Abstract
Acute myeloid leukemia (AML) is a clonal malignant disorder derived from a small number of leukemic stem cells (LSCs). MLL gene rearrangements are found in AML associated with poor prognosis. The upregulation of Hox genes is critical for LSC induction and maintenance, but is unlikely to support malignancy and the high LSC frequency observed in MLL leukemias. The present study shows that MLL fusion proteins interact with the transcription factor PU.1 to activate the transcription of CSF-1R, which is critical for LSC activity. AML is cured by either deletion of PU.1, or ablation of cells expressing CSF-1R. Kinase inhibitors specific for CSF-1R prolong survival time. These findings indicate that PU.1-mediated upregulation of CSF-1R is a critical effector of MLL leukemogenesis.
Keywords: Acute Myeloid Leukemia, CSF-1R, MLL, Spi-1, Stem Cells
Introduction
Acute myeloid leukemia (AML) is a clonal malignant disorder derived from a small number of leukemic stem cells (LSCs)1, 2. LSCs are capable of the limitless self-renewal that is necessary for cancer initiation and maintenance. Conventional chemotherapies are often effective in reducing the total number of leukemia cells, but are not curative in many cases of AML. Since LSCs are often resistant to conventional chemotherapies, residual LSCs are a potential cause of AML relapse. Thus, eradication of LSCs is critical to cure the disease.
Chromosome translocations that involve the mixed lineage leukemia gene (MLL) are frequently observed in human AML and often predict a poor prognosis3–6. Over 60 genes have been identified as MLL fusion partners to date; chromosome rearrangements such as t(9;11), t(11;19), and t(10;11), which express MLL-AF9, MLL-ELL, and MLL-AF10, respectively, are commonly associated with AML5. MLL fusion proteins transform non-self-renewing myeloid progenitors into LSCs7, 8. AML with MLL rearrangements consistently express HOX genes such as HOXA7, HOXA9, and MEIS19–11. The upregulation of Hox genes is critical for LSC induction and maintenance, but does not recapitulate the entire phenotype and biology of MLL leukemias12–15. Moreover it is unlikely to support malignancy and the high LSC levels observed in MLL leukemias16. These facts suggest that unknown critical mediators of leukemogenesis exist.
The present study shows that the upregulation of macrophage colony-stimulating factor receptor (CSF-1R, also called M-CSFR/c-FMS/CD115) is critical for LSC activity in MLL leukemia. AML was cured upon eradication of cells expressing high levels of Csf-1r in mice. MLL fusions were found to regulate CSF-1R transcription through a novel mechanism involving interaction with the transcription factor PU.1. These findings indicate that PU.1-mediated upregulation of CSF-1R is a novel therapeutic target for MLL leukemias.
Materials and Methods
Mice
C57BL/6 mice were purchased from CREA Japan (Tokyo). NGF-FKBP-Fas transgenic mice17 (Jackson Lab.), CSF-1R-deficient mice18, PU.1-null/conditional deficient mice19, and CreERT2 mice (TaconicArtemis GmbH)20 were maintained on a C57BL/6 genetic background. Mouse experiments were performed in a specific pathogen-free environment at the National Cancer Center animal facility according to institutional guidelines and with approval of the National Cancer Center Animal Ethics Committee.
Generation of AML mouse models
MSCV-MLL-AF10-ires-GFP was transfected with PLAT-E21 cells using the FuGENE 6 reagent (Roche Diagnostics), and supernatants containing retrovirus were collected 48 h after transfection. The c-Kit+ cells (1 × 105 cells), which were selected from bone marrow (BM) or fetal liver cells using CD117 MicroBeads (Militenyi Biotec), were incubated with the retrovirus using RetroNectin (Takara Bio) for 24 h in StemPro-34 SFM medium (Invitrogen) containing cytokines (20 ng/mL SCF, 10 ng/mL IL-6, 10 ng/mL IL-3). The infectants were then transplanted together with BM cells (2 × 105) into lethally irradiated (9 Gy) 6- to 8-week-old C57BL/6 mice by intravenous (IV) injection. Secondary transplants were performed by intravenous injection of BM cells from the primary AML mice into sub-lethally irradiated (6 Gy) C57BL/6 mice.
Administration of AP20187, AraC, or Ki20227
AP20187 (gift from Ariad Pharmaceuticals; 10 mg/kg) was administered daily by IV injection for 5 d, and then 1 mg/kg AP20187 was administered every 3 d thereafter as described previously17. Ki2022722 (gift from KIRIN Pharma; 20 mg/kg) were orally administrated daily from 7 days after transplantation. AraC (75 mg/kg) was administered daily by IV injection for 5 days from 7 days after transplantation.
Immunofluorescent staining, flow cytometric analysis, and cell sorting
BM cells from AML mice were preincubated with rat IgG, and then incubated on ice with anti-CD115(CSF-1R)-PE (eBioscience) and anti- c-Kit-APC (2B8)-APC (BD Pharmingen). Flow cytometric analysis and cell sorting were performed using the cell sorter JSAN (Baybioscience), and the results were analyzed using FlowJo software (Tree Star).
Reporter analysis
Csf1r-luciferase constructs were generated by ligation of wild-type and PU.1-lacking Csf1r promoter23 with pGL4. For reporter analysis, SaOS2 cells were transfected with Csf1r-luc and phRL-CMV together with various expression constructs in 24-well plates, and luciferase activity was assayed 24 h after transfection using the microplate luminometer GLOMAX (Promega). Results of reporter assays represent the average values for relative luciferase activity generated from at least three independent experiments that were normalized using the activity of the enzyme from phRL-CMV as an internal control.
Immunoprecipitation and immunoblotting
For immunoprecipitation experiments, cells were lysed in a lysis buffer containing 250 mM NaCl, 20 mM sodium phosphate (pH 7.0), 30 mM sodium pyrophosphate, 10 mM NaF, 0.1% NP-40, 5 mM DTT, 1 mM PMSF, and protease inhibitor. Cell lysates were incubated with anti-FLAG antibody-conjugated agarose beads (Sigma) and gently rotated at 4°C overnight. The absorbed beads were washed 6 times with lysis buffer. Precipitated proteins were eluted from the beads by FLAG peptide and dissolved with the same volume of 2× SDS sample buffer. When immunoprecipitation was not performed, total protein lysates were prepared in 2× SDS sample buffer. Antibodies were detected by chemiluminescence using ECL plus Detection Reagents (Amersham Biosciences, Buckinghamshire, United Kingdom). The primary antibodies used in this study were anti-FLAG (M2) (Sigma), anti-HA (3F10) (Roche), and anti-MLL-N24 antibodies.
Statistical analyses
We performed unpaired two-tailed Student's t-tests for comparisons and a log-rank test for survival data using JMP8 software (SAS Institute).
Colony formation assays
Cells were cultured in 1% methylcellulose in Iscove’s modified Dulbecco’s medium (IMDM) containing 15% fetal bovine serum (FBS), 1% bovine serum albumin (BSA), 10 µg/mL rhInsulin, 200 µg/mL human transferrin, 100 µM 2-mercaptoethanol, 2mML-glutamine, and the following cytokines; 50 ng/mL rm SCF, 10 ng/mL rmIL-3, and 10 ng/mL rh IL-6; or 10 ng/mL mCSF-1. Cultures were maintained at 37°C under humidified conditions with 5% CO2. Colonies containing >50 cells were counted on day 5.
