Abstract
Natural killer cells are controlled by peptide selective inhibitory receptors for MHC class I, including the killer cell immunoglobulin-like receptors (KIRs). Despite having similar ligands, KIR2DL2 and KIR2DL3 confer different levels of protection to infectious disease. To investigate how changes in peptide repertoire may differentially affect NK cell reactivity, NK cells from KIR2DL2 and KIR2DL3 homozygous donors were tested for activity against different combinations of strong inhibitory (VAPWNSFAL), weak inhibitory (VAPWNSRAL) and antagonist peptide (VAPWNSDAL). KIR2DL3-positive NK cells were more sensitive to changes in the peptide content of MHC class I than KIR2DL2-positive NK cells. These differences were observed for the weakly inhibitory peptide VAPWNSRAL in single peptide and double peptide experiments (p<0.01 and p<0.03 respectively). More significant differences were observed in experiments using all three peptides (p<0.0001). Mathematical modeling of the experimental data demonstrated that VAPWNSRAL was dominant over VAPWNSFAL in distinguishing KIR2DL3- from KIR2DL2-positive donors. Donors with different KIR genotypes have different responses to changes in the peptide bound by MHC class I. Differences in the response to the peptide content of MHC class I may be one mechanism underlying the protective effects of different KIR genes against infectious disease.
Keywords: Natural killer cells, killer-cell immunoglobulin-like receptors, MHC class I, peptide, peptide selectivity
Introduction
Natural killer (NK) cells are innate lymphocytes involved in the immune response to viruses and cancer, either through direct interaction with target cells or through interactions with macrophages, dendritic cells and T cells [1, 2]. They express activating and inhibitory receptors on the cell surface and signals transduced from these receptors are integrated to determine whether or not an individual NK cell is activated [3]. Whilst the activating receptors are derived from a number of different gene families and have diverse ligands, the dominant inhibitory receptors expressed on NK cells are either from the killer cell immunoglobulin-like receptor (KIR) family or the C-type lectin-like receptor CD94:NKG2A and have MHC class I ligands [4, 5]. Thus NK cell responses can be fine-tuned by interactions with many different ligands including MHC class I.
A number of immunogenetic studies have highlighted the contribution that the KIR:MHC system makes to human disease including viral infections, pregnancy-associated disorders, autoimmune disease and cancer [6–8]. The HLA-C specific KIR are implicated in the immune response to diseases such as hepatitis C virus (HCV), HIV, malaria and psoriatic arthropathy [9–14]. For instance, in HCV infection, KIR2DL3 in combination with its HLA-C group 1 ligands is protective, whereas KIR2DL2, which also has group 1 HLA-C allotypes as ligands, is not [15–17]. Binding studies have shown that KIR2DL2 is a higher affinity receptor for group 1 HLA-C allotypes than KIR2DL3, suggesting that KIR2DL3-positive NK cells are more easily activated than their KIR2DL2-positive counterparts. However, in functional experiments, no difference has been observed in the inhibition of KIR2DL2- and KIR2DL3-positive NK cells by group 1 HLA-C alleles [18, 19]. Binding of both KIR2DL2 and KIR2DL3 to MHC class I is dependent on the peptide bound by HLA-C [20–22]. However, this is a broad specificity defined by residues 7 and 8 of the bound peptide as distinct from the fine specificity of the T-cell receptor. Recent work has demonstrated an unexpected sensitivity of NK cells to combinations of peptides presented by MHC Class I [23, 24]. Thus, when the peptide VAPWNSDAL is presented by HLA-Cw*0102 it does not alter the reactivity of KIR2DL2- and KIR2DL3-positive NK cells. However, it can significantly reduce the inhibition due to a strong inhibitory peptide VAPWNSFAL in a process termed “peptide antagonism”. This implies that NK cells may be sensitive to changes in peptide repertoire and, based on the immunogenetic findings, suggests that KIR2DL2- and KIR2DL3-positive NK cells may respond differently to these changes.
We originally screened a panel of P7 and P8 variants of VAPWNSLSL, a naturally processed HLA-Cw*0102 epitope [23, 25]. We identified three classes of peptides, strong inhibitory peptides, weak inhibitory peptides and non-inhibitory/antagonist peptides [23]. We postulate that these represent the major classes of peptides presented by MHC class I that can influence NK cell reactivity. We have used peptides representative of these different categories to investigate how NK cells from donors with different genetic backgrounds (KIR2DL3 homozygotes and KIR2DL2 homozygotes) may respond to changes in the peptide content of MHC class I.
