Abstract
Motivation
Mutating residues into alanine (alanine scanning) is one of the fastest experimental means of probing hypotheses about protein function. Alanine scans can reveal functional hot spots, i.e. residues that alter function upon mutation. In vitro mutagenesis is cumbersome and costly: probing all residues in a protein is typically as impossible as substituting by all non-native amino acids. In contrast, such exhaustive mutagenesis is feasible in silico.
Results
Previously, we developed SNAP to predict functional changes due to non-synonymous single nucleotide polymorphisms. Here, we applied SNAP to all experimental mutations in the ASEdb database of alanine scans; we identified 70% of the hot spots (≥1kCal/mol change in binding energy); more severe changes were predicted more accurately. Encouraged, we carried out a complete all-against-all in silico mutagenesis for human glucokinase. Many of the residues predicted as functionally important have indeed been confirmed in the literature, others await experimental verification, and our method is ready to aid in the design of in vitro mutagenesis.
Availability
ASEdb and glucokinase scores are available at http://www.rostlab.org/services/SNAP. For submissions of large/whole proteins for processing please contact the author.
Contact: yb2009@columbia.edu
1 INTRODUCTION
The role of a protein in an interaction pathway is arguably its most important function (Eisenberg, et al., 2000). Thus, protein-protein and protein-substrate interactions are essential for survival. Typically very few residues are essential for any protein interaction interface in the sense that mutating these significantly impacts the reaction (Bogan and Thorn, 1998; Weiss, et al., 2000); these crucial residues are often referred to as protein-protein interaction hot spots. One coarse-grained experimental probe for elucidating the function of a protein is to mutate residues that are hypothesized to be involved in function. Alanine, glycine, proline, and cysteine scanning mutagenesis (individual substitutions of residues by any of the said amino acids) are used to identify functionally important sites (Clackson and Wells, 1995; Gardsvoll, et al., 2006; Konishi, et al., 1999; Kouadio, et al., 2005; Qin, et al., 2003). Because of a variety of biophysical and technical reasons alanine scans dominate. Rarely multiple mutations are tested for the same residue (Xiang, et al., 2006; Yang, et al., 2003). The impact of mutations on function is captured by a variety of probes; one of the more accurate means is the measurement of the change in the binding energy between the wild-type (native sequence) and the mutated protein. Although large energy changes may result from destabilization of the affected proteins and from deformation of the binding sites, such dramatic alterations often indicate that a hot spot was mutated. To illustrate the relevance of hot spots to research: over 400 PubMed records mention hot spots in 2007 alone. One reasonable definition for a hot spot is that its mutation alters the binding energy by ≥1kcal/mol (Kortemme and Baker, 2002).
Computational methods can identify hot spots for proteins of known three-dimensional (3D) structure (DeLano, 2002; Guerois, et al., 2002; Shulman-Peleg, et al., 2007), and more recent attempts even spot these crucial sites from sequence (Gonzalez-Ruiz and Gohlke, 2006; Ofran and Rost, 2007). ISIS (Ofran and Rost, 2007) was the first tool to specifically predict protein-protein interaction hot spots from sequence, but estimates for the effects of single substitutions have long been around (Epstein, 1966; Vegotsky and Fox, 1962; Zuckerkandl and Pauling, 1965). The most recent methods are tailored to predict the effects of non-synonymous single nucleotide polymorphisms (SNPs), i.e. single nucleotide changes that alter the protein sequence (Bromberg and Rost, 2007; Ng and Henikoff, 2003; Ramensky, et al., 2002; Yue, et al., 2006). Such methods have not been assessed in light of large-scale alanine scans and hot spots. One reason might be that while function changes are sensed by such methods, the amount or severity of change is not. Thus, the predicted functional change may just as likely be a hot spot as it may not be.
Here we examined the potential of one particular implementation for in silico mutagenesis, namely SNAP (Bromberg and Rost, 2007), that has been optimized to predict the effect of non-synonymous SNPs on a version of the public database PMD (Kawabata, et al., 1999; Nishikawa, et al., 1994) curated by us. SNAP evaluates functional effects of single amino acid substitutions using neural networks; its output is a value from −100 (no effect) to +100 (effect). First, we established that SNAP correctly captured the effect of alanine scans extracted from ASEdb (Thorn and Bogan, 2001). Then, we assessed substitutions by amino acids other than alanine. Combining these results, we could analyze in silico to which extent alanine scans correlate with all possible mutations. For technical reasons, we confined this analysis to one particular protein with ample experimental data (hexokinase)
To the best of our knowledge this is the first comprehensive study that connects biophysical data from alanine scans with methods optimized to capture the functional effects of SNPs. Making this connection is by itself an important novelty. What makes it even more interesting is that only in silico can we comprehensively address the question as to how representative current alanine scanning is, and only by this means can we comprehensively study the effects of mutagenesis without exorbitant costs. Further large-scale testing of our pilot study is required to establish more clearly that our approach actually captures functionally important residues and hot spots.
