Abstract
Type I interferon (IFN) production is an important host immune response against viral and bacterial infections. However, little is known about the ligands and corresponding host receptors that trigger type I IFN production during bacterial infections. We used a model intracellular pathogen, Francisella novicida to begin characterizing the type I IFN response to bacterial pathogens. F. novicida replicates in the cytosol of host cells and elicits a robust type I IFN response that is largely TLR-independent, but is dependent on the adapter molecule STING, suggesting that the type I IFN stimulus during F. novicida infection is cytosolic. In this study, we report that the cytosolic DNA sensors, cGAS and Ifi204, are both required for the STING-dependent type I IFN response to F. novicida infection in both primary and immortalized murine macrophages. We created cGAS, Ifi204 and Sting functional knockouts in RAW264.7 macrophages and demonstrated that cGAS and Ifi204 cooperate to sense dsDNA and activate the STING-dependent type I IFN pathway. Additionally, we showed that dsDNA from F. novicida is an important type I IFN stimulating ligand. One outcome of cGAS-STING signaling is the activation of the AIM2 inflammasome in response to F. novicida infection. While the AIM2 inflammasome is beneficial to the host during F. novicida infection, type I IFN signaling by STING and IRF3 is detrimental to the host during F. novicida infection. Collectively, our studies indicate that cGAS and Ifi204 cooperate to sense cytosolic dsDNA and F. novicida infection to produce a strong type I IFN response.
Introduction
The innate immune system plays a key role in the early recognition and elimination of invading pathogens. Many recognition systems are in place to detect conserved pathogen-associated molecular patterns (PAMPs), such as nucleic acids and cell wall components (1). Upon PAMP recognition, immune cells initiate signal transduction cascades that trigger a type I interferon (IFN) transcriptional response, which can prompt a broad range of additional responses to infection, including caspase-1-mediated cell death and pro-inflammatory cytokine release (2, 3). While, it is appreciated that many bacterial species, including Francisella tularensis, Listeria monocytogenes, Salmonella typhimurium, Mycobacterium tuberculosis, and Chlamydia trachomatis can initiate a type I IFN response, the mechanism of host recognition remains largely undetermined (4, 5).
Microbes can stimulate the type I IFN response either by activating members of the Toll-like receptor (TLR) family that signal through the endosomal adapter TRIF or by activating cytosolic receptors that activate a type I IFN transcriptional response (6, 7). One important class of cytosolic receptors includes DNA sensors that activate the endoplasmic reticulum membrane protein stimulator of IFN genes (STING, also known as MITA, MPSY, ERIS and TMEM173). STING activation leads to the recruitment of the kinase TBK1, which phophorylates IRF-3, a transcription factor required for the induction of IFN-β1 (8, 9). Recent studies have demonstrated that the host-derived second messenger cyclic GMP-AMP (cGAMP) synthesized from the DNA sensor cyclic GMP-AMP synthase (cGAS) or bacterial-derived cyclic-dinucleotides can directly activate STING (8, 10, 11). In addition to cGAS, a number of cytosolic sensors were identified to bind DNA and trigger the type I IFN response. They include RNA polymerase III, DNA-dependent activator of IFN-regulatory factors (DAI), Lrrfip1, Ifi204 (human: IFI16), Mre11, DNA-dependent protein kinase (DNA-PK), and Ddx41 (7, 12–17). Although many cytosolic DNA sensors have been identified, the role of these sensors during bacterial infections remains unclear.
Francisella novicida is a model organism used to study the cytosolic responses of immune cells to intracellular bacteria (18). Upon phagocytosis by host macrophages, F. novicida rapidly escapes the Francisella containing vacuole (FCV) and replicates in the cytosol (19). Cytosolic F. novicida trigger a pro-inflammatory response characterized by the production of type I IFNs followed by pyroptotic cell death (20, 21). The type I IFN response to F. novicida infection is largely TLR-independent, but STING-dependent, making F. novicida an ideal organism to study the cytosolic responses in macrophages to an intracellular bacterial pathogen (20, 22, 23). To date, the Francisella ligand(s) and corresponding host sensor(s) have not been identified. Ultimately, production of type I IFNs increases protein levels of the DNA sensor, absent in melanoma 2 (AIM2), a protein that binds cytosolic DNA and engages the adaptor protein ASC to form a caspase-1 inflammasome complex (24–27). An active AIM2 inflammasome leads to the secretion of pro-inflammatory cytokines (IL-18 and IL-1β) and caspase-1-dependent cell death (20), which are required for a protective innate immune response in mice (23).
In this study, we identified cGAS and Ifi204 as two important host factors involved in type I IFN signaling in response to F. novicida infection in both BMMs (bone-marrow macrophages) and RAW264.7 macrophages. Using targeted knockouts and complementation vectors, we demonstrated that cGAS and Ifi204 both contribute to STING-dependent type I IFN response to high concentrations of cytosolic dsDNA. Additionally, we show that dsDNA is the primary molecule found in F. novicida lysates that stimulate cGAS- and Ifi204-dependent type I IFN production. Taken together, our results suggest that cGAS and Ifi204 sense dsDNA during a F. novicida infection to elicit STING activation and the type I IFN response.
Materials and Methods
Bacteria, plasmids, primers and generation of RAW264.7 knockout cell lines
Bacterial strains used in this study include wild-type F. novicida U112, ΔFPI (28), and ΔfopA (29) and wild-type L. monocytogenes strain 10403S. The cDNA from cGAS (Clone ID: 40130956, Thermo Scientific) and Ifi204 (Clone ID: 4018506, Thermo Scientific) were amplified with primers listed on Table S1 and cloned into MSCV2.2 retroviral expression construct upstream of an internal ribosome entry site (IRES)-green fluorescent protein (GFP). MSCV2.2 Sting (30) and MSCV2.2 Sting R231A(31) were kindly provided by R. Vance. pCherry and hCas9(32) were kindly provided by J. Carette. pCherry was created by replacing DsRed with mCherry RFP (Pubmed ID: 15558047) in pDsRed C1 (Clontech). cGAS, Ifi204 and Sting knockouts were generated in RAW264.7 cells using the CRISPR/Cas9 system (32). Target gRNA expression constructs were generated from 455bp gene blocks (IDT) containing target RNA sequences listed in Table S2 cloned into pCR-Blunt cloning vector (Invitrogen). 1 X 106 RAW264.7 macrophages were transfected with 2.5 µg hCas9, 2.5 µg Target gRNA and 0.5 µg pCherry in a 6-well tissue culture-treated plate with Targefect-RAW (Targeting Systems). Two days post-transfection, macrophages were single-cell sorted into 96-well tissue culture-treated plates and allowed to grow up ~ 2 weeks. Genomic DNA from macrophage colonies was extracted using QIAamp DNA mini kit (Qiagen). The targeted DNA sequence was amplified with primers (Table S1) flanking the mutant target site and sequenced (Elim Biopharm). qRT-PCR primers are listed in Table S1.
Cell culture, infections and immunofluorescence
Bone marrow-derived macrophages (BMMs) were isolated, differentiated, and cultured as previously described (33). Aim2−/− and ASC−/− BMMs Femurs from C57BL/6-Tmem173gt (Stinggt)(30) and C57BL/6-cGAS−/− (34) were kindly provided by R. Vance and H. Virgin, respectively. Infections with F. novicida were performed as previously described (20). For infections with L. monocytogenes, log-phase bacteria grown in Brain-Heart Infusion (BHI) broth at 37°C, shaking, were washed twice with PBS and infected similarly to F. novicida.
RAW264.7 macrophages and constructed KOs were cultured in DMEM 10% fetal bovine serum (FBS). RAW264.7 macrophages were seeded at a density of 1 X 105 macrophages per well of a 96-well tissue culture treated plate or 2.5 X 105 macrophages per well of a 24-well tissue culture treated plate and allowed to adhere overnight at 37°C, 5% CO2. Infections were conducted as described for BMMs (20). Immunofluorescence microscopy and western blots were performed as previously described (23, 35). A minimum of 50 bacterial cells were quantified for LAMP-1 colocalization. Antibodies used include: caspase-1 p10 (sc514, Santa Cruz Biotech); β-actin (M-2, Santa Cruz Biotech); anti-F. novicida (Monack Laboratory) and anti-LAMP-1 (1D4B, Abcam).
Retroviral transductions and siRNA knockdown
Retroviral constructs were transduced into RAW264.7 macrophages using vesicular stomatitis pseudotyped virus packaged in 293FT cells and sorted by FACS to select for GFP+ macrophages. Gene expression in BMMs was knocked-down using siRNA and TransIT-siQUEST transfection reagent according to manufacture’s recommendations (Mirus). siRNAs used were: Non-targeting (NT) D-001206-13, cGAS D-0555608-01, Ddx41 m-052130-00, lrrfip1 m-047145-01, Ifi204 m-044641-01, RIG-I m-0655328-01 (Dharmacon, GE Healthcare).