Results
Upregulation of CSF-1R is critical for MLL-AF10-induced AML
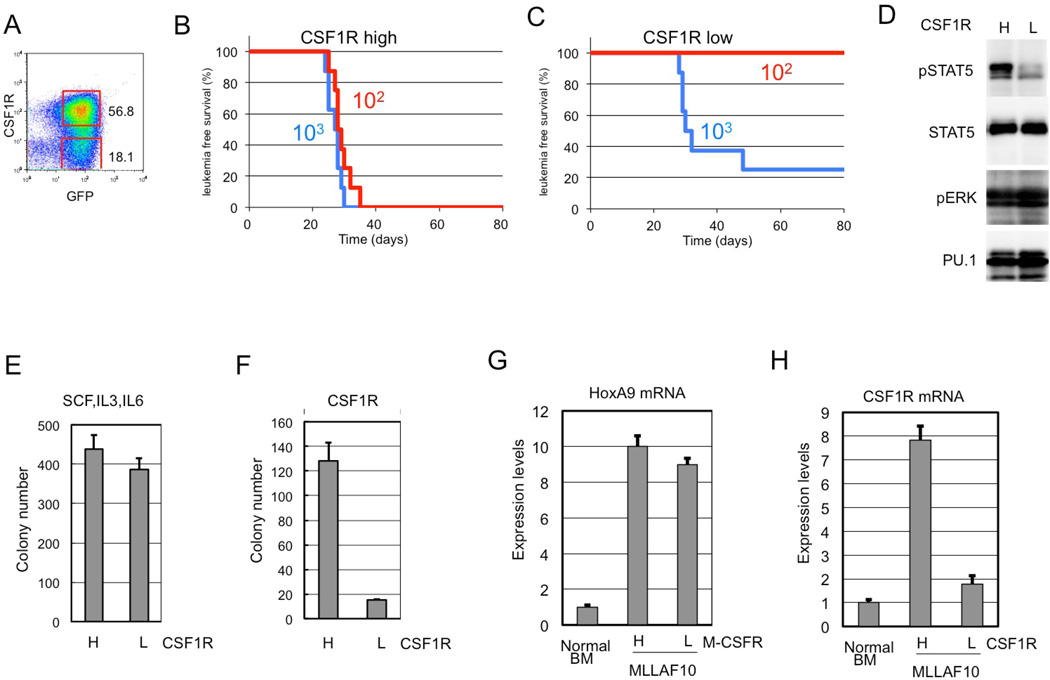
Previous results indicated that the expression of CSF-1R was high in MOZ-TIF2-induced AML25 and human AML26. Csf-1r expression was investigated in MLL-AF10-induced AML in mice. Results showed that Csf-1r expression was high in some AML cell populations (Figure 1A). To assess LSC activity, cells expressing high (Csf-1rhigh) and low (Csf-1rlow/−) levels of Csf-1r were purified and transplanted into irradiated mice. Transplantation of 102 flow-sorted Csf-1r high cells was sufficient to induce AML in all mice transplanted (Figure 1B). Conversely, no mice developed AML upon transplantation of 102 Csf-1rlow/− cells (Figure 1C). Thus, Csf-1rhigh cells displayed stronger LIC activity compared to Csf-1rlow/− cells in MLL-AF10-induced AML.
Figure 1. CSF-1Rhigh cells show potent leukemia-initiating activity.
(A) The bone marrow (BM) cells from MLL-AF10–induced AML mice were analyzed by flow cytometer for expression of GFP and Csf-1r. (B,C) Csf-1r high and Csf-1r low/− cells were sorted by flow cytometry, and the indicated numbers of flow-sorted CSF-1Rhigh (B) and Csf-1r low/− (C) cells were transplanted into sub-lethally irradiated mice, and leukemia-free survival was investigated. n = 8, P < 0.001. (D) Csf-1r high and Csf-1r low/− cells were analyzed for levels of total and phosphorylated Stat5, phosphorylated Erk, and Pu.1 (E,F). Csf-1rhigh and Csf-1rlow/− cells were analyzed for colony-forming activity in methylcellulose medium supplemented with IL3, SCF and IL6 (E) or with M-CSF (F). (G,H) Levels of HoxA9 (G) and Csf1r (H) mRNAs were measured in Csf-1r high and Csf-1r low/− cells prepared from AML mouse BM.
STAT5 and ERK, which are downstream effectors of CSF-1R, are activated in a variety of leukemias and myeloproliferative disorders. The phosphorylation status of these proteins was investigated in Csf-1rhigh and Csf-1rlow/− cells from MLL-AF10-induced AML mice by immunoblot analysis with phospho-specific anti-STAT5 and anti-ERK antibodies. Stat5 was highly phosphorylated in Csf-1rhigh cells but not in Csf-1rlow/− cells (Figure 1D), while Erk1/2 were phosphorylated in both Csf-1rhigh and Csf-1rlow/− cells. Further analyses are required to determine the role(s) of Stat5 during leukemogenesis.
Since MLL-AF10-induced leukemia cells can form colonies in methylcellulose27, flow-sorted Csf-1rhigh and Csf-1rlow/− cells were tested for colony formation in the presence of either M-CSF or multiple cytokines. Csf-1rhigh cells and Csf-1rlow/− formed equivalent numbers of colonies when stimulated with multiple cytokines (Figure 1E). However, Csf-1rlow/− cells showed reduced colony formation when stimulated with M-CSF alone (Figure 1F). Quantitative RT-PCR analysis showed that HoxA9 was upregulated in both Csf-1rhigh and Csf-1rlow/− cells (Figure 1G) and that Csf1r mRNA was appropriately differentially expressed (Figure 1H). Csf-1rhigh and Csf-1rlow/− cells were also observed in normal BM and fetal lever (Supplemental Figure 1). Population of Csf-1rhigh were reduced in Mll−/− Fetal liver cells, suggesting that Csf-1r expression is regulated by wild type Mll as well as by Mll-fusions.