Results
A weak inhibitory peptide discriminates KIR2DL2-poisitve NK cells from KIR2DL3-positive NK cells
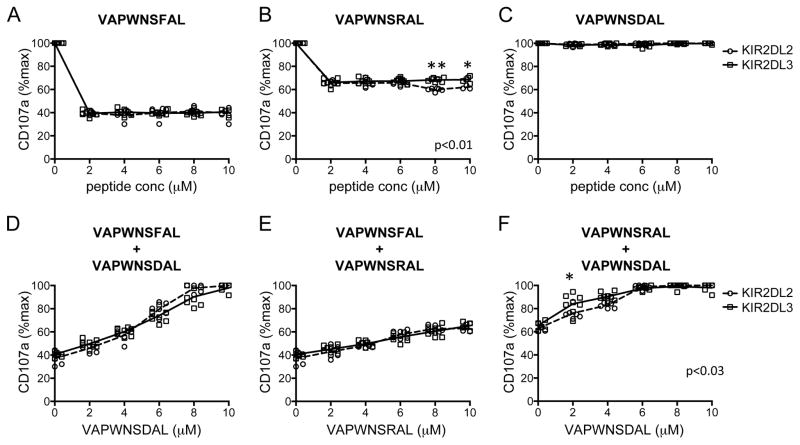
We have used the 721.174 cell line to study the interaction of KIR with group 1 HLA-C. This cell line expresses HLA-C*0102, but as it is deficient in TAP, the HLA on the cell surface is expressed with low affinity peptide, and can therefore be exogenously loaded with peptide. Using this cell line for a peptide screen we identified VAPWNSFAL (VAP-FA) as a high affinity ligand for KIR, VAPWNSDAL (VAP-DA) as a low affinity ligand and VAPWNSRAL (VAP-RA) as having an intermediate affinity for both KIR2DL2 and KIR2DL3 (Fig. 1A–C) [23]. The peptides have similar affinities for HLA-Cw*0102 the allele naturally expressed on 721.174 cells (Fig. 1A and Supporting Information Fig 1). This binding correlates well with function in that VAP-FA is strongly inhibitory, VAP-RA is weakly inhibitory and VAP-DA is non-inhibitory for both KIR2DL2 and KIR2DL3 homozygous donors (Fig. 1D–F). To investigate whether different donors had different responses to single peptides, 721.174 cells were loaded with VAP-FA, VAP-RA or VAP-DA and used as targets for CD158b+ NK cells from eight KIR2DL3 homozygous and six KIR2DL2 homozygous donors in degranulation assays. VAP-FA and VAP-RA inhibited CD158b-positive NK cells from both sets of donors at all peptide concentrations (Fig. 2A–C). This is consistent with our previous data showing that in a single peptide model, inhibition is determined by levels of MHC class I expression, and this starts to decline significantly only at levels of less than 2 μM peptide. However, there was a difference in the level of inhibition between the two donor groups for the weakly inhibitory VAP-RA peptide (p=0.01). This was more evident at higher levels of VAP-RA in which NK cells from the KIR2DL2-positive donors were marginally more inhibited than those from KIR2DL3-positive donors (8 μM VAP-RA [p<0.001] or 10 μM VAP-RA [0.1>p>0.05]) (Fig. 2B). Double peptide experiments were conducted using a final peptide concentration of 10 μM and varying the relative proportions of the two peptides. Consistent with our previous data VAP-DA behaved as a peptide antagonist in that it reduced inhibition due to VAP-FA, despite not binding to KIR2DL2-Fc or KIR2DL3-Fc, or inhibiting KIR2DL2- or KIR2DL3-positive NK cells. For instance, at 4 μM VAP-FA alone CD107a expression was approximately 40% of maximum in the absence of VAP-DA (Fig. 2A), but in its presence (VAP-DA 6 μM) CD107a expression was 80% (Fig. 2D). Overall CD158b-positive NK cells from both KIR2DL2- and KIR2DL3-positive donors were inhibited at similar levels for the VAP-FA/VAP-DA and VAP-FA/VAP-RA ratios tested (Fig. 2D and E). However, we observed small but significantly increased inhibition of NK cells from the KIR2DL2-positive donors for VAP-RA/VAP-DA mixtures overall (p<0.03 (two way ANOVA)) (Fig. 2F). Thus, similar to the single peptide experiments significant differences between the donors were seen with the weak inhibitory peptide VAP-RA, rather than the strongly inhibitory peptide VAP-FA.
Figure 1.
Correlation of KIR binding with NK cell inhibition in KIR2DL2+ and KIR2DL3+ donors. (A) Stabilization of HLA-Cw*0102 on 721.174 by the indicated VAPWNSLSL derivative peptides and a control peptide (VMAPRTLFL) at saturating concentrations 10 μM (filled histograms). The no primary Ab control is indicated in each plot and the titration of the peptides is shown in the far right panel. Binding of (B) KIR2DL2 or (C) KIR2DL3 fusion constructs to 721.174 cells in the presence (filled lines) or absence of the indicated peptide at a concentration of 10 μM. Representative histograms (n=3 samples) are shown. Summaries of the mean fluorescence intensities (MFI) of KIR binding pooled from two independent experiments are shown in the far right panels (**p<0.01, ***p<0.001; Student’s t-test). Gating strategy and degranulation assays of CD158b-positive CD3-CD56+ NK cells from (D) KIR2DL2 homozygous or (E) KIR2DL3 homozygous donors in response to 721.174 cells incubated with the indicated peptides at a final concentration of 10 μM. Representative histograms of six 2DL2 and eight 2DL3 donors, analyzed in duplicate, are shown.