Cytotoxicity, cytokine measurement and cell stimulations
Secreted type I IFNs were measured using the ISRE-L929 reporter cells as previously described (29, 36). IL-1β was measured by ELISA (R&D systems). Cytotoxicity was measured via lactate dehydrogenase release using CytoTox 96 (Non-Radioactive Cytotoxicity Assay, Promega). When specified, RAW264.7 macrophages were stimulated with100 ng/mL lipopolysaccharide (LPS). dsDNA (pCherry) and poly(I:C) (Invivogen) were transfected into RAW264.7 macrophages using Targefect-Raw (Targeting Systems). c-di-GMP (Invivogen) was transfected into RAW264.7 macrophages using Lipofectamine 2000 (Life Technologies) as previously described (22).
F. novicida lysates were prepared similarly to previously described protocols (37). Briefly, a 3 mL overnight culture was pelleted and resuspended in 1 mL PBS supplemented with 1 mg/mL lysozyme. The cells were lysed using multiple freeze-thaw cycles. Remaining debris was removed by centrifugation and cleared extracts were adjusted to contain 2mM MgCl2, 50mM KCl and 20mM Tris, pH 8.0. 100 µl aliquots were treated with 100 U/mL DNase I (Invitrogen) or 100 µg/mL RNase A (Qiagen) for 45 min at 37°C. EDTA was added to 2.5 mM and samples were heated to 70°C for 10 min. DNase I was heat inactivated prior to treating the lysate by heating the enzyme to 70°C for 20 min. Extracts were cleared by centrifugation and 5 µl of lysate was complexed with 1 µl targefect-RAW and transfected into RAW264.7 macrophages.
Mouse infections
Mice between seven- to nine- weeks were used for in vivo experiments comparing C57Bl/6J mice and C57Bl/6J-Tmem173gt/J (Stinggt/gt) (The Jackson Laboratory). David Schneider, with permission from Tadatsugu Taniguchi, generously provided 9 C57Bl/6 and ten IRF3−/− mice (38, 39) eight- to 16-wks-old. Mice were infected by subcutaneous (s.c.) injection with a target dose of 105 CFU of F. novicida strain U112. After 72 h, spleens and livers were harvested, weighed and ground in PBS for CFU determination. For survival experiments mice were monitored twice daily for 15 days for survival.
Results
F. novicida induces a type I IFN response in macrophages that is dependent on cGAS, Ifi204 and STING
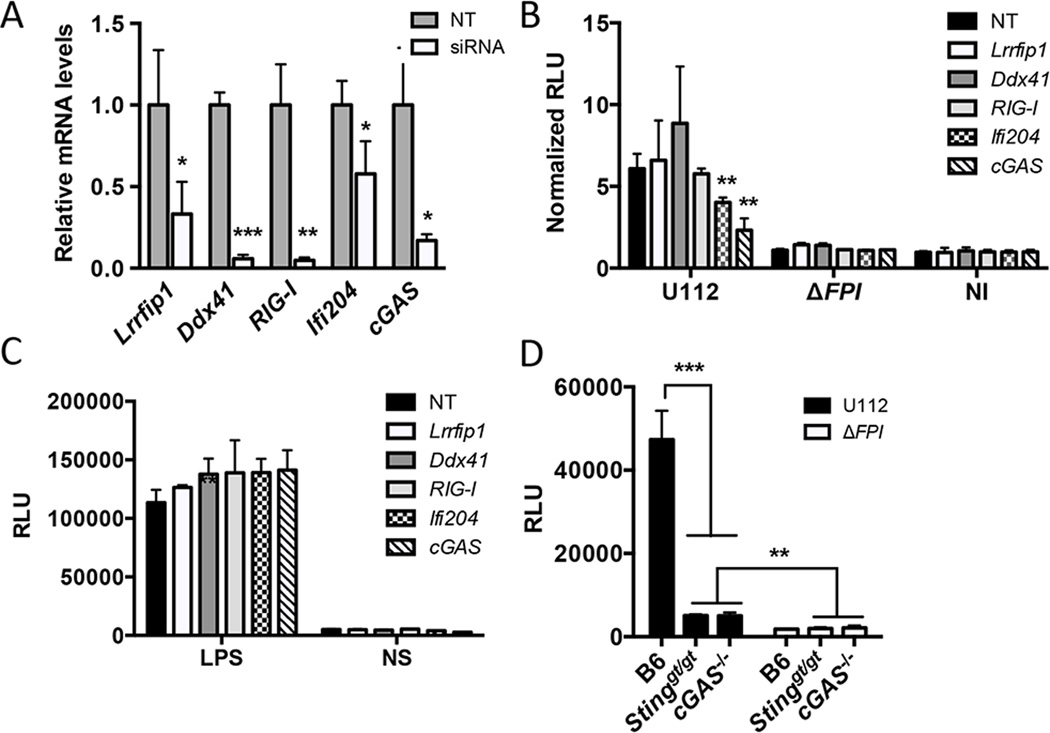
Previous studies showed that F. novicida releases dsDNA into the macrophage cytosol during infection and elicits a type I IFN response that is dependent on STING (22, 23, 29). We hypothesized that a DNA sensor is necessary to mediate the STING-dependent type I IFN response. To test this notion, we knocked down the expression of known DNA sensors that are important for triggering a type I IFN response (10, 13, 14, 17, 40). Small interfering RNAs (siRNAs) were generated for Lrrfip1, RIG-I, Ddx41, Ifi204 and cGAS and transfected into BMMs. Gene expression levels for each siRNA-targeted gene were reduced compared to the non-targeting (NT) control after 36 h (Fig. 1A). Upon F. novicida infection, siRNA knockdown of cGAS and Ifi204 resulted in reduced type I IFN production, while siRNA knockdown of Lrrfip1, RIG-I and Ddx41 did not influence the type I IFN response (Fig. 1B). The type I IFN response was measured using the L929-ISRE reporter cell line (29, 36). A representative standard curve for purified IFN- β and relative luciferase units (RLUs) is shown (Fig. S1A). To confirm that these siRNAs were not indirectly affecting other type I IFN pathways, we stimulated the siRNA knockdowns with LPS and observed similar type I IFN responses across all knockdowns (Fig. 1C). Additionally, we isolated BMMs from the recently described cGAS−/− mice and examined the type I IFN response to F. novicida infection. As expected, the type I IFN response to F. novicida infection was dampened in Sting-deficient (Stinggt/gt) and cGAS−/− BMMs compared to WT B6 BMMs (Fig. 1D and Fig. S1C). Collectively, these results indicate that cGAS and Ifi204 contribute to the type I IFN response to cytosolic F. novicida in BMMs.
Figure 1. cGAS and Ifi204 are required for type I IFN production in response to cytosolic F. novicida in BMMs.
siRNA targeting known cytosolic sensor genes or a non-targeting (NT) control was transfected into BMMs for 36 h (A) qRT-PCR measured mRNA levels for each targeted gene in uninfected BMMs. Gene expression was normalized to GAPDH and the NT control. Type I IFN levels were measured (B) 9 h post-infection with the indicated F. novicida strain at an MOI of 10 or (C) 4 h post-stimulation with LPS. Results are presented as Relative Luciferase Units (RLUs). Data presented as “Normalized RLUs” are type I IFN levels normalized to the un-infected, NT control. (D) Type I IFN levels were measured from un-stimulated primary C57Bl/6 (WT), Stinggt/gt and cGAS−/− BMMs infected with F. novicida strains at an MOI of 10 for 12 hours. *p < 0.05, **p < 0.01, ***p < 0.001. Graphs show the mean ± standard deviation (SD) of triplicate wells and are representative of three independent experiments.
