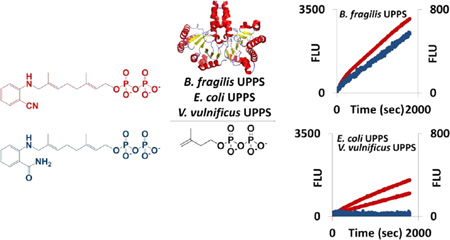
Abstract
Undecaprenyl pyrophosphate synthase (UPPS) is a critical enzyme required for the biosynthesis of polysaccharides essential for bacterial survival. In this report, we have tested the substrate selectivity of UPPS derived from the mammalian symbiont Bacteroides f ragilis, the human pathogen Vibrio vulnif icus, and the typically benign but opportunistic pathogen Escherichia coli. An anthranilamide-containing substrate, 2-amideanilinogeranyl diphosphate (2AA-GPP), was an effective substrate for only the B. f ragilis UPPS protein, yet replacing the amide with a nitrile [2-nitrileanilinogeranyl diphosphate (2CNA-GPP)] led to a compound that was fully functional for UPPS from all three target organisms. These fluorescent substrate analogues were also found to undergo increases in fluorescence upon isoprenoid chain elongation, and this increase in fluorescence can be utilized to monitor the activity and inhibition of UPPS in 96-well plate assays. The fluorescence of 2CNA-GPP increased by a factor of 2.5-fold upon chain elongation, while that of 2AA-GPP increased only 1.2-fold. The 2CNA-GPP compound was therefore more versatile for screening the activity of UPPS from multiple species of bacteria and underwent a larger increase in fluorescence that improved its ability to detect increases in chain length. Overall, this work describes the development of new assay methods for UPPS and demonstrates the difference in substrate utilization between forms of UPPS from different species, which has major implications for UPPS inhibitor development, assay construction, and the development of polysaccharide biosynthesis probes.
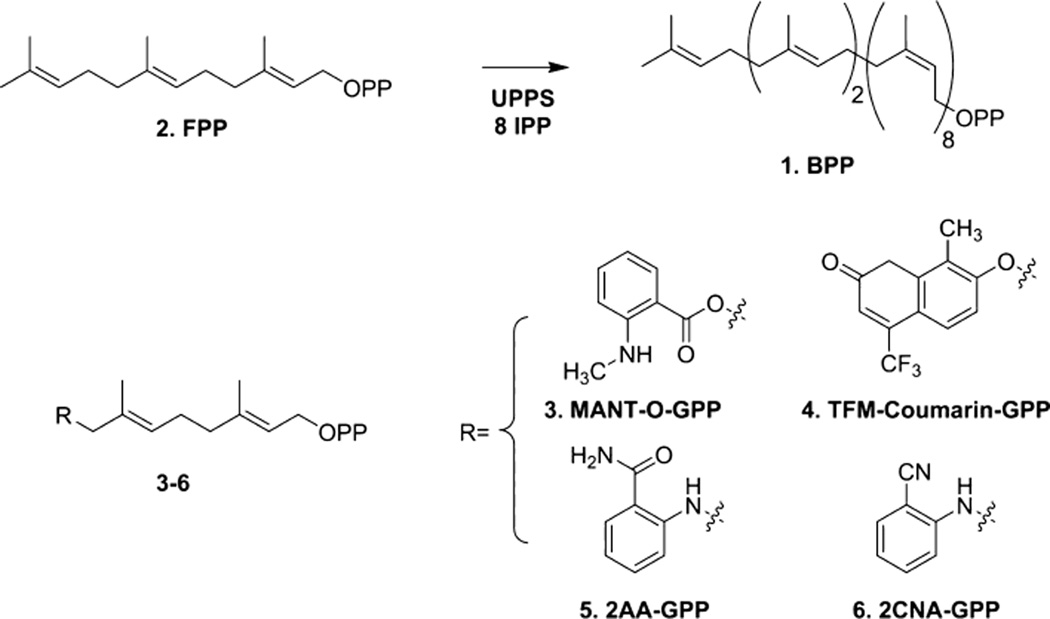
Bactoprenyl diphosphate (BPP 1) is a key platform utilized in the biosynthesis of critical bacterial glycoconjugates, including peptidoglycan and capsular polysaccharides.1–6 Undecaprenyl pyrophosphate synthase (UPPS) catalyzes the formation of BPP 1 from the 15-carbon isoprenoid farnesyl diphosphate (FPP 2) and five-carbon isopentenyl diphosphates (IPPs) (Figure 1).7,8 The critical role this enzyme plays in complex sugar biosynthesis makes it an excellent target for antibiotic development. However, numerous nonpathogenic organisms depend on functional UPPS, and antibacterial agents that target its function would affect these benign and symbiotic organisms as well as pathogens.6 Recently, several classes of compounds that inhibit UPPS function have been discovered. Bisphosphonate derivatives and tetrameric acids have exhibited clear effects on the UPPS-catalyzed biosynthesis of BPP 1.9–12 In addition, several anti-infective lead compounds without a known mechanism of action are thought to target the protein.9 Importantly, while many anti-UPPS compounds appear to have similar activity between bacterial species, several examples in which enhanced activity is observed with one type of bacteria over another have been noted.9,13
Figure 1.
Reaction catalyzed by undecaprenyl pyrophosphate synthase and analogues used to probe UPPS activity.
The function of UPPS from Escherichia coli (UPPSEc) has been extensively characterized using steady-state and single-turnover enzyme kinetics, X-ray crystallography, and site-directed mutagenesis.7,8,13–17 UPPSEc is a homodimer in which each monomer contains two α-helices and four β-sheets that form a hydrophobic tunnel, which is the site of FPP 2.elongation.7,8 Polar residues in the binding site of UPPSEc coordinate to the diphosphate head of FPP 2, while nonpolar residues stabilize the positioning of the isoprenoid chain through hydrophobic interactions. The interaction between the tunnel and isoprenoid is held so firmly that in vitro studies require a surfactant to promote product release.8,18,19 It is presumed that in vivo, longer chain products are removed from the enzyme through membrane interactions that promote turnover.
Several analogues (Figure 1) of the native FPP 2 substrate have been designed to probe the substrate chemical properties required for recognition by the protein.20–24 UPPSEc is competent to catalyze the full extension of fluorescent MANT-O-GPP 3 through the incorporation of seven Z-configuration isoprenes.20 However, a coumarin analogue, 4, accepts the addition of only a single isoprene unit.21 The UPPS from the mammalian symbiont Bacteroides f ragilis (UPPSBf) has also been probed with fluorescent analogue 2AA-GPP 5.23 This analogue was a substrate for UPPSBf, and its optical properties have been utilized to track the biosynthesis of complex bacterial polysaccharides.6,23 Interestingly, while the activity of various inhibitors on a few species of bacteria has been tested, the effect of substrate modification on the ability of various UPPS proteins to recognize different substrates has not.
Traditional methods for quantifying UPPS activity have depended on either the incorporation of radiolabeled isoprenes from IPP or coupled assays that detect the production of diphosphate.17,19A particularly interesting feature of fluorescent analogues designed to probe UPPSEc activity was the ability to use the fluorescence of these compounds to probe both the kinetics and the association of the substrate with the protein.20,21 The fluorescence of MANT-O-GPP 3 increases upon elongation of the isoprenoid chain, which was utilized to develop a continuous assay for UPPSEc activity.20 Our group has recently reported the use of 2AA-GPP 5 as a fluorescent substrate for UPPSBf and the development of a robust HPLC assay to monitor fluorescent product formation.23 It was not known whether substrate recognition restrictions were similar for forms of UPPS from different bacteria or whether this compound could be used for continuous fluorescent monitoring of UPPS activity. Here we have tested forms of UPPS from three species of bacteria and have found clear differences in the ability of the enzymes from different bacterial species to recognize the substrate. The results of this report have implications for the development of new probes for down-stream pathways and the use of particular probes in bacterial UPPS screening platforms and provide new insights into the effect of subtle differences in functionally identical proteins on substrate recognition.
