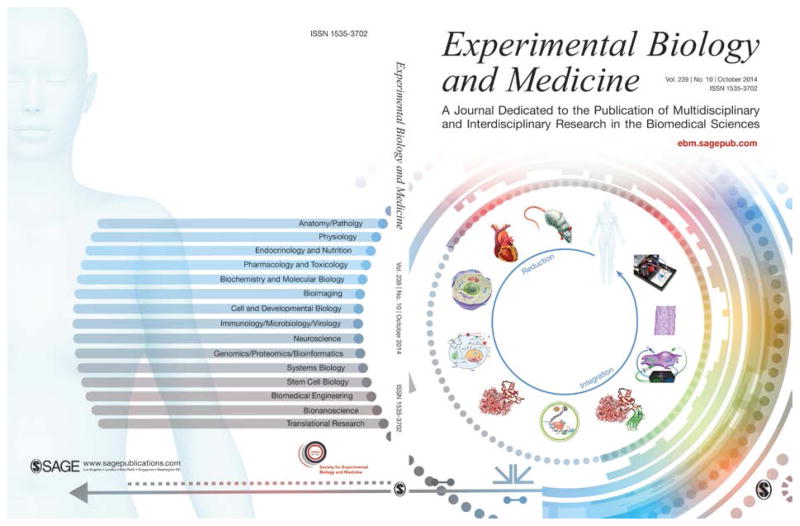
With this issue, Experimental Biology and Medicine (EBM) changes its cover from one that it has used for 8 and 1/2 years to one that is based upon the hermeneutic circle of biology (Figure 1), which was highlighted in the introduction to last year’s Thematic Issue on the Biology and Medicine of Microphysiological Systems.1 The rationale for the new cover is provided in the accompanying letter from EBM Editor-in-Chief, Steven R. Goodman.2 The symbolism of the cover is explained in this commentary by two physicists, one active in biophysics, physiology, and biomedical engineering (JPW), and one who has made the transition from computational physics to philosophical theology and hermeneutics (APP).
Figure 1.
The new cover for Experimental Biology and Medicine (EBM) that shows the hermeneutic circle of biology on the right (front cover) and the topical areas of EBM on the left (back cover). The circle begins with the animal or organism at the top and progresses counterclockwise through organs, cells, metabolic and signaling networks, biomolecules, and finally the genome at the bottom, thereby representing the reductionist limit of biology. Continuing up the right side through the synthesis of molecules, instrumented and controlled cells, engineered tissues, and engineered organs (including organs for regenerative medicine and organ-on-chip microphysiological systems), the circle closes with a representation of the human that integrates these syntheses. It may be necessary to traverse the circle multiple times, and also to employ corresponding circles for the classic biological model organisms that range from bacteria and yeast to zebrafish.
Hermeneutics is the study of interpretation and meaning, originally of texts. In this sense, the hermeneutic circle is a means of exploring the relationship of the parts of a document to its entirety,3 and the concept has been expanded to explore the role of context in a vocabulary within a language, and in history. The basic principle of the hermeneutic circle3 is that “one cannot understand the whole until one understands the parts, and one cannot understand the parts until one understands the whole,” and indeed, the distinction of whole from parts is fundamental to viewing biology as a circle. As a first approximation, one could define a biological hierarchy that spans organism, organ, cell, signaling network, molecule, and genome. Anything below the organism is either parts, or lesser parts. So what constitutes the parts as parts?
In many ways, the identification of whole versus parts reduces to the need to define boundaries, which in living organisms are often neither well defined nor amenable to explanation by a quantitative theory, and support high fluxes and multiple interactions (in the human, for example, there is the microbiome bounded by the lumen of the gastrointestinal tract, and different ones on external surfaces). Of interest to biology are the boundaries within an organism, and boundaries within those boundaries. Whereas in physics, system boundaries may be defined for computational convenience and to minimize the complexity of the problem, in biology they are defined in terms of biological function, which for our purposes means recreating a biochemical and biomechanical environment. At the level of biochemistry, function means the reactions that matter in vitro have to be sufficiently like the same reactions in vivo. At all levels, function means whatever contributes to the life of the organism, whatever makes it live, whatever enables it to cope with changes in its environment, large or small, i.e., contributes to the fitness of the organism. While we need not define life here, in contrast to inanimate objects, a living organism has a stake in its own existence as well as that of other organisms, if only to eat, stay fit, reproduce, and not be eaten. In one form or another, biological systems express intention, whereas physical systems simply respond passively. Having a stake (or equivalently, coping) is not an explanatory category in physics or chemistry, but appears for the first time in biology. It is what allows us to recognize function in the parts of biological systems.