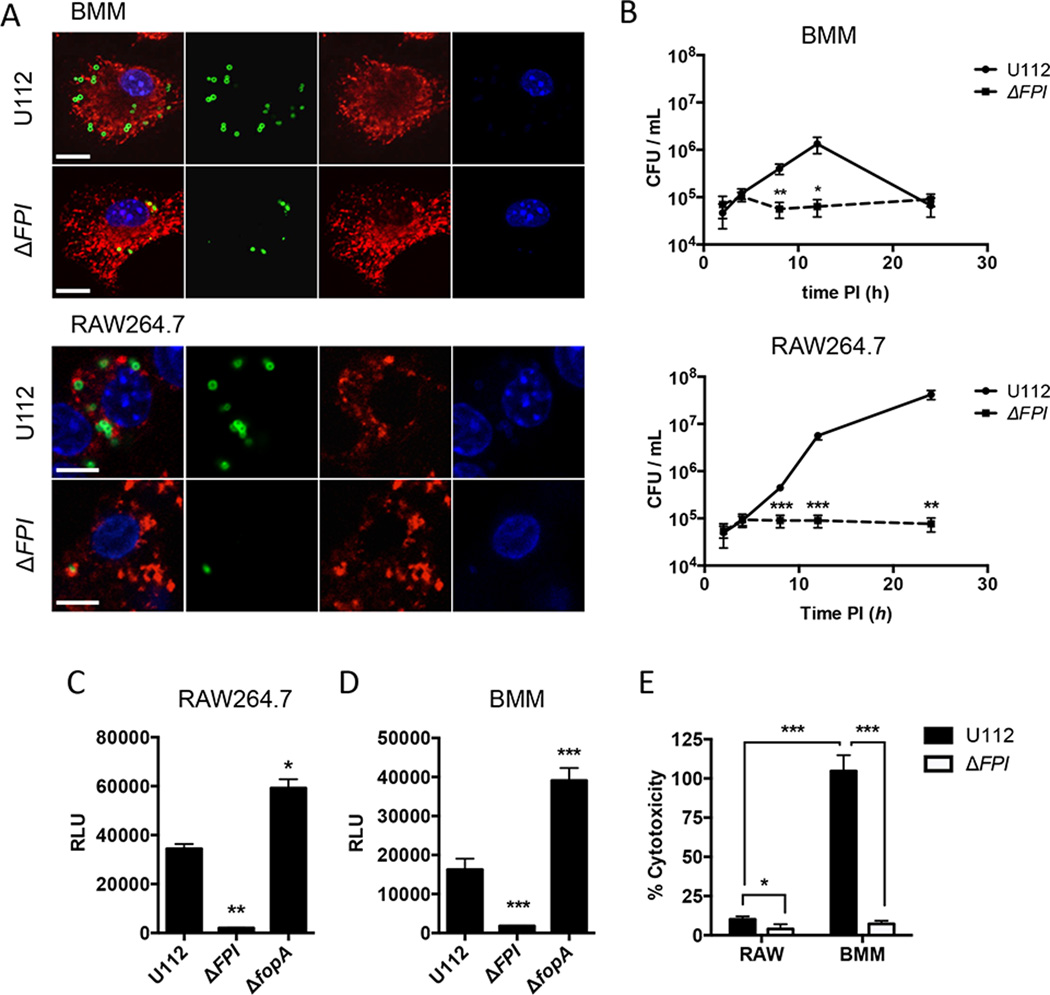
To our knowledge, Ifi204-deficient mice are not available. Additionally, the siRNA knockdown efficiency of Ifi204 was only moderately effective, reducing Ifi204 mRNA levels ~50% (Fig. 1A). Despite this modest siRNA knockdown, type I IFN production was significantly reduced upon infection with F. novicida compared to the NT control (Fig. 1B). To further dissect the roles of Ifi204 and cGAS in the generation of type I IFNs during F. novicida infection of macrophages, we created gene knockouts in immortalized RAW264.7 murine macrophages using the newly described CRISPR/Cas9 system (32). However, we first needed to determine whether F. novicida infection in RAW264.7 macrophages induces type I IFN responses that are similar to infections of BMMs. It has been previously published that Francisella escapes the initial phagosome and replicates in the cytosol of BMMs (41). We confirmed that wild-type F. novicida quickly and efficiently escaped the FCV and entered the cytosol of both C57Bl/6 BMMs and RAW264.7 macrophages. At 8 h post-infection only 15.2% of wild-type F. novicida in BMMs and 14% of wild-type F. novicida in RAW264.7 macrophages co-localized with the lysosomal marker LAMP-1 (Fig. 2A), These levels are similar to previous studies (41). Moreover, F. novicida replicated to high levels in the cytosol of RAW264.7 macrophages, similar to the first 12 h of infected BMMs (Fig. 2B). In contrast, a F. novicida strain lacking the Francisella pathogenicity island (FPI), a predicted type 6 secretion system that is important for escaping the phagosome, co-localized with LAMP-1 at a higher percentage (78% in BMMs and 61% in RAW264.7 macrophages) and did not replicate (Fig. 2A, 2B). The FPI mutant also failed to stimulate type I IFNs in BMMs (Fig. 2D) and dramatically reduced type I IFN stimulation in RAW264.7 macrophages compared to the parental strain (Fig 2C). Additionally, a F. novicida strain lacking FopA, a membrane protein important for maintaining cell wall stability, elicited a higher type I IFN response compared to wild-type F. novicida in both BMMs and RAW264.7 macrophages (Fig. 2C and Fig. 2D)(29). One notable difference was that the type I IFN response pattern in RAW264.7 macrophages was delayed compared to BMMs (Fig. S1B). We measured type I IFN levels at 24 h post-infection of RAW264.7 macrophages and 12 h post-infection of BMMs (Fig. 2C and Fig. 2D). We, and others, have shown that. F. novicida infection of BMMs results in cell death, which requires AIM2, ASC and caspase-1 (23, 24). In contrast, F. novicida infection of RAW264.7 macrophages did not result in high levels of cell death (Fig. 2E), which is consistent with the observation that this cell line does not express ASC (42, 43). Moreover, the induction of cell death in BMMs mirrored a reduction in bacterial CFUs in our intracellular replication assays likely due to bacterial exposure to the extracellular antibiotic gentamicin (Fig. 2B). These results indicate that RAW264.7 macrophages are suitable to study mechanisms of type I IFN production during F. novicida infections.
Figure 2. The type I IFN response to F. novicida is similar in BMM and RAW264.7 macrophages.
BMMs and RAW264.7 macrophages were infected with the indicated F. novicida strains at an MOI of 10. (A) Immunofluorescence microscopy of BMMs and RAW264.7 macrophages stained for F. novicida (green), LAMP-1 (red) and DAPI (blue) 8 hours post-infection. Scale bars, 10 µm. (B) Intracellular survival was assessed by CFU plating. Type I IFN levels from (C) RAW264.7 macrophages or (D) C57Bl/6 BMMs were measured 24 h and 12 h post-infection, respectively. (E) Cytotoxicity was determined by measuring LDH release 24 h post-infection. *p < 0.05, **p < 0.01, ***p < 0.001. Graphs show the mean ± standard deviation (SD) of triplicate wells and are representative of three independent experiments.
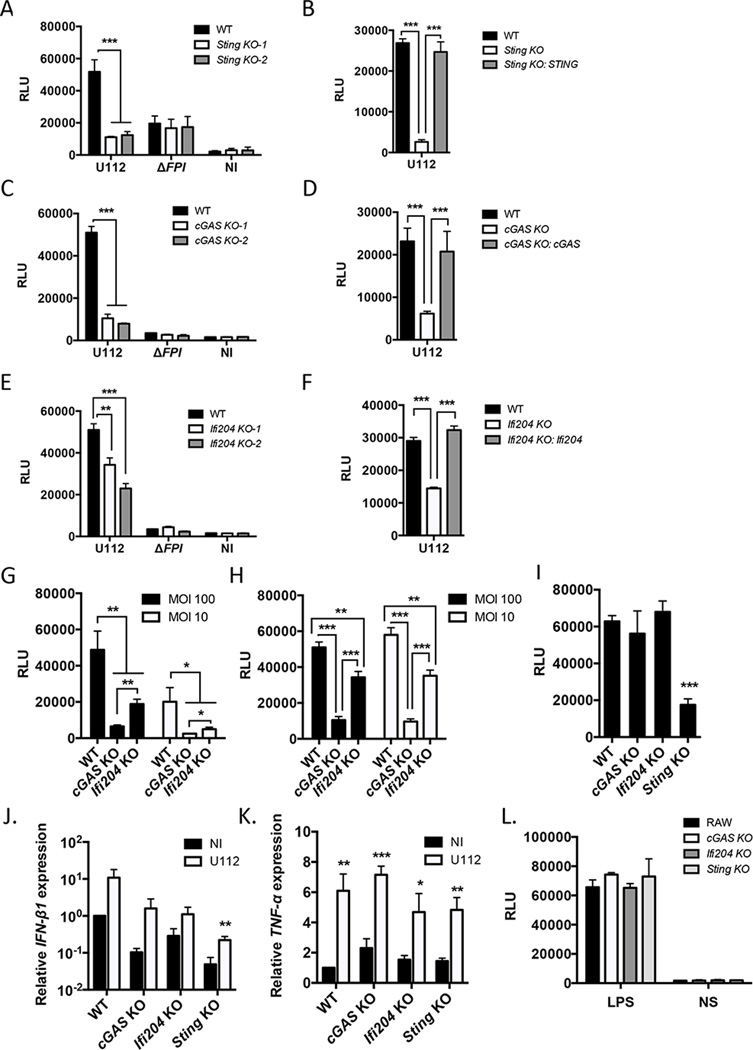
To determine if cGAS, Ifi204 and STING are important factors in mediating the type I IFN response to F. novicida in RAW264.7 macrophages, we created out-of-frame biallelic indel mutations in each gene using the CRISPR/Cas9 genome editing system, resulting in functional knockouts (32). Knockouts were verified by DNA sequencing (Table S2) and two clones were chosen for each gene for further study. As a proof of principle, F. novicida infection of Sting knockout (KO) RAW264.7 macrophages did not produce type I IFNs (Fig. 3A and Fig. S1D), similar to infections with Sting-deficient BMMs (Fig. 1D)(22, 23). To verify that the altered type I IFN response was not due to off target effects of the CRISPR/Cas9 system, we stably expressed STING in Sting KO cells and complemented the type I IFN response to F. novicida infection (Fig. 3B). cGAS KO macrophages and Ifi204 KO macrophages infected with wild-type F. novicida strain U112 produced significantly lower amounts of type I IFN compared to the RAW264.7 parental genotype (Fig 3C, Fig 3E and Fig. S1D). Stable expression of cGAS in cGAS KO macrophages complemented the type I IFN response to F. novicida infection, as did stable expression of Ifi204 in Ifi204 KO macrophages (Fig. 3D and Fig. 3F, respectively). To further evaluate the contribution of cGAS and Ifi204 to the type I IFN response during a F. novicida infection, we infected the knockout cell lines with low and high MOIs and examined two time points: 12 h post-infection, an early time point, and 24 h post-infection. Similar to the siRNA knockdown experiments (Fig. 1B), cGAS-deficiency resulted in a more pronounced reduction in type I IFN response to F. novicida infection compared to Ifi204 KO cells (Fig. 3G and Fig. 3H).