2 METHODS
Alanine scan data
Alanine scanning data was extracted from ASEdb database (Thorn and Bogan, 2001). For each complex we recorded the name of the mutated partner, the position of the mutation, and the change in energy (ΔΔGcomplex) of stability of the given complex due to the mutation. If more than one complex was reported for the given mutant, only the complex resulting in the highest energy change was retained. For the purposes of ASEdb, ΔΔGcomplex is computed as the difference in energy of the wild-type complex (ΔGwild) as compared to the energy of the mutated complex (ΔGmut). Thus, a negative ΔΔG represents a more stable complex (ΔGmut > ΔGwild) and a positive ΔΔGcomplex represents a destabilized complex (ΔGmut < ΔGwild). We used a value of 1kCal/mol change in binding energy as cutoff for determining hot spot residues.
Computing SNAP scores
SNAP outputs a score that ranges from −100 (no effect) to +100 (strong effect). A score cutoff is chosen to classify all mutations into neutral and non-neutral. By default, positive scores define non-neutral mutations; scores ≤0 identify neutral mutations; higher scores yield stronger predictions. For this work, we recorded SNAP predictions for all 19 non-native substitutions for each mutated residue in the by ASEdb data sets. We also compiled the average over all substitution scores at each position. Accuracy (often also referred to as specificity) and coverage (also referred to as sensitivity) of all performances were computed using Eqn. 1, where TP is the number of hot spots predicted to be non-neutral, FP is the number of non-hot spots predicted to be non-neutral, and FN is the number of hot spots predicted to be neutral.
| (1) |
We assumed that all residues predicted and not observed to be functionally important were incorrect predictions (false positives). In particular, we assumed that for each protein in ASEdb there is only one binding site, namely the one probed in that experiment. This is obviously an extreme position that will considerably underestimate our levels of accuracy.
The correlation between score distributions was computed by:
| (2) |
Overlap between PMD and ASEdb
SNAP networks were trained on data from PMD (Kawabata, et al., 1999; Nishikawa, et al., 1994) which slightly overlaps with ASEdb. To avoid over-estimating performance by testing on mutants that were seen in training we aligned all sequences in ASEdb against all proteins in PMD (BLAST at e=0.001). For each of the aligned sequences we collected the mutants found in both databases and recorded their functional effects according to PMD. These were then compared to the corresponding classifications from ASEdb.
Solvent accessibility
We utilized PROFacc (Rost, 2000; Rost, 2005; Rost and Sander, 1994) to predict location of affected residues in ASEdb in protein structure. Residues were split into three classes: buried = <9% exposed surface area, intermediate = >9% and <36%, exposed = > 36%. SNAP prediction accuracy and coverage (Eqn.1) were computed separately for each accessibility class as well as over all classes.
Human hexokinase data
The sequence of human hexokinase (SWISS-PROT identifier HXK4_HUMAN; P35557; 465 amino acids) was taken from SWISS-PROT (Bairoch and Apweiler, 2000; Bairoch, et al., 2005). Four evaluations of residue importance were performed using scores from alanine, glycine, cysteine, and average substitutions. For residues with the native amino acid non-X, the SNAP score of the by-X substitution was recorded; for residues with acid X, the average SNAP score was taken.
3 RESULTS AND DISCUSSION
3.1 Results of alanine scans can be predicted
We extracted 1073 mutants from 48 distinct protein chains from ASEdb. Of these 323 were classified as hot spots at the cutoff of ≥1kcal/mol change. Using this distribution with a random model (probability of observing a hot spot at any given residue is 0.5) to predict hot spots would result in 30% accuracy at 50% coverage (Eqn. 1). With default parameters, accuracy and coverage of SNAP predictions were 36% and 70% respectively. When excluding any overlap between ASEdb and PMD (Methods), these numbers fell to 33% and 67% (Fig.1). While both of these sets of numbers significantly exceeded random, it is unclear which better estimated the method’s performance. Of 174 overlapping mutants 45 (~26%) were annotated differently between PMD and ASEdb (i.e. PMD annotated the mutant as non-neutral when the corresponding ASEdb energy change was <1 kCal/mol, or vice versa). SNAP correctly classified 20 (~44% of 45) of these according to the ASEdb energy change. This implies that SNAP did not “memorize” the training samples, but learned to make decisions based on observed patterns. Arguably, removing the overlapping mutants is therefore unnecessary and artificially reduces performance by decreasing sample diversity in the data set.
