Abstract
Translocating proteins across the double membrane of gram-negative bacteria, Type III secretion systems (T3SS) occur in two evolutionarily related forms: injectisomes, delivering virulence factors into host cells, and the flagellar system, secreting the polymeric filament used for motility. While both systems share related elements of a cytoplasmic sorting platform that facilitates the hierarchical secretion of protein substrates, its assembly and regulation remain unclear. Here we describe a module mediating the assembly of the sorting platform in both secretion systems, and elucidate the structural basis for segregation of homologous components among these divergent T3SS subtypes sharing a common cytoplasmic milieu. These results provide a foundation for the subtype-specific assembly of T3SS sorting platforms and will support further mechanistic analysis and anti-virulence drug design.
INTRODUCTION
Type III secretion systems (T3SS) allow the transport of protein substrates directly across the double membrane of gram-negative bacteria. There are two evolutionarily related, yet functionally distinct subtypes of T3SS: “injectisomes,” which deliver effector proteins into the cytoplasm of eukaryotic host cells1, and the flagellar apparatus, which secretes the polymeric filament used for motility2. Despite their functional divergence, injectisomes and the flagella share a common core of homologous gene products and possess ultrastructural similarities3. For example, both systems share related elements of a “sorting platform” that facilitates the hierarchical secretion of protein substrates4.
Proteomic analyses have identified the major components of the sorting platform for the Salmonella typhimurium SPI-1 injectisome: the AAA+ ATPase InvC, its regulator OrgB, and the proteins SpaO and OrgA4. While SpaO has been shown to be necessary for formation of the sorting platform4, little is known about its molecular structure. In Yersinia, the SpaO homologue is expressed as a full-length protein as well as a carboxy-terminal fragment translated from an internal translation start site5; this carboxy-terminal fragment dimerizes and can interact with the full length protein. The crystal structure of the Yersinia carboxy-terminal dimer is similar to that of its Pseudomonas6 and flagellar7 homologues, together characterizing a structural class known as the surface presentation of antigens (SPOA) domain. Whether other domains within SpaO possess a similar structure, and how these structures correlate with function remains unknown.
In the flagellar apparatus, the SpaO homologues FliM and FliN form a robust, stable ring (the “C-ring”) at the cytoplasmic face of the basal body8. Electron microscopic analyses have similarly localized the SpaO homologue to the cytoplasmic face of the Shigella injectisome9, and recent cryoelectron tomographic studies in the same organism identified SpaO homologue-dependent “pods” of density beneath the injectisome10. In contrast to the flagellar C-ring, this sub-injectisome structure is less robust10, and fluorescence microscopic analysis of the Yersinia SpaO homologue show that there is dynamic exchange between cytoplasmic and injectisome-associated forms11. How SpaO and its homologues interact with other elements of the T3SS has yet to be shown at high resolution, and how homologous flagellar and injectisome components are properly segregated to their cognate secretion systems remains an open question.
Here we show that a novel, heterotypic interaction between SPOA domains serves as a scaffold for sorting platform assembly in both injectisome and flagellar T3SS. Solution NMR data support the crystallographic model, and structure-guided mutagenesis shows that this interaction is necessary for formation of the SpaO-OrgB-InvC complex, the proper localization of SpaO to the bacterial inner membrane, and T3SS function. Structures of the flagellar SpaO-OrgB homologues FliM, FliN and FliH reveal a mechanism for the proper segregation of homologous sorting platform components among T3SS subtypes sharing a common cytoplasmic milieu. Together, these structures define a common module utilized in sorting platform assembly and provide insight into the subtype-specific assembly of T3SS.