Figure 3. The type I IFN response to cytosolic F. novicida requires cGAS, Ifi204 and STING in RAW264.7 macrophages.
Type I IFN levels were measured from RAW264.7 (WT) macrophages and the indicated genotypes (A, C, E) infected with F. novicida strains at an MOI 10 for 24 h or not infected (NI). Type I IFN levels were measured from deficient macrophages stably expressing Sting (B), cGAS (D) or Ifi204 (F) infected with U112 at an MOI 10 for 24 h. Type I IFN levels were measured from RAW264.7 (WT) macrophages and indicated genotypes infected with F. novicida for12 h (G) and 24 h (H) or infected with wild-type L. monocytogenes for 8 h at an MOI of 20 (I). mRNA expression levels of IFN- β1 (J) and TNF-α (K) from uninfected and U112 infected macrophage cell lines at an MOI 10 for 8 h. mRNA levels were normalized to GAPDH and the WT RAW264.7 macrophages. (L) Type I IFN levels were measured from macrophages stimulated with LPS for 8 h. NS: not stimulated *p < 0.05, **p < 0.01, ***p < 0.001. Graphs show the mean ± standard deviation (SD) of triplicate wells and are representative of three independent experiments.
To corroborate our type I IFN data as measured by the L929-ISRE assay, we also measured IFN-β1 gene expression in each RAW264.7 macrophage cell line 8 h after F. novicida infection. IFN-β1 gene expression increased, albeit to different levels, upon F. novicida infection in all macrophage cell lines, but was reduced in expression compared to the RAW264.7 parental genotype in both un-infected and infected conditions (Fig. 3J). To confirm that these cell lines are not altered in type I IFN-independent responses, we measured mRNA expression of TNF-α. Each macrophage cell line expressed similar levels of TNF-α before infection and there were no differences in the levels of TNF-α mRNA between the cell lines after infection (Fig. 3K). We also stimulated type I IFN production through STING-independent mechanisms (44, 45). TLR4-dependent type I IFN production via LPS stimulation resulted in comparable type I IFN levels for all macrophage cell lines (Fig. 3L). Thus, cGAS, Ifi204 and STING specifically influence type I IFNs in response to F. novicida infection.
We next tested whether cGAS and Ifi204 are important for type I IFN signaling during Listeria monocytogenes infection, which is another cytosolic bacterial pathogen known to trigger a robust STING-dependent type I IFN response (30, 46). While L. monocytogenes can directly activate STING through the secretion of c-di-AMP (11), it is unknown if L. monocytogenes DNA is an important ligand during infection and whether cGAS and Ifi204 contribute to the type I IFN response. We infected wild-type, cGAS, Ifi204 and Sting KO macrophages with L. monocytogenes and found that only the Sting KO macrophages had reduced type I IFN levels (Fig. 3I). These results suggest that cGAS and Ifi204 are important for triggering the type I IFN response to F. novicida, but not L. monocytogenes, in RAW264.7 macrophages.
cGAS regulates Ifi204 expression
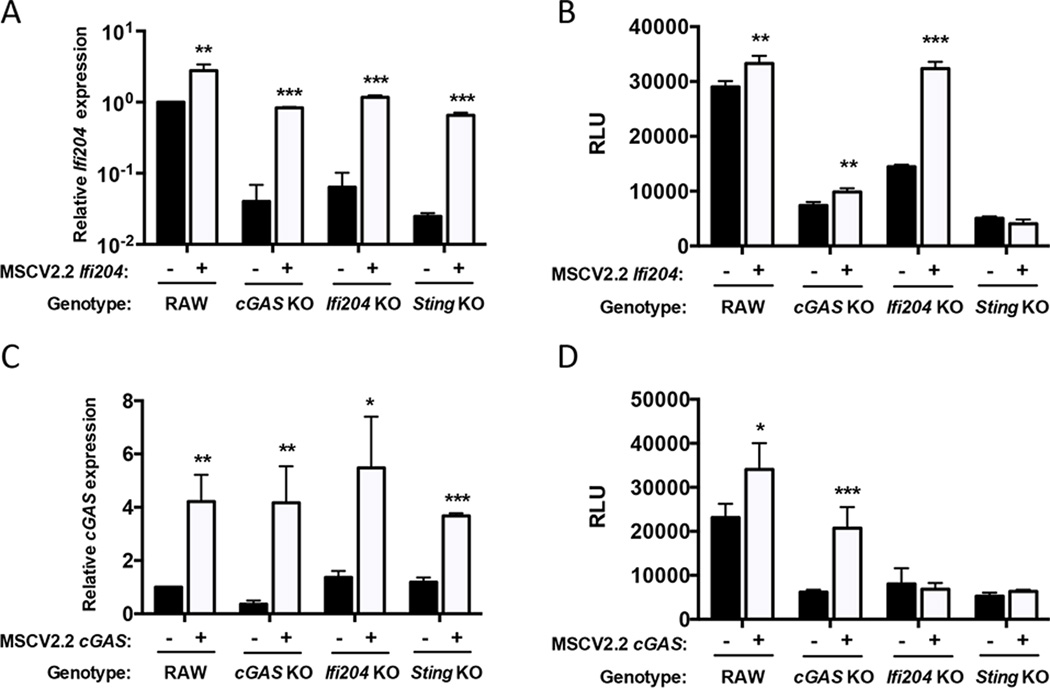
In addition to viruses and bacteria releasing nucleic acids in the cytosol of infected cells, endogenous nucleic acids, such as retroelements, can also trigger a robust type I IFN response (47, 48). Not surprisingly, we found that reducing either cGAS or Ifi204 expression levels resulted in lower endogenous IFN-β1 gene expression and other interferon-stimulated genes (ISGs), including RIG-I (Fig. S2A and Fig. S2B). Ifi204 is an ISG (49), and reduction of cGAS gene expression also resulted in lower Ifi204 gene expression (Fig. S2A and Fig. S2B). In contrast, reduction of Ifi204 did not alter cGAS gene expression and reduced cGAS or Ifi204 levels did not alter Sting or Ddx41 gene expression. We also examined gene expression changes during an infection. Ifi204 gene expression increased in wild-type and cGAS KO RAW264.7 macrophages, but not Sting KO RAW264.7 macrophages (Fig. S2C), while cGAS expression levels were unchanged (Fig. S2E). Interestingly, Sting expression levels were lower in all macrophages during F. novicida infection (Fig. S2D). These data demonstrate that Ifi204 and cGAS are important in regulating type I IFN levels in the absence of infection and indicate that a cGAS KO may, in effect, function as a cGAS and Ifi204 double knockout.
Signaling through both cGAS and Ifi204 is required for the full type I IFN response to F. novicida infection
To determine whether the reduced type I IFN response to F. novicida in cGAS KO macrophages was due to lower expression of the Ifi204 gene, we stably expressed Ifi204 in wild-type, cGAS KO, Ifi204 KO and Sting KO RAW264.7 macrophages. Ectopic expression of Ifi204 led to increased Ifi204 mRNA expression in all cells (Fig. 4A). As expected, expression of Ifi204 in wild-type and Ifi204 KO macrophages complemented the type I IFN phenotype in response to F. novicida infection, but only modestly increased the response in cGAS KO (Fig. 4B). From these data, we hypothesized that cGAS was epistatic to Ifi204. To test this, we ectopically expressed cGAS in an Ifi204 KO and measured type I IFN levels after F. novicida infection. Surprisingly, ectopic expression of cGAS in Ifi204 KO macrophages did not rescue the ability of Ifi204 KO macrophages to secrete type I IFN (Fig. 4D). Importantly, ectopic expression of cGAS increased cGAS mRNA levels in all macrophages (Fig. 4C) and type I IFN production in wild-type and cGAS KO macrophages (Fig. 4D). Ectopic expression of cGAS or Ifi204 in Sting KO macrophages infected with F. novicida did not restore type I IFN production (Fig. 4B and Fig. 4D). These data are consistent with previous studies showing cGAS and Ifi204 signal through STING to facilitate the type I IFN response (10, 13). Collectively, our results suggest that Ifi204 and cGAS are both necessary to fully engage STING-dependent type I IFN responses upon F. novicida infection.