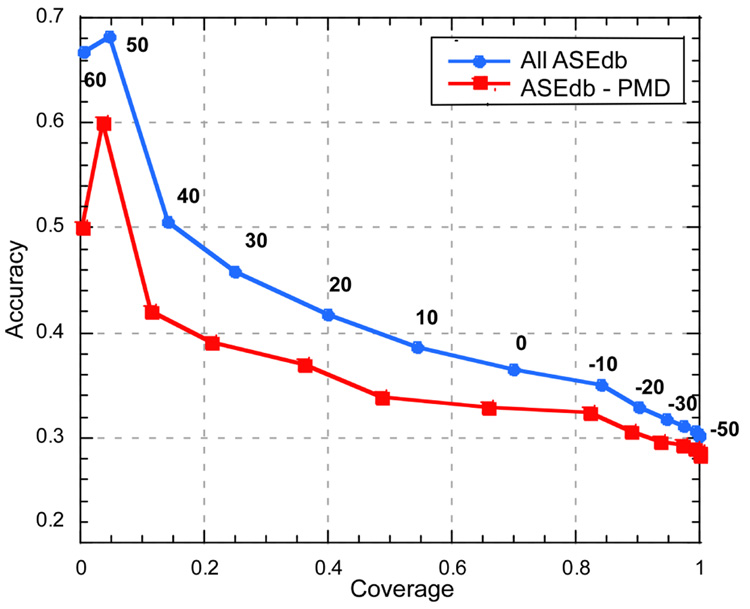
Figure 1. Variation of SNAP cutoff influences performance.
By varying the threshold in the SNAP output (−100 to +100) for considering a mutation as effecting function, we can dial through the ROC curve for interaction hot spots. On the one end, choosing a very low threshold we find all hot spots at very low accuracy (−50 on the lower right), conversely, at high positives we find few hot spots but those we find at high accuracy (50 at top left). Performance is slightly worse for the reduced data set where all mutants overlapping with PMD are removed; it is unclear which data set is better for estimating the method’s performance (Results). For the full ASEdb data set at thresholds >30, we find ~25% of the observed hot spots, and ~45% of the sites predicted at that threshold are hot spots. To compile accuracy we assumed that proteins have only one binding site and that was the one probed in ASEdb; the degree to which this statement is wrong describes the degree to which our method underestimated accuracy.
Increasing the SNAP non-neutrality cutoff (to 5 or 10, i.e. fewer residues predicted as hot spots; Fig. 1) reduced coverage without increasing accuracy correspondingly. Slightly increasing the threshold for considering a residue as a hot spot (from 1 to 2 or 2.5 kcal/mol) slightly increased coverage and decreased accuracy. In contrast, significantly increasing this energy threshold (from 1 to 4 or 4.5 kCal/mol) significant raised coverage (80% and 90% respectively). Overall, more severe (larger) changes in binding energy tended to yield higher SNAP scores. When we considered as neutral only mutations for which the binding energy remained identical between wild-type and mutant, our default method achieved 84% accuracy at 62% coverage.
Extending “no change” to an interval of ±0.2kcal/mol in the change of binding energy (approximation of experimental error in energy change measurement) yielded 68% accuracy at 63% coverage. SNAP predictions were more accurate for residues that were predicted to be buried: 80% buried hot spots were identified, 79% intermediate ones, and only 55% of the exposed hot spots.