Function in biology (and life itself) is an emergent behavior of an ensemble of different molecules – a collective behavior that is not readily predicted from knowledge of the behavior of each individual molecule. Functions are often associated with boundaries, and boundaries contribute to persistence. The cell with its cytoplasmic membrane is a basic unit of biological function and meets most people’s minimum requirements for the definition of “life.” But cells can come together and exhibit emergent behavior that supports a discrete process, whether as the first, rudimentary tubular heart to perfuse an organism, or the endothelial-epithelial interface of the lung alveolus for gas exchange, or the kidney tubules that form the nephron for filtration, absorption, and secretion. Early isolated-organ research that undergirds much of our understanding of physiology benefited from the fact that cells within the boundary provided by an isolated organ were more robust and could sustain their normal function longer than they could alone.1
Were one to remove for study a living organ from an animal, the ability of that organ to function normally in isolation would be determined by the extent to which one can recreate the organ’s environment as seen at the organ boundary. Perfusion is most important, in that nutrients must be delivered across the boundary and waste products removed, but organs often require other types of input (electrical, hormonal, immune, mechanical and neuronal) to support and regulate organ function and avoid cellular dedifferentiation. Similarly, were one to remove a living cell from a tissue to study the cell in isolation, the accuracy with which the behavior of the cell would match that in the tissue would be determined by the nature of the interactions between the cell and its microenvironment. Were the cell normally to interact only with the surrounding fluid medium, then recapitulation of that microenvironment would simply require matching the chemistries and flows. If the cell had mechanical interactions with matrix or neighboring cells, reconstructing the boundary would be much more complicated, in that stresses or strains would need to be controlled. Similar issues arise with electrical signaling, as in skeletal and smooth muscle and the heart and brain. Two-dimensional life in a Petri dish or well plate has been studied in great detail to create many self-consistent biological explanations, but cells in a Petri dish may not have the same experiences as cells in three-dimensional tissue. The issue is one of context and boundaries.
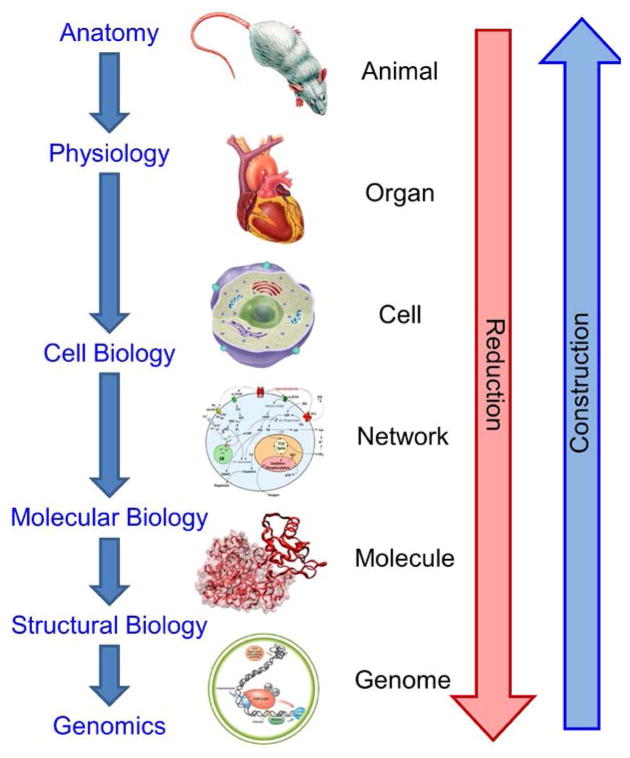
Over the past 2,500 years, physics, physiology, and biology have been highly reductionist, with the goal of finding the most fundamental particles, interactions, or information. Figure 2 illustrates reductionist biology, which began ~200 A.D. with the physiological studies of humans and animals by Galen of Pergamon,4 and benefited greatly from studies of isolated organs beginning in the 1880s.5 The 1950s brought cell biology to the forefront, with our favorite contributions being the development of the yeast chemostat by the organic chemist Aaron Novick and the famous nuclear physicist and inventor Leo Szilard6;7 and the discovery by George Gey of the HeLa immortal cell line isolated from Henrietta Lacks.8 The biochemistry of regulatory networks was the focus of intense research throughout most of the twentieth century.9 X-ray crystallography, for example that of the chemist Rosalind Franklin,10;11 nuclear magnetic resonance, and other techniques led to accurate models of biomolecules and the development of structural biology. Today, we can readily explore what may be the reductionist limit of biology – the rapid sequencing of the genome of an individual human and a myriad of other organisms.