Figure 4. Both cGAS and Ifi204 are required for the full type I IFN response to F. novicida infection.
mRNA expression levels were measured in uninfected RAW264.7 (WT) cell lines stably expressing Ifi204 (A) or cGAS (C). mRNA expression was normalized to GAPDH and wild-type (WT) RAW264.7 macrophages. Macrophages were infected with wild-type F. novicida at an MOI of 10 for 24 h and type I IFN levels measured (B and D, respectively). *p < 0.05, **p < 0.01, ***p < 0.001. Graphs show the mean ± standard deviation (SD) of triplicate wells and are representative of three independent experiments.
Type I IFN induction by cytosolic dsDNA and dsDNA from F. novicida lysates is dependent on cGAS and Ifi204
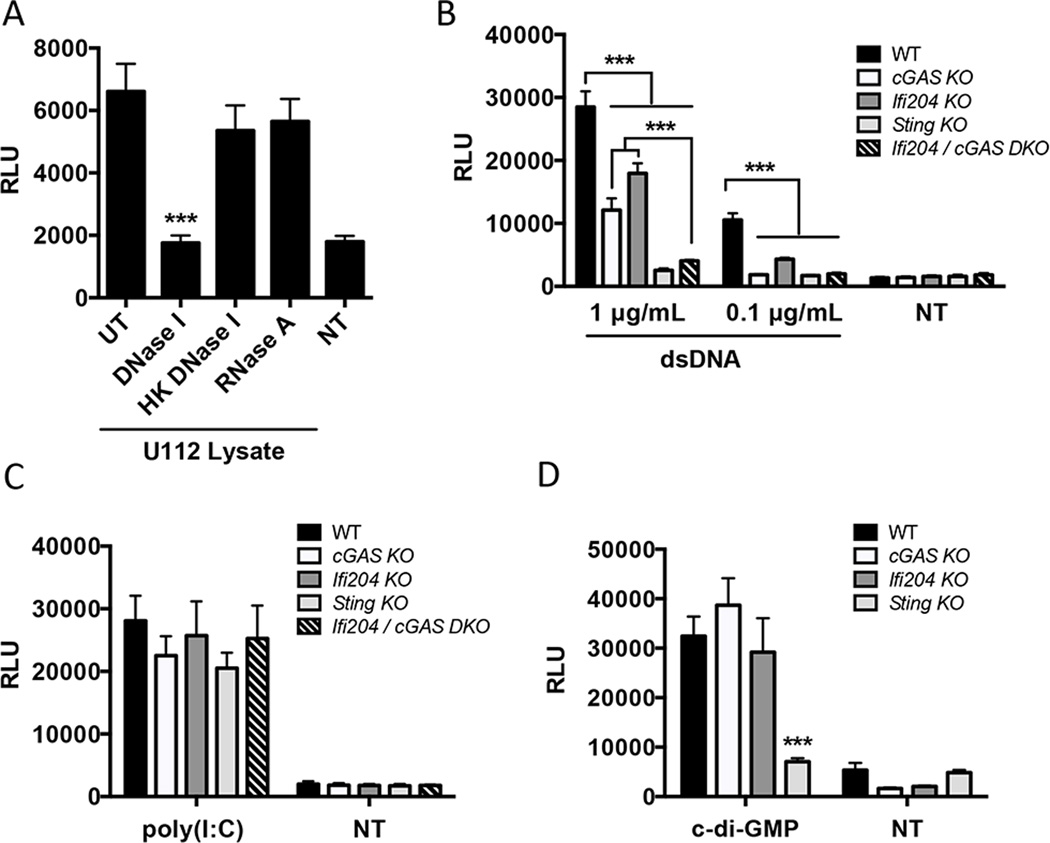
We showed that cGAS and Ifi204 are key factors in the type I IFN response to F. novicida infection (Fig. 3G and Fig. 3H). Since cGAS and Ifi204 have both been implicated in cytosolic DNA sensing, we hypothesized that the type I IFN stimulating PAMP during F. novicida infection is DNA (7). To test this, we transfected RAW264.7 macrophages with untreated F. novicida lysates or with bacterial lysates treated with DNase or RNase. F. novicida lysates stimulated a robust type I IFN response that was abolished when the lysates were treated with DNase I, but not with heat inactivated DNase I or with RNase A (Fig. 5A). These results suggest that DNA from F. novicida lysates is the primary type I IFN stimulus. We next examined the contribution of cGAS, Ifi204 and STING to type I IFN signaling in response to purified dsDNA. Each macrophage cell line was transfected with two different concentrations of dsDNA. Transfection with 0.1 µg/mL of dsDNA resulted in a type I IFN response that was dampened in Ifi204 KO, and undetectable in cGAS KO, Sting KO and cGAS / Ifi204 DKO macrophages (Fig. 5B). Transfecting a higher concentration of dsDNA (1 µg/mL) produced an intermediate type I IFN response for both cGAS KO and Ifi204 KO compared to Sting KO macrophages, which produced a nearly undetectable response (Fig. 5B). Surprisingly, the cGAS / Ifi204 DKO macrophages were equally defective for type I IFN signaling as the Sting KO macrophages (Fig. 5B). These results suggest that cGAS and Ifi204 independently contribute to the cytosolic dsDNA type I IFN response.
Figure 5. cGAS and Ifi204 cooperate to sense cytosolic dsDNA.
(A) Type I IFN levels were measured from wild-type RAW264.7 macrophages transfected for 24 h with U112 lysates either un-treated (UT) or treated with DNase I, heat-killed (HK) DNase I or RNase A. Type I IFN levels were measured from RAW264.7 macrophages transfected for 24 h with (B) endo-free plasmid DNA (pCherry), (C) 1 µg/mL Poly(I:C) or (D) 10 µg/mL c-di-GMP NT: not transfected. *p < 0.05, **p < 0.01, ***p < 0.001. Graphs show the mean ± standard deviation (SD) of triplicate wells and are representative of three independent experiments.
To determine if cGAS and Ifi204 are solely involved in the STING-dependent type I IFN signaling pathway, we stimulated type I IFN production through STING-independent mechanisms in the cytosol (44, 45). MDA5-mediated type I IFN production in response to cytosolic dsRNA stimulation also resulted in comparable type I IFN levels (Fig. 5C). Additionally, we examined whether cGAS or Ifi204 played a role in responding to bacterial cyclic-dinucleotides. Transfection of c-di-GMP led to similar type I IFN responses in wild-type, cGAS KO and Ifi204 KO macrophages (Fig. 5D) In contrast, Sting KO macrophages were defective for signaling in response to c-di-GMP (Fig. 5D), which is consistent with previous studies (31). Our results demonstrate that cGAS and Ifi204 cooperate to signal through the STING-dependent type I IFN pathway in response to cytosolic dsDNA. Additionally, these data support dsDNA as the primary F. novicida PAMP that triggers the type I IFN response.
cGAS increases inflammasome activity in BMMs
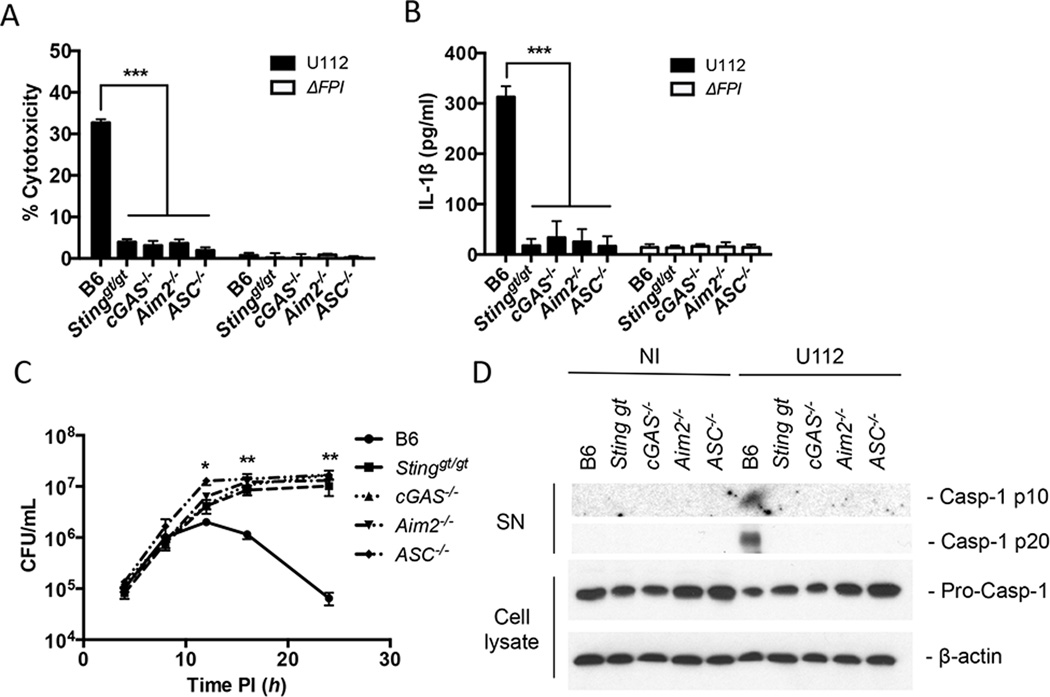
F. novicida infection triggers the activation of the AIM2 inflammasome, a host response pathway that is critical for the defense against F. novicida, resulting in the secretion of pro-inflammatory cytokines (IL-1β and IL-18) and caspase-1-dependent pyroptotic cell death (23, 24). Previous work demonstrated AIM2 protein levels are regulated by type I IFNs and in Sting-deficient BMMs the AIM2 inflammasome is not activated during F. novicida infection (23, 24). Thus, we hypothesized that the AIM2 inflammasome response to F. novicida infection would also be dampened in the absence of either of the DNA sensors that we showed here stimulate STING (cGAS or Ifi204). Using the recently described cGAS−/− mice (34), we examined inflammasome activation by measuring cell death, IL-1β secretion, and caspase-1 processing in response to F. novicida infection. As predicted, when WT, Stinggt/gt, cGAS−/−, Aim2−/− and ASC−/− BMMs were infected with F. novicida, only wild-type cells showed appreciable levels of cell death (Fig. 6A), secreted IL-1β (Fig. 6B) and processed caspase-1 (Fig. 6D). Moreover, F. novicida replicated to higher levels in Stinggt/gt, cGAS−/−, Aim2−/− and ASC−/− BMMs compared to WT BMMs, consistent with the increased cell death of WT BMMs limiting the intracellular niche (Fig. 6C). Collectively, these results demonstrate that cGAS is important for activating the AIM2 inflammasome during F. novicida infection.
