Abstract
Epigenetic influences, such as DNA methylation, histone acetylation and upregulation/downregulation of genes by microRNAs, change the genetic makeup of an individual without affecting DNA base pair sequences. Indeed, epigenetic changes play an integral role in the progression from normal esophageal mucosa to Barrett’s esophagus to esophageal adenocarcinoma via dysplasia- metaplasia-neoplasia sequence. Many genes involved in esophageal adenocarcinoma display hypermethylation, leading to their downregulation. The classes of these genes include cell cycle control, DNA and growth factor repair, tumor suppressors, anti-metastasis, WNT-related genes, and pro-apoptotic genes. Histone acetylation in the pathophysiology of esophageal diseases has not been thoroughly investigated, and its critical role in the development of esophageal adenocarcinoma is less defined. Many microRNAs have been associated with the development of Barrett’s esophagus and esophageal adenocarcinoma. Here, we critically addressed the specific steps most closely influenced by microRNAs in the progression from Barrett’s esophagus to esophageal adenocarcinoma. However, microRNAs can target up to hundreds of genes, making it difficult to correlate directly with a given phenotype of the disease. Esophageal adenocarcinoma progressing from pre-malignant condition of Barrett’s esophagus carries an extremely poor prognosis. Risk stratification for patients based on their epigenetic profiles may be useful in providing more targeted and directed treatment to patients.
Keywords: DNA methylation, epigenetics, Barrett’s esophagus, esophageal adenocarcinoma, esophageal squamous cell carcinoma, histone acetylation, miRNA
Introduction
Esophageal cancer is the 8th most common cancer worldwide, and the 5th leading cause of cancer-related death in men.1,2 There are two main types of cancer that occur in the esophagus: esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC). ESCC typically affects the upper 2/3rd of the esophagus, while EAC typically affects the lower 1/3rd of the esophagus.3 The focus of this review article is esophageal adenocarcinoma. Rates of esophageal adenocarcinoma have been rising over the past four decades particularly in developed countries, and EAC carries a poor prognosis with roughly 25% of patients presenting with metastatic disease4.
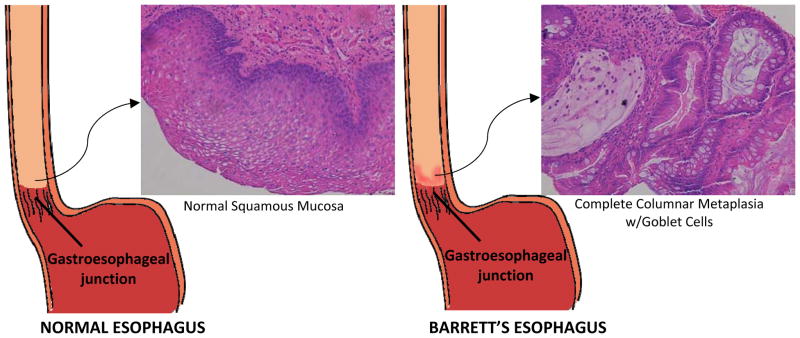
Known risk factors for EAC include gastro-esophageal reflux disease (GERD), Barrett’s esophagus (BE), obesity, Caucasian race, increasing age, and smoking.5,6 It is believed that EAC develops through a metaplasia-dysplasia-neoplasia sequence as a result of chronic GERD.6 Long standing GERD leads to Barrett’s esophagus (Fig. 1), a condition in which the normal squamous epithelium lining the esophagus is replaced by metaplastic columnar epithelium with intestinalization.7 Barrett’s esophagus is a pre-cancerous condition and is the only known precursor of esophageal adenocarcinoma.8 While the molecular mechanism of the transition into adenocarcinoma is not clear, it has been speculated that GERD may cause genetic and epigenetic changes in the epithelium leading to the characteristics seen in Barrett’s esophagus and EAC.6
Figure 1.
Diagram Showing Changes in Epithelium in the Development of Barrett’s esophagus. Stratified squamous epithelium of the esophagus undergoes metaplasia to mimic the columnar epithelium with goblet cells found in the intestines. Modified from Spechler.74
There is a lacunae in our understanding of the mechanisms underlying the progression of BE into EAC as only about 5% of patients with BE EAC. One study found that age might play a role in the development of familial versus non-familial Barrett’s esophagus.9 Multiplex familial Barrett’s esophagus was defined as having at least 2 family members with Barrett’s esophagus.9 This familial aggregation could be caused by either common environmental exposures in family members or a genetic predisposition to the disease or both.9 However, the interpretation of these results is problematic because GERD is symptomatic and associated with the development of Barrett’s esophagus, while patients with BE may not be symptomatic.9 Therefore, it is difficult to establish the age of incidence.9 Endoscopic surveillance of Barrett’s esophagus has not proven to be a very strong preventative measure of Barrett’s esophagus, as only a small percentage (5%) of patients with EAC have a preexisting diagnosis of BE. Additionally, the rate of progression from BE to EAC is only 0.5% per year with a lifetime risk of 5%.9 Therefore, there is a great need for markers to predict which patients with GERD are at risk for developing BE and which patients with BE are at risk of developing EAC. This would allow for the prudent use of resources for screening and surveillance endoscopy, and it would potentially indicate the need for more aggressive ablative treatment of BE. The role of several biomarkers, including DNA methylation in Barrett’s esophagus, has been explored. In this article, we critically reviewed the current status of biomarkers in the GERD → BE → EAC sequence and discussed a potential role of epigenetics in the pathogenesis and progression of the disease process. We also discussed the significance of this knowledge in developing biomarkers that correlate more closely with disease progression.
Genomic instability could be a critical factor to initiate the metaplasia- dysplasia- neoplasia sequence. Several studies have explored the role of p53, APC, CDKN2A, cyclin D1, and Rb genes. Mutations in these genes have been implicated in the progression of many cancers including EAC.10 While some genetic linkages have been found, epigenetics may be more helpful in predicting the progression of esophageal adenocarcinoma in patients with Barrett’s esophagus. Epigenetics is defined as the “study of changes in gene function that are mitotically and/or meiotically heritable that do not entail a change in DNA sequence.”6 The three major processes in epigenetics are DNA methylation, histone acetylation/deacetylation, and miRNA.11 DNA methylation involves modification of the DNA itself, whereas histone acetylation/deacetylation involves modification in the packaging of DNA, and miRNAs are short non-coding molecules that can alter gene expression.6 DNA methylation is the most widely studied area in the context of Barrett’s esophagus and esophageal adenocarcinoma.
DNA Methylation
One epigenetic mechanism is DNA methylation, where methyl groups are added to gene promoter sequences. This methylation primarily occurs on cytosine bases in cytosine-guanine (CpG) dinucleotides, especially when the cytosine and guanine contents are greater than 50% in the DNA sequence.11 Hypermethylation of these CpG islands on the promoter region results in transcriptional silencing, which decreases the expression of genes, while hypomethylation results in increased expression.11 The enzymes, DNA methyltransferases (DNMTs) catalyze DNA methylation. Changes in these enzymes can lead to aberrant methylation of genes. The most extensively studied DNMT in esophageal cancer is O6-Methylguanine-DNA Methyltransferase (MGMT).12 MGMT mutations have been implicated in a number of cancers, including esophageal squamous cell carcinoma.13 Hypermethylation of this gene has been found to be associated with esophageal adenocarcinoma, but not necessarily with a patient’s outcome.12,14 The only other DNMT that seems to have been studied in esophageal cancer is DNMT1. Overexpression of the DNMT1 gene was found to be associated with ESCC and correlated with lymph node metastasis.15 Additional investigation into DNA methyltransferasesis indeed a key area to study in esophageal adenocarcinoma.
