Abstract
Epidermal growth factor receptor (EGFR) is commonly affected in cancer, generally in the form of an increase in DNA copy number and/or as mutation variants [e.g., EGFR variant III (EGFRvIII), an in-frame deletion of exons 2–7]. While detection of EGFR aberrations can be expected to be relevant for glioma patients, such analysis has not yet been implemented in a routine setting, also because feasible and robust assays were lacking.
We evaluated multiplex ligation-dependent probe amplification (MLPA) for detection of EGFR amplification and EGFRvIII in DNA of a spectrum of 216 diffuse gliomas. EGFRvIII detection was verified at the protein level by immunohistochemistry and at the RNA level using the conventionally used endpoint RT-PCR as well as a newly developed quantitative RT-PCR. Compared to these techniques, the DNA-based MLPA assay for EGFR/EGFRvIII analysis tested showed 100% sensitivity and specificity. We conclude that MLPA is a robust assay for detection of EGFR/EGFRvIII aberrations. While the exact diagnostic, prognostic and predictive value of such EGFR testing remains to be seen, MLPA has great potential as it can reliably and relatively easily be performed on routinely processed (formalin-fixed, paraffin-embedded) tumor tissue in combination with testing for other relevant glioma markers.
Keywords: clinical implications, EGFR amplification, EGFRvIII, MLPA, molecular pathology
INTRODUCTION
Epidermal growth factor receptor (EGFR) is a transmembrane tyrosine kinase receptor protein and one of the most commonly affected growth factor receptors in cancer (20). Elevated levels of EGFR are usually induced by amplification of the EGFR gene and correlate with tumorigenicity and an unfavorable prognosis (12, 13, 39, 55, 68, 72). Furthermore, different mutational variants of EGFR have been identified of which EGFR variant III (EGFRvIII) is the most frequent and detected in gliomas (16, 17, 62), as well as in cancers of the lung (50), breast (11), prostate (51) and head and neck (70). EGFRvIII is characterized by an in-frame DNA deletion of exons 2–7, generating a truncated protein that lacks the majority of the extracellular ligand-binding domain (7, 21, 35, 36, 40, 71, 84, 85). EGFRvIII leads to constitutive activation of downstream signaling including second messenger pathways not available to wild-type EGFR (48, 73) and shows decreased protein degradation (88). Expression of EGFRvIII results in increased cell proliferation, resistance to apoptosis, increased cell migration and has been associated with shorter life expectancy (1, 32, 45, 46, 49, 68).
Due to its prominent role in oncogenesis, inhibition of EGFR/EGFRvIII signaling is an attractive therapeutic approach for several types of cancer (43, 47, 58, 60, 81). Especially for the most malignant and common glioma subtype, glioblastoma multiforme (GBM), showing a poor prognosis in spite of aggressive treatment, this approach might offer new therapeutic options as EGFR is frequently affected. Different anti-EGFR agents are available and were proven effective in specific subsets of patients (9, 14, 33, 41, 53, 59, 64, 65). Investigating the molecular background of the tumors will increase our understanding of response to therapy and allow for identification of those patients that are most likely to respond. For example, as agents blocking the ligand-binding site are unlikely to silence signaling through EGFRvIII, in this situation other inhibiting agents such as small molecules affecting the intracellular tyrosine kinase domain might be much more promising. In gliomas, several trials with such small molecules were performed using, for example, Erlotinib (Tarceva®; Roche, Basel, Switzerland) or Gefitineb (Iressa®; AstraZeneca, London, UK) (79). Efficacy was shown in a subset of patients, and although conflicting results have been described, molecular analysis of these responsive cases (EGFRvIII and PTEN (phosphatase and tensin homologue deleted in chromosome 10) positive or high EGFR and low p-AKT (phosphorylated v-akt murine thymoma viral oncogene homolog)) suggests additional clinical value of molecular diagnostics for patient stratification in the (near) future (41, 65).
Molecular analysis in a routine diagnostic setting next to the histopathologic diagnosis is increasingly performed for diffusely infiltrating gliomas providing additional information about, for example, the type and malignancy grade of the tumor, response to chemotherapeutic treatment and prognosis (i.e., diagnostic, therapeutic/predictive and prognostic value, respectively) (22, 38, 74). Such routine molecular diagnostics are DNA based and most commonly evaluate loss of chromosome arms 1p and 19q (1p/19q) as well as methylation of the promotor of the O6-methylguanine-DNA methyltransferase gene (MGMT) (3, 15, 24, 31, 75). Newly identified correlations between molecular characteristics of the tumor and response to specific therapeutic agents, for example, against EGFR/EGFRvIII, or other agents for targeted therapy, can be implemented into the routine molecular diagnostics and thereby contribute to optimal patient stratification, resulting in improved prognosis and quality of life.
Identification of the clinical relevance of, for example, EGFR/ EGFRvIII aberrations as well as implementation in a routine diagnostic setting, requires a feasible and robust assay that can be easily performed, preferably is DNA based so it can be performed in parallel with evaluation of the other relevant molecular markers and ideally is suitable for the analysis of routinely processed, formalin-fixed and paraffin-embedded (FFPE) tissue. We evaluated detection of EGFR amplification and of EGFRvIII by multiplex ligation-dependent probe amplification (MLPA) using tumor DNA and established the specificity and sensitivity of this assay by a parallel analysis of tumor RNA and protein. Analyzing a spectrum of 216 gliomas, we investigated the distribution of EGFR copy number changes and presence of EGFRvIII among the different types of gliomas. We conclude the MLPA assay has great potential for glioma diagnostics in a routine diagnostic setting as it provides additional diagnostic, prognostic and therapeutic information.
MATERIAL AND METHODS
Tumor samples
Diffuse glioma samples for this study were retrieved from the neurooncology archive at the Department of Pathology of the Radboud University Nijmegen Medical Centre, the Netherlands. The use of brain tumor tissue after completing histopathologic diagnosis for research purposes has been approved by the regional ethics committee. Tumors were classified and graded according to the World Health Organization (WHO)-2000 classification as diffuse astrocytoma (A-II), anaplastic astrocytoma (A-III), GBM, oligodendroglioma (O-II), anaplastic oligodendroglioma (O-III), oligoastrocytoma (OA-II) or anaplastic oligoastrocytoma (OA-III) (29). In the current study we analyzed in total 216 tumors: 20 A-IIs, 5 A-IIIs, 104 GBMs, 21 O-IIs, 25 O-IIIs, 13 OA-IIs, 28 OA-IIIs for EGFR copy number changes and the presence of EGFRvIII as detected by MLPA assay P105.
DNA isolation
DNA was isolated from routinely processed, FFPE tumor samples as well as from snap frozen tumor tissue. In case of paraffin-embedded tissue, 50 μm paraffin sections were cut and incubated in P-buffer (50 mM Tris-HCl pH 8.2, 100 mM NaCl, 1 mM EDTA, 0.5% Tween-20, 0.5% NP40, 20 mM DTT) at 90°C for 15 minutes after which a protein digestion was performed using 0.5 mg/mL proteinase K (Roche Diagnostics GmbH, Mannheim, Germany) at 55°C overnight and another 48 h at 37°C (fresh proteinase K being added every 24 h). Subsequently, DNA was isolated using the DNeasy Tissue Kit (Qiagen, Venlo, the Netherlands). DNA from snap-frozen tumor tissue was isolated with this kit as described by the manufacturer. Furthermore, DNA previously isolated using a salting out procedure (26, 42) was purified using the DNeasy Tissue kit (Qiagen). An additional wash step using the AW2 wash buffer was always included prior to elution.
Multiplex ligation-dependent probe amplification (MLPA)
To evaluate copy number changes in EGFR we used MLPA kit P105 (lot-nr 0306 and 0407) prepared by MRC-Holland (Amsterdam, the Netherlands) containing probes specifically recognizing EGFR exon 1 (Probe name MRC-Holland: 2062-L3282), exon 2 (5435-L4851), exon 3 (5436-L4852), exon 4 (5437-L4853), exon 5 (5438-L6027), exon 6 (6121-L6286), exon 7 (5440-L4856), exon 8 (2063-L3283), exon 13 (5441-L5612), exon 17 (2064-L1577) and exon 22 (5968-L5385). Additionally this kit contains 5 CDKN2A, 10 (lot-nr 0306) or 11 (lot-nr 0407) PTEN and 8 TP53 specific probes, whereas the other 6 probes were used as control probes (http://www.mlpa.com) (66).
MLPA was performed as described by the manufacturer with minor modifications. Briefly, 200 ng DNA in 5 μL TE-buffer (10 mM Tris pH 8.2, 1 mM EDTA pH 8.0) or Milli Q was denatured and cooled down to 25°C. After adding the probe mix the sample was denatured, probes were allowed to hybridize (16 h at 60°C) and ligation was performed. PCR was performed in twice the amount as described by the manufacturer (in a volume of 50 μL containing 10 μL of the ligation reaction mixture), which in our experience results in a major improvement of (reliable) copy number analysis (unpublished results using MLPA kit P088 and P105). PCR was performed on a PTC 200 thermal cycler (MJ Research Inc., Waltham, MA, USA) and encompassed 33 cycles of 20 s at 95°C for, 30 s at 60°C and 1 minute at 72°C, with a final extension of 20 minutes at 72°C. Aliquots of 1 μL were combined with 0.3 μL LIZ-labeled internal size standard (LIZ-500 Genescan, ABI 401734) and 8.7 μL deionized formamid. Denatured fragments were separated and quantified (peak height) by electrophoresis on an ABI 3730 capillary sequencer (Applied Biosystems, Nieuwerkerk aan de Ijssel, the Netherlands) and Genemapper analysis (Applied Biosystems) (76).
