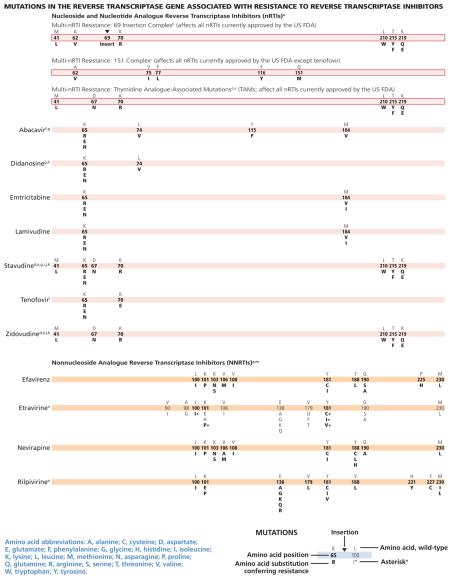
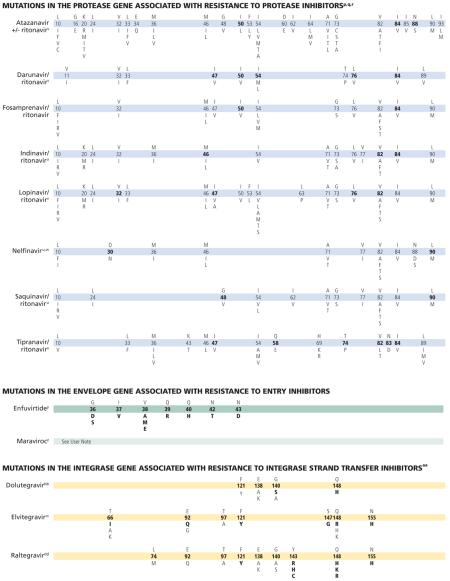
This July 2014 edition of the IAS–USA drug resistance mutations list updates the figures last published in March 2013.1
The following mutations have been added to existing classes or drugs: K65E/N has been added to the bars for the nucleoside and nucleotide analogue reverse transcriptase inhibitors (nRTIs) abacavir, didanosine, emtricitabine, lamivudine, stavudine, and tenofovir2; L100I has been added to the bar for the nonnucleoside analogue reverse transcriptase inhibitor (NNRTI) rilpivirine3,4; and F121Y has been added to the bars for the integrase strand transfer inhibitors (InSTIs) dolutegravir, elvitegravir, and raltegravir.5,6 With regard to protease inhibitors (PIs), it cannot be excluded that drug resistance may be selected for outside the protease encoding region.7,8
Methods
The IAS–USA Drug Resistance Mutations Group is an independent, volunteer panel of experts charged with delivering accurate, unbiased, and evidence-based information on these mutations to HIV clinical practitioners. As with all IAS–USA volunteer panels, members are rotated on a structured, planned basis. The group reviews new data on HIV drug resistance to maintain a current list of mutations associated with clinical resistance to HIV. This list includes mutations that may contribute to a reduced virologic response to a drug.
In addition, the group considers only data that have been published or have been presented at a scientific conference. Drugs that have been approved by the US Food and Drug Administration (FDA) as well as any drugs available in expanded access programs are included (listed in alphabetical order by drug class). User notes provide additional information as necessary. Although the Drug Resistance Mutations Group works to maintain a complete and current list of these mutations, it cannot be assumed that the list presented here is exhaustive.
Identification of Mutations
The mutations listed are those that have been identified by 1 or more of the following criteria: (1) in vitro passage experiments or validation of contribution to resistance by using site-directed mutagenesis; (2) susceptibility testing of laboratory or clinical isolates; (3) nucleotide sequencing of viruses from patients in whom the drug is failing; (4) association studies between genotype at baseline and virologic response in patients exposed to the drug.
The development of more recently approved drugs that cannot be tested as monotherapy precludes assessment of the impact of resistance on antiretroviral activity that is not seriously confounded by activity of other drug components in the background regimen. Readers are encouraged to consult the literature and experts in the field for clarification or more information about specific mutations and their clinical impact. Polymorphisms associated with impaired treatment responses that occur in otherwise wild-type viruses should not be used in epidemiologic analyses to identify transmitted HIV-1 drug resistance.
Clinical Context
The figures are designed for practitioners to use in identifying key mutations associated with antiretroviral drug resistance and in making therapeutic decisions. In the context of making clinical decisions regarding antiretroviral therapy, evaluating the results of HIV-1 genotypic testing includes: (1) assessing whether the pattern or absence of a pattern in the mutations is consistent with the patient’s antiretroviral therapy history; (2) recognizing that in the absence of drug (selection pressure), resistant strains may be present at levels below the limit of detection of the test (analyzing stored samples, collected under selection pressure, could be useful in this setting); and (3) recognizing that virologic failure of the first regimen typically involves HIV-1 isolates with resistance to only 1 or 2 of the drugs in the regimen (in this setting, resistance develops most commonly to lamivudine or emtricitabine or the NNRTIs).
The absence of detectable viral resistance after treatment failure may result from any combination of the following factors: the presence of drug-resistant minority viral populations, a prolonged interval between the time of antiretroviral drug discontinuation and genotypic testing, nonadherence to medications, laboratory error, lack of current knowledge of the association of certain mutations with drug resistance, the occurrence of relevant mutations outside the regions targeted by routine resistance assays, drug-drug interactions leading to subtherapeutic drug levels, and possibly compartmental issues, indicating that drugs may not reach optimal levels in specific cellular or tissue reservoirs.
For more in-depth reading and an extensive reference list, see the 2008 IAS–USA panel recommendations for resistance testing9 and 2014 IAS–USA panel recommendations for antiretroviral therapy.10 Updates are posted periodically at www.iasusa.org.
Comments
Please send your evidence-based comments, including relevant reference citations, to the journal“at”iasusa.org or by fax to 415-544-9401.
Reprint Requests
The Drug Resistance Mutations Group welcomes interest in the mutations figures as an educational resource for practitioners and encourages dissemination of the material to as broad an audience as possible. However, permission is required to reprint the figures and no alterations in format or the content can be made.
Requests to reprint the material should include the name of the publisher or sponsor, the name or a description of the publication in which you wish to reprint the material, the funding organization(s), if applicable, and the intended audience. Requests to make any minimal adaptations of the material should include the former, plus a detailed explanation of the adaptation(s) and, if possible, a copy of the proposed adaptation. To ensure the integrity of the mutations figures, IAS–USA policy is to grant permission for only minor, preapproved adaptations of the figures (eg, an adjustment in size). Minimal adaptations only will be considered; no alterations of the content of the figures or user notes will be permitted.
Permission will be granted only for requests to reprint or adapt the most current version of the mutations figures as they are posted at www.iasusa.org. Because scientific understanding of HIV drug resistance evolves rapidly and the goal of the Drug Resistance Mutations Group is to maintain the most up-to-date compilation of mutations for HIV clinicians and researchers, publication of out-of-date figures is counterproductive. If you have any questions about reprints or adaptations, please contact the IAS–USA.