There are three main mechanisms in which DNA methylation can result in carcinogenesis: base substitution gene mutation, where a 5-methylcytosine is deaminated to thymine; aberrant DNA methylation, which can be associated with allelic loss; and hypermethylation, which may correlate to inactivation of tumor suppressor genes.16 Hypermethylation-associated silencing of tumor suppressor genes is the most recognized epigenetic disruption, first discovered in the retinoblastoma gene (Rb1).16
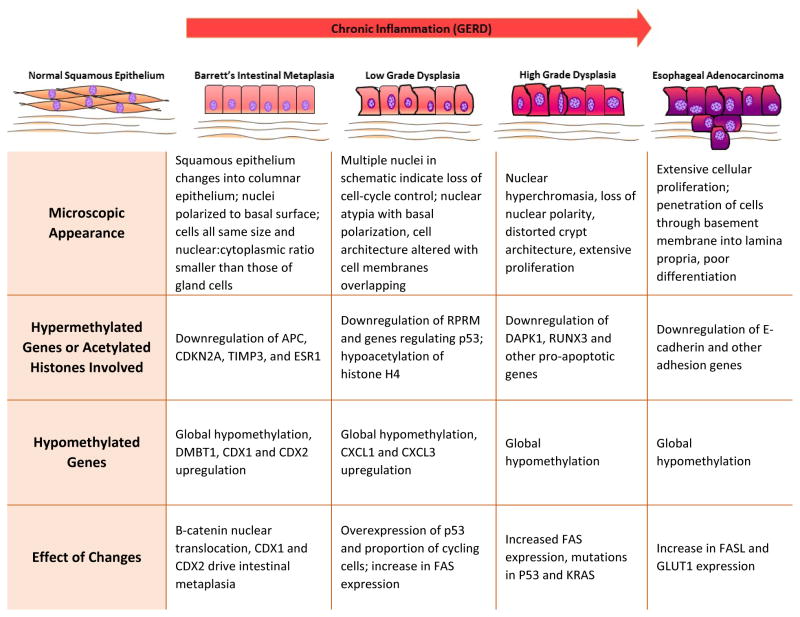
In EAC, hypermethylation of genes has been extensively studied. Table 1 summarizes the results of studies examining hypermethylation in various genes in Barrett’s esophagus and esophageal adenocarcinoma. While these studies characterize hypermethylation, they do not discuss the exact functional loss that correlates with different degrees of hypermethylation. Since one of the goals in studying EAC is to establish a biomarker that indicates prognosis of patients, an important aspect that needs to be studied is the differences in methylation found in normal esophageal mucosa compared to levels found in other tissues and in EAC. It is also important to note that while hypermethylation of these genes correlates with certain stages of EAC, it is not necessarily a causal relationship. In fact, since these studies do not establish a temporal relationship between aberrant methylation patterns and dysplasia, hypermethylation could be a result rather than a cause of the dysplasia. None-the-less, careful and well-designed studies are warranted to establish causal or consequential effect of hypermethylation of genes at various stages of the initiation, progression and chronicity of metaplasia-dysplasia-adenocarcinoma in the esophagus.
TABLE 1.
Genes with Hypermethylation Patterns Reported in the Progression of Barrett’s esophagus and/or Esophageal Adenocarcinoma
| CLASSIFICATION | GENE | FULL NAME | FUNCTION | CITATIONS |
|---|---|---|---|---|
| Cell Cycle Control Gene | CDKN2A | P16 | Cell cycle control | 6,44 |
| DNA repair genes | MGMT | O6-methylguanine DNA methyltransferase | DNA repair | 29,45–47 |
| Growth factor response related genes | CRBP1 | Cellular Retinol Binding Protein 1 | Retinol transport | 6,8,44 |
| IGFBP7 | Insulin-like growth factor binding protein 7 | Modulates binding of insulin-like factors to IGF receptors | 48 | |
| SOCS3 | Suppressor of Cytokine Signaling 3 | Suppression of JAK/STAT pathway | 1,6,8,46 | |
| Metastasis antagonizing genes | CDH1 | E-cadherin | Cell adhesion | 1,6,40,49,50 |
| CDH13 | T-cadherin | Cell adhesion, proliferation, metastasis | 1,6,10–12 | |
| Pro-apoptotic genes | DAPK1 | Death Associated Protein Kinase 1 | Apoptosis | 6,45 |
| RUNX3 | Runt-related transcription factor 1 | Pro-apoptotic factor in TGF-β Pathway | 6,8,49,50 | |
| Tumor Suppressor Gene | AKAP12 | A-Kinase anchoring protein 12 | Controls cell signaling, cell adhesion, mitogenesis and differentiation | 6,8,50 |
| WNT signaling related genes | APC | Adenomatous Polyposis Coli | Involved in cell adhesion through its interaction with beta catenin- cadherin proteins. | 1,8,29,44,48,50,51 |
| SFRP1 | Secreted frizzled-related protein 1 | Antagonist of WNT protein receptors | 6,45,47 | |
| WIF1 | Wnt inhibitory factor 1 | WNT-signaling pathway inhibitor | 6,8,44,50 | |
| Other Genes with Tumor Suppressive Functions | CALCA | Calcitonin | Regulates calcitonin levels through adenylatecyclase | 6,40 |
| ESR1 | Estrogen Receptor α | Hormone receptor in mammary cells | 1,6,8,40 | |
| EYA4 | Eyes absent homolog 4 | Transcriptional activator important for function of the Organ of Corti | 1,8,50 | |
| GPX3 | Glutathione Peroxidase | Catalyzes the reduction of hydrogen peroxide | 1,4,6,8,44,47 | |
| GSTM2 | Glutathione S- transferase Mu 2 | Glutathione transferase activity | 45,50,52 | |
| MYOD1 | Myoblast determination protein 1 | Muscle differentiation | 1,6,40 | |
| NELL1 | Protein kinase C-binding protein NELL1 | Cell growth regulation and differentiation | 6,44,50 | |
| RPRM | REPRIMO | Regulates p53-mediated cell cycle arrest | 6,8,41,50 | |
| SST | Somatostatin | Somatostatin hormone | 44,50 | |
| TAC1 | Protachykinin-1 | Tachykinin peptide hormone | 6,44,50 | |
| TIMP3 | Tissue Inhibitor of Metalloproteinase | Metalloproteinase inhibitor | 1,6,40,44,47,50,53 |
In order to establish these differences, the first step is to differentiate between methylation patterns in normal squamous esophagus versus patterns in other normal tissues. For example, in one study it was found that MT3 gene has a large CpG island, and methylation level of the promoter of this gene was high in normal stomach, but low in normal esophagus.17 This study also found that patients with BE and EAC had hypermethylatedMT3 in the esophagus.17 On the other hand, some genes are not highly methylated in most tissues. Therefore, it is reasonable to speculate that hypermethylation in any tissue could induce pathological lesions. This could be supported by the study of Kwong and colleagues who found that high levels of methylation in the DLEC1 gene were associated with carcinogenesis in the lung, kidney and esophagus.18
In addition to looking at normal mucosa, the ideal way to stratify prognosis of patients who develop GERD is to look at methylation patterns in the different stages of dysplasia (Figure 1). A study by Agarwal and colleagues describes the stages of progression as shown below6:
Normal Squamous Mucosa → Inflammation (GERD) → BE Metaplasia → Low Grade Dysplasia → High Grade Dysplasia → EAC
Most cases of Barrett’s esophagus do not progress to esophageal adenocarcinoma. Currently, the best marker of EAC is high grade dysplasia, but there is no biomarker to predict transition of BE into low or high grade dysplasia.19 One study found that Wnt-related genes, such as APC, SFRP1, and WIF1, are more highly methylated in the development of neoplasia from metaplasia, than in the development of metaplasia.6 Wnt signaling is present in healthy esophagus, and it has been hypothesized that normal Wnt signaling can result in a change in gene expression.20 Trowbridge and colleagues proposed that epigenetic changes, including the methylation of SOX17 promoter may play a role in allowing normal Wnt signaling to result in a change of gene expression.21 However, potentially additional mechanisms cannot be ruled out.