RESULTS
SpaO contains two bona fide SPOA domains
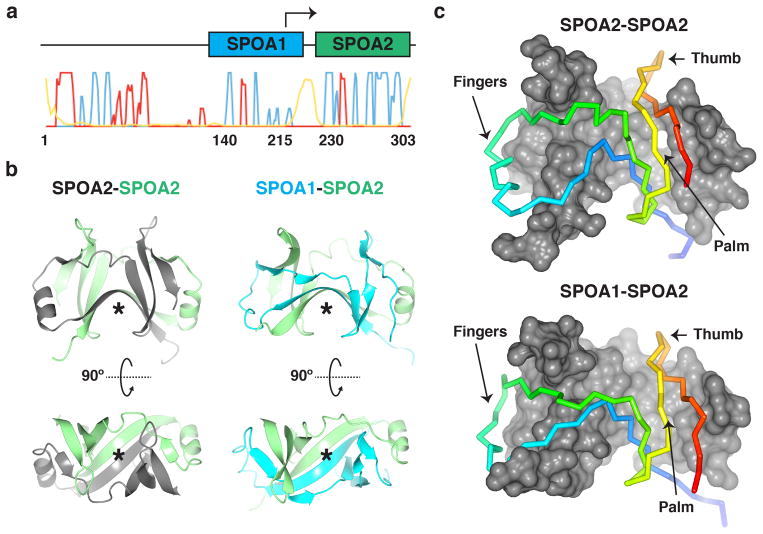
To dissect the structural basis for sorting platform assembly, we determined the structures of individual domains of S. typhimurium SpaO and then characterized their interactions with other sorting platform components. Preliminary bioinformatic analyses suggested the presence of two putative SPOA domains in the carboxy-terminal half of SpaO, which we denote SPOA1 and SPOA2 (Fig. 1a). We first determined the structure of the SPOA2 homodimer to 1.35Å resolution (Fig. 1b,c; Table 1; Supplementary Fig. 1). The SPOA2 homodimer structure is architecturally similar to its homologues5,6: Like two left hands grasping one another, an antiparallel beta-sheet “palm” of each protomer is grasped by the “fingers” of the other, with a “thumb” protruding from the top of the palm and strands from each protomer forming an anti-parallel beta sheet on the “floor” of the assembly (Fig. 1b,c). The Salmonella SPOA2 homodimer superposes on its Yersinia and Pseudomonas homologues with 2.24Å and 3.05Å RMSD, respectively (Supplementary Fig. 1b).
Figure 1. Homotypic and heterotypic SPOA interactions.
(a) Bioinformatic analysis of SpaO. PSIPRED secondary structure predictions and sequence homology suggest the presence of two putative SPOA domains in SpaO. Probability of helical character is plotted in red, strand in blue, and disorder in yellow. The arrow at codon 203 represents a predicted ValGTG internal translation start site, as has been shown for YscQ Met218 in Yersinia pseudotuberculosis5. (b,c) Comparison of homotypic SPOA2-SPOA2 and heterotypic SPOA1-SPOA2 structures from SpaO. (b) Ribbon diagrams show the similar organization of secondary structural elements in both SPOAs. Asterisks denote the antiparallel beta sheet “floor.” (c) Amino- (blue) to carboxy-terminus (red) Cα traces of SPOA2 (top) and SPOA1 (bottom) reveal a similar topology in interaction with SPOA2 (gray surface representation, top and bottom).
Table 1.
Data collection and refinement statistics for injectisome structures
| SpaO(232-297 SeMet) | SpaO(145-213, SeMet) + SpaO(232-297, SeMet) | SpaO(145-213) + SpaO (232-297) | SpaO(145-213) + SpaO (232-297) + OrgB(1-30)::lysozyme | SpaO(145-213, SeMet) + SpaO(232-297, SeMet) + OrgB(1-30)::lysozyme | |
|---|---|---|---|---|---|
| Data collection | |||||
| Space group | P 1 21 1 | P 41 21 2 | P 41 21 2 | P 1 21 1 | P 1 21 1 |
| Cell dimensions | |||||
| a, b, c (Å) | 35, 41.27, 48 | 66.38, 66.38, 95.21 | 65.76, 65.76, 95.65 | 62.092, 89.07, 62.092 | 62.88, 88.5, 63.32 |
| α, β, γ (°) | 90, 103.92, 90 | 90, 90, 90 | 90, 90, 90 | 90, 114.94, 90 | 90, 116.07, 90 |
| Resolution (Å) | 31.26 - 1.35 (1.37 - 1.35) | 46.94 – 3.00 (3.18 – 3.00) | 38.68 - 2.9 (3.08 - 2.9) | 47.59 - 2.0 (2.05 - 2.0) | 45.8 - 2.35 (2.43 - 2.35) |
| Rmerge | 0.146 (1.281) | 0.221 (1.463) | 0.166 (1.447) | 0.102 (0.530) | 0.088 (0.617) |
| I / σI | 8.6 (2.1) | 11.8 (3.1) | 14.4 (2.7) | 10.5 (3.2) | 12.8 (2.6) |
