Abstract
A quinazolinedione-derived screening hit 2 was discovered with cellular antiviral activity against respiratory syncytial virus (CPE EC50 = 2.1 µM), moderate efficacy in reducing viral progeny (4.2 log at 10 µM), and marginal cytotoxic liability (selectivity index, SI ~ 24). Scaffold optimization delivered analogs with improved potency and selectivity profiles. Most notable were compounds 15 and 19 (EC50 = 300–500 nM, CC50 > 50 µM, SI > 100), which significantly reduced viral titer (>400,000-fold), and several analogs were shown to block the activity of the RNA-dependent RNA-polymerase complex of RSV.
INTRODUCTION
Acute bronchiolitis, a common lower respiratory tract infection most seriously affecting infants and the elderly, is predominately caused by the highly infectious human respiratory syncytial virus (hRSV).1–4 The virus belongs to the paramyxovirus family, which also includes mumps and measles viruses;5 however, unlike these related pathogens for which vaccines have been developed, a safe and effective vaccine remains elusive to prevent the contraction and transmission of RSV.6,7 In fact, since the discovery8 of the virus over 50 years ago, the only FDA approved small molecule inhibitor for treatment of the infection is ribavirin, a nucleoside antimetabolite, that is limited to use in critical cases due to its toxicological side effects.9,10 In the United States, the prevalence of RSV infection in adults over the age of 65 results in approximately 170,000 hospitalizations and 10,000 deaths annually11 while the global incidence of RSV infection was estimated in 2005 to result in the hospitalization of 3.4 million children under the age of 5.12 Furthermore, exposure does not impart full immunity from future infection and, in fact, promotes an inflammatory response that can contribute to chronic lung complications such as asthma.13,14 These burdens, coupled with the absence of suitable therapeutic agents for susceptible populations, underscore the importance of identifying effective and safe pharmacological countermeasures for RSV.
The scientific literature is replete with examples from translational development programs aimed at addressing this important need.15–17 Replication inhibitors18–22 have been investigated, along with several compounds that target RSV’s entry-enabling F protein,23–27 though in most cases the compounds were not pursued or clinical development was discontinued.28,29 Despite these efforts, the search continues for RSV inhibitors that offer a superior pharmacological and safety profile compared to that of ribavirin.30
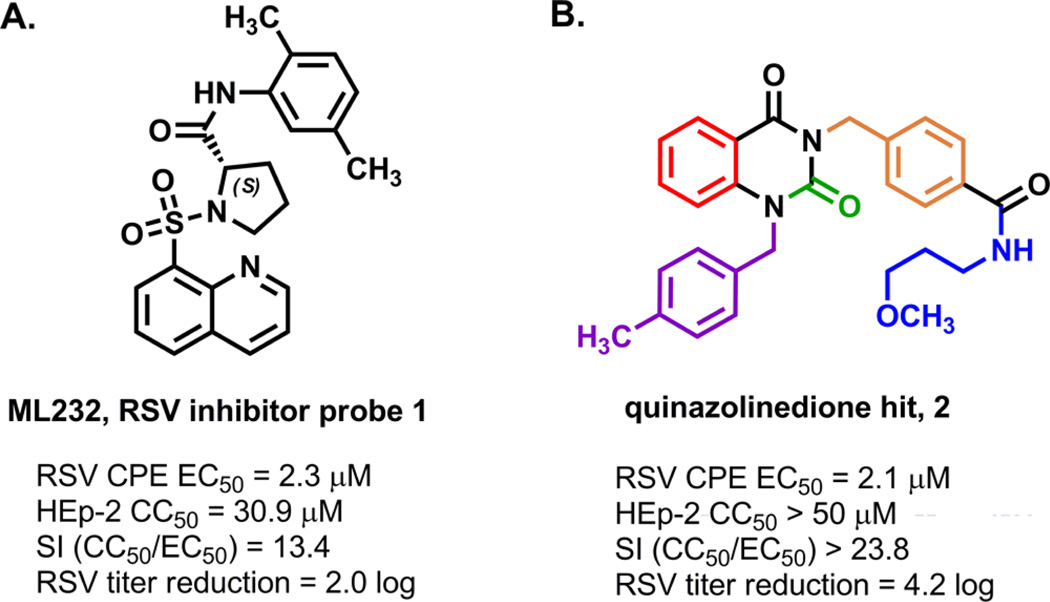
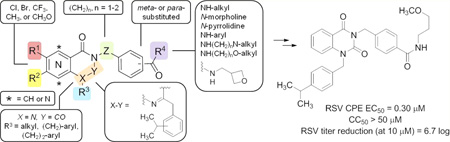
As part of the National Institutes of Health Molecular Libraries Initiative, we pursued a subset of RSV-inhibiting hit scaffolds identified through a high-throughput screen31–33 of the national compound repository.34 Optimization of a screening hit led to compound 1, probe ML232, a sulfonamide-based RSV inhibitor with single-digit micromolar in vitro activity, and a proposed entry-based mechanism of inhibition based on time-of-addition studies (Figure 1A).15,35 In a parallel effort, the team also launched an optimization campaign on a quinazolinedione compound series for which we noted key differences with respect to the breadth of tunable structure–activity and structure–property relationships (SAR and SPR, respectively) and a potentially different mechanism of action as compared to the ML232 compound series. The quinazolinedione hit 2 was determined to inhibit a RSV-induced cytopathic effect with an EC50 of 2.14 µM and showed HEp-2 cellular toxicity with a CC50 > 50 µM, resulting in a selectivity index (CC50/EC50) of >23.8 (Figure 1B). In a titer reduction assay, hit 2 was also found to reduce viral plaques by 4.2 log (~14,000-fold as compared to control) at a concentration of 10 µM. The team undertook an optimization effort that focused on the five colored regions of the scaffold with the primary aims of broadening the selectivity index by enhancing potency and attenuating cellular toxicity, amplifying the plaque reducing effect, and improving solubility (Figure 1B).
Figure 1.
(A) Structure and data for sulfonylpyrrolidine-derived probe 1, ML232. (B) Structure and data for hit quinazolinedione 2 with highlighted regions of structure–activity relationship optimization.
CHEMISTRY
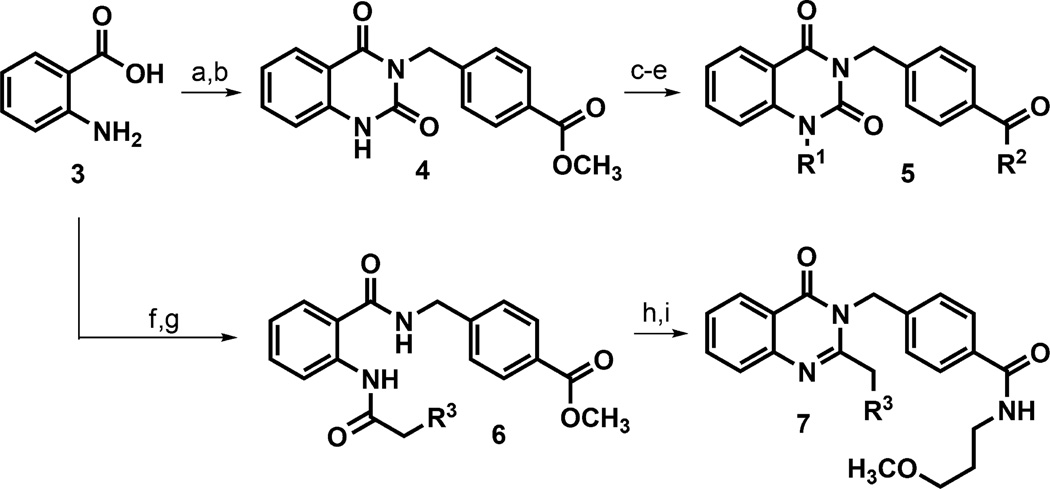
Analogs of hit 2 were generally prepared using standard peptide coupling conditions of 2-amino benzoic acid 3 with methyl 4-(aminomethyl)benzoate to afford an aminobenzamide intermediate (Scheme 1). Subsequent cyclization with CDI generated quinazolinedione core intermediate 4. Most analogs were made by ester hydrolysis of 4, followed by incorporation of the pendent amido alkyl ether (R2) with routine amide coupling, followed by installation of the N-benzyl appendage (R1) to afford products 5. In some cases, it was advantageous to affix the N-benzyl portion prior to revealing the benzoic acid functionality for coupling to the preferred amine component (i.e., shuffling the sequence of R1 vs R2 integration). In either case, the synthetic route was flexible and offered selective, orthogonal, late-stage diversification to prepare the desired analog sets. Quinazolinones 7 were also prepared from anthranillic acid 3. Amide coupling between 3 and methyl 4-(aminomethyl)benzoate afforded a 2-aminobenzamide intermediate that was treated with an isopropylphenylacetic acid chloride. The resulting bisamide 6 was treated with hydroxide base to reveal a benzoic acid that could be further manipulated; however, these conditions fortuitously induced the intended hydrolysis and necessary cyclization to generate the quinazolinone core in one step. Subsequent coupling of the unmasked benzoic acid with 3-methoxypropylamine afforded the desired analogs 7.
Scheme 1. Chemical Synthesis of Quinazolinedione and Quinazolinone Analogsx.
xReagents: (a) DIPEA, HATU, methyl 4-(aminomethyl)benzoate hydrochloride, DMF, rt, 2 h, 68%; (b) DIPEA, 1,1-carbonyldiimidazole, CH2Cl2, reflux, 16 h, 95%; (c) LiOH·H2O, THF, 40 °C, 1 h, 88%; (d) DIPEA, HATU, 3-methoxypropylamine or other amine for R2, DMF, rt, 2 h, 44–76%; (e) K2CO3, 4-isopropylbenzyl bromide other aryl halide for R1, DMF, 40 °C, 16 h, 11–71%; (f) DIPEA, EDCI, HOBt, methyl 4-(aminomethyl)benzoate hydrochloride, CH2Cl2, rt, 74%; (g) 3- or 4-isopropylphenylacetic acid, (COCl)2, cat. DMF, CH2Cl2, then pyridine, CH2Cl2, 1.5 h, rt, 93–100%; (h) LiOH, THF, H2O, 40 °C, 20 h; (i) 3-methoxypropylamine, EDCI, HOBt, DIPEA, CH2Cl2, rt, 10–22% over 2 steps.
RESULTS AND DISCUSSION
Medicinal Chemistry Optimization
For this stage of our program, 73 quinazolinedione-derived analogs were prepared and analyzed. All compounds were evaluated for inhibition of an RSV-induced cytopathic effect and assessment of mammalian cell cytotoxicity. Both assays were performed in a 10-point dose response format using HEp-2 cells (hRSV strain Long), and the data from these screens was employed to drive iterative structural revision. A smaller subset of the most promising analogs was subsequently tested for their ability to reduce viral plaques at a compound concentration of 10 µM. Solubility was determined for selected analogs as their overall activity profiles were improved and as modifications were implemented with an expectation of enhancing this particular parameter.
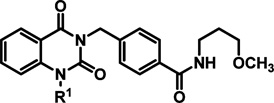
Initially, changes in the quinazolinedione N-benzyl entity (R1) were examined while preserving the hit structure’s 3-methoxypropylamine (R2) component (Table 1). Replacement of the 4-methylbenzyl substituent with a hydrogen atom or use of truncated alkyl replacements such as N-methyl, N-n-propyl, or N-CH2-cyclohexyl were not tolerated, resulting in a loss of potency (EC50 > 50 µM). Similarly, extension of the R1 benzyl linker by one additional methylene unit was not advantageous (N-(CH2)2-4-tolyl, or N-(CH2)2-4-trifluoromethylphenyl, or N-(CH2)2-4-bromophenyl, EC50 > 50 µM). Generally, 4-benzyl substitution was preferred, as 3-substituted benzyl derivatives lost potency compared to hit 2, and 2-substituted benzyl analogs were inactive (Table 1). Increasing steric bulk at the 4-benzyl position (methyl (2) → ethyl (18) → i-propyl (19)) resulted in improved potency. Importantly, cell toxicity was not observed at concentrations exceeding 50 µM for these analogs. Electron withdrawing groups incorporated at this same position were acceptable, and those that mimicked the steric character of small branched aliphatic groups already known to be beneficial were the most promising. Consequently, the N-4-nitrobenzyl derivative 15 and the N-4-isopropylbenzyl analog 19 showed the best potency, viral titer assay efficacy, and cytotoxicity profile in the collection. Heterocyclic variants in this region led to suboptimal potency, cytotoxicity, and reduction in viral plaques (entries 14–17).
Table 1.
hRSV CPE Assay Potency, Cytotoxicity, Selectivity Index, and Logarithmic Reduction in Viral Plaques for Analogs with Structural Variations in the (R1) Region of Hit Compound 2
 | ||||||
|---|---|---|---|---|---|---|
| entry | cmpd | R1 | RSV CPE potency ± standard deviation EC50 (µM)a |
HEp-2 cellular toxicity ± standard deviation CC50 (µM)b |
selectivity index CC50/EC50) |
viral titer reduction at 10 µM (log) |
| 1 | 2 | CH2-4-methylphenyl | 2.1 ± 0.5c | >50.0c | >23.8 | 4.2 |
| 2 | 8 | CH2-2-bromophenyl | >50.0 | 8.2 ± 0.2 | <0.2 | NT |
| 3 | 9 | CH2-3-bromophenyl | 2.2 ± 0.1 | 3.7 ± 0.4 | 1.7 | NT |
| 4 | 10 | CH2-4-bromophenyl | 0.9 ± 0.2 | >50.0 | >55.6 | 4.1 |
| 5 | 11 | CH2-2-fluorophenyl | >50.0 | 7.3 ± 1.0 | <0.2 | NT |
| 6 | 12 | CH2-4-fluorophenyl | 5.1 ± 0.4 | 7.6 ± 0.2 | 1.5 | NT |
| 7 | 13 | CH2-4-chlorophenyl | 6.7 ± 1.9 | >50.0 | >7.5 | 3.1 |
| 8 | 14 | CH2-4-methoxyphenyl | 2.0 ± 0.9 | >50.0 | >25.0 | 2.7 |
| 9 | 15 | CH2-4-nitrophenyl | 0.5 ± 0.05d | >50.0d | >100.0 | 5.6c |
| 10 | 16 | CH2-4-trifluoromethylphenyl | 1.3 ± 0.1 | >50.0 | >38.5 | 2.7 |
| 11 | 17 | CH2-4-nitrilephenyl | 1.3 ± 0.2 | 12.5 ± 1.2 | 7.7 | 4.2 |
| 12 | 18 | CH2-4-ethylphenyl | 1.0 ± 0.05 | >50.0 | >38.5 | 5.9 |
| 13 | 19 | CH2-4-isopropylphenyl | 0.3 ± 0.03c | >50.0c,e | >166.7 | 6.7 |
| 14 | 20 | CH2-5-benzooxadiazole | 4.9 ± 0.7 | >50.0 | >10.2 | 2.1 |
| 15 | 21 | CH2-3-(5-methylisoxazole) | >50.0 | 7.7 ± 0.7 | <0.2 | NT |
| 16 | 22 | CH2-2-pyridyl | >50.0 | 16.0 ± 0.3 | <0.3 | 0.1 |
| 17 | 23 | CH2-3-pyridyl | >50.0 | 8.0 ± 0.3 | <0.2 | 0.9 |
Data were averaged from ≥3 experiments.
Data were averaged from ≥2 experiments.
Data were averaged from two separate compound lots.
Data were averaged from three separate compound lots.
Data were obtained from a 3-day exposure experiment versus 5-day duration due to precipitation of compound after 3 days.
NT = not tested. Data were analyzed using Microsoft Excel 2010.
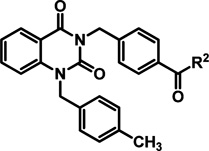
Alterations of the secondary amide (R2) were independently carried out in a parallel effort. Exchange of the methyl ether for the ethyl or isopropyl ether did not appreciably alter potency in the CPE assay; however, the cytotoxic effects associated with those analogs increased relative to the parent hit 2 (Table 2, entries 1–3). Replacement of the methyl ether with a tertiary amine for the purpose of enhancing solubility was inferior in terms of both potency and toxicity (entry 4). Elongation of the alkyl chain by one methylene unit only marginally enhanced potency and reduced the therapeutic window (entry 5); however, truncating the linker afforded analog 28 with comparable potency to hit 2 (entry 6). Introducing an oxetane as a cyclized version of the linear ether chain resulted in a promising potency, cytotoxicity, and viral plaque reduction profile (entry 7). Other modifications were explored to improve solubility or potency without inducing cytotoxicity, but none were found to be more advantageous when considering multiparameter optimization.
