Abstract
GWAS have identified a breast cancer susceptibility locus on 2q35. Here we report the fine mapping of this locus using data from 101,943 subjects from 50 case-control studies. We genotype 276 SNPs using the ‘iCOGS’ genotyping array and impute genotypes for a further 1,284 using 1000 Genomes Project data. All but two, strongly correlated SNPs (rs4442975 G/T and rs6721996 G/A) are excluded as candidate causal variants at odds against > 100:1. The best functional candidate, rs4442975, is associated with oestrogen receptor positive (ER + ) disease with an odds ratio (OR) in Europeans of 0.85 (95% confidence interval = 0.84 – 0.87; P = 1.7 × 10 −43) per t-allele. This SNP flanks a transcriptional enhancer that physically interacts with the promoter of IGFBP5 (encoding insulin-like growth factor-binding protein 5) and displays allele-specific gene expression, FOXA1 binding and chromatin looping. Evidence suggests that the g-allele confers increased breast cancer susceptibility through relative downregulation of IGFBP5, a gene with known roles in breast cell biology.
The 2q35 breast cancer locus was originally identified in an Icelandic genome-wide association study (GWAS)1, and subsequently confirmed in larger European studies. The largest replication study, comprising 25 studies from the Breast Cancer Association Consortium, yielded odds ratio (OR) of 0.89 (95% CI – 0.87 to 0.92) per g-allele for rs13387042 with evidence for association with both oestrogen receptor-positive (ER + ) and ER-negative (ER −) disease2. rs13387042 lies in a 210-kb linkage disequilibrium (LD) block within a gene ‘desert’, bounded centromerically by the transition nuclear protein 1 gene (TNP1—181 kb proximal) and telomerically by the disrupted in renal carcinoma 3 gene (DIRC3—243 kb distal). Additional but more distant centromeric genes are two members of the insulin growth factor-binding protein family, IGFBP5 (345 kb proximal) and IGFBP2 (376 kb proximal).
In the current study, we describe the fine-scale mapping of the 2q35 breast cancer susceptibility locus using 1,560 genotyped and imputed single nucleotide polymorphisms (SNPs) in 101,943 subjects from 50 case-control studies. The strongest candidate for causality, SNP rs4442975, flanks a transcriptional enhancer that physically interacts with the promoter of IGFBP5. Furthermore, we demonstrate that rs4442975 is associated with allele-specific FOXA1 binding, chromatin looping and IGFBP5 expression. Our data suggest that the g-allele of rs4442975 confers increased breast cancer susceptibility through reduced IGFBP5 expression.
Results
Fine-scale mapping identifies two candidate causal variants
Association analyses were performed on 1,560 2q35 SNPs (276 genotyped and 1,284 imputed at r2 > 0.3). Three hundred and fifty-two SNPs are associated with overall breast cancer, 327 with ER + and none with ER − breast cancer (P values <10 −4; Supplementary Data 1) in European-ancestry women. The genotyped SNP rs4442975 displays the strongest association (per-t-allele OR = 0.87; 95% CI − 0.86 to 0.89; P = 3.9 × 10 −46; Fig. 1; Table 1; Supplementary Fig. 1) and this is stronger for ER + disease (OR = 0.85; 95% CI – 0.84 to 0.87; P = 1.69 × 10 −43) than for ER − disease (OR = 0.95; 95% CI − 0.91 to 0.98; P = 0.0043; P heterogeneity = 2.8 × 10 −6; Table 1).
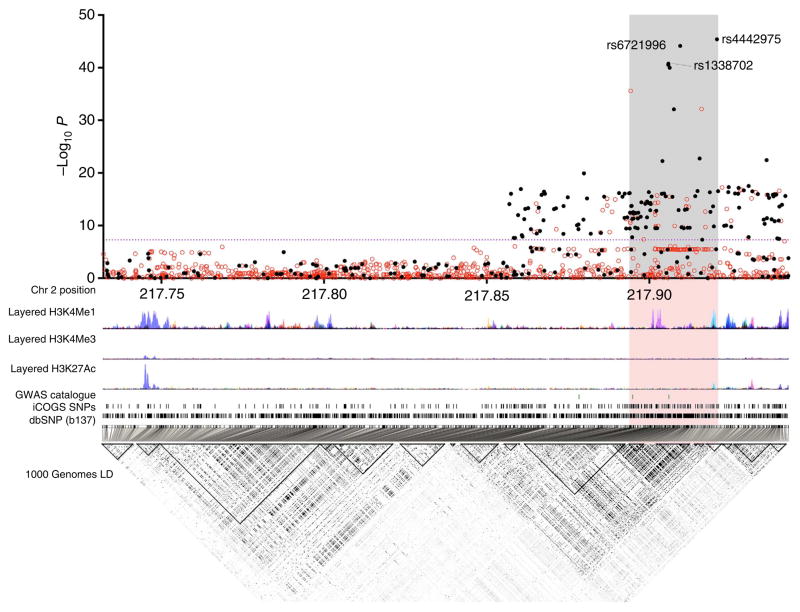
Figure 1. Genetic mapping and epigenetic landscape at the 2q35 locus.
Manhattan plot of the 2q35 breast cancer susceptibility locus. Genotyped (black dots) and imputed (red dots) SNPs are plotted based on their chromosomal position on the x axis and their overall P values (log10 values, likelihood ratio test) from the European BCAC studies (46,451 cases and 42,599 controls) on the y axis. The shaded region represents an area bounded by SNPs correlated with rs4442975 at r2 = 0.8. Data from the UCSC Genome Browser, including epigenetic marks for methylation of histone H3 at lysine 4 (H3K4me1, H3K4me3) and acetylation of H3 at lysine 27 (H3K27ac) in seven cell types from ENCODE28. The positions of all analysed iCOGS SNPs are marked. LD, using data from the BCAC population, is depicted beneath—white represents r2 = 0 and black r2 = 1.
Table 1.
Association of the two most strongly associated SNPs (rs4442975 and rs6721996) and the original GWAS SNP (rs13387042) with breast cancer.
| SNP | Position | Ref | Alt | EAF* | OR_overall | P1df_overall | OR_ER− | P1df_ER− | OR_ER + | P1df_ER + |
|---|---|---|---|---|---|---|---|---|---|---|
| Europeans | ||||||||||
| rs4442975 | 217920769 | G | T | 0.49 | 0.87 (0.86–0.89) | 3.91E −46 | 0.95 (0.91–0.98) | 0.0043 | 0.85 (0.84–0.87) | 1.69E −43 |
| rs6721996 | 217909463 | G | A | 0.49 | 0.87 (0.86–0.89) | 7.09E −45 | 0.94 (0.91–0.98) | 0.0028 | 0.86 (0.84–0.88) | 4.02E −42 |
| rs13387042 | 217905832 | A | G | 0.51 | 0.88 (0.86–0.89) | 1.69E −41 | 0.96 (0.92–0.99) | 0.023 | 0.86 (0.84–0.88) | 5.63E −40 |
| Asians | ||||||||||
| rs4442975 | 217920769 | G | T | 0.13 | 0.94 (0.87–1.02) | 0.12 | 1.01 (0.89–1.14) | 0.90 | 0.93 (0.85–1.02) | 0.11 |
| rs6721996 | 217909463 | G | A | 0.12 | 0.95 (0.88–1.03) | 0.20 | 1 (0.89–1.14) | 0.96 | 0.94 (0.86–1.03) | 0.20 |
| rs13387042 | 217905832 | A | G | 0.12 | 0.95 (0.88–1.03) | 0.21 | 1.01 (0.89–1.14) | 0.89 | 0.94 (0.85–1.03) | 0.18 |
Alt, alternative; ER, oestrogen receptor; GWAS, genome-wide association study; OR, odds ratio; Ref, reference; SNP, single nucleotide polymorphism.
The table displays the per-allele odds ratios for breast cancer in Europeans and Asians, and separately for ER-positive and ER-negative disease. In the Asian studies, the protective/rare alleles for these three SNPs are rarer (minor allele frequencies (MAFs) = 0.13, 0.12 and 0.12, respectively) than in Europeans (MAF = 0.49) but their associated relative risk estimates with overall breast cancer are consistent: per-t-allele OR (rs4442975) = 0.94; 95% confidence interval (CI) −0.87 to 1.02; P = 0.12; per-a-allele OR (rs6721996) = 0.95; 95% CI −0.88 to 1.03; P = 0.20; and per-a-allele OR (rs13387042) = 0.95; 95% CI −0.88 to 1.03; P = 0.21.
EAF, effect allele frequency (frequency of the alternative allele relative to the reference allele).
We next conducted multivariable logistic regression for both overall and ER + breast cancer, examining each SNP with univariate P<10 −4 (N = 330) in an analysis adjusted for the most significant SNP rs4442975. No further variants are strongly associated with overall or ER + disease. The second most strongly associated SNP for overall breast cancer after adjusting for rs4442975 is rs10191184 (OR = 0.96; 95% CI = 0.93 to 0.99; P = 0.0048), consistent with the hypothesis of a single causative variant. We compared the log likelihoods from the ER + univariate regression models for each SNP with the log likelihood for rs4442975. All SNPs except one (rs6721996), which was almost perfectly correlated with rs4442975 (r2 = 0.98), have log likelihoods > 100 times lower than rs4442975 and hence can reasonably be excluded as being causative. The excluded variants include the original GWAS hit, rs13387042, which is strongly correlated with rs4442975 (r2 = 0.93) but has odds of 3300:1 against being causative (Table 1). Haplotype analyses of the five most strongly associated SNPs identified two common and one rarer haplotype (frequency 1.4%: Supplementary Table 1). The rare haplotype (1) carries the cancer-protective alleles at rs4442975 (t-allele) and rs6721996 (a-allele), but not rs13387042, and has a similar risk to haplotype 2, carrying the protective alleles at all five SNPs, which is consistent with the hypothesis of rs4442975 and/or rs6721996 being the causal variant.
In Asian studies, the protective alleles for both candidate causal variants (rs4442975 and rs6721996) are rarer (minor allele frequencies (MAFs) = 0.13 and 0.12, respectively) than in Europeans (MAF = 0.49) but their associated relative risk estimates with overall breast cancer are consistent: per t-allele OR (rs4442975) = 0.94; 95% CI − 0.87 to 1.02; P = 0.12 and per a-allele OR (rs6721996) = 0.95; 95% CI − 0.88 to 1.03; P = 0.20 (Table 1).
rs4442975 resides near a putative regulatory element
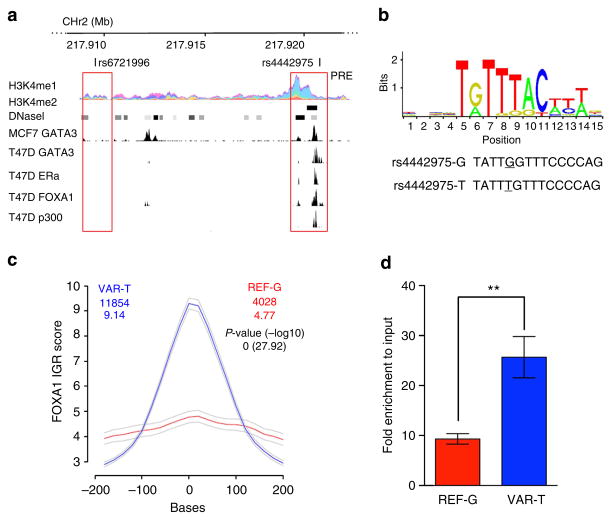
We used available ENCODE chromatin immunoprecipitation-sequencing (ChIP-seq) data to map the candidate causal SNPs relative to transcriptional regulatory elements. SNP rs4442975 lies near a putative regulatory element (PRE) as defined by H3K4Me1 histone modifications in seven cell types from ENCODE, and H3K4Me2 in MCF7 cells (Figs 1 and 2a). This PRE also contains DNaseI-hypersensitive sites in both MCF7 and HMEC cell lines (indicative of regions of open chromatin) and binds several transcription factors (TFs) associated with oestrogen signalling3 (Fig. 2a). By contrast, the region surrounding SNP rs6721996 does not contain specific histone modifications or relevant TF binding in the cell lines analysed (Fig. 2a).