| CC1/2 | 0.994 (0.750) | 0.989 (0.839) | 0.996 (0.856) | 0.994 (0.828) | 0.997 (0.816) |
| Completeness (%) | 99.7 (100) | 100 (100) | 99.3 (99.3) | 99.5 (99.8) | 99.0 (99.6) |
| Redundancy | 7.0 (7.1) | 25.6 (27.2) | 24.6 (26.2) | 5.1 (5.2) | 6.6 (6.6) |
| Refinement | |||||
| No. reflections | 29246 | 4964 | 41183 | 25740 | |
| Rwork / Rfree | 0.1724/0.2053 | 0.2085/0.2795 | 0.1571/0.2096 | 0.1984/0.2618 | |
| No. atoms | 1286 | 1024 | 5769 | 4940 | |
| Protein | 1062 | 1023 | 5112 | 4818 | |
| Ligand/ion | 2 | 1 | 0 | 0 | |
| Water | 222 | 0 | 657 | 122 | |
| B factors | |||||
| Protein | 14.70 | 74.20 | 33.10 | 46.90 | |
| Ligand/ion | 14.20 | 105.00 | |||
| Water | 32.10 | 39.60 | 45.60 | ||
| r.m.s. deviations | |||||
| Bond lengths (Å) | 0.007 | 0.010 | 0.008 | 0.011 | |
| Bond angles (°) | 1.09 | 1.33 | 1.16 | 1.46 | |
While SPOA1 alone was insoluble, constructs containing both SPOA1 and SPOA2 (residues 140-297) were stable and soluble. SpaO(140-297) was analyzed by solution NMR (Supplementary Fig. 2a), and chemical shift deviation analysis of backbone amide resonances suggested a secondary structure pattern similar to that predicted by bioinformatic analyses: two SPOA domains connected by a flexible linker (Supplementary Fig. 2b). We hypothesized that SPOA2 interacts with and stabilizes SPOA1; consistent with this hypothesis, a SPOA1 construct (145-213) could be co-refolded with SPOA2. This complex crystalized, and its structure was determined to 2.9Å resolution (Fig. 1b,c; Table 1; Supplementary Fig. 3). SPOA1 and SPOA2 form a distinct, heterotypic SPOA-SPOA interaction with an overall topology similar to that of the SPOA2 homodimer. The SPOA1 backbone follows that of the prototypical SPOA fold, retaining the antiparallel beta-sheet floor and fingers-to-palm architecture (Fig. 1b,c). In both SPOA1,2 and the SPOA2 homodimer, interacting protomers each bury about 1800Å2 against their binding partner. SPOA1,2 and the SPOA2 homodimer superpose with 2.47Å RMSD (Supplementary Fig. 3b), and the conformation of SPOA2 in association with SPOA1 is grossly similar to that seen in the homodimer, superposing with an RMSD of 1.67Å (Supplementary Fig. 3c).
Further supporting the hypothesis that SPOA1 and SPOA2 interact in solution, a post hoc analysis of the 15N-NOESY-HSQC spectrum for SpaO(140-297) revealed long-range amide proton correlations between SPOA1 and SPOA2 (Supplementary Fig. 4). Given the <20 residue linker connecting SPOA1 and SPOA2, they would experience a low millimolar-range relative concentration and would likely interact in an intramolecular fashion (Fig. 2a). However, at high local SpaO concentrations (e.g. in association with the T3SS), intermolecular heterotypic SPOA interactions might explain the apparent oligomeric nature of the sorting platform (Fig. 2a). Indeed, a similar model of intermolecular domain swapping was recently suggested for the ring-forming injectisome protein PrgK12.