Similarly, P16 hypermethylation is seen in higher frequency in Barrett’s dysplasia than in the Barrett’s metaplasia.22 Another study found that the hypermethylation of CDKN2A, TIMP3, ESR1 genes is associated with the onset of Barrett’s metaplasia.23 Furthermore, the study by Wild and colleagues found that E-cadherin hypermethylation is an important marker in the transition from dysplasia into adenocarcinoma.24 Findings from additional similarly designed studies would help to further characterize the different stages of dysplasia in esophageal adenocarcinoma.23 While some of these genes are known to be hypermethylated in EAC, their exact role is unknown. Also, few of these genes, for example SFRP1, have been characterized in esophageal squamous cell carcinoma, and may also be hypermethylated in EAC.25 This information may be helpful in developing hypermethylation profiles of various genes. Comparing gene profiles at various stages of the esophageal dysplastic sequence could increase the specificity of the results and help clarify which genes may truly be responsible for inducing phenotypic changes.
In addition to the findings of hypermethylation in several loci of the genome, some studies have observed that the entire genome in cells of BE and EAC is hypomethylated when matched with normal squamous mucosa.26 Global hypomethylation of CpG islands is present to some extent in all cancers, and is observed in EAC as early as low grade dysplasia.26 While hypomethylation of specific genes has not been extensively studied, there are only a few potential oncogenes that are upregulated, due to hypomethylation. These genes include Deleted in Malignant Brain Tumor 1 (DMBT1)6, CXCL1 and CXCL3 genes6, and CDX1 and CDX210. CXCL1 and CXCL3 play a role in spinal cord development by inhibiting the migration of oligodendrocyte precursors.27 CDX1 and CDX2 genes are essential for skeletal and intestinal development.28 These genes are usually expressed in the mucosal epithelium from the duodenum to the rectum, but one study demonstrated their presence in EAC tissue.28
Histone Acetylation
Histone acetylation is another epigenetic mechanism that affects chromatin by acetylation of histone proteins. Levels of acetylation/deactylation of histone proteins are determined by two opposing enzymes: histone acetyltransferases (HATs) and histone deacetylases (HDACs).29 Hypoacetylation leads to silencing of gene expression, while hyperacetylation leads to gene activation.29 There are not many studies examining the role of histone acetylation in the carcinogenesis of esophageal adenocarcinoma.
One study on esophageal squamous cell carcinoma (ESCC) found that histone H4 of esophageal carcinoma cells was significantly hyperacetylated in the early stage of cancer and progressively changed into a hypoacetylated state as the cancer progressed (Figure 2).29 The only other study that focused specifically on histone acetylation theorized that this progress may have an effect on the expression of INHBA (activin – a ligand in the TBG-β superfamily) in EAC, but the results were not statistically significant.30 Since there is an overlap between EAC and ESCC, this is a potential important area for future research and additional studies are warranted to dissect the role of histone acetylation in ESCC and EAC.30
Figure 2.
Schematic of Role of Hypermethylated Genes and Acetylated Histones in the Progression from Normal Mucosa to Esophageal Adenocarcinoma. Chronic inflammation due to gastroesophageal reflux disease is understood to be the primary factor that drives this pathogenesis. The figure offers a potential role for hypermethylated genes and altered histones in this progression.23,75–78
MicroRNA
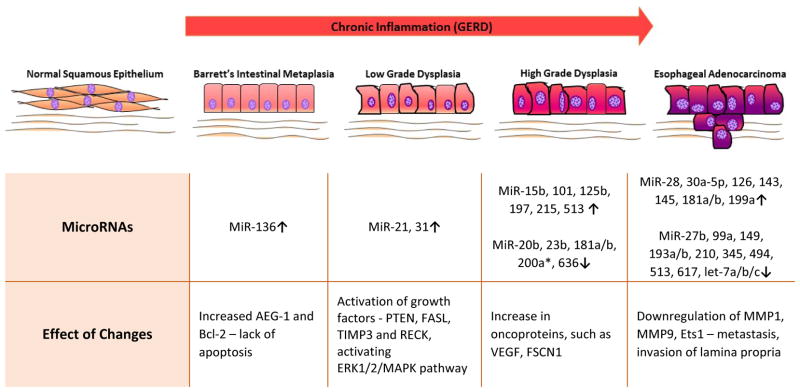
MicroRNAs (miRNA) are small non-coding strands of RNA that are involved in the regulation of transcription and translation by binding to complementary strands of DNA and RNA. They are smaller and perhaps better biomarkers than mRNA. These molecules play an important role in the modification of gene expression, because while not all genes are targeted by miRNA, one miRNA can target multiple genes, leading to large changes in gene expression.26 Some studies suggest that miRNA may be more helpful than methylation in risk stratification for progression of BE to EAC.31 Several miRNAs that have been implicated in EAC (Table 2). This is the first report that summarized the current findings on the type of miRNA involved in various stages of the disease pathogenesis with their expression and the target (Figure 3).
TABLE 2.
