Abstract
Antiviral immunity is initiated upon host recognition of viral products via non-self molecular patterns known as pathogen-associated molecular patterns (PAMPs). Such recognition initiates signaling cascades that induce intracellular innate immune defenses and an inflammatory response that facilitates development of the acquired immune response. The retinoic acid-inducible gene I (RIG-I) and the RIG-I-like receptor (RLR) protein family are key cytoplasmic pathogen recognition receptors that are implicated in the recognition of viruses across genera and virus families, including functioning as major sensors of RNA viruses, and promoting recognition of some DNA viruses. RIG-I, the charter member of the RLR family, is activated upon binding to PAMP RNA. Activated RIG-I signals by interacting with the adapter protein MAVS leading to a signaling cascade that activates the transcription factors IRF3 and NF-κB. These actions induce the expression of antiviral gene products and the production of type I and III interferons that lead to an antiviral state in the infected cell and surrounding tissue. RIG-I signaling is essential for the control of infection by many RNA viruses. Recently, RIG-I crosstalk with other pathogen recognition receptors and components of the inflammasome has been described. In this review, we discuss the current knowledge regarding the role of RIG-I in recognition of a variety of virus families and its role in programming the adaptive immune response through cross-talk with parallel arms of the innate immune system, including how RIG-I can be leveraged for antiviral therapy.
Keywords: RIG-I, RNA virus, RIG-I-like receptor, innate immunity, infection, pathogen-associated molecular pattern
Introduction
The innate immune response provides the first line of defense against virus infection. The innate immune system of vertebrates is multi-pronged and highly differentiated. Cell-associated proteins known as pathogen recognition receptors (PRRs) recognize non-self molecular patterns known as pathogen-associated molecular patterns (PAMPs). PAMPs are viral products, including protein, protein/lipid complexes, and viral nucleic acid, that are specific to the virus or aberrantly located within the cell and typically accumulate during infection. During virus infection PAMPs accumulate at various sites within the infected cell, including at the cell surface, inside endosomal compartments, or free in the cell cytosol. PAMP engagement and activation of PRRs initiates intracellular signaling cascades leading to the expression of genes whose products function to restrict virus replication and spread, attract innate immune cells to the site of infection and induce their antiviral activity, and modulate the adaptive immune response to the viral infection. Secretion of chemokines and cytokines, including type 1 and 3 interferon (IFN), from the infected cell acts to warn neighboring cells of the threat and to attract immune cells from elsewhere in the body to the site of infection. Based on the signals received by the components of the innate and adaptive immune responses, the invading pathogen will be either attacked or tolerated. Programming the immune response against viral infection is highly dependent on nonself discrimination of PAMP versus self molecular patterns found within healthy cells. Aberrant recognition and innate immune signaling induced by self molecular patterns have important consequences that can impart autoimmune disease. Thus, tight regulation of the innate immune response is necessary to maintain immune homeostasis while allowing for quick and robust control of viral infection.
Mammalian PRRs are categorized according to ligand specificity, function, localization and/or evolutionary and structural relationship. PRRs can be secreted, membrane bound, or cytoplasmic proteins. Examples of secreted PRRs include the mannose-binding lectin (MBL) and ficolin which recognize D-mannose and L-fucose carbohydrate residues on a range of microbes including viruses to initiate the complement pathway, a major arm of the host innate immune response (Matsushita, 2010; Murphy et al., 2008). These secreted PRRs are not involved in intracellular signaling cascades but instead trigger soluble protease cascades to complement activation in the blood. Membrane-bound PRRs include the Toll-like receptors (TLRs), which are found on the cell surface or within endosomal compartments in most cells of the body, and C-type lectin receptors which are typically found on dendritic cells or macrophages (Murphy et al., 2008). TLRs recognize a variety of PAMPs relevant to virus infection including unmethylated CpG DNA found in the genome of DNA viruses (TLR9), and double-stranded (ds) or single-stranded (ss) RNA viral products recognized by TLR3 and TLR7/8, respectively (Kawai and Akira, 2010). Cytoplasmic PRRs include NOD-like receptors (NLRs) (Ye and Ting, 2008), the cytosolic DNA sensor cyclic GMP-AMP synthase (cGAS) (Cai et al., 2014), and RIG-I-like receptors (RLRs)(Loo and Gale, 2011). NLRs recognize a wide range of PAMPs and DAMPs (damage-associated molecular patterns produced form damaged host cells). There are several NLR family members, each involved in recognizing intracellular PAMPs or DAMPs that accumulate in response to virus infection. NLR signaling results in formation of the inflammasome, a multicomponent protein complex that catalyzes the downstream processing and activation of latent caspase 1 zymogen and its cleavage and cellular release of pro-IL-1β family members to induce an inflammatory cascade (Wen et al., 2013). cGAS is a recently described cytosolic dsDNA sensor that catalyzes the formation of a heterocyclic dinucleotide, cGAMP (Sun et al., 2013; Xiao and Fitzgerald, 2013; Zhang et al., 2013). cGAMP binding to the adaptor protein STING located on intracellular membranes initiates TBK-1 phosphorylation and IRF3 activation (Shu et al., 2014). Although cGAS senses dsDNA during DNA virus infection, recent reports have implicated this pathway in the sensing of RNA viruses as well (Maringer and Fernandez-Sesma, 2014; Schoggins et al., 2014). RIG-I-like receptors (RLRs) include the cytosolic PRRs retinoic acid-induced gene I (RIG-I), melanoma differentiation-associated gene 5 (MDA5) and laboratory of genetics and physiology 2 (LGP2). In particular, RIG-I is the subject of this review and is described in detail in the following sections. All three RLRs share a common structure which includes a DExD/H-box helicase domain and a C-terminal domain known as the repressor domain (RD). RIG-I and MDA5 also contain N-terminal tandem caspase activation and recruitment domains (CARDs) that function in signaling, which LGP2 lacks. Activation of MDA5 or RIG-I leads to interactions of the CARDs with the CARD of the mitochondrial activator of virus signaling (MAVS) protein, the essential signaling adaptor protein of the RLRs (also known as IPS-1, VISA, or Cardif) (Kawai et al., 2005; Meylan et al., 2005; Xu et al., 2005). CARD-CARD interactions result in initiation of a signaling cascade that includes activation of TBK1 and IKKε protein kinases which phosphorylate and activate the transcription factors interferon regulatory factor (IRF) 3 and NF-κB. Recruitment of tumor necrosis factor receptor type 1 -associated death domain protein (TRADD), Fas-associated protein with death domain (FADD), receptor-interacting protein 1 (RIP1), caspase-8 and caspase-10 leads to activation of the IKK complex consisting of IKKα, IKKβ, and IKKγ, which in turn activate NF-κB (Kawai et al., 2005; Takahashi et al., 2006). Upon activation, IRF3 and NF-κB translocate from the cytosol to the nucleus to induce transcription of a variety of innate immune response genes, including IFNs, direct antiviral genes, and pro-inflammatory genes whose products orchestrate the innate immune response to infection. IFNs then induce hundreds of interferon-stimulated genes (ISGs) whose products have antiviral, immunomodulatory, cell growth regulatory, and metabolic regulatory actions that create an antiviral state. If successful, this response potently restricts virus replication and cell-to-cell spread of infection. However, many, if not all, pathogenic viruses possess strategies to evade the innate immune response and thus cause disease (Chiang et al., 2014).
RIG-I activation and regulation
RIG-I is expressed at a low level in most cells of the body and its abundance increases in response to IFN. In non-infected cells RIG-I is found in a resting state with the RD covering the RNA-binding and helicase domains (Saito et al., 2007). CARDs of “resting” RIG-I are also folded over one another with these interactions governing the signaling activity of RIG-I (Fig. 1.1) (Kowalinski et al., 2011). Following recognition of PAMP RNA, RIG-I hydrolyzes ATP and undergoes a conformational change which opens the RNA binding domain for closer interaction with PAMP RNA while releasing the CARDs for MAVS interaction and signaling (Fig. 1.2) (Jiang et al., 2011; Kowalinski et al., 2011). The hydrolysis of ATP also facilitates translocation of RIG-I along the RNA, thus making room for other RIG-I proteins to oligomerize onto the PAMP molecule (Patel et al., 2013). RIG-I is then modified with ubiquitin at N-terminal sites by the TRIM25 protein thus allowing CARD-CARD tetramers to form in the presence of ubiquitin (Gack et al., 2008; Gack et al., 2007). For sustained activation, RIG-I interacts with the dsRNA binding protein PACT, which binds to the C-terminal domain of RIG-I aiding in ATP hydrolysis to maintain the protein in an active state (Kok et al., 2011). Interactions with zinc antiviral protein short isoform (ZAPS) also maintain RIG-I in an activated state (Hayakawa et al., 2011). As shown in Figure 1, the entire complex then translocates from the cytoplasm to the mitochondrial associated membrane (MAM) facilitated by the 14–3–3ε protein to chaperone the RIG-I “translocase” to the MAM (Liu et al., 2012). At the MAM, RIG-I CARDs interact with the MAVS CARD, catalyzing filament formation of MAVS which in turn activates TBK1 and IKKε to initiate downstream signaling (Fig. 1.4) (Hou et al., 2011). It is still unclear whether the RNA PAMP ligand translocates to the MAM still bound to RIG-I or if it is released upon CARD tetramer formation. Moreover, it should be noted that though MAVS filaments have been identified in cell-free extracts, their formation and presence has not been demonstrated in intact mammalian cells in the context of virus infection (Hou et al., 2011). It is also noted that LGP2, which lacks CARDs is thought to negatively regulate RIG-I though interactions of the RD (Komuro et al., 2008; Komuro and Horvath, 2006; Saito et al., 2007), while other studies indicate a positive regulatory role for LGP2 in RLR signaling (Bruns et al., 2014; Goubau et al., 2014; Satoh et al., 2010). Much work has been focused on understanding the regulation of RLR signaling pathway by various host proteins and is reviewed elsewhere (Chiang et al., 2014; Eisenacher and Krug, 2012; Komuro et al., 2008).
Figure 1.