Acknowledgments
Financial affiliations in the past 12 months: The authors (listed alphabetically) disclose the following affiliations with commercial organizations that may have interests related to the content of this article: Dr Calvez has served on advisory boards for Abbott Laboratories, Bristol-Myers Squibb, Gilead Sciences, Inc, GlaxoSmithKline, Janssen Pharmaceuticals, Inc, Pfizer, Inc, Roche, and ViiV Healthcare. Dr Günthard has served as an advisor and/or consultant for Abbott Laboratories, Boehringer Ingelheim, Bristol-Myers Squibb, Gilead Sciences, Inc, GlaxoSmithKline, Novartis, Pfizer, Inc, Roche, and Tibotec Therapeutics, with all compensation going to his institution, University Hospital of Zurich. He has received unrestricted research and educational grants to his institution from Abbott Laboratories, AstraZeneca, Bristol-Myers Squibb, Gilead Sciences, Inc, GlaxoSmithKline, Merck Sharp & Dohme, and Roche; has served on a data and safety monitoring board for Merck Sharp & Dohme; and has received travel grants from Bristol-Myers Squibb and Gilead Sciences, Inc. Dr Johnson has received research support from Abbott Molecular, Roche Molecular Diagnostics, and Siemens Healthcare Diagnostics, Inc. Dr Paredes has received research grants awarded to IrsiCaixa and Lluita Contra la SIDA Foundations from Gilead Sciences, Inc, and ViiV Healthcare. Dr Pillay has no relevant financial affiliations to disclose. Dr Richman has been a consultant to Bristol-Myers Squibb, Chimerix, Gen-Probe Inc, Gilead Sciences, Inc, Sirenas, Prism, and Monogram Biosciences, Inc. He owns stock from Chimerix. Dr Shafer has served as a consultant or advisor for Celera and has received grants from Bristol-Myers Squibb F. Hoffmann-La Roche, Ltd, Gilead Sciences, Inc, and Merck & Co, Inc. Dr Wensing has served on advisory boards for Bristol-Myers Squibb and Gilead Sciences, Inc; has received grants from Janssen Pharmaceuticals, Inc, and ViiV Healthcare; and has received travel, accommodation, or meeting expenses from Bristol-Myers Squibb and Virology Education. Funding/Support: This work was funded by the IAS–USA. No private sector or government funding was used to support the effort. Panel members are not compensated.
User Notes
a. Some nucleoside (or nucleotide) analogue reverse transcriptase inhibitor (nRTI) mutations, like T215Y and H208Y,1 may lead to viral hypersusceptibility to the nonnucleoside analogue reverse transcriptase inhibitors (NNRTIs), including etravirine,2 in nRTI-treated individuals. The presence of these mutations may improve subsequent virologic response to NNRTI-containing regimens (nevirapine or efavirenz) in NNRTI-naive individuals,3-7 although no clinical data exist for improved response to etravirine in NNRTI-experienced individuals. Mutations at the C-terminal reverse transcriptase domains (amino acids 293-560) outside of regions depicted on the figure bars may prove to be important for nRTI and NNRTI HIV-1 drug resistance. The clinical relevance of these connection domain mutations arises mostly in conjunction with thymidine analogue-associated mutations (TAMs) and M184V and have not been associated with increased rates of virologic failure of etravirine or rilpivirine in clinical trials.8-10 K65E/N variants are increasingly reported in patients experiencing treatment failure with tenofovir, stavudine, or didanosine. K65E usually occurs in mixtures with wild type. K65N gives an approximately 4-fold decrease in susceptibility. Patient-derived viruses with K65E and site-directed mutations replicate very poorly in vitro; as such, no susceptibility testing can be performed.11,12
b. The 69 insertion complex consists of a substitution at codon 69 (typically T69S) and an insertion of 2 or more amino acids (S-S, S-A, S-G, or others). The 69 insertion complex is associated with resistance to all nRTIs currently approved by the US FDA when present with 1 or more TAMs at codons 41, 210, or 215.13 Some other amino acid changes from the wild-type T at codon 69 without the insertion may be associated with broad nRTI resistance.
c. Tenofovir retains activity against the Q151M complex of mutations.13 Q151M is the most important mutation in the complex (ie, the other mutations in the complex [A62V, V75I, F77L, and F116Y] in isolation may not reflect multidrug resistance).
d. Mutations known to be selected by TAMs (ie, M41L, D67N, K70R, L210W, T215Y/F, and K219Q/E) also confer reduced susceptibility to all currently approved nRTIs.14 The degree to which cross-resistance is observed depends on the specific mutations and number of mutations involved.15-18
e. Although reverse transcriptase changes associated with the E44D and V118I mutations may have an accessory role in increased resistance to nRTIs in the presence of TAMs, their clinical relevance is very limited.19-21
f. The M184V mutation alone does not appear to be associated with a reduced virologic response to abacavir in vivo. When associated with TAMs, M184V increases abacavir resistance.22,23
g. As with tenofovir, the K65R mutation may be selected by didanosine, abacavir, or stavudine (particularly in patients with nonsubtype-B clades) and is associated with decreased viral susceptibility to these drugs.22,24,25 Data are lacking on the potential negative impact of K65R on clinical response to didanosine.
h. The presence of 3 of the following mutations—M41L, D67N, L210W, T215Y/F, K219Q/E—is associated with resistance to didanosine.26 The presence of K70R or M184V alone does not decrease virologic response to didanosine.27
i. K65R is selected frequently (4%–11%) in patients with some nonsubtype-B clades for whom stavudine-containing regimens are failing in the absence of tenofovir.28,29
j. The presence of M184V appears to delay or prevent emergence of TAMs.30 This effect may be overcome by an accumulation of TAMs or other mutations.