Table 2.
hRSV CPE Assay Potency, Cytotoxicity, Selectivity Index,and Logarithmic Reduction in Viral Plaques for Analogs with Structural Variations in the (R2) Region of Hit Compound 2
 | ||||||
|---|---|---|---|---|---|---|
| entry | cmpd | R1 | RSV CPE potency ± standard deviation EC50 (µM)a |
HEp-2 cellular toxicity ± standard deviation CC50 (µM)b |
selectivity index (CC50/EC50) |
viral titer reduction at 10 µM (log) |
| 1 | 2 | NH(CH2)3OCH3 | 2.1 ± 0.5c | > 50.0c | >23.8 | 4.2 |
| 2 | 24 | NH(CH2)3OCH2CH3 | 1.9 ± 0.2 | 8.5 ± 0.3 | 4.4 | NT |
| 3 | 25 | NH(CH2)3OCH(CH3)2 | 2.0 ± 0.2 | 19.3 ± 0.9 | 9.8 | NT |
| 4 | 26 | NH(CH2)3N(CH3)2 | 9.5 ± 0.4 | 16.6 ± 1.5 | 1.8 | NT |
| 5 | 27 | NH(CH2)4OCH3 | 0.8 ± 0.05 | 6.5 ± 0.3 | 7.8 | 2.1 |
| 6 | 28 | NH(CH2)2OCH3 | 2.2 ± 1.4 | >50.0d | 22.7 | NT |
| 7 | 29 | NH(CH2)-(3-oxetane) | 0.7 ± 0.1 | 47.0 ± 1.9 | 66.1 | 5.1 |
| 8 | 30 | NH(CH2)-cyclobutane | 1.0 ± 0.08 | 7.6 ± 0.3 | 7.9 | 1.2 |
| 9 | 31 | N-morpholine | >50.0 | 17.5 ± 1.1 | <0.4 | NT |
| 10 | 32 | N-pyrrolidine | >50.0 | 5.4 ± 0.3 | <0.1 | NT |
| 11 | 33 | N-piperidine | >50.0 | 7.3 ± 1.3 | <0.2 | NT |
| 12 | 34 | NHCH3 | 2.2 ± 0.9 | 45.3 ± 1.7 | 21.1 | NT |
| 13 | 35 | N(CH3)2 | >50.0 | 10.1 ± 0.2 | <0.2 | NT |
| 14 | 36 | NH-tert-butyl | 0.7 ± 0.03 | 8.4 ± 0.3 | 12.1 | 5.5 |
| 15 | 37 | NH-cyclohexyl | >50.0 | >50.0 | NA | NT |
| 16 | 38 | NH-phenyl | >50.0 | >50.0 | NA | NT |
| 17 | 39 | NH-benzyl | >50.0 | 41.8 ± 7.9 | NA | NT |
| 18 | 40 | NH-CH2-2-furyl | >50.0 | >50.0 | NA | NT |
| 19 | 41 | NH-2-thiazole | >50.0 | >50.0 | NA | NT |
| 20 | 42 | NH-4-pyridyl | 1.0 ± 0.04 | 1.8 ± 0.4 | 1.8 | 3.5 |
Data were an average of ≥3 experiments.
Data were an average of ≥2 experiments.
Data were an average of outcomes from two separate lots of compound 2.
Data were obtained from a 3-day exposure experiment versus 5-day duration due to precipitation of compound after 3 days.
NT = not tested; NA = not applicable. Data were analyzed using Microsoft Excel 2010.
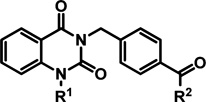
Hybrid analogs derived from the most advantageous individual R1 or R2 modifications were then prepared to assess synergistic effects, and additional SAR data was pursued using these compounds as templates. At this stage, several compounds were also evaluated for improvement in solubility. The initial analog sets (Tables 1 and 2) revealed that analogs bearing the 4-isopropylbenzyl or 4-nitrobenzyl moiety for R1 and the R2 modification of a NHCH2-3-oxetane were independently the most beneficial in terms of combined potency, cytotoxicity, and plaque reduction (compounds 15, 19, and 29, respectively). In pairing the 4-nitrobenzyl functionality (R1) with the NHCH2-3-oxetane (R2) subunit, compound 43 (entry 2, Table 3) showed comparable potency and toxicity to hit 2, but with a 14-fold improvement in PBS solubility and at least a 2 log increase in capacity to reduce viral titer. Combining the 4-isopropylbenzyl group (R1) with the NHCH2-3-oxetane (R2) component resulted in potency comparable to the best analogs in the series (44, entry 3, Table 3); however, the selectivity index was diminished due to unwanted cytotoxicity. Alternatives to the 4-nitrobenzyl element were examined, such as a 4-benzoic acid derivative 45 which imparted a desirable solubility effect but also abrogated activity, which may be the result of reduced cellular permeability. The N-4-dimethylaminobenzyl analog 46 was also found to have improved solubility but did not robustly provide cytoprotection to the extent observed with other architectural combinations (entry 5). Furyl and pyridyl variants in place of the R2 appendage were also determined to be suboptimal overall (entries 7–9 and other analogs not shown). Analogs bearing a substituent (halide, −OCH3, −CH3, or −CF3) at the C6 or C7 position of the quinazolinedione core resulted in loss of potency (EC50 > 50 µM). The same result was determined for analogs with an extra methylene spacer between the core and the benzamide moiety or for compounds with the meta-positioned amide functionality as opposed to the para-arrangement present in the hit (data not shown).
Table 3.
hRSV CPE Assay Potency, Cytotoxicity, Selectivity Index, Logarithmic Reduction in Viral Plaques, and Solubility Assessments for Analogs with Tandem Structural Variations in the (R1) and (R2) Regions of Hit Compound 2
 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| solubility (µM) | |||||||||
| entry | cmpd | R1 | R2 | RSV CPE potency ± standard deviation EC50 (µM)a |
HEp-2 cellular toxicity ± standard deviation CC50 (µM)b |
selectivity index (CC50/ EC50) |
viral titer reduction at 10 µM (log) |
PBSc | mediad |
| 1 | 2 | CH2-4-methylphenyl | NH(CH2)3OCH3 | 2.1 ± 0.5e | >50.0e | >23.8 | 4.2 | 0.1 | 2.5 |
| 2 | 43 | CH2-4-nitrophenyl | NHCH2-3-oxetane | 1.3 ± .05 | .05 > 50.0 | >38.5 | >6.2 | 1.4 | 7.2 |
| 3 | 44 | CH2-4-i-propylphenyl | NHCH2-3-oxetane | 0.4 ± .01 | 3.6 ± .08 | 9.0 | 5.8 | NT | NT |
| 4 | 45 | CH2-4-CO2H-phenyl | NHCH2-3-oxetane | >50.0 | >50.0 | NA | NT | 98.1 | NT |
| 5 | 46 | CH2-4-N(CH3)2 -phenyl | NHCH2-3-oxetane | 1.1 ± 0.2 | 44.7 ± 7.2 | 41.8 | >6.2 | 11.2 | 18.5 |
| 6 | 47 | CH2-4-tert butylphenyl | NHCH2-3-oxetane | 1.6 ± .08 | 3.7 ± 0.3 | 2.3 | 3.4 | NT | NT |
| 7 | 48 | CH2-4-i-propylphenyl | NHCH2-2-furyl | 1.2 ± 0.1 | >50.0 | >41.7 | 2.8 | 0.3 | NT |
| 8 | 49 | CH2-4-chlorophenyl | NHCH2-2-furyl | 0.8 ± .01 | >50.0 | >62.5 | NT | NT | 10.2 |
| 9 | 50 | CH2-4-i-propylphenyl | NH-2-CH3O-pyridyl | 0.5 ± .06 | <1.6 | <3.3 | NT | NT | NT |
Data were an average of ≥3 experiments.
Data are an average of ≥2 experiments.
Kinetic solubility in 1×PBS, pH 7.4.
Kinetic solubility in CPE assay media: (DMEM/F12(r) (Sigma, Cat # D6434)/1×Pen/Strep/Glutamine (Gibco, Cat # 10378)/2% Heat Inactivated FBS (Gibco Cat # 10082)).
NT = not tested; NA = not applicable. Data were analyzed using Microsoft Excel 2010.
Data were an average of outcomes from two separate lots of compound 2.
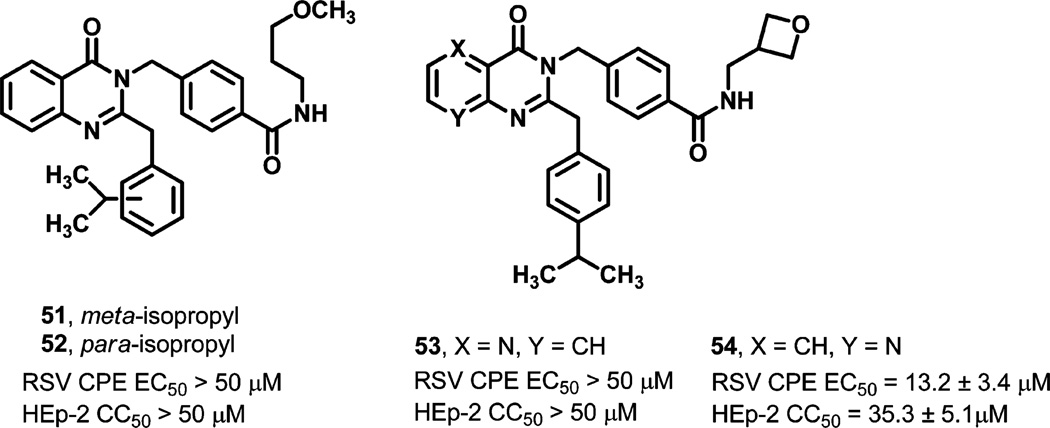
Given the idiosyncractic cytotoxicity profile for several of the compounds in the quinazolinedione series and our experience with a related, antiviral quinazolinone scaffold36,37 that lacked any detectable mammalian cytotoxicity, two quinazolinone analogs were prepared to explore if this modified core was beneficial against RSV (51–52, Figure 2). Quinazolinone analogs 51 and 52 were generated bearing peripheral components that had been shown to possess anti-RSV activity when integrated with the quinazolinedione core. While the compounds were not protective against RSV (>50 µM), the effort underscored the importance of the carbonyl group situated between the core nitrogens to retaining anti-RSV potency. Additionally, the impact of introducing a nitrogen atom into the fused phenyl ring of the quinazolinedione core was studied (53–54, Figure 2). Only analog 54 was weakly active, but the compound also demonstrated some appreciable toxicity. Though limited somewhat in scope, the core alterations represented by these four analogs and those previously discussed in which functional groups were introduced at the C6 or C7 position of the scaffold showed that no core modification made to date was well tolerated.
Figure 2.
Activity of quinazolinones and azaquinazolinediones.
Profiling Assays
The purpose of this program was to identify and develop new compounds with compelling in vitro anti-RSV activity that could be used as a platform for deriving suitable probes for future in vivo efficacy studies. Toward this goal, several analogs emerged from the SAR effort as interesting probe candidates worthy of further characterization based on improvements in CPE potency, solubility, and viral titer. Nonetheless, limitations in aqueous solubility or the presence of functionality with suspected metabolic liability prompted the team to assess passive permeability and hepatocyte toxicity for select analogs. Compounds 15, 19, and 46 were evaluated accordingly (Table 4).
Table 4.
Comparative SAR, Physiochemical, and In Vitro ADME Data for Select Analogs
| solubility (µM)g |
aqueous stability (%)j,k |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| entry | analog | RSV CPE potency ± standard deviation EC50 (µM)a |
HEp-2 cellular toxicity ± standard deviation CC50 (µM)d |
selectivity index (CC50/ EC50) |
viral titer reduction at 10 µM (log) |
PBSh | mediai | PBS | 1:1 PBS/ ACN |
Pe, PAMPA permeability (×10−6 cm/s)g,l |
hepatocyte toxicity LC50 (µM)g,o |
| 1 | 2 | 2.1 ± 0.5 | >50.0 | 23.8 | 4.2 | 0.1 | NT | 88.1 | 94.0 | NT | NT |
| 2 | 15 | 0.5 ± 0.05b | >50.0b | >100 | 5.6f | 1.3 | 2.8 | 83.4 | 94.9 | 364/349/356m | >50 |
| 3 | 19 | 0.3 ± 0.03c | >50.0c,e | >166.7 | 6.7 | 0.4 | 10.2 | 27.0 | 100 | 718/631/703n | >30 |
| 4 | 46 | 1.1 ± 0.2 | 44.7 ± 7.2 | 41.8 | >6.2 | 11.2 | 18.5 | 90.2 | 100 | 780/686/514m | >50 |
Data were an average of ≥3 experiments.
Data were an average of outcomes from three separate lots of compound 15.
Data were an average of outcomes from two separate lots of compound 19.
Data are an average of ≥2 experiments.
Data were obtained from a 3-day exposure experiment versus 5-day duration due to precipitation of compound after 3 days.
Data were an average of outcomes from two separate lots of compound 15.
Data collected by Ms. Arianna Mangravita-Novo at the Conrad Prebys Sanford Burnham Medical Research Institute.
Kinetic solubility in 1× PBS, pH 7.4.
Kinetic solubility in CPE assay media: (DMEM/F12(r) (Sigma, Cat # D6434)/1× Pen/Strep/Glutamine (Gibco, Cat # 10378)/2% Heat Inactivated FBS (Gibco Cat # 10082)).
Data collected by Mr. Patrick Porubsky at the University of Kansas Analytical Chemistry Core, Specialized Chemistry Center.
Results are represented as percent parent remaining after 48 h; Stability assessment was done independently in PBS or with 1:1 PBS and acetonitrile; the latter was used to account for limitations in solubility affecting PBS results.
PAMPA donor pH: 5.0/6.2/7.4, acceptor pH: 7.4; controls: verapamil (222/1097/1936–highly permeable), metoprolol (14/60/472–moderately permeable), ranitidine (<10/<10/<10–poorly permeable).
PAMPA done in PBS, no additives.
PAMPA done with 20% acetontrile added to compensate for PBS solubility. Without acetonitrile, Pe was insignificant at each pH.
Fa2N-4 immortalized human hepatocytes. Data were analyzed using Microsoft Excel 2010;
NT = not tested.
Aqueous solubility for isopropylbenzyl derivative 19 was the most limited of these three compounds, resulting in a skewed result in aqueous stability (27% parent remaining, entry 2, Table 4). The addition of acetonitrile to the stability experiment to account for compound precipitation in PBS alone reflected that compound 19 was stable to degradation. Solubility for each compound in CPE assay media was improved compared to PBS buffer, likely due to protein binding. Passive permeability was negligible for 19 in PBS, as expected from the solubility data. Moderate to good permeability was observed for the nitrobenzyl and oxetanecontaining derivatives 15 and 46, respectively, at each of 3 pH levels. Toxicity in hepatocytes, most concerning for the nitrobenzyl analog 15, was not observed (>50 µM), nor was it significant with the other two analogs tested. While each compound possessed desirable attributes, compound 19 was selected as our lead, ML275, due to the overall profile which included the most improved potency, selectivity window and reduction in viral titer. Probe candidates were routinely assessed in a Eurofins PanLabs (formerly Ricerca) Hit LeadProfiling screen against 67 discrete GPCRs, ion channels and transporters. All assays were performed in duplicate with probe ML275 (19) at a concentration of 10 µM, and >50% inhibition was noted for five targets (Table 5). Inhibition of the human adenosine A3 receptor and the human platelet activating factor was determined to be 44% and 43%, respectively; however, inhibition of all remaining targets did not exceed 34%. A full list of the targets and percent inhibition by compound 19 is provided in the Supporting Information. The liability posed by significant inhibition of any given host target depends on a multitude of factors that includes but is not limited to potency, metabolism, and physiological compartmental exposure (e.g., CNS). These results, while not negligible, were considered informational at this stage of development, as pursuit of individual IC50 values and bioavailability data was cost-prohibitive and revision of the scaffold architecture was expected prior to finding a suitable tool for in vivo assessment. Nonetheless, the profiling outcome served as a useful alert to potential adverse effects associated with the series that will need to be surveyed as development continues toward an advanced lead candidate.
Table 5.
Off Target Profiling Results for Compound 19 at 10 µM (>50% Inhibition)
| entry | biological target | Eurofins Panlabs assay codea |
species | percent inhibition (%)b |
|---|---|---|---|---|
| 1 | calcium channel, L-type, benzothiazepine | 214510 | rat | 52 |
| 2 | calcium channel, L-type, dihydropyridine | 214600 | rat | 70 |
| 3 | cannabinoid CB1 | 217030 | human | 86 |
| 4 | serotonin (5-hydroxytryptamine) 5-HT2B | 271700 | human | 54 |
| 5 | norepinephrine transporter | 204410 | human | 54 |
Detailed assay descriptions can be found at https://www.eurofinspanlabs.com/catalog.
A full list of targets and percent inhibition by compound 19 is provided in the Supporting Information.