METHODS
General
IPP and FPP 2 were synthesized from isopentenol and farnesol as described previously by Poulter and co-workers25 and purified by reverse phase HPLC. Full synthetic procedures of new compounds followed routes previously described by Spielmann and Chehade and are provided in the Supporting Information.26 2AA-GPP 5 was prepared as described previously.23 All analytical HPLC was performed at 1 mL/min in a 65:35 n-propanol/100 mM ammonium bicarbonate mixture using a reverse phase C18 Agilent Eclipse XDB-C18, 5 µm, 4.6 mm × 150 mm column, unless noted otherwise. Anthranilamide-containing compounds were detected with an excitation wavelength 340 nm and an emission wavelength of 450 nm. 2-Nitrileaniline-containing compounds were detected with an excitation wavelength of 340 nm and an emission wavelength of 390 nm. All HPLC was performed on an Agilent 1100 HPLC system equipped with an autosampler, a diode array detector, and a fluorescence detector. All spectrophotometry and plate assays were performed on a Molecular Devices M5 cuvette and plate reader. Electrospray ionization mass spectrometry (ESI-MS) was performed on a Velos ESI-MS system in negative ion mode. Nuclear magnetic resonance (NMR) analysis was performed on a 500 MHz JEOL spectrometer.
UPPS Cloning
The UPPS gene was amplified using the polymerase chain reaction (PCR) from B. f ragilis strain 25285 genomic DNA (ATCC) as previously described.22 Genomic DNA from E. coli was isolated from DH5α cells, and Vibrio vulnif icus genomic DNA was isolated from MO6-24/O cells provided by the J. D. Oliver Laboratory (Department of Biology, University of North Carolina at Charlotte). PCR primers were designed with BamHI and XhoI restriction sites. PCR was performed with a Promega PCR core system kit on a thermocycler with an annealing temperature of 55 °C and 35 cycles. Following amplification, the purified PCR product was doubly digested with BamHI and XhoI (New England Biolabs) and purified from a 1% agarose gel. The doubly digested insert was ligated into the purified doubly digested pET24a vector (Novagen). Ligation mixes containing 1:3 and 1:10 vector:-insert ratios were transformed into chemically competent DH5α cells and plated. Colonies showing resistance to kanamycin (encoded by the intact pET-24a vector) were cultured, doubly digested, and analyzed on a 1% agarose gel to ensure the gene of interest had been successfully incorporated. Sequencing results of the gene of interest also confirmed the successful incorporation of the UPPS gene for E. coli and V. vulnif icus UPPS. Plasmids were isolated from the DH5α cells and transformed into C41 expression cells (Lucigen) for protein expression.
UPPS Expression
A 5 mL starter culture of C41 cells transformed with each UPPS encoding vector was grown overnight. Lysogeny broth (LB, 1 L) was inoculated with 1 mL of starter culture and incubated at 37 °C until an optical density (OD600) of 0.8 was reached. The temperature was reduced to 16 °C for 30 min, and overexpression was induced with 1 mM isopropyl thiogalactoside (IPTG). After an overnight induction at 16 °C while being shaken, the culture was centrifuged for 15 min at 5000g. Pellets were resuspended in 20 mL of lysis buffer [50 mM Tris-HCl (pH 8.0), 300 mM NaCl, and 20 mM imidazole] and lysed by sonication on ice at 25% power for 2 min with a pulse of 1 s on and 1 s off. The lysate was centrifuged under vacuum at 150000g for 75 min at 4 °C, and the resulting supernatant was mixed with 1 mL of 1:1 (v/v) nickel-nitrilotriacetic acid (NI-NTA, PerfectoPro, 5 Prime Inc.) resin in lysis buffer for 30 min. The flow-through was passed twice through the column, and the column was washed thrice with 4 column volumes of wash buffer [50 mM Tris-HCl, 50 mM imidazole, and 300 mM NaCl (pH 8)]. The protein was eluted in six half-column volume fractions (1 mL) with elution buffer [50 mM Tris-HCl, 500 mM imidazole, and 300 mM NaCl (pH 8)]. SDS–PAGE was performed with aliqouts from the flow-through, wash, and elutions followed by Coomassie staining. Additionally, a Ponceau stain and Western blot was performed with an alkaline phosphatase-linked anti-T7 antibody and visualized with NBT/BCIP (Pierce). All fractions that contained UPPS were dialyzed thrice in dialysis buffer [50 mM Tris-HCl, 200 mM NaCl, and 0.5% glycerol (pH 8.0)]. Protein was further purified by gel filtration chromatography performed on a Hi-Load 16/60 Superdex 200 column (GE Healthcare) using an Akta prime plus Fast Protein Liquid Chromatography system (GE Healthcare). Purified protein was stored at −80 °C. The protein concentration was determined with a 1:1 addition of guanidinium hydrochloride and heating at 60 °C for 8 min, followed by quantification at 280 nm on a Thermo Scientific NanoDrop Spectrophotometer.
HPLC Assay General Protocol
Reaction mixtures comprised of 25 mM Bicine (pH 8.3), 0.1% DDM, 5 mM KCl, 0.5 mM MgCl2, and 1 mM IPP were prepared with 5 µM 2CNA-GPP 6 or 2AA-GPP 5 with 170 nM UPPS from each organism in a total volume of 200 µL. Reactions mixtures were incubated at 37 °C for 1 h, and then reactions were quenched with 50 µL of 2-propanol and were analyzed by HPLC as described in General. Nearly stoichiometric enzyme reaction mixtures were prepared under identical buffer conditions with 2.5 µM 2CNA-GPP 6 or 2AA-GPP 5, 0.1 mM IPP, and 1.7 µM UPPS of each type in a total volume of 200 µL. Reaction mixtures were incubated at room temperature and reactions quenched with 100 µL of 1-propanol at 2 min and then analyzed by HPLC.
Extinction Coefficient
The extinction coefficient of 2-nitrileaniline was measured at 310 nm using dilutions of pure weighed 2-nitrileaniline at 11, 33, 55, and 77 µM dissolved in 20% PrOH in 25 mM ammonium bicarbonate. The extinction coefficient was confirmed to be the same for 2CNA-GOAc at 340 nm, using a standard sample with a known concentration based on mass. Each absorbance was measured with triplicate samples.
Solvent Effects
Triplicate solutions of 2CNA-GPP 6 and 2AA-GPP 5 were prepared at 10 µM in water or n-propanol. Solutions were placed in a quartz cuvette with 2AA-GPP 5; the fluorophore was excited at 350 nm, and emissions were scanned from 390 to 500 nm. 2CNA-GPP 6 was excited at 340 nm, and the emission was scanned from 350 to 450 nm. For the comparison of BPP and GPP analogue fluorescence, stock solutions of the 2CNA-B(6)PP (where 6 is the number of Z-isoprenes) and 2CNA-GPP 6 were dissolved in a solution of 40% i-PrOH in 25 mM ammonium bicarbonate at identical concentrations (48 µM) as measured spectrophotometrically at 340 nm. The fluorescence emission from 350 to 450 nm (excitation at 340 nm) was then compared for triplicate preparations of 5 µM BPP or GPP analogue in buffer containing 0.1% n-dodecyl β-d-maltoside, 2 mM MgCl2, 20 mM KCl, and 100 mM HEPES. A similar procedure was followed with 2CNA-B(6–8)PP and 2CNA-GPP at 1 µM dissolved in water.