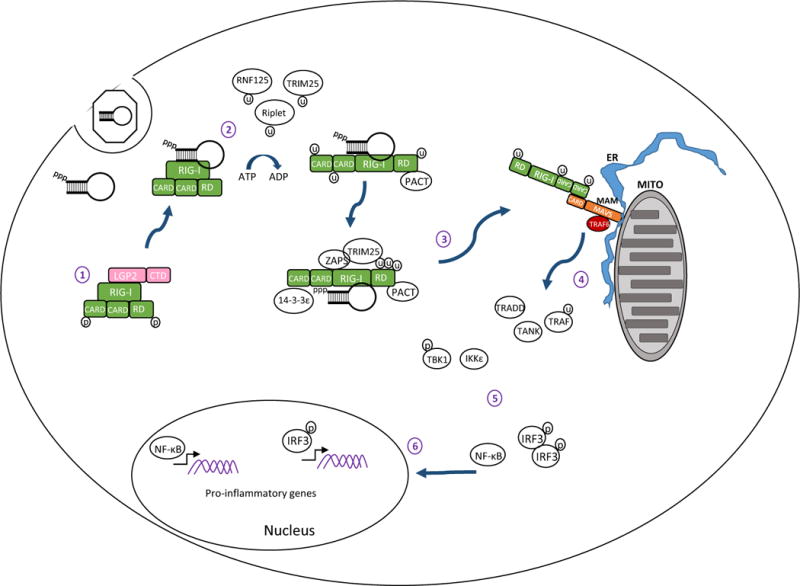
RIG-I signaling pathway. 1) In resting cells RIG-I is maintained in a resting state in the cytosol characterized by a ‘closed fist’ conformation with the RD and CARD domains folded over the helicase domain. Interactions with LGP2 may be important for regulating RIG-I. 2) Upon recognition of an RNA ligand containing PAMP motif and exposed 5′ppp RIG-I is ubiquitinated and hydrolyzes ATP to promote a conformational change that holds the ligand PAMP RNA in the RNA binding groove of the RD. 3) Interactions with PACT, ZAPS, TRIM25 and 14–3–3ε lead to formation of a RIG-I translocase that moves from a soluble compartment in the cell to the mitochondrial associated membranes (MAM) where the complex engages MAVS for downstream signaling. The MAM is a specialized extension of outer mitochondria membrane (Mito) and endoplasmic reticulum (ER) that form a synapse to create an innate immune signaling platform. 4) CARD-CARD interactions between RIG-I and MAVS then initiate activation of the MAVS ‘signalosome’ which includes TRAF2, 3, and 6, TRADD, TANK, the IRF3 kinases TBK1 and/or IKKε, and the NF-κB kinase complex. 5) Signaling through these kinases activates IRF3 and NF-κB. 6) Phosphorylated IRF3 and activated NF-κB translocate to the nucleus and initiate transcription of genes encoding products with antiviral, proinflammatory, and immune-regulatory actions.
PAMP ligands of RIG-I
RIG-I recognizes PAMP RNA ligands based on a level of specificity in terms of sequence composition, PAMP motif length, ss or dsRNA, and presence of 5′ cap versus exposed 5′triphosphate (5′ppp). 5′ capped RNA does not trigger RIG-I signaling nor does RNA lacking exposed 5′ppp or 5′ diphosphosphate (5′pp) (Fig. 2). dsRNA of <300 base pairs with blunt ends containing a 5′-triphosphate (5′ppp) moiety are ideal ligands for RIG-I, although the minimal length has been shown to be 10 base pairs as long as no mismatches exist near the blunt end (Hornung et al., 2006; Kato et al., 2008; Kato et al., 2006; Pichlmair et al., 2006; Schmidt et al., 2009). RNA panhandle or bulge/loop structures are stimulatory if the RNA contains 5′ppp and blunted 5′ end (Marq et al., 2010). However, recent studies have also reported RIG-I binding and activity in the presence of RNA containing 5′pp moiety (Goubau et al., 2014; Lu et al., 2010). It is possible that a significant difference in the on-off rate of the PAMP RNA binding to RIG-I containing different 5′ moieties could explain this discrepancy (Lu et al., 2010; Wang et al., 2010). Two significant crystal structures have been solved to reveal how RIG-I binds to different RNA species and has provided insights into the conformational changes required for signaling (Jiang et al., 2011; Luo et al., 2011). RIG-I has also been shown to recognize specific sequences found in viral genomes, such as the poly-U/UC motif found in the 3′ untranslated region of hepatitis C virus or the mRNA of the N gene of Hantaan virus (Fig. 2) (Lee et al., 2011; Runge et al., 2014; Saito et al., 2008; Schnell et al., 2012). Some controversy exists surrounding the ability of RIG-I to bind to single-stranded RNA (ssRNA) synthesized using the T7 polymerase, which has a tendency to add a short double-stranded copy-back section to 5′ppp ssRNA.(Cazenave and Uhlenbeck, 1994). When one such ssRNA was purified to remove any contaminating dsRNA, it failed to stimulate RIG-I dependent signaling in vitro (Marq et al., 2010; Schlee et al., 2009). However, production of 5′ppp RNA itself by T7 polymerase is not sufficient to produce a PAMP. Such transcripts produced from a variety of templates do not induce RIG-I signaling (Cui et al., 2008; Saito et al., 2008; Schnell et al., 2012). Thus, despite a number of high resolution crystal structures and excellent reports describing structural and sequence determinants for RIG-I ligands, the true definition of a RIG-I ligand remains controversial. The field has collected a large amount of information about what RNA structures or sequences may be beneficial for RIG-I binding and what may be sufficient for RIG-I signaling, but it remains difficult to predict RIG-I binding sequences and stimulatory RNA motifs based on structure and sequence. Defining PAMP ligands of RIG-I thus requires a comprehensive assessment of direct binding to RIG-I, PAMP stimulation of RIG-I ATPase activity and conformation change, as well as ability of PAMP stimulation to drive the cytosol-to-MAM translocation of RIG-I to mediate MAVS interaction and downstream intracellular signaling. The following sections provide an overview of RIG-I interaction with viruses from different genera that reveal shared and unique features of RIG-I interaction among diverse virus families.
Figure 2.
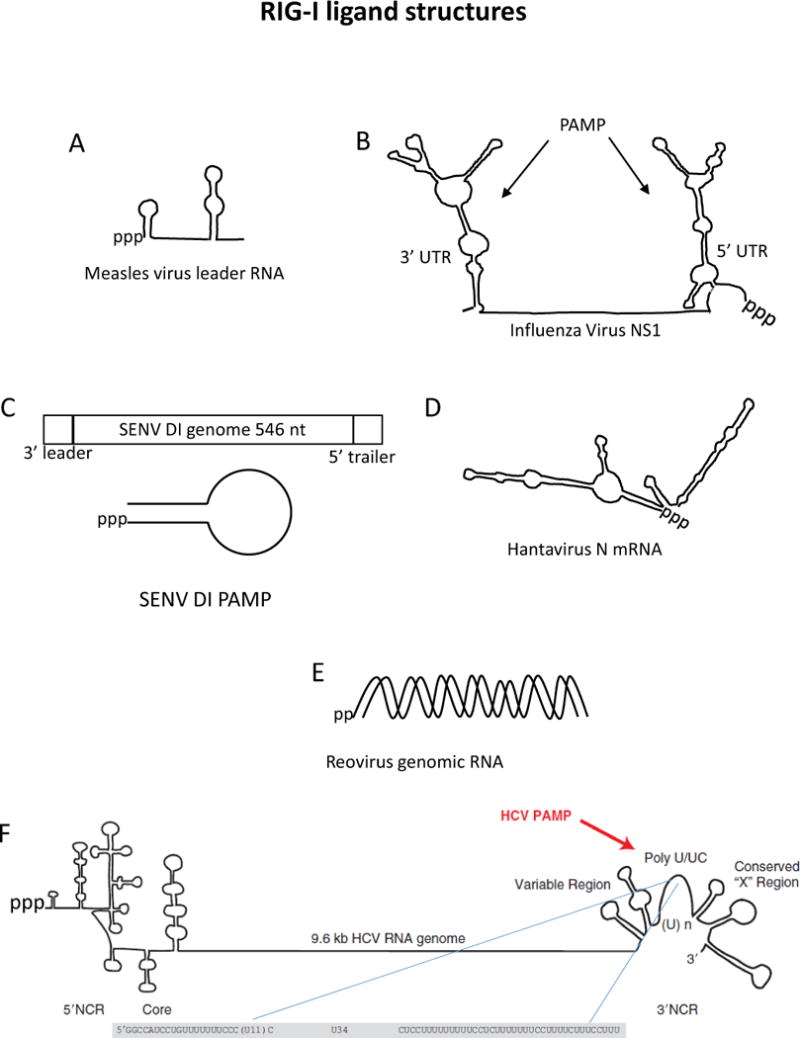
Examples of known RIG-I PAMP RNAs with models of specific structures shown .A) RNAfold predicted structure of measles virus leader RNA using Edmonson B strain sequence. B) Influenza virus PAMPs found at genomic 5′ and 3′ nontranslated regions of the NS1 segment (adapted from (Davis et al., 2012)). C) Sendai virus defective-interfering (DI) particle structure (adapted from (Martinez-Gil et al., 2013)). D) RNAfold predicted structure of Hantavirus nucleoprotein mRNA using TPM strain sequence. E) Reovirus genomic dsRNA with 5′pp motif identified as RIG-I PAMP in (Goubau et al., 2014). F) Hepatitis C virus genome with 3′ nontranslated region and poly-U/UC PAMP highlighted (adapted from (Horner and Gale, 2013)).
RIG-I recognition and regulation of innate immunity
ssRNA viruses Negative-sense
Bornaviridae
Bornaviruses are the only members of the order mononegavirales, viruses with ssRNA negative sense genome, which replicate and transcribe its genome exclusively in the nucleus of infected cells, while assembly occurs in the cytoplasm. Bornaviruses establish persistent infection in the host in the absence of an IFN response. RNA genomes isolated from virus preparations revealed uniformly trimmed 5′ termini, indicating that ‘genome trimming’ occurs efficiently during replication (Schneider et al., 2007). This ‘genome trimming’ removes nucleotides from the 5′ end and could function to regulate transcription and replication during periods of persistence. Importantly, this trimming leaves a terminal 3′ overhang during circularization for replication and also converts 5′ppp to 5′p, thus evading recognition by RIG-I and avoiding the antiviral actions of the innate immune response.
Borna disease virus (BDV) has been shown to actively antagonize the innate immune signaling pathway that leads to IRF3 activation. Unterstab et al. demonstrated that the BDV P protein antagonizes TBK1 to prevent IFNp induction during infection (Unterstab et al., 2005). The viral P protein can bind directly to TBK1 and prevent its kinase activity in vitro. It is hypothesized that the ability to prevent transcription of IRF3-dependent genes during viral infection, thus evading the effects of activating RLR pathway, is a key element to establishing persistent infection in the mammalian and avian host (Reuter et al., 2010).