Data analysis was performed in Microsoft Excel as described previously (25). The “probe fraction” of each peak is calculated by dividing the peak value of each probe amplification product by the combined value of the control probes (n = 6) within the sample (to compensate for differences in PCR efficiency). These “probe fractions” are then divided by the average probe fraction of the normal DNAs included in each experiment, resulting in a probe ratio. Based on our previous experience with MLPA copy number detection, we chose to use thresholds of 1.2 and 0.8 for the detection of gains and losses, respectively (25). Furthermore, ratios below 0.4 were considered homozygous losses, whereas ratios above 2.0 were considered as amplifications and those exceeding 10 as high-copy (HC) number amplifications. Due to the presence of non-neoplastic cells within tumor samples, theoretical thresholds of, for example, 0.5 for a hemizygous loss and 0.00 for a homozygous loss are inadequate. To identify EGFRvIII we determined the average ratio for exons 2–7 probes and compared this with the average ratio of probes for exons 1, 8, 13, 17 and 22 (EGFRvIII ratio). EGFRvIII ratios below 0.8 were considered to harbor the EGFRvIII deletion variant. This EGFRvIII threshold proved to result in a highly specific and sensitive assay (results of the current paper). Next to assessment of the EGFRvIII ratio we inspected the individual probe ratios in order to confirm the presence of EGFRvIII instead of other EGFR variants (e.g., as present in N216, see results).
RNA analysis
Cases selected for RNA analysis included those identified as EGFRvIII positive by MLPA analysis or showing EGFRvIII ratios near the threshold set for detection of EGRFvIII (i.e., 0.8), whereas the remaining cases were selected randomly. Availability of sufficient tumor tissue proved to be a limiting factor for some cases excluding them from further evaluation at the RNA level.
Total RNA was isolated using Trizol (Invitrogen, Breda, the Netherlands). Briefly, 10-μm sections were collected, preferably from snap-frozen tissue or alternatively from FFPE tissue (30–50 or 5–10 sections, respectively). Paraffin from FFPE tissue was removed using 500 μL xylol (3 times) and the tissue was air-dried after three 96% ethanol washes. Lysisbuffer (Biozym TC, Kerensheide, the Netherlands) and Proteinase K (Roche Diagnostics) were added and put overnight at 56°C. Frozen tissue or 250 μL FFPE tissue lysate was dissolved in 1 mL Trizol (Invitrogen) and incubated on ice for 15 minutes after which 200 μL chloroform was added. Total RNA was either isolated by precipitation (pellet dissolved in 25 μL MQ) or using the NucleoSpin® RNA II Columns (Bioké, Leiden, the Netherlands) as described by the manufacturer (eluation in 40 μL MQ) and stored in −80°C.
cDNA was prepared from 500 ng isolated RNA using SuperScript II (Invitrogen) as described by the manufacturer with random hexamers. We performed endpoint RT-PCR as described by Mellinghoff et al (41), the primers generating products of 243 basepairs (bp) and 1044 bp for EGFRvIII and EGFR-wild-type (EGFR-wt), respectively. As these fragments proved to be rather long for the analysis of RNA isolated from FFPE primers for quantitative RT-PCR (q-RT-PCR) were designed, located in exons 1 and 8 and exons 2 and 3, representing EGFRvIII and EGFR-wt and generating products of only 98 and 89 base pairs, respectively. Primers sequences used are 1-for: GCTCTGGAGGAAAAGA AAGGTAATTAT, 8-rev: ACGCCGTCTTCCTCCATCT, 2-for: TACCTATGTGCAGAGGAATTATGATCTTT, and 3-rev: CCAC TGTGTTGAGGGCAATG. For comparison, the housekeeping gene GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used (forward-primer: CCACATCGCTCAGACACCAT and reverse-primer: GCGCCCAATACGACCAAA), generating a 66 bp product. q-RT-PCR was performed using 25 ng cDNA and 15 pmol of each primer in Power SYBR Green PCR Master Mix from Applied Biosystems. PCR was performed on an ABI Prism 7000 Sequence Detection System (Applied Biosystems) (2 minutes at 50°C, 10 minutes at 95°C, 40 subsequent cycles of 15 s at 95°C and 1 minute at 60°C, followed by 15 s at 95°C, 1 minute at 60°C, 15 s at 95°C and 15 s at 60°C).
Immunohistochemical detection of EGFRvIII
Tissue micro-arrays (TMAs) were established from FFPE glioma tissue and included 3–4 representative cores (2 mm in diameter) of each tumor. Gliomas included in these TMAs were previously screened by us for chromosomal imbalances using comparative genomic hybridization (26, 27). Whenever sufficent DNA or tissue was available these cases were included in the present study. These arrays were used to study the expression of EGFRvIII by immunohistochemistry (IHC) and included 76 of the gliomas analyzed by MLPA.
Antigenic localization was performed using the BondMax automated stainer (Leica Microsystems, Bannockburn, IL, USA) IHC was performed using the AbL8A4 antibody that is specific for EGFRvIII but unreactive with wild-type EGFR (83). Five-micrometer sections from TMA blocks were programmed to be processed on the BondMax including paraffin removal, heat-induced epitope retrieval using Bond Epitope Retrieval Solution 2 (Vision BioSystems, Norwell, MA, USA) for 20 minutes and application of the primary antibody. Detection of the bound antibody was accomplished with the use of Bond Polymer Refine Detection (Vision BioSystem). Following application of the Refine Enhancement reagent and labeling with the Refine Polymer, the tissue sections were treated with hydrogen peroxide. The bound immune complex was then visualized with the online application of diaminobenzidine and subsequently counter stained with hematoxylin. Completed slides were dehydrated with alcohol, cleared with xylene and cover slipped with a permanent mounting media. The extensiveness of IHC staining was initially scored semi-quantitatively as − (absent), + [occasionally (groups of) cells positive], ++ (dispersed positivity), or +++ (widespread positivity). For comparison with other EGFRvIII detection, methods cases were classified as either immunohistochemically positive or negative.
RESULTS
Detection of EGFR copy number changes and EGFRvIII in tumor DNA
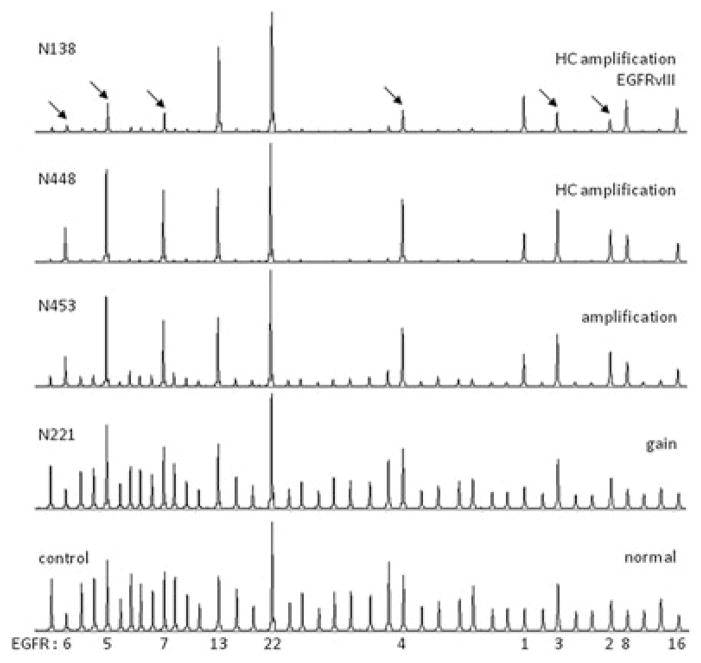
We used MLPA assay P105 to detect copy number changes of EGFR as well as the presence of the deletion variant EGFRvIII (deletion exons 2–7) in 216 diffuse gliomas, encompassing the different histopathologic subtypes and malignancy grades. Copy number changes of the EGFR gene were detected in 102 cases (47%), most frequently involving additional copies of the gene (47 gains, 19 amplifications and 33 HC amplifications), whereas losses were only occasionally detected (3 cases). Additional copies were most frequently found in GBMs (74/104; 71%) or anaplastic glioma (19/67; 28%), more frequently affecting OA-IIIs (12/28; 43%) than O-IIIs (7/25; 28%). Vice versa, the majority of cases with an (HC) amplification (n = 52) was diagnosed as GBM (81%), whereas the remaining cases included anaplastic gliomas (17%) and a single AII (2%). In cases showing a low-copy number gain (n = 47), the percentage diagnosed as GBMs was lower (68%), whereas this aberration was also identified in anaplastic (21%) and low-grade gliomas (11%). These results suggest increase of EGFR copy number in the course of malignant progression of gliomas. Examples of MLPA images reflecting different levels of EGFR copy number (normal, gain, amplification and HC amplification) are shown in Figure 1.
Figure 1.