Figure 2.
Comparison of inhibition of CD158b+ NK cells from KIR2DL2+ and KIR2DL3+ donors by individual peptides and double peptide combinations. 721.174 cells were incubated with the indicated peptides and used as target cells in degranulation assays for CD158b-positive NK cells from eight KIR2DL3 homozygous donors and six KIR2DL2 homozygous donors. The individual responses of KIR2DL2 (circles) and KIR2DL3 (squares) donor NK cells to (A) VAP-FA, (B) VAP-RA and (C) VAP-DA are shown. The dashed line connects the mean of the results from the KIR2DL2 homozygous donors and the full line the KIR2DL3 donors. (D–F) Responses to pairs of peptide are shown with the final concentration of peptides in all mixes being 10 μM. The concentration of each of the peptides is indicated on each x-axis. Data show the mean value for each individual donor, and are from one experiment performed in duplicate. P values for the differences between the donors as determined by ANOVA are indicated. For comparisons at individual peptide concentration *0.05<p<0.1 and **p<0.001. Degranulation data are normalized to 721.174 with no peptide.
Differences between KIR2DL2- and KIR2DL3-positive NK cells are greater at lower levels of inhibition
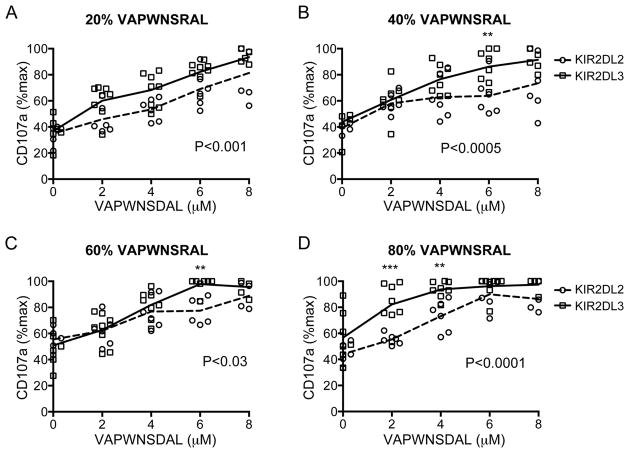
In order to explore the effects of changing class I-bound peptide in more detail we devised experiments using peptides of all three categories: strongly inhibitory, weakly inhibitory, and antagonistic. These experiments were conducted using fixed ratios of weak:strong inhibitory peptides (VAP-RA:VAP-FA) and then gradually increasing the amount of antagonistic VAP-DA in the mix (Supporting Information Table 1). All experiments were performed at a final peptide concentration of 10 μM. Overall there was a highly significant difference between the two groups of donors as determined by two way ANOVA (p<0.0001) (Fig. 3). For each ratio of VAP-RA:VAP-FA overall there were statistically significant differences in the change in inhibition of CD158b-positive NK cells induced by the addition of VAP-DA between the KIR2DL2 and KIR2DL3 homozygotes (Fig. 3). However, the patterns varied with VAP-RA:VAP-FA ratio. When the strongly inhibitory peptide predominated (20%:80% VAP-RA:VAP-FA) there were small, but individually insignificant differences induced by the antagonist between the donor groups at all VAP-RA:VAP-FA ratios (Fig. 3A). For 40%:60% and 60%:40% VAP-RA:VAP-FA the differences were more evident at higher concentrations of VAP-DA, and reached statistical significance at 6 μM VAP-DA (p<0.05 and p<0.01 respectively) (Fig. 3B and C). When the weakly inhibitory peptide predominated (VAP-RA:VAP-FA, 80%:20%), differences induced by the antagonist were more evident at low concentrations of VAP-DA and were significant at both 2 μM and 4 μM (p<0.001 and p<0.01 respectively) (Fig. 3D). Degranulation saturates at higher concentrations of VAP-DA therefore differences are no longer apparent. Thus, in general, differences between the donors were observed at lower levels of inhibition. Analyzing the subgroup of donors with ligands for KIR2DL2 and KIR2DL3 (group 1 HLA-C positive) gave similar results to the overall cohort (Supporting Information Fig. 2).
Figure 3.
Comparison of inhibition of CD158b+ NK cells from KIR2DL2+ and KIR2DL3+ donors to different combinations of VAP-FA, VAP-DA and VAP-RA. (A–D) 721.174 cells were incubated with combinations of three peptides as shown in Table 1 and used as target cells in degranulation assays for CD158b-positive NK cells from KIR2DL2 (circles) and KIR2DL3 (squares) homozygous donors. The mean CD107a expression levels normalised to no peptide are connected by a dashed line (KIR2DL2 homozygous donors) or a full line (KIR2DL3 homozygous donors). The ratios of VAP-RA to VAP-FA were: (A) 20% VAP-RA:80% VAP-FA, (B) 40% VAP-RA:60% VAP-FA, (C) 60% VAP-RA:40% VAP-FA and (D) 80% VAP-RA:20% VAP-FA. In all experiments, the VAP-RA:VAP-FA ratio was kept constant but the total quantity was varied in combination with VAP-DA in order to maintain a total peptide concentration of 10 μM in the peptide mix (Supporting Information Table 1). Data show the mean value for each individual donor, and are from one experiment performed in duplicate. P values for the differences between the donors as determined by ANOVA are shown. For comparisons at individual peptide concentrations *p<0.05, **p<0.01, ***p<0.001.