Altered Expression of Micro RNAs Reported in BE and/or EAC
| STAGE | MICRO RNA* | EXPRESSION | TARGET | CITATIONS |
|---|---|---|---|---|
| NSM to BE | hsa-miR-136* | Increased | AEG-1 and Bcl-2 | 34,37,54 |
| BE to LGD | miR-21 | Increased | PTEN, FASL, TIMP3 and RECK, activating ERK1/2/MAPK pathway | 26,31,34,37,43,55–57 |
| miR-31 | Increased | EMP1 (epithelial membrane protein 1), KSR2 (kinase suppressor of ras 2) and RGS4 (regulator of G-protein signalling 4) – found in Esophageal Squamous Cell carcinoma | 36,56,58 | |
| BE to EAC | miR-25 | Increased | Bcl-2-like protein 11† - apoptosis | 34,59,60 |
| miR-93 | Increased | CDKN1A – cell cycle | 26,60 | |
| miR-106b-25 | Increased | CDKN1A – cell cycle | 34,36,47,60–62 | |
| hsa-miR-136* | Decreased | ‡ | 34,37 | |
| hsa-miR-192 | Increased | DHFR, CDC7, LMNB2, MAD2L1, CUL5 – cell cycle, cell proliferation | 31,34,36,37,56,60,62 | |
| hsa-miR-194* | Increased | EP300 - metastasis | 31,36,37,56,60 | |
| miR-196a/b | Increased | ANXA1, SPRR2C, S1009 and KRT5 - apoptosis | 24,32,36,60–62 | |
| hsa-miR-203 | Decreased | ABL1, TP63 – cell proliferation | 11,26,34,36,37,43,56,60,62,63 | |
| hsa-miR-205 | Decreased | HER3, PRKCD, VEGF-A – cell proliferation, EMT | 11,26,29,34,36,37,43,56,60,62,63 | |
| hsa-miR-223 | Increased | ARTN, a known tumor metastasis-related gene | 34,36,37,61,64 | |
| hsa-miR-424* | Increased | ‡ | 9,11,29,34,36,37 | |
| hsa-miR-450a | Increased | ‡ | 34,37 | |
| LGD to HGD | miR-15b | Increased | Bcl-2 | 36 |
| miR-20b | Decreased | ‡ | 34,36,62 | |
| miR-23b | Decreased | c-Myc | 34,36,65 | |
| hsa-miR-101 | Increased | rap1GAP – tumor suppressor gene | 36,37,65,66 | |
| miR-125b | Increased | CYP24, ERBB2, ERBB3 – cell proliferation | 11,34,36,43,60 | |
| miR-181a/b | Decreased | TIMP- 3† | 32,36,55,67 | |
| miR-197 | Increased | ‡ | 36 | |
| miR-200a* | Decreased | E-cadherin transcriptional repressors ZEB1 and ZEB2 | 36,55–57,68 | |
| miR-215 | Increased | ZEB2 | 26,34,56,62,68 | |
| miR-513 | Increased | ‡ | 36 | |
| miR-636 | Decreased | ‡ | 36 | |
| HGD to EAC | miR-27b | Decreased | ST14, CYP1B1 - Cell proliferation, cell migration, invasion, drug metabolism | 36,43,55,59,60,65 |
| miR-28 | Increased | ‡ | 36 | |
| miR-30a-5p | Increased | ‡ | 34,36 | |
| miR-99a | Decreased | mTOR | 11,29,34,36,59,62,69 | |
| miR-126 | Increased | EGFL7† | 36,57,59,70 | |
| miR-143 | Increased | FSCN1 | 26,34,36,55,56,59,62 | |
| miR-145 | Increased | FSCN1 | 26,34,36,37,56,57,59 | |
| miR-149 | Decreased | FOXM1† - EMT | 36,71 | |
| miR-181a/b | Increased | TIMP- 3† | 32,36,55 | |
| miR-193a/b | Decreased | ‡ | 36 | |
| miR-199a | Increased | Brm | 36,61,72 | |
| miR-210 | Decreased | MNT† – myc antagonist, FGFRL1 | 24,34,36,65,73 | |
| miR-345 | Decreased | ‡ | 36 | |
| miR-494 | Decreased | ‡ | 36 | |
| miR-513 | Decreased | ‡ | 36 | |
| miR-617 | Decreased | ‡ | 36 | |
| let-7a/b/c | Decreased | IL-6, Ras, HMGA2 | 32,34,36,57,60,62,63,65 | |
| EAC | miR-16-2 | Increased | RAR-b2† | 36,57 |
| miR-30e | Increased | ‡ | 55,57 | |
| miR-34a | Decreased | NF-κ B Inhibition of c-Met and cyclin D1 protein expression | 36,55,57 | |
| miR-195p | Increased | ‡ | 36,55,57 | |
| miR-221 | Increased | p27Kip1 and CDX2 | 31,33 | |
| miR-222 | Increased | p27Kip1 and CDX2 | 31,33 | |
| miR-375 | Decreased | PDK1, JAK2 | 29,31,33,34,36,37,44,55,56,61,65 | |
| hsa-miR-518b | Decreased | ‡ | 36,37,55 |
potential target
unknown target,
Abbreviations: BE = Barrett’s esophagus, EAC = Esophageal Adenocarcinoma, HGD = High Grade Dysplasia, LGD = Low Grade Dysplasia, NSM = Normal Squamous Mucosa
Figure 3.
Schematic of Role of MicroRNAs Esophageal Adenocarcinoma Progression. The figure offers a potential role for specific miRNAs in the different stages of pathogenesis.76,77,79
In one study, the expression of four miRNAs (miR-192, miR-194, miR-196a, and miR-196b) was significantly higher in esophageal tissue of patients with progression to esophageal adenocarcinoma than in patients who did not show disease progression.31 Another study found that miR-196a could be a potential biomarker in the progression of EAC – its target genes are SPRR2C, S100A9, and KRT5 whose expression is characteristically decreased or lost during neoplastic transformation of esophageal tissue.32 In this study, miR-196a was found to be 10- to 100-fold higher in precancerous lesions and EAC than in normal squamous mucosa (NSM), and levels of miR- 196a proportionally increase with higher histological grades of dysplasia.32 This is an important point, because all miRNAs do not follow this pattern. For example, miRNA-136, −181 and −513 are included twice in this table, because they have been implicated in various stages with different effects. Up regulation versus downregulation of these miRNAs seems to have a role in different stages of EAC.
MiRNA-21 is one of the most significant miRNAs, because it has been implicated in the carcinogenesis of other tissues, such as breast cancer and lung cancer.11 MiRNA-21 is important for EAC because it has been found to be upregulated in a progressive manner through the Barrett’s metaplasia-dysplasia-adenocarcinoma sequence, making it a useful biomarker in the diagnosis and prognosis of EAC. MiR-221 and miR-222 may also be useful, because a couple of studies found that their expression is higher in EAC than in the surrounding BE. Due to GERD, esophageal cells are exposed to bile acids, which may activate farnesoid × receptor and upregulate levels of miR-221/222.33 The result of this is a reduction in levels of p27Kip1 and degradation of CDX2.33 As a result, levels of p27Kip1 and CDX2 were lower in areas of EAC than in those of BE.31
While most of the miRNAs that have been examined so far appear to correlate with higher levels in the progression of EAC, some significant miRNAs that may be decreased in BE and EAC are let-7c, miR-203 and miR-205.26 MiR-375 was the only miRNA found to be significantly down-regulated in EAC and unchanged in BE metaplasia.34 The most comprehensive study available on the upregulation and downregulation of miRNAs in BE and EAC is the study by Wu et al.34 The most important biomarkers to examine would be those that are significantly different in EAC versus BE metaplasia. While the study by Wu and colleagues.34 describes many miRNAs, it does not identify the target genes of these molecules. This is an important area that needs to be explored.