Examples of representative peak patterns of multiplex ligation-dependent probe amplification (MLPA) analysis using assay P105. On the right, the detected epidermal growth factor receptor (EGFR) copy number changes and the presence of EGFR variant III (EGFRvIII) are indicated, whereas case numbers are indicated on the left side of the images. A reference peak pattern is shown in the lower image with indication of the peaks representing the different EGFR exons (1, 2, 3, 4, 5, 6, 7, 8, 13, 16 and 22). Cases showing increasing amounts of EGFR copy number are shown [normal, gain, amplification and high-copy (HC) amplification], as well as a case containing EGFRvIII in the background of an EGFR HC amplification. Note that peak height of EGFR probes increases relatively to the other peaks with increasing copy number. A clear decrease in peak height for exons 2–7 (indicated by arrows) relative to the other EGFR peaks is seen in the EGFRvIII positive case. Peak patterns may slightly differ when probes are exchanged in newer versions of this assay (images shown were generated using P105-B, lot 0407).
MLPA analysis additionally identified EGFRvIII in 22 tumors [17 GBMs, 4 OA-IIIs, and 1 O-III) (Table 1, Figure 1)]. Interestingly, EGFRvIII was only identified in cases that also showed additional copies of the EGFR gene, most frequently involving an (HC) amplification (21 cases). Not all cases, however, showing such an amplification contain EGFRvIII, suggesting that gene rearrangement takes place during gene amplification as a late event in glioma oncogenesis. Overall, of the 99 gliomas with additional EGFR copies, 22 (22%) contained the mutant EGFRvIII. As shown in Table 1, variation in MLPA ratios among the different probes evaluated is less when ratios are smaller (e.g., within the deletion), whereas they can be high evaluating HC number changes (e.g., outside the deletion). Although some of these differences may represent actual additional EGFR variants (as described below), it is known that the linear correlation between actual copy number and MLPA ratio is affected when evaluating (HC) amplifications.
Table 1.
Identification of EGFRvIII at the DNA, RNA and protein levels.
| Case | Tissue type | Pathology | Exon 1 | Exon 2 | Exon 3 | Exon 4 | Exon 5 | Exon 6 | Exon 7 | Exon 8 | Exon 13 | Exon 17 | Exon 22 | Copy number | EGFRvIII | MLPA | Endpoint RT-PCR | q-RT-PCR | IHC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N31 | p | GBM | 16.4 | 2.6 | 2.7 | 2.7 | 2.3 | 2.1 | 2.4 | 16.8 | 14.7 | 17.0 | 12.1 | 15.4 | 0.2 | + | − | + | + |
| N65 | f | GBM | 21.9 | 12.6 | 12.4 | 12.1 | 11.5 | 11.1 | 11.1 | 20.9 | 17.8 | 21.2 | 17.9 | 19.9 | 0.6 | + | + | + | + |
| N88 | p | OA-III | 37.7 | 9.4 | 18.2 | 19.6 | 11.4 | 17.1 | 12.6 | 53.3 | 36.3 | 33.8 | 30.6 | 38.3 | 0.4 | + | + | + | + |
| N101 | p | OA-III | 12.3 | 8.1 | 6.7 | 7.6 | 9.4 | 8.0 | 9.8 | 11.2 | 14.6 | 12.1 | 14.8 | 13.0 | 0.6 | + | + | + | + |
| N116 | f | OA-III | 11.1 | 3.4 | 2.9 | 3.2 | 3.9 | 3.5 | 3.9 | 10.0 | 11.6 | 10.9 | 11.7 | 11.1 | 0.3 | + | nd | nd | + |
| N125 | p | OA-III | 6.3 | 3.7 | 3.8 | 3.2 | 3.4 | 3.7 | 3.1 | 5.2 | 4.4 | 5.3 | 4.7 | 5.2 | 0.7 | + | + | + | + |
| N138 | f | O-III | 22.4 | 5.2 | 5.4 | 5.0 | 4.7 | 4.4 | 4.5 | 20.1 | 21.8 | 19.7 | 14.6 | 19.7 | 0.3 | + | + | + | nd |
| N156* | f | GBM | 4.3 | 2.9 | 2.9 | 3.0 | 3.2 | 3.6 | 3.3 | 4.4 | 5.2 | 4.2 | 4.5 | 4.5 | 0.7 | + | nd | nd | nd |
| N219 | f | GBM | 4.3 | 1.6 | 1.3 | 1.9 | 3.2 | 3.0 | 3.0 | 4.5 | 7.5 | 4.7 | 5.7 | 5.3 | 0.4 | + | + | + | + |
| N292 | p | GBM-O | 18.0 | 1.6 | 1.9 | 2.0 | 2.1 | 2.6 | 2.4 | 18.4 | 15.4 | 16.7 | 12.6 | 16.2 | 0.1 | + | − | + | + |
| N345 | p | GBM | 4.3 | 1.7 | 1.6 | 1.7 | 1.7 | 1.6 | 1.6 | 3.2 | 3.2 | 3.1 | 2.9 | 3.4 | 0.5 | + | − | + | + |
| N400 | f | GBM | 15.8 | 9.0 | 7.4 | 8.2 | 9.5 | 9.1 | 10.6 | 13.8 | 18.5 | 14.5 | 14.7 | 15.5 | 0.6 | + | + | + | + |
| N455* | f | GBM | 15.2 | 2.8 | 2.9 | 2.7 | 2.9 | 2.8 | 3.0 | 14.3 | 15.2 | 14.3 | 11.1 | 14.0 | 0.2 | + | nd | nd | nd |
| N516 | p | GBM | 32.3 | 21.4 | 21.3 | 16.9 | 18.1 | 18.2 | 15.2 | 24.9 | 25.0 | 24.1 | 21.7 | 25.6 | 0.7 | + | + | + | nd |
| N520 | p | GBM | 29.4 | 17.9 | 17.2 | 16.9 | 16.0 | 17.4 | 15.6 | 22.2 | 20.5 | 19.0 | 16.6 | 21.6 | 0.8 | + | − | + | nd |
| N528 | p | GBM | 35.8 | 26.8 | 27.9 | 26.1 | 30.0 | 29.2 | 29.4 | 33.7 | 35.8 | 34.5 | 38.2 | 35.6 | 0.8 | + | + | + | + |
| N530 | p | GBM | 24.0 | 15.0 | 14.2 | 14.5 | 14.4 | 13.4 | 11.6 | 18.1 | 20.0 | 16.2 | 13.9 | 18.4 | 0.8 | + | + | + | nd |
| N535 | p | GBM | 28.4 | 1.9 | 2.0 | 1.7 | 1.6 | 1.2 | 1.7 | 15.4 | 15.3 | 16.5 | 9.6 | 17.0 | 0.1 | + | + | + | nd |
| N766* | f | GBM | 1.5 | 1.1 | 1.1 | 1.0 | 0.9 | 0.9 | 0.8 | 1.4 | 1.3 | 1.4 | 1.4 | 1.4 | 0.7 | + | nd | nd | nd |
| N771 | p | GBM | 17.8 | 2.7 | 2.6 | 2.5 | 2.4 | 1.9 | 2.1 | 16.9 | 14.2 | 18.3 | 12.9 | 16.0 | 0.2 | + | + | + | nd |
| N775 | p | GBM | 8.0 | 1.1 | 1.0 | 1.0 | 1.2 | 1.0 | 1.0 | 2.4 | 2.6 | 2.9 | 7.1 | 4.6 | 0.2 | + | + | + | nd |
| N793* | f | GBM | 17.8 | 3.7 | 3.2 | 3.2 | 3.0 | 2.1 | 2.3 | 17.3 | 15.6 | 19.4 | 12.7 | 16.6 | 0.2 | + | nd | nd | nd |
| Other EGFR variant | |||||||||||||||||||
| N216† | GBM | 15.9 | 2.3 | 1.8 | 1.9 | 2.2 | 1.8 | 2.0 | 1.9 | 2.1 | 18.6 | 6.3 | 9.0 | 0.2 | − | − | − | nd | |
Due to the absence of additional tissue these cases could not be further evaluated at the RNA or protein levels (IHC). Shown are the ratios as detected using MLPA assay P105 for the EGFR exons included. Exons that are relatively deleted are indicated in gray. EGFR copy number was determined using the average ratio of exons 1, 8, 13, 17 and 22. The presence of EGFRvIII was determined by calculating: (the average MLPA ratios within the deleted exons 2–7 region) / (the average MLPA ratios outside this deleted region).
Using MLPA assay p315 evaluating all EGFR exons N216 was shown to contain a deletion exceeding the EGFRvIII region (exons 2–7) and in fact encompassing exons 2–14. Deletions exceeding exons 2–7 may also result in low EGFRvIII ratios, therefore subsequent evaluation of the individual exon ratios is required for the actual identification of EGFR variants.
Abbreviations: EGFR = epidermal growth factor receptor; EGFRvIII = EGFR variant III; f = fresh frozen tissue; GBM = glioblastoma multiforme; GBM-O = GBM with oligodendroglial features; IHC = immunohistochemistry; MLPA = multiplex ligation-dependent probe amplification; nd = not done; O-III = anaplastic oligodendroglioma; OA-III = anaplastic oligoastrocytoma; p = formalin-fixed and paraffin-embedded tissue; q-RT-PCR = quantitative RT-PCR; RT-PCR = Reverse Transcriptase PCR; + = present; − = absent.