3.2 Accuracy higher than it appears?
SNAP identifies functional effects of single amino acid substitutions. The tool was not explicitly developed to outline residues of functional importance. Surprisingly, it recognized 70% of the hot spots in the ASEdb data set, albeit it did so at very low accuracy. To some extent low accuracy undoubtedly reflected limitations in our method. However, there are three major problems with the data and the way we used them that also contributed to low accuracy. Firstly, a particular mutation may not de-stabilize an interaction enough to pass the chosen threshold. For example, the K110A mutant in the basic fibroblast growth factor (bFGF) is part of a second important binding site (Springer, et al., 1994). Mutation of this residue by an alanine slightly stabilizes the probed complex (ΔΔGcomplex=−0.33 kcal/mol). Secondly, all experiments probe only one particular reaction. A residue not predicted to be a hot spot might be involved in another interaction. For instance, the H114A mutation in angiogenin is known to greatly decrease enzymatic activity of angiogenin with respect to tRNA (Shapiro and Vallee, 1989). However, the change in energy recorded in ASEdb is of the bound angiogenin to ribonuclease inhibitor complex. The mutant described has very little effect on this binding (~0.7 kCal/mol). Thirdly, the precise threshold for considering a residue a hot spot is neither well-defined nor reaction-independent. For instance, the mutation of residue D28 in CD2 to alanine changes the binding energy of its complex with CD48 by >1.7 kcal/mol although this residue has been shown in a more detailed study to contribute little to the actual binding (Davis, et al., 1998). Instead, this particular mutation likely induces local changes in the adjacent binding site. Considering all the possible false assignments of functionality importance using alanine scans it is not surprising that a fair number of non-hot spot residues are assigned to the non-neutral class by SNAP, and vice versa. Nevertheless, as the severity of change correlated fairly well with the SNAPs scores absolutely crucial hot spots (e.g. >4 kCal/mol change) are virtually guaranteed to be included in the prediction at any cutoff.
The observation that buried hot spots are predicted more reliably could be due to the fact that buried residues are, on average, more sequence conserved than exposed residues (Rost and Sander, 1994) and that the success of SNAP is intricately linked to sequence conservation. Another reason might simply be that the experimental results are more reliable for the exceptional buried hot spots.
3.3 Predicting HXK4 functional residues
We used scores for substitutions “by alanine”, “by cysteine”, “by glycine”, and the average over all possible substitutions to high-light residues of importance in the human glucokinase protein. For alanine substitutions, the most conservative of all, a total of 214 of 465 (46%) residues in the Human glucokinase (Hexokinase IV or D; HXK4) sequence were predicted to be functionally important at the default SNAP score cutoff. For cysteine and glycine, the functional residue counts were 254 and 275, respectively. The average substitution by all 19 non-native amino acids outlined 232 residues as functionally important (Fig. 2).
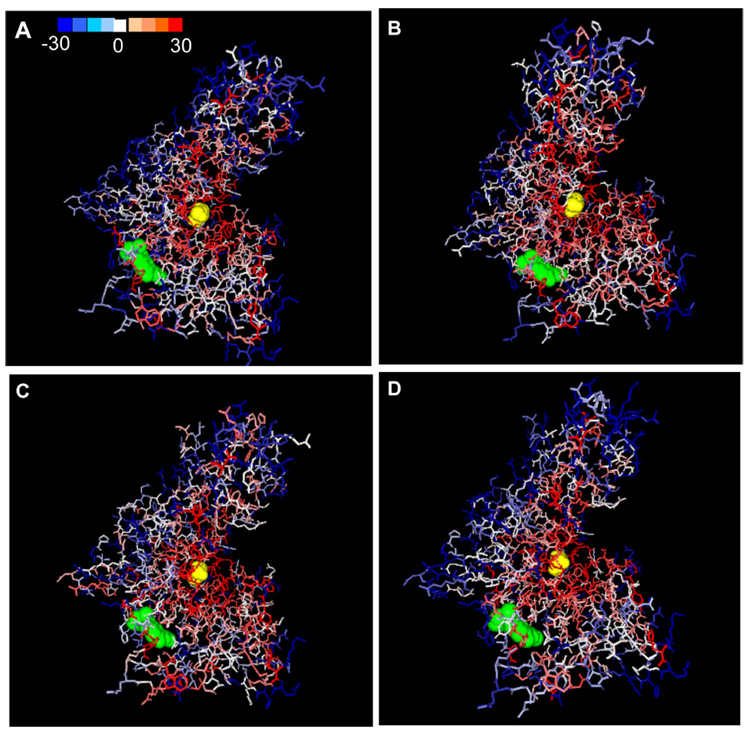
Figure 2. Comprehensive mutagenesis for human glucokinase (HXK4).
The crystal structure of HXK4 was taken from Kamata, et al (PDB: 1v4s; 2004); visualization by GRASP2 (Nichols, etal. 1991). The two ligands in the picture are glucose (yellow spheres) and a synthetic activator (green spheres). The scale of predictions ranges from blue (neutral; SNAP score <−30) to red (strong effect; SNAP score > 30). Blue indeed largely highlights regions that have not been implicated in functional changes, red highlights important residues, and white regions are unknown. Measurements shown reflect SNAP scores of mutation to alanine (A), to glycine (B), to cysteine (C) and to all 19 non-native acids [average score](D).