Response to changes in peptide repertoires discriminates KIR2DL2 from KIR2DL3-positive donors
In order to describe these data quantitatively, we constructed a regression model. The model assumes that the degranulation (A), quantified by CD107a in a population of NK cells, varies with concentrations of FA, DA, and RA peptides as,
| (1) |
First we evaluated the parameters in the model in the absence of any cubic order term, i.e., kDFR = 0. We also set kDA=0 as DA minimally affects activation when NK cells are stimulated with DA peptide alone. When searching for parameter values we restricted the search to only positive values as the peptides have been shown to induce inhibition in single peptide experiments (kF, kD, kR) and antagonize inhibition in two and three peptide experiments (kDF, kFR, kRD, kDFR). The model in Eq. (1) agreed slightly better with the data from KIR2DL3 donors compared with the KIR2DL2 donors. This is reflected in the larger values of χ2 (Table 1) produced for the KIR2DL2 data.
Table 1.
Parameter values obtained by minimizing χ2
| Parameter values | KIR2DL2 donor | KIR2DL3 donor |
|---|---|---|
| kF | 6.4 | 6.05 |
| kD | 0 | 0 |
| kR | 5.27 | 2.36 |
| kDF | 0.003 | 0.0007 |
| kFR | 0.018 | 0.0016 |
| kRD | 0.125 | 0.54 |
| kDFR | 0 | 0 |
| χ2 | 12.6 | 8 |
From the parameter values in Table 1, the [FA]-[DA] and [FA]-[RA] pairs appear to have less impact in inducing activation compared with the [DA]-[RA] pair in the triple peptide experiments. The qualitative features of the model did not change when we added the cubic term in estimating the model parameters. Thus the [RA]-[DA] pair seemed to exert more influence in activation compared with the other two pairs.
The fitted function shows that KIR2DL3 expressing cells are activated more easily than the KIR2DL2 expressing cells (Fig. 4A). This is shown in Figure 4B in which the peptide combinations that produce more than 65% CD107a de-granulation are shown in red. This shows that activation stays below 65% (shown in green) for a greater fraction of peptide combinations for KIR2DL2-positive NK cells, compared with KIR2DL3-positive NK cells. Thus 12 out of 25 peptide combinations for KIR2DL2 donors and 7 out of 25 peptide combinations for KIR2DL3 donors yielded a degranulation value of less than 65%. The role of the [DA]-[RA] pair seems to be greater for KIR2DL3 (KRD=0.54) compared with KIR2DL2 (KRD=0.125). This is also consistent with the single peptide data in which KR for KIR2DL2 is approximately twice that for KIR2DL3. Based on the data in Table 1 the differences in inhibition between KIR2DL3 and KIR2DL2 donors can be given by:
Figure 4.
Regression modeling of experimental data from assays of inhibition using the triple mix peptide combinations. (A) Comparison between the model and the experiments. The activation determined by the model (gray surface) in Eq. (1) for the parameter values noted in Table 1 when the cubic term in Eq. (1) was set to zero. The points show the measured %CD107a degranulation averaged over multiple trails (n ≥ 3). The left panel shows the comparison for KIR2DL2 positive donors and the right panel for KIR2DL3 positive donors. (B) Comparison of the regression models for KIR2DL2 and KIR2DL3-positive donors. The peptide combinations determined by Eq. 1 that produce greater than 65% degranulation as compared with no peptide for KIR2DL2 donors (left panel) and KIR2DL3 donors (right panel). Degranulation of more than 65% is shown in red, and less than 65% is shown in green. Computer generated graphics from data in Figure 3 are shown.
Therefore the [RA] term is dominant over the [FA] term and the [RA][DA] term dominant over [FA][DA]. Thus these data imply that KIR2DL3-positive NK cells respond more readily to changes in peptide repertoire than KIR2DL2-positive NK cells, and the difference in inhibition between donors is determined by the concentrations of the weak agonist peptide and the weak agonist:antagonist peptide combination.
Discussion
We have used a model system to investigate how NK cells may respond to changes in peptide repertoire. We have assumed that there are four broad classes of peptide:MHC class I with respect to KIR: strong inhibitory, weak inhibitory, antagonistic and null, with null peptides not contributing to NK cell signaling. We, therefore, modeled changes in peptide repertoire using representatives of the three classes of peptide that could modulate changes in NK cell activity. It has previously been shown that CD107a degranulation correlates well with binding using KIR-Fc fusion constructs and that the selected peptides are representative of these groups, with VAP-FA being the strongest binder in our peptide screen, VAP-DA being one of the non-binders and VAP-RA being intermediate between the two [23]. Our experimental and modeling data indicate that there are differences in the way that CD158b-positive NK cells from donors with different KIR genotypes respond to changes in peptide repertoire.