Plate Reader Assay General Protocol
Triplicate plate reader assays were performed in a 96-well standard nonbinding surface opaque plate, and the fluorescence was recorded for 60 min with a 20 s interval. Each reaction mixture contained 25 mM Bicine (pH 8.3), 0.1% DDM, 5 mM KCl, 0.5 mM MgCl2, 1 mM IPP, and 5 µM 2CNA-GPP 6 or 2AA-GPP 5. Each reaction mixture was incubated at 37 °C for 5 min prior to initiation with 167 nM UPPS from B. f ragilis, V. vulnif iicus, or E. coli; 50 µL of 2-propanol was used to quench each reaction. Alternatively, matched rate reaction mixtures were prepared under identical conditions except enzyme concentrations were 8.4, 167, and 334 nM for UPPS from B. f ragilis, E. coli, and V. vulnif icus, respectively, with 2CNA-GPP 6 and 0.042, 1.0, and 1.7 µM, respectively, with 2AA-GPP 5. Reactions were quenched with 50 µL of 2-propanol for HPLC analysis.
UPPS Inhibition Assays
Inhibition of 2AA-GPP 5 and 2CNA-GPP 6 with disodium diphosphate was performed via the 96-well plate reader assay. Reaction mixtures contained 0.010–3.5 mM diphosphate and 10 µM 2AA-GPP 5. Identical experiments were performed with 10 µM 2CNA-GPP 6 and 0.010–100 mM diphosphate. Inhibition assays were performed with the plate assay protocols described above with a UPPSBf concentration of 215 nM. The fluorescence was monitored to determine initial rates of product formation and plotted relative to uninhibited rates. The data were fit to the Hill equation to determine IC50 values.
Preparative UPPS Reactions for ESI-MS Analysis
Reaction mixutres were prepared using buffer conditions identical to those described above with 0.24 mM 2AA-GPP 5 or 1 mM 2CNA-GPP 6 with 15 µM UPPSBf and 4 mM IPP. Reactions were monitored using the plate reader assay until no further increase in fluorescence was detected, and then mixtures were purified on a semipreparative C18 HPLC column at 3 mL/min in a 65:35 propanol/100 mM ammonioum bicarbonate mixture. ESI-MS characterization is given in Figure 3 of the Supporting Information. HPLC analysis was performed on the enriched n = 6–8 products derived from each analogue (Figure 4 of the Supporting Information).
RESULTS
Subtle Differences in UPPS from Three Bacterial Species
Previous work from our group has focused on UPPS derived from the symbiotic microbe B. f ragilis.6,22,23 In this work, we were interested in whether the activity, and in particular substrate recognition, of this protein was similar to that of forms of UPPS derived from other organisms. Two species that had amino acid sequences moderately similar to that of UPPSBf were sought. The UPPSs from V. vulnif icus (UPPSVv) and E. coli were chosen, as these proteins were 47% identical and 65 and 67% similar to UPPSBf, respectively. UPPSVv and UPPSEc were 56% identical and 75% similar to each other. UPPS was cloned from genomic DNA isolated from V. vulnif icus strain M06-24/O and E. coli DH5α and then incorporated into pET-24a vectors that encoded an N-terminal T7 tag for highly sensitive Western blot detection and a C-terminal hexahistidine tag for purification. SDS–PAGE and Western Blot analysis confirmed the purity and identity of the proteins overexpressed in C41 E. coli cells (Figure 1 of the Supporting Information). Gel filtration analysis of the purified proteins suggested that UPPSEc and UPPSVv formed homodimers similar to what was observed previously with UPPSBf and UPPSEc (Figure 2 of the Supporting Information).22,27
Major Differences in the Ability of UPPS To Utilize 2AA-GPP
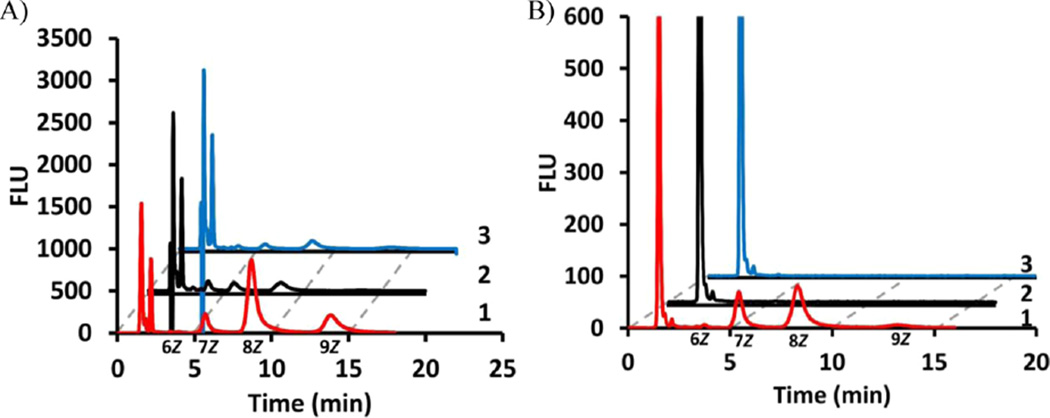
Recently, fluorescent analogue 2AA-GPP 5 has been utilized by UPPSBf to provide fluorescent substrates to track polysaccharide biosynthetic systems.23,28 It was not known whether UPPSEc or UPPSVv was also able to utilize this fluorescent analogue as a substrate. The formation of products in UPPS-catalyzed reactions with a fluorescent analogue can be readily monitored by reverse phase HPLC with fluorescence detection over relatively short analysis times.23 Reaction mixtures were prepared with each isolated UPPS protein, 2AA-GPP 5, and IPP, and then reactions were quenched with 2-propanol after incubation for 1 h. Four major product peaks were observed by HPLC analysis with retention times (tR) of 3.9, 5.6, 8.7, and 13.8 min (Figure 2A). All peaks except the peak at 3.9 min were significantly larger with the B. f ragilis protein, where the amounts of V. vulnif icus and E. coli products were 9 and 4% (tR = 13.8 min), 10 and 10% (tR = 8.7 min), and 24 and 45% (tR = 5.6 min), respectively, of the amount of product formed with the B. f ragilis protein. ESI-MS analysis suggested that these major product peaks corresponded to six to nine Z-configuration isoprenes incorporated by the UPPS proteins (Figure 3A–D of the Supporting Information). The concentration of all three proteins was increased by 1 order of magnitude to nearly stoichiometric levels, and then reactions were quenched after 2 min (Figure 2B). Under these conditions, no product was observed with the E. coli or V. vulnif icus protein, while a substantial amount of product was observed with the B. f ragilis enzyme. Taken together, these data suggested that even though the differences in the UPPS forms among species were not major, significant differences were observed for the activity of the three proteins, with respect to the ability to utilize a specific substrate.
Figure 2.
2AA-GPP is an ineffective substrate for UPPSEc and UPPSVv. HPLC analysis of 2AA-GPP with (1, red) UPPSBf, (2, black) UPPSEc, and (3, blue) UPPSVv at (A) 167 nM and (B) 1.7 µM. Chromatograms 2 and 3 are offset by 2 and 4 min, respectively. Note that the peaks representing flow-through material in panel B (2AA-GPP) are cut to give the best view of long chain isoprenoid products. In all chromatograms, the ESI-MS-identified product is indicated below the time axis with the number of Z-configuration isoprenes associated with each peak.