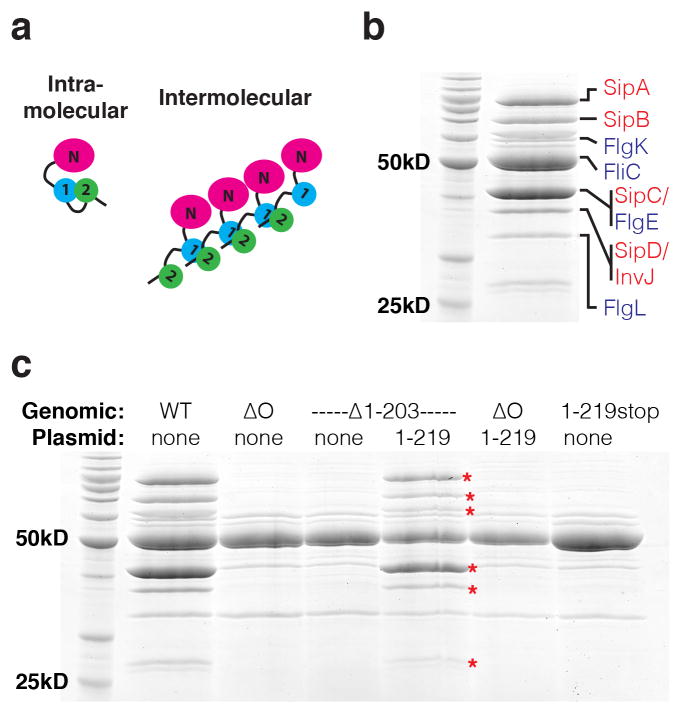
Figure 2. Intermolecular SPOA1-SPOA2 interactions are not necessary for T3SS function.
(a) Schematic models for putative intra- and intermolecular SPOA1-SPOA2 interactions and their implications for the SpaO oligomerization state. 1 and 2 indicate the SpaO SPOA1 and SPOA2, respectively, and N indicates the SpaO amino-terminal domain(s). (b) Coomassie stained PAGE of S. typhimurium culture supernatants grown under T3SS stimulating conditions (0.3M NaCl, strain SB1741). Bands previously identified by Aizawa and colleagues13 are noted and color-coded by T3SS subtype – injectisome in red, flagellar in blue. (c) Coomassie stained PAGE of S. typhimurium culture supernatants grown under T3SS stimulating conditions. Red asterisks indicate injectisome-specific secretion substrates. Abbreviations: WT, wild-type; ΔO, deletion of spaO; Δ1-203, deletion of spaO codons 1-203; 1-219, complementation with SpaO(1-219); 1-219stop, insertion of two stop codons following spaO codon 219. SpaO was 3xFLAG tagged at its amino-terminus in each S. typhimurium strain (except ΔO) and complementation construct. Data shown are representative of three experiments.
Hypothetical SpaO oligomerization driven by intermolecular heterotypic SPOA interactions would be dependent on the covalent linkage of SPOA1 and SPOA2. Thus, we tested whether genomic deletion of the SpaO amino-terminal domain and SPOA1 can be complemented in trans, as assayed by Salmonella T3SS function in vitro. When grown under T3SS stimulating conditions, the culture supernatant of S. typhimurium has a stereotyped protein composition, consisting of both flagellar and injectisome secretion substrates (Fig. 2b, secreted proteins are annotated as per Aizawa and colleagues13). Deletion of spaO results specifically in the loss of injectisome-dependent secretory products from the culture supernatant, and deletion of spaO codons 1-203 phenocopies spaO deletion, indicating that the SpaO amino-terminal domain(s) and/or SPOA1 are necessary for T3SS function (Fig. 2c). Because SpaO(1-219) is able to complement the deletion of spaO codons 1-203 (Fig. 2c, red asterisks), the covalent linkage of SPOA1 and SPOA2 is not necessary for T3SS function. Thus, if intermolecular heterotypic SPOA interactions do occur in vivo, they are not explicitly necessary for secretion. It should be noted that SpaO(1-219) does not complement a full genomic deletion of spaO, demonstrating that SPOA2 is also necessary for T3SS function (Fig. 2c). Similarly, insertion of a double stop codon after spaO codon 219 abrogates T3SS (Fig. 2c).
SpaO SPOA1,2 is a scaffold for interaction with OrgB-InvC
Double hexahistidine-tagged SpaO is able to co-affinity purify the sorting platform components OrgB and InvC when co-expressed in Escherichia coli (Fig. 3a). Formation of the SpaO-OrgB-InvC termary complex is OrgB-dependent, as SpaO alone is insufficient to co-affinity purify InvC (Fig. 3a). We hypothesized that SPOA1,2 might serve as a scaffold for the interaction of SpaO with OrgB-InvC. Indeed, SPOA1,2 is sufficient to co-affinity purify OrgB-InvC (Fig. 3a). This construct contains a Val203GTG to Val203GTT mutation to prevent the duplicitous translation of SPOA2 from its cryptic internal translation start site, demonstrating that the SPOA2 homodimer is dispensable for SpaO-OrgB-InvC complex formation.
