IL-1beta-induced inflammatory response in human lung alveolar epithelial cells. J Immunol. 2008;180:5689–5698. doi: 10.4049/jimmunol.180.8.5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cameron JE, Yin Q, Fewell C, Lacey M, McBride J, Wang X, et al. Epstein–Barr virus latent membrane protein 1 induces cellular MicroRNA miR-146a, a modulator of lymphocyte signaling pathways. J Virol. 2008;82:1946–1958. doi: 10.1128/JVI.02136-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shin VY, Jin H, Ng EK, Cheng AS, Chong WW, Wong CY, et al. NF-κB targets miR-16 and miR-21 in gastric cancer: involvement of prostaglandin E receptors. Carcinogenesis. 2011;32:240–245. doi: 10.1093/carcin/bgq240. [DOI] [PubMed] [Google Scholar]
- 33.Lu Z, Li Y, Takwi A, Li B, Zhang J, Conklin DJ, et al. miR-301a as an NF-κB activator in pancreatic cancer cells. EMBO J. 2011;30:57–67. doi: 10.1038/emboj.2010.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou R, Hu G, Gong AY, Chen XM. Binding of NF-kappaB p65 subunit to the promoter elements is involved in LPS-induced transactivation of miRNA genes in human biliary epithelial cells. Nucleic Acids Res. 2010;38:3222–3232. doi: 10.1093/nar/gkq056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou R, Hu G, Liu J, Gong AY, Drescher KM, Chen XM. NF-kappaB p65-dependent transactivation of miRNA genes following Cryptosporidium parvum infection stimulates epithelial cell immune responses. PLoS Pathog. 2009;5:e1000681. doi: 10.1371/journal.ppat.1000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Hara SP, Splinter PL, Gajdos GB, Trussoni CE, Fernandez-Zapico ME, Chen XM, et al. NFkappaB p50-CCAAT/enhancer-binding protein beta (C/EBPbeta)-mediated transcriptional repression of microRNA let-7i following microbial infection. J Biol Chem. 2010;285:216–225. doi: 10.1074/jbc.M109.041640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu S, Wu LC, Pang J, Santhanam R, Schwind S, Wu YZ, et al. Sp1/NFkappaB/HDAC/ miR-29b regulatory network in KIT-driven myeloid leukemia. Cancer Cell. 2010;17:333–347. doi: 10.1016/j.ccr.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H, Garzon R, Sun H, Ladner KJ, Singh R, Dahlman J, et al. NF-κB-YY1-miR-29 Regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. 2008;14:369–381. doi: 10.1016/j.ccr.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitmarsh AJ, Davis RJ. Transcription factor AP-1 regulation by mitogen-activated protein kinase signal transduction pathways. J Mol Med. 1996;74:589–607. doi: 10.1007/s001090050063. [DOI] [PubMed] [Google Scholar]
- 40.Fujita S, Ito T, Mizutani T, Minoguchi S, Yamamichi N, Sakurai K, et al. miR-21 Gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J Mol Biol. 2008;378:492–504. doi: 10.1016/j.jmb.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 41.Misawa A, Katayama R, Koike S, Tomida A, Watanabe T, Fujita N. AP-1-Dependent miR-21 expression contributes to chemoresistance in cancer stem cell-like SP cells. Oncol Res. 2010;19:23–33. doi: 10.3727/096504010x12828372551759. [DOI] [PubMed] [Google Scholar]
- 42.Shao M, Rossi S, Chelladurai B, Shimizu M, Ntukogu O, Ivan M, et al. PDGF induced microRNA alterations in cancer cells. Nucleic Acids Res. 2011;39:4035–4047. doi: 10.1093/nar/gkq1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yin Q, Wang X, McBride J, Fewell C, Flemington E. B-cell receptor activation induces BIC/miR-155 expression through a conserved AP-1 element. J Biol Chem. 2008;283:2654–2662. doi: 10.1074/jbc.M708218200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oneyama C, Ikeda J, Okuzaki D, Suzuki K, Kanou T, Shintani Y, et al. MicroRNA-mediated downregulation of mTOR/FGFR3 controls tumor growth induced by Src-related oncogenic pathways. Oncogene. 2011 doi: 10.1038/onc.2011.63. in press. [DOI] [PubMed] [Google Scholar]