Mechanism of Action Studies
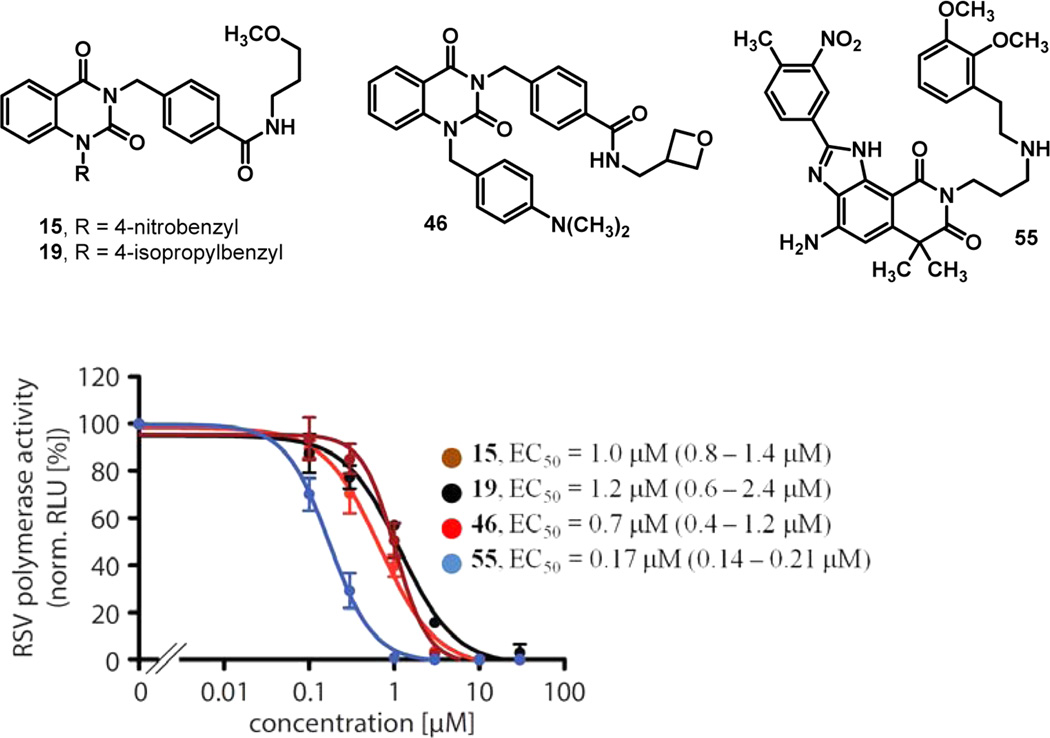
To gain an understanding of how this quinazolinedione class of compounds was inhibiting RSV, a cell-based, time-of-addition study was initially performed to determine the ability of these compounds to inhibit different stages of a single round of viral replication. The assay was performed by infecting cells with RSV and adding compound at a 10 µM concentration at each of several time points up to 24 h postinfection and then tracking cell viability over the course of the experiment. Compounds 15, 19, and 46 were evaluated as a panel alongside ribavirin. While ribavirin treatment protected cells from RSV-induced CPE (approximately 100%) for up to 7 h postinfection, indicating that it targets the period of infection during which viral replication is in progress, none of the three quinazolinediones showed dramatic changes in cell viability between 7 and 24 h postinfection, suggesting that these compounds acted at a later stage of the viral life cycle (data not shown). This time-of-addition profile suggested possible inhibition of the viral RNA-dependent RNA-polymerase (RdRp) activity rather than interference with receptor binding and/or membrane fusion. To test this hypothesis experimentally, compounds 15, 19 and 46 were subjected, along with a known literature-described RdRp inhibitor 55,18 to a plasmid-based RSV minigenome reporter assay that specifically monitored bioactivity of the viral polymerase machinery (Figure 3).38 Luciferase reporter interference was tested by comparing the activity of 19 in assays employing recRSV-luciferase or the equivalent measles virus recombinant (recMeV-luciferase), both of which rely on luciferase activity as the assay readout. While recRSV-luciferase was efficiently inhibited, compound 19 was inactive against recMeV-luciferase, excluding direct reporter interference. All four analogs showed a dose-dependent inhibition of reporter expression with active concentrations similar to those observed against live virus, indicating that the compounds blocked RSV RdRp activity.
Figure 3.
Transient RSV luciferase replicon reporter assay to determine RdRp activity in the presence of the quinazolinedione analogs. Values were normalized for vehicle (DMSO) treated samples and represent averages of three independent experiments, each performed in duplicate. Error bars represent standard deviation; EC50 values and 95% confidence intervals (in parentheses) are shown for each compound.
At this point the exact target with which these compounds interact leading to the observed block of RdRp activity is unknown, but sequencing of RSV-resistant mutant viruses resulting from compound treated cells is currently underway. These studies are expected to pinpoint the location of mutation within the genome that may be responsible for resistance and identify a potential target to investigate further. Attempts to unravel the nature of inhibition are being actively pursued.
CONCLUSION
In summary, we have identified a quinazolinedione class of compounds that show promising cellular activity against RSV. While the SAR for this set of quinazolinediones was relatively limited in texture, we have discovered distinct scaffold regions that tolerate structural change and permit tuning of physiochemical properties and whose modification has led to a 7-fold improvement in selectivity over the hit compound 2. Several compounds in the series inhibited a virus-induced cytopathic effect in the submicromolar range without exhibiting significant cellular toxicity, and importantly, these same compounds also significantly reduced viral plaque formation at a concentration of 10 µM. Furthermore, the mechanism of action for a subset of quinazolinedione analogs was explored and demonstrated to block the activity of the viral RNA-dependent RNA–polymerase complex which is responsible for RSV genome replication and transcription. These results, combined with the insights from structural modifications of the quinazolinedione scaffold and in vitro ADME data of select analogs, suggest that a favorable pharmacological profile can be tuned to produce a lead antiviral compound suitable for in vivo efficacy assessment against RSV. As such, the current set of compounds will be useful tools in establishing baseline pharmacokinetic parameters and further investigating how these agents interact with the RdRp complex to inhibit viral replication.
EXPERIMENTAL SECTION
Chemistry
The purity of all final compounds was >95% and was confirmed by HPLC/MS analysis employing an Agilent 1200 RRL chromatograph with photodiode array UV detection and an Agilent 6224 TOF mass spectrometer. The chromatographic method utilized a Waters Acquity BEH C-18 2.1 × 50 mm, 1.7 µm column; UV detection wavelength = 214 nm; flow rate = 0.4 mL/min; gradient = 5–100% acetonitrile over 3 min with a hold of 0.8 min at 100% acetonitrile; the aqueous mobile phase contained 0.15% ammonium hydroxide (v/v). The mass spectrometer utilized the following parameters: an Agilent multimode source which simultaneously acquires ESI+/APCI+; a reference mass solution consisting of purine and hexakis(1H,1H,3H-tetrafluoropropoxy)phosphazine; and a makeup solvent of 90:10:0.1 MeOH:water:formic acid which was introduced to the LC flow prior to the source to assist ionization. 1H and 13C NMR spectra were recorded on a Bruker AM 400 spectrometer (operating at 400 and 101 MHz, respectively) or a Bruker AVIII spectrometer (operating at 500 and 126 MHz, respectively) in CDCl3 with 0.03% TMS as an internal standard or DMSO-d6. The chemical shifts (δ) reported are given in parts per million (ppm) and the coupling constants (J) are in Hertz (Hz). The spin multiplicities are reported as s = singlet, bs = broad singlet, d = doublet, t = triplet, q = quartet, dd = doublet of doublet, hept = heptet, and m = multiplet. Melting points were determined on a Stanford Research Systems OptiMelt apparatus. Compounds were generally prepared according to Scheme 1 and the protocols detailed for compound 19, below, unless otherwise specified.
Synthesis of 4-((1-(4-isopropylbenzyl)-2,4-dioxo-1,2-dihydroquinazolin- 3(4H)-yl)methyl)-N-(3-methoxypropyl)benzamide (19)
Step 1
To a solution of 2-aminobenzoic acid 3 (5.00 g, 36.50 mmol) in DMF (45 mL) was added methyl 4-(aminomethyl)benzoate hydrochloride (7.35 g, 36.5 mmol), HATU (15.25 g, 40.10 mmol) and N,N-diisopropylethylamine (18.08 mL, 109 mmol). The reaction mixture was stirred for 16 h at room temperature, then diluted with CH2Cl2 (200 mL) and washed sequentially with 1 M HCl (150 mL), sat. aqueous NaHCO3 (150 mL) and water (2 × 800 mL). The separated organic extract was dried (MgSO4), filtered, and concentrated under reduced pressure to afford a crude product which was purified by silica gel flash column chromatography (0–60% v/v EtOAc/Hexane), yielding the product, methyl 4-((2-aminobenzamido)methyl)benzoate, as a white solid (5.00 g, 17.59 mmol, 48% yield). 1H NMR (400 MHz, CDCl3): δ 8.01 (d, J = 8.4 Hz, 2H), 7.41 (d, J = 8.6 Hz, 2H), 7.35 (dd, J = 7.9, 1.4 Hz, 1H), 7.25–7.19 (m, 1H), 6.70 (dd, J = 8.3, 0.9 Hz, 1H), 6.67–6.61 (m, 1H), 6.43 (broad s, 1H), 5.56 (broad s, 2H), 4.66 (d, J = 5.9 Hz, 2H), 3.91 (s, 3H).
Step 2
After stirring a solution of methyl 4-((2-aminobenzamido)-methyl)benzoate (5.00 g, 17.59 mmol) and N,N-diisopropylethylamine (14.53 mL, 88 mmol) in CH2Cl2 (260 mL) for 10 min at room temperature under nitrogen, 1,1′-carbonyldiimidazole (8.55 g, 52.80 mmol) was added, and the reaction mixture was heated 16 h at reflux. The formed precipitate was filtered, dried under vacuum and the desired product, methyl 4-((2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)benzoate 4, was furnished as a white solid without further purification (4.65 g, 14.99 mmol, 85% yield). 1H NMR (400 MHz, DMSO-d6): δ 11.58 (s, 1H), 7.94 (dd, J = 8.3, 1.4 Hz, 1H), 7.90 (d, J = 8.4 Hz, 2H), 7.72–7.64 (m, 1H), 7.43 (d, J = 8.4 Hz, 2H), 7.26–7.18 (m, 2H), 5.15 (s, 2H), 3.83 (s, 3H).
Step 3
To a solution of methyl 4-((2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)benzoate 4 (2.60 g, 8.38 mmol) in THF (50 mL) was added 1 M lithium hydroxide (50 mL, 50.3 mmol). The reaction mixture was stirred at 40 °C for 1 h, at which point TLC confirmed reaction completion. Then 1 M HCl was cautiously added until the reaction mixture reached pH 2, at which point the product precipitated out of solution. The precipitate was collected by filtration, washed with water (2 × 70 mL), dried under high vacuum to afford the desired product, 4-((2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)-benzoic acid, as a white solid (2.19 g, 7.39 mmol, 88% yield). 1H NMR (400 MHz, DMSO-d6): δ 12.90 (broad s, 1H), 11.58 (s, 1H), 7.95 (d, J = 7.3 Hz, 1H), 7.88 (d, J = 8.3 Hz, 2H), 7.74–7.65 (m, 1H), 7.40 (d, J = 8.3 Hz, 2H), 7.28–7.18 (m, 2H), 5.15 (s, 2H).
Step 4
To a solution of 4-((2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)benzoic acid (1.00 g, 3.38 mmol) in DMF (15 mL) was added 3-methoxypropylamine (0.35 mL, 3.38 mmol), HATU (1.41 g, 3.71 mmol) and N,N-diisopropylethylamine (1.67 mL, 10.13 mmol). The reaction mixture was stirred for 16 h at room temperature, then diluted with CH2Cl2 (90 mL) and washed sequentially with 1 M HCl (60 mL), sat. aqueous NaHCO3 (60 mL) and water (2 × 180 mL). The organic extract was separated, dried (MgSO4), filtered, and concentrated under reduced pressure to afford a crude product which was purified by silica gel flash column chromatography (0–5% v/v MeOH/CH2Cl2) yielding the desired product, 4-((2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)-N-(3-methoxypropyl)benzamide, as a white solid (0.91 g, 2.48 mmol, 73% yield), mp 244–246 °C. 1H NMR (400 MHz, DMSO-d6): δ 11.56 (s, 1H), 8.40 (t, J = 5.6 Hz, 1H), 7.95 (d, J = 7.3 Hz, 1H), 7.76 (d, J = 8.4 Hz, 2H), 7.71–7.63 (m, 1H), 7.36 (d, J = 8.4 Hz, 2H), 7.26–7.17 (m, 2H), 5.13 (s, 2H), 3.35 (t, J = 6.3 Hz, 2H), 3.31–3.24 (m, 2H), 3.22 (s, 3H), 1.78–1.67 (m, 2H).
Step 5
To a solution of 4-((2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)-N-(3-methoxypropyl)benzamide (0.050 g, 0.14 mmol) in DMF (2 mL) was added 4-isopropylbenzyl bromide (0.028 mL, 0.16 mmol) and potassium carbonate (0.056 g, 0.41 mmol). The resulting reaction mixture was stirred at 40 °C for 16 h. The formed residue was dissolved in CH2Cl2 (6 mL) and sequentially washed with 1 M HCl (4 mL), water (3 × 20 mL) and brine (8 mL). The organic layer was separated, dried (MgSO4), filtered, and concentrated under reduced pressure to give a crude product which was purified by silica gel flash column chromatography (0–5% v/v MeOH/CH2Cl2) yielding 4-((1-(4-isopropylbenzyl)-2,4-dioxo-1,2-dihydroquinazolin- 3(4H)-yl)methyl)-N-(3-methoxypropyl)benzamide (19) as a white solid (0.030 g, 0.060 mmol, 44% yield), mp 178–180 °C. 1H NMR (500 MHz, CDCl3): δ 8.24 (dd, J = 7.9, 1.6 Hz, 1H), 7.74–7.68 (m, 2H), 7.61–7.52 (m, 3H), 7.25–7.20 (m, 1H), 7.20– 7.12 (m, 5H), 6.89 (t, J = 5.3 Hz, 1H), 5.36 (s, 2H), 5.33 (s, 2H), 3.59–3.52 (m, 4H), 3.37 (s, 3H), 2.87 (h, J = 6.9 Hz, 1H), 1.91–1.83 (m, 2H), 1.21 (d, J = 6.9 Hz, 6H). 13C NMR (126 MHz, CDCl3) δ 167.01, 161.92, 151.48, 148.54, 140.41, 140.16, 135.40, 134.15, 132.89, 129.25, 129.11, 127.19, 127.17, 126.57, 123.29, 115.74, 114.69, 72.59, 59.10, 47.36, 44.94, 39.31, 33.89, 28.95, 24.06. LCMS Retention time: 3.422 min. LCMS purity 98.8%. HRMS (ESI): m/z calcd for C30H33N3O4 [M + H]+ 500.2544, found 500.2540.
N-(3-Methoxypropyl)-4-((1-(4-methylbenzyl)-2,4-dioxo-1,2-dihydroquinazolin- 3(4H)-yl)methyl)benzamide (2)
Isolated as a white solid (36 mg, 56% yield), mp 197–199 °C. 1H NMR (500 MHz, CDCl3): δ 8.23 (dd, J = 7.9, 1.6 Hz, 1H), 7.74–7.68 (m, 2H), 7.60–7.51 (m, 3H), 7.22 (ddd, J = 8.0, 7.3, 0.9 Hz, 1H), 7.16–7.09 (m, 5H), 6.89 (t, J = 5.2 Hz, 1H), 5.36 (s, 2H), 5.33 (s, 2H), 3.59–3.52 (m, 4H), 3.37 (s, 3H), 2.31 (s, 3H), 1.91–1.83 (m, 2H). 13C NMR (126 MHz, CDCl3): δ 167.01, 161.92, 151.49, 140.41, 140.11, 137.59, 135.38, 134.16, 132.59, 129.80, 129.26, 129.11, 127.17, 126.54, 123.30, 115.76, 114.64, 72.58, 59.09, 47.37, 44.95, 39.31, 28.95, 21.23. LCMS Retention time: 1.98 min. LCMS purity 97.8%. HRMS (ESI): m/z calcd for C28H29N3O4 [M + H]+ 472.2158, found 472.2241.
4-((1-(2-Bromobenzyl)-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)-N-(3-methoxypropyl)benzamide (8)
Isolated as a white solid (31 mg, 71% yield), mp 187–189 °C. 1H NMR (500 MHz, CDCl3): δ 8.27 (dd, J = 8.0, 1.6 Hz, 1H), 7.74–7.69 (m, 2H), 7.63 (dd, J = 7.7, 1.5 Hz, 1H), 7.61–7.52 (m, 3H), 7.29–7.22 (m, 1H), 7.21–7.11 (m, 2H), 6.93–6.87 (m, 2H), 6.85–6.79 (m, 1H), 5.41 (s, 2H), 5.37 (s, 2H), 3.60–3.52 (m, 4H), 3.37 (s, 3H), 1.91–1.83 (m, 2H). 13C NMR (126 MHz, CDCl3): δ 166.98, 161.83, 151.39, 140.28, 139.75, 135.65, 134.25, 134.09, 133.30, 129.37, 129.29, 129.21, 128.13, 127.20, 126.69, 123.63, 122.44, 115.76, 114.56, 72.60, 59.10, 48.02, 45.00, 39.33, 28.94. LCMS Retention time: 3.250 min. LCMS purity 97.2%. HRMS (ESI): m/z calcd for C27H26BrN3O4 [M + H]+ 538.1163, found 538.1159.
4-((1-(3-Bromobenzyl)-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)-N-(3-methoxypropyl)benzamide (9)
Isolated as a white solid (28 mg, 64% yield), mp 172–174 °C. 1H NMR (500 MHz, CDCl3): δ 8.26 (dd, J = 8.0, 1.6 Hz, 1H), 7.75–7.69 (m, 2H), 7.61–7.54 (m, 3H), 7.44–7.36 (m, 2H), 7.29–7.23 (m, 1H), 7.20 (apparent t, J = 7.8 Hz, 1H), 7.17–7.12 (m, 1H), 7.06 (d, J = 8.4 Hz, 1H), 6.89 (t, J = 5.2 Hz, 1H), 5.36 (s, 2H), 5.33 (s, 2H), 3.61–3.51 (m, 4H), 3.37 (s, 3H), 1.93–1.81 (m, 2H). 13C NMR (126 MHz, CDCl3): δ 166.98, 161.76, 151.46, 140.25, 139.80, 138.05, 135.55, 134.24, 131.12, 130.74, 129.61, 129.49, 129.10, 127.22, 125.15, 123.60, 123.29, 115.82, 114.31, 72.59, 59.10, 47.02, 45.02, 39.32, 28.95. LCMS Retention time: 3.197 min. LCMS purity 99.4%. HRMS (ESI): m/z calcd for C27H26BrN3O4 [M + H]+ 538.1163, found 538.1159.