MLL fusions activate CSF-1R transcription through interaction with PU.1
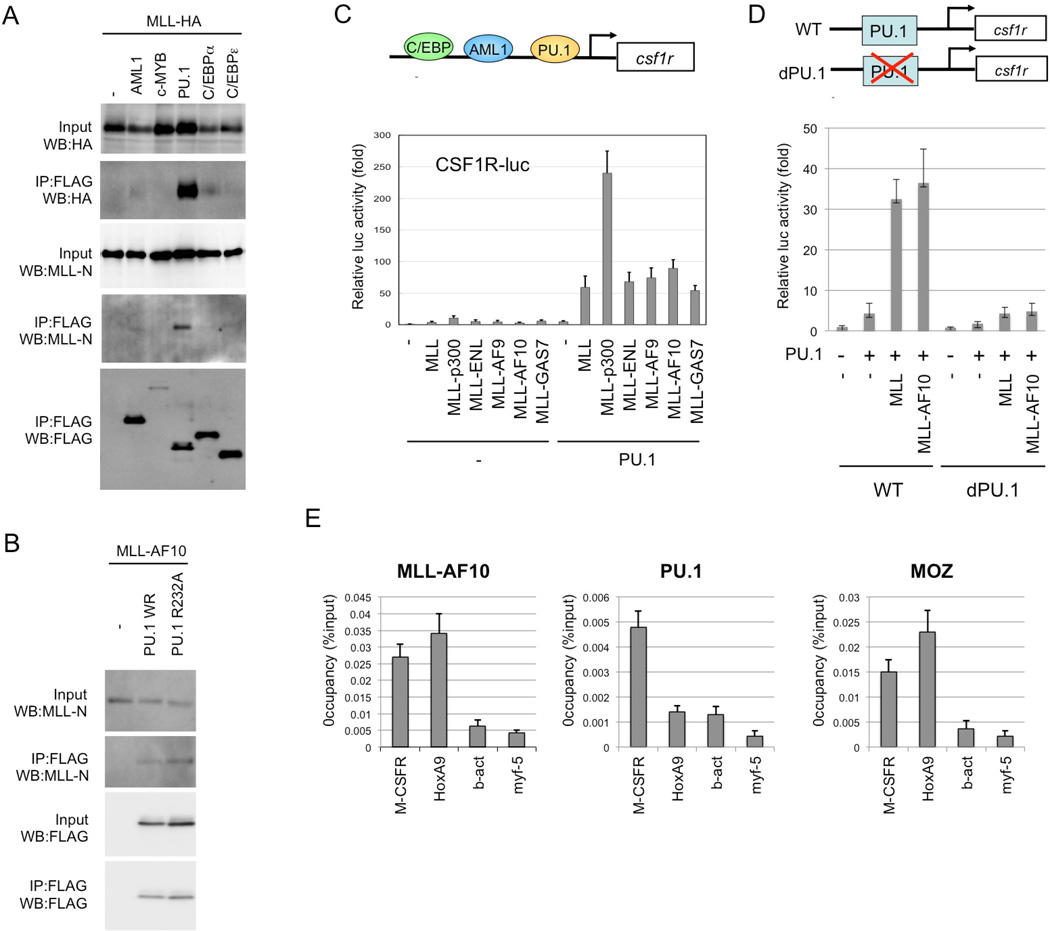
Monocyte-specific expression of CSF-1R is reportedly regulated by transcription factors such as AML1, PU.1, and C/EBP28. To investigate MLL-mediated regulation of CSF-1R transcription, the interaction of MLL with several hematopoietic transcription factors was tested. Results showed that MLL strongly interacts with PU.1 (Figure 2A). MLL-AF10 also interacted with PU.1 (Figure 2B). MLL and MLL fusions very strongly stimulated PU.1-dependent activation of the CSF-1R promoter (Figure 2C). Neither MLL nor MLLAF10 activated a CSF-1R promoter mutant lacking PU.1-binding sites (Figure 2D). Interaction of MLL with AML1/RUNX129 and other factors was less strong, and MLL and MLL fusions did not activate the CSF-1R promoter in the presence of AML1 or C/EBPα (data not shown). Chromatin immunoprecipitation (ChIP) analysis indicated that genomic localizations of MLL-AF10 and PU.1 on Csf-1r (Figure 2E). These results suggest that MLL and MLL fusion proteins interact with PU.1 to activate CSF-1R transcription.
Figure 2. PU.1-dependent upregulation of CSF-1R by MLL and MLL fusions.
(A) Interaction of MLL with PU.1. 293T cells were co-transfected with MLL-HA and the indicated FLAG-tagged transcription factors, including FLAG-PU.1. Anti-FLAG antibody immunoprecipitates (IP:FLAG) or cell lysates (Input) were subjected to immunoblotting with anti-HA, anti-MLL-N or anti-FLAG antibodies. (B) Interaction between MLL-AF10 and PU.1. 293T cells were co-transfected with MLL-AF10 and FLAG-tagged wild-type PU.1 or PU.1/FR232A. Anti-FLAG antibody immunoprecipitates (IP:FLAG) or cell lysates (Input) were subjected to immunoblotting with anti-MLL-N or anti-PU.1 antibodies. (C) Effects of MLL, and MLL fusions on PU.1-mediated Csf1r promoter-driven transcription. SaOS2 cells were co-transfected with the Csf1r–luciferase construct and the indicated effectors. Luciferase activity was analyzed 24 h after transfection. Error bars represent SD (n = 3). (D) PU.1 binding site-dependence of MLL enhancement of Csf1r promoter-driven transcription. SaOS2 cells were transfected with the wild-type Csf1r–luciferase construct or its mutant lacking the PU.1-binding site, together with the indicated effectors. (E) Chromatin immunoprecipitation (ChIP) of MLL-AF10 and PU.1. The BM cells from AML mice induced by Flag-MLL-AF10, were subjected to ChIP analysis using anti-Flag (MLL-AF10), anti-PU.1 and anti-MOZ antibodies. Semiquantitative real-time PCR was performed on the co-precipitated DNAs.
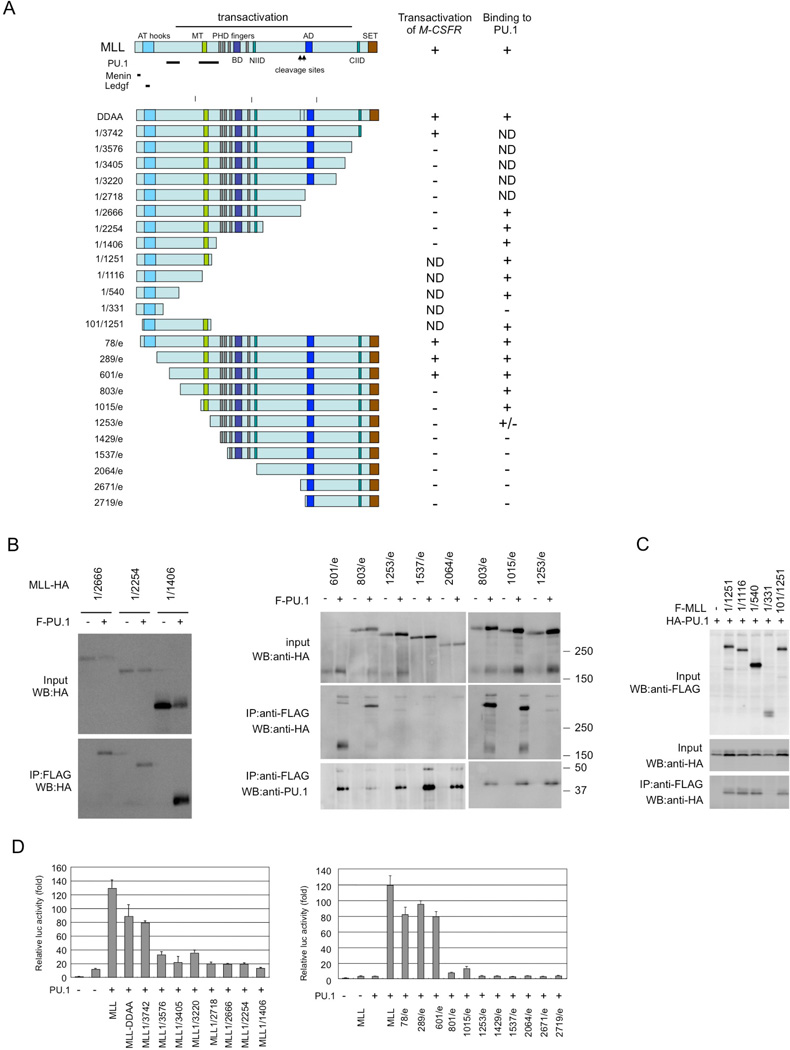
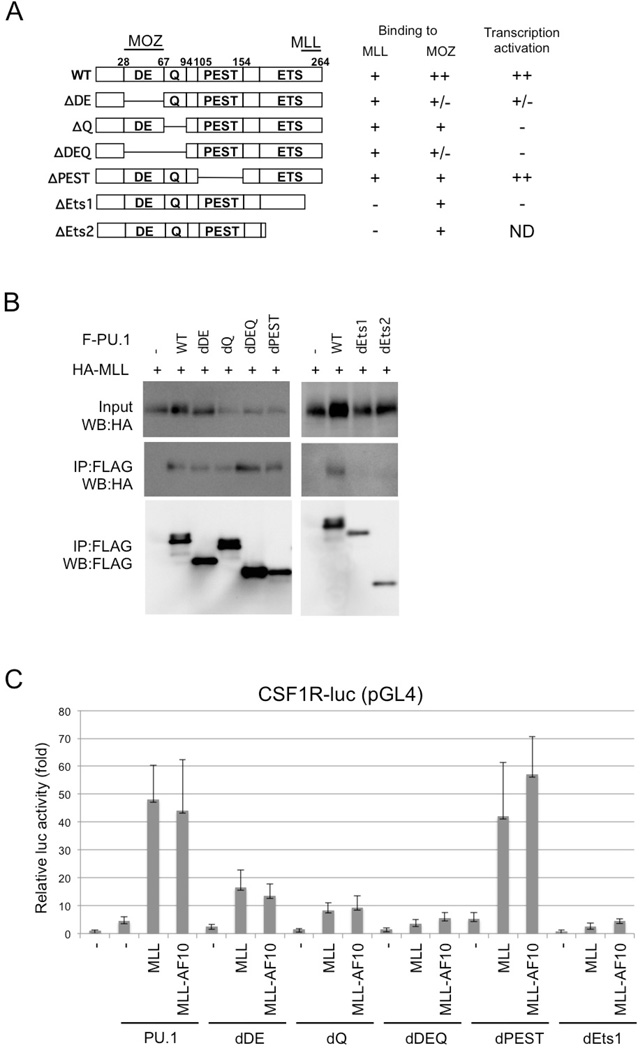