Figure 2. Allele-specific binding of FOXA1 at the rs4442975 site.
(a) Epigenetic and transcriptional landscape of the 2q35 risk interval. Coloured histogram denotes histone modification ChIP-seq data from ENCODE. Data from the UCSC Genome Browser, including epigenetic marks for H3K4me1 in seven cell types from ENCODE28, H3K4me2 from MCF7 cells4, DNaseI hypersensitivity clusters in 125 cell types from ENCODE28, and TF ChIP-seq data from MCF7 and T47D ER + breast cancer cells, which are homozygous for the g-allele of rs4442975 and rs6721996 (ENCODE). The PRE contains SNP rs4442975. (b) Position weight matrix of FOXA1 from JASPAR, with homology to the risk (g) and cancer-protective (t) alleles of rs4442975 coloured below. (c) IGR histogram for SNP rs4442975 predicting the binding intensity of FOXA1 using a seven-nucleotide affinity model5. The top row of coloured numbers shows the number of instances for each K-mer found genome wide within H3K4me2 elements in MCF7 cells. The bottom row shows the averaged binding intensities at the K-mers (50 bp window). Control profiles, shown in grey, are generated by scrambling the probed sequence. (d) Allele-specific FOXA1 ChIP-qPCR results assessed at the rs4442975 SNP in heterozygous BT474 breast cancer cells. Error bars denote s.d. P values were determined with a two-tailed t-test. **P<0.01.
rs4442975 alters FOXA1 DNA binding
Breast cancer susceptibility loci have been shown to be enriched for FOXA1-binding sites at active regulatory elements in breast cancer cells; and the 2q35 locus contains variants predicted to modulate the affinity of FOXA1 (ref. 4). FOXA1 is a pioneer factor and master regulator of ER activity due to its ability to open local chromatin and recruit ER to target gene promoters5,6. SNP rs4442975 is predicted, in silico, to lie in a FOXA1-binding site with the t-allele promoting increased FOXA1 binding compared with the g-allele (Fig. 2b,c; Supplementary Fig. 2). To assess occupancy of FOXA1 in vivo, we conducted ChIP followed by allele-specific quantitative PCR (qPCR) in the heterozygous BT474 breast cancer cell line. We found that FOXA1 is indeed preferentially recruited to the t (cancer-protective) allele of candidate causal SNP rs4442975 (Fig. 2d; Supplementary Fig. 3). Of note, ChIP-seq data from ENCODE identified a second, albeit weaker, FOXA1-binding motif upstream of rs4442975 that may also influence FOXA1 recruitment (Fig. 2a). However, ChIP-qPCR did not detect FOXA1 binding in vivo to this additional site, and due to the limited availability of FOXA1-positive breast cancer cell lines with the relevant genotypes, we are unable to unequivocally discern its affinity for FOXA1. Consequently, while our results support a role for rs4442975 in modulating FOXA1-binding affinity on the site of overlap, we cannot exclude additional cis-effects typical of multi-enhancer variants7 where a rare variant, yet to be identified, would be in LD with rs4442975 and influence the recruitment of FOXA1 or other factors found in the same LD block.
rs4442975 interacts with the IGFBP5 promoter
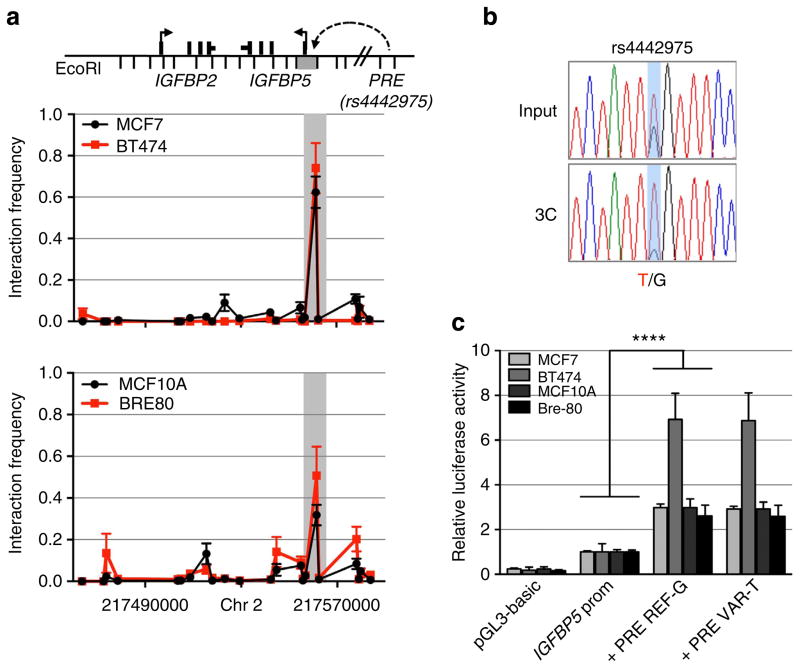
To determine the target gene(s), we used chromatin conformation capture (3C), which revealed that the PRE containing rs4442975 frequently interacts with the IGFBP5 promoter (located 345 kb proximal) in both ER + breast cancer cell lines (MCF7 and BT474) and in normal breast epithelial cells (MCF10A and Bre-80; Fig. 3a). No significant interactions were detected between this PRE and other flanking genes including IGFBP2, XRCC5, TNP1 and DIRC3 (Fig. 3a; Supplementary Figs 4–7). The region surrounding SNP rs6721996 did not interact with any flanking genes including the IGFBP5 promoter (Supplementary Figs 4–7). To assess any potential impact of SNP rs4442975 on this chromatin interaction, allele-specific 3C was performed in heterozygous BT474 cell lines. Sequence profiles indicate that the rs4442975 t-allele is more strongly associated with looping of this PRE to the IGFBP5 promoter than the g-allele (Fig. 3b; Supplementary Fig. 8), suggesting that the cancer-protective t-allele may increase IGFBP5 expression through preferential contact between this element and the IGFBP5 promoter.
Figure 3. Chromatin interactions at the 2q35 risk region with IGFBP5 in breast cell lines.
(a) 3C interaction profiles between the PRE (containing rs4442975) and the IGFBP5 promoter region (grey box). 3C libraries were generated with EcoRI, with the anchor point set at the PRE. A physical map of the region interrogated by 3C is shown above, with the grey bar representing the position of the IGFBP5 promoter (not to scale). Graphs represent three biological replicates assayed in duplicate. Error bars denote s.d. (b) 3C followed by sequencing for the rs4442975-containing region in heterozygous BT474 breast cancer cells shows allele-specific chromatin looping. Chromatograms represent one of the three independent 3C libraries generated and sequenced. (c) Luciferase reporter assays in breast cell lines demonstrating enhancer activity of the PRE at the 2q35 risk locus. The PRE was cloned upstream of an IGFBP5 promoter-driven luciferase reporter with and without SNP rs4442975. Cells were transiently transfected with each of these constructs and assayed for luciferase activity after 24 h. Graphs represent two independent experiments assayed in triplicate. Error bars denote s.d. P values were determined with a two-tailed t-test. ****P<0.0001.
rs4442975 influences IGFBP5 expression
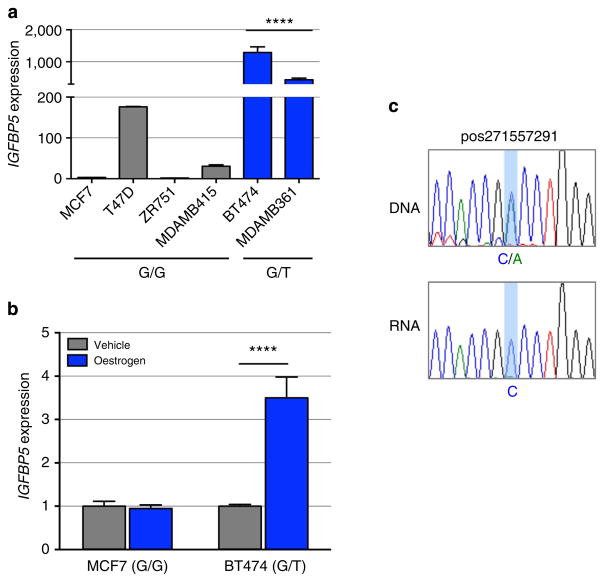
The regulatory capability of the PRE, combined with the effect of SNP rs4442975, was further examined in luciferase reporter assays, using constructs containing the IGFBP5 promoter. The wild-type PRE acts as a transcriptional enhancer, leading to a 2–3 fold increase in IGFBP5 promoter activity (Fig. 3c; PRE REF-G), but inclusion of the rs4442975 t-allele has no significant effect on the PRE enhancer activity (Fig. 3c; PRE REF-T). While this appears to rule out an effect of this SNP on transactivation, it is possible that rs4442975 is influencing gene expression through other regulatory mechanisms. To assess the impact of the rs4442975 alleles on IGFBP5 expression, we measured endogenous levels of IGFBP5 mRNA in ER-positive breast cancer cell lines either homozygous (G/G) or heterozygous (G/T) for SNP rs4442975. While limited in number, the results showed that IGFBP5 mRNA was significantly increased in heterozygous cell lines (Fig. 4a). Furthermore, given the importance of FOXA1 in oestrogen–ER activity, we also measured endogenous levels of IGFBP5 mRNA in MCF7 (G/G) and BT474 (G/T) cells following oestrogen induction and found that IGFBP5 mRNA was significantly increased but only in the heterozygous BT474 cells (Fig. 4b; Supplementary Fig. 9). To evaluate allele-specific IGFBP5 expression, we identified a heterozygous variant (pos271557291) in the first intron of IGFBP5 in BT474 cells. Sequencing of the 3C product showed that the t-allele of rs4442975 is physically linked to the variant c-allele of pos271557291 (Supplementary Fig. 10). Allele-specific expression assays revealed that the c-allele of variant pos271557291 is preferentially expressed, supporting our conclusion that the protective t-allele of rs4442975 is associated with an increase in IGFBP5 expression (Fig. 4c; Supplementary Fig. 11).
Figure 4. IGFBP5 expression in breast cancer cell lines and normal breast tissue.