4-((1-(4-Bromobenzyl)-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)-N-(3-methoxypropyl)benzamide (10)
Isolated as a white solid (23 mg, 49% yield), mp 179–181 °C. 1H NMR (500 MHz, CDCl3): δ 8.25 (dd, J = 7.9, 1.6 Hz, 1H), 7.74–7.68 (m, 2H), 7.60–7.53 (m, 3H), 7.48–7.42 (m, 2H), 7.28–7.21 (m, 1H), 7.14–7.09 (m, 2H), 7.05 (d, J = 8.4 Hz, 1H), 6.89 (t, J = 5.4 Hz, 1H), 5.35 (s, 2H), 5.31 (s, 2H), 3.60–3.52 (m, 4H), 3.37 (s, 3H), 1.91–1.83 (m, 2H). 13C NMR (126 MHz, CDCl3): δ 166.94, 161.76, 151.44, 140.24, 139.82, 135.48, 134.72, 134.26, 132.29, 129.46, 129.15, 128.32, 127.20, 123.55, 121.78, 115.80, 114.34, 72.62, 59.11, 47.06, 44.99, 39.34, 28.95. LCMS Retention time: 3.208 min. LCMS purity 99%. HRMS (ESI): m/z calcd for C27H26BrN3O4 [M + H]+ 538.1163, found 538.1191.
4-((1-(4-Fluorobenzyl)-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)- methyl)-N-(3-methoxypropyl)benzamide (12)
Isolated as a white solid (27 mg, 70% yield), mp 173–175 °C. 1H NMR (500 MHz, CDCl3): δ 8.25 (dd, J = 7.9, 1.6 Hz, 1H), 7.75–7.69 (m, 2H), 7.61– 7.54 (m, 3H), 7.26–7.19 (m, 3H), 7.10 (d, J = 8.4 Hz, 1H), 7.05–6.98 (m, 2H), 6.90 (t, J = 5.4 Hz, 1H), 5.36 (s, 2H), 5.33 (s, 2H), 3.60– 3.53 (m, 4H), 3.37 (s, 3H), 1.91–1.83 (m, 2H). 13C NMR (126 MHz, CDCl3): δ 166.97, 162.34 (d, J = 246.5 Hz), 161.80, 151.46, 140.28, 139.90, 135.44, 134.22, 131.38 (d, J = 3.2 Hz), 129.42, 129.14, 128.35 (d, J = 8.1 Hz), 127.19, 123.49, 116.11 (d, J = 21.7 Hz), 115.80, 114.38, 72.59, 59.09, 46.95, 44.98, 39.32, 28.94. LCMS Retention time: 3.067 min. LCMS purity 99.4%. HRMS (ESI): m/z calcd for C27H26FN3O4 [M + H]+ 476.1980, found 476.1972.
4-((1-(4-Chlorobenzyl)-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)-N-(3-methoxypropyl)benzamide (13)
Isolated as a white solid (37 mg, 55% yield), mp 185–187 °C. 1H NMR (500 MHz, CDCl3): δ 8.25 (dd, J = 8.0, 1.6 Hz, 1H), 7.75–7.68 (m, 2H), 7.61– 7.53 (m, 3H), 7.34–7.28 (m, 2H), 7.28–7.21 (m, 1H), 7.20–7.14 (m, 2H), 7.06 (d, J = 8.5 Hz, 1H), 6.91 (t, J = 5.3 Hz, 1H), 5.35 (s, 2H), 5.33 (s, 2H), 3.60–3.52 (m, 4H), 3.37 (s, 3H), 1.92–1.83 (m, 2H). 13C NMR (126 MHz, CDCl3): δ 166.94, 161.77, 151.44, 140.25, 139.83, 135.47, 134.25, 134.18, 133.73, 129.45, 129.34, 129.15, 127.99, 127.19, 123.54, 115.80, 114.35, 72.61, 59.10, 47.01, 44.99, 39.33, 28.95. LCMS Retention time: 3.171 min. LCMS purity 100%. HRMS (ESI): m/z calcd for C27H26ClN3O4 [M + H]+ 492.1685, found 492.1700.
4-((1-(4-Methoxybenzyl)-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)-N-(3-methoxypropyl)benzamide (14)
Isolated as a white solid (33 mg, 50% yield), mp 171–173 °C. 1H NMR (500 MHz, CDCl3): δ 8.24 (dd, J = 7.9, 1.6 Hz, 1H), 7.75–7.68 (m, 2H), 7.60– 7.53 (m, 3H), 7.22 (apparent t, J = 7.1 Hz, 1H), 7.20–7.14 (m, 3H), 6.90 (t, J = 5.2 Hz, 1H), 6.87–6.82 (m, 2H), 5.36 (s, 2H), 5.31 (s, 2H), 3.77 (s, 3H), 3.60–3.52 (m, 4H), 3.37 (s, 3H), 1.93–1.82 (m, 2H). 13C NMR (126 MHz, CDCl3): δ 166.99, 161.89, 159.22, 151.48, 140.40, 140.07, 135.35, 134.16, 129.28, 129.10, 128.00, 127.62, 127.17, 123.29, 115.77, 114.60, 114.51, 72.58, 59.09, 55.42, 47.06, 44.93, 39.30, 28.95. LCMS Retention time: 3.002 min. LCMS purity 100%. HRMS (ESI): m/z calcd for C28H29N3O5 [M + H]+ 488.2180, found 488.2190.
N-(3-Methoxypropyl)-4-((1-(4-nitrobenzyl)-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)benzamide (15)
Isolated as an off-white solid (21 mg, 48% yield), mp 170–173 °C. 1H NMR (500 MHz, CDCl3): δ 8.29 (dd, J = 7.9, 1.6 Hz, 1H), 8.23–8.17 (m, 2H), 7.75– 7.69 (m, 2H), 7.61–7.54 (m, 3H), 7.43–7.37 (m, 2H), 7.31–7.24 (m, 1H), 6.98 (d, J = 8.4 Hz, 1H), 6.91 (t, J = 5.3 Hz, 1H), 5.46 (s, 2H), 5.36 (s, 2H), 3.60–3.53 (m, 4H), 3.38 (s, 3H), 1.91–1.84 (m, 2H). 13C NMR (126 MHz, CDCl3): δ 166.89, 161.61, 151.41, 147.69, 143.14, 140.06, 139.57, 135.63, 134.37, 129.74, 129.21, 127.39, 127.23, 124.47, 123.86, 115.88, 113.97, 72.65, 59.11, 47.12, 45.07, 39.38, 28.94. LCMS Retention time: 2.973 min. LCMS purity 97.8%. HRMS (ESI): m/z calcd for C27H26N4O6 [M + H]+ 503.1925, found 503.1951.
4-((2,4-Dioxo-1-(4-(trifluoromethyl)benzyl)-1,2-dihydroquinazolin-3(4H)-yl)methyl)-N-(3-methoxypropyl)benzamide (16)
Isolated as a white solid (27 mg, 59% yield), mp 174–177 °C. 1H NMR (500 MHz, CDCl3): δ 8.27 (dd, J = 7.9, 1.6 Hz, 1H), 7.75–7.69 (m, 2H), 7.62–7.53 (m, 5H), 7.34 (d, J = 7.9 Hz, 2H), 7.29–7.23 (m, 1H), 7.03 (d, J = 8.3 Hz, 1H), 6.90 (t, J = 5.3 Hz, 1H), 5.42 (s, 2H), 5.36 (s, 2H), 3.63–3.51 (m, 4H), 3.37 (s, 3H), 1.93–1.82 (m, 2H). 13C NMR (126 MHz, CDCl3): δ 166.92, 161.72, 151.45, 140.18, 139.76, 135.57, 134.30, 130.26 (q, J = 32.6 Hz), 129.56, 129.18, 127.21, 126.83, 126.18 (q, J = 3.8 Hz), 125.10, 123.67, 122.94, 115.83, 114.22, 72.63, 59.10, 47.21, 45.02, 39.36, 28.95. LCMS Retention time: 3.207 min. LCMS purity 97.8%. HRMS (ESI): m/z calcd for C28H26F3N3O4 [M + H]+ 526.1948, found 526.1973.
4-((1-(4-Cyanobenzyl)-2,4-dioxo-1,2-dihydroquinazolin-3(4H)- yl)methyl)-N-(3-methoxypropyl)benzamide (17)
Isolated as a white solid (43.4 mg, 66% yield), mp 187–189 °C. 1H NMR (500 MHz, CDCl3): δ 8.28 (dd, J = 7.9, 1.6 Hz, 1H), 7.75–7.69 (m, 2H), 7.66– 7.61 (m, 2H), 7.61–7.53 (m, 3H), 7.34 (d, J = 8.3 Hz, 2H), 7.30–7.24 (m, 1H), 6.98 (d, J = 8.4 Hz, 1H), 6.92 (t, J = 5.2 Hz, 1H), 5.41 (s, 2H), 5.35 (s, 2H), 3.62–3.50 (m, 4H), 3.38 (s, 3H), 1.93–1.82 (m, 2H). 13C NMR (126 MHz, CDCl3): δ 166.90, 161.63, 151.40, 141.16, 140.08, 139.60, 135.61, 134.32, 133.00, 129.66, 129.19, 127.23, 127.21, 123.80, 118.50, 115.83, 114.02, 111.96, 72.62, 59.09, 47.27, 45.04, 39.35, 28.92. LCMS Retention time: 2.910 min. LCMS purity 99.6%. HRMS (ESI): m/z calcd for C28H26N4O4 [M + H]+ 483.2027, found 483.2024.
4-((1-(4-Ethylbenzyl)-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)-methyl)-N-(3-methoxypropyl)benzamide (18)
Isolated as a white solid (39 mg, 59% yield) mp 170–172 °C. 1H NMR (500 MHz, CDCl3): δ 8.24 (dd, J = 7.9, 1.6 Hz, 1H), 7.75–7.68 (m, 2H), 7.61–7.52 (m, 3H), 7.22 (ddd, J = 8.0, 7.3, 0.9 Hz, 1H), 7.18–7.12 (m, 5H), 6.91 (t, J = 5.4 Hz, 1H), 5.36 (s, 2H), 5.34 (s, 2H), 3.60–3.52 (m, 4H), 3.37 (s, 3H), 2.61 (q, J = 7.6 Hz, 2H), 1.91–1.83 (m, 2H), 1.20 (t, J = 7.6 Hz, 3H). 13C NMR (126 MHz, CDCl3): δ 167.01, 161.91, 151.48, 143.91, 140.40, 140.12, 135.38, 134.14, 132.78, 129.24, 129.09, 128.60, 127.17, 126.58, 123.28, 115.74, 114.66, 72.56, 59.08, 47.36, 44.93, 39.29, 28.95, 28.59, 15.61. LCMS Retention time: 3.323 min. LCMS purity 99.1%. HRMS (ESI): m/z calcd for C29H31N3O4 [M + H]+ 486.2387, found 486.2383.
4-((1-(Benzo[c][1,2,5]oxadiazol-5-ylmethyl)-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)-N-(3-methoxypropyl)benzamide (20)
Isolated as an off-white solid (25 mg, 58% yield), mp 70–74 °C. 1H NMR (500 MHz, CDCl3): δ 8.31 (dd, J = 8.0, 1.6 Hz, 1H), 7.87 (dd, J = 9.3, 1.0 Hz, 1H), 7.75–7.69 (m, 2H), 7.63–7.54 (m, 4H), 7.36 (dd, J = 9.3, 1.5 Hz, 1H), 7.33–7.27 (m, 1H), 7.05 (d, J = 8.3 Hz, 1H), 6.91 (t, J = 5.2 Hz, 1H), 5.44 (s, 2H), 5.37 (s, 2H), 3.61–3.52 (m, 4H), 3.37 (s, 3H), 1.92–1.84 (m, 2H). 13C NMR (126 MHz, CDCl3): δ 166.91, 161.57, 151.37, 149.12, 148.82, 140.02, 139.76, 139.49, 135.73, 134.39, 130.85, 129.82, 129.19, 127.25, 123.99, 117.97, 115.88, 113.88, 113.04, 72.61, 59.11, 47.41, 45.12, 39.35, 28.95. LCMS Retention time: 2.955 min. LCMS purity 97.9%. HRMS (ESI): m/z calcd for C27H25N5O5 [M + H]+ 500.1928, found 500.1955.
N-(3-Methoxypropyl)-4-((1-((5-methylisoxazol-3-yl)methyl)-2,4-dioxo-1,2 dihydroquinazolin-3(4H)-yl)methyl)benzamide (21)
Isolated as a white solid (37 mg, 59% yield), mp 164–166 °C. 1H NMR (500 MHz, CDCl3): δ 8.23 (dd, J = 7.9, 1.6 Hz, 1H), 7.74–7.69 (m, 2H), 7.65 (ddd, J = 8.7, 7.3, 1.6 Hz, 1H), 7.59–7.53 (m, 2H), 7.46 (d, J = 8.4 Hz, 1H), 7.30–7.24 (m, 1H), 6.91 (t, J = 5.1 Hz, 1H), 5.97 (apparent d, J = 1.0 Hz, 1H), 5.34 (s, 2H), 5.33 (s, 2H), 3.60–3.52 (m, 4H), 3.37 (s, 3H), 2.37 (apparent d, J = 0.8 Hz, 3H), 1.92–1.82 (m, 2H). 13C NMR (126 MHz, CDCl3): δ 170.89, 166.95, 161.75, 159.51, 151.19, 140.17, 139.66, 135.68, 134.24, 129.32, 129.07, 127.19, 123.69, 115.69, 114.28, 101.28, 72.62, 59.09, 44.93, 39.78, 39.34, 28.94, 12.45. LCMS Retention time: 2.779 min. LCMS purity 100%. HRMS (ESI): m/z calcd for C25H26N4O5 [M + H]+ 463.1976, found 463.1994.
4-((2,4-Dioxo-1-(pyridin-2-ylmethyl)-1,2-dihydroquinazolin-3(4H)-yl)methyl)-N-(3-methoxypropyl)benzamide (22)
Isolated as a white solid (23 mg, 53% yield), mp 191–193 °C. 1H NMR (500 MHz, CDCl3): δ 8.57 (dt, J = 4.7, 1.5 Hz, 1H), 8.24 (dd, J = 7.9, 1.6 Hz, 1H), 7.75–7.69 (m, 2H), 7.63 (td, J = 7.7, 1.8 Hz, 1H), 7.61–7.53 (m, 3H), 7.30 (d, J = 8.5 Hz, 1H), 7.26–7.18 (m, 3H), 6.90 (t, J = 5.1 Hz, 1H), 5.48 (s, 2H), 5.37 (s, 2H), 3.62–3.51 (m, 4H), 3.37 (s, 3H), 1.93–1.82 (m, 2H). 13C NMR (126 MHz, CDCl3): δ 167.00, 161.92, 155.77, 151.54, 149.69, 140.35, 140.15, 137.32, 135.46, 134.18, 129.19, 129.09, 127.18, 123.45, 122.94, 121.64, 115.73, 114.93, 72.60, 59.10, 49.51, 44.98, 39.32, 28.95. LCMS Retention time: 1.610 min. LCMS purity 100%. HRMS (ESI): m/z calcd for C26H26N4O4 [M + H]+ 459.2030, found 459.2100.
4-((2,4-Dioxo-1-(pyridin-3-ylmethyl)-1,2-dihydroquinazolin-3(4H)-yl)methyl)-N-(3-methoxypropyl)benzamide (23)
Isolated as a white solid (4.8 mg, 11% yield). 1H NMR (500 MHz, CDCl3): δ 8.62 (s, 1H), 8.55 (d, J = 4.9 Hz, 1H), 8.27 (dd, J = 7.9, 1.6 Hz, 1H), 7.75– 7.69 (m, 2H), 7.63–7.52 (m, 4H), 7.30–7.23 (m, 2H), 7.10 (d, J = 8.4 Hz, 1H), 6.90 (t, J = 5.2 Hz, 1H), 5.39 (s, 2H), 5.36 (s, 2H), 3.61– 3.52 (m, 4H), 3.38 (s, 3H), 1.93–1.82 (m, 2H). 13C NMR (126 MHz, CDCl3): δ 166.96, 161.68, 151.48, 149.48, 148.54, 140.16, 139.64, 135.59, 134.61, 134.29, 131.49, 129.63, 129.14, 127.23, 124.01, 123.69, 115.85, 114.05, 72.59, 59.10, 45.24, 45.03, 39.32, 28.95. LCMS Retention time: 1.510 min. LCMS purity 99%. HRMS (ESI): m/z calcd for C26H26N4O4 [M + H]+ 459.2030, found 459.2000.