2CNA-GPP Is a Substrate for B. f ragilis UPPS
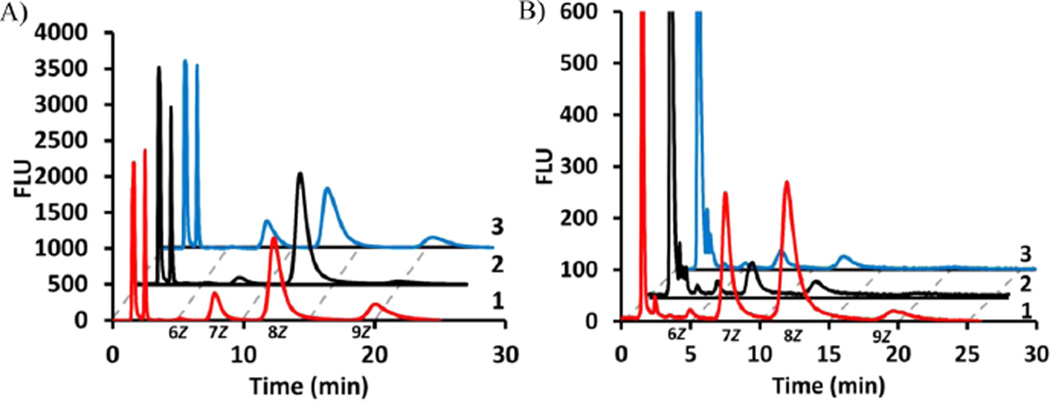
It was suspected that the large 2-amide functionality on the aniline ring may be responsible for the difference in activity between species. To test this, a new analogue was designed that was fluorescent but had a smaller nitrile at position 2 of the aniline. The 2CNA-GPP 6 (Figure 1) analogue was synthesized (Scheme 1 of the Supporting Information) using methods similar to those previously described by our group and others23,25,26,29,30 and then quantified on the basis of a measured extinction coefficient (E340 = 2700 ± 600 M−1 cm−1). To determine if the new analogue was a substrate for B. f ragilis UPPS, 2CNA-GPP 6 was mixed with IPP and UPPSBf, and then the reaction was analyzed by HPLC after incubation for 1 h (excitation at 340 nm and emission at 390 nm). Presumed long chain products were formed with HPLC tR values of 5.1, 7.7, 12.3, and 20 min (Figure 3A). ESI-MS analysis of isolated peaks (Figure 3E–H of the Supporting Information) suggested that the peaks corresponded to 2-nitrileanilinobactoprenyl diphosphates [2CNA-B(n)PP] with six to nine Z-configuration isoprenes incorporated. ESI-MS analysis of the anthranilamide and nitrileaniline products was performed on the purified product isolated from reactions with 0.2 and 1 mM substrate analogue. In several cases, mono-phosphate and diphosphate were present in these purified samples. This was surprising considering that we have previously shown significant differences in the HPLC retention of these types of materials. HPLC analysis of the isolated products (Figure 4A,B of the Supporting Information) indicated that the samples were primarily diphosphate but there was some decomposition to the monophosphate. Because the isoprenoid length was consistent with the diphosphate and monophosphate in the MS analysis and only one peak was isolated by HPLC, the decomposition likely occurred during the removal of the HPLC solvent after purification or during the MS analysis.
Figure 3.
2CNA-GPP is an effective substrate for all three UPPS proteins. HPLC analysis of 2CNA-GPP with (1, red) UPPSBf, (2, black) UPPSEc, and (3, blue) UPPSVv at (A) 167 nM and (B) 1.7 µM. Chromatograms 2 and 3 are offset by 2 and 4 min, respectively. Note that the peaks representing flow-through material are cut to give the best view of long chain isoprenoid products.
V. vulnif icus UPPS and E. coli UPPS Are Competent To Catalyze the Elongation of 2CNA-GPP
To test whether replacing the amide of 2AA-GPP 5 with the smaller nitrile in 2CNA-GPP 6 led to a compound that could be utilized by all three UPPS proteins, 2CNA-GPP 6 was tested for activity with UPPSVv and UPPSEc. HPLC analysis of the reaction mixtures demonstrated that both of these enzymes utilized the new fluorescent analogue under conditions in which very little product was observed with 2AA-GPP 5 (Figure 3A). The product profiles of the V. vulnif icus and B. f ragilis proteins were very similar, while the E. coli protein appeared to primarily produce the 8Z-isoprene product. The percentages of products with the V. vulnif icus and E. coli proteins relative to the B. f ragilis enzyme were 68 and 18% (tR = 20 min), 73 and 133% (tR = 12.3 min), and 108 and 27% (tR = 7.7 min), respectively. The enzyme concentration was increased to nearly stoichiometric concentrations, and unlike 2AA-GPP 5, significant product was formed under these conditions in just 2 min with all three enzymes, although UPPSBf was clearly more active (Figure 3B). These results suggested that just this small change in the 2-substituted aniline had a drastic effect on substrate recognition by these similar but not identical enzymes.
2AA-GPP and 2CNA-GPP Fluorescence
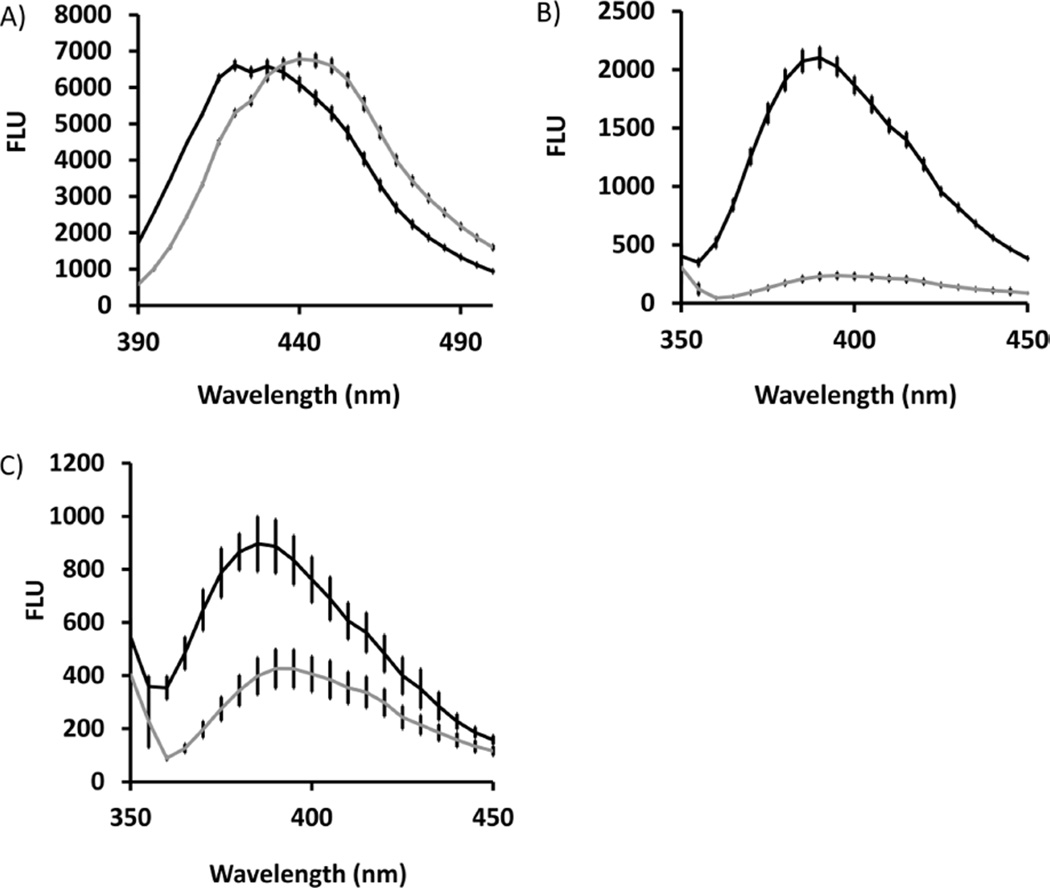
Continuous assay methods for UPPS could allow for simpler methods for monitoring the reaction rates of the UPPS proteins for further analysis of the differences between the enzymes. The previously reported assay with MANT-O-GPP 3 (Figure 1) relied on an increase in fluorescence of the analogue upon chain elongation.20 To test this solvatochromic effect with 2AAGPP 5 and 2CNA-GPP 6, each was diluted into an aqueous buffer solution or 1-propanol to determine whether there was a significant change in fluorescence in these different environments. Under these conditions, the 2AA-GPP 5 fluorescence emission spectrum shifted by 20 nm without a significant change in fluorescence intensity (Figure 4A). The 2CNA-GPP 6 fluorescence emission maximum shifted very little but increased 9-fold in the nonpolar solvent (Figure 4B). To determine if there was a significant change in fluorescence from 2CNA-GPP 6 to the bactoprenyl isoprenoids, the concentration of the substrate and potential products were verified by spectrophotometry, and then 1 µM solutions were prepared with 2CNA-GPP 6 and the six to eight Z-configuration [2CNA-B(6–8)PP] isoprenoids. Initially, the fluorescence difference was tested in water (Figure 5 of the Supporting Information) where an 11-fold increase was observed for the six to eight Z-configuration isoprenoids relative to 2CNA-GPP 6, with very little difference between the fluorescence values of the larger isoprenoids. Next, the difference in fluorescence was tested under typical reaction conditions in the presence of the n-dodecyl β-d-maltoside (DDM) surfactant and other reaction components excluding IPP and enzyme (Figure 4C). The baseline fluorescence was increased for 2CNA-GPP 6 as would be expected in the presence of the surfactant. Importantly, a 2-fold increase in fluorescence was observed even under these conditions with the 6Z-configuration isoprenoid 2CNA-B(6)PP.