Filoviridae
Members of the filovirus family include the human pathogens Ebola and Marburg viruses that can cause severe hemorrhagic fevers and high case-fatality ratios. The importance of the proinflammatory response in controlling Ebola virus (EBOV) infection was demonstrated by pre-activation of RIG-I in cells by treatment with RIG-I agonist RNA prior to virus challenge. These cells were then resistant to EBOV infection (Spiropoulou et al., 2009). Because stimulation of RIG-I leads to a general pro-inflammatory response through induction of IFNs, it is not clear what specific cellular antiviral effector mechanisms were responsible for the protection against EBOV observed in this study. Moreover, EBOV has been shown to encode an innate immune antagonist protein, VP35. This viral protein was first identified as an antagonist of IFN induction (Basler et al., 2000). Subsequent reports have suggested possible mechanisms for this inhibition including sequestration of viral dsRNA through interactions with the VP35 RNA binding domain that prevent PAMP recognition by RIG-I (Cardenas et al., 2006), inhibition of phosphorylation of IRF3/7 through interactions with TBK1 (Basler et al., 2003; Hartman et al., 2008; Prins et al., 2009), and direct interaction with RIG-I to suppress RIG-I signaling activity (Luthra et al., 2013). While these studies suggest that EBOV dsRNA might harbor PAMP motifs for RIG-I recognition and innate immune signaling, the potential for RIG-I actually sensing the EBOV genome has yet to be directly examined. It is possible that the dsRNA sequestered by VP35 would trigger RLR signaling activation during infection but the absolute nature of this putative PAMP is not known (Ramanan et al., 2012).
Paramyxoviridae
RIG-I, as well as MDA5, has been reported to induce IFN following infection of human cells with measles virus (Ikegame et al., 2010). The viral leader transcript of the measles virus has been identified as a potential PAMP ligand of RIG-I (Fig. 2) (Plumet et al., 2007). However, during viral infection, limited IFN is detected in the supernatant of infected cells, suggesting that measles virus may antagonize RIG-I signaling actions. Originally, the V proteins of paramyxoviruses were identified as MDA5 antagonists, preventing its activation and thereby suppressing innate immune signaling during infection (Childs et al., 2007; Childs et al., 2009; Lo et al., 2012; Motz et al., 2013). Interestingly, a recent study shows that a single amino acid difference between RLRs determines the ability of paramyxovirus V proteins to suppress MDA5 versus RIG-I (Rodriguez and Horvath, 2013). In 2012, Childs et al. reported that the V protein of the related parainfluenzavirus type 5 (PIV5) interacts with LGP2 to regulate the activation of RIG-I (Childs et al., 2012). They reported a synergism between PIV5 V protein and LGP2 to inhibit RIG-I activation when cells were exposed to known RIG-I PAMP ligands. Childs et al. demonstrated that interaction between LGP2 and RIG-I occurred only in the presence of PIV5 V protein and not the P protein, suggesting that LGP2 inhibition of RIG-I was a result of V protein interference (Childs et al., 2012). However, others reported no inhibitor effect on RIG-I activity in the presence of physiologically relevant levels of V proteins from measles virus and PIV5 (Rodriguez and Horvath, 2014). Because LGP2 can regulate RIG-I and MDA5 in opposing ways, it has been difficult to tease out the importance of LGP2 interference with RIG-I activation and innate immune stimulation by paramyxovirus V protein interactions. Thus, further studies are warranted to define the role of LGP2 for RLR and innate immune regulation in paramyxovirus infection.
Sendai virus (SenV) activates RIG-I signaling and is often used in the field as a model virus to stimulate robust type I interferon responses through RIG-I activation. Recently, defective interfering (DI) particles produced in SenV infection have been characterized to robustly stimulate RIG-I activation and innate immune signaling, likely explaining the nature of the viral agonists of RIG-I as products of DI particles (Baum et al., 2010). The dsRNA present in particles derived from the Cantell SenV strain is 546 nucleotides in length and contains perfectly complementary ends, creating a hairpin structure with a 5′ppp that can function as a RIG-I-stimulatory RNA (Fig. 2) (Kolakofsky, 1976). A recent report describes the use of this stimulatory viral RNA as a vaccine adjuvant that can enhance the immunogenicity of influenza A virus vaccination in a mouse model, further demonstrating that RIG-I signaling drives immune-enhancing actions against virus infection (Martinez-Gil et al., 2013).
Rhabdoviridae
The rhabdovirus vesicular stomatitis virus (VSV) has been demonstrated to trigger RIG-I specific innate immune signaling during infection (Furr et al., 2010; Kato et al., 2006). In RIG-I−/− mouse embryonic fibroblasts (MEFs), infection with VSV failed to induce IFN-β expression compared to WT MEFs (Kato et al., 2006). Similar results in RIG-I−/− primary astrocytes indicate that RIG-I recognition of viral RNA is essential for innate immune signaling in cells of the central nervous system (Furr et al., 2010). While the exact RNA PAMP of VSV that is recognized and bound by RIG-I to initiate the innate immune response is not known, sequence similarity of the 5′ leader segment of the VSV genome to other known viral RIG-I ligands suggests that this may be a stimulatory RNA motif (Saito et al., 2008).
Arteriviridae
Arteriviruses establish infections in horses, pigs and non-human primates causing a wide-range of disease states including edema, respiratory distress, and hemorrhagic fever. Arteriviruses were shown to be primarily recognized by MDA5 in MEFs (van Kasteren et al., 2012). However Arteriviruses encode a deubiquitinase (DUB) activity within their nonstructural protein 2 (nsp2) that serves to catalyze the removal of ubiquitin chains from MDA5 (and RIG-I) that otherwise facilitate RLR activation. This process results in attenuation of MDA5 signaling and overall suppression of downstream IRF3 nuclear localization and IFN-β expression, thus supporting viral replication and spread (Luo et al., 2008). Innate immune evasion was linked to the nsp2 of porcine respiratory and reproductive syndrome virus (PRRSV), which was sufficient to inhibit virus-induced IRF3 activation when expressed in target cells (Li et al., 2010a). Viral DUB activity is conserved among arterivirus strains and is linked with pathogenesis (van Kasteren et al., 2012).
Arenaviridae
Arenaviruses contain a bipartite ambisense genome in which the two termini are either exactly complimentary or contain two mismatches at nucleotide positions 6 and 8 (Albarino et al., 2009). Interestingly, arenavirus genome synthesis initiates with a GTP at position +2 and the initiating 5′pppG is realigned to form 5′pppGpCOH before the replication primer is extended (Garcin and Kolakofsky, 1992). Marq et al. demonstrated that the 5′ RNA overhang resulting from this ‘prime and realign’ mechanism of replication significantly decreases IFN-β expression compared to RNA with no overhang (Marq et al., 2010). Further, the mismatched base pairings could also contribute to a reduction in innate immune signaling and elimination of the 5′ppp completely eliminated IFN-β expression (Marq et al., 2010). Ligand competition assays revealed that the 5′ppp overhang RNA acted as a sink for RIG-I, holding it tightly, but not allowing for subsequent signaling (Marq et al., 2011). In this way, it is thought that arenaviruses are recognized by RIG-I and that they are able to actively antagonize RIG-I signaling to evade the innate immune response.
New world arenaviruses have evolved the ability to prevent RIG-I association with MAVS during infection. Huang et al. reported a significant decrease in IFN-β production in RIG-I knockout cells infected with Junin Virus, with complete abrogation of ISG expression (Huang et al., 2012). The Z proteins of new world arenaviruses, but not old world arenaviruses, were shown to co-localize with RIG-I and could co-immunoprecipitate RIG-I in vitro (Fan et al., 2010). This association with RIG-I was sufficient to block RIG-I-MAVS interactions and NF-κB and IRF3 activation. However, most human infections with pathogenic arenaviruses are known to involve high levels of IFN expression and pro-inflammatory cytokines. Thus, RIG-I antagonism must be incomplete during the course of in vivo infection. Understanding the mechanisms underlying pathogenic arenavirus infection and RIG-I antagonism will be essential to development of novel therapeutics to treat increasingly common zoonotic infections in humans.
Bunyaviridae
Hantavirus
Hantaviruses are members of the Bunyaviridae family, which is comprised of more than 350 viruses, of which Hantaviruses and Phleboviruses, as well as other genera are significant human or agricultural pathogens. Hantaviruses are transmitted by respiratory route while phleboviruses are transmitted by the bite of an insect vector (Horne and Vanlandingham, 2014). The hantavirus genome consist of three negative-sense, single-stranded genome segments which form blunt-ended panhandle structures containing a 5′ monophosphate (5′p) motif (Garcin et al., 1995; Wang et al., 2011). Studies of hantavirus revealed that this 5′p motif prevents RIG-I recognition of the genomic RNA (Habjan et al., 2008; Wang et al., 2011). However, during infection, RIG-I is required for innate immune signaling (Lee et al., 2011). Lee et al. demonstrated that the viral N gene from Hantaan virus (an Old World hantavirus) could serve as a ligand for RIG-I when transfected in a mammalian expression vector, suggesting that viral mRNAs could serve as the stimulatory RNA in hantavirus infection (Lee et al., 2011). The authors concluded that the mRNA from the N gene contained double-stranded RNA recognized by RIG-I because co-transfection of the N gene with the E3 gene from Vaccinia virus, which sequesters dsRNA, resulted in ablation of innate signaling (Fig. 2) (Lee et al., 2011). Further elucidation of the structural motif recognized by RIG-I should complement similar studies to determine whether other hantavirus mRNAs are also recognized by RIG-I, and the elucidate the role of 5′ phosphates in this process.
RIG-I activation is inhibitory to hantavirus replication, and hantaviruses have evolved mechanisms to inhibit the inflammatory response induced by RIG-I signaling. Interestingly, the N protein from Andes virus (a New World hantavirus) has been implicated in suppressing the RIG-I signaling pathway (Cimica et al., 2014). Cimica et al. demonstrated that expression of the N protein inhibited IFN-β and ISG expression in the presence of a constitutively active RIG-I protein and that this inhibition occurs by blocking downstream activation of TBK1 and phosphorylation of IRF3 (Cimica et al., 2014). In addition to the N protein, Alff et al. showed that the cytoplasmic tail of the viral G2 protein could bind cellular protein kinases and prevent phosphorylation of TBK1 (Alff et al., 2006). Importantly, the G2 cytoplasmic tail of human pathogenic hantaviruses was found to exhibit this function, whereas the G2 cytoplasmic tail of non-pathogenic hantaviruses did not (Alff et al., 2006). The presence of redundant mechanisms for inhibition of RIG-I-dependent signaling suggests that this pathway presents a strong selective pressure on the virus.
Phlebovirus
The specific roles of RIG-I and MDA5 in phlebovirus recognition are not well defined but the importance of this RLR pathway is exemplified in the active antagonism by the nonstructural protein (NSs) of severe fever with thrombocytopenia syndrome virus (SFTSV) (Ning et al., 2014). Ning et al. demonstrated that NSs binds and sequesters the components of the TBK1/IKKε complex to prevent activation of IRF3 for downstream innate immune signaling induced by the RLRs (Ning et al., 2014). Therefore, RLR recognition and activation must be a strong pressure on phleboviruses leading to the evolution of specific evasion strategies. Further, research must focus on understanding how each RLR is involved in recognition and what viral RNA species is recognized as PAMP by each RLR.