It has been shown previously that KIR2DL2 has a greater affinity for HLA class I than KIR2DL3 due to a difference in the hinge angle between the two domains of the receptors [18, 19]. However, functional experiments did not detect differences in inhibition by group 1 HLA-C alleles between KIR2DL2 and KIR2DL3 positive NK cells. These experiments were performed using transfectant cell lines expressing single HLA-C alleles, which are strongly inhibitory, and this is consistent with our observations that VAP-FA is not discriminatory between the two donor genotypes. Our data show that functional differences are more subtle and related to the peptides presented by MHC class I. Thus, in single peptide experiments both sets of donors made similar responses to VAP-DA and VAP-FA, but not to VAP-RA. In the triple mix experiments, the ratio of VAP-RA to VAP-DA appeared to be the most discriminatory between the donors, but in the mathematical analysis the concentration of VAP-RA appeared dominant over the combination of the two.
The observed differences between the donors are small. However, given that KIR2DL2 and KIR2DL3 differ on average by only 4 amino acids in the D1 and D2 domains this is not surprising. Additionally, large immunogenetic studies have been required to define the protective effects of KIR against infection, arguing against individual KIR having dramatic effects and demonstrating the subtly of functional polymorphisms within the KIR system.
Our data show that in this model system changes in peptide repertoire lead more readily to activation of KIR2DL3-positve NK cells than KIR2DL2-positive NK cells. Testing of these data with additional peptide:HLA combinations would assist in establishing the broad applicability of these findings. Furthermore, the in vivo significance of this work needs further exploration. However, changes in peptide repertoire may occur during viral infection or tumorigenesis through the presentation of viral antigens, tumor antigens or self-peptides [26]. The formation of the MHC class I peptide repertoire is complex and appears to depend on a combination of the degradation of mature proteins, and also recently synthesized misfolded proteins (defective ribosomal products “DRiPs”) [27, 28]. Viral infection can readily change the MHC class I peptide repertoire either by presentation of viral peptides, but also by changing the self-peptides presented by MHC class I. Moreover, the induction of the immunoproteosome by proinflammatory signals during an active immune response is likely to significantly alter the peptide landscape presented by MHC when compared with non-inflammatory conditions. At an early time point during vaccinia infection, nearly 50% of the peptide repertoire of MHC class I is composed of viral peptides [29]. Therefore, changes in peptide repertoire during viral infection can be substantial and, based on our data, potentially sufficient to activate an NK cell. Peptide antagonism is a mechanism that permits detection of changes in peptide repertoire and this would be more readily accomplished in the presence of weaker inhibitory peptides. Based on our peptide binding screen, the subset constituting the inhibitory peptides form a continuum of which VAP-RA is at the weaker KIR-binding and VAP-FA at the stronger KIR-binding ends. Thus, although other peptides may have subtly different effects the peptide combinations tested broadly represent changes in MHC class I peptide repertoire.
The modeling data provide a template for how the inhibition of NK cells may change in response to the peptide content of MHC class I and the better fit of the model in Eq. (1) with the data for KIR2DL3 compared with KIR2DL2, could indicate the presence of additional factors, that modulate NK cell function for only KIR2DL2 donors. One such factor could be KIR2DS2, which is also bound by the CD158b antibody used in this study. KIR2DS2 is in strong linkage disequilibrium with KIR2DL2, therefore all our KIR2DL2 donors also had this activating receptor. It has recently been shown that HLA-A11 is a ligand for KIR2DS2 and that KIR2DS2-positive NK cell clones can recognize HLA-Cw*0304 [30, 31]. However, we were unable to confirm or refute a role for KIR2DS2 as excluding these individuals meant that numbers were too small for statistical analysis. However, the affinity of KIR2DS2 for HLA class I is much lower than for KIR2DL2 [22] and in our experiments NK cells from the KIR2DL2 donors were inhibited to a greater extent than those from the KIR2DL3 donors. Taken together, this suggests that the effect of KIR2DS2 in these experiments is likely to be small, if any.
KIR2DL3, but not KIR2DL2 has been shown to be protective against viral infection such as hepatitis C virus and also against fulminant malaria. It is thought that the selection for the KIR A haplotype, containing KIR2DL3, is related to defense against pathogens. Our data suggest that fine tuning of NK cell activity by MHC class I peptide may discriminate KIR2DL3, from KIR2DL2, positive donors in a manner that allows marginally greater reactivity of KIR2DL3 positive NK cells. These subtle differences may be important for understanding how KIR contribute to the genetic susceptibility to infectious disease.