- 45.Ding XC, Weiler J, Grosshans H. Regulating the regulators: mechanisms controlling the maturation of microRNAs. Trends Biotechnol. 2009;27:27–36. doi: 10.1016/j.tibtech.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 46.Fukuda T, Yamagata K, Fujiyama S, Matsumoto T, Koshida I, Yoshimura K, et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat Cell Biol. 2007;9:604–611. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- 47.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guil S, Caceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 49.Michlewski G, Guil S, Semple CA, Caceres JF. Posttranscriptional regulation of miRNAs harboring conserved terminal loops. Mol Cell. 2008;32:383–393. doi: 10.1016/j.molcel.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shimada N, Rios I, Moran H, Sayers B, Hubbard K. p38 MAP kinase dependent regulation of the expression level and subcellular distribution of heterogeneous nuclear ribonucleoprotein A1 and its involvement in cellular senescence in normal human fibroblasts. RNA Biol. 2009;6:293–304. doi: 10.4161/rna.6.3.8497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, et al. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruggiero T, Trabucchi M, de Santa F, Zupo S, Harfe BD, McManus MT, et al. LPS induces KH-type splicing regulatory protein-dependent processing of microRNA-155 precursors in macrophages. FASEB J. 2009;23:2898–2908. doi: 10.1096/fj.09-131342. [DOI] [PubMed] [Google Scholar]
- 53.Dean JL, Sully G, Clark AR, Saklatvala J. The involvement of AU-rich element-binding proteins in p38 mitogen-activated protein kinase pathway-mediated mRNA stabilisation. Cell Signal. 2004;16:1113–1121. doi: 10.1016/j.cellsig.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 54.Melo SA, Ropero S, Moutinho C, Aaltonen LA, Yamamoto H, Calin GA, et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet. 2009;41:365–370. doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55.Paroo Z, Ye X, Chen S, Liu Q. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell. 2009;139:112–122. doi: 10.1016/j.cell.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Melo S, Villanueva A, Moutinho C, Davalos V, Spizzo R, Ivan C, et al. Small molecule enoxacin is a cancer-specific growth inhibitor that acts by enhancing TAR RNA-binding protein 2-mediated microRNA processing. Proc Natl Acad Sci USA. 2011;108:4394–4399. doi: 10.1073/pnas.1014720108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Asirvatham AJ, Gregorie CJ, Hu Z, Magner WJ, Tomasi TB. MicroRNA targets in immune genes and the Dicer/Argonaute and ARE machinery components. Mol Immunol. 2008;45:1995–2006. doi: 10.1016/j.molimm.2007.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNFalpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 59.Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, et al. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 60.Witwer KW, Sisk JM, Gama L, Clements JE. MicroRNA regulation of IFN-beta protein expression: rapid and sensitive modulation of the innate immune response. J Immunol. 2010;184:2369–2376. doi: 10.4049/jimmunol.0902712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ueda R, Kohanbash G, Sasaki K, Fujita M, Zhu X, Kastenhuber ER, et al. Dicer-regulated microRNAs 222 and 339 promote resistance of cancer cells to cytotoxic T-lymphocytes by down-regulation of ICAM-1. Proc Natl Acad Sci USA. 2009;106:10746–10751. doi: 10.1073/pnas.0811817106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kawahigashi Y, Mishima T, Mizuguchi Y, Arima Y, Yokomuro S, Kanda T, et al. MicroRNA profiling of human intrahepatic cholangiocarcinoma cell lines reveals biliary epithelial cell-specific microRNAs. J Nippon Med Sch. 2009;76:188–197. doi: 10.1272/jnms.76.188. [DOI] [PubMed] [Google Scholar]
- 63.Hu G, Gong AY, Liu J, Zhou R, Deng C, Chen XM. miR-221 suppresses ICAM-1 translation and regulates interferon-gamma-induced ICAM-1 expression in human cholangiocytes. Am J Physiol. 2010;298:542–550. doi: 10.1152/ajpgi.00490.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suárez Y, Wang C, Manes TD, Pober JS. Cutting edge: TNF-induced microRNAs regulate TNF-induced expression of E-selectin and intercellular adhesion molecule-1 on human endothelial cells: feedback control of inflammation. J Immunol. 2010;184:21–25. doi: 10.4049/jimmunol.0902369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Natl Acad Sci USA. 2008;105:1516–1521. doi: 10.1073/pnas.0707493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gong AY, Zhou R, Hu G, Li X, Splinter PL, O’Hara SP, et al. MicroRNA-513 regulates B7-H1 translation and is involved in interferon-gamma-induced B7-H1 expression in cholangiocytes. J Immunol. 