We chose this example because HXK4 is experimentally well studied; it is an enzyme that functions in glucose metabolism (Kamata, et al., 2004). Variants of the glucokinase encoding gene are implicated in type 2 diabetes (MODY-2 maturity onset diabetes of the young) (Vionnet, et al., 1992). The enzyme exists in three forms super-open, open, and closed. It has at least two functional sites: the glucose binding site (including residues E256, E290, T168, K169, N204 and D205) and the allosteric binding site (incl. V455, A456 and Y214, mutations of which cause a metabolic disease persistent hyperinsulemic hypoglycemia (Christesen, et al., 2002; Glaser, et al., 1998))). Kamata, et al. (2004) describe a synthetic glucokinase activator which binds the allosteric site and interacts with residues R63, M210, I211, Y215, M235, and V452. Allosteric binding is facilitated by the flexibility of connecting region I (residues 64–72), which, although not responsible for binding itself, is very important to proper function. In the super-open form glucokinase has reduced affinity for glucose and no allosteric binding site. A slow, energetically costly, conformational change transforms the protein into the open form upon glucose binding; this form has higher affinity for glucose binding, and is capable to rapidly transform into the closed form.
Binding of the allosteric regulator prevents glucokinase from going into its super-open form and thus contributes to continuous glucose metabolism (Kamata, et al., 2004). The crystal structure of HXK4 was solved by Kamata, et al in 2004 (PDB: 1v4s, Fig. 2); it captures the closed (glucose bound) conformation of HXK4. The synthetic activator loosely bound to the allosteric site is also seen. In all SNAP evaluations the glucose binding site is very well highlighted with red (implying sites predicted to be functionally important). Neighboring internal regions also shown in red somewhat correspond to the stretches of sequence involved in facilitating transformation changes. Some of the residues interacting with the synthetic compound are also lit up. Quantitative predictions for the binding residues discussed here are given in Table 1.
Table 1.
Evaluation of human glucokinase (HXK4) functional cites.
| SNAP scores | |||||
|---|---|---|---|---|---|
| Residue | Interactionsite** | Ala | Cys | Gly | Average |
| R63 | A | −7 | −21 | 0 | −18 |
| T168 | G | 26 | 29 | 27 | 28 |
| K169 | G | 35 | 44 | 37 | 38 |
| N204 | G | 26 | 28 | 26 | 28 |
| D205 | G | 28 | 29 | 27 | 29 |
| M210 | A | 21 | 26 | 23 | 23 |
| I211 | A | −17 | 0 | 8 | −2 |
| Y214 | A | −16 | −16 | −12 | −2 |
| Y215 | A | 25 | 29 | 27 | 23 |
| M235 | A | 7 | 9 | 11 | 5 |
| E256 | G | 30 | 34 | 30 | 32 |
| E290 | G | 26 | 29 | 27 | 27 |
| V452 | A | −6 | −27 | −1 | 2 |
| V455 | A | 15 | 17 | 19 | 11 |
| A456 | A | 0 | −9 | −11 | 7 |
Using SNAP scores of by alanine, cysteine, glycine and average over all possible substitutions at a given location we predicted HXK4 sites of importance. Zero and negative scores indicate neutral predictions, while positive scores are non-neutral. Higher absolute value of a given score indicates better reliability of the prediction. The glucose binding site residues were correctly identified by all methods. The allosteric interaction residues were predicted somewhat worse. Arguably, this is due to the fact that the synthetic molecule interactions do not exactly mimic the natural allosteric regulator binding patterns.
‘A’ stands for allosteric site and ‘G’ for glucose binding site.
When considering the four images of glucokinase (Fig. 2), it is intuitively clear that for this example by-alanine substitutions appear to be best in identifying functionally important residues (red predictions limited to potential functional sites and there is a higher resolution of color; i.e. very few residues for which prediction is made with low confidence). However, a more detailed study/comparison is required to determine which, if any one (as opposed to a few), substitution scoring is best at finding all functionally important residues.