Immunoprecipitation analysis using MLL deletion mutants indicated that PU.1 interacts with at least two regions in the N-terminus of MLL (Figures 3A and S1). The menin and LEGDF-interacting domains30 and the C-terminal SET domain, which is needed for histone methyltransferase activity31, are not required for interaction with PU.1 (Figure3B and 3C) or the PU.1-dependent activation of CSF-1R by MLL (Figure 3D), suggesting that interaction with menin and LEGDF and histone methyltransferase activity are not required for MLL-mediated transactivation of CSF-1R. PU.1 deletion analysis indicated that the ETS domain of PU.1 was required for the interaction of PU.1 with MLL (Figures 4A and 4B). Since the ETS domain is a DNA-binding domain, it is possible that the interaction between MLL/MLL fusions and PU.1 is DNA-dependent. However, this seems unlikely because MLL-AF10 also interacted with PU.1/R232A, which lacks DNA-binding capacity (Figure 2B). Both the DEQ region and the ETS domain of PU.1 were required to activate PU.1-mediated transcription by MLL and MLL-AF10 (Figure 4C).
Figure 3. Functional domains of MLL required for interaction with PU.1 and for PU.1-mediated activation of Csf1r promoter.
(A) PU.1 binding and PU.1-mediated Csf1r promoter activity of MLL deletion mutants. The PU.1-, menin-, and LEDGF-interacting domains and the results for interaction with PU.1 and PU.1-mediated transactivation of Csf1r-luc are indicated. ND, not determined (B–C) Pu.1 binding. 293T cells were co-transfected with wild-type or mutants of HA-tagged MLL and FLAG-tagged PU.1 (B), or with wild-type or mutants of FLAG-tagged MLL and HA-tagged PU.1 (C). Anti-FLAG antibody immunoprecipitates (IP:FLAG) or cell lysates (Input) were subjected to immunoblotting with anti-HA or anti-FLAG antibodies. (D) PU.1-mediated Csf1r promoter-driven transcription. SaOS2 cells were transfected with the Csf1r–luciferase construct and PU.1, together with deletion mutants of MLL. Luciferase activity was analyzed 24 h after transfection. Error bars represent SD (n = 3).
Figure 4. Functional domains of PU.1.
(A) Deletion mutants of PU.1. The MOZ- and MLL-interacting domains and the results for interaction with PU.1 and PU.1-mediated transactivation of Csf1r-luc are indicated. ND, not determined (B) Interaction of PU.1 mutants with MLL. 293T cells were co-transfected with HA-tagged MLL and wild-type or mutants of FLAG-tagged PU.1. Anti-FLAG antibody immunoprecipitates (IP:FLAG) or cell lysates (Input) were subjected to immunoblotting with anti-HA or anti-FLAG antibodies. (C) SaOS2 cells were co-transfected with the Csf1r–luciferase construct and the indicated effectors. Luciferase activity was analyzed 24 h after transfection. Error bars represent SD (n = 3).
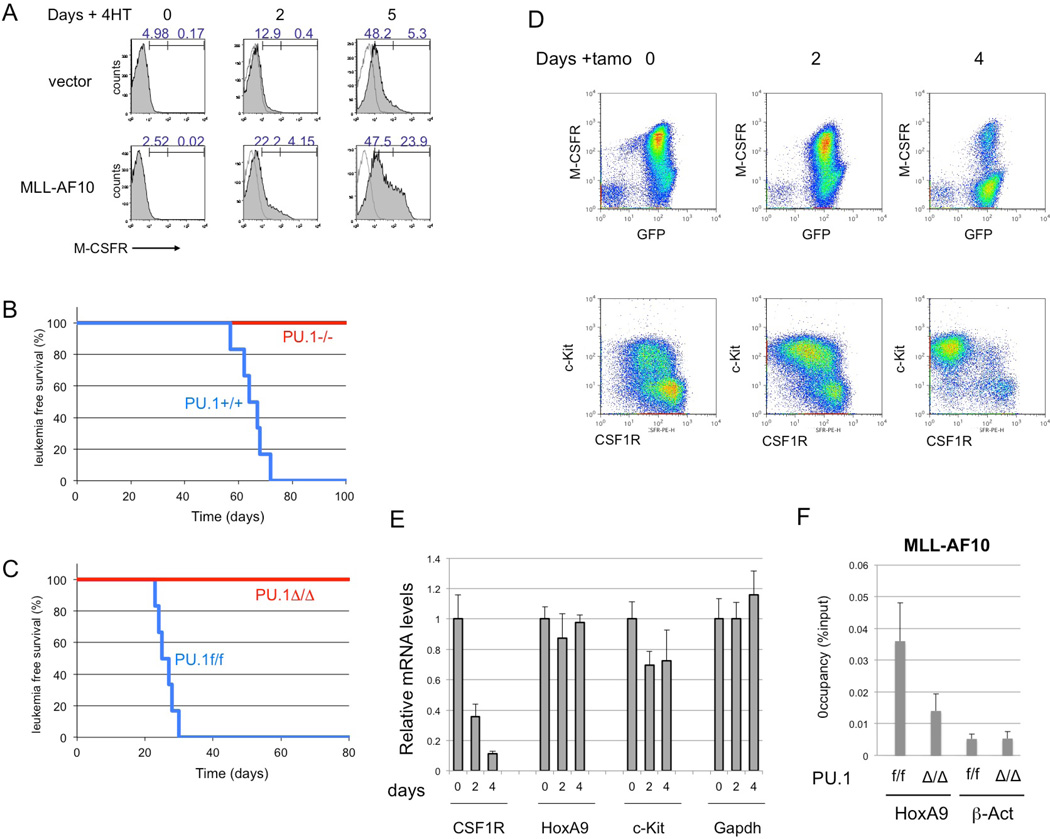
To test whether MLL-AF10 stimulates PU.1-dependent induction of endogenous Csf-1r, Pu.1−/− myeloid progenitors expressing the PU.1-estrogen receptor fusion protein (PUER) were used. These cells can differentiate into macrophages upon restoration of PU.1 activity by exposure to 4-hydroxytamoxifen (4-HT)32. PUER cells were infected with MSCV-MLL-AF10-ires-GFP or control retroviruses. GFP+ cells were sorted and cultured in the presence of 4-HT. Five days after the addition of 4-HT, flow cytometry analysis indicated a strong increase in Csf-1r expression by cells expressing MLL-AF10, but only a slight increase in cells infected with the control vector (Figure 5A). Thus, MLL-AF10 induces expression of endogenous Csf-1r in a PU.1-dependent manner.