k. The T215A/C/D/E/G/H/I/L/N/S/V substitutions are revertant mutations at codon 215 that confer increased risk of virologic failure of zidovudine or stavudine in antiretroviralnaive patients.31,32 The T215Y mutant may emerge quickly from one of these mutations in the presence of zidovudine or stavudine.33
l. The presence of K65R is associated with a reduced virologic response to tenofovir.13 A reduced response also occurs in the presence of 3 or more TAMs inclusive of either M41L or L210W.13 The presence of TAMs or combined treatment with zidovudine prevents the emergence of K65R in the presence of tenofovir.34-36
m. There is no evidence for the utility of efavirenz, nevirapine, or rilpivirine in patients withNNRTIresistance.37
n. Resistance to etravirine has been extensively studied only in the context of coadministration with darunavir/ritonavir. In this context, mutations associated with virologic outcome have been assessed and their relative weights (or magnitudes of impact) assigned. In addition, phenotypic cutoff values have been calculated, and assessment of genotype-phenotype correlations from a large clinical database have determined relative importance of the various mutations. These 2 approaches are in agreement for many, but not all, mutations and weights.38-40 Asterisks (*) are used to emphasize higher relative weights with regard to reduced susceptibility and reduced clinical response compared with other etravirine mutations.41 The single mutations L100I*, K101P*, and Y181C*/I*/V* reduce clinical utility. The presence of K103N alone does not affect etravirine response.42 Accumulation of several mutations results in greater reductions in susceptibility and virologic response than do single mutations.43-45
o. Fifteen mutations have been associated with decreased rilpivirine susceptibility (K101E/P, E138A/G/K/Q/R, V179L, Y181C/I/V, H221Y, F227C, and M230I/L).46-48 A 16th mutation, Y188L, reduces rilpivirine susceptibility 6-fold.49 K101P and Y181I/V reduce rilpivirine susceptibility about 50-fold and 15-fold, respectively, but are uncommonly observed in patients receiving rilpivirine.50-52 K101E, E138K, and Y181C, each of which reduces rilpivirine susceptibility 2.5-fold to 3-fold, occur commonly in patients receiving rilpivirine. E138K and to a lesser extent K101E usually occur in combination with the nRTI resistance mutation M184I, which alone does not reduce rilpivirine susceptibility. When M184I is combined with E138K or K101E, rilpivirine susceptibility is reduced about 7-fold and 4.5-fold, respectively.52-55 The combinations of reverse transcriptase mutations L100I + K103N/S and L100I + K103R + V179D were strongly associated with reduced susceptibility to rilpivirine. However, for isolates harboring the L100I/ K103N/R/S or V179D as single mutations, no reduction in susceptibility was detected.48,56
p. Often, numerous mutations are necessary to substantially impact virologic response to a ritonavir-boosted protease inhibitor (PI).57 In some specific circumstances, atazanavir might be used unboosted. In such cases, the mutations that are selected are the same as with ritonavir-boosted atazanavir, but the relative frequency of mutations may differ.
q. Resistance mutations in the protease gene are classified as “major” or “minor.”
Major mutations in the protease gene (positions in bold type) are defined as those selected first in the presence of the drug or those substantially reducing drug susceptibility. These mutations tend to be the primary contact residues for drug binding.
Minor mutations generally emerge later than major mutations and by themselves do not have a substantial effect on phenotype. They may improve replication of viruses containing major mutations. Some minor mutations are present as common polymorphic changes in HIV-1 nonsubtype-B clades.
Mutations in the cytoplasmic tail of gp41 of env or mutations in gag cleavage sites may confer resistance to all protease inhibitors and may emerge before mutations in protease do.58,59 A large proportion of virus samples from patients with confirmed virologic failure on a PI-containing regimen is not found to have PI resistance mutations. Preliminary data from recent studies suggest that several mutations in the Gag protein60 and in the cytoplasmic tail of the Env protein59 may be responsible for reduced PI susceptibility in a subset of these patients.
r. Ritonavir is not listed separately, as it is currently used only at low dose as a pharmacologic booster of other PIs.
s. Many mutations are associated with atazanavir resistance. Their impacts differ, with I50L, I84V, and N88S having the greatest effect. Higher atazanavir levels obtained with ritonavir boosting increase the number of mutations required for loss of activity. The presence of M46I plus L76V might increase susceptibility to atazanavir when no other related mutations are present.61
t. HIV-1 RNA response to ritonavir-boosted darunavir correlates with baseline susceptibility and the presence of several specific PI mutations. Reductions in response are associated with increasing numbers of the mutations indicated in the figure bar. The negative impact of the protease mutations I47V, I54M, T74P, and I84V and the positive impact of the protease mutation V82A on virologic response to darunavir/ritonavir were shown in 2 data sets independently.62,63 Some of these mutations appear to have a greater effect on susceptibility than others (eg, I50V vs V11I). A median darunavir phenotypic fold-change greater than 10 (low clinical cutoff) occurs with 3 or more of the 2007 IAS–USA mutations listed for darunavir64 and is associated with a diminished virologic response.65
u. The mutations depicted on the figure bar cannot be considered comprehensive because little relevant research has been reported in recent years to update the resistance and cross-resistance patterns for this drug.
v. In PI-experienced patients, the accumulation of 6 or more of the mutations indicated on the figure bar is associated with a reduced virologic response to lopinavir/ritonavir.66,67 The product information states that accumulation of 7 or 8 mutations confers resistance to the drug.68 However, there is emerging evidence that specific mutations, most notably I47A (and possibly I47V) and V32I, are associated with high-level resistance.69-71 The addition of L76V to 3 PI resistance–associated mutations substantially increases resistance to lopinavir/ritonavir.61
w. In some nonsubtype-B HIV-1, D30N is selected less frequently than are other PI mutations.72
x. Clinical correlates of resistance to tipranavir are limited by the paucity of clinical trials and observational studies of the drug. The available genotypic scores have not been validated on large, diverse patient populations. The presence of mutations L24I, I50L/V, F53Y/L/W, I54L, and L76V have been associated with improved virologic response to tipranavir in some studies.73-75
y. Resistance to enfuvirtide is associated primarily with mutations in the first heptad repeat (HR1) region of the gp41 envelope gene. However, mutations or polymorphisms in other regions of the envelope (eg, the HR2 region or those yet to be identified) as well as coreceptor usage and density may affect susceptibility to enfuvirtide.76-78
z. The activity of CC chemokine receptor 5 (CCR5) antagonists is limited to patients with virus that uses only CCR5 for entry (R5 virus). Viruses that use both CCR5 and CXC chemokine receptor 4 (CXCR4; termed dual/mixed [D/M] virus) or only CXCR4 (X4 virus) do not respond to treatment with CCR5 antagonists. Virologic failure of these drugs frequently is associated with outgrowth of D/M or X4 virus from a preexisting minority population present at levels below the limit of assay detection. Mutations in HIV-1 gp120 that allow the virus to bind to the drug-bound form of CCR5 have been described in viruses from some patients whose virus remained R5 after virologic failure of a CCR5 antagonist. Most of these mutations are found in the V3 loop, the major determinant of viral tropism. There is as yet no consensus on specific signature mutations for CCR5 antagonist resistance, so they are not depicted in the figure. Some CCR5 antagonist-resistant viruses selected in vitro have shown mutations in gp41 without mutations in V3;79 the clinical significance of such mutations is not yet known.
aa. In site-directed mutants and clinical isolates, the mutation F121Y has a profound effect on susceptibility to elvitegravir and raltegravir and to a lesser extent to dolutegravir.59,60
bb. Cross-resistance studies with raltegravir- and elvitegravir-resistant viruses indicate that Q148H and G140S in combination with mutations L74I/M, E92Q, T97A, E138A/K, G140A, or N155H are associated with 5-fold to 20-fold reduced dolutegravir susceptibility80 and reduced virologic suppression in patients.81-84 Results of the phase III dolutegravir study in antiretroviral treatment-naive patients are expected to provide additional resistance information.