Other studies focused on miRNAs that are specifically involved in various stages of EAC. One study found that 3 oncogenic miRNAs, miR-25, miR-93 and miR-106b, are up-regulated in EAC relative to BE, indicating a potential progressive correlation of these miRNAs to the neoplastic process.35 Another study looked specifically at the progression from low grade dysplasia to high grade dysplasia, and found that this progression was associated with up-regulation of miR-200a*, miR-513, miR-125b, miR-101 and miR-197, as well as down-regulation of miR-23b, miR-20b, miR-181b, miR-203, miR-193b and miR-636.36 In the transition from high grade dysplasia to EAC, miR-126, miR-143, miR-145, miR-181a, miR-181b, miR-199a, miR-28 and miR-30a-5p appear to be upregulated.36 Let-7a/b/c, miR-193a, miR-345 and miR-494 are down-regulated.36
In addition to the involvement of miRNAs at various stages, miRNA could serve as a prognostic marker. A study by Huang and colleagues36 found a correlation between miR-126 expression and tumor cell differentiation and lymph node metastasis (LNM), and that miR-16-2 and miR-195p are associated with LNM and higher tumor stage.36 Huang and colleagues suggest that miRNAs could potentially be good targets for therapy, because most miRNAs exert their effects on multiple target genes. However, since a single miRNA could target many genes, such as those reported by Wang and colleagues,37 where a single miRNA targeted up to 50 genes, there is a possibility that upregulation of one gene is regulated by downregulaiton of other genes. This could result in globally adverse and unpredictable effects where the role of a specific miRNA in the pathogenesis of esophageal carcinoma would remain unclear.
LONG NON-CODING RNA
Long non-coding RNA (lncRNA) is similar to miRNA and snoRNA in that these are non-coding strands of RNA. They are usually longer than miRNA (longer than 200 bp). Also, unlike other non-coding RNA molecules, such as miRNA and snoRNAs, lncRNA molecules are not strongly conserved across diverse species.38 While this lack of conservation has been cited as a reason for non-functionality, it could also be an evidence to support that non-coding RNAs are subject to different selection pressures. This is a far less studied area than DNA methylation and miRNA, but this could also be a potential epigenetic mechanism.
One published study on lncRNA by Wu and colleagues39 found a role of lncRNA in the development of BE. These investigators selected AFAP1-AS1 as the lncRNA to study because it was significantly and aberrantly hypomethylated in BE.39 AFAP1 modulates actin filament integrity and serves as an adaptor protein linking Src family members and other signaling proteins to actin filaments.39 AFAP1 plays a role in breast cancer, because it is required for actin stress fiber formation and cell adhesion in breast cancer cells.39 While this was the only study that looked at lncRNA, this field is wide open to explore in regards to epigenetic mechanisms involved in the pathogenesis of esophageal adenocarcinoma.
EXPERT COMMENTARY AND FIVE YEAR REVIEW
One goal in studying epigenetics is to develop risk stratification that is helpful in delivering a clearer prediction of developing EAC in patients with BE. DNA hypermethylation studies are most helpful in this regard, because different genes have been found to be hypermethylated in different stages of the BE to EAC progression.40 One study found that 19 CpG islands segregate into six classes of genes silenced in cancer. Each class undergoes unique epigenetic changes at different steps of disease progression to EAC. The classes were defined as sharing similar epigenetic behaviors. Now that these changes have been characterized, the next step in studying these patterns will be to identify the molecular mechanisms and factors affecting the various CpG island clusters.40
In studying epigenetics and its role in esophageal adenocarcinoma, we encounter a number of obstacles. The clinical application of methylated DNA biomarkers for both diagnosis and prognosis of BE and esophageal cancer is limited because there are not enough clinical trials for validation (such as phase 2–3 biomarker studies).8 Also, many of the studies that have been done involve the characterization of expression without any link to function. Prospective studies would be helpful to achieve this goal. There is a need for longitudinal studies between patients with non progressive BE and patients with progressive BE. However, these studies could be challenging to complete due to the amount of time and resources that are required.
In addition, significant studies have been done on the hypermethylation and miRNA expression, but more focus on functional studies of the gene is required to see exactly what effect miRNAs have.26 Chronic inflammation in the esophagus clearly has an effect on the progression of this cancer, so studying the association between inflammatory processes and inflammatory alterations in miRNA expression is a critical area of research.26 This is difficult to achieve since miRNA molecules often exert effects on multiple genes.
Since the goal of these studies is to eventually find a useful biomarker, it is important to characterize markers that are as noninvasive as possible. Circulating biomarkers would be less invasive and less expensive. However, the problem with looking at these biomarkers is that the plasma levels might show different methylation patterns based on the severity of the progression. A study by Shah and colleagues found that the APC gene in the plasma was hypermethylated in late-stage EAC, but not in BE; tissue APC gene showed hypermethylation even in early stages.41 Thus, plasma levels may lag tissue levels in showing hypermethylation patterns, making them less than ideal despite being less invasive. The use of biopsy tissue would be best for such studies. However, this could involve a more invasive procedure for the patient, if additional biopsy tissue is collected.
In regard to hypermethylation, most of the published studies focus on the role of tumor suppressor genes in the development of BE and EAC, but few studies examined the inflammatory processes in the development of BE. Damage from GERD leads to increased activity of COX-2, suggesting a potentially important role in the methylation of genes regulating inflammation.42 This may be an important area for future investigation, because it could yield more helpful findings on the molecular mechanisms leading to dysplasia. These biomarkers could also be more useful in earlier stages of the disease. Most of the studies that show some sort of risk stratification do not discuss the progression of BE into low grade dysplasia and further progression into higher grades of dysplasia. These inflammatory mechanisms may hold the key to finding the likelihood of cancer at much earlier stages.
While there are a number of studies focusing on the upregulation and downregulation of miRNAs in EAC and BE, there are limited studies that aim to determine the target genes of these miRNA molecules. The problem with determining target genes is that there are too many targets to be able to realistically pinpoint the cellular mechanism that leads to neoplasia. For example, a study by Wang and colleagues found the two most significant miRNA markers wheremiR-21 was upregulated and miR-203 was downregulated.37 MiRNA profiles of BE and EAC are more similar to each other than to the profiles of any squamous epithelial tissue. Perhaps this makes sense, since EAC arises from columnar epithelium, rather than squamous epithelium.43
Although miRNAs do have the potential to be useful biomarkers, with the ease of accessibility, their function is not fully understood since current studies have only characterized expression levels. It is important to perform more studies that examine the target genes of these molecules. However, this is a challenging goal, since some miRNAs have over 100 target genes. This is critical from a treatment standpoint, because altering the levels of miRNAs can have large global effects. At the same time, the large number of target genes poses a problem in identifying specific genes that are affected.
CONCLUSION
Overall, significant research has been done in hypermethylation, and these studies may show the best biomarkers for the future, but prospective studies would provide stronger evidence for correlation. Histone acetylation seems to play an important role in esophageal squamous cell carcinoma, and may show good results in EAC. Studies on miRNAs is promising, but more work needs to be done on the effect of gene function. While presence of, or lack thereof, many miRNA molecules have been found, but, without any link to the function of these miRNAs. Studies on lncRNA are very limited, and there is not enough information to support whether it will be a useful biomarker to predict the development of EAC. An interesting area for future directions would be to examine the relative levels of hypermethylation and miRNA and compare these to the response of the patients to neoadjuvant therapy – if there is a difference in these levels, the method of neoadjuvant therapy may need to be accordingly adjusted.