Figure 5. PU.1 is critical for MLL-AF10-induced AML.
(A) PUER cells infected with MSCV-GFP or MSCV-FLAG-MLL-AF10-ires-GFP retroviruses were exposed to 100 nM 4-hydroxytamoxifen (4-HT) for 0, 2, or 5 days and analyzed by FACS for CSF-1R expression. (B) Fetal liver cells of E12.5 PU.1+/+ and PU.1−/− mouse embryo littermates were infected with either MLL-AF10, and transplanted into irradiated mice. Leukemia-free survivals of the mice were analyzed. n = 6, P < 0.001 (C) The fetal liver cells of E14.5 PU.1flox/flox with ER-Cre were infected with MLL-AF10, and transplanted into irradiated mice. The BM cells of the primary AML mice were transplanted into sub-lethally irradiated wild-type mice. Tamoxifene (PU.1Δ/Δ or solvent (PU.1f/f) was administered to the secondary AML mice every 2 d by intravenous injection 17 d after transplantation, when GFP+ cells were detected in peripheral blood. Leukemia-free survivals of the secondary mice were investigated. n = 6, P < 0.001 (D, E) The BM cells were prepared 0, 2, or 4 days after injection of Tamoxifen and analysed for expression of CSF-1R and c-Kit proteins (D) and for Csf1r, HoxA9, c-Kit, Meis1 and Gapdh mRNAs (E). (F) The BM cells, untreated (f/f) or Tamoxifen-treated for 4 days (Δ/Δ), were subjected to ChIP analysis using anti-Flag (MLL-AF10) antibodies. Semiquantitative real-time PCR was performed on the co-precipitated DNAs.
To determine whether PU.1 is essential for initiation of MLL-AF10-induced AML, the wild-type and Pu.1−/− fetal liver cells of E12.5 litter mates were infected with MLL-AF10 retrovirus and transplanted into irradiated mice. Although the mice with wild-type cells expressing MLL-AF10 developed AML 2–3 months after transplantation, mice with Pu.1−/− cells were quite healthy for at least 6 months (Figure 5B).
To determine if PU.1 is required for maintenance of MLL-AF10-induced AML, AML mice were generated using fetal liver cells of Pu.1 conditional KO mice (Pu.1flox/flox ERT2-Cre). The bone marrow (BM) cells of the AML mice were transplanted into secondary recipient mice and deletion of the Pu.1 gene was induced 3 weeks after transplantation. All the control mice died within 1 month, while none of the mice with deletion of Pu.1 developed AML or died (Figure 5C). The population of Csf-1rhigh cells in BM decreased within 4 days after deletion of Pu.1 (Figure 5D). By contrast, c-Kit-positive cells were still remained. These results indicate that PU.1 is required for both development and maintenance of MLL-AF10-induced AML. RT-PCR analysis indicated that levels of Csf-1r mRNAs were decreased after Pu.1 deletion but levels of HoxA9, c-Kit and Gapdh mRNAs were stable at least 4 days after Tamoxifen treatment (Figure 5E). Chromatin immunoprecipitation (ChIP) analysis indicated that MLL-AF10 enrichment at the CSF-1R locus was reduced by deleting Pu.1 (Figure 5F).
CSF-1R is a promising target for AML therapy
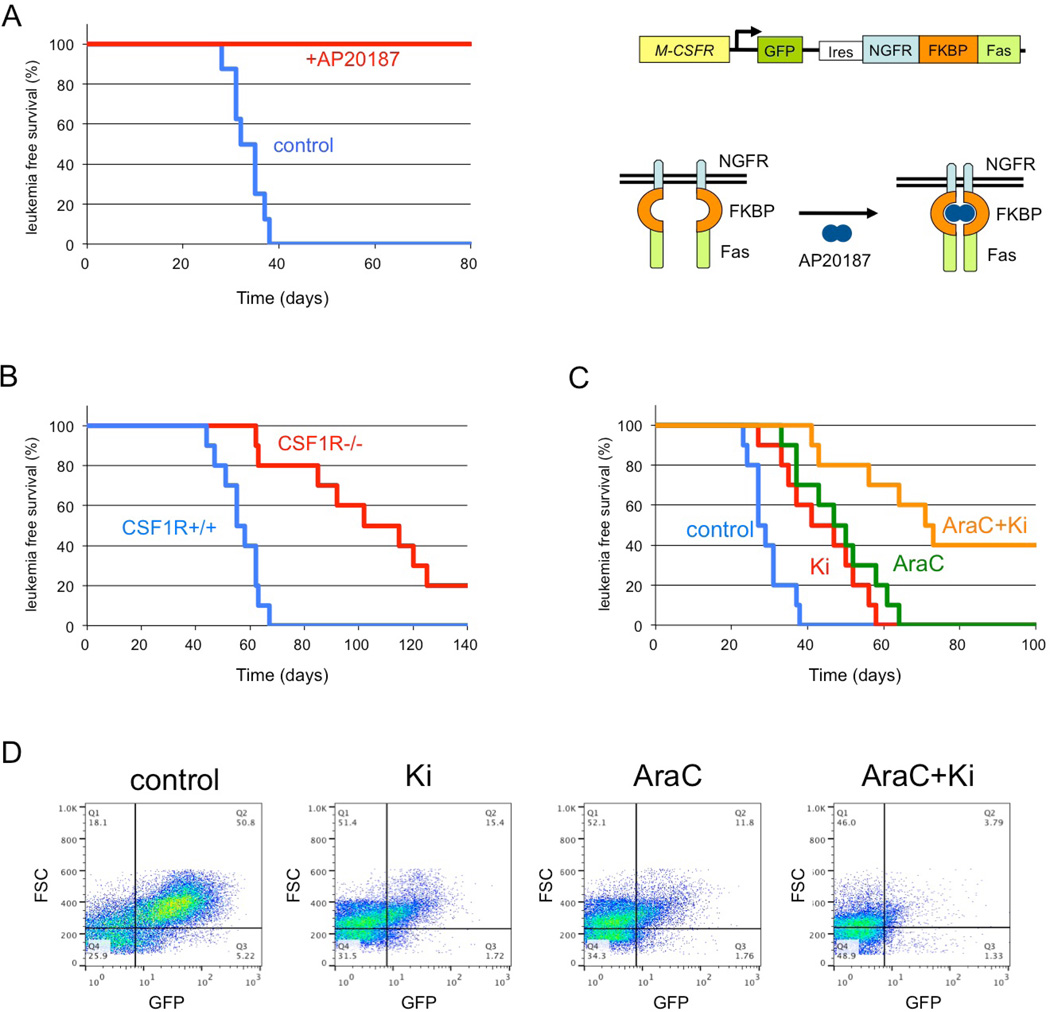
To determine whether a high level of CSF-1R expression is an essential element of LICs, transgenic mice expressing drug-inducible FKBP-Fas suicide gene and EGFP under the control of the Csf-1r promoter were used17 (Figure 6A). In these mice, conditional ablation of Csf-1r-expressing cells can be induced by injection of the AP20187 dimerizer17. c-Kit+ BM cells of transgenic mice were infected with MLL-AF10 retrovirus and transplanted into lethally irradiated wild-type mice. These mice developed AML about 2 months after transplantation, and their BM cells were transplanted into secondary recipient mice. Seven days after transplantation, the mice were injected with AP20187 as described previously17. All untreated mice, and none of the AP20187-treated mice, developed AML 4–6 weeks after transplantation (Figure 6A), indicating that a high level of CSF-1R expression is a key LIC functional element in MLL-AF10-induced AML mice.