Figure 2.
A simplified, linear view of the history of biology and medicine, showing the directions of reduction and construction.
One might ask, “What do you do when you get to the bottom?” You can look for a new understanding of the unanswered questions. You can continue to dig deeper and figure out how atoms make genes, and then how elementary particles make atoms, etc., on the way to the theory of everything. You might identify a distinct biological “layer” below the genome. Or you can work your way back up toward the animal or organism, in accord with Anderson’s constructionist approach,12 building upon the knowledge gained at the bottom. One possibility would be to use the computational models of systems biology for the upward construction; however, a fully computational model from genome to organism might, if ever attempted, require dealing with a mole of differential equations, termed a Leibniz.13;14 The intercalation of computational modeling and experiments may enable the crossing of emergent “boundaries” that defy calculation, such as the need to “solve” the protein folding problem to get from genome to protein. But in any case, the linear map in Figure 2 of the ongoing exploration of biology is by far overly simplistic and is provided merely as a heuristic tool.
Reductionism by definition ignores the whole and focuses on the parts. In a post-reductionist era, one might also ask, “What is the role of a specific part in the whole?” That is a question of both function and integration and construction, and it presents clear differences between physics and biology. Physics describes dynamic interactions in terms of fundamental or phenomenological laws that govern the state of the matter being studied. In 1949, the physicist Max Delbrück provided a concise distinction between physics and biology: “… any living cell carries with it the experiences of a billion years of experimentation by its ancestors. You cannot expect to explain so wise an old bird in a few simple words.”15 Biology has laws, but the operation of every living organism is determined not only by the laws of biology, physics, and chemistry, but also by historic instructions within the genome and its design for molecules that may be specific to each individual organism. Just as in our studies of literature, as we seek to deepen our understanding of human physiology it is important not to overlook the historical context, which for biology reaches back to the origins of life.
Returning to the top along the left side of the circle in Figure 1 is topologically equivalent to the linear Figure 2 and might be accomplished with some mix of computation and experiment. But with the circle, we have the alternative possibility of returning from the genome to the organism by a different path – one that allows through construction precise control of the boundaries to the problem, and hence the right side of Figure 1: it is now possible to engineer biomolecules for particular functions, including reporting the state of the molecule or that of its environment, which in turn leads to the ability to perform closed-loop control of cellular function.16–18 It is now possible to engineer tissues, either for basic science, repair, or replacement, wherein the cells within the engineered tissue are enjoying microenvironments (internal boundaries) that are much more representative of those in vivo than those provided by two-dimensional life on plastic. The development of engineered tissue constructs, often with human cells, is ushering in tissues on chips, the subject of the 2014 EBM Thematic Issue1;19–34 and other reviews.35–39 Ultimately, these novel constructions will lead to organs on chips that will be connected together to create coupled organ systems40–43 that will allow us to generate microphysiological models that are simpler than the complete organism. The net result is that we can now choose to close the hermeneutic circle on the right not with an exhaustively accurate representation of biology, but with a synthetic one that allows us to insert sensors and actuators and more cleanly define the boundaries of the biological subsystem under study. Organs on chips can play a central role in this process, but there are many other ways to close the circle. Human physiology and medicine have been advanced by studies of biological model organisms such as bacteria, bacteriophage, yeast, C. elegans, zebrafish, mice in general and particularly humanized mice.44 Each of these can be explored with their own hermeneutic circles that overlay that of the human. They may allow us to traverse the circle at many different levels of reduction and construction.