Acknowledgments
This work was supported by research grants from the LB692 State of Nebraska to SKM and grants from the National Institutes of Health, USA to DK Agrawal.
Footnotes
The nomenclature of miRNA is included as found in the original studies. Hsa- prefix indicates that the miRNA is of human origin; mir- refers to an immature transcript, compared to a mature (miR) transcript.11 The number refers to order in which these molecules were discovered (i.e. miR-21 was the 21stmiRNA molecule to be discovered).11 A letter after the number indicates an identical sequence found from a different part of the genome.11 Also, since miRNAs are processed from miRNA hairpins, multiple miRNAs may be obtained from a single pre-transcript, so 5p or 3p is included to indicate whether the molecule came from the 3′ or 5′ end.11 Finally, an asterisk indicates that this particular miRNA is found in lower levels than another transcript processed from the same pre-miRNA without an asterisk.11
DISCLAIMER
The content of this review is solely the responsibility of the authors and does not necessarily represent the official views of the State of Nebraska or NIH.
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
- 1.Kaz AM, et al. DNA methylation profiling in Barrett’s esophagus and esophageal adenocarcinoma reveals unique methylation signatures and molecular subclasses. Epigenetics. 2011;6:1403–12. doi: 10.4161/epi.6.12.18199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Jonge PJF, van Blankenstein M, Grady WM, Kuipers EJ. Barrett’s oesophagus: epidemiology, cancer risk and implications for management. Gut. 2014;63:191–202. doi: 10.1136/gutjnl-2013-305490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abedi-Ardekani Behnoush, Hainaut P. Cancers of the Upper Gastro-intestinal Tract: A Review of Somatic Mutation Distributions. Arch Iran Med. 2014;17:286–292. [PubMed] [Google Scholar]
- 4.Reid BJ, Li X, Vaughan T. Barrett’s oesophagus and oesophageal adenocarcinoma: time for a new synthesis The natural history of Barrett’s oesophagus Epidemiology and etiology Oesophageal adenocarcinoma. Nat Rev Cancer. 2011;10:87–101. doi: 10.1038/nrc2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chak A, et al. Assessment of Familiality, Obesity, and Other Risk Factors for Early Age of Cancer Diagnosis in Adenocarcinomas of the Esophagus and Gastro-esophageal Junction Amitabh. Am J Gastroenterol. 2009;104:1913–1921. doi: 10.1038/ajg.2009.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agarwal A, Polineni R, Hussein Z, Vigoda I, Bhagat TD. Role of epigenetic alterations in the pathogenesis of Barrett’s esophagus and esophageal adenocarcinoma. Int J Clin Exp Pathol. 2012;5:382–396. [PMC free article] [PubMed] [Google Scholar]
- 7.Maley CC, Reid BJ. Natural selection in neoplastic progression of Barrett’s esophagus. Semin Cancer Biol. 2005;15:474–83. doi: 10.1016/j.semcancer.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Kaz AM, Grady WM. Epigenetic biomarkers in esophageal cancer. Cancer Lett. 2014;342:193–9. doi: 10.1016/j.canlet.2012.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chak A, et al. Variation in age at cancer diagnosis in familial versus non familial Barrett’s esophagus. Cancer Epidemiol Biomarkers Prev. 2012;21:376–83. doi: 10.1158/1055-9965.EPI-11-0927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conteduca V, et al. Barrett’s esophagus and esophageal cancer: an overview. Int J Oncol. 2012;41:414–24. doi: 10.3892/ijo.2012.1481. [DOI] [PubMed] [Google Scholar]
- 11.Trowbridge RM, Pittelkow MR. Epigenetics in the pathogenesis and pathophysiology of psoriasis vulgaris. J Drugs Dermatol. 2014;13:111–8. [PubMed] [Google Scholar]
- 12.Baumann S, et al. The prognostic impact of O6-Methylguanine-DNA Methyltransferase (MGMT) promotor hypermethylation in esophageal adenocarcinoma. Int J Cancer. 2006;119:264–8. doi: 10.1002/ijc.21848. [DOI] [PubMed] [Google Scholar]
- 13.Zhang L, et al. Inactivation of DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation and its relation to p53 mutations in esophageal squamous cell carcinoma. Carcinogenesis. 2003;24:1039–44. doi: 10.1093/carcin/bgg062. [DOI] [PubMed] [Google Scholar]
- 14.Su Y, et al. Malignant progression in O6-methylguanine-DNA methyltransferase-deficient esophageal cancer cells is associated with Ezrin protein. DNA Cell Biol. 2012;31:856–66. doi: 10.1089/dna.2011.1318. [DOI] [PubMed] [Google Scholar]
- 15.Zhao SL, Zhu ST, Hao X, Li P, Zhang ST. Effects of DNA methyltransferase 1 inhibition on esophageal squamous cell carcinoma. Dis Esophagus. 2011;24:601–10. doi: 10.1111/j.1442-2050.2011.01199.x. [DOI] [PubMed] [Google Scholar]
- 16.Baba Y, Watanabe M, Baba H. Review of the alterations in DNA methylation in esophageal squamous cell carcinoma. Surg Today. 2013;43:1355–64. doi: 10.1007/s00595-012-0451-y. [DOI] [PubMed] [Google Scholar]
- 17.Peng D, et al. Location-specific epigenetic regulation of the metallothionein 3 gene in esophageal adenocarcinomas. PLoS One. 2011;6:e22009. doi: 10.1371/journal.pone.0022009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwong J, et al. Epigenetic Inactivation of the Deleted in Lung and Esophageal Cancer 1 Gene in Nasopharyngeal Carcinoma. Genes Chromosomes Cancer. 2007;46:171–180. doi: 10.1002/gcc.20398. [DOI] [PubMed] [Google Scholar]
- 19.Clemons Nicholas J, Koh Shze Yung, Phillips WA. Advances in Understanding the Pathogenesis of Barrett’s Esophagus. Discov Med. 2014;17:7–14. [PubMed] [Google Scholar]
- 20.Ali I, et al. Dickkopf homologs in squamous mucosa of esophagitis patients are overexpressed compared with Barrett’s patients and healthy controls. Am J Gastroenterol. 2006;101:1437–48. doi: 10.1111/j.1572-0241.2006.00584.x. [DOI] [PubMed] [Google Scholar]
- 21.Trowbridge R, Kizer RT, Mittal SK, Agrawal DK. 1,25-dihydroxyvitamin D in the pathogenesis of Barrett’s esophagus and esophageal adenocarcinoma. Expert Rev Clin Immunol. 2013;9:517–533. doi: 10.1586/eci.13.38. [DOI] [PubMed] [Google Scholar]
- 22.Hong J, Li D, Wands J, Souza R, Cao W. Role of NADPH oxidase NOX5-S, NF-κB, and DNMT1 in acid-induced p16 hypermethylation in Barrett’s cells. Am J Physiol Cell Physiol. 2013;305:C1069–79. doi: 10.1152/ajpcell.00080.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wild Christopher P, Hardie LJ. Molecular events associated with the development of Barrett’s oesophagus and oesophageal adenocarcinoma. Nat Rev Cancer. 2003;3:676–684. doi: 10.1038/nrc1166. [DOI] [PubMed] [Google Scholar]