Figure 4.
Solvatochromic properties of 2AA and 2CNA-GPP. (A) 2AA-GPP in water (gray line) or 1-propanol (black line) and (B) 2CNA-GPP in water (gray line) or 1-propanol (black line). (C) Difference in fluorescence between matched concentrations of 2CNA-GPP and 2CNA-B(6)PP under typical reaction conditions without enzyme or IPP. Error bars represent one standard deviation from the average value for three samples.
2CNA-GPP Is a General and Effective Continuous Probe for UPPS Activity
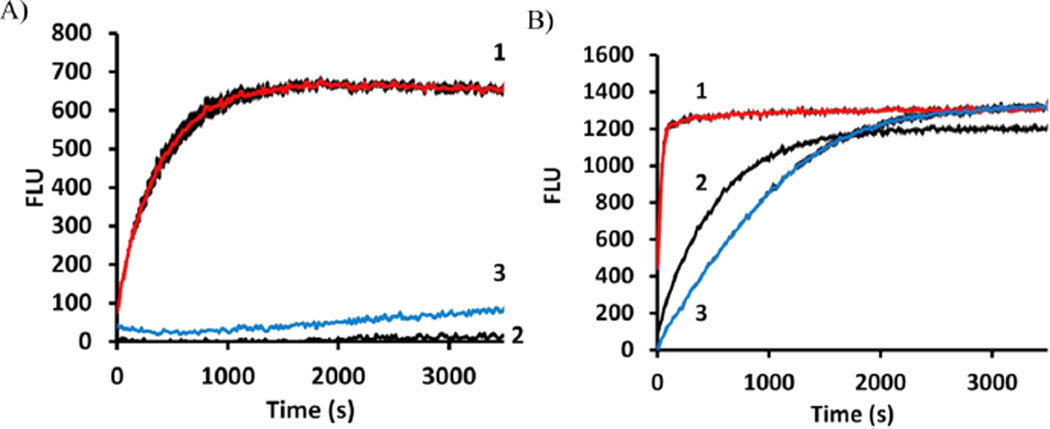
The solvatochromic changes in fluorescence cited above suggested that both 2CNA-GPP 6 and 2AA-GPP 5 could be effective for continuous monitoring of UPPS activity. The ability of these two compounds to undergo a change in fluorescence as the isoprenoid is elongated was tested in a 96-well plate format with each of the UPPS proteins isolated (Figure 5A,B). The fluorescence (excitation at 350 nm and emission at 405 nm) of 2AA-GPP 5 did increase over time with the addition of UPPSBf, but as expected, it did not substantially increase with UPPSVv or UPPSEc (Figure 5A). Importantly, the fluorescence increased with all three enzymes when 2CNA-GPP 6 was the substrate (Figure 5B). No fluorescence increase was observed when UPPS or IPP was omitted from the reaction mixture. The increase in fluorescence with 2AA-GPP 5 was only 1.2-fold, while the fluorescence of 2CNA-GPP 6 increased 2.5-fold, which was consistent with the model systems described above. These results suggested that 2CNA-GPP 6 was a more effective probe than 2AA-GPP 5 because of the lower signal-to-noise ratio associated with the larger fold enhancement in fluorescence. In addition, nitrile analogue 6 was more versatile than anthranilamide 5 because of our ability to use this analogue with forms of UPPS from several species.
Figure 5.
2AA-GPP and 2CNA-GPP are effective probes for monitoring UPPS activity. Plate monitoring of reactions of (1, red) UPPSBf, (2, black) UPPSEc, and (3, blue) UPPSVv with (A) 2AA-GPP and (B) 2CNA-GPP. Error bars represent one standard deviation from the average of three samples. Product distributions upon quenching at 3500 s are shown in Figures 2A and 3A.
Analogue Specific Difference in UPPS Activity
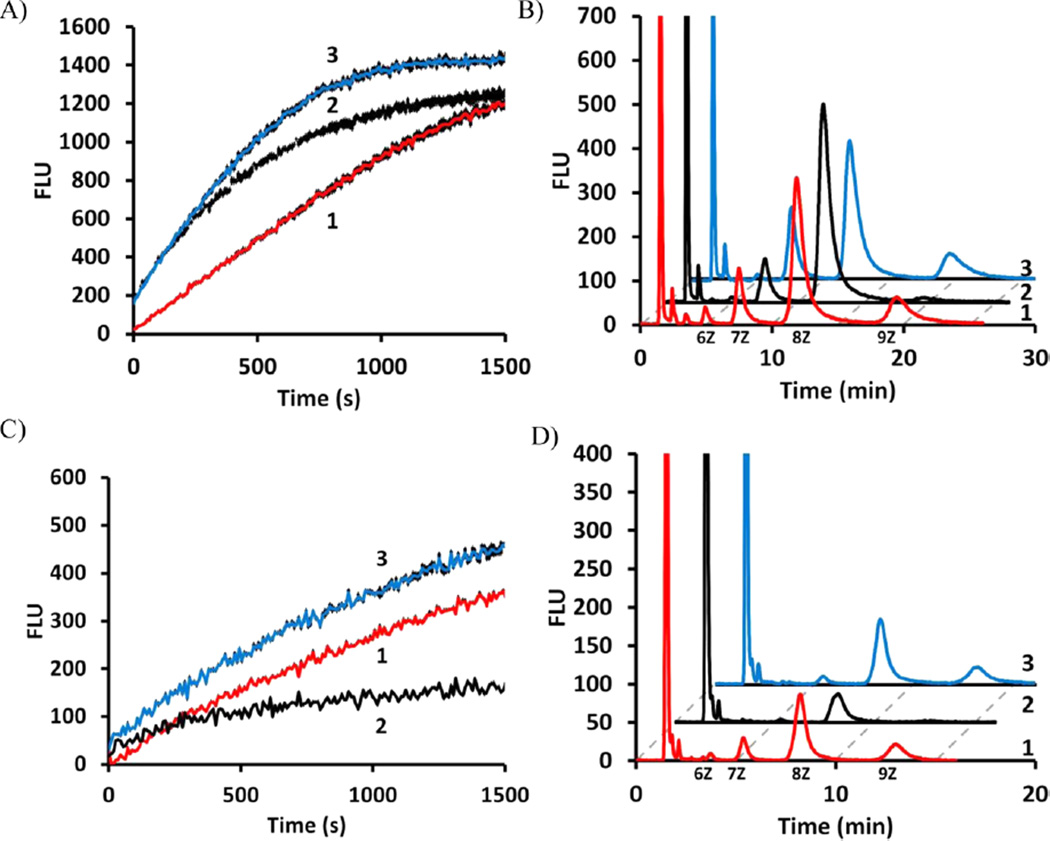
The plate assay results suggested that the UPPSEc and UPPSVv reactions were slower than the UPPSBf reactions with 2CNA-GPP 6. It was possible that 2AA-GPP 5 was a less effective substrate for all three enzymes and that the lack of activity observed with the E. coli and V. vulnif icus proteins did not represent a difference between their ability to use the analogue but instead a lack of ability to detect the slower turnover product with 2AA-GPP 5 as the substrate. To test this, reactions were prepared with UPPS concentrations that doubled the rate of 2CNA-GPP 6 utilization by UPPSEc and UPPSVv relative to that by UPPSBf, and these conditions were then tested with 2AA-GPP 5 (Figure 6A–D). These reaction conditions led to a very similar distribution of products among the three proteins with 