Mathematical models can help us along the way: biology has a growing number of theories and empirical (or effective) models that simplify upward construction at appropriate scales. It is generally more efficient to invoke Michaelis-Menton kinetics than compute enzyme molecular dynamics from first principles, just as functional models of integrated circuits and larger electronic modules allow us to build supercomputers without knowledge of the individual electronic gates. Given the control of boundary conditions and variables offered by the constructionist approach, it may be easier to calculate our way upwards on the right side of the circle than in either direction on the left. Mathematics can provide biology with rigorous predictions that can be tested quantitatively in a controlled, reproducible setting.45
Is such a synthetic system (and its models) complex enough to be of any value? Clearly, no one would attempt in the immediate future to create a multi-organ microphysiological system with a hundred organs – it would be too complicated to understand. To quote the physician and physiologist Arturo Rosenblueth and the mathematician and philosopher Norbert Wiener, “[t]he best material model for a cat is another, or preferably the same cat.”46 Little new information could be extracted from a cumbersome “[m]ap of the Empire that was of the same Scale as the Empire and that coincided with it point for point.”47 Instead, each of the stages along the right side of the circle might best be viewed as a toy model, as exemplified by the work of the physicist Freeman Dyson on the origin of life.48 Such toy models serve as a framework in which questions about biology can be posed with some degree of precision, with answers that build intuition, rather than exact reconstructions of in vivo biology. Models become particularly interesting when they are of sufficient complexity that they can produce emergent phenomena, which for an in vitro neural network would be some form of biocomputation and in coupled microphysiological systems would be organ-organ interactions that lead to unanticipated actions of drugs. Anderson anticipates as a general principle what we might learn from the constructionist approach of assembling simplified in vitro biological models into more complicated in vitro systems: “We expect to encounter fascinating and, I believe, very fundamental questions at each stage in fitting together less complicated pieces into the more complicated system and understanding the basically new types of behavior which can result.”12 Ultimately, at each stage of mirroring tissue and organ-level function(s), the fundamental question must be whether the details included in the model are both necessary and sufficient.
We can now return, full circle, to Figure 1 and the new cover of EBM. The hermeneutical circle of understanding, in its original sense of interpreting texts, is an iterative process. It is like a mathematical iterative process in that it may or may not converge, and unlike mathematical iterative processes in that it is not metrical. In biology, convergence gets tested in another way: the functioning of the parts must support the functioning of the whole. Any explanation that does not exhibit that coherence of function has failed to converge. Gadamer3 points out that more than a single passage around a hermeneutic circle will be required: “the repeated return from the whole to the parts, and vice versa, is essential. Moreover, this circle is constantly expanding, since the concept of the whole is relative, and being integrated into larger contexts always affects the understanding of the individual part.” Hence the new cover.
Acknowledgments
We thank Steven Goodman for the opportunity to provide the new cover for Experimental Biology and Medicine and to write this commentary; Frank E. Block III, Dominic Doyle, Dmitry Markov, and Virginia Pensabene for their contributions to the figures; Jeff Davidson, Kristin Fabre, Rosemarie Hunziker, Shane Hutson, Ilya Nemenman, Lans Taylor and Marija Zanic for their comments and suggestions; and Don Berry and Allison Price for their bibliographic and editorial support. The preparation of this editorial has been supported in part by the National Center for Advancing Translational Sciences of the National Institutes of Health (NIH) under Award No. UH2/UH3 TR000491 and the Defense Advanced Research Projects Administration (DARPA) under grant W911NF-12-2-0036, but its content reflects the views of the authors and not those of either agency.