- 24.David S, Meltzer SJ. MicroRNA involvement in esophageal carcinogenesis. Curr Opin Pharmacol. 2011;11:612–6. doi: 10.1016/j.coph.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meng Y, et al. Epigenetic inactivation of the SFRP1 gene in esophageal squamous cell carcinoma. Dig Dis Sci. 2011;56:3195–203. doi: 10.1007/s10620-011-1734-7. [DOI] [PubMed] [Google Scholar]
- 26.Fang Y, et al. Cellular origins and molecular mechanisms of Barrett’s esophagus and esophageal adenocarcinoma. Ann N Y Acad Sci. 2013;1300:187–99. doi: 10.1111/nyas.12249. [DOI] [PubMed] [Google Scholar]
- 27.Tsai H, et al. The Chemokine Receptor CXCR2 Controls Positioning of Oligodendrocyte Precursors in Developing Spinal Cord by Arresting Their Migration of Cleveland. Cell. 2002;110:373–383. doi: 10.1016/s0092-8674(02)00838-3. [DOI] [PubMed] [Google Scholar]
- 28.Makita K, et al. Cdx2 expression and its promoter methylation during metaplasia-dysplasia-carcinoma sequence in Barrett’s esophagus. World J Gastroenterol. 2013;19:536–41. doi: 10.3748/wjg.v19.i4.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toh Y, Egashira A, Yamamoto M. Epigenetic alterations and their clinical implications in esophageal squamous cell carcinoma. Gen Thorac Cardiovasc Surg. 2013;61:262–9. doi: 10.1007/s11748-013-0235-3. [DOI] [PubMed] [Google Scholar]
- 30.Seder CW, Hartojo W, Lin L, Silvers AL. INHBA Overexpression Promotes Cell Proliferation and May Be Epigenetically Regulated in Esophageal Adenocarcinoma. J Thorac Oncol. 2009;4:455–462. doi: 10.1097/JTO.0b013e31819c791a. [DOI] [PubMed] [Google Scholar]
- 31.Matsuzaki Juntaro, Suzuki H. MicroRNA expression profiling in human Barrett’s carcinogenesis. Int J Cancer. 2011;129:1661–70. doi: 10.1002/ijc.25823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maru DM, et al. MicroRNA-196a is a potential marker of progression during Barrett’s metaplasia-dysplasia-invasive adenocarcinoma sequence in esophagus. Am J Pathol. 2009;174:1940–8. doi: 10.2353/ajpath.2009.080718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuzaki J, et al. Bile acids increase levels of microRNAs 221 and 222, leading to degradation of CDX2 during esophageal carcinogenesis. Gastroenterology. 2013;145:1300–11. doi: 10.1053/j.gastro.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 34.Wu X, et al. MicroRNA expression signatures during malignant progression from Barrett’s esophagus to esophageal adenocarcinoma. Cancer Prev Res (Phila) 2013;6:196–205. doi: 10.1158/1940-6207.CAPR-12-0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Soest EM, Dieleman JP, Siersema PD, Sturkenboom MCJM, Kuipers EJ. Increasing incidence of Barrett’s oesophagus in the general population. Gut. 2005;54:1062–6. doi: 10.1136/gut.2004.063685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang J, et al. MicroRNAs as oncogenes or tumour suppressors in oesophageal cancer: potential biomarkers and therapeutic targets. Cell Prolif. 2014 doi: 10.1111/cpr.12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang H Bin, Jiang ZB, Li M. Research on the Typical miRNA and Target Genes in Squamous Cell Carcinoma and Adenocarcinoma of Esophagus Cancer with DNA Microarray. Pathol Oncol Res. 2014:245–252. doi: 10.1007/s12253-013-9688-z. [DOI] [PubMed] [Google Scholar]
- 38.Brosius J. Waste not, want not – transcript excess in multicellular eukaryotes. TRENDS Genet. 2005;21:287–288. doi: 10.1016/j.tig.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 39.Wu W, et al. Hypomethylation of Noncoding DNA Regions and Overexpression of the Long Noncoding RNA, AFAP1-AS1, in Barrett’s Esophagus and Esophageal Adenocarcinoma Wenjing. Gastroenterology. 2013;144:956–966. doi: 10.1053/j.gastro.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eads CA, et al. Epigenetic Patterns in the Progression of Esophageal Adenocarcinoma Epigenetic Patterns in the Progression of Esophageal Adenocarcinoma 1. Cancer Res. 2001;61:3410–3418. [PubMed] [Google Scholar]
- 41.Shah AK, Saunders Na, Barbour AP, Hill MM. Early diagnostic biomarkers for esophageal adenocarcinoma--the current state of play. Cancer Epidemiol Biomarkers Prev. 2013;22:1185–209. doi: 10.1158/1055-9965.EPI-12-1415. [DOI] [PubMed] [Google Scholar]
- 42.Yang Zhengduo, Guan Baoxiang, Men Taoyan, Fujimoto Junya, Xu X. Comparable Molecular Alterations in 4-Nitroquinoline 1-Oxide- induced Oral and Esophageal Cancer in Mice and in Human Esophageal Cancer, Associated with Poor Prognosis of Patients. In Vivo (Brooklyn) 2013;27:473–484. [PMC free article] [PubMed] [Google Scholar]
- 43.Feber A, et al. MicroRNA expression profiles of esophageal cancer. J Thorac Cardiovasc Surg. 2008;135:255–60. doi: 10.1016/j.jtcvs.2007.08.055. discussion 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li JS, et al. Review TSGs Silenced by Promoter Methylation in ESCC We briefly summarized the epigenetically silenced Cell cycle control genes. Chin J Cancer. 2013;32:3–11. [Google Scholar]
- 45.Herceg Z, Hainaut P. Genetic and epigenetic alterations as biomarkers for cancer detection, diagnosis and prognosis. Mol Oncol. 2007;1:26–41. doi: 10.1016/j.molonc.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaz AM, Luo Y, Grady WM. Aberrantly methylated PKP1 in the progression of Barrett ’ s esophagus to esophageal adenocarcinoma Materials and Methods Tissue samples and cell lines. Genes Chromosomes Cancer. 2011;51:384–393. doi: 10.1002/gcc.21923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soutto M, et al. Epigenetic and genetic silencing of CHFR in esophageal adenocarcinomas. Cancer. 2010;116:4033–42. doi: 10.1002/cncr.25151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith E, Ruszkiewicz aR, Jamieson GG, Drew Pa. IGFBP7 is associated with poor prognosis in oesophageal adenocarcinoma and is regulated by promoter DNA methylation. Br J Cancer. 2014;110:775–82. doi: 10.1038/bjc.2013.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guo M, et al. Accumulation of promoter methylation suggests epigenetic progression in squamous cell carcinoma of the esophagus. Clin Cancer Res. 2006;12:4515–22. doi: 10.1158/1078-0432.CCR-05-2858. [DOI] [PubMed] [Google Scholar]
- 50.Zhang XM, Guo MZ. The value of epigenetic markers in esophageal cancer. Front Med China. 2010;4:378–84. doi: 10.1007/s11684-010-0230-3. [DOI] [PubMed] [Google Scholar]