Nairovirus
Nairoviruses are members of the family Bunyaviridae that contain negative-sense, single-stranded circular genomes. The viral RNA is not recognized by RIG-I due to the lack of a free 5′ or 3′ end. However, nairoviruses, like arterviruses, encode DUB enzymes that deubiquitinate RIG-I to prevent innate immune signaling activation (van Kasteren et al., 2012), suggesting that during nairovirus infection/replication a PAMP, likely a viral transcription product, is produced that can activate RIG-I. The RIG-I-stimulatory PAMP RNA involved in triggering RIG-I signaling remains undefined.
Orthomyxoviridae
The Orthomyxoviridae include Influenza A, B, and C viruses. The viruses harbor a negative sense multi-segmented single stranded RNA genome. Influenza A virus (IAV) encodes 8 genome segments and represents the major epidemic and pandemic virus of past and recent time (Iwasaki and Pillai, 2014; Palese, 2004). IAV is maintained in water fowl and other avian species and is typically transmitted among encounters of domestic water fowl or chickens with pigs and humans wherein reassortment of viral genome segments in pigs co-infected with multiple IAV strains can result in new viruses. When exposed to humans, these new viruses cause contemporary seasonal epidemics or in the worst outcome creates an IAV pandemic strain against which the human population has little or no pre-existing immunity (Palese, 2004). Induction of an innate immune response by IAV occurs differentially among strains owing to the fact that the virus directs a variety of host response evasion strategies that impose limitations on host cell gene expression, including 5′ cap snatching activity, transcriptional regulation, and evasion of RIG-I activation of innate immune signaling (Schmolke and Garcia-Sastre, 2010). Early studies revealed that the viral nonstructural protein NS1 could direct evasion of IFN induction and host defenses during early infection through direct interference or indirectly via interference with host mRNA processing (Garcia-Sastre et al., 1998). Since then Pichlmair et al. demonstrated that RIG-I recognizes 5′ppp genomic ssRNA from IAV during infection (Fig. 2) (Pichlmair et al., 2006). This was the first demonstration, that RIG-I could recognize both dsRNA and ssRNA in a virus infection albeit via 5′ppp. Further studies identified the 3′ nontranslated region of the viral genome as a ligand for RIG-I independent of the 5′-ppp wherein lariat structure of the viral RNA end provides a substrate for recognition by RIG-I (Davis et al., 2012). The IAV non-structural protein NS1 was shown to antagonize RIG-I signaling (Mibayashi et al., 2007) and further shown to form a complex with RIG-I to suppress its function (Pichlmair et al., 2006). This complex formation blocks TRIM25 ubiquitin E3 ligase-mediated Lys63-linked ubiquitination of RIG-I, which is required for downstream signaling (Rajsbaum et al., 2012). Interestingly, this inhibition by NS1 is species specific, suggesting that this activity may be one of many required adaptations for host-species jump of influenza viruses (Rajsbaum et al., 2012). Mular et al. showed that NS1 from IAV strains that induce RIG-I activation can co-opt LGP2 for inhibition of RIG-I signaling (Malur et al., 2012).
Other studies suggest that the proinflammatory response triggered by RIG-I and TLR7 could actually be essential for efficient IAV replication (Pang et al., 2013). Pang et al. observed that viral replication in mice deficient for both MAVS and TLR7 was significantly reduced in the lung compared to wild-type mice. This replication could be enhanced through addition of bronchial lavages from wild-type infected mice, indicating that a robust interferon and pro-inflammatory responses may be helpful for viral replication. Indeed, the authors demonstrated that TLR7 and MAVS-dependent signaling is required to recruit monocyte-derived DCs to the lung and that these cells serve as target cells for viral infection (Pang et al., 2013). Thus, a careful balance between inflammatory signals that restrict viral infection and sustained inflammation that enhances viral replication may determine IAV infection outcome in humans.
Positive Sense RNA viruses
Coronaviridae
Both RIG-I and MDA5-dependent signaling has been shown to play a role in innate immune defenses against murine hepatitis virus infection (Li et al., 2010b). Experiments to knock down expression of either MDA5 or RIG-I revealed that lack of either imposes a block in virus-induced IFN-α/β production in murine oligodendrocyte cells (Li et al., 2010b). However, the respective roles of each RLR in recognition and innate immune activation were not defined. It remains possible that each RLR is essential for recognition of viral RNA at distinct stages during the infection as has been described for West Nile virus (WNV) infection (Errett et al., 2013). Because the coronavirus genomic RNA contains a 5′ cap moiety known to block RIG-I recognition of RNA, it is likely that replication intermediates or negative sense subgenomic RNAs and not genome/virion RNA serve as the stimulatory RNA for RIG-I during infection while MDA5 may recognize the replicating genome dsRNA products.
As large RNA viruses, coronaviruses contain the genetic space to encode for multiple proteins that interact with various factors in the innate immune signaling pathway to evade the host response to infection. Similar to arteriviruses, coronaviruses encode a protease capable of inhibiting ubiquitination of important innate signaling molecules to prevent induction of IFNs. For example, the SARS coronavirus has been shown to encode a Papin-like protease (PLpro) which inhibits IRF3 activation through deubiquitination of RIG-I, STING, TRAF and TBK1 (Chen et al., 2014; Sun et al., 2012). MERS-CoV protein 4a suppresses the RIG-I activator PACT in an RNA-dependent manner to prevent RIG-I signaling (Siu et al., 2014b). The M protein of SARS CoV has also been shown to interact with RIG-I, TRAF, and TBK1 to prevent the interaction of TRAF with other downstream effectors for signaling IFN induction (Siu et al., 2014a). Thus, coronaviruses encode multiple factors that direct a variety of innate immune evasion strategies to support infection.
Flaviviridae
Members of the Flaviviridae are enveloped, positive sense ssRNA viruses. The family includes flaviviruses, hepacivurses, and pestiviruses that cause disease in humans or agriculturally important animals (Fields et al., 2013).
Flavivirus
Flaviviruses are arboviruses that are transmitted between mosquito to birds within enzootic cycles that include humans and other animals as dead-end hosts (Fields et al., 2013). Various studies have shown that sustained IRF3 activity though MAVS-dependent RLR signaling is essential for host protection from WNV, an emerging flavivirus (Errett et al., 2013; Suthar et al., 2010). WNV is recognized by both RIG-I and MDA5, although the PAMP that includes 5′ppp for RIG-I recognition and dsRNA for MDA5 recognition is not yet fully charactered (Errett et al., 2013). Of interest is that the flaviviruses are recognized as nonself in the vertebrate host through the actions of both RIG-I and MDA5. Errett et al. described distinct temporal activity of each PRR during WNV infection, with RIG-I recognition and activity occurring early (less than12 hours post infection) and MDA5 activity beginning late (greater than 24 hours post-infection) (Errett et al., 2013) wherein both RLRs were important for host survival in vivo. The WNV virion RNA contains a 5′cap which is expected to mask the 5′ppp required for RIG-I recognition (Fields et al., 2013). Thus the likely stimulatory RNA for RIG-I includes either a 5′ppp replication intermediate which may form dsRNA complexes or non-coding subgenomic RNAs found during infection (Roby et al., 2014).
RIG-I activity is essential for survival of mice infected with the Japanese encephalitis virus (JEV), a flavivirus related to WNV (Kato et al., 2006). RIG-I was also shown to mediate the inflammatory response of neurons during infection which leads to the release of pro-inflammatory cytokines that recruit immune cells to the brain (Nazmi et al., 2011). The RNA ligand for RIG-I recognition of JEV is unknown but, based on similarity to WNV, it is likely a replication intermediate or subgenomic RNA marked by the presence of a 5′ppp for RIG-I recognition.
Innate immune responses to Dengue virus (DENV) are thought to involve RIG-I, MDA5, and TLR3 (Loo et al., 2008; Nasirudeen et al., 2011). Loo et al. describe incomplete signal suppression in cells lacking either RIG-I or MDA5, while DENV-induced innate immune signaling was completely lost in cells lacking both RLRs. Thus, both PRRs could be responsible for innate immune signaling during acute DENV infection, perhaps operating in temporally distinct fashion as in WNV infection (Loo et al., 2008). Similarly, Nasirudeen et al. observed measurable IFNβ production in individual knockdowns for RIG-I, MDA5, and TLR3, but triple knockdown cells completely lost all innate immune signal during DENV infection (Nasirudeen et al., 2011). DENV, like the other flaviviruses, has not been shown to antagonize the RLR signaling pathway directly but has been shown to antagonize the IFN signaling JAK-STAT pathway through degradation of STAT2 and other means recently reviewed in (Green et al., 2014).
Hepacivirus
Like their flavivirus cousins, hepaciviruses are enveloped ssRNA viruses. However, hepaciviruses are not arboviruses but instead are transmitted among humans via parenteral exposure to blood or blood products (Hajarizadeh et al., 2013). Hepatitis C virus (HCV) is the major human pathogen in this virus family, which also includes the related GB viruses (GBV), GBV-C through GBV-D, that do not cause disease in humans (Fields et al., 2013; Stapleton et al., 2011). In 2005 RIG-I was identified as an essential host factor that mediates the recognition of HCV RNA in hepatoma cell lines wherein RIG-I serves to recognize and bind to the 3′ nontranslated region of the HCV RNA (Sumpter et al., 2005). This discovery was followed with identification of the viral poly-uridine/cytosine (poly-U/UC) motif in the 3′ nontranslated region together with 5′ppp of the viral genome as the HCV PAMP (Fig. 2) (Saito et al., 2008; Uzri and Gehrke, 2009). The poly-U/UC PAMP motif is marked by a U-core region that typically includes 17 or more poly-U nucleotides that are essential for PAMP recognition such that U-core length and 5′ppp are critical determinants of PAMP activity and recognition of HCV RNA by RIG-I (Schnell et al., 2012). When introduced in cells the poly-U/UC motif was shown to trigger RIG-I signaling of innate immune defenses that could suppress HCV infection (Saito et al., 2008). The induction of innate immunity by RIG-I signaling is countered however during HCV infection through the actions of the viral nonstructural (NS)3/4A protease that targets and cleaves MAVS to suppress RIG-I signaling (Foy et al., 2003; Loo et al., 2006). HCV most often mediates a chronic infection in people acutely exposed to the virus, and NS3/4A cleavage of MAVS to inactivate the RIG-I pathway has been linked to chronic infection progression in models of HCV infection (Horner et al., 2012). The structural requirements of RIG-I recognition of the HCV poly-U/UC and the viral mechanisms of innate immune evasion have been thoroughly reviewed previously (Horner, 2014; Horner and Gale, 2013).