(a) Endogenous IGFBP5 expression measured by qPCR in untreated ER + human breast cancer cell lines and (b) oestrogen-stimulated breast cancer cell lines. Graphs represent three independent experiments. Error bars denote s.e.m. P values were determined by a two-tailed t-test. ****P<0.0001. (c) Allele-specific IGFBP5 expression measured by allelic amplification of intronic marker variant pos271557291. Chromatograms represent one of the three independent experiments performed and sequenced.
Gene expression analyses in breast tissue
Finally, we examined the associations of rs4442975 with expression levels of genes within 1 Mb of the SNPs, in 123 normal breast tissue samples and 254 breast tumour samples in the Norwegian Breast Cancer Study (NBCS), and additionally in 135 normal breast tissue samples from the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) study. In normal breast tissue from NBCS, SNP rs4442975 is associated with expression levels of the IGFBP5 probe, A_23_P154115 (P = 0.045), and similarly in METABRIC with the IGFBP5 probe, ilmn_1750324 (P = 0.026; Supplementary Table 2), but there are no associations with other IGFBP5 probes used in these studies. In both studies, the protective t-allele of rs4442975 was associated with slightly increased IGFBP5 levels (Supplementary Fig. 12). However, for each tested IGFBP5 probe there are other more strongly expression-associated SNPs (eSNPs) at this locus, none of which are significantly correlated with the breast cancer risk candidate SNP, rs4442975 (r2<0.001; Supplementary Table 2). No significant associations were observed between rs4442975 and expression of any other genes in NBCS normal breast tissues or breast tumours, nor in METABRIC normal breast samples (Supplementary Table 3).
Discussion
In this study, we have conducted a comprehensive analysis of all known common variants within a 210-kb interval of the original 2q35 locus. We identified one independent set of correlated, highly trait-associated variants (iCHAV)8 for ER-positive breast cancer. Our data are consistent with a single disease-associated variant, with no evidence for further SNPs being associated with breast cancer risk after adjustment for the candidate causal SNP, rs4442975. However, we recently identified another iCHAV for breast cancer > 300 Kb telomeric to rs4442975 (ref. 9). These two iCHAVs are separated by several recombination hotspots, and their tagging SNPs are uncorrelated (r2 = 0.002). This observation fits the general pattern that multiple independent cancer susceptibility variants fall within GWAS-identified loci7,10, and raises the possibility that both associations are mediated through the same target gene.
Our allele-specific 3C and expression analyses provided evidence that rs4442975 contributes to changes in IGFBP5 expression. Although not robustly supported by our expression quantitative trait locus (eQTL) studies, two independent data sets showed that the protective t-allele of rs4442975 was associated with slightly increased IGFBP5 levels, which is consistent with our functional results. However, we also identified other eSNPs in the region that are more strongly associated with IGFBP5 expression in normal breast tissue, but do not drive breast cancer risk. This situation is not dissimilar to other loci we have studied, where we have not found that the causal risk SNPs are strong eQTLs for the gene they regulate11–13. This disparity may at least partly be explained by the fact that eSNPs are acting in multiple tissues, but risk-associated SNPs may only act in one specific cell type. Given that normal breast tissue is so heterogeneous, any eQTL effect that is specific to one cell type (such as stem cells) is going to be significantly diluted. In addition, eQTLs are very context dependent, so might only be expressed in breast tissue under particular stimuli or stages of development. It is also possible that the relevant cells for the analysis are luminal progenitor cells in adolescence, when the human breast seems susceptible to environmental and hormonal influences, but we have no access to data from them.
The best understood activity of the IGFBPs is sequestration of extracellular IGFs to control their growth-promoting actions. IGFBP5, which is expressed in both normal and cancer tissues, is a key member of this IGF axis—regulating cellular growth, differentiation and apoptosis14,15, but IGF-independent actions of IGFBP5 have also been demonstrated in various cell types16,17. The roles of IGFBP5 in human breast cancer are complex and there are many contradictory findings: some lines of evidence suggest that IGFBP5 acts as an inhibitor of tumour growth. For example, Butt et al.18 reported that increased expression of IGFBP5 inhibits human breast cancer cell growth. Consistent with a pro-apoptotic effect, transgenic mice, expressing IGFBP5 in mammary gland, have impaired mammary development and increased apoptotic cell death19. Other evidence indicates, conversely, that IGFBP5 has anti-apoptotic and tumour-promoting actions; Perks et al.20 reported that exogenous IGFBP5 inhibits apoptosis of breast cancer cells in vitro. Very low IGFBP5 expression has been detected in benign breast epithelium with high expression levels in adjacent breast tumour tissue21,22.
We propose that the g-allele of SNP rs4442975 (associated with increased risk) reduces FOXA1 binding and hence results in reduced chromatin accessibility, cofactor recruitment and long-range chromatin interactions. Taken together, all these lines of evidence point to increased breast cancer risk, associated with the rs4442975 g-allele, being mediated through reduced IGFBP5 expression. The IGF axis is already an important therapeutic target in other human cancers23, and our findings suggest further studies on IGFBP5 and breast cancer prevention may be merited.
Methods
Study populations and genotyping
Epidemiological data were obtained from 50 breast cancer case-control studies participating in the Breast Cancer Association Consortium; these comprised 41 studies from populations of European ancestry and 9 studies from populations of East Asian ancestry9. Genotyping was conducted using the iCOGS array, a custom array comprising ~200,000 SNPs. Details of the participating studies, genotyping calling and quality control are given elsewhere9. After quality control exclusions, we analysed data from 46,451 cases and 42,599 controls of European ancestry and 6,269 cases and 6,624 controls of Asian ancestry. ER status of the primary tumour was available for 34,539 European and 4,972 Asian cases; of these 7465 (22%) European and 1610 (32%) Asian cases were ER negative9.
SNP selection and genetic mapping
We first defined a mapping interval of 210,596 bp (positions 217, 732, 119–217, 942, 715; NCBI build 37 assembly) based on the LD block that included rs13387042 in Hapmap (CEU). We catalogued 1,578 variants in the region using the 1000 Genomes Project (March 2010 Pilot version 60 CEU project data), of which 751 variants had a MAF > 2%. Of these, we selected all SNPs correlated with the rs13387042 at r2 > 0.1 (N = 150), plus a set of SNPs designed to tag all remaining SNPs with r2 > 0.9 (N = 137). All but 11 SNPs passed a designability score (DS) provided by Illumina (DS > 0.9) and were included on the iCOGS array. The 276 SNPs included on the array all passed quality control and were included in this analysis. The genotype data were then used to impute genotypes at all additional known SNPs in the interval using IMPUTE version 2.0 and the 1000 Genome Project data (March 2012 version) as a reference panel. One thousand two hundred and eighty-four variants were successfully imputed, with imputation r2 > 0.3 in Europeans.
Statistical analysis
Per-allele ORs and s.e. were estimated for each SNP using logistic regression, separately for subjects of European and Asian ancestry, and separately for overall, ER-positive and ER-negative breast cancer. The association between each SNP and breast cancer risk was tested using a one-degree-of-freedom trend test adjusted for study and seven principal components. The statistical significance of each SNP was derived using a Wald test. To evaluate evidence for multiple association signals, we performed conditional analyses, in which the association for each SNP was re-evaluated after including other associated SNPs in the model. SNPs with a P value <10 −4 and MAF > 2% in the single SNP analysis were included in this analysis9. Differences in the OR between ER-positive and ER-negative disease were assessed using a case-only analysis, with ER status as the dependent variable. Haplotype-specific ORs and confidence limits were estimated using haplo.stats24.
Cell lines and treatments
Breast cancer cell lines MCF7 (ER + ; ATCC #HTB22), T47D (ER + ; ATCC #HTB-133), ZR751 (ER + ; ATCC #CRL-1500), MDAMB415 (ER + ; ATCC #HTB-128) and BT474 (ER + ; ATCC #HTB20) were grown in RPMI medium with 10% fetal calf serum and antibiotics. MDAMB361 (ER + ; kindly provided by Sunil Lakhani, UQCCR, Brisbane) were grown in DMEM with 20% fetal calf serum and antibiotics. Normal breast epithelial cell lines MCF10A (ATCC #CRL 10317) and Bre-80 (kindly provided by Roger Reddel, CMRI, Sydney) were grown in DMEM/F12 medium with 5% horse serum, 10 μg ml −1 insulin, 0.5 μg ml −1 hydrocortisone, 20 ng ml −1 epidermal growth factor and 100 ng ml −1 cholera toxin and antibiotics. For oestrogen induction, 24 h after plating MCF7 or BT474 cells into 24-well plates, medium was replaced with that containing 10 nM fulvestrant. Cells were incubated for 48 h and then fresh medium containing either 10 nM oestrogen or DMSO (dimethylsulphoxide; as vehicle control) was added25. All cell lines were maintained under standard conditions, routinely tested for Mycoplasma and identity profiled with short tandem repeat markers.
Chromatin conformation capture (3C)
Breast cancer cell lines were grown to 80% confluence, then crosslinked with 1% formaldehyde at 37 °C for 10 min, quenched with ice-cold 125 mM glycine and collected by cell scraping. Cells were then washed twice in ice-cold phosphate-buffered saline (PBS), lysed for 30 min on ice in 10 ml lysis buffer (10 mM Tris-HCl, pH 7.5, 10 mM NaCl, 0.2% Igepal, 1 × protease inhibitor cocktail) and homogenized with 15 strokes in a Dounce homogenizer. Nuclei were then pelleted for 10 min (800g at 4 °C), washed in PBS and resuspended in 1 ml 1.2 × EcoRI restriction buffer and 0.3% SDS for 1 h at 37 °C with shaking. Triton X-100 (2%) was added to sequester SDS, and then each tube was digested with 1,500 U EcoRI for 24 h at 37 °C with shaking. One aliquot of digested cells was set aside to assess restriction enzyme efficiency by real-time PCR (qPCR), the rest was ligated with 4,000 U of T4 DNA ligase for 4 h at 16 °C. Crosslinks were reversed by proteinase K digestion overnight, and then the 3C DNA template was purified by phenol–chloroform extraction followed by four rounds of ethanol precipitation. The final DNA pellet was dissolved in 10 mM Tris (pH 7.5) overnight at 4 °C, purified through Amicon Ultra 0.5 ml columns (EMD Millipore) and quantitated by qPCR. 3C interactions were quantitated by qPCR using primers designed within EcoRI restriction fragments (Supplementary Table 4). All qPCRs were performed on a RotorGene 6000 using MyTaq HS DNA polymerase with the addition of 5 mM of Syto9, annealing temperature of 66 °C and extension of 30 s. 3C analyses were performed in three independent experiments with each experiment quantified in duplicate. BAC clones (RP11-96E20, RP11-944D16, RP11-14F16, RP11-639B13, RP11-43F9, RP11-22K2) covering the 2q35 region were used to create artificial libraries of ligation products to normalize for PCR efficiency. Data were normalized to the signal from the BAC clone library and, between cell lines, by reference to a region within GAPDH. All qPCR products were electrophoresed on 2% agarose gels, gel purified and sequenced to verify the 3C product.