References
- 1.Wikswo JP. The relevance and potential roles of microphysiological systems in biology and medicine. Exp Biol Med. 2014;239:1061–72. doi: 10.1177/1535370214542068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goodman SR. Letter from the Editor-in-Chief: Launching our new EBM cover. Exp Biol Med. 2015 doi: 10.1177/1535370214564539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weinsheimer J, Marshall DG. In: Truth and method. 2. Gadamer HG, translator; New York: Continuum; 2000. revised ed. [Google Scholar]
- 4.Nutton V. The chronology of Galen’s early career. Classical Quarterly (New Series) 1973;23:158–71. doi: 10.1017/s0009838800036600. [DOI] [PubMed] [Google Scholar]
- 5.Martin HN. The direct influence of gradual variations of temperature upon the rate of beat of the dog’s heart. Philos Trans R Soc London. 1883;174:663–88. [Google Scholar]
- 6.Novick A, Szilard L. Experiments with the chemostat on spontaneous mutations of bacteria. PNAS. 1950;36:708–19. doi: 10.1073/pnas.36.12.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Novick A, Szilard L. Description of the chemostat. Science. 1950;112:715–6. doi: 10.1126/science.112.2920.715. [DOI] [PubMed] [Google Scholar]
- 8.Scherer WF, Syverton JT, Gey GO. Studies on the propagation in vitro of poliomyelitis viruses: IV. Viral multiplication in a stable strain of human malignant epithelial cells (strain HeLa) derived from an epidermoid carcinoma of the cervix. J Exper Med. 1953;97:695–710. doi: 10.1084/jem.97.5.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Exton JH. Crucible of science: the story of the Cori laboratory. Oxford: Oxford University Press; 2013. [Google Scholar]
- 10.Franklin RE, Gosling RG. Molecular configuration in sodium thymonucleate. Nature. 1953;171:740–1. doi: 10.1038/171740a0. [DOI] [PubMed] [Google Scholar]
- 11.Franklin RE. Structure of tobacco mosaic virus. Nature. 1955;175:379–81. doi: 10.1038/175379a0. [DOI] [PubMed] [Google Scholar]
- 12.Anderson PW. More is different: broken symmetry and the nature of the hierarchical structure of science. Science. 1972;177:393–6. doi: 10.1126/science.177.4047.393. [DOI] [PubMed] [Google Scholar]
- 13.Huang S, Wikswo J. Dimensions of systems biology. In: Amara SG, Bamberg E, Gudermann T, Hebert SC, Jahn R, Lederer WJ, Lill R, Miyajima A, Offermanns S, editors. Reviews of Physiology, Biochemistry and Pharmacology. 157. 2007. pp. 81–104. [DOI] [PubMed] [Google Scholar]
- 14.Wikswo JP, Prokop A, Baudenbacher F, Cliffel D, Csukas B, Velkovsky M. Engineering challenges of BioNEMS: the integration of microfluidics, and micro- and nanodevices, models, and external control for systems biology. IEE Proc -Nanobiotechnol. 2006;153:81–101. doi: 10.1049/ip-nbt:20050045. [DOI] [PubMed] [Google Scholar]
- 15.Delbruck M. A physicist looks at biology. Trans Conn Acad Arts Sci. 1949;38:173–90. [Google Scholar]
- 16.Miesenbock G. Optogenetic control of cells and circuits. Annu Rev Cell Dev Biol. 2011;27:731–58. doi: 10.1146/annurev-cellbio-100109-104051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung K, Wallace J, Kim SY, Kalyanasundaram S, Andalman AS, Davidson TJ, Mirzabekov JJ, Zalocusky KA, Mattis J, Denisin AK, Pak S, Bernstein H, Ramakrishnan C, Grosenick L, Gradinaru V, Deisseroth K. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–7. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LeDuc PR, Messner WC, Wikswo JP. How do control-based approaches enter into biology? Annu Rev Biomed Engr. 2011;13:369–96. doi: 10.1146/annurev-bioeng-071910-124651. [DOI] [PubMed] [Google Scholar]