N-(3-Ethoxypropyl)-4-((1-(4-methylbenzyl)-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)benzamide (24)
Isolated as a white solid (24 mg, 44% yield), mp 174–176 °C. 1H NMR (500 MHz, CDCl3): δ 8.24 (dd, J = 7.9, 1.6 Hz, 1H), 7.75–7.70 (m, 2H), 7.61–7.51 (m, 3H), 7.22 (ddd, J = 8.1, 7.3, 0.9 Hz, 1H), 7.16–7.11 (m, 5H), 7.08 (t, J = 5.1 Hz, 1H), 5.36 (s, 2H), 5.33 (s, 2H), 3.64–3.54 (m, 4H), 3.51 (q, J = 7.0 Hz, 2H), 2.31 (s, 3H), 1.91–1.83 (m, 2H), 1.24 (t, J = 7.0 Hz, 3H). 13C NMR (126 MHz, CDCl3): δ 166.83, 161.90, 151.49, 140.38, 140.10, 137.57, 135.36, 134.14, 132.59, 129.79, 129.26, 129.09, 127.15, 126.54, 123.28, 115.76, 114.63, 70.71, 66.76, 47.36, 44.93, 39.63, 28.95, 21.23, 15.55. LCMS Retention time: 3.278 min. LCMS purity 99.7%. HRMS (ESI): m/z calcd for C29H31N3O4 [M + H]+ 486.2387, found 486.2389.
N-(3-Isopropoxypropyl)-4-((1-(4-methylbenzyl)-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)benzamide (25)
Isolated as a white solid (35 mg, 62% yield), mp 163–165 °C. 1H NMR (500 MHz, CDCl3): δ 8.24 (dd, J = 7.9, 1.6 Hz, 1H), 7.76–7.70 (m, 2H), 7.61–7.51 (m, 3H), 7.22 (ddd, J = 8.0, 7.3, 0.9 Hz, 1H), 7.16 (t, J = 4.5 Hz, 1H), 7.15–7.10 (m, 5H), 5.36 (s, 2H), 5.33 (s, 2H), 3.65–3.53 (m, 5H), 2.31 (s, 3H), 1.91–1.81 (m, 2H), 1.18 (d, J = 6.1 Hz, 6H). 13C NMR (126 MHz, CDCl3): δ 166.77, 161.90, 151.49, 140.34, 140.10, 137.56, 135.36, 134.16, 132.59, 129.79, 129.26, 129.07, 127.17, 126.54, 123.28, 115.77, 114.62, 72.14, 68.29, 47.36, 44.93, 39.85, 29.15, 22.36, 21.23. LCMS Retention time: 3.374 min. LCMS purity 99.6%. HRMS (ESI): m/z calcd for C30H33N3O4 [M + H]+ 500.2544, found 500.2541.
N-(3-(Dimethylamino)propyl)-4-((1-(4-methylbenzyl)-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)benzamide (26)
Isolated as a white solid (12 mg, 22% yield), mp 185–187 °C. 1H NMR (500 MHz, CDCl3): δ 8.44 (t, J = 4.8 Hz, 1H), 8.24 (dd, J = 7.9, 1.6 Hz, 1H), 7.76–7.69 (m, 2H), 7.61–7.51 (m, 3H), 7.22 (ddd, J = 8.1, 7.3, 0.9 Hz, 1H), 7.17–7.09 (m, 5H), 5.36 (s, 2H), 5.33 (s, 2H), 3.58–3.51 (m, 2H), 2.52–2.46 (m, 2H), 2.31 (s, 3H), 2.28 (s, 6H), 1.80–1.70 (m, 2H). 13C NMR (126 MHz, CDCl3): δ 166.75, 161.93, 151.49, 140.19, 140.11, 137.58, 135.37, 134.16, 132.59, 129.79, 129.26, 129.06, 127.18, 126.55, 123.29, 115.77, 114.63, 59.64, 47.36, 45.62, 44.95, 40.87, 25.31, 21.23. LCMS Retention time: 3.187 min. LCMS purity 98.7%. HRMS (ESI): m/z calcd for C29H32N4O3 [M + H]+ 485.2547, found 485.2545.
N-(4-Methoxybutyl)-4-((1-(4-methylbenzyl)-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)benzamide (27)
Isolated as a white solid (21 mg, 57% yield), mp 167–169 °C. 1H NMR (500 MHz, CDCl3): δ 8.23 (dd, J = 7.9, 1.6 Hz, 1H), 7.75–7.69 (m, 2H), 7.62–7.51 (m, 3H), 7.22 (ddd, J = 8.1, 7.3, 0.9 Hz, 1H), 7.17–7.09 (m, 5H), 6.51 (t, J = 5.9 Hz, 1H), 5.36 (s, 2H), 5.33 (s, 2H), 3.51–3.39 (m, 4H), 3.34 (s, 3H), 2.31 (s, 3H), 1.75–1.64 (m, 4H). 13C NMR (126 MHz, CDCl3): δ 167.27, 161.90, 151.48, 140.46, 140.10, 137.58, 135.38, 134.24, 132.58, 129.80, 129.26, 129.13, 127.18, 126.54, 123.30, 115.75, 114.64, 72.58, 58.83, 47.37, 44.93, 39.93, 27.29, 26.61, 21.23. LCMS Retention time: 3.207 min. LCMS purity 96.5%. HRMS (ESI): m/z calcd for C29H31N3O4 [M + H]+ 486.2387, found 486.2383.
N-(2-Methoxyethyl)-4-((1-(4-methylbenzyl)-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)benzamide (28)
Isolated as a white solid (26 mg, 53% yield), mp 194–196 °C. 1H NMR (500 MHz, CDCl3): δ 8.23 (dd, J = 7.9, 1.6 Hz, 1H), 7.77–7.70 (m, 2H), 7.62– 7.57 (m, 2H), 7.55 (ddd, J = 8.7, 7.3, 1.7 Hz, 1H), 7.22 (ddd, J = 8.1, 7.3, 0.9 Hz, 1H), 7.17–7.08 (m, 5H), 6.48 (t, J = 5.5 Hz, 1H), 5.37 (s, 2H), 5.33 (s, 2H), 3.68–3.60 (m, 2H), 3.58–3.52 (m, 2H), 3.37 (s, 3H), 2.31 (s, 3H). 13C NMR (126 MHz, CDCl3): δ 167.31, 161.90, 151.49, 140.66, 140.10, 137.59, 135.38, 133.93, 132.59, 129.81, 129.27, 129.14, 127.30, 126.54, 123.31, 115.75, 114.64, 71.34, 58.99, 47.37, 44.94, 39.79, 21.23. LCMS Retention time: 3.088 min. LCMS purity 99.3%. HRMS (ESI): m/z calcd for C27H27N3O4 [M + H]+ 458.2074, found 458.2088.
4-((1-(4-Methylbenzyl)-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)-N-(oxetan-3-ylmethyl)benzamide (29)
Isolated as a white solid (24 mg, 67% yield), mp 200–202 °C. 1H NMR (500 MHz, CDCl3): δ 8.23 (dd, J = 7.9, 1.6 Hz, 1H), 7.75–7.68 (m, 2H), 7.61– 7.55 (m, 2H), 7.55 (ddd, J = 8.7, 7.3, 1.7 Hz, 1H), 7.22 (ddd, J = 8.1, 7.3, 0.9 Hz, 1H), 7.17–7.08 (m, 5H), 6.36 (t, J = 5.9 Hz, 1H), 5.36 (s, 2H), 5.33 (s, 2H), 4.82 (dd, J = 7.7, 6.3 Hz, 2H), 4.46 (apparent t, J = 6.1 Hz, 2H), 3.73 (apparent t, J = 6.1 Hz, 2H), 3.33–3.23 (m, 1H), 2.31 (s, 3H). 13C NMR (126 MHz, CDCl3): δ 167.80, 161.90, 151.47, 140.91, 140.09, 137.61, 135.42, 133.64, 132.54, 129.80, 129.24, 129.20, 127.23, 126.52, 123.34, 115.71, 114.66, 75.19, 47.38, 44.92, 42.56, 35.14, 21.23. LCMS Retention time: 3.038 min. LCMS purity 96.9%. HRMS (ESI): m/z calcd for C28H27N3O4 [M + H]+ 470.2074, found 470.2070.
N-(Cyclobutylmethyl)-4-((1-(4-methylbenzyl)-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)benzamide (30)
Isolated as a white solid (22 mg, 63% yield), mp 218–220 °C. 1H NMR (500 MHz, CDCl3): δ 8.23 (dd, J = 7.9, 1.6 Hz, 1H), 7.73–7.67 (m, 2H), 7.61– 7.55 (m, 2H), 7.54 (ddd, J = 8.7, 7.3, 1.6 Hz, 1H), 7.25–7.18 (m, 1H), 7.17–7.09 (m, 5H), 6.02 (t, J = 5.7 Hz, 1H), 5.36 (s, 2H), 5.33 (s, 2H), 3.46 (dd, J = 7.3, 5.7 Hz, 2H), 2.62–2.51 (m, 1H), 2.31 (s, 3H), 2.13–2.03 (m, 2H), 1.98–1.84 (m, 2H), 1.79–1.68 (m, 2H). 13C NMR (126 MHz, CDCl3): δ 167.47, 161.90, 151.48, 140.53, 140.10, 137.59, 135.39, 134.27, 132.57, 129.80, 129.25, 129.16, 127.17, 126.53, 123.31, 115.74, 114.64, 47.37, 45.31, 44.94, 35.20, 25.83, 21.23, 18.47. LCMS Retention time: 3.422 min. LCMS purity 96.0%. HRMS (ESI): m/z calcd for C29H29N3O3 [M + H]+ 468.2282, found 468.2274.
1-(4-Methylbenzyl)-3-(4-(morpholine-4-carbonyl)benzyl)-quinazoline-2,4(1H,3H)-dione (31)
Isolated as a white solid (35 mg, 60% yield), mp 172–174 °C. 1H NMR (500 MHz, CDCl3): δ 8.23 (dd, J = 7.9, 1.6 Hz, 1H), 7.61–7.57 (m, 2H), 7.55 (ddd, J = 8.7, 7.3, 1.7 Hz, 1H), 7.39–7.34 (m, 2H), 7.22 (ddd, J = 8.1, 7.3, 0.9 Hz, 1H), 7.17–7.10 (m, 5H), 5.35 (s, 2H), 5.33 (s, 2H), 3.97–3.25 (m, 8H), 2.31 (s, 3H). 13C NMR (126 MHz, CDCl3): δ 170.35, 161.89, 151.49, 140.09, 139.02, 137.60, 135.39, 134.68, 132.57, 129.79, 129.30, 129.22, 127.45, 126.55, 123.31, 115.75, 114.64, 67.03, 47.37, 44.92, 21.23. LCMS Retention time: 3.126 min. LCMS purity 99.3%. HRMS (ESI): m/z calcd for C28H27N3O4 [M + H]+ 470.2074, found 470.2086.
1-(4-Methylbenzyl)-3-(4-(pyrrolidine-1-carbonyl)benzyl)-quinazoline-2,4(1H,3H)-dione (32)
Isolated as a white solid (34 mg, 67% yield), mp 193–196 °C. 1H NMR (500 MHz, CDCl3): δ 8.23 (dd, J = 7.9, 1.6 Hz, 1H), 7.59–7.51 (m, 3H), 7.50–7.45 (m, 2H), 7.21 (ddd, J = 8.0, 7.2, 0.9 Hz, 1H), 7.16–7.10 (m, 5H), 5.35 (s, 2H), 5.33 (s, 2H), 3.63 (t, J = 7.0 Hz, 2H), 3.41 (t, J = 6.6 Hz, 2H), 2.31 (s, 3H), 1.99–1.91 (m, 2H), 1.88–1.80 (m, 2H). 13C NMR (126 MHz, CDCl3): δ 169.58, 161.88, 151.48, 140.09, 138.82, 137.55, 136.64, 135.32, 132.61, 129.78, 129.22, 129.00, 127.44, 126.55, 123.26, 115.78, 114.61, 49.75, 47.35, 46.28, 44.95, 26.52, 24.58, 21.23. LCMS Retention time: 3.241 min. LCMS purity 100%. HRMS (ESI): m/z calcd for C28H27N3O3 [M + H]+ 454.2125, found 454.2144.
1-(4-Methylbenzyl)-3-(4-(piperidine-1-carbonyl)benzyl)-quinazoline-2,4(1H,3H)-dione (33)
Isolated as a white solid (36 mg, 69% yield), mp 171–173 °C. 1H NMR (500 MHz, CDCl3): δ 8.23 (dd, J = 7.9, 1.6 Hz, 1H), 7.60–7.51 (m, 3H), 7.37–7.32 (m, 2H), 7.21 (ddd, J = 8.0, 7.3, 0.9 Hz, 1H), 7.16–7.10 (m, 5H), 5.39–5.30 (m, 4H), 3.69 (apparent broad s, 2H), 3.32 (apparent broad s, 2H), 2.31 (s, 3H), 1.72–1.57 (m, 4H), 1.48 (apparent broad s, 2H). 13C NMR (126 MHz, CDCl3): δ 170.20, 161.89, 151.49, 140.09, 138.39, 137.55, 135.91, 135.33, 132.61, 129.78, 129.21, 129.17, 127.14, 126.55, 123.26, 115.78, 114.61, 48.88, 47.36, 44.94, 43.23, 26.69, 25.73, 24.73, 21.23. LCMS Retention time: 3.412 min. LCMS purity 98.7%. HRMS (ESI): m/z calcd for C29H29N3O3 [M + H]+ 468.2282, found 468.2305.
N-Methyl-4-((1-(4-methylbenzyl)-2,4-dioxo-1,2-dihydroquinazolin- 3(4H)-yl)methyl)benzamide (34)
Isolated as a white solid (15 mg, 29% yield), mp 236–238 °C. 1H NMR (500 MHz, CDCl3): δ 8.23 (dd, J = 7.9, 1.6 Hz, 1H), 7.73–7.68 (m, 2H), 7.60–7.51 (m, 3H), 7.22 (ddd, J = 8.0, 7.3, 0.9 Hz, 1H), 7.16–7.09 (m, 5H), 6.13 (q, J = 4.7 Hz, 1H), 5.36 (s, 2H), 5.33 (s, 2H), 2.99 (d, J = 4.9 Hz, 3H), 2.31 (s, 3H). 13C NMR (126 MHz, CDCl3): δ 168.02, 161.91, 151.48, 140.54, 140.10, 137.59, 135.39, 134.01, 132.56, 129.80, 129.25, 129.15, 127.15, 126.53, 123.31, 115.74, 114.64, 47.37, 44.92, 26.97, 21.23. LCMS Retention time: 3.047 min. LCMS purity 97.6%. HRMS (ESI): m/z calcd for C25H23N3O3 [M + H]+ 414.1812, found 414.1820.
N,N-Dimethyl-4-((1-(4-methylbenzyl)-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)benzamide (35)
Isolated as a white solid (21 mg, 39% yield), mp 153–155 °C. 1H NMR (500 MHz, CDCl3): δ 8.24 (dd, J = 7.9, 1.6 Hz, 1H), 7.60–7.51 (m, 3H), 7.40–7.35 (m, 2H), 7.22 (ddd, J = 8.0, 7.3, 0.9 Hz, 1H), 7.16–7.10 (m, 5H), 5.35 (s, 2H), 5.33 (s, 2H), 3.09 (s, 3H), 2.96 (s, 3H), 2.31 (s, 3H). 13C NMR (126 MHz, CDCl3): δ 171.53, 161.90, 151.49, 140.10, 138.57, 137.57, 135.74, 135.34, 132.61, 129.79, 129.23, 129.13, 127.40, 126.55, 123.27, 115.78, 114.62, 47.37, 44.94, 39.74, 35.47, 21.23. LCMS Retention time: 3.147 min. LCMS purity 99.3%. HRMS (ESI): m/z calcd for C26H25N3O3 [M + H]+ 428.1969, found 428.1982.
N-(tert-Butyl)-4-((1-(4-methylbenzyl)-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)benzamide (36)
Isolated as a white solid (36 mg, 79% yield), mp 157–159 °C. 1H NMR (500 MHz, CDCl3): δ 8.23 (dd, J = 7.9, 1.6 Hz, 1H), 7.69–7.64 (m, 2H), 7.59–7.51 (m, 3H), 7.21 (ddd, J = 8.1, 7.3, 0.9 Hz, 1H), 7.16–7.09 (m, 5H), 5.89 (s, 1H), 5.35 (s, 2H), 5.32 (s, 2H), 2.31 (s, 3H), 1.45 (s, 9H). 13C NMR (126 MHz, CDCl3): δ 166.79, 161.88, 151.47, 140.26, 140.09, 137.57, 135.36, 132.58, 129.79, 129.24, 129.09, 127.01, 126.52, 123.28, 115.74, 114.63, 51.71, 47.35, 44.92, 29.00, 21.23. LCMS Retention time: 3.424 min. LCMS purity 100%. HRMS (ESI): m/z calcd for C28H29N3O3 [M + H]+ 456.2282, found 456.2274.