Plasmid construction and luciferase assays
The IGFBP5 promoter-driven luciferase reporter construct was generated by inserting a 1,071-bp fragment containing the IGFBP5 promoter into the KpnI and XhoI sites of pGL3-basic. To assist cloning, AgeI and SbfI sites were inserted into the BamHI and SalI sites downstream of luciferase. A 1,296-bp fragment containing the PRE was inserted into the AgeI and SbfI sites downstream of luciferase. SNP rs4442975 was incorporated into the PRE using overlap extension PCR. All constructs were sequenced to confirm variant incorporation (AGRF, Australia). Primers used to generate all constructs are listed in Supplementary Table 4. MCF7, BT474, MCF10A and Bre-80 breast cells were transfected with equimolar amounts of luciferase reporter plasmids and 50 ng of pRLTK using Lipofectamine 2000. The total amount of transfected DNA was kept constant per experiment by adding carrier plasmid (pUC19). Luciferase activity was measured 24 h post transfection using the Dual-Glo Luciferase Assay System on a Beckman-Coulter DTX-880 plate reader. To correct for any differences in transfection efficiency or cell lysate preparation, Firefly luciferase activity was normalized to Renilla luciferase. The activity of each test construct was calculated relative to IGFBP5 promoter construct, the activity of which was arbitrarily defined as 1.
Intragenomic replicates
Intragenomic replicates (IGR) predicts the modulation in affinity produced by a SNP at a TF-binding site4. The affinity of a TF for a particular DNA sequence of length K (K-mer) is obtained by averaging binding data across a ChIP-seq data set for that TF. IGR accounts for displacement effects by computing affinity models over a sliding window of K-mers around the SNP of interest. Through this process, the collection of affinity models for increasing values of K is placed in a lattice structure that connects K-mers, which are 1 bp apart. Two lattices are constructed, one for each of the variants alleles. The maxima among the affinity models in the lattices is used to calculate the IGR score. T-tests are used to assess the statistical significance of the affinity modulation between the two K-mers with the maximum affinities.
Allele-specific ChIP-qPCR
Breast cancer cell lines were grown to 95% confluence, crosslinked with 1% formaldehyde at 37 °C for 10 min, cells were rinsed with ice-cold PBS plus 5% bovine serum albumin and then with PBS and collected with PBS plus 1 × protease inhibitor cocktail (Roche Molecular Biochemicals, Indianapolis, IN). Collected cells were centrifuged for 2 min at 3,000 r.p.m. Cell pellet was then resuspended in 0.35 ml of lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 8.1, 1 × protease inhibitor cocktail) and sonicated 20 times in 30 s on 30 s off cycles at the maximum setting (Diagenode Biorupter 300) followed by centrifugation at maximum speed for 15 min. Supernatants were collected and diluted in dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl, pH 8.1). Four micrograms of FOXA1 antibody (Acris, AP16139PU-N) was pre-bound for 6 h to protein A and protein G Dynal magnetic beads (Dynal Biotech, Norway) and washed three times with ice-cold PBS plus 5% bovine serum albumin and then added to the diluted chromatin for overnight immunoprecipitation. The magnetic bead–chromatin complexes were collected and washed six times in RIPA buffer (50 mM HEPES (pH 7.6), 1 mM EDTA, 0.7% Na deoxycholate, 1% NP-40, 0.5 M LiCl), then washed twice with Tris-EDTA buffer. To reverse the formaldehyde crosslinking, decrosslinking buffer (1% SDS, 0.1 M NaHCO3) was added to the complexes overnight at 65 °C. DNA fragments were purified with a QIAquick Spin Kit (Qiagen, CA). For PCR, 2.5 μl from a 125-μl immunoprecipitated chromatin extraction and 250-μl input extraction, and 40 cycles of amplification were used. To assess differential FOXA1 binding at the heterozygous alleles, the MAMA (Mismatch Amplification Mutation Assays) PCR-based technique was used26. Reverse MAMA primers specific to each allele were designed with one mismatched nucleotide at the 3′ end26. The primers are listed in Supplementary Table 4.
Gene expression analysis
MCF7 and BT474 total RNA was extracted using Trizol (Life Technologies) from untreated, oestrogen (10 nM)- or vehicle (DMSO)-treated cells. Residual DNA contaminants were removed by DNAse treatment (Ambion) and complementary DNA was synthesized using random primers as per manufacturers’ instructions (Life Technologies). All qPCRs were performed on a RotorGene 6000 (Corbett Research) with TaqMan Gene Expression assays (Hs00181213_m1 for IGFBP5 and Hs00907239_m1 for TFF1) and TaqMan Universal PCR master mix. All reactions were normalized against B-glucuronidase (MIM 611499; Catalogue No. 4326320E). For in vivo allele-specific gene expression, a primer outside of the rs4442975 SNP and its closest EcoRI restriction enzyme site and a primer outside of the SNP pos271557291 and its closest EcoRI site were first used to PCR amplify the EcoRI 3C product from BT474 cells. PCR-amplified products were cloned into pBLUNT empty vector (Life Technologies), then sequenced using the Sanger sequencing, which revealed the linkage between the two alleles (Supplementary Fig. 10). BT474 genomic DNA was extracted using Qiagen DNeasy blood and tissue kit. BT474 total nuclear RNA was extracted using Trizol and cDNA synthesized using a gene-specific primer. PCR-amplified sequences from BT474 genomic DNA or cDNA were gel purified (Qiagen) and Sanger sequenced to measure the DNA and RNA levels of each allele. All experiments were conducted in biological triplicates and qPCR reactions as technical duplicates. The primers are listed in Supplementary Table 4.
eQTL analysis
eQTL analyses were conducted in two studies: 123 normal breast tissue and 254 breast tumours from women in the Norwegian Breast Cancer Study (NBCS); all women were of Caucasian origin. The 123 normal breast tissue is a cohort of expression data from normal breasts biopsy (n = 74), reduction plastic surgery (n = 37) and adjacent normal (n = 12) (adjacent to tumour). Correlations between the two most likely causative SNPs (rs4442975 and rs6721996) and expression levels of nearby genes (500 kb upstream and downstream of the SNPs) were assessed using a linear regression model in which an additive effect on expression level was assumed for each copy of the rare allele. Calculations were carried out using the eMap library in R (www.bios.unc.edu/~weisun/software/eMap).
The second eQTL analysis was based on 135 adjacent normal breast samples from women of Caucasian origin in the METABRIC study27. Matched gene expression (Illumina HT-12 v3 microarray) and germline SNP data that were either genotyped (Affymetrix SNP 6.0) or imputed (1000 Genomes Project, March 2012 data using IMPUTE version 2.0) were used. Statistical methods were identical to the NBCS analysis.
Supplementary Material
Acknowledgments
BCAC thanks all the individuals who took part in these studies and all the researchers, clinicians, technicians and administrative staff who have enabled this work to be carried out. BCAC is funded by Cancer Research UK (C1287/A10118, C1287/A12014) and by the European Community’s Seventh Framework Programme under grant agreement number 223175 (grant number HEALTH-F2-2009-223175; COGS). Meetings of the BCAC have been funded by the European Union COST programme (BM0606). The COGS study would not have been possible without the contributions of the following: Andrew Berchuck (OCAC), Rosalind A. Eeles, Ali Amin Al Olama, Zsofia Kote-Jarai, Sara Benlloch (PRACTICAL), Antonis Antoniou, Lesley McGuffog, Ken Offit (CIMBA), Andrew Lee, the staff of the Centre for Genetic Epidemiology Laboratory in Cambridge, the staff of the CNIO genotyping unit, Sylvie LaBoissière and Frederic Robidoux and the staff of the McGill University and Génome Québec Innovation Centre, the staff of the Copenhagen DNA laboratory, and Julie M. Cunningham, Sharon A. Windebank, Christopher A. Hilker, Jeffrey Meyer and the staff of Mayo Clinic Genotyping Core Facility. Genotyping of the iCOGS array was funded by the European Union (HEALTH-F2-2009-223175), Cancer Research UK (C1287/A10710), the Canadian Institutes of Health Research for the ‘CIHR Team in Familial Risks of Breast Cancer’ program—grant #CRN-87521 and the Ministry of Economic Development, Innovation and Export Trade of Quebec—grant #PSR-SIIRI-701. The QIMR Berghofer group was supported by a National Health and Medical Research Council of Australia project grant (1021731). The Princess Margaret Cancer Centre-University Health Network group was supported by the US National Institutes of Health (NIH; R01CA155004 to M.L.) and OICR Young Investigator Award (M.L.). D.F.E. is a Principal Research Fellow of CR-UK. G.C.-T. is an NHMRC Senior Principal Research Fellow. S.L.E. and J.D.F. are supported by Fellowships from the National Breast Cancer Foundation (NBCF) Australia. The funders have no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. The Australian Breast Cancer Family Study (ABCFS) would like to thank Maggie Angelakos, Judi Maskiell and Gillian Dite. ABCFS was supported by grant UM1 CA164920 from the National Cancer Institute (USA).