Next to EGFRvIII, an additional EGFR variant was detected in GBM N216 (Table 1) characterized by a deletion spanning not only exons 2–7 but extending to exons 8 and 13. Further evaluation of this tumor DNA using MLPA assay P315 (evaluating all EGFR exons) revealed that the deletion encompasses exons 2–14 (data not shown). Relatively decreased or increased ratios for individual exons was occasionally (4 cases) detected and affected different exons [relatively low ratios for exon 2, 3 and 4 in N473 (GBM, EGFR gain) or for exon 2 in N12 (O-III, EGFR HC-amplification) and N88 (EGFR HC-amplification, EGFRvIII); relatively high ratios for exon 1 and 22 in N775 (GBM, EGFR amplification, EGFRvIII) or exon 1 in N535 (GBM, EGFR HC-amplification, EGFRvIII)]. Whether these reflect alternative EGFR variants requires further investigation and is beyond the scope of the current study.
Detection of EGFRvIII protein or RNA
Part of the gliomas were additionally screened for the presence of EGFRvIII protein by IHC (76 cases) and/or EGFRvIII transcripts (61 cases) by endpoint RT-PCR as well as q-RT-PCR.
IHC (Figure 2) was performed using tissue arrays including 3–4 representative cores of 2 mm for each tumor. All 11 EGFRvIII positive cases as identified by MLPA included in these arrays were shown to contain the EGFRvIII protein (Table 1). As exemplified in Figure 2, a considerable amount of heterogeneity in EGFRvIII staining was detected. While in some cores homogeneous staining of tumor cells was found (Figure 2A, N292), clear tumor heterogeneity was detected among the different cores (N292: 2 cores negative, 2 cores homogenous staining) and frequently heterogeneity was present within cores, either as regional staining (Figure 2B, N65) or as dispersed positive cells intermingled with a majority of negative tumor cells (Figure 2C, N88). The fact that no clear correlation between the MLPA EGFRvIII ratio and extent of IHC staining (semi-quantitatively scored as −, +, ++ and +++ for this purpose) could be established might be explained by this tumor heterogeneity (e.g., a few cells with very high levels of EGFR/ EGFRvIII or a lot of cells with increased levels of EGFR/ EGFRvIII), the use of micro-arrays for EGFRvIII IHC and the use of different samples of the tumor for MLPA and IHC analysis.
Figure 2.
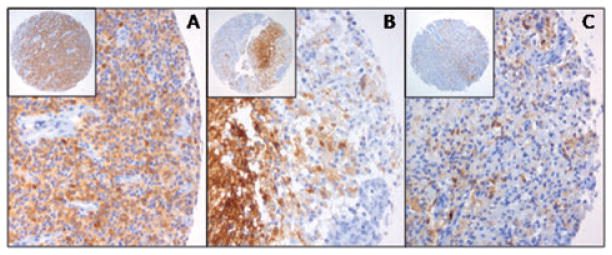
Immunohistochemical detection of epidermal growth factor receptor variant III (EGFRvIII) using tissue microarrays. Shown are cores from the EGFRvIII-positive tumors N292 (A), N65 (B) and N88 (C). EGFRvIII staining can be homogenous within a core (A), but still heterogeneous among the different cores of the same tumor (N292: only 2 out of 4 cores positive; negative cores not shown). Furthermore, within a core heterogeneity can be detected with regional differences in staining intensity (B) or even staining of individual cells in between a majority of negative tumor cells (C).
Evaluating RNA of the EGFRvIII positive cases (n = 17), differences were detected using either endpoint RT-PCR (Figure 3) or q-RT-PCR (Figure 4). By endpoint RT-PCR, the EGFRvIII fragment (234 bp) was detected in 13 of the 17 cases (Table 1), whereas the wild-type fragment (1044 bp) could only be detected in RNA isolated from frozen tumor tissue. In contrast, EGFRvIII (98 bp) and EGFR-wt (89 bp) could be identified in all positive cases using q-RT-PCR (Table 1). The detected discrepancies (only present when using FFPE tissue) most likely result from degradation of the RNA, which can be better evaluated using q-RT-PCR generating short PCR products. In order to validate the threshold set for EGFRvIII detection by MLPA we selected a relative high number of cases showing an EGFRvIII ratio close to 0.8. As shown in Figure 4, using q-RT-PCR two groups can be distinguished, either with or without EGFRvIII, which clearly correlates with MLPA EGFRvIII ratios of <0.8 or >0.8 respectively. So although the amount of EGFRvIII present in a tumor can be variable (among tumors, tumor regions or possibly even among individual tumor cells) as is reflected by EGFRvIII ratios ranging from 0 to 1, using the 0.8 threshold in MLPA analysis results in the same detection sensitivity as RNA analysis. Overall, in all 18 evaluable cases, EGFRvIII was also detected at the RNA and/or protein level, confirming the high sensitivity of this DNA-based MLPA assay. The fact that none of the 83 EGFRvIII negative cases was positive at the RNA level (n = 40) and/or at the protein level (n = 69) underlines the high specificity of the MLPA approach.
Figure 3.

Detection of epidermal growth factor receptor (EGFR) variant III (EGFRvIII) and EGFR-wild-type (EGFR-wt) using endpoint RT-PCR. Shown are representative examples of endpoint RT-PCR analysis using RNA isolated from frozen (f) or formalin-fixed and paraffin-embedded (FFPE) (p) tissue, either with (+) or without (−) EGFRvIII as identified by multiplex ligation-dependent probe amplification (MLPA). Note that due to RNA fragmentation, EGFR-wt was only detected when using frozen tissue and never using FFPE. Similarly, EGFRvIII was not always detected in EGFRvIII positive cases (N345, N31 and N520). When using frozen tissue, EGFR-wt [1044 basepairs (bp) fragment] is not as clearly detected as EGFRvIII (234 bp fragment) due to more efficient PCR amplification of the short fragments.
Figure 4.
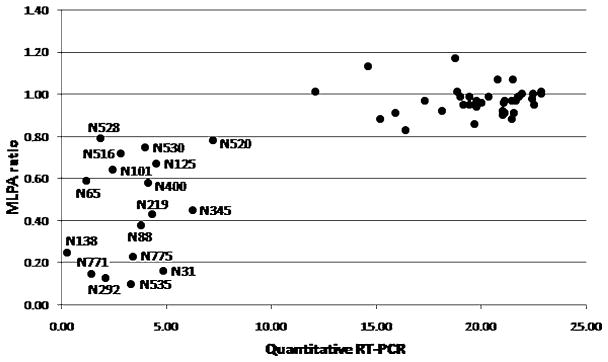
Detection of epidermal growth factor receptor variant III (EGFRvIII) by quantitative RT-PCR (q-RT-PCR). X-axis shows the cycle number when threshold is reached adjusted for PCR efficiency evaluating the housekeeping gene GAPDH. Y-axis shows the EGFRvIII ratio as detected by multiplex ligation-dependent probe amplification (MLPA). A clear correlation exists between q-RT-PCR and the EGFRvIII ratio as identified by MLPA (threshold for EGFRvIII detection: 0.8). Two different groups were identified either with (lower left) or without (upper right) EGFRvIII. In order to validate the threshold set, a relatively high number of cases with EGFRvIII ratios around 0.8 were evaluated.
DISCUSSION
Detection of EGFR/EGFRvIII aberrations
In diffuse, high-grade gliomas EGFR is frequently overexpressed, and in a significant subset of these tumors a variant of EGFR (EGFRvIII) is expressed that is constitutively active (88). EGFRvIII has originally been described as a DNA deletion of exons 2–7 (71), but is sometimes referred to as an alternative splice variant, implicating that exons 2–7 are present in the DNA but excluded from the mRNA together with the introns during splicing. This misconception is probably caused by the fact that so far EGFRvIII was generally identified at the RNA level, also because convenient assays evaluating the tumor DNA were not readily available.
Expression of EGFR-wt can be detected using IHC. However, as immunopositivity proved to be dependent on the type of antibody and the scoring scheme used, it has been suggested that clinical testing of EGFR signaling is not yet mature enough (86). EGFRvIII has been identified at the protein level by, for example, IHC (83). The EGFRvIII deletion is in-frame and generates a protein missing the majority of its extracellular domain. Antibodies have been produced that specifically recognize this truncated protein. Although IHC can easily be implemented in a routine histopathologic diagnostic setting, existing patents currently prohibit the use of an antibody recognizing EGFRvIII for clinical detection (87). To confirm the presence or absence of EGFRvIII by IHC, part of our cases were stained and heterogeneous staining patterns were detected throughout the tumor. Interestingly, in our study the correlation between the results obtained by MLPA and IHC analysis of the presence of EGFRvIII was perfect.
RNA analysis for EGFRvIII is commonly performed by endpoint RT-PCR using primers located in exons 1 and 9 (41), generating two products representing either the EGFR-wt (long fragment) or EGFRvIII (801 bp shorter). In the literature several primer sets have been described generating PCR fragments ranging from approximately 200 to 350 bp for EGFRvIII (41, 50, 56). Here we show that when using high molecular weight (undegraded) RNA isolated from snap frozen tumor tissue, EGFR-wt (1044 bp) was readily detected. In EGFRvIII negative cases the EGFR-wt fragment was clearly identified, whereas it is less abundantly present in the EGFRvIII positive cases due to more efficient amplification of shorter (i.e., EGFRvIII, 234 bp) fragments during PCR. The EGFRvIII fragment (243 bp) was found in 4 cases, all identified as EGFRvIII positive using other approaches (DNA/RNA/protein). However, such analysis becomes more complex when using degraded RNA from, for example, FFPE tissue, the type of tissue that is commonly available from routine diagnostics. In all 14 FFPE cases, EGFR-wt could not be detected, whereas EGFRvIII was detected in 9 cases, all confirmed positive at the DNA or protein level. However, for those cases in which no EGFRvIII fragment is generated it cannot be determined whether this is due to an inadequate experiment (N31, N292, N345 and N520; Table 1, Figure 2) or due to the absence of EGFRvIII (N599; Figure 2). We therefore developed new primer sets generating smaller fragments (<100 bp) and suitable for q-RT-PCR. Primer sets spanned exons 1–8 and 2–3 representing EGFRvIII (98 bp) and EGFR-wt (89 bp), respectively. Identification of EGFRvIII in all positive cases confirmed that fragment length of the RNA caused the underestimation of EGFRvIII when using the conventional endpoint RT-PCR.