3.4 Alanine scans correlated with average over all possible scans
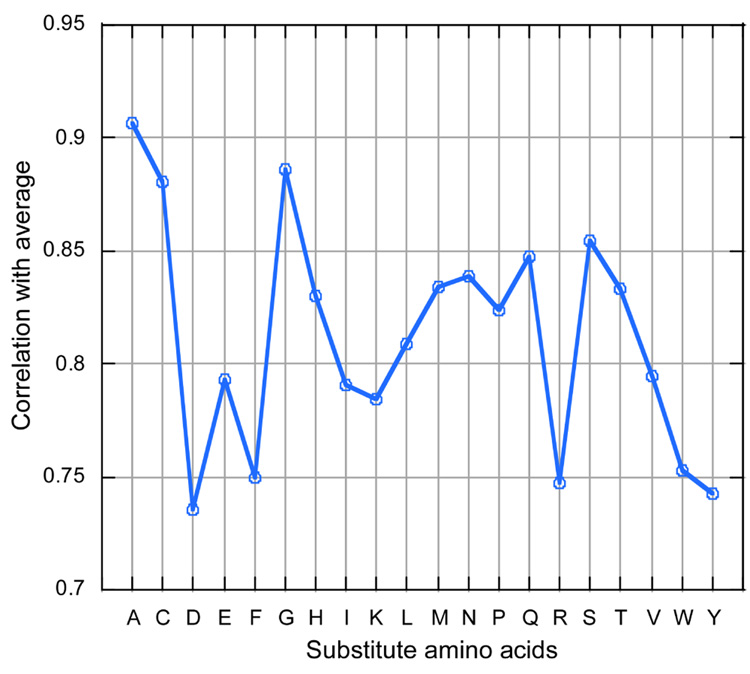
Because in silico mutagenesis is so much cheaper than its experimental sister, we could comprehensively analyze the degree to which alanine scans are representative of all possible mutations. We found that SNAP prediction scores for by-alanine substitutions correlated strongly with the average SNAP scores over all possible substitutions (for both, reported ASEdb mutant locations (Fig.3) and over all glucokinase residues (data not shown)). This suggested that using alanine scans aimed at estimating functional importance of residues may likely be just as informative as sequentially substituting each of the other 18 amino acids. For ASEdb mutagenesis sites, the average correlated also significantly with by-cysteine and by-glycine substitutions.
Figure 3. Average substitution effect correlated with single amino acid substitutions.
Among all single amino acid substitutions (at ASEdb mutant sequence positions), the distribution of predictions that best estimated the average was that of alanine, followed by cysteine, and glycine. These are also the amino acids that are often used in experimental mutagenesis studies to define functional sites.
3.5 Computational mutagenesis is a good first step toward annotating protein active sites
ASEdb data is likely skewed with regard to interface residues; i.e. most alanine scanning mutagenesis experiments are performed on suspected sets of binders. When considering entire protein sequences, however, other notions become important. For instance, core residues may be predicted as functionally important due to their utter necessity for maintenance of protein stability. There currently is no simple automated way to separate out the reasons behind functional importance annotations. However, as the example with HXK4 shows, there is validity in filtering entire sequences.
First, the ability to consider all possible substitutions at each residue may aid experimentalists in choosing the optimal site for mutagenesis. Second, in this particular sequence, and likely in many others, over half of the residues are excluded from functional considerations by almost any measure. This significantly narrows down the number of suspects. Third, SNAP scores have a scale meaning; i.e. substitutions that have severe effects are more likely to have higher scores. This suggests priorities for processing mutations of interest. While in silico mutagenesis may not yet be good enough to do the experiment, we challenge that tools of the type we used have finally come sufficiently of age to aid experimental mutagenesis in their design and prioritization. In other words, comprehensive in silico mutagenesis is not ready to be an end, but certainly it is ready to make for a good beginning.
4 CONCLUSION
Alanine scans aid the experimental elucidation of protein function. We demonstrated that SNAP, a method developed for a very different purpose, namely to predict the effects of non-synonymous single nucleotide polymorphisms (SNPs), correctly identified over 70% of the functionally important sites in ASEdb. As an example for a comprehensive in silico mutagenesis, we presented a demi-formal, graphical and intuitive evaluation of predictions made for all possible substitutions in the human glucokinase. This exercise highlighted the potential value in using SNAP predictions to guide experiments. Our work also suggested that alanine scans may be surprisingly representative of what could be found if we had the means to experimentally test the mutation of all residues by all non-native amino acids in say all human proteins.
ACKNOWLEDGEMENTS
Thanks to Rudolph L. Leibel, Marco Punta, Ta-tsen Soong, and Chani Weinreb (all Columbia) for helpful discussions. Particular thanks to Guy Yachdav (Columbia) for all his help with setting up and maintaining the SNAP web-server. Last not least, thanks to all those who deposit experimental data into databases and to all of those who make their carefully evaluated tools available.