Figure 6. cGAS increases inflammasome activity in BMMs.
(A) cytotoxicity, (B) IL-1β secretion and (C) intracellular survival were measured from un-stimulated primary C57Bl/6 (WT), Stinggt/gt, cGAS−/−, Aim2−/− and ASC−/− BMMs infected with F. novicida strains at an MOI of 10 for 12 hours unless otherwise indicated. (D) Release of processed caspase-1 (casp-1 p10 and p20) into the supernatants (SN) was measured by western blot from cells either not infected (NI) or infected with wild-type F. novicida at an MOI of 100 for 12 hours. Corresponding cell lysates were probed for pro-caspase-1 and β-actin. *p < 0.05, **p < 0.01, ***p < 0.001. Graphs show the mean ± standard deviation (SD) of triplicate wells and are representative of three independent experiments.
STING and IRF3 deficiency enhances host survival during F. novicida infection
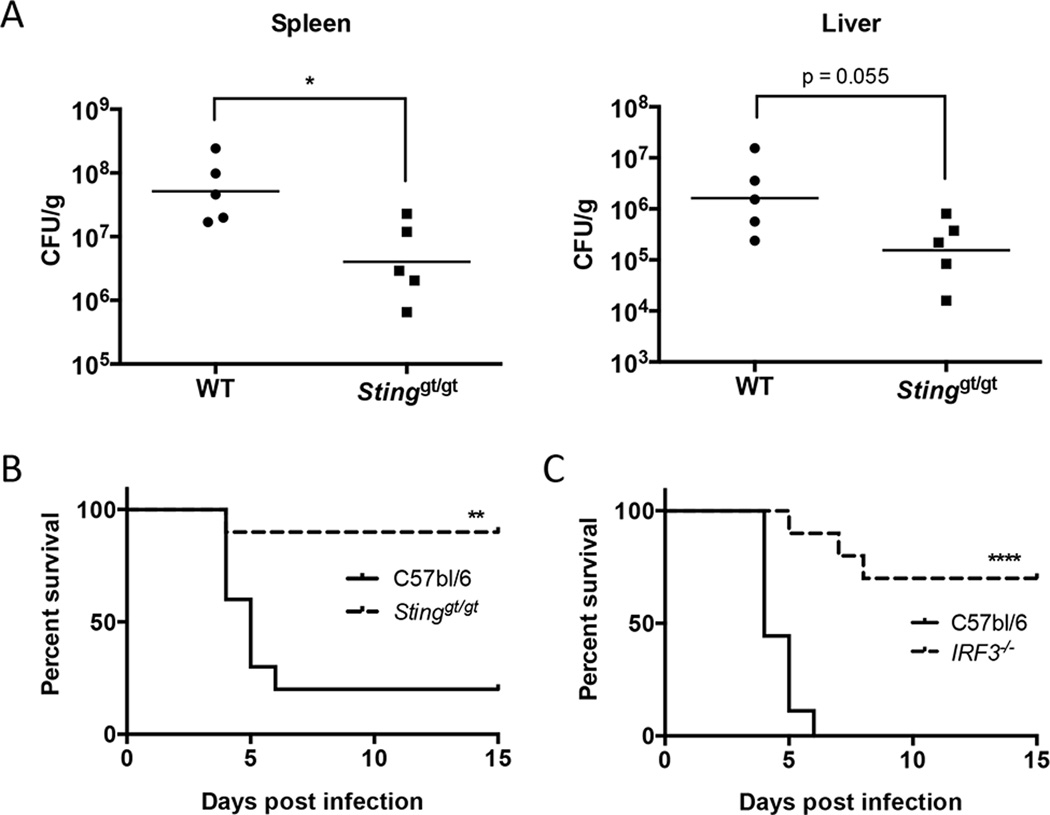
While type I IFN production during viral infections can lead to increased host survival, there are many examples in which type I IFN production in the context of a bacterial infection is associated with decreased host survival (50–52). Indeed, we have previously shown that mice deficient in the type I IFN signaling receptor, IFNAR, are more resistant to F. novicida infection (20, 52). We have shown in this study that both cGAS and Ifi204, which signal through STING, are required to produce the maximum type I IFN levels in response to F. novicida infection. Thus, to test the role of the cGAS/Ifi204/STING axis in mice, we infected Stinggt/gt mice with F. novicida. Mice were infected s.c. with 105 F. novicida CFU and evaluated for bacterial burdens in the spleen and liver 3 d PI. The bacterial loads in the spleen were significantly higher in the wild-type mice compared to the Stinggt/gt mice and similar trends occurred in the liver (Fig. 7A). Mice were also evaluated for their relative susceptibility to F. novicida as assessed in a survival experiment (Fig. 7B). The median time to death of wild-type mice was 5 d and 80% of this group did not survive the F. novicida challenge. In contrast, the median time to death of Stinggt/gt mice was >15 d and 90% of this group survived the infection. To corroborate our findings that type I IFN-dependent STING signaling was detrimental to the host during F. novicida infection, we also infected IRF3-deficient mice with F. novicida. Interferon regulatory factor 3 (IRF3) is the transcription factor activated by STING to induce the transcription of IFN-β and other ISGs (9, 53). Similar to Stinggt/gt mice, IRF3−/− mice infected s.c. with 2x105 F. novicida CFU were significantly more resistant to F. novicida infection compared to wild-type mice (Fig. 7C). The median time to death for wild-type mice and IRF3−/− mice was 4 d and > 15 d, respectively. These results demonstrate that STING- and IRF3-dependent signaling is detrimental to the host during a F. novicida infection.
Figure 7. STING and IRF3 are detrimental to host survival during F. novicida infection.
WT and Stinggt/gt mice were infected s.c. with 105 CFU of U112. (A) The spleen and liver from five infected WT and Stinggt/gt mice were harvested and plated for CFU/g 3 d post-infection, geometric mean shown. (B) 10 WT and 10 Stinggt/gt mice were monitored twice daily over 15 d for survival. (C) 9 WT and 10 IRF3−/− mice were infected s.c. with 2x105 CFU of U112 and were monitored twice daily over 15 d for survival. *p < 0.05, **p < 0.01, ***p < 0.001.
Discussion
Recognition of microbial pathogens is essential to initiate an effective immune response. Host cells have developed numerous strategies to identify infection and tissue injury. One mode of detection is surveying the cytosol for the presence of nucleic acids. While the importance of recognizing and responding to nucleic acids in the cytosol is well-appreciated for viral infections, the role of nucleic acids triggering the type I IFN response during bacterial infections is not well understood. To complicate matters, recent studies have identified numerous cytosolic receptors important for type I IFN signaling in response to dsDNA. We were interested in identifying the host sensors important for type I IFN signaling in response to bacterial infections. In this study we show that during infection with the cytosolic bacterium, F. novicida, the primary type I IFN response is likely dependent on the presence of F. novicida DNA in the cytosol. Moreover, we demonstrate that two proposed host DNA sensors, cGAS and Ifi204, are required to generate type I IFNs during F. novicida infection and in response to cytosolic dsDNA in murine macrophages.