The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centres in the Breast Cancer Family Registry (BCFR), nor does it mention trade names, commercial products or organizations that imply endorsement by the USA Government or the BCFR. The ABCFS was also supported by the National Health and Medical Research Council of Australia, the New South Wales Cancer Council, the Victorian Health Promotion Foundation (Australia) and the Victorian Breast Cancer Research Consortium. J.L.H. is a National Health and Medical Research Council (NHMRC) Australia Fellow and a Victorian Breast Cancer Research Consortium Group Leader. M.C.S. is a NHMRC Senior Research Fellow and a Victorian Breast Cancer Research Consortium Group Leader. The ABCS study was supported by the Dutch Cancer Society (grants NKI 2007-3839; 2009 4363); BBMRI-NL, which is a Research Infrastructure financed by the Dutch government (NWO 184.021.007); and the Dutch National Genomics Initiative. The ACP study wishes to thank the participants in the Thai Breast Cancer study. Special thanks also go to the Thai Ministry of Public Health (MOPH), doctors and nurses who helped with the data collection process. Finally, the study would like to thank Dr Prat Boonyawongviroj, the former Permanent Secretary of MOPH and Dr Pornthep Siriwanarungsan, the Department Director-General of Disease Control who have supported the study throughout. The ACP study is funded by the Breast Cancer Research Trust, UK. The BBCC study would like to thank Matthias Rübner, Alexander Hein and Michael Schneider. The work of the BBCC was partly funded by ELAN-Fond of the University Hospital of Erlangen. The BBCS would like to thank Eileen Williams, Elaine Ryder-Mills and Kara Sargus. The BBCS is funded by Cancer Research, UK, and Breakthrough Breast Cancer and acknowledges NHS funding to the NIHR Biomedical Research Centre, and the National Cancer Research Network (NCRN). The BIGGS would like to thank Niall McInerney, Gabrielle Colleran, Andrew Rowan and Angela Jones. E.J.S. is supported by NIHR Comprehensive Biomedical Research Centre, Guy’s & St Thomas’ NHS Foundation Trust in partnership with King’s College London, UK. I.T. is supported by the Oxford Biomedical Research Centre. The BSUCH would like to thank Peter Bugert, Medical Faculty Mannheim. The BSUCH study was supported by the Dietmar-Hopp Foundation, the Helmholtz Society and the German Cancer Research Center (DKFZ). The CECILE study was funded by Fondation de France, Institut National du Cancer (INCa), Ligue Nationale contre le Cancer, Ligue contre le Cancer Grand Ouest, Agence Nationale de Sécurité Sanitaire (ANSES) and Agence Nationale de la Recherche (ANR). The CGPS study would like to thank staff and participants of the Copenhagen General Population Study. It would also like to thank Dorthe Uldall Andersen, Maria Birna Arnadottir, Anne Bank and Dorthe Kjeldgård Hansen for the excellent technical assistance. The Danish Breast Cancer Group (DBCG) is acknowledged for the tumour information. The CGPS was supported by the Chief Physician Johan Boserup and Lise Boserup Fund, the Danish Medical Research Council and Herlev Hospital. The CNIO-BCS would like to thank Guillermo Pita, Charo Alonso, Daniel Herrero, Nuria Álvarez, Pilar Zamora, Primitiva Menendez and the Human Genotyping-CEGEN Unit (CNIO). The CNIO-BCS was supported by the Genome Spain Foundation, the Red Temática de Investigación Cooperativa en Cáncer and grants from the Asociación Española Contra el Cáncer and the Fondo de Investigación Sanitario (PI11/00923 and PI081120). The Human Genotyping-CEGEN Unit (CNIO) is supported by the Instituto de Salud Carlos III. The CTS study would like to thank the CTS steering committee: Leslie Bernstein, Susan Neuhausen, James Lacey, Sophia Wang, Huiyan Ma, Yani Lu and Jessica Clague DeHart at the Beckman Research Institute of the City of Hope; Dennis Deapen, Rich Pinder, Eunjung Lee and Fred Schumacher at the University of Southern California; Pam Horn-Ross, Peggy Reynolds, Christina Clarke and David Nelson at the Cancer Prevention Institute of California; and Hoda Anton-Culver, Hannah Park and Al Ziogas at the University of California, Irvine. The CTS was initially supported by the California Breast Cancer Act of 1993 and the California Breast Cancer Research Fund (contract 97-10500) and is currently funded through the National Institutes of Health (R01 CA77398). Collection of cancer incidence data was supported by the California Department of Public Health as part of the statewide cancer reporting program mandated by California Health and Safety Code Section 103885. H.A.-C. receives support from the Lon V Smith Foundation (LVS39420). The ESTHER study would like to thank Hartwig Ziegler, Sonja Wolf and Volker Hermann. The ESTHER study was supported by a grant from the Baden Württemberg Ministry of Science, Research and Arts. Additional cases were recruited in the context of the VERDI study, which was supported by a grant from the German Cancer Aid (Deutsche Krebshilfe). The GENICA Network would like to thank Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart; and University of Tübingen, Germany (H.B., Wing-Yee Lo, Christina Justenhoven); Department of Internal Medicine, Evangelische Kliniken Bonn gGmbH, Johanniter Krankenhaus, Bonn, Germany (Yon-Dschun Ko, Christian Baisch); Institute of Pathology, University of Bonn, Germany (Hans-Peter Fischer); Molecular Genetics of Breast Cancer, Deutsches Krebsforschungszentrum (DKFZ), Heidelberg, Germany (Ute Hamann); Institute for Prevention and Occupational Medicine of the German Social Accident Insurance, Institute of the Ruhr University Bochum (IPA), Bochum, Germany (T.B., Beate Pesch, Sylvia Rabstein, Anne Lotz); and Institute of Occupational Medicine and Maritime Medicine, University Medical Center Hamburg-Eppendorf, Germany (Volker Harth). The GENICA was funded by the Federal Ministry of Education and Research (BMBF) Germany grants 01KW9975/5, 01KW9976/8, 01KW9977/0 and 01KW0114; the Robert Bosch Foundation, Stuttgart; Deutsches Krebsforschungszentrum (DKFZ), Heidelberg; the Institute for Prevention and Occupational Medicine of the German Social Accident Insurance, Institute of the Ruhr University Bochum (IPA), Bochum; as well as the Department of Internal Medicine, Evangelische Kliniken Bonn gGmbH, Johanniter Krankenhaus, Bonn, Germany. The HEBCS would like to thank Kirsimari Aaltonen, Karl von Smitten, Sofia Khan, Tuomas Heikkinen and Irja Erkkilä. The HEBCS was financially supported by the Helsinki University Central Hospital Research Fund, Academy of Finland (266528), the Finnish Cancer Society, The Nordic Cancer Union and the Sigrid Juselius Foundation. The HERPACC was supported by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports, Culture and Technology of Japan, by a Grant-in-Aid for the Third Term Comprehensive 10-Year Strategy for Cancer Control from Ministry Health, Labour and Welfare of Japan, by Health and Labour Sciences Research Grants for Research on Applying Health Technology from Ministry Health, Labour and Welfare of Japan and by National Cancer Center Research and Development Fund. The HMBCS would like to thank Natalia Antonenkova, Peter Hillemanns, Hans Christiansen and Johann H. Karstens.
The HMBCS was supported by a grant from the Friends of Hannover Medical School and by the Rudolf Bartling Foundation. Financial support for KARBAC was provided through the regional agreement on medical training and clinical research (ALF) between Stockholm County Council and Karolinska Institutet, the Swedish Cancer Society, The Gustav V Jubilee foundation and Bert von Kantzows foundation. The KBCP would like to thank Eija Myöhänen and Helena Kemiläinen. The KBCP was financially supported by the special Government Funding (EVO) of Kuopio University Hospital grants, Cancer Fund of North Savo, the Finnish Cancer Organizations, the Academy of Finland and by the strategic funding of the University of Eastern Finland. The kConFab/AOCS study would like to thank Heather Thorne, Eveline Niedermayr, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics and the Clinical Follow Up Study (which has received funding from the NHMRC, the National Breast Cancer Foundation, Cancer Australia and the National Institute of Health (USA)) for their contributions to this resource, and the many families who contributed to kConFab. kConFab is supported by a grant from the National Breast Cancer Foundation and previously by the National Health and Medical Research Council (NHMRC), the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia, and the Cancer Foundation of Western Australia. The LAABC study would like to thank all the study participants and the entire data collection team, especially Annie Fung and June Yashiki. LAABC is supported by grants (1RB-0287, 3PB-0102, 5PB-0018, 10PB-0098) from the California Breast Cancer Research Program. Incident breast cancer cases were collected by the USC Cancer Surveillance Program (CSP), which is supported under subcontract by the California Department of Health. The CSP is also part of the National Cancer Institute’s Division of Cancer Prevention and Control Surveillance, Epidemiology, and End Results Program, under contract number N01CN25403. The LMBC would like to thank Gilian Peuteman, Dominiek Smeets, Thomas Van Brussel and Kathleen Corthouts. LMBC is supported by the ‘Stichting tegen Kanker’ (232-2008 and 196-2010). Diether Lambrechts is supported by the FWO and the KULPFV/10/016-SymBioSysII. The MARIE study would like to Judith Heinz, Nadia Obi, Alina Vrieling, Sabine Behrens, Ursula Eilber, Muhabbet Celik, Til Olchers and Stefan Nickels. The MARIE study was supported by the Deutsche Krebshilfe e.V. (70-2892-BR I), the Hamburg Cancer Society, the German Cancer Research Center and the genotype work in part by the Federal Ministry of Education and Research (BMBF) Germany (01KH0402). The MBCSG would like to thank Siranoush Manoukian, Bernard Peissel and Daniela Zaffaroni of the Fondazione IRCCS Istituto Nazionale dei Tumori (INT); Monica Barile and Irene Feroce of the Istituto Europeo di Oncologia (IEO); and Loris Bernard and the personnel of the Cogentech Cancer Genetic Test Laboratory. MBCSG is supported by grants from the Italian Association for Cancer Research (AIRC) and by funds from the Italian citizens who allocated the 5/1,000 share of their tax payment in support of the Fondazione IRCCS Istituto Nazionale Tumori, according to Italian laws (INT-Institutional strategic projects ‘5 × 1000’). The MCBCS was supported by the NIH grants CA128978, CA116167, CA176785, an NIH Specialized Program of Research Excellence (SPORE) in Breast Cancer (CA116201), and the Breast Cancer Research Foundation and a generous gift from the David F. and Margaret T. Grohne Family Foundation and the Ting Tsung and Wei Fong Chao Foundation. MCCS cohort recruitment was funded by VicHealth and Cancer Council Victoria. The MCCS was further supported by Australian NHMRC grants 209057, 251553 and 504711 and by infrastructure provided by Cancer Council Victoria. The MEC was support by NIH grants CA63464, CA54281, CA098758 and CA132839. The MTLGEBCS would like to thank Martine Tranchant (Cancer Genomics Laboratory, CHU de Québec Research Center), Marie-France Valois, Annie Turgeon and Lea Heguy (McGill University Health Center, Royal Victoria Hospital, McGill University) for DNA extraction, sample management and skilful technical assistance. J.S. is Chairholder of the Canada Research Chair in Oncogenetics. The work of MTLGEBCS was supported by the Quebec Breast Cancer Foundation, the Canadian Institutes of Health Research for the ‘CIHR Team in Familial Risks of Breast Cancer’ program—grant #CRN-87521—and the Ministry of Economic Development, Innovation and Export Trade—grant #PSR-SIIRI-701. The MYBRCA study would like to thank Phuah Sze Yee, Peter Kang, Kang In Nee, Kavitta Sivanandan, Shivaani Mariapun, Yoon Sook-Yee, Teh Yew Ching and Nur Aishah Mohd Taib for DNA Extraction and patient recruitment. MYBRCA is funded by research grants from the Malaysian Ministry of Science, Technology and Innovation (MOSTI), Malaysian Ministry of Higher Education (UM.C/HlR/MOHE/06) and Cancer Research Initiatives Foundation (CARIF). Additional controls were recruited by the Singapore Eye Research Institute, which was supported by a grant from the Biomedical Research Council (BMRC08/1/35/19/550), Singapore and the National medical Research Council, Singapore (NMRC/CG/SERI/2010). The NBCS was supported by grants from the Norwegian Research council, 155218/V40, 175240/S10 to A.-L.B.-D., FUGE-NFR 181600/V11 to V.N.K. and a Swizz Bridge Award to A.-L.B.-D. The NBHS_TN study would like to thank participants and research staff for their contributions and commitment to this study. The NBHS was supported by NIH grant R01CA100374. Biological sample preparation was conducted by the Survey and Biospecimen Shared Resource, which is supported by P30 CA68485. The OBCS study would like to thank Meeri Otsukka, Kari Mononen, Mervi Grip and Saila Kauppila. The OBCS was supported by research grants from the Finnish Cancer Foundation, the Academy of Finland (grant number 250083, 122715 and Center of Excellence grant number 251314), the Finnish Cancer Foundation, the Sigrid Juselius Foundation, the University of Oulu, the University of Oulu Support Foundation and the special Governmental EVO funds for Oulu University Hospital-based research activities. The OFBCR study would like to thank Teresa Selander and Nayana Weerasooriya. The Ontario Familial Breast Cancer Registry (OFBCR) was supported by grant UM1 CA164920 from the National Cancer Institute (USA).