2009;182:1325–1333. doi: 10.4049/jimmunol.182.3.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gong AY, Zhou R, Hu G, Liu J, Sosnowska D, Drescher KM, et al. Cryptosporidium parvum induces B7-H1 expression in cholangiocytes by down-regulating microRNA-513. J Infect Dis. 2010;201:160–169. doi: 10.1086/648589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 69.Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kesimer M, Scull M, Brighton B, DeMaria G, Burns K, O’Neal W, et al. Characterization of exosome-like vesicles released from human tracheobronchial ciliated epithelium: a possible role in innate defense. FASEB J. 2009;23:1858–1868. doi: 10.1096/fj.08-119131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ohshima K, Inoue K, Fujiwara A, Hatakeyama K, Kanto K, Watanabe Y, et al. Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. PLoS One. 2010;5:e13247. doi: 10.1371/journal.pone.0013247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Otsuka M, Jing Q, Georgel P, New L, Chen J, Mols J, et al. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity. 2007;27:123–134. doi: 10.1016/j.immuni.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 73.Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, et al. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ahluwalia JK, Khan SZ, Soni K, Rawat P, Gupta A, Hariharan M, et al. Human cellular microRNA hsa-miR-29a interferes with viral nef protein expression and HIV-1 replication. Retrovirology. 2008;5:117. doi: 10.1186/1742-4690-5-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang J, Wang F, Argyris E, Chen K, Liang Z, Tian H, et al. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat Med. 2007;13:1241–1247. doi: 10.1038/nm1639. [DOI] [PubMed] [Google Scholar]
- 76.Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, et al. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- 77.Wang P, Hou J, Lin L, Wang C, Liu X, Li D, et al. Inducible microRNA-155 feedback promotes type I IFN signaling in antiviral innate immunity by targeting suppressor of cytokine signaling 1. Immunology. 2010;185:6226–6233. doi: 10.4049/jimmunol.1000491. [DOI] [PubMed] [Google Scholar]
- 78.Chen XM, Splinter PL, O’Hara SP, LaRusso NF. A cellular micro-RNA, let-7i, regulates Toll-like receptor 4 expression and contributes to cholangiocyte immune responses against Cryptosporidium parvum infection. J Biol Chem. 2007;282:28929–28938. doi: 10.1074/jbc.M702633200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 80.Linnstaedt SD, Gottwein E, Skalsky RL, Luftig MA, Cullen BR. Virally induced cellular microRNA miR-155 plays a key role in B-cell immortalization by Epstein–Barr virus. J Virol. 2010;84:11670–11678. doi: 10.1128/JVI.01248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lagos D, Pollara G, Henderson S, Gratrix F, Fabani M, Milne RS, et al. miR-132 regulates antiviral innate immunity through suppression of the p300 transcriptional co-activator. Nat Cell Biol. 2010;12:513–519. doi: 10.1038/ncb2054. [DOI] [PubMed] [Google Scholar]
- 82.Triboulet R, Mari B, Lin YL, Chable-Bessia C, Bennasser Y, Lebrigand K, et al. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science. 2007;315:1579–1582. doi: 10.1126/science.1136319. [DOI] [PubMed] [Google Scholar]
- 83.Ceppi M, Pereira PM, Dunand-Sauthier I, Barras E, Reith W, Santos MA, et al. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc Natl Acad Sci USA. 2009;106:2735–2740. doi: 10.1073/pnas.0811073106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tang B, Xiao B, Liu Z, Li N, Zhu ED, Li BS, et al. Identification of MyD88 as a novel target of miR-155, involved in negative regulation of Helicobacter pylori-induced inflammation. FEBS Lett. 2010;584:1481–1486. doi: 10.1016/j.febslet.2010.02.063. [DOI] [PubMed] [Google Scholar]
- 85.Sheedy FJ, Palsson-McDermott E, Hennessy EJ, Martin C, O’Leary JJ, Ruan Q, et al. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol. 2010;11:141–147. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
- 86.Hu G, Zhou R, Liu Jiu, Gong AY, Eischeid AN, Dittman JW, et al. MicroRNA-98 and let-7 confer cholangiocyte expression of cytokine-inducible Src homology 2-containing protein in response to microbial challenge. J Immunol. 2009;183:1617–1624. doi: 10.4049/jimmunol.0804362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhu QY, Liu Q, Chen JX, Lan K, Ge BX. MicroRNA-101 targets MAPK phosphatase-1 to regulate the activation of MAPKs in macrophages. J Immunol. 2010;185:7435–7442. doi: 10.4049/jimmunol.1000798. [DOI] [PubMed] [Google Scholar]