Materials and methods
Peptide stabilization assay
The HLA-Cw*0102-positive, TAP deficient 721.174 cell line was cultured in RPMI medium 1640 supplemented with 1% penicillin/streptomycin (Invitrogen, Paisley, UK) and 10% fetal calf serum (FCS) (Lonza). For peptide stabilization, 2 × 105 721.174 cells were incubated with or without synthetic peptides (GL Biochem, Shanghai Ltd, China) overnight at 26°C. After 16 h, cells were washed and stained with a primary HLA-Cw*0102-specific antibody VP6G3 for 1 h at room temperature, followed by a secondary FITC-conjugated goat anti-human IgM antibody (AbD Serotec) for 30 min at room temperature. Cells were then washed and fixed in 1% PFA/FCS. Cells were analyzed by flow cytometry (BD Biosciences, Oxford, UK). The mean intensity of fluorescence of 10 000 events was collected.
KIR2DL2/3-Fc binding assay
KIR2D2L2-IgG and KIR2DL3-IgG fusion constructs (KIR2DL2-Fc & KIR2DL3-Fc Chimera; R&D Systems) were conjugated with protein A Alexa Fluor 488 (Invitrogen) at a molar ratio of 6:1. 1 × 106 721.174 cells were incubated with 10 μM peptide as described above and then stained with KIR-Fc. After fixation in 4% (w/v) paraformaldehyde cells were analyzed by flow cytometry.
CD107a NK Cell Degranulation Assay
Human peripheral blood mononuclear cells (PBMC) were isolated from the blood of 14 healthy donors using Hypaque-Ficoll (GE Healthcare, Amersham, UK) density centrifugation, with informed consent and full ethical approval (REC ref 06/Q1701/120). 3 × 105 PBMCs were stimulated overnight with 1 ng/mL recombinant human IL-15. Peptide pulsed 721.174 targets were prepared as for the stabilization assays. Target cells were resuspended with PBMCs at an effector-to-target (E:T) ratio of 5:1 in fresh R10 medium containing peptide and anti-CD107a-Alexa-fluor-647 antibody (eBioscience, Hatfield, UK). Cells were incubated for 1 h at 26°C, then 6 μg/mL Golgi-Stop™ (BD Biosciences) was added, and incubated for a further 3 h at 26°C. Cells were washed in wash buffer (1% BSA/0.1% NaN3) and blocked with blocking buffer (10% human serum in PBS) for 30 min and then stained with the following antibodies: anti-CD3-PerCP (Biolegend, San Diego, US), anti-human CD56-PE and anti-human CD158b-FITC (both BD Biosciences). Cells were fixed in 1% PFA and analyzed by flow cytometry. Individual assays for each donor were performed once in duplicate and the mean value used for subsequent analysis.
KIR and HLA Genotyping
All donors were typed for KIR using a quantitative PCR comparative Ct method [3232]. Donors were typed for HLA-B and HLA-C at the Anthony Nolan Institute (London, UK) using Luminex xMAP® technology
Statistics
These were calculated using GraphPad Prism by two-way ANOVA to compare KIR2DL2 and KIR2DL3 homozygous donors, with a post Student’s t-test and Bonferroni correction to compare individual data points.
Data Modeling
We constructed a regression model using the experimental data. The model assumes that the degranulation (A), quantified by CD107a in a population of NK cells, varies with concentrations of FA, DA, and RA peptides as:
| (1) |
In the above expression, [x] denotes the concentration of a peptide, x; kF, kD, kR, kDF, kFR, kRD, and kDFR are model parameters that are evaluated by fitting the above expression to the data characterizing NK cell activation stimulated by mixtures of the three peptides. The terms linear in the concentrations in Eq. (1) describe inhibition of NK cell activation when NK cells are stimulated individually by the peptides. The terms proportional to the product of two peptide concentrations describe the antagonistic effect induced by the peptides. The last term describes the effect of a triple peptide stimulation that cannot be captured by pair-wise stimulations of the peptides. NK cell activation is assumed to be 100% in the absence of any DA, FA or RA peptide. We determine the parameters in Eq. (1) by fitting the expression with the data from the three-peptide mixture experiments. Then we use Eq. (1) to address two principal issues:
To quantify differences between the data generated from the KIR2DL2 and the KIR2DL3 donors in terms of the parameter values.
To quantify differences between the ability of the NK cells from the KIR2DL2 and the KIR2DL3 donors to respond when the peptide mixtures stimulate the cells.
We calculated the parameters in Eq. (1) by minimizing χ2 as defined below. χ2 quantifies the difference between the model and the experimental data.
| (2) |
where, i refers to an experiment with any mixture of the three peptides, FA, DA, and RA. Aiexpt denotes the average value of the % of CD107a degranulation measured over multiple trials (n>3) for a particular mixture of peptides indexed by i. σiexpt denotes the variance in the measured % of CD107a degranulation in the trials done in the ith experiment. In the experiments, the peptide combinations were chosen such that the total concentration of the peptides was held fixed at 10 μM, i.e., [FA]+[DA]+[RA]=10 μM. Aimodel represents the % of CD107 degranulation calculated from Eq. (1) for the peptide concentrations corresponding to the ith experiment. We chose the parameters that minimized χ2 in Eq. (2). The minimization was carried out by the routine lsqcurvefit in MATLAB.