N-Cyclohexyl-4-((1-(4-methylbenzyl)-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)benzamide (37)
Isolated as a white solid (18 mg, 33% yield), mp 246–248 °C. 1H NMR (500 MHz, CDCl3): δ 8.23 (dd, J = 7.9, 1.6 Hz, 1H), 7.73–7.67 (m, 2H), 7.61–7.56 (m, 2H), 7.54 (ddd, J = 8.7, 7.3, 1.7 Hz, 1H), 7.22 (ddd, J = 8.1, 7.3, 0.9 Hz, 1H), 7.16–7.09 (m, 5H), 5.90 (d, J = 8.1 Hz, 1H), 5.36 (s, 2H), 5.33 (s, 2H), 4.02–3.90 (m, 1H), 2.31 (s, 3H), 2.05–1.97 (m, 2H), 1.78–1.69 (m, 2H), 1.68–1.61 (m, 1H), 1.48–1.36 (m, 2H), 1.28–1.14 (m, 3H). 13C NMR (126 MHz, CDCl3): δ 166.46, 161.89, 151.48, 140.45, 140.10, 137.59, 135.38, 134.53, 132.59, 129.80, 129.25, 129.15, 127.14, 126.53, 123.30, 115.75, 114.64, 48.75, 47.37, 44.95, 33.38, 25.71, 25.03, 21.24. LCMS Retention time: 2.320 min. LCMS purity 100%. HRMS (ESI): m/z calcd for C30H31N3O3 [M + H]+ 482.2448, found 482.2440.
4-((1-(4-Methylbenzyl)-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)-N-phenylbenzamide (38)
Isolated as a white solid (18 mg, 30% yield) mp 247–249 °C. 1H NMR (500 MHz, CDCl3): δ 8.24 (dd, J = 7.9, 1.7 Hz, 1H), 7.86–7.77 (m, 3H), 7.63 (d, J = 8.4 Hz, 4H), 7.55 (ddd, J = 8.7, 7.3, 1.6 Hz, 1H), 7.40–7.32 (m, 2H), 7.22 (apparent t, J = 7.2 Hz, 1H), 7.18–7.09 (m, 6H), 5.38 (s, 2H), 5.34 (s, 2H), 2.31 (s, 3H). 13C NMR (126 MHz, CDCl3): δ 165.57, 161.92, 151.49, 141.12, 140.11, 138.06, 137.62, 135.44, 134.39, 132.54, 129.81, 129.34, 129.26, 129.22, 127.36, 126.53, 124.65, 123.36, 120.23, 115.72, 114.68, 47.39, 44.94, 21.23. LCMS Retention time: 3.456 min. LCMS purity 98.9%. HRMS (ESI): m/z calcd for C30H25N3O3 [M + H]+ 476.1969, found 476.1988.
N-Benzyl-4-((1-(4-methylbenzyl)-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)benzamide (39)
Isolated as a white solid (39 mg, 71% yield) mp 218–220 °C. 1H NMR (500 MHz, CDCl3): δ 8.22 (dd, J = 7.9, 1.6 Hz, 1H), 7.78–7.72 (m, 2H), 7.61–7.56 (m, 2H), 7.54 (ddd, J = 8.7, 7.3, 1.6 Hz, 1H), 7.37–7.31 (m, 4H), 7.31–7.27 (m, 1H), 7.21 (ddd, J = 8.0, 7.3, 0.9 Hz, 1H), 7.16–7.09 (m, 5H), 6.39 (t, J = 5.7 Hz, 1H), 5.35 (s, 2H), 5.32 (s, 2H), 4.63 (d, J = 5.7 Hz, 2H), 2.31 (s, 3H). 13C NMR (126 MHz, CDCl3): δ 167.13, 161.89, 151.47, 140.80, 140.09, 138.30, 137.59, 135.39, 133.74, 132.56, 129.80, 129.25, 129.20, 128.91, 128.01, 127.74, 127.29, 126.53, 123.31, 115.73, 114.64, 47.37, 44.92, 44.23, 21.23. LCMS Retention time: 3.387 min. LCMS purity 100%. HRMS (ESI): m/z calcd for C31H27N3O3 [M + H]+ 490.2125, found 490.2149.
N-(Furan-2-ylmethyl)-4-((1-(4-methylbenzyl)-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)benzamide (40)
Isolated as a white solid (29 mg, 54% yield), mp 217–219 °C. 1H NMR (500 MHz, CDCl3): δ 8.23 (dd, J = 8.0, 1.6 Hz, 1H), 7.76–7.71 (m, 2H), 7.60–7.56 (m, 2H), 7.54 (ddd, J = 8.7, 7.2, 1.7 Hz, 1H), 7.36 (dd, J = 1.9, 0.8 Hz, 1H), 7.21 (ddd, J = 8.0, 7.3, 0.9 Hz, 1H), 7.17–7.08 (m, 5H), 6.41 (t, J = 5.5 Hz, 1H), 6.33 (dd, J = 3.2, 1.9 Hz, 1H), 6.28 (dd, J = 3.2, 0.9 Hz, 1H), 5.35 (s, 2H), 5.32 (s, 2H), 4.62 (d, J = 5.5 Hz, 2H), 2.31 (s, 3H). 13C NMR (126 MHz, CDCl3): δ 166.99, 161.89, 151.47, 151.28, 142.43, 140.86, 140.09, 137.59, 135.39, 133.53, 132.56, 129.80, 129.25, 129.18, 127.33, 126.52, 123.31, 115.73, 114.64, 110.64, 107.79, 47.36, 44.92, 37.13, 21.23. LCMS Retention time: 3.280 min. LCMS purity 100%. HRMS (ESI): m/z calcd for C29H25N3O4 [M + H]+ 480.1918, found 480.1936.
4-((1-(4-Methylbenzyl)-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)-N-(thiazol-2-yl)benzamide (41)
Isolated as a colorless oil (8 mg, 17% yield). 1H NMR (500 MHz, CDCl3): δ 10.02 (s, 1H), 8.25 (dd, J = 7.9, 1.6 Hz, 1H), 7.95–7.88 (m, 2H), 7.70–7.64 (m, 2H), 7.56 (ddd, J = 8.7, 7.3, 1.7 Hz, 1H), 7.39 (d, J = 3.6 Hz, 1H), 7.30– 7.20 (m, 1H), 7.19–7.09 (m, 5H), 6.99 (d, J = 3.6 Hz, 1H), 5.41 (s, 2H), 5.35 (s, 2H), 2.31 (s, 3H). 13C NMR (126 MHz, CDCl3): δ 164.29, 161.92, 158.65, 151.49, 142.38, 140.13, 137.74, 137.67, 135.50, 132.52, 131.37, 129.84, 129.53, 129.32, 127.82, 126.55, 123.41, 115.71, 114.70, 114.05, 47.41, 44.92, 21.24. LCMS Retention time: 3.278 min. LCMS purity 100%. HRMS (ESI): m/z calcd for C27H22N4O3S [M + H]+ 483.1485, found 483.1482.
4-((1-(4-Methylbenzyl)-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)-N-(pyridin-4-yl)benzamide (42)
Isolated as a colorless oil (7 mg, 14% yield). 1H NMR (500 MHz, DMSO-d6): δ 11.55 (s, 1H), 8.74 (d, J = 6.6 Hz, 2H), 8.30 (d, J = 6.9 Hz, 2H), 8.11 (dd, J = 7.9, 1.7 Hz, 1H), 8.04–7.98 (m, 2H), 7.70 (ddd, J = 8.7, 7.2, 1.7 Hz, 1H), 7.55 (d, J = 8.3 Hz, 2H), 7.36–7.27 (m, 2H), 7.21 (d, J = 8.0 Hz, 2H), 7.14 (d, J = 8.0 Hz, 2H), 5.36 (s, 2H), 5.31 (s, 2H), 2.26 (s, 3H). 13C NMR (126 MHz, DMSO): δ 167.07, 161.22, 150.88, 142.80, 142.55, 139.74, 136.48, 135.50, 133.13, 131.96, 129.29, 128.62, 128.14, 127.39, 126.48, 123.12, 115.24, 115.07, 115.04, 106.96, 46.23, 44.41, 20.66. LCMS Retention time: 3.159 min. LCMS purity 98%. HRMS (ESI): m/z calcd for C29H24N4O3 [M + H]+ 477.1921, found 477.1917.
4-((1-(4-Nitrobenzyl)-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)-methyl)-N-(oxetan-3-ylmethyl)benzamide (43)
Isolated as a white solid (45 mg, 53% yield), mp 170–173 °C. 1H NMR (500 MHz, CDCl3): δ 8.28 (dd, J = 7.9, 1.6 Hz, 1H), 8.23–8.17 (m, 2H), 7.75– 7.69 (m, 2H), 7.62–7.55 (m, 3H), 7.43–7.37 (m, 2H), 7.31–7.24 (m, 1H), 6.99 (d, J = 8.2 Hz, 1H), 6.37 (t, J = 5.9 Hz, 1H), 5.46 (s, 2H), 5.36 (s, 2H), 4.83 (dd, J = 7.7, 6.3 Hz, 2H), 4.46 (apparent t, J = 6.1 Hz, 2H), 3.74 (apparent t, J = 6.3 Hz, 2H), 3.33–3.24 (m, 1H). 13C NMR (126 MHz, CDCl3): δ 167.74, 161.60, 151.40, 147.69, 143.10, 140.58, 139.55, 135.69, 133.82, 129.72, 129.31, 127.37, 127.29, 124.47, 123.91, 115.83, 113.99, 75.16, 47.12, 45.04, 42.53, 35.14. LCMS Retention time: 2.935 min. LCMS purity 98.4%. HRMS (ESI): m/z calcd for C27H24N4O6 [M + H]+ 501.1769, found 501.1765.
4-((1-(4-Isopropylbenzyl)-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)-N-(oxetan-3-ylmethyl)benzamide (44)
Isolated as a white solid (50.4 mg, 83% yield), mp 178–180 °C. 1H NMR (500 MHz, CDCl3): δ 8.23 (dd, J = 7.9, 1.6 Hz, 1H), 7.71 (d, J = 8.3 Hz, 2H), 7.60–7.53 (m, 3H), 7.24–7.12 (m, 6H), 6.33 (brt, 1H), 5.36 (s, 2H), 5.33 (s, 2H), 4.82 (dd, J = 7.7, 6.3 Hz, 2H), 4.46 (apparent t, J = 6.1 Hz, 2H), 3.73 (apparent t, J = 6.1 Hz, 2H), 3.33–3.22 (m, 1H), 2.87 (h, J = 7.0 Hz, 1 H), 1.21 (d, J = 6.9 Hz, 6H). 13C NMR (126 MHz, CDCl3): δ 167.64, 161.75, 151.32, 148.42, 140.78, 140.01, 135.27, 133.50, 132.71, 129.08, 129.06, 129.20, 127.08, 126.04, 126.40, 123.17, 115.56, 114.55, 75.03, 47.22, 44.77, 42.42, 35.02, 33.73, 23.90. LCMS Retention time: 3.278 min. LCMS purity 97.9%. HRMS (ESI): m/z calcd for C30H31N3O4 [M + H]+ 498.2387, found 498.2387.
4-((1-(4-(Dimethylamino)benzyl)-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)-N-(oxetan-3-ylmethyl)benzamide (45)
Step 1
Synthesis of methyl 4-((1-(4-dimethylamino)benzyl)-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)benzoate. To a stirred suspension of sodium hydride (0.076 g, 1.89 mmol) in dry DMF (2.5 mL) at 0 °C was added methyl 4-((2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)-methyl)benzoate 4 (0.24 g, 0.76 mmol) and the mixture was stirred for 15 min at 0 °C. After adding 4-(dimethylamino)benzyl 4-methylbenzenesulfonate (0.25 g, 0.83 mmol) in DMF (2.5 mL), the suspension was heated at 80 °C for 16 h and then was then cooled to 0 °C. Water was added (5 mL), and the mixture was extracted with CH2Cl2 (3 × 8 mL). The organic extracts were washed with water (3 × 25 mL), brine (12 mL) and dried (MgSO4), filtered and concentrated under reduced pressure to afford the crude product which was purified by reverse phase column chromatography (0– 100% v/v MeCN/H2O) yielding the desired product as a pale green solid (0.036 g, 0.081 mmol, 11% yield). 1H NMR (400 MHz, CDCl3): δ 8.22 (dd, J=7.9, 1.5 Hz, 1H), 8.04–7.95 (m, 2H), 7.61–7.52 (m, 3H), 7.26–7.18 (m, 2H), 7.14 (d, J = 8.8 Hz, 2H), 6.66 (d, J = 8.8 Hz, 2H), 5.37 (s, 2H), 5.27 (broad s, 2H), 3.90 (s, 3H), 2.91 (s, 6H).
Step 2
Synthesis of 4-((1-(4-dimethylamino)benzyl)-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)benzoic acid. To a solution of methyl 4-((1-(4-(dimethylamino)benzyl)-2,4-dioxo-1,2-dihydroquinazolin- 3(4H)-yl)methyl)benzoate (0.060 g, 0.14 mmol) in THF (1 mL) was added 1 M lithium hydroxide (0.81 mL, 0.81 mmol). The reaction mixture was stirred at 40 °C for 3 h, at which point TLC confirmed completion of reaction. Then 1 M HCl was cautiously added until the reaction mixture was at pH 7 (isoelectric point), at which point the product precipitated out of solution. The precipitate was filtered, washed with H2O (2 × 6 mL), collected and dried under high vacuum to afford the desired product as a pale green solid (0.026 g, 0.061 mmol, 45% yield). 1H NMR (400 MHz, DMSO-d6): δ 12.90 (s, 1H), 8.08 (d, J = 7.5 Hz, 1H), 7.90 (d, J = 8.2 Hz, 2H), 7.70 (apparent t, J = 7.6 Hz, 1H), 7.44 (d, J = 8.2 Hz, 2H), 7.40 (d, J = 8.5 Hz, 1H), 7.28 (apparent t, J = 7.5 Hz, 1H), 7.14 (d, J = 8.6 Hz, 2H), 6.66 (d, J = 8.6 Hz, 2H), 5.27 (s, 4H), 2.83 (s, 6H).
Step 3
Synthesis of 4-((1-(4-(dimethylamino)benzyl)-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)-N-(oxetan-3-ylmethyl)- benzamide 46. To a solution of 4-((1-(4-dimethylamino)benzyl)-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)benzoic acid (1.0 equiv) in DMF was added 3-aminomethyl-oxetane (1.0 equiv), HATU (1.1 equiv) and N,N-diisopropylethylamine. The reaction mixture was stirred for 16 h at room temperature, then diluted with CH2Cl2 and washed sequentially with 1 M HCl, sat. aqueous NaHCO3, and water. The organic extract was separated, dried (MgSO4), filtered, and concentrated under reduced pressure to afford a crude product which was purified by silica gel flash column chromatography (0–5% v/v MeOH/CH2Cl2) yielding 46 as a white solid (33 mg, 36% yield). 1H NMR (500 MHz, CDCl3): δ 8.21 (dd, J = 7.9, 1.6 Hz, 1H), 7.74–7.68 (m, 2H), 7.62–7.52 (m, 3H), 7.25–7.18 (m, 2H), 7.17–7.10 (m, 2H), 6.70–6.63 (m, 2H), 6.35 (t, J = 5.8 Hz, 1H), 5.36 (s, 2H), 5.27 (s, 2H), 4.82 (dd, J = 7.7, 6.3 Hz, 2H), 4.46 (apparent t, J = 6.0 Hz, 2H), 3.75–3.71 (m, 2H), 3.34–3.21 (m, 1H), 2.91 (s, 6H). 13C NMR (126 MHz, CDCl3): δ 167.83, 161.97, 151.49, 150.18, 141.01, 140.23, 135.35, 133.58, 129.18, 129.15, 127.88, 127.22, 123.17, 123.02, 115.71, 114.79, 112.85, 75.20, 47.15, 44.88, 42.58, 40.64, 35.15. LCMS Retention time: 1.82 min. LCMS purity 100% (method used 0.05% formic acid). HRMS (ESI): m/z calcd for C29H30N4O4 [M + H]+ 499.2340, found 499.2338.
4-((1-(4-tert-Butylbenzyl)-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)-N-(oxetan-3-ylmethyl)benzamide (47)
Prepared in sequence as described for 46. Isolated as a white solid (40.0 mg, 40% yield). 1H NMR (500 MHz, CDCl3): δ 8.23 (dd, J = 7.9, 1.6 Hz, 1H), 7.71 (d, J = 8.3 Hz, 2H), 7.59 (d, J = 8.3 Hz, 2H), 7.56 (ddd, J = 8.7, 7.3, 1.7 Hz, 1H), 7.33 (d, J = 8.4 Hz, 2H), 7.22 (ddd, J = 8.7, 7.3, 1.7 Hz, 1H), 7.17–7.14 (m, 3H), 6.35 (brt, 1H), 5.36 (s, 2H), 5.33 (s, 2H), 4.82 (dd, J = 7.7, 6.3 Hz, 2H), 4.46 (apparent t, J = 6.1 Hz, 2H), 3.73 (apparent t, J = 6.1 Hz, 2H), 3.33–3.24 (m, 1H), 1.28 (s, 9H). 13C NMR (126 MHz, CDCl3): δ 167.66, 161.75, 151.30, 150.69, 140.77, 140.02, 135.28, 133.49, 132.33, 129.08, 129.05, 127.08, 126.13, 125.90, 123.16, 115.55, 114.56, 75.03, 47.14, 44.77, 42.42, 35.01, 34.50, 31.26. LCMS Retention time: 3.346 min. LCMS purity 98.7%. HRMS (ESI): m/z calcd for C31H33N3O4 [M + H]+ 512.2527, found 512.2527.