2CNA-GPP 6 (Figure 6B). However, with 2AA-GPP 5, the reaction rates with UPPSEc and UPPSVv were 0.7 and 1.2 times the rate with UPPSBf, respectively (Figure 6C). In addition, the distributions of products from these reactions were consistent with the fluorescence enhancement (Figure 6D). Interestingly, the E. coli protein again did not appear to form the 9Z-configuration isoprenoid while both of the other two proteins did. Another assay was prepared in which the reaction rates were matched, in which the E. coli and V. vulnif icus UPPS reaction rates were 1.1 times the rate of the B. f ragilis protein (Figure 6A,B of the Supporting Information) with 2CNA-GPP 6, yet the rates of 2AA-GPP 5 utilization by the E. coli and V. vulnif icus enzymes under these same conditions were 0.3 and 0.7 times the reaction rate with the B. f ragilis enzyme, respectively (Figure 6c,d of the Supporting Information). These results suggested that even with reaction rates increased relative to that of the B. f ragilis UPPS with 2CNA-GPP 6 the rate of 2AA-GPP 5 utilization was not proportional. 2AA-GPP 5 was therefore a less effective substrate for all three enzymes and was less well utilized by the V. vulnif icus and E. coli proteins beyond just differences in the general activity of the enzymes.
Figure 6.
2AA-GPP utilization is impaired with V. vulnif icus UPPS and E. coli UPPS. (A) UPPS concentrations that doubled the 2CNA-GPP UPPSBf rate with the E. coli and V vulnif icus proteins were used. (B) Distribution of products form reactions in panel A quenched at 1500 s. (C) Change in fluorescence with 2AA-GPP with all enzyme concentrations used in panel A increased by 3.5-fold. (D) Distribution of products in panel C after quenching at 1500 s: (1, red) UPPSBf, (2, black) UPPSEc, and (3, blue) UPPSVv. Chromatograms 2 and 3 are offset by 2 and 4 min, respectively. Note that the peaks representing flow-through material are cut to give the best view of long chain isoprenoid products.
Monitoring UPPS Inhibition by a Plate Assay
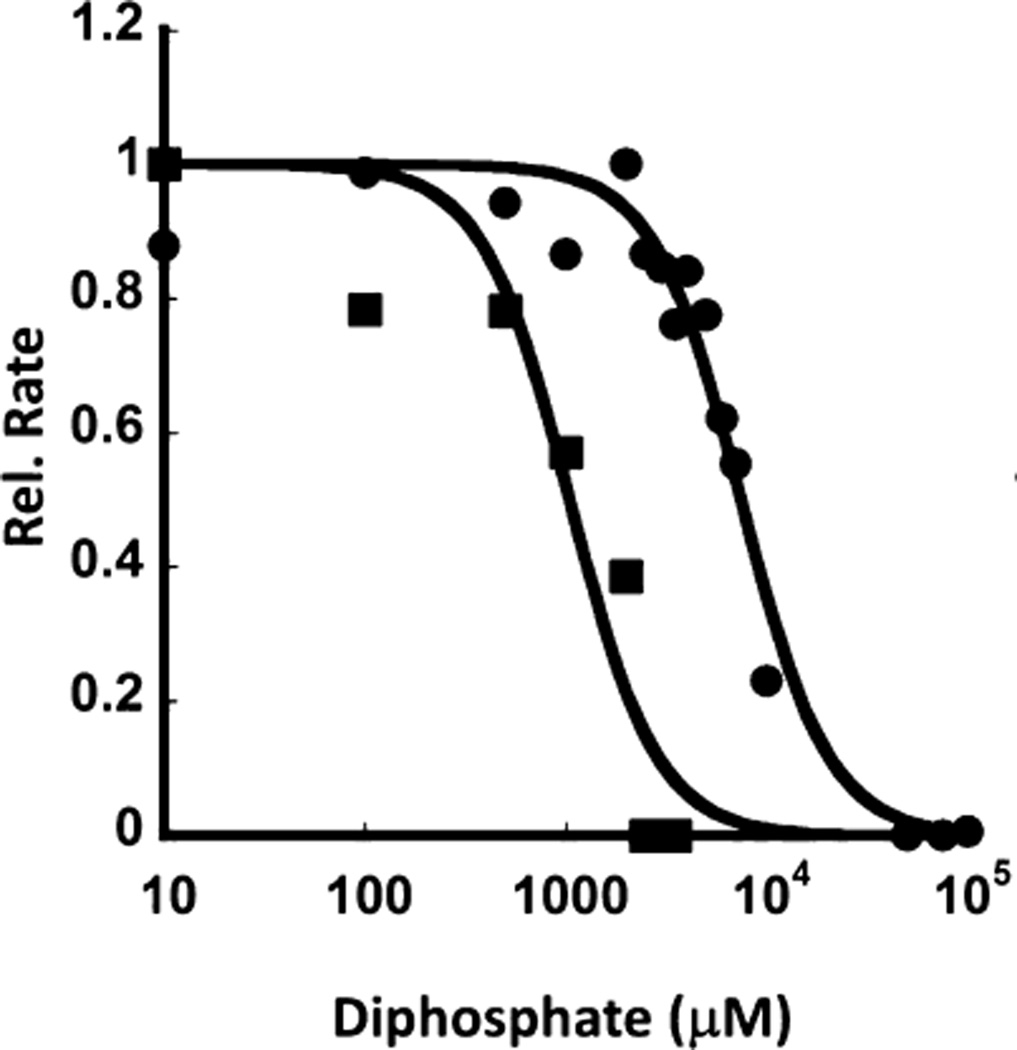
We next tested whether the difference in UPPS activity for the analogues affected their ability to be inhibited. The ability of diphosphate to act as an inhibitor was tested using the 96-well plate assay described above (Figure 7). We found that with 10 µM 2AA-GPP 5 and varying concentrations of diphosphate, the diphosphate IC50 with UPPSBf was 1.0 ± 0.2 mM. In parallel inhibition assays containing 10 µM 2CNA-GPP 6, the IC50 of diphosphate was found to be 7.4 ± 0.5 mM with UPPSBf. These results suggest that more diphosphate is required in competitions with 2CNA-GPP 6 than with 2AA-GPP 6, again suggesting that nitrile analogue 6 is a more effective substrate with UPPSBf than anthranilamide analogue 5.
Figure 7.
UPPS inhibition by diphosphate. 2CNA-GPP (●) and 2AA-GPP (■) inhibition by diphosphate. Reaction mixtures containing each analogue and varying concentrations of diphosphate were monitored using the 96-well plate assay. Rates are reported relative to that of the uninhibited reaction.