- 51.Kuroki T, Trapasso F, Yendamuri S. Allele Loss and Promoter Hypermethylation of VHL, RAR - β, RASSF1A, and FHIT Tumor Suppressor Genes on Chromosome 3p in Esophageal Squamous Cell Carcinoma Tumor Suppressor Genes on Chromosome 3p in Esophageal Squamous. Cancer Res. 2003;63:3724–3728. [PubMed] [Google Scholar]
- 52.Talukdar FR, Ghosh SK, Laskar RS, Mondal R. Epigenetic, genetic and environmental interactions in esophageal squamous cell carcinoma from northeast India. PLoS One. 2013;8:e60996. doi: 10.1371/journal.pone.0060996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sato F, et al. Three-tiered risk stratification model to predict progression in Barrett’s esophagus using epigenetic and clinical features. PLoS One. 2008;3:e1890. doi: 10.1371/journal.pone.0001890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang Y, et al. MiR-136 promotes apoptosis of glioma cells by targeting AEG-1 and Bcl-2. FEBS Lett. 2012;586:3608–12. doi: 10.1016/j.febslet.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 55.Fu C, et al. The expression of miR-21 and miR-375 predict prognosis of esophageal cancer. Biochem Biophys Res Commun. 2014;446:1197–203. doi: 10.1016/j.bbrc.2014.03.087. [DOI] [PubMed] [Google Scholar]
- 56.Saad R, et al. Deciphering the unique microRNA signature in human esophageal adenocarcinoma. PLoS One. 2013;8:1–11. doi: 10.1371/journal.pone.0064463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu Y, et al. Prognostic significance of differentially expressed miRNAs in esophageal cancer. Int J Cancer. 2011;128:132–43. doi: 10.1002/ijc.25330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang T, et al. The oncogenetic role of microRNA-31 as a potential biomarker in oesophageal squamous cell carcinoma. Clin Sci (Lond) 2011;121:437–47. doi: 10.1042/CS20110207. [DOI] [PubMed] [Google Scholar]
- 59.Sakai NS, Samia-Aly E, Barbera M, Fitzgerald RC. A review of the current understanding and clinical utility of miRNAs in esophageal cancer. Semin Cancer Biol. 2013;23:512–21. doi: 10.1016/j.semcancer.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 60.Kan T, Meltzer SJ. MicroRNAs in Barrett’s esophagus and esophageal adenocarcinoma. Curr Opin Pharmacol. 2009;9:727–32. doi: 10.1016/j.coph.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Streppel MM, et al. MicroRNA 223 is upregulated in the multistep progression of Barrett’s esophagus and modulates sensitivity to chemotherapy by targeting PARP1. Clin Cancer Res. 2013;19:4067–78. doi: 10.1158/1078-0432.CCR-13-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fassan M, et al. MicroRNA expression profiling in human Barrett’s carcinogenesis. Int J Cancer. 2011;129:1661–70. doi: 10.1002/ijc.25823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee KH, et al. MicroRNA-373 (miR-373) post-transcriptionally regulates large tumor suppressor, homolog 2 (LATS2) and stimulates proliferation in human esophageal cancer. Exp Cell Res. 2009;315:2529–38. doi: 10.1016/j.yexcr.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 64.Li S, et al. miR-223 regulates migration and invasion by targeting Artemin in human esophageal carcinoma. J Biomed Sci. 2011;18:24. doi: 10.1186/1423-0127-18-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang P, et al. MicroRNA 23b regulates autophagy associated with radioresistance of pancreatic cancer cells. Gastroenterology. 2013;145:1133–1143. e12. doi: 10.1053/j.gastro.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 66.Banerjee R, et al. The tumor suppressor gene rap1GAP is silenced by miR-101-mediated EZH2 overexpression in invasive squamous cell carcinoma. Oncogene. 2011;30:4339–49. doi: 10.1038/onc.2011.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guo J-X, et al. miR-181b as a potential molecular target for anticancer therapy of gastric neoplasms. Asian Pac J Cancer Prev. 2012;13:2263–7. doi: 10.7314/apjcp.2012.13.5.2263. [DOI] [PubMed] [Google Scholar]
- 68.Smith CM, et al. miR-200 family expression is downregulated upon neoplastic progression of Barrett’s esophagus. World J Gastroenterol. 2011;17:1036–44. doi: 10.3748/wjg.v17.i8.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun J, et al. MicroRNA-99a/100 promotes apoptosis by targeting mTOR in human esophageal squamous cell carcinoma. Med Oncol. 2013;30:411. doi: 10.1007/s12032-012-0411-9. [DOI] [PubMed] [Google Scholar]
- 70.Meister J, Schmidt MHH. miR-126 and miR-126*: new players in cancer. Sci World J. 2010;10:2090–100. doi: 10.1100/tsw.2010.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ke Y, Zhao W, Xiong J, Cao R. miR-149 Inhibits Non-Small-Cell Lung Cancer Cells EMT by Targeting FOXM1. Biochem Res Int. 2013;2013:506731. doi: 10.1155/2013/506731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sakurai K, et al. MicroRNAs miR-199a-5p and -3p target the Brm subunit of SWI/SNF to generate a double-negative feedback loop in a variety of human cancers. Cancer Res. 2011;71:1680–9. doi: 10.1158/0008-5472.CAN-10-2345. [DOI] [PubMed] [Google Scholar]
- 73.Cho WCS. MicroRNAs in cancer - from research to therapy. Biochim Biophys Acta. 2010;1805:209–17. doi: 10.1016/j.bbcan.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 74.Spechler SJ. Clinical Practice: Barrett’s Esophagus. N Engl J Med. 2002;346:836–842. doi: 10.1056/NEJMcp012118. [DOI] [PubMed] [Google Scholar]
- 75.Kerkhof M, et al. Grading of dysplasia in Barrett’s oesophagus: substantial interobserver variation between general and gastrointestinal pathologists. Histopathology. 2007;50:920–7. doi: 10.1111/j.1365-2559.2007.02706.x. [DOI] [PubMed] [Google Scholar]
- 76.Shi HY, Zhu SC, Shen WB, Liu ML. Pathological characteristics of esophageal cancer. Oncol Lett. 2014;8:533–538. doi: 10.3892/ol.2014.2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tolone S, et al. The patterns of reflux can affect regression of non-dysplastic and low-grade dysplastic Barrett. s esophagus after medical and surgical treatment: a prospective case-control study. Surg Endosc. 2014 doi: 10.1007/s00464-014-3713-5. [DOI] [PubMed] [Google Scholar]
- 78.Kumar Vinay, Abbas Abul, Fausto N. Robbins and Cotran Pathologic Basis of Disease. Elsevier, Inc; 2005. pp. 808–809. [Google Scholar]
- 79.Wadhwa R, et al. Gastric cancer-molecular and clinical dimensions. Nat Rev Clin Oncol. 2013;10:643–55. doi: 10.1038/nrclinonc.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]