N-(Furan-2-ylmethyl)-4-((1-(4-isopropylbenzyl)-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)benzamide (48)
Isolated as a white solid (70 mg, 74% yield), mp 194–197 °C. 1H NMR (500 MHz, CDCl3): δ 8.23 (dd, J = 7.9, 1.7 Hz, 1H), 7.77–7.70 (m, 2H), 7.61–7.52 (m, 3H), 7.36 (dd, J = 1.9, 0.9 Hz, 1H), 7.22 (ddd, J = 8.0, 7.2, 0.9 Hz, 1H), 7.20–7.11 (m, 5H), 6.43 (t, J = 5.5 Hz, 1H), 6.32 (dd, J = 3.2, 1.9 Hz, 1H), 6.28 (d, J = 3.1 Hz, 1H), 5.35 (s, 2H), 5.33 (s, 2H), 4.62 (d, J = 5.5 Hz, 2H), 2.87 (h, J = 6.9 Hz, 1H), 1.21 (d, J = 6.9 Hz, 6H). 13C NMR (126 MHz, CDCl3): δ 167.00, 161.89, 151.45, 151.28, 148.54, 142.42, 140.85, 140.14, 135.41, 133.52, 132.87, 129.24, 129.17, 127.33, 127.18, 126.55, 123.30, 115.70, 114.69, 110.63, 107.79, 47.35, 44.91, 37.12, 33.88, 24.05. LCMS Retention time: 3.591 min. LCMS purity 97.4%. HRMS (ESI): m/z calcd for C31H29N3O4 [M + H]+ 508.2231, found 508.2223.
4-((1-(4-Isopropylbenzyl)-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)methyl)-N-(2-methoxypyridin-4-yl)benzamide (50)
Isolated as a white solid (14.3 mg, 23% yield). 1H NMR (500 MHz, CDCl3): δ 8.24 (dd, J = 7.9, 1.6 Hz, 1H), 8.10 (d, J = 6.3 Hz, 1H), 7.84–7.74 (m, 3H), 7.65 (d, J = 8.2 Hz, 2H), 7.57 (ddd, J = 8.7, 7.3, 1.7 Hz, 1H), 7.25–7.21 (m, 1H), 7.20–7.09 (m, 7H), 5.39 (s, 2H), 5.34 (s, 2H), 3.94 (s, 3H), 2.87 (h, J = 6.9 Hz, 1H), 1.21 (d, J = 6.9 Hz, 6H). 13C NMR (126 MHz, CDCl3): δ 165.63, 165.48, 161.78, 151.32, 148.46, 147.71, 146.95, 141.64, 140.01, 135.36, 133.40, 132.67, 129.33, 129.11, 127.30, 127.06, 123.24, 115.52, 114.59, 108.25, 99.67, 53.62, 47.24, 44.76, 33.74, 23.90. LCMS Retention time: 3.53 min. LCMS purity 100%. HRMS (ESI): m/z calcd for C32H30N4O4 [M + H]+ 535.2345, found 535.2345.
4-((2-(3-Isopropylbenzyl)-4-oxoquinazolin-3(4H)-yl)methyl)-N-(3-methoxypropyl) benzamide (51)
Step 1: Synthesis of methyl-4-((2-aminobenzamido)methyl)benzoate
Anthranilic acid (3.63 g, 26.5 mmol) was dissolved in dry CH2Cl2 (200 mL) and to this solution methyl-4-(aminomethyl)benzoate hydrochloride (6.43 g, 32 mmol), DIPEA (14 mL, 80 mmol), HOBt (4.34 g, 32 mmol), and EDCI (6.17 g, 32 mmol) were added. The mixture was stirred at rt for 24 h. The reaction mixture was washed with 1 M aq. HCl (2 × 200 mL) and saturated aq. NaHCO3 (2 × 200 mL) and dried with Na2SO4 to give the title compound (5.6 g, 74%) as a white solid. 1H NMR (500 MHz, acetone-d6) δ 8.17 (t, J = 6.0 Hz, 1H), 7.99–7.94 (m, 2H), 7.60 (dd, J = 8.0, 1.5 Hz, 1H), 7.52–7.47 (m, 2H), 7.16 (ddd, J = 8.4, 7.1, 1.5 Hz, 1H), 6.77 (dd, J = 8.2, 1.2 Hz, 1H), 6.54 (ddd, J = 8.1, 7.1, 1.2 Hz, 1H), 6.30 (s, 2H), 4.65 (d, J = 6.0 Hz, 2H), 3.86 (s, 3H).
Step 2: Synthesis of ethyl-2-(3-(prop-1-en-2-yl)phenyl)acetate
A MW vial was charged with ethyl-2-(3-bromophenyl)acetate (122 mg, 0.50 mmol), Pd(OAc)2 (5.6 mg, 0.025 mmol), RuPhos (23 mg, 0.05 mmol), Cs2CO3 (492 mg, 1.51 mmol), and potassium isopropenyltrifluoroborate (97 mg, 0.66 mmol). The vial was capped and evacuated/refilled with Ar (3 times). Degassed toluene (3 mL) and water (0.5 mL) were added to the vial and the mixture was heated in a MW reactor at 100 °C for 15 min. The biphasic layers were separated and the aq. layer was extracted with EtOAc (2 × 4 mL). The combined organic extracts were concentrated and the product was purified by flash chromatography (0–15% EtOAc/hexanes) to give the title compound (91 mg, 89%) as a clear, pale-yellow oil. 1H NMR (400 MHz, CDCl3) δ 7.38–7.36 (m, 1H), 7.34 (dt, J = 7.7, 1.5 Hz, 1H), 7.25 (t, J = 7.6 Hz, 1H), 7.17 (dt, J = 7.5, 1.4 Hz, 1H), 5.35 (dq, J = 1.6, 0.8 Hz, 1H), 5.06 (p, J = 1.5 Hz, 1H), 4.12 (q, J = 7.1 Hz, 2H), 3.58 (s, 2H), 2.12 (dd, J = 1.5, 0.8 Hz, 3H), 1.22 (t, J = 7.1 Hz, 3H).
Step 3: Synthesis of ethyl-2-(3-isopropylphenyl)acetate
Palladium on carbon (10% wt., 72 mg, 0.07 mmol) was added to a solution of ethyl-2-(3-(prop-1-en-2-yl)phenyl)acetate (273 mg, 1.34 mmol) in EtOH (9 mL). The mixture was stirred under 1 atm of H2 (balloon) at rt for 2 h. The reaction mixture was diluted with CH2Cl2 (10 mL), filtered through Celite and rinsed with CH2Cl2 (2 × 10 mL) to give the title compound (267 mg, 97%) as a pleasant-smelling, clear, colorless oil. 1H NMR (400 MHz, CDCl3) δ 7.21 (t, J = 7.6 Hz, 1H), 7.15–7.06 (m, 3H), 4.12 (q, J = 7.1 Hz, 2H), 3.56 (s, 2H), 2.87 (hept, J = 6.9 Hz, 1H), 1.26–1.18 (m, 9H).
Step 4: Syntheis of 2-(3-isopropylphenyl)acetic acid
A solution of LiOH (217 mg, 9.1 mmol) in water (5 mL) was added to a solution of ethyl-2-(3-isopropylphenyl)acetate (267 mg, 1.3 mmol) in THF (5 mL). The mixture was stirred at rt for 4.5 h. The reaction mixture was quenched with 1 M aq. HCl (20 mL). The layers were separated and the aq. layer was extracted with CH2Cl2 (2 × 20 mL). The combined organic extracts were dried with Na2SO4 to give the title compound (234 mg, 100%) as a clear, colorless oil. 1H NMR (400 MHz, CDCl3) δ 11.75 (s, 1H), 7.25–7.19 (m, 1H), 7.15–7.05 (m, 3H), 3.58 (s, 2H), 2.86 (hept, J = 6.9 Hz, 1H), 1.22 (d, J = 7.0 Hz, 6H).
Step 5: Syntheis of methyl-4-((2-(2-(3-isopropylphenyl)-acetamido)benzamido)methyl)benzoate
A catalytic amount of dry DMF (4 drops) was added to a solution of 2-(3-isopropylphenyl)-acetic acid (219 mg, 1.23 mmol) in dry CH2Cl2 (3 mL) under Ar. Oxalyl chloride (0.11 mL, 1.30 mmol) was added dropwise to the mixture at rt. The reaction mixture was stirred at rt for 30 min and then added dropwise to a mixture of methyl-4-((2-aminobenzamido)-methyl)benzoate (384 mg, 1.35 mmol) and pyridine (0.20 mL, 2.5 mmol) in dry CH2Cl2 (3 mL). After stirring at rt for 1.5 h, the mixture was diluted with CH2Cl2 (10 mL) and successively washed with saturated aq. NaHCO3 (10 mL) and 1 M aq. HCl (10 mL) to give the title compound (550 mg, 100%) as a forrest-green solid. 1H NMR (400 MHz, CDCl3) δ 11.07 (s, 1H), 8.49 (d, J = 8.4 Hz, 1H), 7.98 (d, J = 8.3 Hz, 2H), 7.48 (d, J = 7.8 Hz, 1H), 7.39–7.32 (m, 3H), 7.29– 7.21 (m, 2H), 7.15 (t, J = 8.1 Hz, 2H), 7.11–7.04 (m, 1H), 6.96 (t, J = 7.5 Hz, 1H), 4.59 (d, J = 5.9 Hz, 2H), 3.91 (s, 3H), 3.67 (s, 2H), 2.89 (hept, J = 6.9 Hz, 1H), 1.24 (d, J = 6.9 Hz, 6H).
Step 6: Synthesis of 4-((2-(3-isopropylbenzyl)-4-oxoquinazolin-3(4H)-yl)methyl)benzoic acid
A solution of LiOH (237 mg, 9.9 mmol) in water (10 mL) was added to a solution of methyl-4-((2-(2-(3-isopropylphenyl)acetamido)benzamido)methyl)benzoate (440 mg, 0.99 mmol) in THF (10 mL). The mixture was stirred at 40 °C for 16 h. At rt, the reaction was quenched with 1 M aq. HCl (25 mL) and the product was extracted with CH2Cl2 (3 × 40 mL). The combined extracts were dried with Na2SO4 and the product was purified by flash chromatography (0–100% EtOAc/hexanes) to yield an inseparable mixture of 4-((2-(2-(3-isopropylphenyl)acetamido)benzamido)-methyl)benzoic acid and the title compound (258 mg, 63%) as a pale-yellow solid. It was used in the next step without further purification.
Step 7: Syntheis of 4-((2-(3-isopropylbenzyl)-4-oxoquinazolin-3(4H)-yl)methyl)-N-(3-methoxypropyl)benzamide (51)
A mixture of 4-((2-(3-isopropylbenzyl)-4-oxoquinazolin-3(4H)-yl)methyl)benzoic acid (113 mg, 0.27 mmol) and 4-((2-(2-(3-isopropylphenyl)-acetamido)benzamido)methyl)benzoic acid was dissolved in dry CH2Cl2 (3 mL) and to this solution 3-methoxypropylamine (0.04 mL, 0.4 mmol), DIPEA (0.07 mL, 0.4 mmol), HOBt (45 mg, 0.33 mmol), and EDCI (64 mg, 0.33 mmol) were added. The mixture was stirred at rt for 2.5 h and then concentrated. The remaining residue was purified by flash chromatography (0–100% EtOAc/hexanes). Single-spot fractions (by TLC) were combined and concentrated to give the title compound (13 mg, 10%) as a clear, colorless oil. 1H NMR (500 MHz, CDCl3) δ 8.33 (dt, J = 8.0, 1.0 Hz, 1H), 7.88–7.80 (m, 2H), 7.74–7.70 (m, 2H), 7.58–7.51 (m, 1H), 7.25 (t, J = 7.6 Hz, 1H), 7.18–7.13 (m, 3H), 7.07–7.05 (m, 1H), 7.04–7.01 (m, 1H), 6.97 (t, J = 4.5 Hz, 1H), 5.29 (s, 2H), 4.09 (s, 2H), 3.59–3.54 (m, 4H), 3.37 (s, 3H), 2.86 (hept, J = 6.9 Hz, 1H), 1.91–1.84 (m, 2H), 1.22 (d, J = 6.9 Hz, 6H). 13C NMR (126 MHz, CDCl3) δ 166.7, 162.5, 155.9, 150.2, 146.8, 139.3, 135.0, 134.6, 134.4, 129.4, 127.7, 127.39, 127.35, 127.0, 126.5, 126.4, 125.7, 125.5, 120.4, 72.7, 59.1, 46.3, 42.3, 39.5, 34.2, 28.9, 24.1. LC-MS: t R = 3.40 min, purity =100%. HRMS (m/z): calcd for C30H34N3O3 (M + H)+ 484.2595; found 484.2593.
4-((2-(4-Isopropylbenzyl)-4-oxoquinazolin-3(4H)-yl)methyl)-N-(3-methoxypropyl)benzamide (52)
Step 1: Synthesis of methyl-4-((2-(2-(4-isopropylphenyl)acetamido)benzamido)methyl)benzoate
A catalytic amount of dry DMF (7 drops) was added to a solution of 2-(4-isopropylphenyl)acetic acid (0.891 g, 5.0 mmol) in dry CH2Cl2 (10 mL) under Ar. Oxalyl chloride (2 M in CH2Cl2, 2.8 mL, 5.6 mmol) was slowly added to the mixture at rt. The reaction mixture was stirred at rt for 30 min and then slowly added to a mixture of methyl-4-((2-aminobenzamido)methyl)benzoate (1.563 g, 5.5 mmol) and pyridine (0.81 mL, 10.0 mmol) in dry CH2Cl2 (10 mL). After stirring at rt for 3 h, the mixture was successively washed with 1 M aq. HCl (2 × 20 mL) and saturated aq. NaHCO3 (2 × 20 mL) and dried with Na2SO4 to give the title compound (2.072 g, 93%) as a yellow solid. 1H NMR (400 MHz, CDCl3) δ 11.05 (s, 1H), 8.47 (dd, J = 8.5, 1.1 Hz, 1H), 8.00–7.95 (m, 2H), 7.44 (dd, J = 7.9, 1.4 Hz, 1H), 7.36– 7.30 (m, 3H), 7.28–7.23 (m, 2H), 7.20–7.16 (m, 2H), 7.11 (t, J = 6.0 Hz, 1H), 6.93 (td, J = 7.6, 1.2 Hz, 1H), 4.56 (d, J = 5.9 Hz, 2H), 3.90 (s, 3H), 3.64 (s, 2H), 2.87 (hept, J = 6.8 Hz, 1H), 1.22 (d, J = 6.9 Hz, 6H).
Step 2: Synthesis of 4-((2-(4-isopropylbenzyl)-4-oxoquinazolin-3(4H)-yl)methyl)benzoic acid
A solution of LiOH (0.285 g, 11.9 mmol) in water (12 mL) was added to a solution of methyl-4-((2-(2-(4-isopropylphenyl)acetamido)benzamido)methyl)benzoate (1.056 g, 2.38 mmol) in THF (12 mL). The mixture was stirred at 40 °C for 2 h and a second portion of LiOH (0.285 g, 11.9 mmol) was added to the mixture. The mixture was stirred at 40 °C for an additional 19 h. At rt, the reaction was quenched with 1 M aq. HCl (25 mL) and the product was extracted with CH2Cl2 (2 × 50 mL). The extracts were dried with Na2SO4 to yield a mixture of 4-((2-(2-(4-isopropylphenyl)acetamido)-benzamido)methyl)benzoic acid and the title compound (0.985 g, 100%) as a pale-yellow solid. It was used in the next step without further purification.
Step 3: Synthesis of 4-((2-(4-isopropylbenzyl)-4-oxoquinazolin-3(4H)-yl)methyl)-N-(3-methoxypropyl)benzamide (52)
A mixture of 4-((2-(4-isopropylbenzyl)-4-oxoquinazolin-3(4H)-yl)methyl)benzoic acid (0.985 g, 2.4 mmol) and 4-((2-(2-(4-isopropylphenyl)-acetamido)benzamido)methyl)benzoic acid was dissolved in dry CH2Cl2 (20 mL) and to this solution 3-methoxypropylamine (0.28 mL, 2.75 mmol), DIPEA (0.60 mL, 3.4 mmol), HOBt (0.371 g, 2.75 mmol), and EDCI (0.527 g, 2.75 mmol) were added. The mixture was stirred at rt for 62 h and then concentrated. The remaining residue was purified by flash chromatography (0–10% MeOH/CH2Cl2), followed by mass-directed fractionation to give the title compound (0.252 g, 22%) as a white solid. mp 52–55 °C. 1H NMR (400 MHz, CDCl3) δ 8.31 (ddd, J = 8.0, 1.5, 0.7 Hz, 1H), 7.83–7.70 (m, 4H), 7.51 (ddd, J = 8.1, 6.8, 1.6 Hz, 1H), 7.20–7.09 (m, 7H), 5.29 (s, 2H), 4.03 (s, 2H), 3.58–3.51 (m, 4H), 3.36 (s, 3H), 2.88 (hept, J = 6.9 Hz, 1H), 1.91– 1.83 (m, 2H), 1.23 (d, J = 6.9 Hz, 6H). 13C NMR (101 MHz, CDCl3) δ 166.6, 162.6, 155.4, 148.2, 147.3, 139.5, 134.5, 134.2, 132.2, 128.0, 127.6, 127.3, 127.2, 127.1, 126.9, 126.2, 120.4, 72.2, 58.9, 46.1, 42.0, 39.0, 33.7, 28.9, 23.9. LC-MS: tR = 3.74 min, purity =99.7%. HRMS (m/z): calcd for C30H34N3O3 (M + H)+ 484.2595; found 484.2611.