The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centres in the Breast Cancer Family Registry (BCFR), nor does it mention trade names, commercial products or organizations that imply endorsement by the USA Government or the BCFR. The ORIGO study would like to thank E. Krol-Warmerdam, and J. Blom for patient accrual, administering questionnaires and managing clinical information. The LUMC survival data were retrieved from the Leiden hospital-based cancer registry system (ONCDOC) with the help of Dr J. Molenaar. The ORIGO study was supported by the Dutch Cancer Society (RUL 1997-1505) and the Biobanking and Biomolecular Resources Research Infrastructure (BBMRI-NL CP16). The PBCS study would like to thank Louise Brinton, Mark Sherman, Neonila Szeszenia-Dabrowska, Beata Peplonska, Witold Zatonski, Pei Chao and Michael Stagner. The PBCS was funded by Intramural Research Funds of the National Cancer Institute, Department of Health and Human Services, USA. The pKARMA study would like to thank The Swedish Medical Research Counsel. The pKARMA study was supported by Märit and Hans Rausings Initiative Against Breast Cancer. The RBCS study would like to thank Petra Bos, Jannet Blom, Ellen Crepin, Elisabeth Huijskens, Annette Heemskerk and the Erasmus MC Family Cancer Clinic. The RBCS was funded by the Dutch Cancer Society (DDHK 2004-3124, DDHK 2009-4318). The SASBAC would like to thank The Swedish Medical Research Counsel. The SASBAC study was supported by funding from the Agency for Science, Technology and Research of Singapore (A*STAR), the US National Institute of Health (NIH) and the Susan G. Komen Breast Cancer Foundation. The SBCGS would like to thank participants and research staff for their contributions and commitment to this study. The SBCGS was supported primarily by NIH grants R01CA64277, R01CA148667 and R37CA70867. Biological sample preparation was conducted by the Survey and Biospecimen Shared Resource, which is supported by P30 CA68485. The scientific development and funding of this project were, in part, supported by the Genetic Associations and Mechanisms in Oncology (GAME-ON) Network U19 CA148065. The SBCS would like to thank Sue Higham, Helen Cramp, Sabapathy Balasubramanian, Ian Borck and Dan Connley. The SBCS was supported by Yorkshire Cancer Research S295, S299 and S305PA. The SEARCH study would like to thank The SEARCH and EPIC teams. SEARCH is funded by a programme grant from Cancer Research UK (C490/A10124) and supported by the UK National Institute for Health Research Biomedical Research Centre at the University of Cambridge. SEBCS was supported by the BRL (Basic Research Laboratory) program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (2012-0000347). The SGBCC study would like to thank the participants and research coordinator Kimberley Chua. SGBCC is funded by the National Medical Research Council start-up Grant and Centre Grant (NMRC/CG/NCIS/2010). Additional controls were recruited by the Singapore Consortium of Cohort Studies-Multi-ethnic cohort (SCCS-MEC), which was funded by the Biomedical Research Council, grant number: 05/1/21/19/425. The SKKDKFZS study thanks all study participants, clinicians, family doctors, researchers and technicians for their contributions and commitment to this study. SKKDKFZS is supported by the DKFZ. The SZBCS was supported by Grant PBZ_KBN_122/P05/2004. The TBCS was funded by The National Cancer Institute, Thailand. The TNBCC was supported by: a Specialized Program of Research Excellence (SPORE) in Breast Cancer (CA116201), a grant from the Breast Cancer Research Foundation, a generous gift from the David F. and Margaret T. Grohne Family Foundation and the Ting Tsung and Wei Fong Chao Foundation, the Stefanie Spielman Breast Cancer fund and the OSU Comprehensive Cancer Center, DBBR (a CCSG Share Resource by National Institutes of Health Grant P30 CA016056), the Hellenic Cooperative Oncology Group research grant (HR R_BG/04) and the Greek General Secretary for Research and Technology (GSRT) Program, Research Excellence II, the European Union (European Social Fund—ESF), and Greek national funds through the Operational Program ‘Education and Lifelong Learning’ of the National Strategic Reference Framework (NSRF)—ARISTEIA. The TWBCS is supported by the Taiwan Biobank project of the Institute of Biomedical Sciences, Academia Sinica, Taiwan. The UKBGS study would like to thank Breakthrough Breast Cancer and the Institute of Cancer Research for support and funding of the Breakthrough Generations Study, and the study participants, study staff, and the doctors, nurses and other health care providers and health information sources who have contributed to the study. We acknowledge NHS funding to the Royal Marsden/ICR NIHR Biomedical Research Centre. The UKBGS is funded by Breakthrough Breast Cancer and the Institute of Cancer Research (ICR), London. ICR acknowledges NHS funding to the NIHR Biomedical Research Centre.
Kyriaki Michailidou4, Silje Nord5, Richard Cowper-Sal · lari6, Kinjal Desai6,7, Siddhartha Kar4, Kristine M Hillman2,3, Susanne Kaufmann2,3, Dylan M. Glubb2, Jonathan Beesley2, Joe Dennis4, Manjeet K. Bolla4, Qin Wang4, Ed Dicks1, Qi Guo1, Marjanka K. Schmidt8, Mitul Shah1, Robert Luben4, Judith Brown4, Kamila Czene9, Hatef Darabi9, Mikael Eriksson9, Daniel Klevebring9, Stig E. Bojesen10,11,12, Børge G. Nordestgaard10,11,12, Sune F. Nielsen10,11, Henrik Flyger13, Diether Lambrechts14,15, Bernard Thienpont15,16, Patrick Neven17,18, Hans Wildiers17,18, Annegien Broeks8, Laura J. Van’t Veer8, Emiel J. Th Rutgers8, Fergus J. Couch19, Janet E. Olson20, Emily Hallberg20, Celine Vachon20, Jenny Chang-Claude21, Anja Rudolph21, Petra Seibold21, Dieter Flesch-Janys22, Julian Peto23, Isabel dos-Santos-Silva23, Lorna Gibson23, Heli Nevanlinna24, Taru A. Muranen24, Kristiina Aittomäki25, Carl Blomqvist26, Per Hall9, Jingmei Li27, Jianjun Liu27, Keith Humphreys9, Daehee Kang28, 29, 30, Ji-Yeob Choi28, 29, Sue K. Park28, 29, 30, Dong-Young Noh31, Keitaro Matsuo32, Hidemi Ito33, Hiroji Iwata34, Yasushi Yatabe35, Pascal Guénel36, 37, Thérèse Truong36, 37, Florence Menegaux36, 37, Marie Sanchez36, 37, Barbara Burwinkel38, 39, Frederik Marme38, 40, Andreas Schneeweiss38, 40, Christof Sohn38, Anna H. Wu41, Chiu-chen Tseng41, David Van Den Berg41, Daniel O. Stram41, Javier Benitez42, 43, M Pilar Zamora44, Jose Ignacio Arias Perez45, Primitiva Menéndez46, Xiao-Ou Shu47, Wei Lu48, Yu-Tang Gao49, Qiuyin Cai47, Angela Cox50, Simon S. Cross51, Malcolm W.R. Reed50, Irene L. Andrulis52, 53, Julia A. Knight54, 55, Gord Glendon56, Sandrine Tchatchou52, Elinor J. Sawyer57, Ian Tomlinson58, Michael J. Kerin59, Nicola Miller59, Australian Ovarian Cancer Management Group†, Christopher A. Haiman61, Brian E. Henderson61, Fredrick Schumacher61, Loic Le Marchand62, Annika Lindblom63, Sara Margolin64, Soo Hwang TEO65, 66, Cheng Har YIP65, Daphne SC Lee66, Tien Y. Wong67, Maartje J. Hooning68, John W.M. Martens68, J Margriet Collée69, Carolien H.M. van Deurzen70, John L. Hopper71, Melissa C. Southey72, Helen Tsimiklis72, Miroslav K. Kapuscinski71, Chen-Yang Shen73, 74, Pei-Ei Wu75, Jyh-Cherng Yu76, Shou-Tung Chen77, Grethe Grenaker Alnæs5, Anne-Lise Borresen-Dale5, 78, Graham G. Giles79, 80, Roger L. Milne80, 81, Catriona McLean82, Kenneth Muir83, 84, Artitaya Lophatananon83, Sarah Stewart-Brown83, Pornthep Siriwanarangsan85, Mikael Hartman86, 87, Hui Miao87, Shaik Ahmad Bin Syed Buhari88, Yik Ying Teo87, 89, Peter A. Fasching90, 91, Lothar Haeberle91, Arif B. Ekici92, Matthias W. Beckmann91, Hermann Brenner93, 94, Aida Karina Dieffenbach93, 94, Volker Arndt93, Christa Stegmaier95, Anthony Swerdlow96, 97, Alan Ashworth98, Nick Orr98, Minouk J. Schoemaker97, Montserrat García-Closas98, 99, Jonine Figueroa100, Stephen J. Chanock100, Jolanta Lissowska101, Jacques Simard102, Mark S. Goldberg103, 104, France Labrèche105, Martine Dumont102, Robert Winqvist106, Katri Pylkäs106, Arja Jukkola-Vuorinen107, Hiltrud Brauch108, 109, 110, Thomas Brüning111, Yon-Dschun Koto112, Paolo Radice113, Paolo Peterlongo114, Bernardo Bonanni115, Sara Volorio114, 116, Thilo Dörk117, Natalia V. Bogdanova118, Sonja Helbig117, Arto Mannermaa119, 120, 121, Vesa Kataja119, 121, Veli-Matti Kosma119, 120, 121, Jaana M. Hartikainen119, 120, 121, Peter Devilee122, Robert A.E.M. Tollenaar123, Caroline Seynaeve124, Christi J. Van Asperen69, Anna Jakubowska125, Jan Lubinski125, Katarzyna Jaworska-Bieniek125, Katarzyna Durda125, Susan Slager20, Amanda E. Toland126, Christine B. Ambrosone127, Drakoulis Yannoukakos128, Suleeporn Sangrajrang129, Valerie Gaborieau130, Paul Brennan130, James McKay130, Ute Hamann131, Diana Torres131, 132, Wei Zheng47,Jirong Long47, Hoda Anton-Culver133, Susan L. Neuhausen134, Craig Luccarini1, Caroline Baynes1, Shahana Ahmed1, Mel Maranian1, Catherine S. Healey1, Anna González-Neira135, Guillermo Pita135, M. Rosario Alonso135, Nuria Álvarez135, Daniel Herrero135, Daniel C. Tessier136, Daniel Vincent137, Francois Bacot137, Ines de Santiago138, Jason Carroll138, Carlos Caldas138, Melissa A. Brown3, Mathieu Lupien6, 56, 139, Vessela N. Kristensen5, 78, 140, Paul D.P. Pharoah1,4, Georgia Chenevix-Trench2, Juliet D. French2,3, Douglas F. Easton1,4 & Alison M. Dunning1
Australian Ovarian Cancer Management Group
Georgia Chenevix-Trench2, Penny Webb141, David Bowtell60, Anna De Fazio142, 143
Footnotes
Centre for Cancer Genetic Epidemiology, Department of Oncology, University of Cambridge, Cambridge CB1 8RN, UK.