cc. Six elvitegravir codon mutations have been observed in integrase strand transfer inhibitor treatment-naive and -experienced patients in whom therapy is failing.85-91 T97A results in only a 2-fold change in elvitegravir susceptibility and may require additional mutations for resistance.88,89 The sequential use of elvitegravir and raltegravir (in either order) is not recommended because of cross-resistance between these drugs.88
dd. Raltegravir failure is associated with integrase mutations in at least 3 distinct, but not exclusive, genetic pathways defined by 2 or more mutations including (1) a signature (major) mutation at Q148H/K/R, N155H, or Y143R/H/C; and (2) 1 or more additional minor mutations. Minor mutations described in the Q148H/K/R pathway include L74M plus E138A, E138K, or G140S. The most common mutational pattern in this pathway is Q148H plus G140S, which also confers the greatest loss of drug susceptibility. Mutations described in the N155H pathway include this major mutation plus either L74M, E92Q, T97A, E92Q plus T97A, Y143H, G163K/R, V151I, or D232N.92 The Y143R/H/C mutation is uncommon.93-97 E92Q alone reduces susceptibility to elvitegravir more than 20-fold and causes limited (<5-fold) cross resistance to raltegravir.87,98-100 N155H mutants tend to predominate early in the course of raltegravir failure, but are gradually replaced by viruses with higher resistance, often bearing mutations G140S+Q148H/R/K, with continuing raltegravir treatment.93
References
- 1.Johnson VA, Calvez V, Gunthard HF, et al. Update of the Drug Resistance Mutations in HIV-1: March 2013. Top Antivir Med. 2013;21(1):6–14. [PMC free article] [PubMed] [Google Scholar]
- 2.Fourati S, Visseaux B, Armenia D, et al. Identification of a rare mutation at reverse transcriptase Lys65 (K65E) in HIV-1–infected patients failing on nucleos(t)ide reverse transcriptase inhibitors. J Antimicrob Chemother. 2013;68(10):2199–2204. doi: 10.1093/jac/dkt200. [DOI] [PubMed] [Google Scholar]
- 3.Haddad M, Napolitano LA, Frantzell A, et al. Combinations of HIV-1 reverse transcriptase mutations L100I + K103N/S and L100I + K103R + V179D reduce susceptibility to rilpivirine [Abstract H-677]. 53rd Interscience Conference on Antimicrobial Agents and Chemotherapy (ICAAC); Denver, Colorado. Sep 10-13, 2013. [Google Scholar]
- 4.Picchio GR, Rimsky LT, Van E, Haddad M, Napolitano LA, Vingerhoets J. Prevalence in the USA of rilpivirine resistance-associated mutations in clinical samples and effects on phenotypic susceptibility to rilpivirine and etravirine. Antivir Ther. 2014 doi: 10.3851/IMP2771. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 5.Malet I, Gimferrer AL, Artese A, et al. New raltegravir resistance pathways induce broad cross-resistance to all currently used integrase inhibitors. J Antimicrob Chemother. 2014 doi: 10.1093/jac/dku095. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 6.Souza CJ, Minhoto LA, de Paula Ferreira JL, da EM, de Souza Dantas DS, de Macedo Brigido LF. In-vivo selection of the mutation F121Y in a patient failing raltegravir containing salvage regimen. Antiviral Res. 2012;95(1):9–11. doi: 10.1016/j.antiviral.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 7.Rabi SA, Laird GM, Durand CM, et al. Multi-step inhibition explains HIV-1 protease inhibitor pharmacodynamics and resistance. J Clin Invest. 2013;123(9):3848–3860. doi: 10.1172/JCI67399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fun A, Wensing AM, Verheyen J, Nijhuis M. Human Immunodeficiency Virus Gag and protease: partners in resistance. Retrovirology. 2012;9:63. doi: 10.1186/1742-4690-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirsch MS, Günthard HF, Schapiro JM, et al. Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an International AIDS Society-USA panel. Clin Infect Dis. 2008;47(2):266–285. doi: 10.1086/589297. [DOI] [PubMed] [Google Scholar]
- 10.Gunthard HF, Aberg JA, Eron JJ, et al. Antiretroviral treatment of adult HIV infection: 2014 recommendations of the International Antiviral Society-USA panel. JAMA. 2014;312(4):410–425. doi: 10.1001/jama.2014.8722. [DOI] [PubMed] [Google Scholar]
References to the User Notes
- 1.Clark SA, Shulman NS, Bosch RJ, Mellors JW. Reverse transcriptase mutations 118I, 208Y, and 215Y cause HIV-1 hypersusceptibility to non-nucleoside reverse transcriptase inhibitors. AIDS. 2006;20(7):981–984. doi: 10.1097/01.aids.0000222069.14878.44. [DOI] [PubMed] [Google Scholar]
- 2.Picchio G, Vingerhoets J, Parkin N, Azijn H, de Bethune MP. Nucleoside-associated mutations cause hypersusceptibility to etravirine. Antivir Ther. 2008;13(Suppl 3):A25. [Google Scholar]
- 3.Shulman NS, Bosch RJ, Mellors JW, Albrecht MA, Katzenstein DA. Genetic correlates of efavirenz hypersusceptibility. AIDS. 2004;18(13):1781–1785. doi: 10.1097/00002030-200409030-00006. [DOI] [PubMed] [Google Scholar]
- 4.Demeter LM, DeGruttola V, Lustgarten S, et al. Association of efavirenz hypersusceptibility with virologic response in ACTG 368, a randomized trial of abacavir (ABC) in combination with efavirenz (EFV) and indinavir (IDV) in HIV-infected subjects with prior nucleoside analog experience. HIV Clin Trials. 2008;9(1):11–25. doi: 10.1310/hct0901-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haubrich RH, Kemper CA, Hellmann NS, et al. The clinical relevance of non-nucleoside reverse transcriptase inhibitor hypersusceptibility: a prospective cohort analysis. AIDS. 2002;16(15):F33–F40. doi: 10.1097/00002030-200210180-00001. [DOI] [PubMed] [Google Scholar]
- 6.Tozzi V, Zaccarelli M, Narciso P, et al. Mutations in HIV-1 reverse transcriptase potentially associated with hypersusceptibility to nonnucleoside reverse-transcriptase inhibitors: effect on response to efavirenz-based therapy in an urban observational cohort. J Infect Dis. 2004;189(9):1688–1695. doi: 10.1086/382960. [DOI] [PubMed] [Google Scholar]
- 7.Katzenstein DA, Bosch RJ, Hellmann N, et al. Phenotypic susceptibility and virological outcome in nucleoside-experienced patients receiving three or four antiretroviral drugs. AIDS. 2003;17(6):821–830. doi: 10.1097/00002030-200304110-00007. [DOI] [PubMed] [Google Scholar]
- 8.von Wyl V, Ehteshami M, Demeter LM, et al. HIV-1 reverse transcriptase connection domain mutations: dynamics of emergence and implications for success of combination antiretroviral therapy. Clin Infect Dis. 2010;51(5):620–628. doi: 10.1086/655764. [DOI] [PubMed] [Google Scholar]