We observed that during a F. novicida infection both cGAS and Ifi204 were necessary to mediate a full type I IFN response (Fig. 4). In addition, we show that both cGAS and Ifi204 need to be eliminated (e.g., cGAS / Ifi204 DKO in RAW264.7 cells) to prevent a type I IFN response to high concentrations of transfected DNA in RAW264.7 macrophages. This result was surprising given that cGAS alone was previously demonstrated to bind cytosolic DNA and generate the STING-activating second messenger, cGAMP, resulting in type I IFN production (8, 10, 54, 55). This discrepancy in findings may be attributed to fundamental variances in cell types, including differences in transfection efficiency and/or alterations in the Ifi204 expression signature. In particular, we observe that transfecting RAW264.7 macrophages using standard transfection reagents (i.e. Lipofectamine 2000 and Targefect-RAW) results in a considerably higher percentage of cells being transfected compared to BMMs and much lower cytotoxicity (data not shown). These factors may permit detection of intermediate to low type I IFN responses that are not seen in cGAS−/− BMMs (54). However, further studies in a variety of cell types are needed to assess the role and/or expression of Ifi204 in the absence of cGAS. It also remains to be determined how Ifi204, a member of AIM2-like receptors (ALRs) (49), fits into the cGAS/STING/type I IFN pathway. To date, 13 ALR genes have been identified within the mouse genome (49). They are all encoded within a single continuous locus and contain a Pyrin and/or HIN domain. One possible model for the requirement of Ifi204 in the cGAS/STING/type I IFN pathway is that Ifi204 either due to higher affinity for dsDNA or to appropriate subcellular localization, may initially recognize F. novicida DNA and transport the DNA ligand to cGAS. This would result in the production of cGAMP and subsequent activation of STING to trigger type I IFN production. Evidence for this model is supported by previous studies, which demonstrate that the human orthologue of Ifi204, IFI16, interacts with STING upon DNA stimulation (13). Additionally, Ifi204 and STING were found to co-localize in the cytosol when transfected in HeLa cells (49). Future studies are required to resolve the mechanism behind the necessity of Ifi204 and determine if other ALR proteins are important in facilitating the cGAS-dependent signaling response to intracellular bacteria and potentially other stimuli.
Access of F. novicida DNA to the host cytosol is a requirement to activate two independent host responses. First, F. novicida DNA is likely recognized by cGAS and Ifi204 to trigger a STING-dependent type I IFN response. Through autocrine and paracrine type I IFN signaling, AIM2 protein levels increase and subsequently associates with F. novicida DNA and ASC to activate caspase-1-mediated cell death and secretion of pro-inflammatory cytokines (23). Though the mechanism of DNA release by F. novicida into the host cytosol is not fully known, F. novicida mutants that are prone to increased bacterial lysis trigger higher type I IFNs and inflammasome responses compared to the parental strain (29). This observation suggests that one mechanism of facilitating access of the DNA sensors to F. novicida DNA is a low level of bacterial lysis in the cytosol. One notable difference between the two cell types we used in this study is that the FPI mutant elicits a type I IFN response that is higher than the levels produced by uninfected RAW264.7 macrophages (Fig. S1D). In contrast, the levels of type I IFN produced by BMMs infected with the FPI mutant are not higher than uninfected BMMs (Fig. S1D). These results may be attributed to the higher proportion of FPI mutant bacteria found in the cytosol of RAW264.7 macrophages compared to BMMs. These studies underscore the importance of DNA recognition during a bacterial infection. This is particularly important for a bacterial pathogen like Francisella that is a stealth invader and does not elicit a TLR-mediated type I IFN response (20).
Many intracellular bacterial pathogens elicit a type I IFN host response and several of these have been demonstrated to require STING, including L. monocytogenes, M. tuberculosis, and C. trachomatis (5, 11, 56). In addition to STING, the type I IFN response in immortalized BMMs to M. tuberculosis infection was also demonstrated to require Ifi204 (56). Herein, we demonstrated that the STING-dependent type I IFN response elicited by L. monocytogenes did not require the DNA sensors, cGAS or Ifi204 (Fig. 3I). These results are consistent with previous findings demonstrating that L. monocytogenes secretes cyclic-di-AMP to directly stimulated STING in murine cells (11). Notably, a recent study showed L. monocytogenes infection in human macrophages induced the type I IFN response that is dependent on cGAS, IFI16 (Ifi204 homologue) and STING (57). Further studies are needed to elucidate the roles of cGAS and Ifi204/IFI16 in recognizing other intracellular bacterial pathogens and potential differences between human and mouse type I IFN signaling pathways.
The role of type I IFNs in controlling bacterial infections is complex. Wild-type mice infected with L. monocytogenes, F. novicida and M. tuberculosis support increased bacterial burdens than mice deficient for the type I IFN receptor (IFNAR1−/−) (50–52). While these studies were initially surprising, it is now known that type I IFN production leads to the transcription of hundreds of ISGs, which modulate a variety of factors in both innate and adaptive immunity (3). In the case of a F. novicida infection, IFNAR1−/− mice are more resistant to infection largely due to an increase expansion of IL-17A+ γδ T cells and increased splenic neutrophils (52). The role of cGAS and Ifi204 during a F. novicida infection in vivo is unknown. Since, cGAS and Ifi204 both require STING for type I IFN signaling, we sought to evaluate Stinggt/gt mice for their susceptibility to F. novicida infection. Previous studies using a different Francisella subspecies, F. tularensis subspecies holactica live vaccine strain (LVS), and different infection route (i.p.) did not identify a difference in splenic bacterial loads between STING (MPYS)-deficient mice and wild-type mice 48 h PI (22). This finding is similar to our previous findings using IFNAR−/− mice, in which there is no difference in bacterial burdens 1 and 2 d PI (52). Importantly, we show here that Stinggt/gt mice carried lower bacterial burdens in the liver and spleen 3 d PI and were significantly more resistant to F. novicida infection compared to wild type mice (Fig. 7A). Furthermore, we show that mice deficient for the downstream transcription factor, IRF3, were also significantly more susceptible to F. novicida infection. We postulate that both Stinggt/gt mice and Irf3−/− mice are more resistant to infection due to similar mechanisms described for IFNAR1−/− mice (52). Despite lower bacterial burdens found in Stinggt/gt mice and IFNAR1−/− mice, the type I IFN response is important in activating specific host responses important for bacterial control, including the AIM2 inflammasome during F. novicida infections (23).
Our results begin to illuminate the mechanisms involved in type I IFN production during intracellular bacterial infections. Our data strongly support the hypothesis that F. novicida DNA is sensed by cGAS and Ifi204 to trigger the STING-dependent type I IFN response. Future experiments investigating the cross talk between TLR-mediated type I IFN production and cytosolic DNA-mediated type I IFN production may discover unique signaling signatures depending on the host receptor activated. Understanding how type I IFN production is regulated and what factors are involved may aid in our therapeutic efforts to prevent and treat auto-inflammatory and infectious diseases.
Supplementary Material
Acknowledgements
We thank Jan Carette and Caleb Marceau for technical support with CRISPR/Cas9 mutagenesis, Bianca Gomez for technical support on the flow cytometer, Russell Vance for Stinggt/gt femurs, Herbert Virgin for cGAS−/− femurs, Damian Trujillo and David Schneider for C57Bl/6 mice and IRF3−/− mice and all members of the Monack lab and Jenny Lumb for valuable comments.
This work was supported by NIH-NIAID grants AI063302 and AI065359 to DM and F32 AI108089 to K.M.S..