DISCUSSION
UPPS is a critical protein for the biosynthesis of complex bacterial polysaccharides and has become an important potential target for new antibacterial agents.9,31 The ability to differentiate between specific types of bacteria could lead to methods for targeting bacterial species selectively. In this study, we have found that while forms of UPPS from a variety of species catalyze identical reactions, differences in these proteins can alter their ability to utilize specific substrates. Importantly, we have observed that the nitrile in 2CNA-GPP 6 has little effect on the utilization of UPPS from three different bacteria, yet the amide of 2AA-GPP 5 does appear to affect which species can utilize this alternative substrate effectively. We propose that this difference in the ability of these enzymes to utilize these analogues is due to the size of the nitrile relative to the amide. However, other properties of the functional group could also play a role, including electronics and polarity. While this report identifies a species specific difference, current work is focused on the structure–activity relationships associated with this difference.
While it is clear that very subtle changes in analogue structure affect the ability of three different UPPS enzymes to utilize them, it is not clear from the protein perspective how these differences come about. It was surprising that three proteins that evolved to have the same biological function would show such a dramatic difference in activity generally and with the anthranilamide substrate analogue. Homology models have been constructed for UPPSBf and UPPSVv to compare with the X-ray crystal structure of UPPSEc (data not shown). On the basis of these models and sequence alignments, several amino acid residues located near the enzyme active site could be responsible for differences in selectivity. However, site-directed mutagenesis work, thus far, has not positively identified residues that alter the selectivity of these proteins (data not shown). Ongoing work is focused on discovering the precise differences in these proteins that lead to the observed variation in activity. While specific residues could be responsible, the selectivity of the proteins may also be governed by several residues rather than a few discriminating amino acids. Both phylogenetic and rational site-directed mutagenesis approaches could be keys to identifying specific amino acids that are responsible for the differences in activity between the proteins.
In our previous work, we have focused on the development of probes to track the biosynthesis of complex polysaccharides in bacteria.6,22,23,28 The strongly fluorescent anthranilamide has been extremely useful for this application. The nitrile analogue developed in this work as a fluorophore is not as potent as the anthranilamide. However, this work highlights a key advantage to this less potent fluorophore, which instead has enhanced sensitivity to environmental changes. The development of the solvatochromic fluorophores, MANT-O-GPP 3,20 2AA-GPP 5,23 and 2CNA-GPP 6, allows continuous monitoring of the UPPS-catalyzed reaction and can in turn provide rapid insight into kinetics and inhibition. Coupling this system to an HPLC assay allows the relatively rapid analysis of the UPPS product profile, as well. Here we have shown that the development of these analogues as UPPS probes should take into account the species from which the UPPS is derived. It is important to note that selectivity may differ with other organisms, as it is possible that forms of UPPS from other species may more readily utilize the amide-containing analogue over the nitrile. In addition, the results described in this report have important implications for the design of new probes for polysaccharide biosynthetic systems. The results demonstrate that forms of UPPS from other organisms may be an alternative source for the development of new probes that cannot be elongated by the E. coli or B. f ragilis UPPS protein.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. James D. Oliver for providing V. vulnif icus genomic DNA, Jennifer Stanziale for the expression of UPPSBf, and Katelyn Erickson for collection of the ESI-MS data.
Funding
This work was supported by National Institutes of Health AREA Grant R15GM100402 (J.M.T.) and National Science Foundation Instrumentation Grant 1337873 (Department of Chemistry, University of North Carolina at Charlotte).
ABBREVIATIONS
- UPPS
undecaprenyl pyrophosphate synthase
- 2AA-GPP
2-amideanilinogeranyl diphosphate
- 2CNA-GPP
2-nitrileanilinogeranyl diphosphate
- BPP
bactoprenyl diphosphate
- FPP
farnesyl diphosphate
- IPP
isopentenyl diphosphate
- MANT-O-GPP
(2E,6E)-8-O-(N-methyl-2-aminobenzoyl)-3,7-dimethyl-2,6-octadien-1-diphosphate
- 4NA-GPP
4-nitroanilinogeranyl diphosphate
- HPLC
high-performance liquid chromatography
- DDM
n-dodecyl β-d-maltoside
- PCR
polymerase chain reaction
- LB
lysogeny broth
- OD
optical density
- IPTG
isopropyl thiogalactoside
- NI-NTA
nickel-nitrilotriacetic acid
- SDS–PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- NBT/BCIP
nitro-blue tetrazolium chloride/5-bromo-4-chloro-3′-indolyl phosphate p-toluidine salt.
Footnotes
ASSOCIATED CONTENT
S Supporting Information
SDS–PAGE and Western blot analysis of all UPPS proteins used, SEC results with standards and each UPPS expressed, ESI-MS data of bactoprenyl isoprenoids, differences in fluorescence between 2CNA-GPP 6 and 2CNA-B(6–9)PP in water, matched rate 2CNA-GPP 6 and 2AA-GPP 5 plate assays and HPLC product profiles, full descriptions of synthetic procedures for 2CNA-GPP 6, synthesis scheme for 2CNA-GPP 6, and 1H NMR, 13C NMR, 31P NMR, and ESI-MS data on all new compounds. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Huang SH, Wu WS, Huang LY, Huang WF, Fu WC, Chen PT, Fang JM, Cheng WC, Cheng TJ, Wong CH. New continuous fluorometric assay for bacterial transglycosylase using Forster resonance energy transfer. J. Am. Chem. Soc. 2013;135:17078–17089. doi: 10.1021/ja407985m. [DOI] [PubMed] [Google Scholar]
- 2.Faridmoayer A, Fentabil MA, Haurat MF, Yi W, Woodward R, Wang PG, Feldman MF. Extreme Substrate Promiscuity of the Neisseria Oligosaccharyl Transferase Involved in Protein O-Glycosylation. J. Biol. Chem. 2008;283:34596–34604. doi: 10.1074/jbc.M807113200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perlstein DL, Zhang Y, Wang TS, Kahne DE, Walker S. The direction of glycan chain elongation by peptidoglycan glycosyltransferases. J. Am. Chem. Soc. 2007;129:12674–12675. doi: 10.1021/ja075965y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen MM, Weerapana E, Ciepichal E, Stupak J, Reid CW, Swiezewska E, Imperiali B. Polyisoprenol specificity in the Campylobacter jejuni N-linked glycosylation pathway. Biochemistry. 2007;46:14342–14348. doi: 10.1021/bi701956x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.James DB, Gupta K, Hauser JR, Yother J. Biochemical activities of Streptococcus pneumoniae serotype 2 capsular glycosyltransferases and significance of suppressor mutations affecting the initiating glycosyltransferase Cps2E. J. Bacteriol. 2013;195:5469–5478. doi: 10.1128/JB.00715-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mostafavi AZ, Troutman JM. Biosynthetic assembly of the Bacteroides fragilis capsular polysaccharide A precursor bactoprenyl diphosphate-linked acetamido-4-amino-6-deoxygalactopyranose. Biochemistry. 2013;52:1939–1949. doi: 10.1021/bi400126w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang SY, Ko TP, Chen APC, Wang AHJ, Liang PH. Substrate binding mode and reaction mechanism of undecaprenyl pyrophosphate synthase deduced from crystallographic studies. Protein Sci. 2004;13:971–978. doi: 10.1110/ps.03519904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang SY, Ko TP, Liang PH, Wang AHJ. Catalytic mechanism revealed by the crystal structure of undecaprenyl pyrophosphate synthase in complex with sulfate, magnesium, and triton. J. Biol. Chem. 2003;278:29298–29307. doi: 10.1074/jbc.M302687200. [DOI] [PubMed] [Google Scholar]