- 9.Gupta S, Vingerhoets J, Fransen S, et al. Connection domain mutations in HIV-1 reverse transcriptase do not impact etravirine susceptibility and virologic responses to etravirine-containing regimens. Antimicrob Agents Chemother. 2011;55(6):2872–2879. doi: 10.1128/AAC.01695-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rimsky L, Van Eygen V, Vingerhoets J, Leijskens E, Picchio G. Reverse transcriptase connection domain mutations were not associated with virological failure or phenotypic resistance in rilpivirine-treated patients from the ECHO and THRIVE Phase III trials (week 96 analysis) Antivir Ther. 2012;17(Suppl 1):A36. [Google Scholar]
- 11.Fourati S, Visseaux B, Armenia D, et al. Identification of a rare mutation at reverse transcriptase Lys65 (K65E) in HIV-1–infected patients failing on nucleos(t)ide reverse transcriptase inhibitors. J Antimicrob Chemother. 2013;68(10):2199–2204. doi: 10.1093/jac/dkt200. [DOI] [PubMed] [Google Scholar]
- 12.Chunduri H, Crumpacker C, Sharma PL. Reverse transcriptase mutation K65N confers a decreased replication capacity to HIV-1 in comparison to K65R due to a decreased RT processivity. Virology. 2011;414(1):34–41. doi: 10.1016/j.virol.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 13.Miller MD, Margot N, Lu B, et al. Genotypic and phenotypic predictors of the magnitude of response to tenofovir disoproxil fumarate treatment in antiretroviral-experienced patients. J Infect Dis. 2004;189(5):837–846. doi: 10.1086/381784. [DOI] [PubMed] [Google Scholar]
- 14.Whitcomb JM, Parkin NT, Chappey C, Hellman NS, Petropoulos CJ. Broad nucleoside reverse-transcriptase inhibitor cross-resistance in human immunodeficiency virus type 1 clinical isolates. J Infect Dis. 2003;188(7):992–1000. doi: 10.1086/378281. [DOI] [PubMed] [Google Scholar]
- 15.Larder BA, Kemp SD. Multiple mutations in HIV-1 reverse transcriptase confer highlevel resistance to zidovudine (AZT) Science. 1989;246(4934):1155–1158. doi: 10.1126/science.2479983. [DOI] [PubMed] [Google Scholar]
- 16.Kellam P, Boucher CA, Larder BA. Fifth mutation in human immunodeficiency virus type 1 reverse transcriptase contributes to the development of high-level resistance to zidovudine. Proc Natl Acad Sci USA. 1992;89(5):1934–1938. doi: 10.1073/pnas.89.5.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calvez V, Costagliola D, Descamps D, et al. Impact of stavudine phenotype and thymidine analogues mutations on viral response to stavudine plus lamivudine in ALTIS 2 ANRS trial. Antivir Ther. 2002;7(3):211–218. [PubMed] [Google Scholar]
- 18.Kuritzkes DR, Bassett RL, Hazelwood JD, et al. Rate of thymidine analogue resistance mutation accumulation with zidovudine- or stavudine-based regimens. JAIDS. 2004;36(1):600–603. doi: 10.1097/00126334-200405010-00008. [DOI] [PubMed] [Google Scholar]
- 19.Romano L, Venturi G, Bloor S, et al. Broad nucleoside-analogue resistance implications for human immunodeficiency virus type 1 reverse-transcriptase mutations at codons 44 and 118. J Infect Dis. 2002;185(7):898–904. doi: 10.1086/339706. [DOI] [PubMed] [Google Scholar]
- 20.Walter H, Schmidt B, Werwein M, Schwingel E, Korn K. Prediction of abacavir resistance from genotypic data: impact of zidovudine and lamivudine resistance in vitro and in vivo. Antimicrob Agents Chemother. 2002;46(1):89–94. doi: 10.1128/AAC.46.1.89-94.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mihailidis C, Dunn D, Pillay D, Pozniak A. Effect of isolated V118I mutation in reverse transcriptase on response to first-line antiretroviral therapy. AIDS. 2008;22(3):427–430. doi: 10.1097/QAD.0b013e3282f3744f. [DOI] [PubMed] [Google Scholar]
- 22.Harrigan PR, Stone C, Griffin P, et al. Resistance profile of the human immunodeficiency virus type 1 reverse transcriptase inhibitor abacavir (1592U89) after monotherapy and combination therapy. CNA2001 Investigative Group. J Infect Dis. 2000;181(3):912–920. doi: 10.1086/315317. [DOI] [PubMed] [Google Scholar]
- 23.Lanier ER, Ait-Khaled M, Scott J, et al. Antiviral efficacy of abacavir in antiretroviral therapy-experienced adults harbouring HIV-1 with specific patterns of resistance to nucleoside reverse transcriptase inhibitors. Antivir Ther. 2004;9(1):37–45. doi: 10.1177/135965350400900102. [DOI] [PubMed] [Google Scholar]
- 24.Winters MA, Shafer RW, Jellinger RA, Mamtora G, Gingeras T, Merigan TC. Human immunodeficiency virus type 1 reverse transcriptase genotype and drug susceptibility changes in infected individuals receiving dideoxyinosine monotherapy for 1 to 2 years. Antimicrob Agents Chemother. 1997;41(4):757–762. doi: 10.1128/aac.41.4.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Svarovskaia ES, Margot NA, Bae AS, et al. Low-level K65R mutation in HIV-1 reverse transcriptase of treatment-experienced patients exposed to abacavir or didanosine. JAIDS. 2007;46(2):174–180. doi: 10.1097/QAI.0b013e31814258c0. [DOI] [PubMed] [Google Scholar]
- 26.Marcelin AG, Flandre P, Pavie J, et al. Clinically relevant genotype interpretation of resistance to didanosine. Antimicrob Agents Chemother. 2005;49(5):1739–1744. doi: 10.1128/AAC.49.5.1739-1744.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molina JM, Marcelin AG, Pavie J, et al. Didanosine in HIV-1-infected patients experiencing failure of antiretroviral therapy: a randomized placebo-controlled trial. J Infect Dis. 2005;191(6):840–847. doi: 10.1086/428094. [DOI] [PubMed] [Google Scholar]
- 28.Hawkins CA, Chaplin B, Idoko J, et al. Clinical and genotypic findings in HIV-infected patients with the K65R mutation failing first-line antiretroviral therapy in Nigeria. JAIDS. 2009;52(2):228–234. doi: 10.1097/QAI.0b013e3181b06125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wallis CL, Mellors JW, Venter WD, Sanne I, Stevens W. Varied patterns of HIV-1 drug resistance on failing first-line antiretroviral therapy in South Africa. JAIDS. 2010;53(4):480–484. doi: 10.1097/QAI.0b013e3181bc478b. [DOI] [PubMed] [Google Scholar]
- 30.Kuritzkes DR, Quinn JB, Benoit SL, et al. Drug resistance and virologic response in NUCA 3001, a randomized trial of lamivudine (3TC) versus zidovudine (ZDV) versus ZDV plus 3TC in previously untreated patients. AIDS. 1996;10(9):975–981. doi: 10.1097/00002030-199610090-00007. [DOI] [PubMed] [Google Scholar]
- 31.Violin M, Cozzi-Lepri A, Velleca R, et al. Risk of failure in patients with 215 HIV-1 revertants starting their first thymidine analog-containing highly active antiretroviral therapy. AIDS. 2004;18(2):227–235. doi: 10.1097/00002030-200401230-00012. [DOI] [PubMed] [Google Scholar]