Togaviridae
The family Togaviridae includes the Alphaviruses, which are mosquito-transmitted viruses, and the Rubiviruses which are aerosol acquired viruses. Togaviridae members are enveloped, positive sense ssRNA viruses. Transcriptional profiling of human cells infected with alphaviruses, such as Chikungunya (CHIKV) and Sindbis (SINV) viruses, has shown a significant transcriptional upregulation of innate immune signaling genes, while host translation is shut off by the nsP2 viral protein to prevent protein production (Frolova et al., 2002) To determine the source of pathogen recognition, Burke et al. investigated IFN-α/β production in murine embryonic fibroblasts (MEFs) following infection with a mutant SINV unable to prevent host protein synthesis (Burke et al., 2009). Infection in WT MEFs with the mutant SINV led to IFN production. MDA5−/− MEFs produced 2-fold less IFN following infection with mutant SINV compared to WT MEFs (Burke et al., 2009). In contrast, RIG−/− MEFs produced significantly increased levels of IFN following mutant SINV infection. While Burke et al. conclude that these results indicate an important role for MDA5 in SINV recognition and signaling, they acknowledge the possibility that RIG-I exerts a significant restriction on SINV replication which would also explain these results (Burke et al., 2009). White et al. demonstrated that infection of human fibroblasts with CHIKV induces IRF3 dimerization and nuclear localization that is MAVS-dependent, consistent with MDA5 or RIG-I activation during infection (White et al., 2011). Further elucidation of the viral region recognized by either RIG-I or MDA5 and finer resolution of the roles of each PRR are necessary to determine the nature of pathogen recognition for the Togaviridae.
dsRNA viruses
Reoviridae
Members of this virus family are non-enveloped viruses that harbor multiple segments of dsRNA as their genome. The ten genomic segments of the reovirus genome form blunt-ended hairpin structures but were thought to evade recognition by RIG-I by encoding a phosohohydrolase that trims the 5′ppp to 5′ diphosphate (pp)(Fields et al., 2013). However, a recent report by Goubau et al. describes RIG-I recognition and activation induced by reovirus RNA in vitro, and reveal that MAVS signaling is important for viral control in vivo(Goubau et al., 2014). This study demonstrated that 5′ pp can serve as a recognition motif of PAMP RNA (Fig 2). Although RIG-I signaling was evident in response to 5′pp RNA treatment, the magnitude of RIG-I-induced IFN production was significantly greater following treatment with 5′ppp compared to 5′pp RNA. Thus, while viral phosohohydrolase activity to render 5′pp reduces RIG-I signaling of innate immunity during reovirus infection, it does not prevent PAMP recognition such that innate immune signaling remains active but attenuated. The physiological relevance of differential RIG-I signaling in response to 5′ppp vs 5′pp motifs remains unclear in terms of immune response induction and host control of virus infection.
Interestingly, resistance to grass carp reovirus (GCRV) has been mapped to specific insertions and deletions found in the Carp RIG-I gene homolog, further underscoring the importance of RIG-I protein in reovirus recognition (Wan et al., 2013). These observations raise the interesting possibility that changes in RIG-I protein composition/structure between species has allowed for changes in PAMP recognition profile of RIG-I orthologs, thus reflecting differential evolutionary pressures by reoviruses across species.
Setting the tone: RLR crosstalk leads to specific adaptive response
PAMP recognition and pathogen recognition receptor signaling and pathways inside the infected cell have profound effects on cell survival, the control of cell to cell virus spread, and the onset of effective host immunity. Often PRR signaling directs major alterations in gene expression of the infected cell, thus programming the cell for virus restriction or cell death. Additionally, PRR signaling does not occur in isolation, but instead viral products trigger a variety of cell signaling pathways that mediate crosstalk with one another to direct synergistic or antagonistic actions toward controlling infection (Loo and Gale, 2011). For example, RLR activation can function together with TLR3 signaling leading to enhanced HLA-I cross-presentation of antigen by dendritic cells (Lalwani et al., 2013). In contrast, activation of the RLR pathway by poly(I:C) or 5′ppp RNA was shown to suppress TLR-mediated transcription of IL 12b during bacterial infection (Negishi et al., 2012). This RLR signaling attenuated TH1 and TH17 responses leading to decreased survival of mice treated with a sublethal dose of Lysteria monocytogenes. Such crosstalk may explain how viral infection can exacerbate the symptoms of other microbial infections. Other in vivo models of RNA virus infection have also revealed the ability of the RLR pathway to program adaptive T cell responses. Utilizing various RLR pathway knockout animals has shown that CD4 and CD8 T cell responses can be severely dysregulated in both a cell extrinsic and cell intrinsic manner (Lazear et al., 2013; Suthar et al., 2010). Moreover, RNA virus infection has been shown to induce inflammasome activation through MAVS (Delaloye et al., 2009; Ermler et al., 2014; Pothlichet et al., 2013). Poeck et al. revealed that RIG-I may interact in a MAVS-dependent manner with CARD9 and Bcl-10 to activate NF-κB for transcription of IL-1β and other factors involved in inflammasome activation and onset of the inflammatory response to virus infection (Poeck et al., 2010). In addition, MAVS has been shown to interact with NLRP3 and direct association with the MAM and facilitate NLRP3 oligomerization leading to caspase-1 activation and release of mature IL-1β to drive the inflammatory response (Park et al., 2013). Regulation of apoptosis and inflammasome-induced cell death or pyroptosis through RLR/MAVS-dependent activation of the inflammasome may have evolved as a mechanism to prevent viral spread and induce a robust pro-inflammatory immune response, thus linking innate antiviral immunity with inflammatory signaling and responses that control virus infection.
Further studies to understand the outcome of PRR crosstalk and RLR signaling will direct the design of novel adjuvants or therapeutics that could be used to trigger specific innate responses to fight viral infection. A number of groups have already explored the use of RIG-I agonists to prevent viral infection. RIG-I activation using 5′ppp RNA can restrict viral infection when treatment occurs before or after virus challenge in vitro and in vivo (Chen et al., 2013; Han et al., 2011; Spiropoulou et al., 2009). Others have already developed RIG-I agonists which can activate the pro-inflammatory response in the absence of stimulatory RNA (Goulet et al., 2013; Han et al., 2011), and small molecule RIG-I agonists are being developed as antivirals to suppress RNA virus infection (Bedard et al., 2012). These studies present proof of concept that stimulation of a specific innate immune responses alone or combined with the presentation of viral antigen or targeted vaccine design could lead to broad spectrum antiviral strategies as therapy for RNA virus infection, as well as providing adjuvant actions for enhancement of RNA virus vaccines.
Conclusions
Detection of non-self nucleic acid PAMPs is essential for cellular and systemic host defense against viral pathogens. The innate antiviral immune response initiated by RIG-I activation serves to program a specific adaptive immune response effective against RNA viruses. In addition, cross-talk between the RLR pathway and other cellular PRR pathways can skew the host response towards tolerance or defense against invading pathogens. RIG-I plays a key role in the defense against RNA viruses from broad genera. RNA viruses have evolved mechanisms for either evading recognition by RIG-I or active antagonism of RIG-I signaling. However, beyond 5′ppp the characteristics of RIG-I ligands remain poorly understood. Future studies must focus on defining the specific RNA species, sequence and structure recognized by RIG-I during virus infection, and on assessing the conservation of such properties among RNA virus genera. Defining the characteristics of RIG-I ligands and the diverse effects on the innate and adaptive immune response when multiple PRR pathways interact is essential for informing design of novel therapeutics or adjuvants tailored to specific RNA virus pathogens.
Table 1.
Summary of RIG-I recognition and antagonism by RNA viruses.
| Virus | Predicted RIG-I ligand(s) | Evasion mechanism(s) | References |
|---|---|---|---|
| ssRNA viruses | |||
| Negative sense | |||
| Bornaviridae | Viral mRNA or replication intermediates | 5′ genome trimming TBK1 antagonism | (Schneider et al., 2007) (Unterstab et al., 2005) |
| Filoviridae | virion dsRNA | Sequestration of dsRNA by VP35 | (Basler et al., 2003; Cardenas et al., 2006) |
| Paramyxoviridae | 5′ trailer sequence | Facilitates LGP2 interaction with RIG-I | (Childs et al., 2012; Plumet et al., 2007) |
| Rhabdoviridae | Unknown | Unknown | |
| Arteriviridae | Unknown | Deubiquitination of RIG-I | (van Kasteren et al., 2012) |
| Arenaviridae | Unknown | 5′ppp overhang RNA may be RIG-I sink Z protein-RIG-I antagonism | (Marq et al., 2011) ((Fan et al., 2010) |
| Bunyaviridae | |||
| Hantavirus | Viral mRNAs | TBK1/IRF3 dephosphorylation | (Alff et al., 2006; Cimica et al., 2014; Lee et al., 2011) |
| Nairovirus | Unknown | Deubiquitination of RIG-I | (van Kasteren et al., 2012) |
| Phlebovirus | Unknown | TBK1/IKKε sequestration | (Ning et al., 2014) |
| Orthomyxoviridae | Genomic RNA (3′ NTR) | Prevents RIG-I Ubiquitination Cap-snatching host Translational shutoff | (Davis et al., 2012; Pichlmair et al., 2006) |
| Positive sense | |||
| Coronaviridae | Unknown | Deubiquitination of RIG-I, STING, TBK1, TRAF Suppression of PACT | (Chen et al., 2014; Sun et al., 2012) (Siu et al., 2014b) |
| Flaviviridae | |||
| Hepacivirus | Genomic RNA (3′ NTR) | MAVS cleavage | (Meylan et al., 2005; Saito et al., 2008) |
| Flavivirus | Unknown | Interferes with I FN pathway | (Nasirudeen et al., 2011) |
| Togaviridae | Unknown | Host translational shutoff | (Frolova et al., 2002) |
| dsRNA viruses | |||
| Reoviridae | 5′pp genomic RNAs | Phosphohydrolase trimming of 5′ppp | (Goubau et al., 2014) |
Research Highlights.
RIG-I is a cytosolic pathogen recognition receptor
RIG-I binds to PAMP RNA
RIG-I initiates the immune response to RNA virus infection
Acknowledgments
We would like to thank Drs. Kwan Chow, Amy Stone, Sowmya Pattabhi, Amina Negash, and John Errett for their helpful discussions and comments on this topic. MG Jr. and AMK are supported by National Institutes of Health grants AI104002–01, AI88778, and AI083019.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interests:
The authors disclose no competing interests.
Author Contributions:
AMK and MG Jr. contributed to the writing and approved the final manuscript.