Figure 6. Cure of MLL-AF10-induced AML by ablation of CSF-1Rhigh cells.
(A) The BM cells from the transgenic (CSF-1R-EGFP-NGFR/ FKBP1A/ TNFRSF6) mice were infected with MSCV-MLL-AF10-ires-GFP and were transplanted into lethally irradiated C57BL/6 mice to induce AML. Bone marrow cells (1 × 104) of primary AML mice were transplanted into sub-lethally irradiated C57BL/6 mice. Administration of AP20187 or solvent (control) to the secondary AML mice was started by IV injection three weeks after transplantation. Leukemia-free survivals of the untreated (n = 8) and AP20187-treated (n = 8) secondary transplanted mice were investigated. P < 0.001. Right panel shows the structure of genes for the Csf1r promoter, EGFP, the NGFR–FKBP–Fas suicide construct, and activation of NGFR–FKBP–Fas. Note that in the transgenic mice, conditional ablation of cells expressing high levels of CSF-1R can be induced by exposure to the AP20187 dimerizer. (B) Fetal liver cells of E16.5 Csf1r+/+ and Csf1r−/− mice littermate embryos were infected with MLL-AF10-ires-GFP and transplanted into irradiated mice. The leukemia-free survivals of the mice were analyzed. n = 10, P < 0.001 (C, D). BM cells (105) from AML mice with MLL-AF10 were transplanted into non-irradiated mice. Vehicle, Ki20227, Ara-C, or Ki20227 plus Ara-C were administrated as described in EXPERIMENTAL PROCEDURES Leukemia-free survivals of the mice were analyzed (C). n = 10, P < 0.01 (control vs +Ki20227, AraC vs AraC+Ki) Peripheral blood cells were prepared 21 days after transplantation and analysed for expression of GFP (D).
To determine if Csf-1r is essential for the development of MLL-AF10-induced AML, AML mice were generated using E16.5 fetal liver cells from Csf-1r −/− 18 and Csf-1r+/+ littermate embryos. The mice transplanted with the wild-type cells developed AML 6–9 weeks after transplantation while those transplanted with Csf-1r −/− cells developed AML 9–18 weeks after transplantation (Figure 6B). Thus, the CSF-1R is required for efficient induction of AML by MLL-AF10.
The present results suggest that signaling through CSF-1R may be a suitable therapeutic target for kinase inhibitors in MLL fusion-induced leukemogenesis. The effect of the CSF-1R-specific inhibitor Ki20227 was tested with or without AraC in MLL-AF10-induced AML in mice. Ki20227. Ki20227 and AraC slowed the onset of AML (Figure 6C) and inhibited the increase in GFP+ leukemic cells (Figure 6D). The combination of Ki20227 plus AraC was more effective than either agent alone.
Discussion
CSF-1R is a potential target for AML therapy
AML is a highly malignant disease. Numerous genetic abnormalities are known in AML, among which chromosome translocations involving the MLL gene are associated with poor prognosis. Conventional chemotherapies are often effective in reducing the total number of leukemia cells, but are not curative in many cases of AML. LSCs are capable of the limitless self-renewal necessary for cancer initiation and maintenance. Since residual LSCs are a potential cause of AML relapse, eradication of LSCs is critical to cure the disease. The present results showed that LSCs are enriched in cells expressing high levels of CSF-1R. Relevant to our observations, a viral integration site of the Friend murine leukemia virus that is utilized in approximately 20% of virus-induced primary myeloid leukemias, was shown to be at the 5’ end of the Csf-1r gene and to result in high expression of a normal-sized Csf-1r mRNA33. Using a mouse model expressing a drug-inducible suicide gene controlled by the Csf-1r promoter, ablation of Csf-1rhigh cells was shown to prevent AML mice from dying of the disease. Moreover MLL-AF10-induced leukemia was suppressed by deletion of the Csf-1r gene. These results clearly show that CSF-1R is a promising target for novel AML therapy. CSF-1R is a receptor tyrosine kinase that regulates the survival, proliferation and/or differentiation of macrophages, osteoclasts and Paneth cells34,35,36. A tyrosine kinase inhibitor specific for CSF-1R slowed the progress of MLL-AF10-induced leukemia. CSF-1R upregulation has been detected in LSCs in MOZ-TIF2-induced AML25 and human AML patients26.
CSF-1R expression is critical for AML initiation but not for immortalization in vitro
MLL leukemias are invariably associated with the expression of Hox genes. Upregulation of Hox genes in MLL leukemias is critical for LSC maintenance; however, Hox upregulation alone does not recapitulate all the biological and clinical features of MLL leukemias and is unlikely to support malignancy and the high LSC frequency observed in MLL leukemias. Forced expression of the HOXA9 gene can immortalize myeloid progenitors in vitro, but is not sufficient to initiate AML in vivo. By contrast, expression of MLL fusions such as MLL-AF10, MLL-AF9, and MLL-ENL is sufficient for both immortalization in vitro and initiation of AML in vivo. Our results demonstrate that cells expressing high levels of Csf-1r show strong AML initiation in vivo (Figures 1B and 1C) while cells expressing high and low levels of Csf-1r showed equivalent colony formation in vitro (Figure 1E). These findings suggest that CSF-1R expression is not important for immortalization in vitro but is critical for initiation of AML in vivo.
The LSC activity of MLL leukemia is known to be reduced after culture in vitro37, suggesting that a certain in vivo microenvironment is required for LSC maintenance, as is also the case for hematopoietic stem cells. Our results show that the expression of Csf-1r is greatly reduced after culture in vitro (data not shown), suggesting that expression of CSF-1R is regulated by microenvironment-dependent epigenetics.