- 32.Chappey C, Wrin T, Deeks S, Petropoulos CJ. Evolution of amino acid 215 in HIV-1 reverse transcriptase in response to intermittent drug selection. Antivir Ther. 2003;8:S37. [Google Scholar]
- 33.Garcia-Lerma JG, MacInnes H, Bennett D, Weinstock H, Heneine W. Transmitted human immunodeficiency virus type 1 carrying the D67N or K219Q/E mutation evolves rapidly to zidovudine resistance in vitro and shows a high replicative fitness in the presence of zidovudine. J Virol. 2004;78(14):7545–7552. doi: 10.1128/JVI.78.14.7545-7552.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parikh UM, Zelina S, Sluis-Cremer N, Mellors JW. Molecular mechanisms of bidirectional antagonism between K65R and thymidine analog mutations in HIV-1 reverse transcriptase. AIDS. 2007;21(11):1405–1414. doi: 10.1097/QAD.0b013e3281ac229b. [DOI] [PubMed] [Google Scholar]
- 35.Parikh UM, Barnas DC, Faruki H, Mellors JW. Antagonism between the HIV-1 reverse-transcriptase mutation K65R and thymidine-analogue mutations at the genomic level. J Infect Dis. 2006;194(5):651–660. doi: 10.1086/505711. [DOI] [PubMed] [Google Scholar]
- 36.von Wyl V, Yerly S, Böni J, et al. Factors associated with the emergence of K65R in patients with HIV-1 infection treated with combination antiretroviral therapy containing tenofovir. Clin Infect Dis. 2008;46(8):1299–1309. doi: 10.1086/528863. [DOI] [PubMed] [Google Scholar]
- 37.Antinori A, Zaccarelli M, Cingolani A, et al. Cross-resistance among nonnucleoside reverse transcriptase inhibitors limits recycling efavirenz after nevirapine failure. AIDS Res Hum Retroviruses. 2002;18(12):835–838. doi: 10.1089/08892220260190308. [DOI] [PubMed] [Google Scholar]
- 38.Benhamida J, Chappey C, Coakley E, Parkin NT. HIV-1 genotype algorithms for prediction of etravirine susceptibility: novel mutations and weighting factors identified through correlations to phenotype. Antivir Ther. 2008;13(Suppl 3):A142. [Google Scholar]
- 39.Coakley E, Chappey C, Benhamida J, et al. Biological and clinical cut-off analyses for etravirine in the PhenoSense HIV assay. Antivir Ther. 2008;13(Suppl 3):A134. [Google Scholar]
- 40.Vingerhoets J, Tambuyzer L, Azijn H, et al. Resistance profile of etravirine: combined analysis of baseline genotypic and phenotypic data from the randomized, controlled Phase III clinical studies. AIDS. 2010;24(4):503–514. doi: 10.1097/QAD.0b013e32833677ac. [DOI] [PubMed] [Google Scholar]
- 41.Haddad M, Stawiski E, Benhamida J, Coakley E. Improved genotypic algorith for predicting etravirine susceptibility: comprehensive list of mutations identified through correlation with matched phenotype. 17th Conference on Retroviruses and Opportunistic Infections (CROI); San Francisco, CA. Feb 16-19, 2010. [Google Scholar]
- 42.Etravirine . Vol. 8. Tibotec Therapeutics; Bridgewater, NJ: 2008. package insert. [Google Scholar]
- 43.Scherrer AU, Hasse B, Von Wyl V, et al. Prevalence of etravirine mutations and impact on response to treatment in routine clinical care: the Swiss HIV Cohort Study (SHCS) HIV Med. 2009;10(10):647–656. doi: 10.1111/j.1468-1293.2009.00756.x. [DOI] [PubMed] [Google Scholar]
- 44.Tambuyzer L, Nijs S, Daems B, Picchio G, Vingerhoets J. Effect of mutations at position E138 in HIV-1 reverse transcriptase on phenotypic susceptibility and virologic response to etravirine. JAIDS. 2011;58(1):18–22. doi: 10.1097/QAI.0b013e3182237f74. [DOI] [PubMed] [Google Scholar]
- 45.Tudor-Williams G, Cahn P, Chokephaibulkit K, et al. Etravirine in treatment-experienced, HIV-1–infected children and adolescents: 48-week safety, efficacy and resistance analysis of the phase II PIANO study. HIV Med. 2014 doi: 10.1111/hiv.12141. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 46.Rilpivirine . Tibotec Therapeutics; Titusville, NJ: 2011. package insert. [Google Scholar]
- 47.Azijn H, Tirry I, Vingerhoets J, et al. TMC278, a next-generation nonnucleoside reverse transcriptase inhibitor (NNRTI), active against wild-type and NNRTI-resistant HIV-1. Antimicrob Agents Chemother. 2010;54(2):718–727. doi: 10.1128/AAC.00986-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Picchio GR, Rimsky LT, Van E, Haddad M, Napolitano LA, Vingerhoets J. Prevalence in the USA of rilpivirine resistance-associated mutations in clinical samples and effects on phenotypic susceptibility to rilpivirine and etravirine. Antivir Ther. 2014 doi: 10.3851/IMP2771. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 49.Haddad M, Napolitano LA, Paquet AC, et al. Impact of HIV-1 reverse transcriptase E138 mutations on rilpivirine drug susceptibility. Antivir Ther. 2011;16(Suppl 1):A18. [Google Scholar]
- 50.Cohen CJ, Andrade-Villanueva J, Clotet B, et al. Rilpivirine versus efavirenz with two background nucleoside or nucleotide reverse transcriptase inhibitors in treatment-naive adults infected with HIV-1 (THRIVE): a phase 3, randomised, non-inferiority trial. Lancet. 2011;378(9787):229–237. doi: 10.1016/S0140-6736(11)60983-5. [DOI] [PubMed] [Google Scholar]
- 51.Molina JM, Cahn P, Grinsztejn B, et al. Rilpivirine versus efavirenz with tenofovir and emtricitabine in treatment-naive adults infected with HIV-1 (ECHO): a phase 3 randomised double-blind active-controlled trial. Lancet. 2011;378(9787):238–246. doi: 10.1016/S0140-6736(11)60936-7. [DOI] [PubMed] [Google Scholar]
- 52.Rimsky L, Vingerhoets J, Van Eygen V, et al. Genotypic and phenotypic characterization of HIV-1 isolates obtained from patients on rilpivirine therapy experiencing virologic failure in the phase 3 ECHO and THRIVE studies: 48-week analysis. JAIDS. 2012;59(1):39–46. doi: 10.1097/QAI.0b013e31823df4da. [DOI] [PubMed] [Google Scholar]
- 53.Kulkarni R, Babaoglu K, Lansdon EB, et al. The HIV-1 reverse transcriptase M184I mutation enhances the E138K-associated resistance to rilpivirine and decreases viral fitness. JAIDS. 2012;59(1):47–54. doi: 10.1097/QAI.0b013e31823aca74. [DOI] [PubMed] [Google Scholar]