FUNDING
The work of YB and BR was supported by the grant RO1-LM07329-01 from the National Library of Medicine (NLM).
REFERENCES
- Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Research. 2000;28:45–48. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A, Apweiler R, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Natale DA, O'Donovan C, Redaschi N, Yeh LS. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. J Mol Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- Bromberg Y, Rost B. SNAP: predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 2007;35:3823–3835. doi: 10.1093/nar/gkm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christesen HB, Jacobsen BB, Odili S, Buettger C, Cuesta-Munoz A, Hansen T, Brusgaard K, Massa O, Magnuson MA, Shiota C, Matschinsky FM, Barbetti F. The second activating glucokinase mutation (A456V): implications for glucose homeostasis and diabetes therapy. Diabetes. 2002;51:1240–1246. doi: 10.2337/diabetes.51.4.1240. [DOI] [PubMed] [Google Scholar]
- Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- Davis SJ, Davies EA, Tucknott MG, Jones EY, van der Merwe PA. The role of charged residues mediating low affinity protein-protein recognition at the cell surface by CD2. Proc Natl Acad Sci U S A. 1998;95:5490–5494. doi: 10.1073/pnas.95.10.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. Unraveling hot spots in binding interfaces: progress and challenges. Curr Opin Struct Biol. 2002;12:14–20. doi: 10.1016/s0959-440x(02)00283-x. [DOI] [PubMed] [Google Scholar]
- Eisenberg D, Marcotte EM, Xenarios I, Yeates TO. Protein function in the post-genomic era. Nature. 2000;405:823–826. doi: 10.1038/35015694. [DOI] [PubMed] [Google Scholar]
- Epstein CJ. Role of the amino acid 'code' and of selection for conformation in the evolution of proteins. Nature. 1966;210:25–28. doi: 10.1038/210025a0. [DOI] [PubMed] [Google Scholar]
- Gardsvoll H, Gilquin B, Le Du MH, Menez A, Jorgensen TJ, Ploug M. Characterization of the functional epitope on the urokinase receptor. Complete alanine scanning mutagenesis supplemented by chemical cross-linking. J Biol Chem. 2006;281:19260–19272. doi: 10.1074/jbc.M513583200. [DOI] [PubMed] [Google Scholar]
- Glaser B, Kesavan P, Heyman M, Davis E, Cuesta A, Buchs A, Stanley CA, Thornton PS, Permutt MA, Matschinsky FM, Herold KC. Familial hyperinsulinism caused by an activating glucokinase mutation. N Engl J Med. 1998;338:226–230. doi: 10.1056/NEJM199801223380404. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Ruiz D, Gohlke H. Targeting protein-protein interactions with small molecules: challenges and perspectives for computational binding epitope detection and ligand finding. Curr Med Chem. 2006;13:2607–2625. doi: 10.2174/092986706778201530. [DOI] [PubMed] [Google Scholar]
- Guerois R, Nielsen JE, Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J Mol Biol. 2002;320:369–387. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- Kamata K, Mitsuya M, Nishimura T, Eiki J, Nagata Y. Structural basis for allosteric regulation of the monomeric allosteric enzyme human glucokinase. Structure. 2004;12:429–438. doi: 10.1016/j.str.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Kawabata T, Ota M, Nishikawa K. The protein mutant database. Nucleic Acids Research. 1999;27:355–357. doi: 10.1093/nar/27.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi S, Iwaki S, Kimura-Someya T, Yamaguchi A. Cysteine-scanning mutagenesis around transmembrane segment VI of Tn10-encoded metal-tetracycline/ H(+) antiporter. FEBS Lett. 1999;461:315–318. doi: 10.1016/s0014-5793(99)01490-8. [DOI] [PubMed] [Google Scholar]
- Kortemme T, Baker D. A simple physical model for binding energy hot spots in protein-protein complexes. Proc Natl Acad Sci U S A. 2002;99:14116–14121. doi: 10.1073/pnas.202485799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouadio JL, Horn JR, Pal G, Kossiakoff AA. Shotgun alanine scanning shows that growth hormone can bind productively to its receptor through a drastically minimized interface. J Biol Chem. 2005;280:25524–25532. doi: 10.1074/jbc.M502167200. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Research. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa K, Ishino S, Takenaka H, Norioka N, Hirai T, Yao T, Seto Y. Constructing a protein mutant database. Protein Engineering. 1994;7:773. doi: 10.1093/protein/7.5.733. [DOI] [PubMed] [Google Scholar]