- 19.Alexander PG, Gottardi R, Lin H, Lozito TP, Tuan RS. Three dimensional osteogenic and chondrogenic systems to model osteochondral physiology and degenerative joint diseases. Exp Biol Med. 2014;239:1080–95. doi: 10.1177/1535370214539232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bale SS, Vernetti L, Senutovitch N, Jindal R, Hegde M, Gough A, McCarty WJ, Bakan A, Bhushan A, Shun TY, Golberg I, DeBiasio R, Usta BO, Taylor DL, Yarmush ML. In vitro platforms for evaluating liver toxicity. Exp Biol Med. 2014;239:1180–91. doi: 10.1177/1535370214531872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng CS, Davis BNJ, Maddan L, Bursac N, Truskey GA. Physiology and metabolism of tissue engineered skeletal muscle. Exp Biol Med. 2014;239:1203–14. doi: 10.1177/1535370214538589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cimetta E, Vunjak-Novakovic G. Microscale technologies for regulating human stem cell differentiation. Exp Biol Med. 2014;239:1255–63. doi: 10.1177/1535370214530369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clark AM, Wheeler SE, Taylor DP, Pillai VC, Young CL, Prantil-Baun R, Nguyen T, Stolz DB, Borenstein JT, Lauffenburger DA, Venkataramanan R, Griffith LG, Wells A. A microphysiological system model of therapy for liver micrometastases. Exp Biol Med. 2014;239:1170–9. doi: 10.1177/1535370214532596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eddie SL, Kim JJ, Woodruff TK, Burdette JE. Microphysiological modeling of the reproductive tract: a fertile endeavor. Exp Biol Med. 2014;239:1192–202. doi: 10.1177/1535370214529387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eungdamrong NJ, Higgins C, Guo Z, Wen-han L, Gillette B, Sia S, Christiano AM. Challenges and promises in modeling dermatologic disorders with bioengineered skin. Exp Biol Med. 2014;239:1215–24. doi: 10.1177/1535370214538747. [DOI] [PubMed] [Google Scholar]
- 26.Fabre K, Livingston C, Tagle DA. Organs-on-chips (microphysiological systems): Tools to expedite efficacy and toxicity testing in human tissue. Exp Biol Med. 2014;239:1073–7. doi: 10.1177/1535370214538916. [DOI] [PubMed] [Google Scholar]
- 27.Foulke-Abel J, In J, Kovbasnjuk O, Zachos NC, Ettayebi K, Blutt SE, Hyser JM, Zeng X-L, Crawford SE, Broughman JR, Estes MK, Donowitz M. Human enteroids as an ex vivo model of host-pathogen interactions in the GI tract. Exp Biol Med. 2014;239:1124–34. doi: 10.1177/1535370214529398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hartman KG, Bortner JD, Jr, Falk GW, Ginsberg GG, Jhala N, Jian Yu MGM, Rustgi AK, Lynch JP. Modeling human gastrointestinal inflammatory diseases using microphysiological culture systems. Exp Biol Med. 2014;239:1108–23. doi: 10.1177/1535370214529388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heylman C, Sobrino A, Shirure VS, Hughes CCW, George SC. A strategy for integrating essential three-dimensional microphysiological systems of human organs for realistic anticancer drug screening. Exp Biol Med. 2014;239:1240–54. doi: 10.1177/1535370214525295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nichols JE, Niles JA, Vega SP, Argueta LB, Eastaway A, Cortiella J. Modeling the lung: Design and development of tissue engineered macro- and micro-physiologic lung models for research use. Exp Biol Med. 2014;239:1135–69. doi: 10.1177/1535370214536679. [DOI] [PubMed] [Google Scholar]