Differential regulation of CSF-1R and Hox
MLL and MLL fusion proteins form a complex with MENIN and LEDGF to regulate the expression of Hox genes30, 38. Our results show that PU.1 mediates the regulation of Csf1r transcription by MLL/MLL fusion proteins. The MENIN and LEGDF-interacting domains of MLL are not required for interaction with PU.1 or transactivation at the Csf1r promoter, suggesting that MENIN and LEGDF are unlikely to be involved in the regulation of CSF-1R expression. While expression of Csf-1r rapidly decreased after deletion of the Pu.1 gene (Figures 5D and 5E), HoxA9 mRNA levels were stable at least 4 days after Pu.1 deletion (Figure 5E). Moreover, HoxA9 mRNA levels were equivalent in AML cells expressing high and low levels of CSF-1R (Figure 1H). These results suggest that expression of the Csf1r and Hox genes is independently regulated by MLL fusion proteins.
The MLL-AF10-induced AML was demonstrated to be cured by deletion of Pu.1 (Figure 5C). However, deletion of Csf-1r prolonged survival time but did not cure the AML completely. These facts suggest that CSF-1R is not the sole critical PU.1-dependent mediator of leukemogenesis, and that there are other PU.1-target genes critical for maintenance of MLL fusion-induced AML. Such genes may be involved in the leukemogenesis in collaboration with CSF-1R and HOX genes.
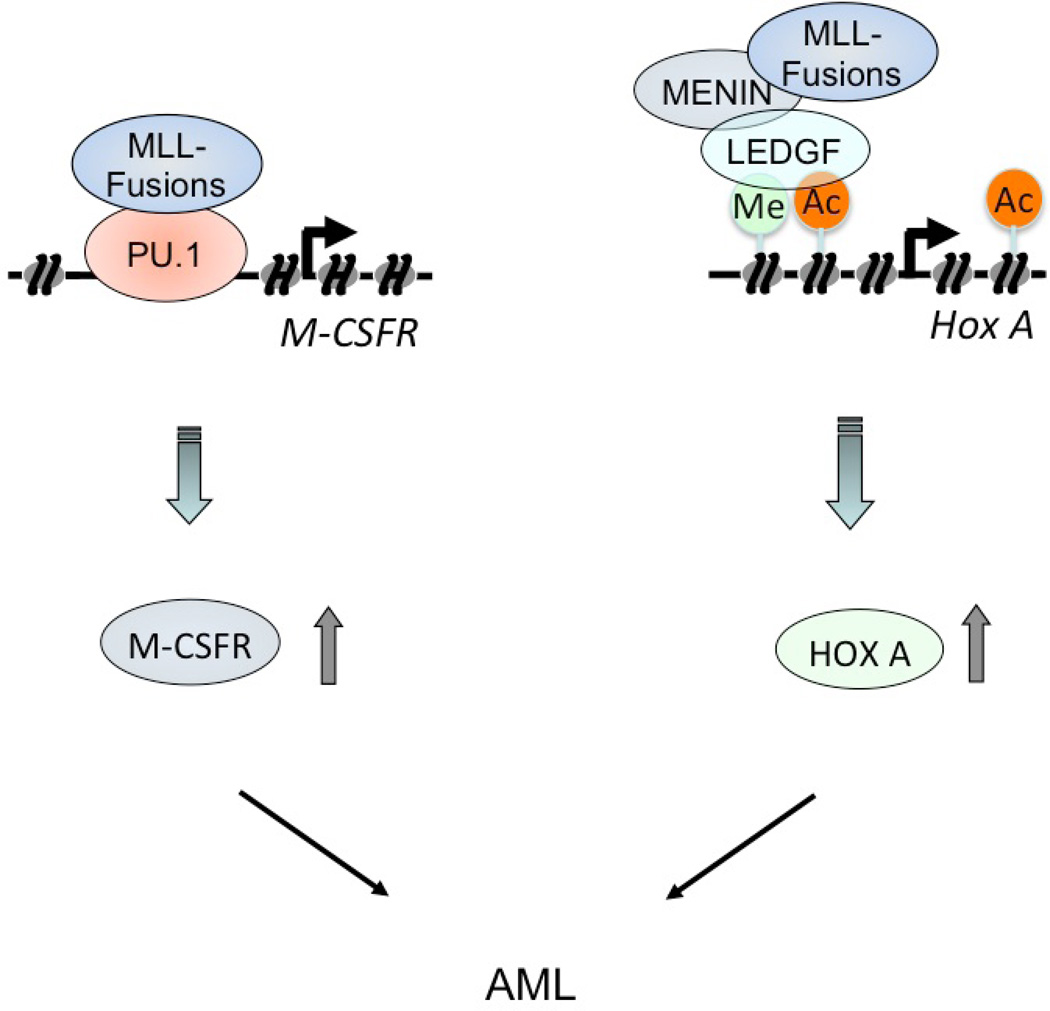
It has been suggested that second mutations, such as activating point mutations, in receptor tyrosine kinases (e.g., FLT3 and c-KIT) are required for fusion genes such as AML1-ETO, PML-RARa, or CBFb-MYH11 to induce acute leukemia39. By contrast, MLL fusions alone can induce the rapid onset of AML40. Our data suggest that MLL fusions induce the upregulation of the receptor tyrosine kinase Csf-1r and Hox genes, thereby inducing the rapid onset of AML by activating two classes of pathways (Figure 7). These pathways provide potential molecular targets for new approaches in the treatment of these forms of leukemia.
Figure 7.
Model for transcriptional regulation of Csf1r and Hox genes by MLL fusion proteins. MLL fusions induce the rapid onset of AML by activating two classes of pathways, CSF-1R pathway and Hox pathway.. MLL-fusions stimulated constitutive CSF-1R expression by binding to PU.1 to induce leukemia (left panel). MLL fusion proteins form a complex with menin and LEDGF to regulate the expression of Hox genes (right panel).
Supplementary Material
Acknowledgments
We would like to thank Dr. D. E. Zhang for Csf-1r promoter mutant lacking PU.1-binding sites and Dr. Harinder Singh for PUER cells. This work was supported in part by Grants-in-Aid from the Ministry of Health, Labor and Welfare; the Ministry of Education, Culture, Sports, Science and Technology; National Cancer Center Research and Development Fund; and NIH grants HL112719, CA32551 and 5P30-CA13330.
Footnotes
Author Contributions
YA, MS, KY, TK and YS performed research and analyzed data. ERS, MLK, KA, and DGT contributed vital new reagents. IK designed the research, analyzed data and wrote the paper.
Disclosure Statement
The authors have no conflicts of interest.