- 88.Ørom UA, Lund AH. Experimental identification of microRNA targets. Gene. 2010;451:1–5. doi: 10.1016/j.gene.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 89.Leung AK, Young AG, Bhutkar A, Zheng GX, Bosson AD, Nielsen CB, et al. Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nat Struct Mol Biol. 2011;18:237–244. doi: 10.1038/nsmb.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kota J, Chivukula RR, O’Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sharma AD, Narain N, Händel EM, Iken M, Singhal N, Cathomen T, et al. MicroRNA-221 regulates FAS-induced fulminant liver failure. Hepatology. 2011;53:1651–1661. doi: 10.1002/hep.24243. [DOI] [PubMed] [Google Scholar]
- 92.Androulidaki A, Iliopoulos D, Arranz A, Doxaki C, Schworer S, Zacharioudaki V, et al. The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs. Immunity. 2009;31:220–231. doi: 10.1016/j.immuni.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Alsaleh G, Suffert G, Semaan N, Juncker T, Frenzel L, Gottenberg JE, et al. Bruton’s tyrosine kinase is involved in miR-346-related regulation of IL-18 release by lipopolysaccharide-activated rheumatoid fibroblast-like synoviocytes. J Immunol. 2009;82:5088–5097. doi: 10.4049/jimmunol.0801613. [DOI] [PubMed] [Google Scholar]
- 94.El Gazzar M, McCall CE. MicroRNAs distinguish translational from transcriptional silencing during endotoxin tolerance. J Biol Chem. 2010;285:20940–20951. doi: 10.1074/jbc.M110.115063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huang TH, Wu F, Loeb GB, Hsu R, Heidersbach A, Brincat A, et al. Up-regulation of miR-21 by HER2/neu signaling promotes cell invasion. J Biol Chem. 2009;284:18515–18524. doi: 10.1074/jbc.M109.006676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Martinez-Nunez RT, Louafi F, Sanchez-Elsner T. The interleukin 13 (IL-13) pathway in human macrophages is modulated by microRNA-155 via direct targeting of interleukin 13 receptor alpha1 (IL13Ralpha1) J Biol Chem. 2011;286:1786–1794. doi: 10.1074/jbc.M110.169367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yin Q, McBride J, Fewell C, Lacey M, Wang X, Lin Z, et al. MicroRNA-155 is an Epstein– Barr virus-induced gene that modulates Epstein–Barr virus-regulated gene expression pathways. J Virol. 2008;82:5295–5306. doi: 10.1128/JVI.02380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Worm J, Stenvang J, Petri A, Frederiksen KS, Obad S, Elmén J, et al. Silencing of microRNA-155 in mice during acute inflammatory response leads to derepression of C/EBPβ and down-regulation of G-CSF. Nucleic Acids Res. 2009;37:5784–5792. doi: 10.1093/nar/gkp577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schembri F, Sridhar S, Perdomo C, Gustafson AM, Zhang X, Ergun A, et al. MicroRNAs as modulators of smoking-induced gene expression changes in human airway epithelium. Proc Natl Acad Sci USA. 2009;106:2319–2324. doi: 10.1073/pnas.0806383106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sonkoly E, Wei T, Janson PC, Sääf A, Lundeberg L, Tengvall-Linder M, et al. MicroRNAs: novel regulators involved in the pathogenesis of Psoriasis? PLoS One. 2007;2:e610. doi: 10.1371/journal.pone.0000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wu F, Zikusoka M, Trindade A, Dassopoulos T, Harris ML, Bayless TM, et al. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology. 2008;135:1624–1635. doi: 10.1053/j.gastro.2008.07.068. [DOI] [PubMed] [Google Scholar]
- 102.Hu G, Zhou R, Liu J, Gong AY, Chen XM. MicroRNA-98 and let-7 regulate expression of suppressor of cytokine signaling 4 in biliary epithelial cells in response to Cryptosporidium parvum infection. J Infect Dis. 2010;202:125–135. doi: 10.1086/653212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Oglesby IK, Bray IM, Chotirmall SH, Stallings RL, O’Neill SJ, McElvaney NG, et al. miR-126 is downregulated in cystic fibrosis airway epithelial cells and regulates TOM1 expression. J Immunol. 2010;184:1702–1709. doi: 10.4049/jimmunol.0902669. [DOI] [PubMed] [Google Scholar]