References
- Albarino CG, Bergeron E, Erickson BR, Khristova ML, Rollin PE, Nichol ST. Efficient reverse genetics generation of infectious junin viruses differing in glycoprotein processing. Journal of virology. 2009;83:5606–5614. doi: 10.1128/JVI.00276-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alff PJ, Gavrilovskaya IN, Gorbunova E, Endriss K, Chong Y, Geimonen E, Sen N, Reich NC, Mackow ER. The pathogenic NY-1 hantavirus G1 cytoplasmic tail inhibits RIG-I- and TBK-1-directed interferon responses. Journal of virology. 2006;80:9676–9686. doi: 10.1128/JVI.00508-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler CF, Mikulasova A, Martinez-Sobrido L, Paragas J, Muhlberger E, Bray M, Klenk HD, Palese P, GarciaSastre A. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. Journal of virology. 2003;77:7945–7956. doi: 10.1128/JVI.77.14.7945-7956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler CF, Wang X, Muhlberger E, Volchkov V, Paragas J, Klenk HD, Garcia-Sastre A, Palese P. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:12289–12294. doi: 10.1073/pnas.220398297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A, Sachidanandam R, Garcia-Sastre A. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:16303–16308. doi: 10.1073/pnas.1005077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedard KM, Wang ML, Proll SC, Loo YM, Katze MG, Gale M, Jr, Iadonato SP. Isoflavone agonists of IRF-3 dependent signaling have antiviral activity against RNA viruses. Journal of virology. 2012;86:7334–7344. doi: 10.1128/JVI.06867-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns AM, Leser GP, Lamb RA, Horvath CM. The innate immune sensor LGP2 activates antiviral signaling by regulating MDA5-RNA interaction and filament assembly. Molecular cell. 2014;55:771–781. doi: 10.1016/j.molcel.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke CW, Gardner CL, Steffan JJ, Ryman KD, Klimstra WB. Characteristics of alpha/beta interferon induction after infection of murine fibroblasts with wild-type and mutant alphaviruses. Virology. 2009;395:121–132. doi: 10.1016/j.virol.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Chiu YH, Chen ZJ. The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Molecular cell. 2014;54:289–296. doi: 10.1016/j.molcel.2014.03.040. [DOI] [PubMed] [Google Scholar]
- Cardenas WB, Loo YM, Gale M, Jr, Hartman AL, Kimberlin CR, Martinez-Sobrido L, Saphire EO, Basler CF. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. Journal of virology. 2006;80:5168–5178. doi: 10.1128/JVI.02199-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazenave C, Uhlenbeck OC. RNA template-directed RNA synthesis by T7 RNA polymerase. Proceedings of the National Academy of Sciences of the United States of Africa. 1994;91:6972–6976. doi: 10.1073/pnas.91.15.6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Qian Y, Yan F, Tu J, Yang X, Xing Y, Chen Z. 5′-triphosphate-siRNA activates RIG-I-dependent type I interferon production and enhances inhibition of hepatitis B virus replication in HepG2.2.15 cells. European journal of pharmacology. 2013;721:86–95. doi: 10.1016/j.ejphar.2013.09.050. [DOI] [PubMed] [Google Scholar]
- Chen X, Yang X, Zheng Y, Yang Y, Xing Y, Chen Z. SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex. Protein & cell. 2014;5:369–381. doi: 10.1007/s13238-014-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang JJ, Davis ME, Gack MU. Regulation of RIG-I-like receptor signaling by host and viral proteins. Cytokine & growth factor reviews. 2014 doi: 10.1016/j.cytogfr.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs K, Randall R, Goodbourn S. Paramyxovirus V proteins interact with the RNA Helicase LGP2 to inhibit RIG-I-dependent interferon induction. Journal of virology. 2012;86:3411–3421. doi: 10.1128/JVI.06405-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs K, Stock N, Ross C, Andrejeva J, Hilton L, Skinner M, Randall R, Goodbourn S. mda-5, but not RIG-I, is a common target for paramyxovirus V proteins. Virology. 2007;359:190–200. doi: 10.1016/j.virol.2006.09.023. [DOI] [PubMed] [Google Scholar]
- Childs KS, Andrejeva J, Randall RE, Goodbourn S. Mechanism of mda-5 Inhibition by paramyxovirus V proteins. Journal of virology. 2009;83:1465–1473. doi: 10.1128/JVI.01768-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimica V, Dalrymple NA, Roth E, Nasonov A, Mackow ER. An innate immunity-regulating virulence determinant is uniquely encoded by the Andes virus nucleocapsid protein. mBio. 2014;5 doi: 10.1128/mBio.01088-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui S, Eisenacher K, Kirchhofer A, Brzozka K, Lammens A, Lammens K, Fujita T, Conzelmann KK, Krug A, Hopfner KP. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Molecular cell. 2008;29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- Davis WG, Bowzard JB, Sharma SD, Wiens ME, Ranjan P, Gangappa S, Stuchlik O, Pohl J, Donis RO, Katz JM, Cameron CE, Fujita T, Sambhara S. The 3′ untranslated regions of influenza genomic sequences are 5′PPP-independent ligands for RIG-I. PloS one. 2012;7:e32661. doi: 10.1371/journal.pone.0032661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaloye J, Roger T, Steiner-Tardivel QG, Le Roy D, Knaup Reymond M, Akira S, Petrilli V, Gomez CE, Perdiguero B, Tschopp J, Pantaleo G, Esteban M, Calandra T. Innate immune sensing of modified vaccinia virus Ankara (MVA) is mediated by TLR2-TLR6, MDA-5 and the NALP3 inflammasome. PLoS pathogens. 2009;5:e1000480. doi: 10.1371/journal.ppat.1000480. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Eisenacher K, Krug A. Regulation of RLR-mediated innate immune signaling–it is all about keeping the balance. European journal of cell biology. 2012;91:36–47. doi: 10.1016/j.ejcb.2011.01.011. [DOI] [PubMed] [Google Scholar]
- Ermler ME, Traylor Z, Patel K, Schattgen SA, Vanaja SK, Fitzgerald KA, Hise AG. Rift Valley fever virus infection induces activation of the NLRP3 inflammasome. Virology. 2014;449:174–180. doi: 10.1016/j.virol.2013.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errett JS, Suthar MS, McMillan A, Diamond MS, Gale M., Jr The essential, nonredundant roles of RIG-I and MDA5 in detecting and controlling West Nile virus infection. Journal of virology. 2013;87:11416–11425. doi: 10.1128/JVI.01488-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Briese T, Lipkin WI. Z proteins of New World arenaviruses bind RIG-I and interfere with type I interferon induction. Journal of virology. 2010;84:1785–1791. doi: 10.1128/JVI.01362-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields BN, Knipe DM, Howley PM. Fields virology. 6. Wolters Kluwer Health/Lippincott Williams & Wilkins; Philadelphia: 2013. p. 1. online resource (2 v. (xx, 2456, I–2482 p.)) [Google Scholar]
- Foy E, Li K, Wang C, Sumpter R, Jr, Ikeda M, Lemon SM, Gale M., Jr Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science. 2003;300:1145–1148. doi: 10.1126/science.1082604. [DOI] [PubMed] [Google Scholar]
- Frolova EI, Fayzulin RZ, Cook SH, Griffin DE, Rice CM, Frolov I. Roles of nonstructural protein nsP2 and Alpha/Beta interferons in determining the outcome of Sindbis virus infection. Journal of virology. 2002;76:11254–11264. doi: 10.1128/JVI.76.22.11254-11264.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furr SR, Moerdyk-Schauwecker M, Grdzelishvili VZ, Marriott I. RIG-I mediates nonsegmented negative-sense RNA virus-induced inflammatory immune responses of primary human astrocytes. Glia. 2010;58:1620–1629. doi: 10.1002/glia.21034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack MU, Kirchhofer A, Shin YC, Inn KS, Liang C, Cui S, Myong S, Ha T, Hopfner KP, Jung JU. Roles of RIG-I N-terminal tandem CARD and splice variant in TRIM25-mediated antiviral signal transduction. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16743–16748. doi: 10.1073/pnas.0804947105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- Garcin D, Kolakofsky D. Tacaribe arenavirus RNA synthesis in vitro is primer dependent and suggests an unusual model for the initiation of genome replication. Journal of virology. 1992;66:1370–1376. doi: 10.1128/jvi.66.3.1370-1376.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcin D, Lezzi M, Dobbs M, Elliott RM, Schmaljohn C, Kang CY, Kolakofsky D. The 5′ ends of Hantaan virus (Bunyaviridae) RNAs suggest a prime-and-realign mechanism for the initiation of RNA synthesis. Journal of virology. 1995;69:5754–5762. doi: 10.1128/jvi.69.9.5754-5762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubau D, Schlee M, Deddouche S, Pruijssers AJ, Zillinger T, Goldeck M, Schuberth C, Van der Veen AG, Fujimura T, Rehwinkel J, Iskarpatyoti JA, Barchet W, Ludwig J, Dermody TS, Hartmann G, Reis e Sousa C. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5′-diphosphates. Nature. 2014;514:372–375. doi: 10.1038/nature13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulet ML, Olagnier D, Xu Z, Paz S, Belgnaoui SM, Lafferty EI, Janelle V, Arguello M, Paquet M, Ghneim K, Richards S, Smith A, Wilkinson P, Cameron M, Kalinke U, Qureshi S, Lamarre A, Haddad EK, Sekaly RP, Peri S, Balachandran S, Lin R, Hiscott J. Systems analysis of a RIG-I agonist inducing broad spectrum inhibition of virus infectivity. PLoS pathogens. 2013;9:e1003298. doi: 10.1371/journal.ppat.1003298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green AM, Beatty PR, Hadjilaou A, Harris E. Innate immunity to dengue virus infection and subversion of antiviral responses. Journal of molecular biology. 2014;426:1148–1160. doi: 10.1016/j.jmb.2013.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habjan M, Andersson I, Klingstrom J, Schumann M, Martin A, Zimmermann P, Wagner V, Pichlmair A, Schneider U, Muhlberger E, Mirazimi A, Weber F. Processing of genome 5′ termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction. PloS one. 2008;3:e2032. doi: 10.1371/journal.pone.0002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajarizadeh B, Grebely J, Dore GJ. Epidemiology and natural history of HCV infection. Nature reviews. Gastroenterology & hepatology. 2013;10:553–562. doi: 10.1038/nrgastro.2013.107. [DOI] [PubMed] [Google Scholar]
- Han Q, Zhang C, Zhang J, Tian Z. Reversal of hepatitis B virus-induced immune tolerance by an immunostimulatory 3p-HBx-siRNAs in a retinoic acid inducible gene I-dependent manner. Hepatology. 2011;54:1179–1189. doi: 10.1002/hep.24505. [DOI] [PubMed] [Google Scholar]
- Hartman AL, Bird BH, Towner JS, Antoniadou ZA, Zaki SR, Nichol ST. Inhibition of IRF-3 activation by VP35 is critical for the high level of virulence of ebola virus. Journal of virology. 2008;82:2699–2704. doi: 10.1128/JVI.02344-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa S, Shiratori S, Yamato H, Kameyama T, Kitatsuji C, Kashigi F, Goto S, Kameoka S, Fujikura D, Yamada T, Mizutani T, Kazumata M, Sato M, Tanaka J, Asaka M, Ohba Y, Miyazaki T, Imamura M, Takaoka A. ZAPS is a potent stimulator of signaling mediated by the RNA helicase RIG-I during antiviral responses. Nature immunology. 2011;12:37–44. doi: 10.1038/ni.1963. [DOI] [PubMed] [Google Scholar]