Department of Genetics, QIMR Berghofer Medical Research Institute, Brisbane, Queensland 4029, Australia.
School of Chemistry and Molecular Biosciences, The University of Queensland, Brisbane, Queensland 4072, Australia.
Centre for Cancer Genetic Epidemiology, Department of Public Health and Primary Care, University of Cambridge, Cambridge CB1 8RN, UK.
Department of Genetics, Institute for Cancer Research, Oslo University Hospital, Radiumhospitalet, N-0310 Oslo, Norway.
The Princess Margaret Cancer Centre, University Health Network, Toronto, Ontario, Canada M5T 2M9.
Geisel School of Medicine, Dartmouth College, Hanover, New Hampshire 03755, USA.
Netherlands Cancer Institute, Antoni van Leeuwenhoek hospital, 1066 CX Amsterdam, The Netherlands.
Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Stockholm SE-17177, Sweden.
Copenhagen General Population Study, Herlev Hospital, 2730 Herlev, Copenhagen, Denmark.
Department of Clinical Biochemistry, Herlev Hospital, Copenhagen University Hospital, 2730 Herlev, Copenhagen, Denmark.
Faculty of Health and Medical Sciences, University of Copenhagen, 2200 Copenhagen, Denmark.
Department of Breast Surgery, Herlev Hospital, Copenhagen University Hospital, 2730 Herlev, Copenhagen, Denmark.
Laboratory for Translational Genetics, Department of Oncology, University of Leuven, 3000 Leuven, Belgium.
Vesalius Research Center (VRC), VIB, 3000 Leuven, Belgium.
Vesalius Research Center, University of Leuven, 3000 Leuven, Belgium.
Department of Oncology, University of Leuven, 3000 Leuven, Belgium.
Multidisciplinary Breast Center, Department of General Medical Oncology, University Hospitals Leuven, 3000 Leuven, Belgium.
Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, Minnesota 55905, USA.
Department of Health Sciences Research, Mayo Clinic, Rochester, Minnesota 55905, USA.
Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), 69120 Heidelberg, Germany.
Department of Cancer Epidemiology/Clinical Cancer Registry and Institute for Medical Biometrics and Epidemiology, University Clinic Hamburg-Eppendorf, 20246 Hamburg, Germany.
Department of Non-Communicable Disease Epidemiology, London School of Hygiene and Tropical Medicine, London WC1E 7HT, UK.
Department of Obstetrics and Gynecology, Helsinki University Central Hospital, Helsinki, FI-00029 HUS, Finland.
Department of Clinical Genetics, University of Helsinki, Helsinki University Central Hospital, Helsinki, FI-00029 HUS, Finland.
Department of Oncology, University of Helsinki, Helsinki University Central Hospital, Helsinki, FI-00029 HUS, Finland.
Human Genetics Division, Genome Institute of Singapore, Singapore 138672, Singapore.
Cancer Research Institute, Seoul National University College of Medicine, Seoul 110-799, Korea.
Department of Biomedical Sciences, Seoul National University Graduate School, Seoul 151-742, Korea.
Department of Preventive Medicine, Seoul National University College of Medicine, Seoul 110-799, Korea.
Department of Surgery, Seoul National University College of Medicine, Seoul 110-799, Korea.
Department of Preventive Medicine, Kyushu University Faculty of Medical Sciences, Fukuoka 812-8582, Japan.
Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi 464-8681, Japan.
Department of Breast Oncology, Aichi Cancer Center Hospital, Nagoya 484-8681, Japan.
Department of Pathology and Molecular Diagnostics, Aichi Cancer Center Hospital, Nagoya 484-8681, Japan.
Inserm (National Institute of Health and Medical Research), CESP (Center for Research in Epidemiology and Population Health), U1018, Environmental Epidemiology of Cancer, 94807 Villejuif, France.
University Paris-Sud, UMRS 1018, 94807 Villejuif, France.
Department of Obstetrics and Gynecology, University of Heidelberg, 69120 Heidelberg, Germany.
Molecular Epidemiology Group, German Cancer Research Center (DKFZ), 69120 Heidelberg, Germany.
National Center for Tumor Diseases, University of Heidelberg, 69120 Heidelberg, Germany.
Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles, California 90033, USA.
Centro de Investigación en Red de Enfermedades Raras (CIBERER), 46010 Valencia, Spain.
Human Genetics Group, Human Cancer Genetics Program, Spanish National Cancer Research Centre (CNIO), 28029 Madrid, Spain.
Servicio de Oncología Médica, Hospital Universitario La Paz, 28046 Madrid, Spain.
Servicio de Cirugía General y Especialidades, Hospital Monte Naranco, 33012 Oviedo, Spain.
Servicio de Anatomía Patológica, Hospital Monte Naranco, 33013 Oviedo, Spain.
Division of Epidemiology, Department of Medicine, Vanderbilt Epidemiology Center, Vanderbilt-Ingram Cancer Center, Vanderbilt University School of Medicine, Nashville, Tennessee 37203, USA.
Shanghai Center for Disease Control and Prevention, Shanghai 200336, China.
Department of Epidemiology, Shanghai Cancer Institute, Shanghai 200032, China.
CRUK/YCR Sheffield Cancer Research Centre, Department of Oncology, University of Sheffield, Sheffield S10 2RX, UK.
Academic Unit of Pathology, Department of Neuroscience, University of Sheffield, Sheffield S10 2HQ, UK.
Lunenfeld-Tanenbaum Research Institute of Mount Sinai Hospital, Toronto, Ontario, Canada M5G 1X5.
Department of Molecular Genetics, University of Toronto, Toronto, Ontario, Canada M5S 1A8.
Division of Epidemiology, Dalla Lana School of Public Health, University of Toronto, Toronto, Ontario, Canada M5T 3M7.
Prosserman Centre for Health Research, Lunenfeld-Tanenbaum Research Institute of Mount Sinai Hospital, Toronto, Ontario, Canada M5G 1X5.
Ontario Cancer Genetics Network, Lunenfeld-Tanenbaum Research Institute of Mount Sinai Hospital, Toronto, Ontario, Canada M5G 1X5.
Division of Cancer Studies, NIHR Comprehensive Biomedical Research Centre, Guy’s & St Thomas’ NHS Foundation Trust in partnership with King’s College London, London SE1 9RT, UK.
Wellcome Trust Centre for Human Genetics, Oxford Biomedical Research Centre, University of Oxford, Oxford OX3 7BN, UK.
Clinical Science Institute, University Hospital Galway, Galway, Ireland.
Peter MacCallum Cancer Center, Melbourne, Victoria 3002, Australia.
Department of Preventive Medicine, Keck School of Medicine, University of Southern California Norris Comprehensive Cancer Center, Los Angeles, California 90033, USA.
Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu, Hawaii 96813, USA.
Department of Molecular Medicine and Surgery, Karolinska Institutet, Stockholm SE-17177, Sweden.
Department of Oncology—Pathology, Karolinska Institutet, Stockholm SE-17177, Sweden.
Breast Cancer Research Unit, University Malaya Cancer Research Institute, University Malaya Medical Centre, 59100 Kuala Lumpur, Malaysia.
Cancer Research Initiatives Foundation, Sime Darby Medical Centre, Subang Jaya 47500 Selangor, Malaysia.
Singapore Eye Research Institute, National University of Singapore, Singapore 168751, Singapore.
Department of Medical Oncology, Erasmus MC Cancer Institute, 3008 AE Rotterdam, The Netherlands.
Department of Clinical Genetics, Erasmus University Medical Center, 3000 CA Rotterdam, The Netherlands.
Department of Pathology, Erasmus University Medical Center, 3075 EA Rotterdam, The Netherlands.
Centre for Molecular, Environmental, Genetic and Analytical Epidemiology, Melbourne School of Population Health, University of Melbourne, Melbourne, Victoria 3010, Australia.
Department of Pathology, The University of Melbourne, Melbourne, Victoria 3010, Australia.
College of Public Health, China Medical University, Taichung 40402, Taiwan, China.
Institute of Biomedical Sciences, Academia Sinica, Taipei 115, Taiwan, China.
Taiwan Biobank, Institute of Biomedical Sciences, Academia Sinica, Taipei 115, Taiwan, China.
Department of Surgery, Tri-Service General Hospital, Taipei 114, Taiwan, China.
Department of Surgery, Changhua Christian Hospital, Changhua City 500, Taiwan, China.
Institute of Clinical Medicine, University of Oslo (UiO), 0318 Oslo, Norway.
Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Victoria 3004, Australia.
Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Melbourne, Victoria 3010, Australia.
Cancer Epidemiology Centre, The Cancer Council Victoria, Melbourne, Victoria 3053, Australia.
Anatomical Pathology, The Alfred Hospital, Melbourne, Victoria 3004, Australia.
Division of Health Sciences, Warwick Medical School, Warwick University, Coventry CV4 7AL, UK.
Institute of Population Health, University of Manchester, Manchester M13 9PL, UK.
Ministry of Public Health, Bangkok 10400, Thailand.
Department of Surgery, Yong Loo Lin School of Medicine, National University of Singapore and National University Health System, Singapore 119228, Singapore.
Saw Swee Hock School of Public Health, National University of Singapore and National University Health System, Singapore 117597, Singapore.
Division of General Surgery, National University Health System, Singapore 117597, Singapore.
Department of Statistics and Applied Probability, National University of Singapore, Singapore 117546, Singapore.
Division of Hematology and Oncology, Department of Medicine, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, California 90095, USA.
Department of Gynecology and Obstetrics, University Breast Center Franconia, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-EMN, 91054 Erlangen, Germany.
Institute of Human Genetics, University Hospital Erlangen, Friedrich Alexander University Erlangen-Nuremberg, 91054 Erlangen, Germany.
Division of Clinical Epidemiology and Aging Research, German Cancer Research Center (DKFZ), 69120 Heidelberg, Germany.
German Cancer Consortium (DKTK), 69120 Heidelberg, Germany.
Saarland Cancer Registry, 66119 Saarbrücken, Germany.
Division of Breast Cancer Research, Institute of Cancer Research, London SM2 5NG, UK.
Division of Genetics and Epidemiology, Institute of Cancer Research, London SM2 5NG, UK.
Breakthrough Breast Cancer Research Centre, Division of Breast Cancer Research, The Institute of Cancer Research, London SW3 6JB, UK.
Division of Genetics and Epidemiology, Institute of Cancer Research, Sutton, Surrey SM2 5NG, UK.
Division of Cancer Epidemiology and Genetics, National Cancer Institute, Rockville, Maryland 20850, USA.
Department of Cancer Epidemiology and Prevention, M. Sklodowska-Curie Memorial Cancer Center and Institute of Oncology, 02-781 Warsaw, Poland.
Cancer Genomics Laboratory, Centre Hospitalier Universitaire de Québec Research Center, Laval University, Quebec, Canada G1V 4G2.
Department of Medicine, McGill University, Montreal, Quebec, Canada H3G 2M1.