- Ofran Y, Rost B. ISIS: interaction sites identified from sequence. Bioinformatics. 2007;23:e13–e16. doi: 10.1093/bioinformatics/btl303. [DOI] [PubMed] [Google Scholar]
- Ofran Y, Rost B. Protein-Protein Interaction Hotspots Carved into Sequences. PLoS Comput Biol. 2007;3:e119. doi: 10.1371/journal.pcbi.0030119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L, Cai S, Zhu Y, Inouye M. Cysteine-scanning analysis of the dimerization domain of EnvZ, an osmosensing histidine kinase. J Bacteriol. 2003;185:3429–3435. doi: 10.1128/JB.185.11.3429-3435.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramensky V, Bork P, Sunyaev SR. Human non-synonymous SNPs: server and survey. Nucleic Acids Research. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B. PROF: predicting one-dimensional protein structure by profile based neural networks. 2000. unpublished. [DOI] [PubMed] [Google Scholar]
- Rost B. In: The Proteomics Protocols Handbook. Walker JE, editor. Totowa, NJ: Humana; 2005. pp. 875–901. [Google Scholar]
- Rost B, Sander C. Conservation and prediction of solvent accessibility in protein families. Proteins: Structure, Function, and Genetics. 1994;20:216–226. doi: 10.1002/prot.340200303. [DOI] [PubMed] [Google Scholar]
- Shapiro R, Vallee BL. Site-directed mutagenesis of histidine-13 and histidine-114 of human angiogenin. Alanine derivatives inhibit angiogenin-induced angiogenesis. Biochemistry. 1989;28:7401–7408. doi: 10.1021/bi00444a038. [DOI] [PubMed] [Google Scholar]
- Shulman-Peleg A, Shatsky M, Nussinov R, Wolfson HJ. Spatial chemical conservation of hot spot interactions in protein-protein complexes. BMC Biol. 2007;5:43. doi: 10.1186/1741-7007-5-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer BA, Pantoliano MW, Barbera FA, Gunyuzlu PL, Thompson LD, Herblin WF, Rosenfeld SA, Book GW. Identification and concerted function of two receptor binding surfaces on basic fibroblast growth factor required for mitogenesis. J Biol Chem. 1994;269:26879–26884. [PubMed] [Google Scholar]
- Thorn KS, Bogan AA. ASEdb: a database of alanine mutations and their effects on the free energy of binding in protein interactions. Bioinformatics. 2001;17:284–285. doi: 10.1093/bioinformatics/17.3.284. [DOI] [PubMed] [Google Scholar]
- Vegotsky A, Fox SW. Protein molecules: intraspecific and interspecific variations. In: Florkin M, Mason HS, editors. Comparative Biochemistry IV. New York, NY: Academic Press; 1962. pp. 185–244. [Google Scholar]
- Vionnet N, Stoffel M, Takeda J, Yasuda K, Bell GI, Zouali H, Lesage S, Velho G, Iris F, Passa P, et al. Nonsense mutation in the glucokinase gene causes early-onset non-insulin-dependent diabetes mellitus. Nature. 1992;356:721–722. doi: 10.1038/356721a0. [DOI] [PubMed] [Google Scholar]
- Weiss GA, Watanabe CK, Zhong A, Goddard A, Sidhu SS. Rapid mapping of protein functional epitopes by combinatorial alanine scanning. Proc Natl Acad Sci U S A. 2000;97:8950–8954. doi: 10.1073/pnas.160252097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Z, Litherland SA, Sorensen NB, Proneth B, Wood MS, Shaw AM, Millard WJ, Haskell-Luevano C. Pharmacological characterization of 40 human melanocortin-4 receptor polymorphisms with the endogenous proopiomelano-cortin- derived agonists and the agouti-related protein (AGRP) antagonist. Biochemistry. 2006;45:7277–7288. doi: 10.1021/bi0600300. [DOI] [PubMed] [Google Scholar]
- Yang Y, Chen M, Lai Y, Gantz I, Yagmurlu A, Georgeson KE, Harmon CM. Molecular determination of agouti-related protein binding to human melanocortin-4 receptor. Mol Pharmacol. 2003;64:94–103. doi: 10.1124/mol.64.1.94. [DOI] [PubMed] [Google Scholar]
- Yue P, Melamud E, Moult J. SNPs3D: candidate gene and SNP selection for association studies. BMC Bioinformatics. 2006;7:166. doi: 10.1186/1471-2105-7-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling L. Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ, editors. Evolving Genes And Proteins. New York and London: Academic Press; 1965. pp. 97–166. [Google Scholar]