References
- 1.Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009;6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hardy MP, Owczarek CM, Jermiin LS, Ejdeback M, Hertzog PJ. Characterization of the type I interferon locus and identification of novel genes. Genomics. 2004;84:331–345. doi: 10.1016/j.ygeno.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 4.Perry AK, Chen G, Zheng D, Tang H, Cheng G. The host type I interferon response to viral and bacterial infections. Cell Res. 2005;15:407–422. doi: 10.1038/sj.cr.7290309. [DOI] [PubMed] [Google Scholar]
- 5.Barker JR, Koestler BJ, Carpenter VK, Burdette DL, Waters CM, Vance RE, Valdivia RH. STING-dependent recognition of cyclic di-AMP mediates type I interferon responses during Chlamydia trachomatis infection. mBio. 2013;4:e00018-00013. doi: 10.1128/mBio.00018-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trinchieri G. Type I interferon: friend or foe? J Exp Med. 2010;207:2053–2063. doi: 10.1084/jem.20101664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu J, Chen ZJ. Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol. 2014;32:461–488. doi: 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]
- 8.Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, Chen ZJ. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Molecular cell. 2013;51:226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Science signaling. 2012;5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, Ohba Y, Taniguchi T. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 13.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang P, An H, Liu X, Wen M, Zheng Y, Rui Y, Cao X. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a beta-catenin-dependent pathway. Nat Immunol. 2010;11:487–494. doi: 10.1038/ni.1876. [DOI] [PubMed] [Google Scholar]
- 15.Kondo T, Kobayashi J, Saitoh T, Maruyama K, Ishii KJ, Barber GN, Komatsu K, Akira S, Kawai T. DNA damage sensor MRE11 recognizes cytosolic double-stranded DNA and induces type I interferon by regulating STING trafficking. Proc Natl Acad Sci U S A. 2013;110:2969–2974. doi: 10.1073/pnas.1222694110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X, Brann TW, Zhou M, Yang J, Oguariri RM, Lidie KB, Imamichi H, Huang DW, Lempicki RA, Baseler MW, Veenstra TD, Young HA, Lane HC, Imamichi T. Cutting edge: Ku70 is a novel cytosolic DNA sensor that induces type III rather than type I IFN. J Immunol. 2011;186:4541–4545. doi: 10.4049/jimmunol.1003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones CL, Napier BA, Sampson TR, Llewellyn AC, Schroeder MR, Weiss DS. Subversion of host recognition and defense systems by Francisella spp. Microbiol Mol Biol Rev. 2012;76:383–404. doi: 10.1128/MMBR.05027-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chong A, Wehrly TD, Nair V, Fischer ER, Barker JR, Klose KE, Celli J. The early phagosomal stage of Francisella tularensis determines optimal phagosomal escape and Francisella pathogenicity island protein expression. Infect Immun. 2008;76:5488–5499. doi: 10.1128/IAI.00682-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henry T, Brotcke A, Weiss DS, Thompson LJ, Monack DM. Type I interferon signaling is required for activation of the inflammasome during Francisella infection. J Exp Med. 2007;204:987–994. doi: 10.1084/jem.20062665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mariathasan S, Weiss DS, Dixit VM, Monack DM. Innate immunity against Francisella tularensis is dependent on the ASC/caspase-1 axis. J Exp Med. 2005;202:1043–1049. doi: 10.1084/jem.20050977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin L, Hill KK, Filak H, Mogan J, Knowles H, Zhang B, Perraud AL, Cambier JC, Lenz LL. MPYS is required for IFN response factor 3 activation and type I IFN production in the response of cultured phagocytes to bacterial second messengers cyclic-di-AMP and cyclic-di-GMP. J Immunol. 2011;187:2595–2601. doi: 10.4049/jimmunol.1100088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones JW, Kayagaki N, Broz P, Henry T, Newton K, O'Rourke K, Chan S, Dong J, Qu Y, Roose-Girma M, Dixit VM, Monack DM. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc Natl Acad Sci U S A. 2010;107:9771–9776. doi: 10.1073/pnas.1003738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, Superti-Furga G. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 27.Landolfo S, Gariglio M, Gribaudo G, Lembo D. The Ifi 200 genes: an emerging family of IFN-inducible genes. Biochimie. 1998;80:721–728. doi: 10.1016/s0300-9084(99)80025-x. [DOI] [PubMed] [Google Scholar]
- 28.Weiss DS, Brotcke A, Henry T, Margolis JJ, Chan K, Monack DM. In vivo negative selection screen identifies genes required for Francisella virulence. Proc Natl Acad Sci U S A. 2007;104:6037–6042. doi: 10.1073/pnas.0609675104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng K, Broz P, Jones J, Joubert LM, Monack D. Elevated AIM2-mediated pyroptosis triggered by hypercytotoxic Francisella mutant strains is attributed to increased intracellular bacteriolysis. Cell Microbiol. 2011;13:1586–1600. doi: 10.1111/j.1462-5822.2011.01643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sauer JD, Sotelo-Troha K, von Moltke J, Monroe KM, Rae CS, Brubaker SW, Hyodo M, Hayakawa Y, Woodward JJ, Portnoy DA, Vance RE. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect Immun. 2011;79:688–694. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Broz P, Newton K, Lamkanfi M, Mariathasan S, Dixit VM, Monack DM. Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. J Exp Med. 2010;207:1745–1755. doi: 10.1084/jem.20100257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, Dabelic R, Manicassamy B, Aitchison JD, Aderem A, Elliott RM, Garcia-Sastre A, Racaniello V, Snijder EJ, Yokoyama WM, Diamond MS, Virgin HW, Rice CM. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014;505:691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Broz P, von Moltke J, Jones JW, Vance RE, Monack DM. Differential requirement for Caspase-1 autoproteolysis in pathogen-induced cell death and cytokine processing. Cell Host Microbe. 2010;8:471–483. doi: 10.1016/j.chom.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang Z, Georgel P, Du X, Shamel L, Sovath S, Mudd S, Huber M, Kalis C, Keck S, Galanos C, Freudenberg M, Beutler B. CD14 is required for MyD88-independent LPS signaling. Nat Immunol. 2005;6:565–570. doi: 10.1038/ni1207. [DOI] [PubMed] [Google Scholar]
- 37.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 38.Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, Nakaya T, Katsuki M, Noguchi S, Tanaka N, Taniguchi T. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity. 2000;13:539–548. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- 39.Daffis S, Suthar MS, Szretter KJ, Gale M, Jr, Diamond MS. Induction of IFN-beta and the innate antiviral response in myeloid cells occurs through an IPS-1-dependent signal that does not require IRF-3 and IRF-7. PLoS Pathog. 2009;5:e1000607. doi: 10.1371/journal.ppat.1000607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parvatiyar K, Zhang Z, Teles RM, Ouyang S, Jiang Y, Iyer SS, Zaver SA, Schenk M, Zeng S, Zhong W, Liu ZJ, Modlin RL, Liu YJ, Cheng G. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat Immunol. 2012;13:1155–1161. doi: 10.1038/ni.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmerk CL, Duplantis BN, Howard PL, Nano FE. A Francisella novicida pdpA mutant exhibits limited intracellular replication and remains associated with the lysosomal marker LAMP-1. Microbiology. 2009;155:1498–1504. doi: 10.1099/mic.0.025445-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bryan NB, Dorfleutner A, Kramer SJ, Yun C, Rojanasakul Y, Stehlik C. Differential splicing of the apoptosis-associated speck like protein containing a caspase recruitment domain (ASC) regulates inflammasomes. Journal of inflammation. 2010;7:23. doi: 10.1186/1476-9255-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pelegrin P, Barroso-Gutierrez C, Surprenant A. P2X7 receptor differentially couples to distinct release pathways for IL-1beta in mouse macrophage. J Immunol. 2008;180:7147–7157. doi: 10.4049/jimmunol.180.11.7147. [DOI] [PubMed] [Google Scholar]
- 44.Uematsu S, Akira S. Toll-like receptors and Type I interferons. J Biol Chem. 2007;282:15319–15323. doi: 10.1074/jbc.R700009200. [DOI] [PubMed] [Google Scholar]
- 45.Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M, Jr, Akira S, Yonehara S, Kato A, Fujita T. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 46.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Volkman HE, Stetson DB. The enemy within: endogenous retroelements and autoimmune disease. Nat Immunol. 2014;15:415–422. doi: 10.1038/ni.2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brunette RL, Young JM, Whitley DG, Brodsky IE, Malik HS, Stetson DB. Extensive evolutionary and functional diversity among mammalian AIM2-like receptors. J Exp Med. 2012;209:1969–1983. doi: 10.1084/jem.20121960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stanley SA, Johndrow JE, Manzanillo P, Cox JS. The Type I IFN response to infection with Mycobacterium tuberculosis requires ESX-1-mediated secretion and contributes to pathogenesis. J Immunol. 2007;178:3143–3152. doi: 10.4049/jimmunol.178.5.3143. [DOI] [PubMed] [Google Scholar]
- 51.Auerbuch V, Brockstedt DG, Meyer-Morse N, O'Riordan M, Portnoy DA. Mice lacking the type I interferon receptor are resistant to Listeria monocytogenes. J Exp Med. 2004;200:527–533. doi: 10.1084/jem.20040976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Henry T, Kirimanjeswara GS, Ruby T, Jones JW, Peng K, Perret M, Ho L, Sauer JD, Iwakura Y, Metzger DW, Monack DM. Type I IFN signaling constrains IL-17A/F secretion by gammadelta T cells during bacterial infections. J Immunol. 2010;184:3755–3767. doi: 10.4049/jimmunol.0902065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hiscott J. Triggering the innate antiviral response through IRF-3 activation. J Biol Chem. 2007;282:15325–15329. doi: 10.1074/jbc.R700002200. [DOI] [PubMed] [Google Scholar]
- 54.Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ablasser A, Hemmerling I, Schmid-Burgk JL, Behrendt R, Roers A, Hornung V. TREX1 deficiency triggers cell-autonomous immunity in a cGAS-dependent manner. J Immunol. 2014;192:5993–5997. doi: 10.4049/jimmunol.1400737. [DOI] [PubMed] [Google Scholar]
- 56.Manzanillo PS, Shiloh MU, Portnoy DA, Cox JS. Mycobacterium tuberculosis activates the DNA-dependent cytosolic surveillance pathway within macrophages. Cell Host Microbe. 2012;11:469–480. doi: 10.1016/j.chom.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hansen K, Prabakaran T, Laustsen A, Jorgensen SE, Rahbaek SH, Jensen SB, Nielsen R, Leber JH, Decker T, Horan KA, Jakobsen MR, Paludan SR. Listeria monocytogenes induces IFNbeta expression through an IFI16-, cGAS- and STING-dependent pathway. The EMBO journal. 2014;33:1654–1666. doi: 10.15252/embj.201488029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.