- 9.Zhu W, Zhang Y, Sinko W, Hensler ME, Olson J, Molohon KJ, Lindert S, Cao R, Li K, Wang K, Wang Y, Liu YL, Sankovsky A, de Oliveira CA, Mitchell DA, Nizet V, McCammon JA, Oldfield E. Antibacterial drug leads targeting isoprenoid biosynthesis. Proc. Natl. Acad. SciU.SA. 2013;110:123–128. doi: 10.1073/pnas.1219899110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo RT, Cao R, Liang PH, Ko TP, Chang TH, Hudock MP, Jeng WY, Chen CKM, Zhang YH, Song YC, Kuo CJ, Yin FL, Oldfield E, Wang AHJ. Bisphosphonates target multiple sites in both cis- and transprenyltransferases. Proc. Natl. Acad. SciU.SA. 2007;104:10022–10027. doi: 10.1073/pnas.0702254104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durrant JD, Cao R, Gorfe AA, Zhu W, Li J, Sankovsky A, Oldfield E, McCammon JA. Non-bisphosphonate inhibitors of isoprenoid biosynthesis identified via computer-aided drug design. Chem. Biol. Drug Des. 2011;78:323–332. doi: 10.1111/j.1747-0285.2011.01164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peukert S, Sun Y, Zhang R, Hurley B, Sabio M, Shen X, Gray C, Dzink-Fox J, Tao J, Cebula R, Wattanasin S. Design and structure-activity relationships of potent and selective inhibitors of undecaprenyl pyrophosphate synthase (UPPS): Tetramic, tetronic acids and dihydropyridin-2-ones. Bioorg. Med. Chem. Lett. 2008;18:1840–1844. doi: 10.1016/j.bmcl.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 13.Kuo CJ, Guo RT, Lu IL, Liu HG, Wu SY, Ko TP, Wang AHJ, Liang PH. Structure-based inhibitors exhibit differential activities against Helicobacter pylori and Escherichia coli undecaprenyl pyrophosphate synthases. J. Biomed. Biotechnol. 2008 doi: 10.1155/2008/841312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang SY, Chen YK, Wang AH, Liang PH. Identification of the active conformation and the importance of length of the flexible loop 72–83 in regulating the conformational change of undecaprenyl pyrophosphate synthase. Biochemistry. 2003;42:14452–14459. doi: 10.1021/bi035283x. [DOI] [PubMed] [Google Scholar]
- 15.Guo RT, Ko TP, Chen AP, Kuo CJ, Wang AH, Liang PH. Crystal structures of undecaprenyl pyrophosphate synthase in complex with magnesium, isopentenyl pyrophosphate, and farnesyl thiopyrophosphate: Roles of the metal ion and conserved residues in catalysis. J. Biol. Chem. 2005;280:20762–20774. doi: 10.1074/jbc.M502121200. [DOI] [PubMed] [Google Scholar]
- 16.Teng KH, Liang PH. Structures, mechanisms and inhibitors of undecaprenyl diphosphate synthase: A cisprenyltransferase for bacterial peptidoglycan biosynthesis. Bioorg. Chem. 2012;43:51–57. doi: 10.1016/j.bioorg.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 17.Pan JJ, Chiou ST, Liang PH. Product distribution and pre-steady-state kinetic analysis of Escherichia coli undecaprenyl pyrophosphate synthase reaction. Biochemistry. 2000;39:10936–10942. doi: 10.1021/bi000992l. [DOI] [PubMed] [Google Scholar]
- 18.Pan JJ, Ramamoorthy G, Poulter CD. Dependence of the Product Chain-Length on Detergents for Long-Chain E-Polyprenyl Diphosphate Synthases. Biochemistry. 2013;52:5002–5008. doi: 10.1021/bi400681d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, Huang JZ, Jiang XH, Seefeld M, McQueney M, Macarron R. The effect of triton concentration on the activity of undecaprenyl pyrophosphate synthase inhibitors. J. Biomol. Screening. 2003;8:712–715. doi: 10.1177/1087057103258185. [DOI] [PubMed] [Google Scholar]
- 20.Teng KH, Chen APC, Kuo CJ, Li YC, Liu HG, Chen CT, Liang PH. Fluorescent substrate analog for monitoring chain elongation by undecaprenyl pyrophosphate synthase in real time. Anal. Biochem. 2011;417:136–141. doi: 10.1016/j.ab.2011.05.043. [DOI] [PubMed] [Google Scholar]
- 21.Chen APC, Chen YH, Liu HP, Li YC, Chen CT, Liang PH. Synthesis and application of a fluorescent substrate analogue to study ligand interactions for undecaprenyl pyrophosphate synthase. J. Am. Chem. Soc. 2002;124:15217–15224. doi: 10.1021/ja020937v. [DOI] [PubMed] [Google Scholar]
- 22.Lujan DK, Stanziale JA, Mostafavi AZ, Sharma S, Troutman JM. Chemoenzymatic synthesis of an isoprenoid phosphate tool for the analysis of complex bacterial oligosaccharide biosynthesis. Carbohydr. Res. 2012;359:44–53. doi: 10.1016/j.carres.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mostafavi AZ, Lujan DK, Erickson KM, Martinez CD, Troutman JM. Fluorescent probes for investigation of isoprenoid configuration and size discrimination by bactoprenolutilizing enzymes. Bioorg. Med. Chem. 2013;21:5428–5435. doi: 10.1016/j.bmc.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujikura K, Maki Y, Ohya N, Satoh M, Koyama T. Kinetic studies of Micrococcus luteus B-P 26 undecaprenyl diphosphate synthase reaction using 3-desmethyl allylic substrate analogs. Biosci., Biotechnol., Biochem. 2008;72:851–855. doi: 10.1271/bbb.70723. [DOI] [PubMed] [Google Scholar]
- 25.Davisson VJ, Woodside AB, Neal TR, Stremler KE, Muehlbacher M, Poulter CD. Phosphorylation of Isoprenoid Alcohols. J. Org. Chem. 1986;51:4768–4779. [Google Scholar]
- 26.Chehade KA, Andres DA, Morimoto H, Spielmann HP. Design and synthesis of a transferable farnesyl pyrophosphate analogue to Ras by protein farnesyltransferase. J. Org. Chem. 2000;65:3027–3033. doi: 10.1021/jo991735t. [DOI] [PubMed] [Google Scholar]
- 27.Guo RT, Ko TP, Chen APC, Kuo CJ, Wang AHJ, Liang PH. Crystal structures of undecaprenyl pyrophosphate synthase in complex with magnesium, isopentenyl pyrophosphate, and farnesyl thiopyrophosphate: Roles of the metal ion and conserved residues in catalysis. J. Biol. Chem. 2005;280:20762–20774. doi: 10.1074/jbc.M502121200. [DOI] [PubMed] [Google Scholar]
- 28.Troutman JM, Sharma S, Erickson KM, Martinez CD. Functional identification of a galactosyltransferase critical to Bacteroides fragilis capsular polysaccharide A biosynthesis. Carbohydr. Res. 2014;395:19–28. doi: 10.1016/j.carres.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chehade KAH, Kiegiel K, Isaacs RJ, Pickett JS, Bowers KE, Fierke CA, Andres DA, Spielmann HP. Photoaffinity analogues of farnesyl pyrophosphate transferable by protein farnesyl transferase. J. Am. Chem. Soc. 2002;124:8206–8219. doi: 10.1021/ja0124717. [DOI] [PubMed] [Google Scholar]
- 30.Labadie GR, Viswanathan R, Poulter CD. Farnesyl diphosphate analogues with omega-bioorthogonal azide and alkyne functional groups for protein farnesyl transferase-catalyzed ligation reactions. J. Org. Chem. 2007;72:9291–9297. doi: 10.1021/jo7017747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crunkhorn S. New antibiotics on the horizon? Nat. Rev. Drug Discovery. 2013;12:99–99. doi: 10.1038/nrd3940. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.