Time of Addition Assay
HEp-2 cells were plated in 96 well black tissue culture plates at 10,000 cells per well in 100 µL and incubated 24 h at 37 °C, 5% CO2. The compounds were diluted in media to give a final concentration of 25 µM and added to plates in triplicate at –1, 0, 1, 2, 3, 5, 7, 14, 17, 21, and 24 h post-infection (p.i.). Cells were infected with RSV strain Long at an MOI of 1.0 and incubated at 37 °C, 5% CO2. Following a six day incubation period, the assay plates were equilibrated to room temperature for 30 min. An equal volume (100 µL) of Cell Titer-Glo reagent (Promega Inc.) was added to each well using a MultiFlo Microplate Dispenser (BioTek, Winooski, VT), and the plates were incubated for an additional 10 min at room temperature. At the end of the incubation, luminescence was measured using a multilabel reader (BioTek Synergy 4, Winooski, VT).
Cytotoxicity Assay
Compound cytotoxicity was measured as described elsewhere36 with some modifications. Briefly, for a dose response study, test compounds were solubilized in DMSO at 20 mM, and then 2-fold serially diluted in DMSO for 8 concentrations. Compounds diluted in 45 µL of assay media (final DMSO concentration of 0.5%) were added to each well in 96-well microtiter plates in which HEp-2 cells were grown overnight (seeding density of 12,000 cells/well in a volume of 45 µL). The cells were incubated for 5 days unless otherwise noted. To measure cell viability, Cell Titer-Glo reagent (Promega) was added to each well and luminescence was measured using a multilabel reader. Experiments were done at least in triplicate. For CC50 calculation, relative cell viability compared to the DMSO control cells was plotted using XLfit (IBDS), and CC50 were calculated using the 4 Parameter Logistic Model or the Sigmoidal Dose–response Model.
Minireplicon Reporter Assay.38
A recently described firefly luciferase-based RSV minigenome construct under the control of the cellular pol I promoter was used to monitor viral polymerase activity. Cells were cotransfected with this plasmid and plasmids pRSV-L, pRSV-M2-1, pRSV-N, and pRSV-P, respectively. Compounds were added in serial dilutions and luciferase reporter activities determined 40 h post-transfection. If possible, 50 percent effective concentrations (EC50 values) were calculated based on four-parameter variable-slope nonlinear regression modeling of mean values using the Prism5 (GraphPad) software package.
Supplementary Material
ACKNOWLEDGMENTS
The authors gratefully acknowledge funding from the National Institutes of Health. Chemistry efforts and pharmacokinetic analyses at the University of Kansas Specialized Chemistry Center were supported by NIH U54HG005031 to J.A. Support for the University of Kansas NMR instrumentation was provided by NIH Shared Instrumentation Grant number S10RR024664 and NSF Major Research Instrumentation Grant number 0320648. High throughput screening performed at the Southern Research Specialized Biocontainment Screening Center was supported by Grant NIH U54HG005034-01, and W.S. acknowledges support from Grant Number R03 MH082403-01A1. Work in R.P.’s laboratory was supported, in part, by Public Health service grants R01 AI071002 and R01 HD079327 from the NIH/NIAID and NIH/NICHD, respectively (to R.K.P.). The authors also wish to thank Mr. Patrick Porubsky at the University of Kansas SCC and Ms. Arianna Mangravita-Novo at the Conrad Prebys Sanford Burnham Medical Research Institute for performing aqueous stability and in vitro ADME experiments, respectively.
ABBREVIATIONS
- ACN
acetonitrile
- ADME
absorption, distribution, metabolism, and excretion
- CC50
concentration that reduced the cell viability by 50% when compared to untreated controls
- CDI
carbonyldiimidazole
- CPE
cytopathic effect
- DIPEA
N, N-diisopropylethylamine
- EDCI
1-ethyl-3-(3-(dimethylamino)-propyl)carbodiimide
- GPCR
G-protein coupled receptor
- HATU
(1-[bis(dimethylamino)methylene]-1H-1,2,3-triazolo-[4,5-b]pyridinium 3-oxid hexafluorophosphate)
- hRSV
human respiratory syncytial virus
- MOI
multiplicity of infection
- PAMPA
parallel artificial membrane permeability assay
- PBS
phosphate buffered saline
- RdRp
RNA-dependent RNA-polymerase
- SAR
structure–activity relationship
- SI
selectivity index
- SPR
structure–property relationship
Footnotes
ASSOCIATED CONTENT
S Supporting Information
Solubility assessment protocols and Eurofins profiling data for compound 19. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interests.
References
- 1.Greenough A. Respiratory syncytial virus infection: clinical features, management, and prophylaxis. Curr. Opin. Pulm. Med. 2002;8:214–217. doi: 10.1097/00063198-200205000-00011. [DOI] [PubMed] [Google Scholar]
- 2.Wright PF, Cutts FT. Generic protocol to examine the incidence of lower respiratory infection due to respiratory syncytial virus in children less than 5 years of age. Geneva: World Health Organization; 2000. [Google Scholar]
- 3.Wright M, Piedimonte G. Respiratory syncytial virus prevention and therapy: Past, present, and future. Pediatr. Pulmonol. 2011;46:324–347. doi: 10.1002/ppul.21377. [DOI] [PubMed] [Google Scholar]
- 4.Falsey AR, Walsh EE. Respiratory syncytial virus infection in adults. Clin. Microbiol. Rev. 2000;13:371–384. doi: 10.1128/cmr.13.3.371-384.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enders G. In: Medical Microbiology. 4th ed. Baron S, editor. University of Texas Medical Branch at Galveston; 1996. [PubMed] [Google Scholar]
- 6.Blanco JCG, Boukhvalova MS, Shirey KA, Prince GA, Vogel SN. New insights for development of a safe and protective RSV vaccine. Hum. Vaccines. 2010;6:482–492. doi: 10.4161/hv.6.6.11562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graham BS. Biological challenges and technological opportunities for respiratory syncytial virus vaccine development. Immunol. Rev. 2011;239:149–166. doi: 10.1111/j.1600-065X.2010.00972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morris JA, Blount RE, Savage RE. Recovery of cytopathic agent from chimpanzees with coryza. Proc. Soc. Exp. Biol. Med. 1956;92:544–549. doi: 10.3181/00379727-92-22538. [DOI] [PubMed] [Google Scholar]
- 9.REBETOL Product Information. Schering Corporation; 1998. [Google Scholar]
- 10.Ben Venue Laboratories, I. Virazole, aerosolized ribavirin. Drug Insert. 2012 http://www.drugs.com/pro/virazole.html. [Google Scholar]
- 11.Murata Y, Falsey AR. Respiratory syncytial virus infection in adults. Antivir. Ther. 2007;12:659–670. [PubMed] [Google Scholar]
- 12.Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, O’Brien KL, Roca A, Wright PF, Bruce N, Chandran A, Theodoratou E, Sutanto A, Sedyaningsih ER, Ngama M, Munywoki PK, Kartasasmita C, Simões EAF, Rudan I, Weber MW, Campbell H. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet. 2010;375:1545–1555. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sigurs N. A cohort of children hospitalised with acute RSV bronchiolitis: impact on later respiratory disease. Paediatr. Respir. Rev. 2002;3:177–183. doi: 10.1016/s1526-0542(02)00191-4. [DOI] [PubMed] [Google Scholar]
- 14.Sigurs N, Bjarnason R, Sigurbergsson F, Kjellman B. Respiratory syncytial virus bronchiolitis in infancy is an important risk factor for asthma and allergy at age 7. Am. J. Respir. Crit. Care Med. 2000;161:1501–1507. doi: 10.1164/ajrccm.161.5.9906076. [DOI] [PubMed] [Google Scholar]
- 15.Moore BP, Chung DH, Matharu DS, Golden JE, Maddox C, Rasmussen L, Noah JW, Sosa MI, Ananthan S, Tower NA, White EL, Jia F, Prisinzano TE, Aubé J, Jonsson CB, Severson WE. (S-N-(2,5-Dimethylphenyl)-1-(quinoline-8-ylsulfonyl)pyrrolidine-2-carboxamide as a small molecule inhibitor probe for the study of respiratory syncytial virus infection. J. Med. Chem. 2012;55:8582–8587. doi: 10.1021/jm300612z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Douglas JL. In search of a small-molecule inhibitor for respiratory syncytial virus. Expert Rev. Anti Infect. Ther. 2004;2:625–639. doi: 10.1586/14787210.2.4.625. [DOI] [PubMed] [Google Scholar]
- 17.Bonavia A, Franti M, Pusateri Keaney E, Kuhen K, Seepersaud M, Radetich B, Shao J, Honda A, Dewhurst J, Balabanis K, Monroe J, Wolff K, Osborne C, Lanieri L, Hoffmaster K, Amin J, Markovits J, Broome M, Skuba E, Cornella-Taracido I, Joberty G, Bouwmeester T, Hamann L, Tallarico JA, Tommasi R, Compton T, Bushell SM. Identification of broad-spectrum antiviral compounds and assessment of the druggability of their target for efficacy against respiratory syncytial virus (RSV) Proc. Natl. Acad. Sci. U. S. A. 2011;108:6739–6744. doi: 10.1073/pnas.1017142108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liuzzi M, Mason SW, Cartier M, Lawetz C, McCollum RS, Dansereau N, Bolger G, Lapeyre N, Gaudette Y, Lagace L, Massariol MJ, Do F, Whitehead P, Lamarre L, Scouten E, Bordeleau J, Landry S, Rancourt J, Fazal G, Simoneau B. Inhibitors of respiratory syncytial virus replication target cotranscriptional mRNA guanylylation by viral RNA-dependent RNA polymerase. J. Virol. 2005;79:13105–13115. doi: 10.1128/JVI.79.20.13105-13115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapman J, Abbott E, Alber DG, Baxter RC, Bithell SK, Henderson EA, Carter MC, Chambers P, Chubb A, Cockerill GS, Collins PL, Dowdell VCL, Keegan SJ, Kelsey RD, Lockyer MJ, Luongo C, Najarro P, Pickles RJ, Simmonds M, Taylor D, Tyms S, Wilson LJ, Powell KL. RSV604, a novel inhibitor of respiratory syncytial virus replication. Antimicrob. Agents Chemother. 2007;51:3346–3353. doi: 10.1128/AAC.00211-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiong H, Foulk M, Aschenbrenner L, Fan J, Tiong-Yip C-L, Johnson KD, Moustakas D, Fleming PR, Brown DG, Zhang M, Ferguson D, Wu D, Yu Q. Discovery of a potent respiratory syncytial virus RNA polymerase inhibitor. Bioorg. Med. Chem. Lett. 2013;23:6789–6793. doi: 10.1016/j.bmcl.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 21.Sudo K, Miyazaki Y, Kojima N, Kobayashi M, Suzuki H, Shintani M, Shimizu Y. YM-53403, a unique anti-respiratory syncytial virus agent with a novel mechanism of action. Antiviral Res. 2005;65:125–131. doi: 10.1016/j.antiviral.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 22.Carter MC, Alber DG, Baxter RC, Bithell SK, Budworth J, Chubb A, Cockerill GS, Dowdell VC, Henderson EA, Keegan SJ, Kelsey RD, Lockyer MJ, Stables JN, Wilson LJ, Powell KL. 1,4-Benzodiazepines as inhibitors of respiratory syncytial virus. J. Med. Chem. 2006;49:2311–2319. doi: 10.1021/jm051185t. [DOI] [PubMed] [Google Scholar]
- 23.Sun Z, Pan Y, Jiang S, Lu L. Respiratory syncytial virus entry inhibitors targeting the F-protein. Viruses. 2013;5:211–225. doi: 10.3390/v5010211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huntley CC, Weiss WJ, Gazumyan A, Buklan A, Feld B, Hu W, Jones TR, Murphy T, Nikitenko AA, O’Hara B, Prince G, Quartuccio S, Raifeld YE, Wyde P, O’Connell JF. RFI-641, a potent respiratory syncytial virus inhibitor. Antimicrob. Agents Chemother. 2002;46:841–847. doi: 10.1128/AAC.46.3.841-847.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Douglas JL, Panis ML, Ho E, Lin KY, Krawczyk SH, Grant DM, Cai R, Swaminathan S, Chen X, Cihlar T. Small molecules VP-14637 and JNJ-2408068 inhibit respiratory syncytial virus fusion by similar mechanisms. Antimicrob. Agents Chemother. 2005;49:2460–2466. doi: 10.1128/AAC.49.6.2460-2466.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonfanti JF, Doublet F, Fortin J, Lacrampe J, Guillemont J, Muller P, Queguiner L, Arnoult E, Gevers T, Janssens P, Szel H, Willebrords R, Timmerman P, Wuyts K, Janssens F, Sommen C, Wigerinck P, Andries K. Selection of a respiratory syncytial virus fusion inhibitor clinical candidate, part 1: improving the pharmacokinetic profile using the structure-property relationship. J. Med. Chem. 2007;50:4572–4584. doi: 10.1021/jm070143x. [DOI] [PubMed] [Google Scholar]
- 27.Cianci C, Genovesi EV, Lamb L, Medina I, Yang Z, Zadjura L, Yang H, D’Arienzo C, Sin N, Yu KL, Combrink K, Li Z, Colonno R, Meanwell N, Clark J, Krystal M. Oral efficacy of a respiratory syncytial virus inhibitor in rodent models of infection. Antimicrob. Agents Chemother. 2004;48:2448–2454. doi: 10.1128/AAC.48.7.2448-2454.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meanwell NA, Cianci CW, Krystal MR. Antiviral Drugs. John Wiley & Sons, Inc.; 2011. Discovery and development of orally active RSV fusion inhibitors; pp. 353–366. [Google Scholar]
- 29.Meanwell N, Langley D. Inhibitors of Protein-Protein Interactions in Paramyxovirus Fusion: A Focus on Respiratory Syncytial Virus. In: Wendt MD, editor. Protein–Protein Interactions. Vol. 8. Berlin Heidelberg: Springer; 2012. pp. 167–196. [Google Scholar]
- 30. Taken from: http://clinicaltrials.gov/show/NCT01756482.
- 31.Chung D-H, Moore B, Matharu D, Golden JE, Maddox C, Rasmussen L, Sosa M, Ananthan S, White EL, Jia F, Jonsson C, Severson W. A cell based high-throughput screening approach for the discovery of new inhibitors of respiratory syncytial virus. Virol. J. 2013;10:1–12. doi: 10.1186/1743-422X-10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.See PubChem Summary AID 2440.
- 33.Noah JW, Severson WE, Chung DH, Moore B, Jia F, Xu X, Maddox C, Rasmussen L, Sosa MI, Tower NA, Ananthan S, Evans CW, White EL, Jonsson C, Matharu DS, Flaherty DP, Simpson DS, Golden JE, Aubé J. Probe Reports from the NIH Molecular Libraries Program [Internet] National Center for Biotechnology Information (US); 2011. Identification of a series of quinazolinediones as potent, selective, post-entry inhibitors of human respiratory syncytial virus (hRSV) via a cell-based high throughput screen and chemical optimization. http://www.ncbi.nlm.nih.gov/books/NBK133445/. [PubMed] [Google Scholar]
- 34.NIH Molecular Libraries Small Molecule Repository (MLSMR) http://mli.nih.gov/mli/compound-repository/.
- 35.Noah JW, Severson W, Chung DH, Moore B, Jia F, Xu X, Maddox C, Rasmussen L, Sosa MI, Tower NA, Ananthan S, White EL, Jonsson C, Matharu DS, Golden JE, Prisinzano TE, Aubé J. NIH Bookshelf. National Center for Biotechnology Information; 2011. A cell based HTS approach for the discovery of new inhibitors of RSV. http://www.ncbi.nlm.nih.gov/books/NBK133442/. [PubMed] [Google Scholar]
- 36.Chung D-H, Jonsson CB, Tower NA, Chu Y-K, Sahin E, Golden JE, Noah JW, Schroeder CE, Sotsky JB, Sosa MI, Cramer DE, McKellip SN, Rasmussen L, White EL, Schmaljohn CS, Julander JG, Smith JM, Filone CM, Connor JH, Sakurai Y, Davey RA. Discovery of a novel compound with anti-Venezuelan equine encephalitis virus activity that targets the nonstructural protein 2. PLoS Pathog. 2014;10:e1004213. doi: 10.1371/journal.ppat.1004213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schroeder CE, Yao T, Sotsky JB, Smith RA, Roy S, Chu YK, Guo H, Tower NA, Noah JW, McKellip S, Sosa M, Rasmussen L, Smith LH, White EL, Aubé J, Jonsson CB, Chung D-H, Golden JE. Development of (E)-2-((1,4-dimethylpiperazin-2-ylidene)amino)-5-nitro-N-phenylbenzamide, ML336: novel 2-amidinophenylbenzamides as potent inhibitors of Venezuelan equine encephalitis virus. J. Med. Chem. 2014;57:8608. doi: 10.1021/jm501203v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan D, Lee S, Thakkar VD, Luo M, Moore ML, Plemper RK. Cross-resistance mechanism of respiratory syncytial virus against structurally diverse entry inhibitors. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E3441–E3449. doi: 10.1073/pnas.1405198111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.