Division of Clinical Epidemiology, McGill University Health Centre, Royal Victoria Hospital, Montreal, Quebec, Canada H3A 1A8.
Département de médecine sociale et préventive, Département de santé environnementale et santé au travail, Université de Montréal, Montreal, Quebec, Canada H3T 1A8.
Laboratory of Cancer Genetics and Tumor Biology, Department of Clinical Chemistry and Biocenter Oulu, NordLab Oulu/Oulu University Hospital, University of Oulu, FI-90220 Oulu, Finland.
Department of Oncology, Oulu University Hospital, University of Oulu, FI-90220 Oulu, Finland.
Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, 70376 Stuttgart, Germany.
University of Tübingen, 72074 Tübingen, Germany.
German Cancer Consortium (DKTK) and German Cancer Research Center (DKFZ), 69120 Heidelberg, Germany.
Institute for Prevention and Occupational Medicine of the German Social Accident Insurance, Institute of the Ruhr University Bochum (IPA), 44789 Bochum, Germany.
Department of Internal Medicine, Evangelische Kliniken Bonn gGmbH, Johanniter Krankenhaus, 53113 Bonn, Germany.
Unit of Molecular Bases of Genetic Risk and Genetic Testing, Department of Preventive and Predictive Medicine, Fondazione IRCCS Istituto Nazionale dei Tumori (INT), 20133 Milan, Italy.
IFOM, Fondazione Istituto FIRC di Oncologia Molecolare, 20139 Milan, Italy.
Division of Cancer Prevention and Genetics, Istituto Europeo di Oncologia (IEO), 20141 Milan, Italy.
Cogentech Cancer Genetic Test Laboratory, 20133 Milan, Italy.
Department of Obstetrics and Gynaecology, Hannover Medical School, 30625 Hannover, Germany.
Department of Radiation Oncology, Hannover Medical School, 30625 Hannover, Germany.
Cancer Center of Eastern Finland, University of Eastern Finland, FI-70211 Kuopio, Finland.
Imaging Center, Department of Clinical Pathology, Kuopio University Hospital, FI-70210 Kuopio, Finland.
School of Medicine, Institute of Clinical Medicine, Oncology, University of Eastern Finland, FI-70211 Kuopio, Finland.
Department of Human Genetics & Department of Pathology, Leiden University Medical Center, 2300 RC Leiden, The Netherlands.
Department of Surgical Oncology, Leiden University Medical Center, 2300 RC Leiden, The Netherlands.
Family Cancer Clinic, Department of Medical Oncology, Erasmus MC-Daniel den Hoed Cancer Centre, 3075 EA Rotterdam, The Netherlands.
Department of Genetics and Pathology, Pomeranian Medical University, 70-115 Szczecin, Poland.
Department of Molecular Virology, Immunology and Medical Genetics, Comprehensive Cancer Center, The Ohio State University, Columbus, Ohio 43210, USA.
Roswell Park Cancer Institute, Buffalo, New York 14263, USA.
Molecular Diagnostics Laboratory, IRRP, National Centre for Scientific Research ‘Demokritos’, Aghia Paraskevi Attikis, Athens 15310, Greece.
National Cancer Institute, Bangkok 10400, Thailand.
International Agency for Research on Cancer, Lyon, Cedex 08, France.
Molecular Genetics of Breast Cancer, German Cancer Research Center (DKFZ), 69120 Heidelberg, Germany.
Institute of Human Genetics, Pontificia University Javeriana, Bogota, DC 11001000, Colombia.
Department of Epidemiology, University of California Irvine, Irvine, California 92697, USA.
Department of Population Sciences, Beckman Research Institute of City of Hope, Duarte, California 92697, USA.
Human Genotyping-CEGEN Unit, Human Cancer Genetics Program, Spanish National Cancer Research Centre (CNIO), 28029 Madrid, Spain.
Centre d’innovation Génome Québec et Université McGill, Montréal, Quebec, Canada H3A OG1.
McGill University, Montreal, Quebec, Canada H3A OG1.
Cancer Research UK, Cambridge Institute, University of Cambridge, Robinson Way, Cambridge CB2 0RE, UK.
Department of Medical Biophysics, University of Toronto, Toronto, Ontario, Canada M5G 1L7.
Department of Clinical Molecular Biology (EpiGen), University of Oslo (UiO), 0450 Oslo, Norway.
Department of Population Health, QIMR Berghofer Medical Research Institute, Brisbane, Queensland 4029, Australia.
Center for Cancer Research, University of Sydney at Westmead Millennium Institute, Sydney, New South Wales 2006, Australia.
Department of Gynaecological Oncology, Westmead Hospital, Sydney, New South Wales 2145, Australia.
A full list of authors and their affiliations appears below.
Author contributions
Manuscript writing group: M.G., S.L.E. and A.M.D. M.G., E.D. and A.M.D. helped in locus SNP selection. iCOGS genotyping, calling and QC was performed by S.E.B., S.F.N., A.G.-N., M.R.A., D.H., J.B., D.C.T., D.V., F.B., F.R., S.A., M.J.M., C.L., C.B., D.C., J.C., J.D., C.S.H., J.S., A.M.D., G.C.-T. and D.F.E.; imputation by K.M. and D.F.E.; statistical analyses and programming by K.M., M.G., D.B., E.D., J.T., S.K. and D.F.E.; functional analysis and bioinformatics by S.L.E., J.D.F., K.D., R.C.-S., M.L., J.B., K.M.H., S.K., S.N., V.N.K., J.C., I.D.S., J.A.B., P.J.B., M.A.B., M.O’R., T.C., A.-T.M., B.A.J.P., S.K., H.N., K.A. and P.H.; COGS coordination by P.H., D.F.E., J.B. and A.M.D.; BCAC coordination by D.F.E., G.C.-T. and P.D.P.; and BCAC data management by M.K.B. and Q.W. All other authors provided participant samples and phenotype information, and read and approved the manuscript.
Supplementary Information accompanies this paper at http://www.nature.com/naturecommunications
Competing financial interests: The authors declare no competing financial interests.
References
- 1.Stacey SN, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39:865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 2.Milne RL, et al. Risk of estrogen receptor-positive and -negative breast cancer and single-nucleotide polymorphism 2q35-rs13387042. J Natl Cancer Inst. 2009;101:1012–1018. doi: 10.1093/jnci/djp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kong SL, Li G, Loh SL, Sung WK, Liu ET. Cellular reprogramming by the conjoint action of ERalpha, FOXA1, and GATA3 to a ligand-inducible growth state. Mol Syst Biol. 2011;7:526. doi: 10.1038/msb.2011.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cowper-Sal lari R, et al. Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat Genet. 2012;44:1191–1198. doi: 10.1038/ng.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cirillo LA, et al. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 6.Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet. 2011;43:27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corradin O, et al. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res. 2014;24:1–13. doi: 10.1101/gr.164079.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edwards SL, Beesley J, French JD, Dunning AM. Beyond GWASs: illuminating the dark road from association to function. Am J Hum Genet. 2013;93:779–797. doi: 10.1016/j.ajhg.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michailidou K, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45:353–361. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghoussaini M, et al. Multiple loci with different cancer specificities within the 8q24 gene desert. J Natl Cancer Inst. 2008;100:962–966. doi: 10.1093/jnci/djn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.French JD, et al. Functional variants at the 11q13 risk locus for breast cancer regulate cyclin D1 expression through long-range enhancers. Am J Hum Genet. 2013;92:489–503. doi: 10.1016/j.ajhg.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bojesen SE, et al. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat Genet. 2013;45:371–384. doi: 10.1038/ng.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyer KB, et al. Fine-scale mapping of the FGFR2 breast cancer risk locus: putative functional variants differentially bind FOXA1 and E2F1. Am J Hum Genet. 2013;93:1046–1060. doi: 10.1016/j.ajhg.2013.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beattie J, Allan GJ, Lochrie JD, Flint DJ. Insulin-like growth factor-binding protein-5 (IGFBP-5): a critical member of the IGF axis. Biochem J. 2006;395:1–19. doi: 10.1042/BJ20060086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gullu G, Karabulut S, Akkiprik M. Functional roles and clinical values of insulin-like growth factor-binding protein-5 in different types of cancers. Chin J Cancer. 2012;31:266–280. doi: 10.5732/cjc.011.10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sureshbabu A, et al. IGFBP5 induces cell adhesion, increases cell survival and inhibits cell migration in MCF-7 human breast cancer cells. J Cell Sci. 2012;125:1693–1705. doi: 10.1242/jcs.092882. [DOI] [PubMed] [Google Scholar]
- 17.Tripathi G, et al. IGF-independent effects of insulin-like growth factor binding protein-5 (Igfbp5) in vivo. FASEB J. 2009;23:2616–2626. doi: 10.1096/fj.08-114124. [DOI] [PubMed] [Google Scholar]
- 18.Butt AJ, Dickson KA, McDougall F, Baxter RC. Insulin-like growth factor-binding protein-5 inhibits the growth of human breast cancer cells in vitro and in vivo. J Biol Chem. 2003;278:29676–29685. doi: 10.1074/jbc.M301965200. [DOI] [PubMed] [Google Scholar]
- 19.Tonner E, et al. Insulin-like growth factor binding protein-5 (IGFBP-5) induces premature cell death in the mammary glands of transgenic mice. Development. 2002;129:4547–4557. doi: 10.1242/dev.129.19.4547. [DOI] [PubMed] [Google Scholar]
- 20.Perks CM, Bowen S, Gill ZP, Newcomb PV, Holly JM. Differential IGF-independent effects of insulin-like growth factor binding proteins (1–6) on apoptosis of breast epithelial cells. J Cell Biochem. 1999;75:652–664. doi: 10.1002/(sici)1097-4644(19991215)75:4<652::aid-jcb11>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 21.Huynh H. In vivo regulation of the insulin-like growth factor system of mitogens by human chorionic gonadotropin. Int J Oncol. 1998;13:571–575. doi: 10.3892/ijo.13.3.571. [DOI] [PubMed] [Google Scholar]
- 22.Pekonen F, Nyman T, Ilvesmaki V, Partanen S. Insulin-like growth factor binding proteins in human breast cancer tissue. Cancer Res. 1992;52:5204–5207. [PubMed] [Google Scholar]
- 23.Bruchim I, Werner H. Targeting IGF-1 signaling pathways in gynecologic malignancies. Expert Opin Ther Targets. 2013;17:307–320. doi: 10.1517/14728222.2013.749863. [DOI] [PubMed] [Google Scholar]
- 24.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prall OW, Sarcevic B, Musgrove EA, Watts CK, Sutherland RL. Estrogen-induced activation of Cdk4 and Cdk2 during G1-S phase progression is accompanied by increased cyclin D1 expression and decreased cyclin-dependent kinase inhibitor association with cyclin E-Cdk2. J Biol Chem. 1997;272:10882–10894. doi: 10.1074/jbc.272.16.10882. [DOI] [PubMed] [Google Scholar]
- 26.Li B, Kadura I, Fu DJ, Watson DE. Genotyping with TaqMAMA. Genomics. 2004;83:311–320. doi: 10.1016/j.ygeno.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 27.Curtis C, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.