- 31.Pamies D, Hartung T, Hogberg HT. Biological and medical applications of a brain-on-chip. Exp Biol Med. 2014;239:1096–107. doi: 10.1177/1535370214537738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Slikker W., Jr Of human-on-a-chip and humans: Considerations for creating and using microphysiological systems. Exp Biol Med. 2014;239:1078–9. doi: 10.1177/1535370214537754. [DOI] [PubMed] [Google Scholar]
- 33.Sung JH, Srinivasan B, Esch MB, McLamb WT, Bernabini C, Shuler ML, Hickman JJ. Using PBPK guided “Body-on-a-Chip” systems to predict mammalian response to drug and chemical exposure. Exp Biol Med. 2014;239:1225–39. doi: 10.1177/1535370214529397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tourovskaia A, Fauver M, Kramer G, Simonson S, Neumann T. Tissue-engineered microenvironment systems modeling the human vasculature. Exp Biol Med. 2014;239:1264–71. doi: 10.1177/1535370214539228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van der Meer AD, van den Berg A. Organs-on-chips: Breaking the in vitro impasse. Integr Biol. 2012;4:461–70. doi: 10.1039/c2ib00176d. [DOI] [PubMed] [Google Scholar]
- 36.Huh D, Torisawa YS, Hamilton GA, Kim HJ, Ingber DE. Microengineered physiological biomimicry: Organs-on-chips. Lab Chip. 2012;12:2156–64. doi: 10.1039/c2lc40089h. [DOI] [PubMed] [Google Scholar]
- 37.Ghaemmaghami AM, Hancock MJ, Harrington H, Kaji H, Khademhosseini A. Biomimetic tissues on a chip for drug discovery. Drug Discov Today. 2012;17:173–81. doi: 10.1016/j.drudis.2011.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moraes C, Mehta G, Lesher-Perez SC, Takayama S. Organs-on-a-chip: A focus on compartmentalized microdevices. Ann Biomed Eng. 2012;40:1211–27. doi: 10.1007/s10439-011-0455-6. [DOI] [PubMed] [Google Scholar]
- 39.Wikswo J, Curtis EL, Eagleton ZE, Evans BC, Kole A, Hofmeister LH, Matloff WJ. Scaling and systems biology for integrating multiple organs-on-a-chip. Lab Chip. 2013;13:3496–511. doi: 10.1039/c3lc50243k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Imura Y, Sato K, Yoshimura E. Micro total bioassay system for ingested substances: Assessment of intestinal absorption, hepatic metabolism, and bioactivity. Anal Chem. 2010;82:9983–8. doi: 10.1021/ac100806x. [DOI] [PubMed] [Google Scholar]
- 41.Shuler ML, Esch MB. Body-on-a chip: Using microfluidic systems to predict human responses to drugs. Pure Appl Chem. 2010;82:1635–45. [Google Scholar]
- 42.Esch MB, King TL, Shuler ML. The role of body-on-a-chip devices in drug and toxicity studies. Annu Rev Biomed Engr. 2011;13:55–72. doi: 10.1146/annurev-bioeng-071910-124629. [DOI] [PubMed] [Google Scholar]
- 43.Sung JH, Esch MB, Prot JM, Long CJ, Smith A, Hickman JJ, Shuler ML. Microfabricated mammalian organ systems and their integration into models of whole animals and humans. Lab Chip. 2013;13:1201–12. doi: 10.1039/c3lc41017j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shultz LD, Ishikawa F, Greiner DL. Humanized mice in translational biomedical research. Nat Rev Immunol. 2007;7:118–30. doi: 10.1038/nri2017. [DOI] [PubMed] [Google Scholar]
- 45.Wigner EP. The unreasonable effectiveness of mathematics in the natural sciences. Richard Courant Lecture in Mathematical Sciences delivered at New York University, May 11, 1959. Comm Pure Appl Math. 1960 Feb 1;13:1–14. [Google Scholar]
- 46.Rosenblueth A, Wiener N. The role of models in science. Philos Sci. 1945;12:316–21. [Google Scholar]
- 47.Di Giovanni NT. Of exactitude in science. In: Borges JL, translator; A universal history of infamy. London: Penguin; 1975. [Google Scholar]
- 48.Dyson FJ. A model for the origin of life. J Mol Evol. 1982;18:344–50. doi: 10.1007/BF01733901. [DOI] [PubMed] [Google Scholar]