So far EGFRvIII is usually not investigated at the DNA level probably because available approaches were not suitable for screening of large sets of tumors. Primer sets used for RT-PCR cannot be used for DNA analysis due to the presence of the introns. Alternatively, southern blotting is quite laborious and requires a substantial amount of intact DNA and is therefore not suitable for analysis of DNA isolated from routinely processed FFPE tissue. MLPA analysis, however, allows simultaneous copy number analysis of multiple small DNA fragments and therefore opens new opportunities for EGFRvIII detection. Here we show that by evaluating copy number changes of several EGFR exons within (exons 2, 3, 4, 5, 6 and 7) as well as outside (exons 1, 8, 13, 17 and 22) the deleted area (MLPA assay p105), cases harboring EGFRvIII can unequivocally be identified not only in snap frozen or fresh tissue, but also when using routinely processed FFPE tissue. Furthermore, the generated results provide information on the EGFR copy number status, information not directly available when IHC or (q-) RT-PCR is used for EGFRvIII evaluation. The fact that all 18 EGFRvIII positive cases as detected by MLPA indeed showed this variant at the RNA and/or protein level, whereas EGFRvIII was not detected at the protein or RNA level in cases not found to contain EGFRvIII by MLPA (n = 83), confirms the high sensitivity and specificity of the MLPA approach. Furthermore, this assay proved to be very robust as all DNAs evaluated could be reliably analyzed, and duplicate experiments performed randomly throughout the study showed similar results. Using P105B lot 0407 and more recent versions, a single analysis was sufficient for reliable analysis, whereas with earlier versions of the assay (e.g., lot 0306) occasionally a second analysis needed to be performed. Finally, in contrast to the previously established detection threshold for analysis of hemizygous 1p/19q losses (50% of tumor cells) (25), a detection limit cannot be determined for EGFR/EGFRvIII as this not only depends on the amount of tumor cells present in the biopsy, but on the amount of cells containing the aberration, the actual copy number (low level gain vs. HC amplification), and the ratios between EGFR and EGFRvIII.
Diagnostic and prognostic implications
Additional copies of EGFR were most frequently identified in high grade gliomas [74/104 GBMs (WHO grade IV) and 19/67 anaplastic (WHO grade III) gliomas], whereas EGFRvIII was detected in 22 of these cases including 17 GBMs (68%) and 5 anaplastic gliomas (22%). Our results are in line with previous reports showing that identification of EGFRvIII and/or increase of EGFR copy number is highly indicative for gliomas of high-grade malignancy and can therefore be of diagnostic use (38, 74, 78). Histopathologic revision of our EGFRvIII positive anaplastic gliomas revealed that all four OA-IIIs (at the time classified according to the WHO-2000 classification (29) would now be classified as GBM with oligodendroglial component based on the presence of necrosis as described in the current WHO-2007 classification (38). Interestingly, while the only EGFRvIII positive O-III was, also after revision, histopathologically diagnosed as pure anaplastic oligodendroglioma (without necrosis), in this tumor the characteristic genetic features of combined complete loss of 1p and 19q was not present. It has been reported for A-IIIs that half of the EGFRvIII-positive tumors when reviewed were classified as GBM (1). These results show that EGFR/EGFRvIII analysis may be helpful for identification of tumors with more aggressive behavior than suggested based on histopathology alone.
Conflicting results have been reported about the prognostic value of EGFR amplifications and the presence of EGFRvIII in gliomas. These discrepancies may not only be caused by the different techniques and tissue types used but also by the group of tumors evaluated. As discussed above, identification of EGFR/EGFRvIII in anaplastic or even low-grade gliomas may be indicative for the highest malignancy grade, and an unfavorable impact on the prognosis for patients with anaplastic gliomas including A-IIIs, OA-IIIs and O-IIIs has been described (1, 26, 27, 42, 69). In a group of gliomas already identified as highly malignant, such correlations may become less obvious (1, 37, 54, 67), although even in this group identification of EGFR/EGFRvIII aberrations has been reported by some to be a poor prognostic factor (6, 10, 16, 19, 23, 44, 52, 68). Furthermore, as amplification of EGFR has been associated with “de novo” primary GBMs that carry a worse prognosis than secondary GBMs arising from progression of a less malignant lesion (30, 77), the prognostic impact of EGFR/EGFRvIII may be dependent on the distribution of primary and secondary GBMs within the group evaluated. Applying robust analysis that is suitable for routinely processed FFPE tissue to characterize well-defined groups of gliomas encompassing all histopathologic as well as genetic subtypes will reveal the actual prognostic value of EGFR copy number changes and EGFRvIII.
Therapeutic implications
Therapeutic implications for the detection of EGFR aberrations depend on the type of therapy used. First, for the currently used (combined) radiation and chemotherapy, detection of EGFRvIII might serve as a negative predictor as introduction of EGFRvIII into cells confers resistance to radiation therapy and chemotherapy (8, 49, 82). Furthermore, overexpression of EGFR has been also been associated with resistance to radiation in vitro (4) and poor response to radiation therapy in GBM patients (2). Since deregulation of EGFR plays a role in therapy resistance, inhibition of EGFR signaling may sensitize tumors and is of particular clinical interest.
Secondly, as EGFR is frequently aberrant in (high-grade) gliomas (by gene amplification or mutational variants) and EGFR activation leads to increased proliferation and survival of (tumor) cells as well as resistance to apoptosis, EGFR-targeted therapy is a promising therapeutic approach, especially since the currently used therapies only result in a limited increase in survival. A selected group of (high-grade) glioma patients may benefit from EGFR kinase inhibitors Erlotinib (Tarceva®) or Gefitineb (Iressa®) (5, 14, 53, 57, 80). Studies aiming to identify the molecular underpinnings of responsive GBMs unfortunately did not shown consistent results. This may be due to the fact that different approaches were used investigating different biomarkers (5, 33, 41, 59, 80). Interestingly, it was clearly shown that GBMs harboring EGFRvIII in combination with PTEN expression (41) or with high levels of EGFR and low levels of p-AKT (65) are likely to respond to EGFR tyrosine kinase inhibitors. Both studies suggest that an intact PTEN–AKT pathway is necessary for clinical response to the inhibitors underlining the relevance of combined analysis of multiple markers. In addition to small molecule inhibitors, a monoclonal antibody against EGFR (Cetuximab; Erbitux, ImClone Systems, Branchbury, NJ, USA), has demonstrated (pre)clinical antitumor activity against GBMs (9, 61). Although a correlation with EGFR amplification was identified in a preclinical setting this could not (yet) be confirmed in GBM patients. Since EGFRvIII is not present in normal tissue it is an attractive target for immunotherapy and promising results were shown in (pre)clinical studies (34, 64). For example, the anti-EGFRvIII peptide vaccine CDX-110 increased (doubled) progression-free survival and overall survival in EGFRvIII-positive GBM patients when added to the conventional therapeutic regimen of TMZ (Temozolomide, Temodal®, Schering-Plough, Kenilworth, NJ, USA) and radiation therapy (18, 63).
So many different approaches/agents have been used for anti-EGFR/EGFRvIII therapies in gliomas, most of them showing efficacy in a subset of patients. The described positive correlations between the molecular background and therapy response may serve as a rational basis for stratification of patients for EGFR-targeted therapies after validation of these biomarkers in large prospective trials. The fact that up till now the value of the EGFR status as a predictive marker for such targeted therapy is not clear might partly be explained by problems with standardization of methods for assessment of biomarkers in routinely processed FFPE tissue and by the limited accessibility of a well-characterized antibody for immunohistochemical detection of EGFRvIII expression. Further molecular evaluation of larger sets of anti-EGFR-treated glioma patients, investigating different types of EGFR aberrations (amplification and mutation variants), as well as molecules involved in downstream pathways using techniques best suitable for the type of tissue available will increase our understanding of the response to these anti-EGFR agents and thereby help to build a firm basis for tailored therapy of future glioma patients.
In summary
In summary, we show that EGFRvIII can be detected with high sensitivity and specificity at the DNA, RNA and protein levels using MLPA, q-RT-PCR and IHC, respectively. The conventionally used RNA-based RT-PCR proved to be less sensitive when evaluating degraded RNA as isolated from routinely processed FFPE tissue (resulting in underestimation of EGFRvIII). Using MLPA analysis, EGFRvIII and EGFR amplification could reliably and relatively easily be detected in routinely processed FFPE glioma tissue. The major advantage of EGFR/EGFRvIII analysis at the DNA level for gliomas is that parallel analysis can be performed of other molecular markers already proven to be of clinical value and that are currently being introduced in a routine diagnostic setting.