- Horne KM, Vanlandingham DL. Bunyavirus-Vector Interactions. Viruses. 2014;6:4373–4397. doi: 10.3390/v6114373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner SM. Activation and evasion of antiviral innate immunity by hepatitis C virus. Journal of molecular biology. 2014;426:1198–1209. doi: 10.1016/j.jmb.2013.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner SM, Gale M., Jr Regulation of hepatic innate immunity by hepatitis C virus. Nature medicine. 2013;19:879–888. doi: 10.1038/nm.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner SM, Park HS, Gale M., Jr Control of innate immune signaling and membrane targeting by the Hepatitis C virus NS3/4A protease are governed by the NS3 helix alphaO. Journal of virology. 2012;86:3112–3120. doi: 10.1128/JVI.06727-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, Endres S, Hartmann G. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Kolokoltsova OA, Yun NE, Seregin AV, Poussard AL, Walker AG, Brasier AR, Zhao Y, Tian B, de la Torre JC, Paessler S. Junin virus infection activates the type I interferon pathway in a RIG-I-dependent manner. PLoS neglected tropical diseases. 2012;6:e1659. doi: 10.1371/journal.pntd.0001659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegame S, Takeda M, Ohno S, Nakatsu Y, Nakanishi Y, Yanagi Y. Both RIG-I and MDA5 RNA helicases contribute to the induction of alpha/beta interferon in measles virus-infected human cells. Journal of virology. 2010;84:372–379. doi: 10.1128/JVI.01690-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A, Pillai PS. Innate immunity to influenza virus infection. Nature reviews. Immunology. 2014;14:315–328. doi: 10.1038/nri3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Ramanathan A, Miller MT, Tang GQ, Gale M, Jr, Patel SS, Marcotrigiano J. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479:423–427. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. The Journal of experimental medicine. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nature immunology. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nature immunology. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- Kok KH, Lui PY, Ng MH, Siu KL, Au SW, Jin DY. The double-stranded RNA-binding protein PACT functions as a cellular activator of RIG-I to facilitate innate antiviral response. Cell host & microbe. 2011;9:299–309. doi: 10.1016/j.chom.2011.03.007. [DOI] [PubMed] [Google Scholar]
- Kolakofsky D. Isolation and characterization of Sendai virus DI-RNAs. Cell. 1976;8:547–555. doi: 10.1016/0092-8674(76)90223-3. [DOI] [PubMed] [Google Scholar]
- Komuro A, Bamming D, Horvath CM. Negative regulation of cytoplasmic RNA-mediated antiviral signaling. Cytokine. 2008;43:350–358. doi: 10.1016/j.cyto.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komuro A, Horvath CM. RNA- and virus-independent inhibition of antiviral signaling by RNA helicase LGP2. Journal of virology. 2006;80:12332–12342. doi: 10.1128/JVI.01325-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunei J, Grigorov B, Gerlier D, Cusack S. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- Lalwani P, Raftery MJ, Kobak L, Rang A, Giese T, Matthaei M, van den Elsen PJ, Wolff T, Kruger DH, Schonrich G. Hantaviral mechanisms driving HLA class I antigen presentation require both RIG-I and TRIF. European journal of immunology. 2013;43:2566–2576. doi: 10.1002/eji.201243066. [DOI] [PubMed] [Google Scholar]
- Lazear HM, Pinto AK, Ramos HJ, Vick SC, Shrestha B, Suthar MS, Gale M, Jr, Diamond MS. Pattern recognition receptor MDA5 modulates CD8+ T cell-dependent clearance of West Nile virus from the central nervous system. Journal of virology. 2013;87:11401–11415. doi: 10.1128/JVI.01403-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Lalwani P, Raftery MJ, Matthaei M, Lutteke N, Kirsanovs S, Binder M, Ulrich RG, Giese T, Wolff T, Kruger DH, Schonrich G. RNA helicase retinoic acid-inducible gene I as a sensor of Hantaan virus replication. The Journal of general virology. 2011;92:2191–2200. doi: 10.1099/vir.0.032367-0. [DOI] [PubMed] [Google Scholar]
- Li H, Zheng Z, Zhou P, Zhang B, Shi Z, Hu Q, Wang H. The cysteine protease domain of porcine reproductive and respiratory syndrome virus non-structural protein 2 antagonizes interferon regulatory factor 3 activation. The Journal of general virology. 2010a;91:2947–2958. doi: 10.1099/vir.0.025205-0. [DOI] [PubMed] [Google Scholar]
- Li J, Liu Y, Zhang X. Murine coronavirus induces type I interferon in oligodendrocytes through recognition by RIG-I and MDA5. Journal of virology. 2010b;84:6472–6482. doi: 10.1128/JVI.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HM, Loo YM, Horner SM, Zornetzer GA, Katze MG, Gale M., Jr The mitochondrial targeting chaperone 14-3-3epsilon regulates a RIG-I translocon that mediates membrane association and innate antiviral immunity. Cell host & microbe. 2012;11:528–537. doi: 10.1016/j.chom.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo MK, Peeples ME, Bellini WJ, Nichol ST, Rota PA, Spiropoulou CF. Distinct and overlapping roles of Nipah virus P gene products in modulating the human endothelial cell antiviral response. PloS one. 2012;7:e47790. doi: 10.1371/journal.pone.0047790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM, Fornek J, Crochet N, Bajwa G, Perwitasari 0, Martinez-Sobrido L, Akira S, Gill MA, Garcia-Sastre A, Katze MG, Gale M., Jr Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. Journal of virology. 2008;82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM, Gale M., Jr Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM, Owen DM, Li K, Erickson AK, Johnson CL, Fish PM, Carney DS, Wang T, Ishida H, Yoneyama M, Fujita T, Saito T, Lee WM, Hagedorn CH, Lau DT, Weinman SA, Lemon SM, Gale M., Jr Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:6001–6006. doi: 10.1073/pnas.0601523103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Xu H, Ranjith-Kumar CT, Brooks MT, Hou TY, Hu F, Herr AB, Strong RK, Kao CC, Li P. The structural basis of 5′ triphosphate double-stranded RNA recognition by RIG-I C-terminal domain. Structure. 2010;18:1032–1043. doi: 10.1016/j.str.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, Ding SC, Vela A, Kohlway A, Lindenbach BD, Pyle AM. Structural insights into RNA recognition by RIG-I. Cell. 2011;147:409–422. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, Xiao S, Jiang Y, Jin H, Wang D, Liu M, Chen H, Fang L. Porcine reproductive and respiratory syndrome virus (PRRSV) suppresses interferon-beta production by interfering with the RIG-I signaling pathway. Molecular immunology. 2008;45:2839–2846. doi: 10.1016/j.molimm.2008.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthra P, Ramanan P, Mire CE, Weisend C, Tsuda Y, Yen B, Liu G, Leung DW, Geisbert TW, Ebihara H, Amarasinghe GK, Basler CF. Mutual antagonism between the Ebola virus VP35 protein and the RIG-I activator PACT determines infection outcome. Cell host & microbe. 2013;14:74–84. doi: 10.1016/j.chom.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malur M, Gale M, Jr, Krug RM. LGP2 downregulates interferon production during infection with seasonal human influenza A viruses that activate interferon regulatory factor 3. Journal of virology. 2012;86:10733–10738. doi: 10.1128/JVI.00510-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maringer K, Fernandez-Sesma A. Message in a bottle: lessons learned from antagonism of STING signalling during RNA virus infection. Cytokine & growth factor reviews. 2014 doi: 10.1016/j.cytogfr.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marq JB, Hausmann S, Veillard N, Kolakofsky D, Garcin D. Short double-stranded RNAs with an overhanging 5′ ppp-nucleotide, as found in arenavirus genomes, act as RIG-I decoys. The Journal of biological chemistry. 2011;286:6108–6116. doi: 10.1074/jbc.M110.186262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marq JB, Kolakofsky D, Garcin D. Unpaired 5′ ppp-nucleotides, as found in arenavirus double-stranded RNA panhandles, are not recognized by RIG-I. The Journal of biological chemistry. 2010;285:18208–18216. doi: 10.1074/jbc.M109.089425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Gil L, Goff PH, Hai R, Garcia-Sastre A, Shaw ML, Palese P. A Sendai virus-derived RNA agonist of RIG-I as a virus vaccine adjuvant. Journal of virology. 2013;87:1290–1300. doi: 10.1128/JVI.02338-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita M. Ficolins: complement-activating lectins involved in innate immunity. Journal of innate immunity. 2010;2:24–32. doi: 10.1159/000228160. [DOI] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- Mibayashi M, Martinez-Sobrido L, Loo YM, Cardenas WB, Gale M, Jr, Garcia-Sastre A. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. Journal of virology. 2007;81:514–524. doi: 10.1128/JVI.01265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motz C, Schuhmann KM, Kirchhofer A, Moldt M, Witte G, Conzelmann KK, Hopfner KP. Paramyxovirus V proteins disrupt the fold of the RNA sensor MDA5 to inhibit antiviral signaling. Science. 2013;339:690–693. doi: 10.1126/science.1230949. [DOI] [PubMed] [Google Scholar]
- Murphy K, Travers P, Walport M, Janeway C. Janeway’s immunobiology. 7. Garland Science; New York: 2008. [Google Scholar]
- Nasirudeen AM, Wong HH, Thien P, Xu S, Lam KP, Liu DX. RIG-I, MDA5 and TLR3 synergistically play an important role in restriction of dengue virus infection. PLoS neglected tropical diseases. 2011;5:e926. doi: 10.1371/journal.pntd.0000926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazmi A, Dutta K, Basu A. RIG-I mediates innate immune response in mouse neurons following Japanese encephalitis virus infection. PloS one. 2011;6:e21761. doi: 10.1371/journal.pone.0021761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negishi H, Yanai H, Nakajima A, Koshiba R, Atarashi K, Matsuda A, Matsuki K, Miki S, Doi T, Aderem A, Nishio J, Smale ST, Honda K, Taniguchi T. Cross-interference of RLR and TLR signaling pathways modulates antibacterial T cell responses. Nature immunology. 2012;13:659–666. doi: 10.1038/ni.2307. [DOI] [PubMed] [Google Scholar]
- Ning YJ, Wang M, Deng M, Shen S, Liu W, Cao WC, Deng F, Wang YY, Hu Z, Wang H. Viral suppression of innate immunity via spatial isolation of TBK1/IKKepsilon from mitochondrial antiviral platform. Journal of molecular cell biology. 2014;6:324–337. doi: 10.1093/jmcb/mju015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palese P. Influenza: old and new threats. Nature medicine. 2004;10:S82–87. doi: 10.1038/nm1141. [DOI] [PubMed] [Google Scholar]