- 54.Hu Z, Kuritzkes DR. Interaction of reverse transcriptase (RT) mutations conferring resistance to lamivudine and etravirine: effects on fitness and RT activity of human immunodeficiency virus type 1. J Virol. 2011;85(21):11309–11314. doi: 10.1128/JVI.05578-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu HT, Asahchop EL, Oliveira M, et al. Compensation by the E138K mutation in HIV-1 reverse transcriptase for deficits in viral replication capacity and enzyme processivity associated with the M184I/V mutations. J Virol. 2011;85(21):11300–11308. doi: 10.1128/JVI.05584-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haddad M, Napolitano LA, Frantzell A, et al. Combinations of HIV-1 reverse transcriptase mutations L100I + K103N/S and L100I + K103R + V179D reduce susceptibility to rilpivirine [Abstract H-677]. 53rd Interscience Conference on Antimicrobial Agents and Chemotherapy (ICAAC); Denver, Colorado. Sep 10-13, 2013. [Google Scholar]
- 57.Hirsch MS, Günthard HF, Schapiro JM, et al. Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an International AIDS Society-USA panel. Clin Infect Dis. 2008;47(2):266–285. doi: 10.1086/589297. [DOI] [PubMed] [Google Scholar]
- 58.Malet I, Gimferrer AL, Artese A, et al. New raltegravir resistance pathways induce broad cross-resistance to all currently used integrase inhibitors. J Antimicrob Chemother. 2014;69(8):2118–2122. doi: 10.1093/jac/dku095. [DOI] [PubMed] [Google Scholar]
- 59.Rabi SA, Laird GM, Durand CM, et al. Multi-step inhibition explains HIV-1 protease inhibitor pharmacodynamics and resistance. J Clin Invest. 2013;123(9):3848–3860. doi: 10.1172/JCI67399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fun A, Wensing AM, Verheyen J, Nijhuis M. Human Immunodeficiency Virus Gag and protease: partners in resistance. Retrovirology. 2012;9:63. doi: 10.1186/1742-4690-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Young TP, Parkin NT, Stawiski E, et al. Prevalence, mutation patterns, and effects on protease inhibitor susceptibility of the L76V mutation in HIV-1 protease. Antimicrob Agents Chemother. 2010;54(11):4903–4906. doi: 10.1128/AAC.00906-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.De Meyer S, Descamps D, Van Baelen B, et al. Confirmation of the negative impact of protease mutations I47V, I54M, T74P and I84V and the positive impact of protease mutation V82A on virological response to darunavir/ ritonavir. Antivir Ther. 2009;14(Suppl 1):A147. [Google Scholar]
- 63.Descamps D, Lambert-Niclot S, Marcelin AG, et al. Mutations associated with virological response to darunavir/ritonavir in HIV-1-infected protease inhibitor-experienced patients. J Antimicrob Chemother. 2009;63(3):585–592. doi: 10.1093/jac/dkn544. [DOI] [PubMed] [Google Scholar]
- 64.Johnson VA, Brun-Vézinet F, Clotet B, et al. Update of the drug resistance mutations in HIV-1: 2007. Top HIV Med. 2007;15(4):119–125. [PubMed] [Google Scholar]
- 65.De Meyer S, Dierynck I, Lathouwers E, et al. Phenotypic and genotypic determinants of resistance to darunavir: analysis of data from treatment-experienced patients in POWER 1, 2, 3 and DUET-1 and 2. Antivir Ther. 2008;13(Suppl 3):A33. [Google Scholar]
- 66.Masquelier B, Breilh D, Neau D, et al. Human immunodeficiency virus type 1 genotypic and pharmacokinetic determinants of the virological response to lopinavir-ritonavir-containing therapy in protease inhibitor experienced patients. Antimicrob Agents Chemother. 2002;46(9):2926–2932. doi: 10.1128/AAC.46.9.2926-2932.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kempf DJ, Isaacson JD, King MS, et al. Identification of genotypic changes in human immunodeficiency virus protease that correlate with reduced susceptibility to the protease inhibitor lopinavir among viral isolates from protease inhibitor-experienced patients. J Virol. 2001;75(16):7462–7469. doi: 10.1128/JVI.75.16.7462-7469.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lopinavir/ritonavir . Abbott Laboratories; Abbott Park, IL: 2010. package insert. [Google Scholar]
- 69.Mo H, King MS, King K, Molla A, Brun S, Kempf DJ. Selection of resistance in protease inhibitor-experienced, human immunodeficiency virus type 1-infected subjects failing lopinavir- and ritonavir-based therapy: mutation patterns and baseline correlates. J Virol. 2005;79(6):3329–3338. doi: 10.1128/JVI.79.6.3329-3338.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Friend J, Parkin N, Liegler T, Martin JN, Deeks SG. Isolated lopinavir resistance after virological rebound of a ritonavir/lopinavir-based regimen. AIDS. 2004;18(14):1965–1966. doi: 10.1097/00002030-200409240-00016. [DOI] [PubMed] [Google Scholar]
- 71.Kagan RM, Shenderovich M, Heseltine PN, Ramnarayan K. Structural analysis of an HIV-1 protease I47A mutant resistant to the protease inhibitor lopinavir. Protein Sci. 2005;14(7):1870–1878. doi: 10.1110/ps.051347405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gonzalez LM, Brindeiro RM, Aguiar RS, et al. Impact of nelfinavir resistance mutations on in vitro phenotype, fitness, and replication capacity of human immunodeficiency virus type 1 with subtype B and C proteases. Antimicrob Agents Chemother. 2004;48(9):3552–3555. doi: 10.1128/AAC.48.9.3552-3555.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rhee SY, Taylor J, Fessel WJ, et al. HIV-1 protease mutations and protease inhibitor cross-resistance. Antimicrob Agents Chemother. 2010;54(10):4253–4261. doi: 10.1128/AAC.00574-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schapiro JM, Scherer J, Boucher CA, et al. Improving the prediction of virological response to tipranavir: the development and validation of a tipranavir-weighted mutation score. Antivir Ther. 2010;15(7):1011–1019. doi: 10.3851/IMP1670. [DOI] [PubMed] [Google Scholar]
- 75.Marcelin AG, Masquelier B, Descamps D, et al. Tipranavir-ritonavir genotypic resistance score in protease inhibitor-experienced patients. Antimicrob Agents Chemother. 2008;52(9):3237–3243. doi: 10.1128/AAC.00133-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reeves JD, Gallo SA, Ahmad N, et al. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc Natl Acad Sci USA. 2002;99(25):16249–16254. doi: 10.1073/pnas.252469399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Reeves JD, Miamidian JL, Biscone MJ, et al. Impact of mutations in the coreceptor binding site on human immunodeficiency virus type 1 fusion, infection, and entry inhibitor sensitivity. J Virol. 2004;78(10):5476–5485. doi: 10.1128/JVI.78.10.5476-5485.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xu L, Pozniak A, Wildfire A, et al. Emergence and evolution of enfuvirtide resistance following long-term therapy involves heptad repeat 2 mutations within gp41. Antimicrob Agents Chemother. 2005;49(3):1113–1119. doi: 10.1128/AAC.49.3.1113-1119.