Diagnostic, prognostic and therapeutic implications have been suggested for molecular analysis investigating EGFR copy numbers changes and the presence of EGFRvIII. Discrepancies, however, have been reported, which may reflect differences in techniques and tissue types used, biomarkers investigated, as well as characteristics of the group of tumors investigated. Using robust MLPA analysis, we investigated a broad spectrum of 216 gliomas for EGFR/EGFRvIII aberrations as these can be of additional diagnostic and prognostic use, increasing the accuracy of identification of high grade malignancy. Predictive implications for different anti-EGFR therapies are to be expected based on reports clearly describing specific molecular characteristics for the responsive gliomas.
Using robust assays investigating different EGFR aberrations simultaneously (copy number changes and the presence of EGFRvIII) and preferably in combination with other relevant aberrations, in a clinically well-documented group of patients will further elucidate the true diagnostic, prognostic and therapeutic values of this analysis. As the MLPA platform can also be used for detection of other relevant biomarkers in gliomas (1p/19q loss, PTEN loss, MGMT promotor hypermethylation) (26, 28, 42), MLPA analysis is a promising and versatile tool for focused clinical implementation of molecular diagnostics (and thereby improving tailor-made decision making) of individual glioma patients.
Acknowledgments
This project was sponsored by the Dutch Cancer Society (KWF: KUN 2004-3143, 2008-4214), the Pauline van Everdingen Foundation, the Pediatric Brain Tumor Foundation (USA) and NIH 5-P50-CA108786. We thank James Burchette and Kathryn Perkinson for the immunologic detection of EGFRvIII, and Lonneke Wigman, Martijn Kramer and Marjolijn Klomp for their contribution to the MLPA analysis.
References
- 1.Aldape KD, Ballman K, Furth A, Buckner JC, Giannini C, Burger PC, et al. Immunohistochemical detection of EGFRvIII in high malignancy grade astrocytomas and evaluation of prognostic significance. J Neuropathol Exp Neurol. 2004;63:700–707. doi: 10.1093/jnen/63.7.700. [DOI] [PubMed] [Google Scholar]
- 2.Barker FG, 2nd, Simmons ML, Chang SM, Prados MD, Larson DA, Sneed PK, et al. EGFR overexpression and radiation response in glioblastoma multiforme. Int J Radiat Oncol Biol Phys. 2001;51:410–418. doi: 10.1016/s0360-3016(01)01609-1. [DOI] [PubMed] [Google Scholar]
- 3.Cairncross JG, Ueki K, Zlatescu MC, Lisle DK, Finkelstein DM, Hammond RR, et al. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J Natl Cancer Inst. 1998;90:1473–1479. doi: 10.1093/jnci/90.19.1473. [DOI] [PubMed] [Google Scholar]
- 4.Chakravarti A, Delaney MA, Noll E, Black PM, Loeffler JS, Muzikansky A, Dyson NJ. Prognostic and pathologic significance of quantitative protein expression profiling in human gliomas. Clin Cancer Res. 2001;7:2387–2395. [PubMed] [Google Scholar]
- 5.Cloughesy TF, Yung A, Vredenberg J, Aldape K, Eberhard D, Prados M, et al. Phase II study of erlotinib in recurrent GBM: molecular predictors of outcome (abstract# 1507) Proc Am Soc Clin Oncol. 2005;41:115. [Google Scholar]
- 6.Dehais C, Laigle-Donadey F, Marie Y, Kujas M, Lejeune J, Benouaich-Amiel A, et al. Prognostic stratification of patients with anaplastic gliomas according to genetic profile. Cancer. 2006;107:1891–1897. doi: 10.1002/cncr.22211. [DOI] [PubMed] [Google Scholar]
- 7.Ekstrand AJ, James CD, Cavenee WK, Seliger B, Pettersson RF, Collins VP. Genes for epidermal growth factor receptor, transforming growth factor alpha, and epidermal growth factor and their expression in human gliomas in vivo. Cancer Res. 1991;51:2164–2172. [PubMed] [Google Scholar]
- 8.Ekstrand AJ, Longo N, Hamid ML, Olson JJ, Liu L, Collins VP, James CD. Functional characterization of an EGF receptor with a truncated extracellular domain expressed in glioblastomas with EGFR gene amplification. Oncogene. 1994;9:2313–2320. [PubMed] [Google Scholar]
- 9.Eller JL, Longo SL, Kyle MM, Bassano D, Hicklin DJ, Canute GW. Anti-epidermal growth factor receptor monoclonal antibody cetuximab augments radiation effects in glioblastoma multiforme in vitro and in vivo. Neurosurgery. 2005;56:155–162. doi: 10.1227/01.neu.0000145865.25689.55. [DOI] [PubMed] [Google Scholar]
- 10.Feldkamp MM, Lala P, Lau N, Roncari L, Guha A. Expression of activated epidermal growth factor receptors, Rasguanosine triphosphate, and mitogen-activated protein kinase in human glioblastoma multiforme specimens. Neurosurgery. 1999;45:1442–1453. doi: 10.1097/00006123-199912000-00034. [DOI] [PubMed] [Google Scholar]
- 11.Ge H, Gong X, Tang CK. Evidence of high incidence of EGFRvIII expression and coexpression with EGFR in human invasive breast cancer by laser capture microdissection and immunohistochemical analysis. Int J Cancer. 2002;98:357–361. doi: 10.1002/ijc.10224. [DOI] [PubMed] [Google Scholar]
- 12.Haas-Kogan DA, Prados MD, Tihan T, Eberhard DA, Jelluma N, Arvold ND, et al. Epidermal growth factor receptor, protein kinase B/Akt, and glioma response to erlotinib. J Natl Cancer Inst. 2005;97:880–887. doi: 10.1093/jnci/dji161. [DOI] [PubMed] [Google Scholar]
- 13.Halatsch ME, Gehrke E, Borhani FA, Efferth T, Werner C, Nomikos P, et al. EGFR but not PDGFR-beta expression correlates to the antiproliferative effect of growth factor withdrawal in glioblastoma multiforme cell lines. Anticancer Res. 2003;23:2315–2320. [PubMed] [Google Scholar]
- 14.Halatsch ME, Schmidt U, Behnke-Mursch J, Unterberg A, Wirtz CR. Epidermal growth factor receptor inhibition for the treatment of glioblastoma multiforme and other malignant brain tumours. Cancer Treat Rev. 2006;32:74–89. doi: 10.1016/j.ctrv.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 15.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 16.Heimberger AB, Hlatky R, Suki D, Yang D, Weinberg J, Gilbert M, et al. Prognostic effect of epidermal growth factor receptor and EGFRvIII in glioblastoma multiforme patients. Clin Cancer Res. 2005;11:1462–1466. doi: 10.1158/1078-0432.CCR-04-1737. [DOI] [PubMed] [Google Scholar]
- 17.Heimberger AB, Suki D, Yang D, Shi W, Aldape K. The natural history of EGFR and EGFRvIII in glioblastoma patients. J Transl Med. 2005;3:38. doi: 10.1186/1479-5876-3-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heimberger AB, Hussain SF, Aldape KD, Sawaya R, Archer G, Friedman H, et al. Tumor-specific peptide vaccination in newly diagnosed patients with GBM (abstract) Proc Am Soc Clin Oncol. 2006;25:2529. [Google Scholar]
- 19.Hoang-Xuan K, He J, Huguet S, Mokhtari K, Marie Y, Kujas M, et al. Molecular heterogeneity of oligodendrogliomas suggests alternative pathways in tumor progression. Neurology. 2001;57:1278–1281. doi: 10.1212/wnl.57.7.1278. [DOI] [PubMed] [Google Scholar]
- 20.Holbro T, Civenni G, Hynes NE. The ErbB receptors and their role in cancer progression. Exp Cell Res. 2003;284:99–110. doi: 10.1016/s0014-4827(02)00099-x. [DOI] [PubMed] [Google Scholar]
- 21.Humphrey PA, Wong AJ, Vogelstein B, Friedman HS, Werner MH, Bigner DD, Bigner SH. Amplification and expression of the epidermal growth factor receptor gene in human glioma xenografts. Cancer Res. 1988;48:2231–2238. [PubMed] [Google Scholar]
- 22.Idbaih A, Marie Y, Pierron G, Brennetot C, Hoang-Xuan K, Kujas M, et al. Two types of chromosome 1p losses with opposite significance in gliomas. Ann Neurol. 2005;58:483–487. doi: 10.1002/ana.20607. [DOI] [PubMed] [Google Scholar]
- 23.Idbaih A, Marie Y, Lucchesi C, Pierron G, Manie E, Raynal V, et al. BAC array CGH distinguishes mutually exclusive alterations that define clinicogenetic subtypes of gliomas. Int J Cancer. 2008;122:1778–1786. doi: 10.1002/ijc.23270. [DOI] [PubMed] [Google Scholar]
- 24.Jenkins RB, Blair H, Ballman KV, Giannini C, Arusell RM, Law M, et al. A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res. 2006;66:9852–9861. doi: 10.1158/0008-5472.CAN-06-1796. [DOI] [PubMed] [Google Scholar]