- Pang IK, Pillai PS, Iwasaki A. Efficient influenza A virus replication in the respiratory tract requires signals from TLR7 and RIG-I. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:13910–13915. doi: 10.1073/pnas.1303275110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Juliana C, Hong S, Datta P, Hwang I, Fernandes-Alnemri T, Yu JW, Alnemri ES. The mitochondrial antiviral protein MAVS associates with NLRP3 and regulates its inflammasome activity. J Immunol. 2013;191:4358–4366. doi: 10.4049/jimmunol.1301170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel JR, Jain A, Chou YY, Baum A, Ha T, Garcia-Sastre A. ATPase-driven oligomerization of RIG-I on RNA allows optimal activation of type-I interferon. EMBO reports. 2013;14:780–787. doi: 10.1038/embor.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Plumet S, Herschke F, Bourhis JM, Valentin H, Longhi S, Gerlier D. Cytosolic 5′-triphosphate ended viral leader transcript of measles virus as activator of the RIG I-mediated interferon response. PloS one. 2007;2:e279. doi: 10.1371/journal.pone.0000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poeck H, Bscheider M, Gross O, Finger K, Roth S, Rebsamen M, Hannesschlager N, Schlee M, Rothenfusser S, Barchet W, Kato H, Akira S, Inoue S, Endres S, Peschel C, Hartmann G, Hornung V, Ruland J. Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1 beta production. Nature immunology. 2010;11:63–69. doi: 10.1038/ni.1824. [DOI] [PubMed] [Google Scholar]
- Pothlichet J, Meunier I, Davis BK, Ting JP, Skamene E, von Messling V, Vidal SM. Type I IFN triggers RIG-I/TLR3/NLRP3-dependent inflammasome activation in influenza A virus infected cells. PLoS pathogens. 2013;9:e1003256. doi: 10.1371/journal.ppat.1003256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins KC, Cardenas WB, Basler CF. Ebola virus protein VP35 impairs the function of interferon regulatory factor-activating kinases IKKepsilon and TBK-1. Journal of virology. 2009;83:3069–3077. doi: 10.1128/JVI.01875-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajsbaum R, Albrecht RA, Wang MK, Maharaj NP, Versteeg GA, Nistal-Villan E, Garcia-Sastre A, Gack MU. Species-specific inhibition of RIG-I ubiquitination and IFN induction by the influenza A virus NS1 protein. PLoS pathogens. 2012;8:e1003059. doi: 10.1371/journal.ppat.1003059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan P, Edwards MR, Shabman RS, Leung DW, Endlich-Frazier AC, Borek DM, Otwinowski Z, Liu G, Huh J, Basler CF, Amarasinghe GK. Structural basis for Marburg virus VP35-mediated immune evasion mechanisms. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:20661–20666. doi: 10.1073/pnas.1213559109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter A, Ackermann A, Kothlow S, Rinder M, Kaspers B, Staeheli P. Avian bornaviruses escape recognition by the innate immune system. Viruses. 2010;2:927–938. doi: 10.3390/v2040927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roby JA, Pijlman GP, Wilusz J, Khromykh AA. Noncoding subgenomic flavivirus RNA: multiple functions in West Nile virus pathogenesis and modulation of host responses. Viruses. 2014;6:404–427. doi: 10.3390/v6020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez KR, Horvath CM. Amino acid requirements for MDA5 and LGP2 recognition by paramyxovirus V proteins: a single arginine distinguishes MDA5 from RIG-I. Journal of virology. 2013;87:2974–2978. doi: 10.1128/JVI.02843-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez KR, Horvath CM. Paramyxovirus V protein interaction with the antiviral sensor LGP2 disrupts MDA5 signaling enhancement but is not relevant to LGP2-mediated RLR signaling inhibition. Journal of virology. 2014;88:8180–8188. doi: 10.1128/JVI.00737-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runge S, Sparrer KM, Lassig C, Hembach K, Baum A, Garcia-Sastre A, Soding J, Conzelmann KK, Hopfner KP. In vivo ligands of MDA5 and RIG-I in measles virus-infected cells. PLoS pathogens. 2014;10:e1004081. doi: 10.1371/journal.ppat.1004081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Hirai R, Loo YM, Owen D, Johnson CL, Sinha SC, Akira S, Fujita T, Gale M., Jr Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:582–587. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K, Tsujimura T, Fujita T, Akira S, Takeuchi O. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:1512–1517. doi: 10.1073/pnas.0912986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, Coch C, Janke M, Mihailovic A, Wardle G, Juranek S, Kato H, Kawai T, Poeck H, Fitzgerald KA, Takeuchi O, Akira S, Tuschl T, Latz E, Ludwig J, Hartmann G. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Schwerd T, Hamm W, Hellmuth JC, Cui S, Wenzel M, Hoffmann FS, Michallet MC, Besch R, Hopfner KP, Endres S, Rothenfusser S. 5′-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:12067–12072. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmolke M, Garcia-Sastre A. Evasion of innate and adaptive immune responses by influenza A virus. Cellular microbiology. 2010;12:873–880. doi: 10.1111/j.1462-5822.2010.01475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider U, Martin A, Schwemmle M, Staeheli P. Genome trimming by Borna disease viruses: viral replication control or escape from cellular surveillance? Cellular and molecular life sciences: CMLS. 2007;64:1038–1042. doi: 10.1007/s00018-007-6545-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell G, Loo YM, Marcotrigiano J, Gale M., Jr Uridine composition of the poly-U/UC tract of HCV RNA defines non-self recognition by RIG-I. PLoS pathogens. 2012;8:e1002839. doi: 10.1371/journal.ppat.1002839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, Dabelic R, Manicassamy B, Aitchison JD, Aderem A, Elliott RM, Garcia-Sastre A, Racaniello V, Snijder EJ, Yokoyama WM, Diamond MS, Virgin HW, Rice CM. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014;505:691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu C, Li X, Li P. The mechanism of double-stranded DNA sensing through the cGAS-STING pathway. Cytokine & growth factor reviews. 2014 doi: 10.1016/j.cytogfr.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu KL, Chan CP, Kok KH, Chiu-Yat Woo P, Jin DY. Suppression of innate antiviral response by severe acute respiratory syndrome coronavirus M protein is mediated through the first transmembrane domain. Cellular & molecular immunology. 2014a;11:141–149. doi: 10.1038/cmi.2013.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu KL, Yeung ML, Kok KH, Yuen KS, Kew C, Lui PY, Chan CP, Tse H, Woo PC, Yuen KY, Jin DY. Middle east respiratory syndrome coronavirus 4a protein is a double-stranded RNA-binding protein that suppresses PACT-induced activation of RIG-I and MDA5 in the innate antiviral response. Journal of virology. 2014b;88:4866–4876. doi: 10.1128/JVI.03649-13. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Spiropoulou CF, Ranjan P, Pearce MB, Sealy TK, Albarino CG, Gangappa S, Fujita T, Rollin PE, Nichol ST, Ksiazek TG, Sambhara S. RIG-I activation inhibits ebolavirus replication. Virology. 2009;392:11–15. doi: 10.1016/j.virol.2009.06.032. [DOI] [PubMed] [Google Scholar]
- Stapleton JT, Foung S, Muerhoff AS, Bukh J, Simmonds P. The GB viruses: a review and proposed classification of GBV-A, GBV-C (HGV), and GBV-D in genus Pegivirus within the family Flaviviridae. The Journal of general virology. 2011;92:233–246. doi: 10.1099/vir.0.027490-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumpter R, Jr, Loo YM, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, Gale M., Jr Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. Journal of virology. 2005;79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Xing Y, Chen X, Zheng Y, Yang Y, Nichols DB, Clementz MA, Banach BS, Li K, Baker SC, Chen Z. Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling. PloS one. 2012;7:e30802. doi: 10.1371/journal.pone.0030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suthar MS, Ma DY, Thomas S, Lund JM, Zhang N, Daffis S, Rudensky AY, Bevan MJ, Clark EA, Kaja MK, Diamond MS, Gale M., Jr IPS-1 is essential for the control of West Nile virus infection and immunity. PLoS pathogens. 2010;6:e1000757. doi: 10.1371/journal.ppat.1000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Kawai T, Kumar H, Sato S, Yonehara S, Akira S. Roles of caspase-8 and caspase-10 in innate immune responses to double-stranded RNA. J Immunol. 2006;176:4520–4524. doi: 10.4049/jimmunol.176.8.4520. [DOI] [PubMed] [Google Scholar]
- Unterstab G, Ludwig S, Anton A, Planz O, Dauber B, Krappmann D, Heins G, Ehrhardt C, Wolff T. Viral targeting of the interferon-{beta}-inducing Traf family member-associated NF-{kappa}B activator (TANK)-binding kinase-1. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:13640–13645. doi: 10.1073/pnas.0502883102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzri D, Gehrke L. Nucleotide sequences and modifications that determine RIG-I/RNA binding and signaling activities. Journal of virology. 2009;83:4174–4184. doi: 10.1128/JVI.02449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kasteren PB, Beugeling C, Ninaber DK, Frias-Staheli N, van Boheemen S, Garcia-Sastre A, Snijder EJ, Kikkert M. Arterivirus and nairovirus ovarian tumor domain-containing Deubiquitinases target activated RIG-I to control innate immune signaling. Journal of virology. 2012;86:773–785. doi: 10.1128/JVI.06277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Q, Su J, Wang L, Peng L, Chen L. Correlation between grass carp (Ctenopharyngodon idella) resistance to grass carp reovirus and the genetic insert-deletion polymorphisms in promoter and intron of RIG-I gene. Gene. 2013;516:320–327. doi: 10.1016/j.gene.2012.12.069. [DOI] [PubMed] [Google Scholar]
- Wang H, Vaheri A, Weber F, Plyusnin A. Old World hantaviruses do not produce detectable amounts of dsRNA in infected cells and the 5′ termini of their genomic RNAs are monophosphorylated. The Journal of general virology. 2011;92:1199–1204. doi: 10.1099/vir.0.029405-0. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ludwig J, Schuberth C, Goldeck M, Schlee M, Li H, Juranek S, Sheng G, Micura R, Tuschl T, Hartmann G, Patel DJ. Structural and functional insights into 5′-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nature structural & molecular biology. 2010;17:781–787. doi: 10.1038/nsmb.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen H, Miao EA, Ting JP. Mechanisms of NOD-like receptor-associated inflammasome activation. Immunity. 2013;39:432–441. doi: 10.1016/j.immuni.2013.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White LK, Sali T, Alvarado D, Gatti E, Pierre P, Streblow D, Defilippis VR. Chikungunya virus induces IPS-1-dependent innate immune activation and protein kinase R-independent translational shutoff. Journal of virology. 2011;85:606–620. doi: 10.1128/JVI.00767-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao TS, Fitzgerald KA. The cGAS-STING pathway for DNA sensing. Molecular cell. 2013;51:135–139. doi: 10.1016/j.molcel.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Molecular cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Ye Z, Ting JP. NLR, the nucleotide-binding domain leucine-rich repeat containing gene family. Current opinion in immunology. 2008;20:3–9. doi: 10.1016/j.coi.2008.01.003. [DOI] [PubMed] [Google Scholar]
- Zhang X, Shi H, Wu J, Sun L, Chen C, Chen ZJ. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Molecular cell. 2013;51:226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]