Supplementary Material
Acknowledgments
This work was supported by NIH grant 1R56AI090115-01A1 to JD and grants WT089883MA, WT076991MA and WT093465MA from The Wellcome Trust to SC, SIK and MAP.
Abbreviations
- KIR
Killer cell immunoglobulin-like receptors
- VAP-FA
VAPWNSFAL
- VAP-RA
VAPWNSRAL
- VAP-DA
VAPWNSDAL
Footnotes
Conflict of interest
The authors declare no commercial or financial conflict of interest.
References
- 1.Crome SQ, Lang PA, Lang KS, Ohashi PS. Natural killer cells regulate diverse T cell responses. Trends Immunol. 2013;34:342–349. doi: 10.1016/j.it.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 2.Moretta A. The dialogue between human natural killer cells and dendritic cells. Curr Opin Immunol. 2005;17:306–311. doi: 10.1016/j.coi.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 3.Long EO, Kim HS, Liu D, Peterson ME, Rajagopalan S. Controlling natural killer cell responses: integration of signals for activation and inhibition. Annu Rev Immunol. 2013;31:227–258. doi: 10.1146/annurev-immunol-020711-075005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol. 2008;9:495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long EO. Negative signaling by inhibitory receptors: the NK cell paradigm. Immunol Rev. 2008;224:70–84. doi: 10.1111/j.1600-065X.2008.00660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parham P, Moffett A. Variable NK cell receptors and their MHC class I ligands in immunity, reproduction and human evolution. Nat Rev Immunol. 2013;13:133–144. doi: 10.1038/nri3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kulkarni S, Martin MP, Carrington M. The Yin and Yang of HLA and KIR in human disease. Semin Immunol. 2008;20:343–352. doi: 10.1016/j.smim.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jamil KM, Khakoo SI. KIR/HLA interactions and pathogen immunity. J Biomed Biotechnol. 2011;2011:298348. doi: 10.1155/2011/298348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahlenstiel G, Martin MP, Gao X, Carrington M, Rehermann B. Distinct KIR/HLA compound genotypes affect the kinetics of human antiviral natural killer cell responses. J Clin Invest. 2008;118:1017–1026. doi: 10.1172/JCI32400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moesta AK, Parham P. Diverse functionality among human NK cell receptors for the C1 epitope of HLA-C: KIR2DS2, KIR2DL2, and KIR2DL3. Front Immunol. 2012;3:336. doi: 10.3389/fimmu.2012.00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alter G, Altfeld M. NK cell function in HIV-1 infection. Curr Mol Med. 2006;6:621–629. doi: 10.2174/156652406778195035. [DOI] [PubMed] [Google Scholar]
- 12.Tiemessen CT, Paximadis M, Minevich G, Winchester R, Shalekoff S, Gray GE, Sherman GG, Coovadia AH, Kuhn L. Natural killer cell responses to HIV-1 peptides are associated with more activating KIR genes and HLA-C genes of the C1 allotype. J Acquir Immune Defic Syndr. 2011;57:181–189. doi: 10.1097/QAI.0b013e3182174a76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirayasu K, Ohashi J, Kashiwase K, Hananantachai H, Naka I, Ogawa A, Takanashi M, Satake M, Nakajima K, Parham P, Arase H, Tokunaga K, Patarapotikul J, Yabe T. Significant Association of KIR2DL3-HLA-C1 Combination with Cerebral Malaria and Implications for Co-evolution of KIR and HLA. PLoS Pathog. 2012;8:e1002565. doi: 10.1371/journal.ppat.1002565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin MP, Nelson G, Lee JH, Pellett F, Gao X, Wade J, Wilson MJ, Trowsdale J, Gladman D, Carrington M. Cutting edge: susceptibility to psoriatic arthritis: influence of activating killer Ig-like receptor genes in the absence of specific HLA-C alleles. J Immunol. 2002;169:2818–2822. doi: 10.4049/jimmunol.169.6.2818. [DOI] [PubMed] [Google Scholar]
- 15.Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, Cheng J, Goedert JJ, Vlahov D, Hilgartner M, Cox S, Little AM, Alexander GJ, Cramp ME, O’Brien SJ, Rosenberg WM, Thomas DL, Carrington M. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 16.Romero V, Azocar J, Zuniga J, Clavijo OP, Terreros D, Gu X, Husain Z, Chung RT, Amos C, Yunis EJ. Interaction of NK inhibitory receptor genes with HLA-C and MHC class II alleles in Hepatitis C virus infection outcome. Mol Immunol. 2008;45:2429–2436. doi: 10.1016/j.molimm.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vidal-Castineira JR, Lopez-Vazquez A, Diaz-Pena R, Alonso-Arias R, Martinez-Borra J, Perez R, Fernandez-Suarez J, Melon S, Prieto J, Rodrigo L, Lopez-Larrea C. Effect of killer immunoglobulin-like receptors in the response to combined treatment in patients with chronic hepatitis C virus infection. J Virol. 2010;84:475–481. doi: 10.1128/JVI.01285-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moesta AK, Norman PJ, Yawata M, Yawata N, Gleimer M, Parham P. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J Immunol. 2008;180:3969–3979. doi: 10.4049/jimmunol.180.6.3969. [DOI] [PubMed] [Google Scholar]