- 25.Jeuken J, Cornelissen S, Boots-Sprenger S, Gijsen S, Wesseling P. Multiplex ligation-dependent probe amplification: a diagnostic tool for simultaneous identification of different genetic markers in glial tumors. J Mol Diagn. 2006;8:433–443. doi: 10.2353/jmoldx.2006.060012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeuken JW, Sprenger SH, Wesseling P, Macville MV, von Deimling A, Teepen HL, et al. Identification of subgroups of high-grade oligodendroglial tumors by comparative genomic hybridization. J Neuropathol Exp Neurol. 1999;58:606–612. doi: 10.1097/00005072-199906000-00005. [DOI] [PubMed] [Google Scholar]
- 27.Jeuken JW, Sprenger SH, Boerman RH, von Deimling A, Teepen HL, van Overbeeke JJ, Wesseling P. Subtyping of oligoastrocytic tumours by comparative genomic hybridization. J Pathol. 2001;194:81–87. doi: 10.1002/path.837. [DOI] [PubMed] [Google Scholar]
- 28.Jeuken JW, Cornelissen SJ, Vriezen M, Dekkers MM, Errami A, Sijben A, et al. MS-MLPA: an attractive alternative laboratory assay for robust, reliable, and semiquantitative detection of MGMT promoter hypermethylation in gliomas. Lab Invest. 2007;87:1055–1065. doi: 10.1038/labinvest.3700664. [DOI] [PubMed] [Google Scholar]
- 29.Kleihues P, Cavenee WK. Pathology and Genetics of Tumours of the Nervous System. IARC Press; Lyon: 2000. [Google Scholar]
- 30.Kleihues P, Ohgaki H. Primary and secondary glioblastomas: from concept to clinical diagnosis. Neurooncol. 1999;1:44–51. doi: 10.1093/neuonc/1.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kouwenhoven MC, Kros JM, French PJ, Biemondter Stege EM, Graveland WJ, Taphoorn MJ, et al. 1p/9q loss within oligodendroglioma is predictive for response to first line temozolomide but not to salvage treatment. Eur J Cancer. 2006;42:2499–2503. doi: 10.1016/j.ejca.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 32.Lal A, Glazer CA, Martinson HM, Friedman HS, Archer GE, Sampson JH, Riggins GJ. Mutant epidermal growth factor receptor up-regulates molecular effectors of tumor invasion. Cancer Res. 2002;62:3335–3339. [PubMed] [Google Scholar]
- 33.Lassman AB, Rossi MR, Raizer JJ, Abrey LE, Lieberman FS, Grefe CN, et al. Molecular study of malignant gliomas treated with epidermal growth factor receptor inhibitors: tissue analysis from North American Brain Tumor Consortium Trials 01–03 and 00–01. Clin Cancer Res. 2005;11:7841–7850. doi: 10.1158/1078-0432.CCR-05-0421. [DOI] [PubMed] [Google Scholar]
- 34.Li G, Wong AJ. EGF receptor variant III as a target antigen for tumor immunotherapy. Expert Rev Vaccines. 2008;7:977–985. doi: 10.1586/14760584.7.7.977. [DOI] [PubMed] [Google Scholar]
- 35.Libermann TA, Razon N, Bartal AD, Yarden Y, Schlessinger J, Soreq H. Expression of epidermal growth factor receptors in human brain tumors. Cancer Res. 1984;44:753–760. [PubMed] [Google Scholar]
- 36.Libermann TA, Nusbaum HR, Razon N, Kris R, Lax I, Soreq H, et al. Amplification, enhanced expression and possible rearrangement of EGF receptor gene in primary human brain tumours of glial origin. Nature. 1985;313:144–147. doi: 10.1038/313144a0. [DOI] [PubMed] [Google Scholar]
- 37.Liu L, Backlund LM, Nilsson BR, Grander D, Ichimura K, Goike HM, Collins VP. Clinical significance of EGFR amplification and the aberrant EGFRvIII transcript in conventionally treated astrocytic gliomas. J Mol Med. 2005;83:917–926. doi: 10.1007/s00109-005-0700-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol (Berl) 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lund-Johansen M, Bjerkvig R, Humphrey PA, Bigner SH, Bigner DD, Laerum OD. Effect of epidermal growth factor on glioma cell growth, migration, and invasion in vitro. Cancer Res. 1990;50:6039–6044. [PubMed] [Google Scholar]
- 40.Malden LT, Novak U, Kaye AH, Burgess AW. Selective amplification of the cytoplasmic domain of the epidermal growth factor receptor gene in glioblastoma multiforme. Cancer Res. 1988;48:2711–2714. [PubMed] [Google Scholar]
- 41.Mellinghoff IK, Wang MY, Vivanco I, Haas-Kogan DA, Zhu S, Dia EQ, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 42.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mineo JF, Bordron A, Quintin-Roue I, Loisel S, Ster KL, Buhe V, et al. Recombinant humanised anti-HER2/neu antibody (Herceptin) induces cellular death of glioblastomas. Br J Cancer. 2004;91:1195–1199. doi: 10.1038/sj.bjc.6602089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murat A, Migliavacca E, Gorlia T, Lambiv WL, Shay T, Hamou MF, et al. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26:3015–3024. doi: 10.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- 45.Nagane M, Coufal F, Lin H, Bogler O, Cavenee WK, Huang HJ. A common mutant epidermal growth factor receptor confers enhanced tumorigenicity on human glioblastoma cells by increasing proliferation and reducing apoptosis. Cancer Res. 1996;56:5079–5086. [PubMed] [Google Scholar]
- 46.Nagane M, Levitzki A, Gazit A, Cavenee WK, Huang HJ. Drug resistance of human glioblastoma cells conferred by a tumor-specific mutant epidermal growth factor receptor through modulation of Bcl-XL and caspase-3-like proteases. Proc Natl Acad Sci U S A. 1998;95:5724–5729. doi: 10.1073/pnas.95.10.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Natale RB. Biologically targeted treatment of non-small-cell lung cancer: focus on epidermal growth factor receptor. Clin Lung Cancer. 2003;5 (Suppl 1):S11–S17. doi: 10.3816/clc.2003.s.010. [DOI] [PubMed] [Google Scholar]
- 48.Nicholas MK, Lukas RV, Jafri NF, Faoro L, Salgia R. Epidermal growth factor receptor—mediated signal transduction in the development and therapy of gliomas. Clin Cancer Res. 2006;12:7261–7270. doi: 10.1158/1078-0432.CCR-06-0874. [DOI] [PubMed] [Google Scholar]
- 49.Nishikawa R, Ji XD, Harmon RC, Lazar CS, Gill GN, Cavenee WK, Huang HJ. A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc Natl Acad Sci U S A. 1994;91:7727–7731. doi: 10.1073/pnas.91.16.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okamoto I, Kenyon LC, Emlet DR, Mori T, Sasaki J, Hirosako S, et al. Expression of constitutively activated EGFRvIII in non-small cell lung cancer. Cancer Sci. 2003;94:50–56. doi: 10.1111/j.1349-7006.2003.tb01351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Olapade-Olaopa EO, Moscatello DK, MacKay EH, Horsburgh T, Sandhu DP, Terry TR, et al. Evidence for the differential expression of a variant EGF receptor protein in human prostate cancer. Br J Cancer. 2000;82:186–194. doi: 10.1054/bjoc.1999.0898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pelloski CE, Ballman KV, Furth AF, Zhang L, Lin E, Sulman EP, et al. Epidermal growth factor receptor variant III status defines clinically distinct subtypes of glioblastoma. J Clin Oncol. 2007;25:2288–2294. doi: 10.1200/JCO.2006.08.0705. [DOI] [PubMed] [Google Scholar]
- 53.Preusser M, Gelpi E, Rottenfusser A, Dieckmann K, Widhalm G, Dietrich W, et al. Epithelial growth factor receptor inhibitors for treatment of recurrent or progressive high grade glioma: an exploratory study. J Neurooncol. 2008;89:211–218. doi: 10.1007/s11060-008-9608-3. [DOI] [PubMed] [Google Scholar]
- 54.Quan AL, Barnett GH, Lee SY, Vogelbaum MA, Toms SA, Staugaitis SM, et al. Epidermal growth factor receptor amplification does not have prognostic significance in patients with glioblastoma multiforme. Int J Radiat Oncol Biol Phys. 2005;63:695–703. doi: 10.1016/j.ijrobp.2005.03.051. [DOI] [PubMed] [Google Scholar]
- 55.Quaranta M, Divella R, Daniele A, Di Tardo S, Venneri MT, Lolli I, Troccoli G. Epidermal growth factor receptor serum levels and prognostic value in malignant gliomas. Tumori. 2007;93:275–280. doi: 10.1177/030089160709300308. [DOI] [PubMed] [Google Scholar]
- 56.Rae JM, Scheys JO, Clark KM, Chadwick RB, Kiefer MC, Lippman ME. EGFR and EGFRvIII expression in primary breast cancer and cell lines. Breast Cancer Res Treat. 2004;87:87–95. doi: 10.1023/B:BREA.0000041585.26734.f9. [DOI] [PubMed] [Google Scholar]