References
- 1.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 2.Warner JK, Wang JC, Hope KJ, Jin L, Dick JE. Concepts of human leukemic development. Oncogene. 2004;23:7164–7177. doi: 10.1038/sj.onc.1207933. [DOI] [PubMed] [Google Scholar]
- 3.Ayton PM, Cleary ML. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene. 2001;20:5695–5707. doi: 10.1038/sj.onc.1204639. [DOI] [PubMed] [Google Scholar]
- 4.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nature reviews Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 5.Dou Y, Hess JL. Mechanisms of transcriptional regulation by MLL and its disruption in acute leukemia. Int J Hematol. 2008;87:10–18. doi: 10.1007/s12185-007-0009-8. [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez-Perales S, Cano F, Lobato MN, Rabbitts TH. MLL gene fusions in human leukaemias: in vivo modelling to recapitulate these primary tumourigenic events. Int J Hematol. 2008;87:3–9. doi: 10.1007/s12185-007-0001-3. [DOI] [PubMed] [Google Scholar]
- 7.Cozzio A, Passegue E, Ayton PM, Karsunky H, Cleary ML, Weissman IL. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev. 2003;17:3029–3035. doi: 10.1101/gad.1143403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huntly BJ, Shigematsu H, Deguchi K, et al. MOZ-TIF2, but not BCR-ABL, confers properties of leukemic stem cells to committed murine hematopoietic progenitors. Cancer Cell. 2004;6:587–596. doi: 10.1016/j.ccr.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 9.Armstrong SA, Staunton JE, Silverman LB, et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- 10.Rozovskaia T, Feinstein E, Mor O, et al. Upregulation of Meis1 and HoxA9 in acute lymphocytic leukemias with the t(4: 11) abnormality. Oncogene. 2001;20:874–878. doi: 10.1038/sj.onc.1204174. [DOI] [PubMed] [Google Scholar]
- 11.Yeoh EJ, Ross ME, Shurtleff SA, et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1:133–143. doi: 10.1016/s1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- 12.Ayton PM, Cleary ML. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes Dev. 2003;17:2298–2307. doi: 10.1101/gad.1111603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar AR, Hudson WA, Chen W, Nishiuchi R, Yao Q, Kersey JH. Hoxa9 influences the phenotype but not the incidence of Mll-AF9 fusion gene leukemia. Blood. 2004;103:1823–1828. doi: 10.1182/blood-2003-07-2582. [DOI] [PubMed] [Google Scholar]
- 14.Wong P, Iwasaki M, Somervaille TC, So CW, Cleary ML. Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes Dev. 2007;21:2762–2774. doi: 10.1101/gad.1602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar AR, Li Q, Hudson WA, et al. A role for MEIS1 in MLL-fusion gene leukemia. Blood. 2009;113:1756–1758. doi: 10.1182/blood-2008-06-163287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Somervaille TC, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 2006;10:257–268. doi: 10.1016/j.ccr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 17.Burnett SH, Kershen EJ, Zhang J, et al. Conditional macrophage ablation in transgenic mice expressing a Fas-based suicide gene. J Leukoc Biol. 2004;75:612–623. doi: 10.1189/jlb.0903442. [DOI] [PubMed] [Google Scholar]
- 18.Dai XM, Ryan GR, Hapel AJ, et al. Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood. 2002;99:111–120. doi: 10.1182/blood.v99.1.111. [DOI] [PubMed] [Google Scholar]
- 19.Iwasaki H, Somoza C, Shigematsu H, et al. Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood. 2005;106:1590–1600. doi: 10.1182/blood-2005-03-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seibler J, Zevnik B, Kuter-Luks B, et al. Rapid generation of inducible mouse mutants. Nucleic Acids Res. 2003;31:e12. doi: 10.1093/nar/gng012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- 22.Ohno H, Kubo K, Murooka H, et al. A c-fms tyrosine kinase inhibitor, Ki20227, suppresses osteoclast differentiation and osteolytic bone destruction in a bone metastasis model. Mol Cancer Ther. 2006;5:2634–2643. doi: 10.1158/1535-7163.MCT-05-0313. [DOI] [PubMed] [Google Scholar]
- 23.Zhang DE, Hetherington CJ, Chen HM, Tenen DG. The macrophage transcription factor PU.1 directs tissue-specific expression of the macrophage colony-stimulating factor receptor. Mol Cell Biol. 1994;14:373–381. doi: 10.1128/mcb.14.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yokoyama A, Kitabayashi I, Ayton PM, Cleary ML, Ohki M. Leukemia proto-oncoprotein MLL is proteolytically processed into 2 fragments with opposite transcriptional properties. Blood. 2002;100:3710–3718. doi: 10.1182/blood-2002-04-1015. [DOI] [PubMed] [Google Scholar]
- 25.Aikawa Y, Katsumoto T, Zhang P, et al. PU.1-mediated upregulation of CSF1R is crucial for leukemia stem cell potential induced by MOZ-TIF2. Nat Med. 2010;16:580–585. doi: 10.1038/nm.2122. 1p following 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kikushige Y, Shima T, Takayanagi S, et al. TIM-3 is a promising target to selectively kill acute myeloid leukemia stem cells. Cell stem cell. 2010;7:708–717. doi: 10.1016/j.stem.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 27.DiMartino JF, Ayton PM, Chen EH, Naftzger CC, Young BD, Cleary ML. The AF10 leucine zipper is required for leukemic transformation of myeloid progenitors by MLL-AF10. Blood. 2002;99:3780–3785. doi: 10.1182/blood.v99.10.3780. [DOI] [PubMed] [Google Scholar]
- 28.Zhang DE, Hetherington CJ, Meyers S, et al. CCAAT enhancer-binding protein (C/EBP) and AML1 (CBF alpha2) synergistically activate the macrophage colony-stimulating factor receptor promoter. Mol Cell Biol. 1996;16:1231–1240. doi: 10.1128/mcb.16.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang G, Zhao X, Wang L, et al. The ability of MLL to bind RUNX1 and methylate H3K4 at PU.1 regulatory regions is impaired by MDS/AML-associated RUNX1/AML1 mutations. Blood. 2011;118:6544–6552. doi: 10.1182/blood-2010-11-317909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yokoyama A, Cleary ML. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell. 2008;14:36–46. doi: 10.1016/j.ccr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milne TA, Briggs SD, Brock HW, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Molecular cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 32.Walsh JC, DeKoter RP, Lee HJ, et al. Cooperative and antagonistic interplay between PU.1 and GATA-2 in the specification of myeloid cell fates. Immunity. 2002;17:665–676. doi: 10.1016/s1074-7613(02)00452-1. [DOI] [PubMed] [Google Scholar]
- 33.Gisselbrecht S, Fichelson S, Sola B, et al. Frequent c-fms activation by proviral insertion in mouse myeloblastic leukaemias. Nature. 1987;329:259–261. doi: 10.1038/329259a0. [DOI] [PubMed] [Google Scholar]
- 34.Chitu V, Stanley ER. Colony-stimulating factor-1 in immunity and inflammation. Current opinion in immunology. 2006;18:39–48. doi: 10.1016/j.coi.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 35.Huynh D, Dai XM, Nandi S, et al. Colony stimulating factor-1 dependence of paneth cell development in the mouse small intestine. Gastroenterology. 2009;137:136–144. 44 e1–44 e3. doi: 10.1053/j.gastro.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pixley FJ, Stanley ER. CSF-1 regulation of the wandering macrophage: complexity in action. Trends in cell biology. 2004;14:628–638. doi: 10.1016/j.tcb.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 37.Krivtsov AV, Twomey D, Feng Z, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 38.Yokoyama A, Wang Z, Wysocka J, et al. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol. 2004;24:5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gilliland DG, Griffin JD. The roles of FLT3 in hematopoiesis and leukemia. Blood. 2002;100:1532–1542. doi: 10.1182/blood-2002-02-0492. [DOI] [PubMed] [Google Scholar]
- 40.Lavau C, Szilvassy SJ, Slany R, Cleary ML. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. Embo J. 1997;16:4226–4237. doi: 10.1093/emboj/16.14.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.