- 19.Winter CC, Gumperz JE, Parham P, Long EO, Wagtmann N. Direct binding and functional transfer of NK cell inhibitory receptors reveal novel patterns of HLA-C allotype recognition. J Immunol. 1998;161:571–577. [PubMed] [Google Scholar]
- 20.Boyington JC, Motyka SA, Schuck P, Brooks AG, Sun PD. Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand. Nature. 2000;405:537–543. doi: 10.1038/35014520. [DOI] [PubMed] [Google Scholar]
- 21.Maenaka K, Juji T, Nakayama T, Wyer JR, Gao GF, Maenaka T, Zaccai NR, Kikuchi A, Yabe T, Tokunaga K, Tadokoro K, Stuart DI, Jones EY, van der Merwe PA. Killer cell immunoglobulin receptors and T cell receptors bind peptide-major histocompatibility complex class I with distinct thermodynamic and kinetic properties. J Biol Chem. 1999;274:28329–28334. doi: 10.1074/jbc.274.40.28329. [DOI] [PubMed] [Google Scholar]
- 22.Vales-Gomez M, Reyburn HT, Erskine RA, Strominger J. Differential binding to HLA-C of p50-activating and p58-inhibitory natural killer cell receptors. Proc Natl Acad Sci U S A. 1998;95:14326–14331. doi: 10.1073/pnas.95.24.14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fadda L, Borhis G, Ahmed P, Cheent K, Pageon SV, Cazaly A, Stathopoulos S, Middleton D, Mulder A, Claas FH, Elliott T, Davis DM, Purbhoo MA, Khakoo SI. Peptide antagonism as a mechanism for NK cell activation. Proc Natl Acad Sci U S A. 2010;107:10160–10165. doi: 10.1073/pnas.0913745107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borhis G, Ahmed PS, Mbiribindi B, Naiyer MM, Davis DM, Purbhoo MA, Khakoo SI. A Peptide Antagonist Disrupts NK Cell Inhibitory Synapse Formation. J Immunol. 2013;190:2924–30. doi: 10.4049/jimmunol.1201032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barber LD, Percival L, Valiante NM, Chen L, Lee C, Gumperz JE, Phillips JH, Lanier LL, Bigge JC, Parekh RB, Parham P. The inter-locus recombinant HLA-B*4601 has high selectivity in peptide binding and functions characteristic of HLA-C. J Exp Med. 1996;184:735–740. doi: 10.1084/jem.184.2.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yewdell JW, Reits E, Neefjes J. Making sense of mass destruction: quantitating MHC class I antigen presentation. Nat Rev Immunol. 2003;3:952–961. doi: 10.1038/nri1250. [DOI] [PubMed] [Google Scholar]
- 27.Schubert U, Anton LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- 28.Yewdell JW. DRiPs solidify: progress in understanding endogenous MHC class I antigen processing. Trends Immunol. 2011;32:548–558. doi: 10.1016/j.it.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Croft NP, Smith SA, Wong YC, Tan CT, Dudek NL, Flesch IE, Lin LC, Tscharke DC, Purcell AW. Kinetics of antigen expression and epitope presentation during virus infection. PLoS Pathog. 2013;9:e1003129. doi: 10.1371/journal.ppat.1003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J, Xiao Z, Ko HL, Shen M, Ren EC. Activating killer cell immunoglobulin-like receptor 2DS2 binds to HLA–A*11. Proc Natl Acad Sci U S A. 2014;111:2662–2667. doi: 10.1073/pnas.1322052111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.David G, Djaoud Z, Willem C, Legrand N, Rettman P, Gagne K, Cesbron A, Retiere C. Large spectrum of HLA-C recognition by killer Ig-like receptor (KIR)2DL2 and KIR2DL3 and restricted C1 SPECIFICITY of KIR2DS2: dominant impact of KIR2DL2/KIR2DS2 on KIR2D NK cell repertoire formation. J Immunol. 2013;191:4778–4788. doi: 10.4049/jimmunol.1301580. [DOI] [PubMed] [Google Scholar]
- 32.Jiang W, Johnson C, Jayaraman J, Simecek N, Noble J, Moffatt MF, Cookson WO, Trowsdale J, Traherne JA. Copy number variation leads to considerable diversity for B but not A haplotypes of the human KIR genes encoding NK cell receptors. Genome Res. 2012;22:1845–1854. doi: 10.1101/gr.137976.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.