- 57.Raizer JJ, Abrey LE, Wen P, Cloughesy T, Robins I, Fine H, et al. A phase II trial of Erlotinib (OSI-774) in patients with recurrent malignant gliomas not on EIAEDs (abstract 1502) Proc Am Soc Clin Oncol. 2004;40:107. [Google Scholar]
- 58.Raymond E, Faivre S, Armand JP. Epidermal growth factor receptor tyrosine kinase as a target for anticancer therapy. Drugs. 2000;60 (Suppl 1):15–23. doi: 10.2165/00003495-200060001-00002. discussion 41–42. [DOI] [PubMed] [Google Scholar]
- 59.Rich JN, Reardon DA, Peery T, Dowell JM, Quinn JA, Penne KL, et al. Phase II trial of gefitinib in recurrent glioblastoma. J Clin Oncol. 2004;22:133–142. doi: 10.1200/JCO.2004.08.110. [DOI] [PubMed] [Google Scholar]
- 60.Rocha-Lima CM, Soares HP, Raez LE, Singal R. EGFR targeting of solid tumors. Cancer Control. 2007;14:295–304. doi: 10.1177/107327480701400313. [DOI] [PubMed] [Google Scholar]
- 61.Sadones JCC, Sadones J, Chaskis C, Joosens E, Dhondt L, Baurain J, In ’t Veld P, et al. A stratified phase II study of cetuximab for the treatment of recurrent glioblastoma multiforme: preliminary results (abstract) Proc Am Soc Clin Oncol. 2006;24:1558. [Google Scholar]
- 62.Saikali S, Avril T, Collet B, Hamlat A, Bansard JY, Drenou B, et al. Expression of nine tumour antigens in a series of human glioblastoma multiforme: interest of EGFRvIII, IL-13Ralpha2, gp100 and TRP-2 for immunotherapy. J Neurooncol. 2007;81:139–148. doi: 10.1007/s11060-006-9220-3. [DOI] [PubMed] [Google Scholar]
- 63.Sampson JH, Aldape KD, Gilbert MR, Hassenbusch SJ, Sawaya R, Schmittling B, et al. Temozolomide as a vaccine adjuvant in GBM (abstract) Proc Am Soc Clin Oncol. 2007;25:2020. [Google Scholar]
- 64.Sampson JH, Archer GE, Mitchell DA, Heimberger AB, Bigner DD. Tumor-specific immunotherapy targeting the EGFRvIII mutation in patients with malignant glioma. Semin Immunol. 2008;20:267–275. doi: 10.1016/j.smim.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sathornsumetee S, Reardon DA, Desjardins A, Quinn JA, Vredenburgh JJ, Rich JN. Molecularly targeted therapy for malignant glioma. Cancer. 2007;110:13–24. doi: 10.1002/cncr.22741. [DOI] [PubMed] [Google Scholar]
- 66.Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shih HA, Betensky RA, Dorfman MV, Louis DN, Loeffler JS, Batchelor TT. Genetic analyses for predictors of radiation response in glioblastoma. Int J Radiat Oncol Biol Phys. 2005;63:704–710. doi: 10.1016/j.ijrobp.2005.03.059. [DOI] [PubMed] [Google Scholar]
- 68.Shinojima N, Tada K, Shiraishi S, Kamiryo T, Kochi M, Nakamura H, et al. Prognostic value of epidermal growth factor receptor in patients with glioblastoma multiforme. Cancer Res. 2003;63:6962–6970. [PubMed] [Google Scholar]
- 69.Smith JS, Tachibana I, Passe SM, Huntley BK, Borell TJ, Iturria N, et al. PTEN mutation, EGFR amplification, and outcome in patients with anaplastic astrocytoma and glioblastoma multiforme. J Natl Cancer Inst. 2001;93:1246–1256. doi: 10.1093/jnci/93.16.1246. [DOI] [PubMed] [Google Scholar]
- 70.Sok JC, Coppelli FM, Thomas SM, Lango MN, Xi S, Hunt JL, et al. Mutant epidermal growth factor receptor (EGFRvIII) contributes to head and neck cancer growth and resistance to EGFR targeting. Clin Cancer Res. 2006;12:5064–5073. doi: 10.1158/1078-0432.CCR-06-0913. [DOI] [PubMed] [Google Scholar]
- 71.Sugawa N, Ekstrand AJ, James CD, Collins VP. Identical splicing of aberrant epidermal growth factor receptor transcripts from amplified rearranged genes in human glioblastomas. Proc Natl Acad Sci U S A. 1990;87:8602–8606. doi: 10.1073/pnas.87.21.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thomas C, Ely G, James CD, Jenkins R, Kastan M, Jedlicka A, et al. Glioblastoma-related gene mutations and over-expression of functional epidermal growth factor receptors in SKMG-3 glioma cells. Acta Neuropathol (Berl) 2001;101:605–615. doi: 10.1007/s004010000332. [DOI] [PubMed] [Google Scholar]
- 73.Toth J, Egervari K, Klekner A, Bognar L, Szanto J, Nemes Z, Szollosi Z. Analysis of EGFR gene amplification, protein over-expression and tyrosine kinase domain mutation in recurrent glioblastoma. Pathol Oncol Res. 2009;15:225–229. doi: 10.1007/s12253-008-9082-4. [DOI] [PubMed] [Google Scholar]
- 74.van den Bent MJ, Kros JM. Predictive and prognostic markers in neurooncology. J Neuropathol Exp Neurol. 2007;66:1074–1081. doi: 10.1097/nen.0b013e31815c39f1. [DOI] [PubMed] [Google Scholar]
- 75.van den Bent MJ, Looijenga LH, Langenberg K, Dinjens W, Graveland W, Uytdewilligen L, et al. Chromosomal anomalies in oligodendroglial tumors are correlated with clinical features. Cancer. 2003;97:1276–1284. doi: 10.1002/cncr.11187. [DOI] [PubMed] [Google Scholar]
- 76.van Dijk MC, Rombout PD, Boots-Sprenger SH, Straatman H, Bernsen MR, Ruiter DJ, Jeuken JW. Multiplex ligation-dependent probe amplification for the detection of chromosomal gains and losses in formalin-fixed tissue. Diagn Mol Pathol. 2005;14:9–16. doi: 10.1097/01.pas.0000146701.98954.47. [DOI] [PubMed] [Google Scholar]
- 77.Van Meir EG, Kikuchi T, Tada M, Li H, Diserens AC, Wojcik BE, et al. Analysis of the p53 gene and its expression in human glioblastoma cells. Cancer Res. 1994;54:649–652. [PubMed] [Google Scholar]
- 78.Viana-Pereira M, Lopes JM, Little S, Milanezi F, Basto D, Pardal F, et al. Analysis of EGFR overexpression, EGFR gene amplification and the EGFRvIII mutation in Portuguese high-grade gliomas. Anticancer Res. 2008;28:913–920. [PubMed] [Google Scholar]
- 79.Voelzke WR, Petty WJ, Lesser GJ. Targeting the epidermal growth factor receptor in high-grade astrocytomas. Curr Treat Options Oncol. 2008;9:23–31. doi: 10.1007/s11864-008-0053-5. [DOI] [PubMed] [Google Scholar]
- 80.Vogelbaum MA, Peereboom D, Stevens GHJ, Barnett GH, Brewer C. Response rate to single agent therapy with the EGFR tyrosine kinase inhibitor erlotinib in recurrent glioblastoma multiforme: results of a phase II study (abstract TA-59) Neurooncol. 2004;6:384. [Google Scholar]
- 81.von Eyben FE. Epidermal growth factor receptor inhibition and non-small cell lung cancer. Crit Rev Clin Lab Sci. 2006;43:291–323. doi: 10.1080/10408360600728369. [DOI] [PubMed] [Google Scholar]
- 82.Weppler SA, Li Y, Dubois L, Lieuwes N, Jutten B, Lambin P, et al. Expression of EGFR variant vIII promotes both radiation resistance and hypoxia tolerance. Radiother Oncol. 2007;83:333–339. doi: 10.1016/j.radonc.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 83.Wikstrand CJ, Hale LP, Batra SK, Hill ML, Humphrey PA, Kurpad SN, et al. Monoclonal antibodies against EGFRvIII are tumor specific and react with breast and lung carcinomas and malignant gliomas. Cancer Res. 1995;55:3140–3148. [PubMed] [Google Scholar]
- 84.Wong AJ, Bigner SH, Bigner DD, Kinzler KW, Hamilton SR, Vogelstein B. Increased expression of the epidermal growth factor receptor gene in malignant gliomas is invariably associated with gene amplification. Proc Natl Acad Sci U S A. 1987;84:6899–6903. doi: 10.1073/pnas.84.19.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yamazaki H, Fukui Y, Ueyama Y, Tamaoki N, Kawamoto T, Taniguchi S, Shibuya M. Amplification of the structurally and functionally altered epidermal growth factor receptor gene (c-erbB) in human brain tumors. Mol Cell Biol. 1988;8:1816–1820. doi: 10.1128/mcb.8.4.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yip S, Iafrate AJ, Louis DN. Molecular diagnostic testing in malignant gliomas: a practical update on predictive markers. J Neuropathol Exp Neurol. 2008;67:1–15. doi: 10.1097/nen.0b013e31815f65fb. [DOI] [PubMed] [Google Scholar]
- 87.Yoshimoto K, Dang J, Zhu S, Nathanson D, Huang T, Dumont R, et al. Development of a real-time RT-PCR assay for detecting EGFRvIII in glioblastoma samples. Clin Cancer Res. 2008;14:488–493. doi: 10.1158/1078-0432.CCR-07-1966. [DOI] [PubMed] [Google Scholar]
- 88.Zandi R, Larsen AB, Andersen P, Stockhausen MT, Poulsen HS. Mechanisms for oncogenic activation of the epidermal growth factor receptor. Cell Signal. 2007;19:2013–2023. doi: 10.1016/j.cellsig.2007.06.023. [DOI] [PubMed] [Google Scholar]