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Anastassopoulou CG, Ketas TJ, Sanders RW, Klasse PJ, Moore JP. Effects of sequence changes in the HIV-1 gp41 fusion peptide on CCR5 inhibitor resistance. Virology. 2012;428(2):86–97. doi: 10.1016/j.virol.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kobayashi M, Yoshinaga T, Seki T, et al. In vitro antiretroviral properties of S/ GSK1349572, a next-generation HIV integrase inhibitor. Antimicrob Agents Chemother. 2011;55(2):813–821. doi: 10.1128/AAC.01209-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Eron JJ, Clotet B, Durant J, et al. Safety and efficacy of dolutegravir in treatment-experienced subjects with raltegravir-resistant HIV type 1 infection: 24-week results of the VI-KING Study. J Infect Dis. 2013;207(5):740–748. doi: 10.1093/infdis/jis750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Raffi F, Rachlis A, Stellbrink HJ, et al. Once-daily dolutegravir versus raltegravir in antiretroviral-naive adults with HIV-1 infection: 48 week results from the randomised, double-blind, non-inferiority SPRING-2 study. Lancet. 2013;381(9868):735–743. doi: 10.1016/S0140-6736(12)61853-4. [DOI] [PubMed] [Google Scholar]
- 83.Hightower KE, DeAnda F, Wang R, Vavro CL, Underwood MR. Combinations of primary and secondary integrase mutations in the VIKING Pilot Study: effects on and rationale for dolutegravir dissociation; 21st International Workshop on HIV & Hepatitis Virus Drug Resistance and Curative Strategies. Sitges, Spain. Jun 5-9, 2012. [Google Scholar]
- 84.Seki T, Kobayashi M, Miki S, et al. A high barrier to resistance for dolutegravir (DTG, S/GSK1349572) against raltegravir resistant Y143 mutants: an in vitro passage study. 21st International Workshop on HIV & Hepatitis Virus Drug Resistance and Curative Strategies; Sitges, Spain. Jun 5-9, 2012. [Google Scholar]
- 85.Goodman D, Hluhanich R, Waters J, et al. Integrase inhibitor resistance involves complex interactions among primary and second resistance mutations: a novel mutation L68V/I associates with E92Q and increases resistance. Antivir Ther. 2008;13(Suppl 3):A15. [Google Scholar]
- 86.Waters J, Margot N, Hluhanich R, et al. Evolution of resistance to the HIV integrase inhibitor (INI) elvitegravir can involve genotypic switching among primary INI resistance patterns. Antivir Ther. 2009;14(Supp 1):A137. [Google Scholar]
- 87.Geretti AM, Fearnhill E, Ceccherini-Silberstein F, et al. Prevalence and patterns of raltegravir resistance in treated patients in Europe. Antivir Ther. 2010;159(Suppl 2):A62. [Google Scholar]
- 88.Abram ME, Hluhanich RM, Goodman DD, et al. Impact of primary elvitegravir resistance-associated mutations in HIV-1 integrase on drug susceptibility and viral replication fitness. Antimicrob Agents Chemother. 2013;57(6):2654–2663. doi: 10.1128/AAC.02568-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.White KL, Kulkarni R, Szwareberg J, Quirk E, Cheng AK, Miles AJG. Integrated analysis of emergent drug resistance from the HIV-1 Phase 3 QUAD studies through week 48. Antivir Ther. 2012;17(Suppl 1):A11. [Google Scholar]
- 90.Sax PE, DeJesus E, Mills A, et al. Co-formulated elvitegravir, cobicistat, emtricitabine, and tenofovir versus co-formulated efavirenz, emtricitabine, and tenofovir for initial treatment of HIV-1 infection: a randomised, double-blind, phase 3 trial, analysis of results after 48 weeks. Lancet. 2012;379(9835):2439–2448. doi: 10.1016/S0140-6736(12)60917-9. [DOI] [PubMed] [Google Scholar]
- 91.DeJesus E, Rockstroh J, Henry K, et al. Co-formulated elvitegravir, cobicistat, emtricitabine, and tenofovir disoproxil fumarate versus ritonavir-boosted atazanavir plus co-formulated emtricitabine and tenofovir disoproxil fumarate for initial treatment of HIV-1 infection: a randomised, double-blind, phase 3, non-inferiority trial. Lancet. 2012;379(9835):2429–2438. doi: 10.1016/S0140-6736(12)60918-0. [DOI] [PubMed] [Google Scholar]
- 92.Hazuda DF, Miller MD, Nguyen BY, Zhao J. Resistance to the HIV-integrase inhibitor raltegravir: analysis of protocol 005, a phase II study in patients with triple-class resistant HIV-1 infection. Antivir Ther. 2007;12:S10. for the P005 Study Team. [Google Scholar]
- 93.Gatell JM, Katlama C, Grinsztejn B, et al. Long-term efficacy and safety of the HIV integrase inhibitor raltegravir in patients with limited treatment options in a Phase II study. JAIDS. 2010;53(4):456–463. doi: 10.1097/qai.0b013e3181c9c967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fransen S, Gupta S, Danovich R, et al. Loss of raltegravir susceptibility by human immunodeficiency virus type 1 is conferred via multiple nonoverlapping genetic pathways. J Virol. 2009;83(22):11440–11446. doi: 10.1128/JVI.01168-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hatano H, Lampiris H, Fransen S, et al. Evolution of integrase resistance during failure of integrase inhibitor-based antiretroviral therapy. J Acquir Immune Defic Syndr. 2010;54(4):389–393. doi: 10.1097/QAI.0b013e3181c42ea4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wittkop L, Breilh D, Da Silva D, et al. Virological and immunological response in HIV-1-infected patients with multiple treatment failures receiving raltegravir and optimized background therapy, ANRS CO3 Aquitaine Cohort. J Antimicrob.Chemother. 2009;63(6):1251–1255. doi: 10.1093/jac/dkp114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Armenia D, Vandenbroucke I, Fabeni L, et al. Study of genotypic and phenotypic HIV-1 dynamics of integrase mutations during raltegravir treatment: a refined analysis by ultra-deep 454 pyrosequencing. J Infect Dis. 2012;205(4):557–567. doi: 10.1093/infdis/jir821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cooper DA, Steigbigel RT, Gatell JM, et al. Subgroup and resistance analyses of raltegravir for resistant HIV-1 infection. N Engl J Med. 2008;359(4):355–365. doi: 10.1056/NEJMoa0708978. [DOI] [PubMed] [Google Scholar]
- 99.Malet I, Delelis O, Valantin MA, et al. Mutations associated with failure of raltegravir treatment affect integrase sensitivity to the inhibitor in vitro. Antimicrob Agents Chemother. 2008;52(4):1351–1358. doi: 10.1128/AAC.01228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Blanco JL, Varghese V, Rhee SY, Gatell JM, Shafer RW. HIV-1 integrase inhibitor resistance and its clinical implications. J Infect Dis. 2011;203(9):1204–1214. doi: 10.1093/infdis/jir025. [DOI] [PMC free article] [PubMed] [Google Scholar]