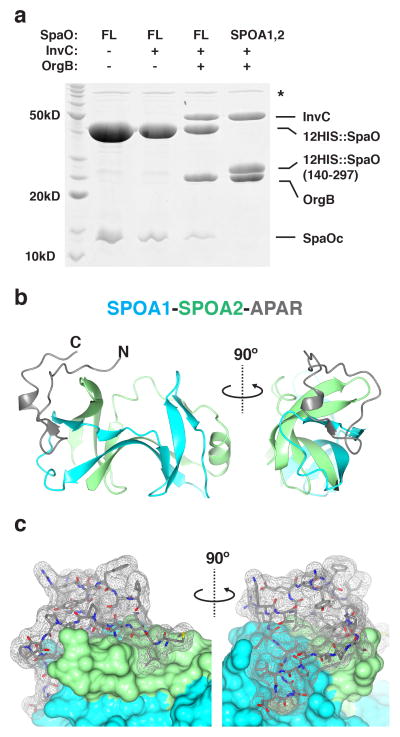
Figure 3. Architecture of the SPOA1,2-APAR interaction.
(a) Coomassie stained gel of protein elution from NiNTA resin shows that double hexahistidine (12HIS)-tagged SpaO (“FL,” full length) and SpaO(140-297, Val203GTT) are each sufficient to co-affinity purify InvC-OrgB when co-expressed in Escherichia coli. Note that OrgB is necessary for ternary complex formation. Asterisk denotes nonspecific co-purifying E. coli proteins, likely chaperones. SpaOc indicates the cryptically expressed SPOA2-containing carboxy-terminal fragment. Data shown are representative of three experiments. (b) Ribbon diagram of the SpaO-OrgB crystal structure. For simplicity, the T4 lysozyme crystallization chaperone has been omitted and only one of the two constituent complexes from the crystallographic asymmetric unit is shown. The amino- and carboxy-termini of the OrgB APAR are denoted as “N” and “C,” respectively. (c) Surface representation of the complex in (b). The OrgB APAR (grey mesh) contacts both SpaO SPOA1 (cyan) and SPOA2 (green).
OrgB and its homologues are predicted to share a common amino-terminal organization: a disordered region followed by a coiled coil. In the flagellar system, the unstructured region at the amino-terminus of the OrgB homologue FliH is necessary for its interaction with the SpaO homologues FliM and FliN14. We solublized the pre-coiled-coil region of OrgB (residues 1-30) by genetic fusion to T4 lysozyme and found that it bound to SpaO SPOA1,2. Herein, we will refer to the SPOA1,2-binding region at the amino-terminus of OrgB and its homologues as the adaptor peptide of the ATPase regulator (APAR).
The SPOA1,2-OrgB(APAR)::lysozyme complex was crystalized and its structure solved to 2.0Å resolution (Fig. 3b; Table 1; Supplementary Figs. 5,6). The OrgB APAR forms a lariat-like structure, contacting the thumb of SPOA2 and fingers of SPOA1 (Fig. 3b,c). OrgB makes substantial contact with both SPOA1 and SPOA2 of SpaO, burying 570Å2 against SPOA1 and 470Å2 against SPOA2. In the APAR-bound structure, there is little change in the conformation of SpaO (Supplementary Fig. 5d, 1.01Å RMSD).
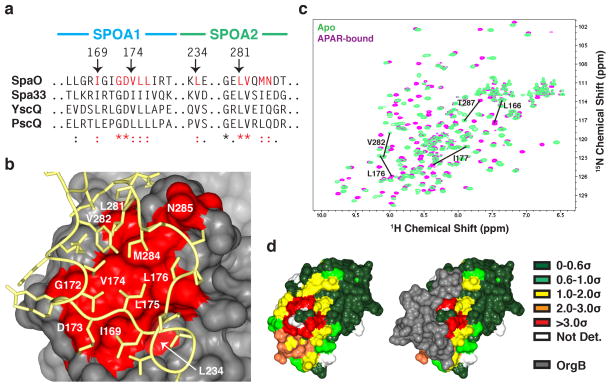
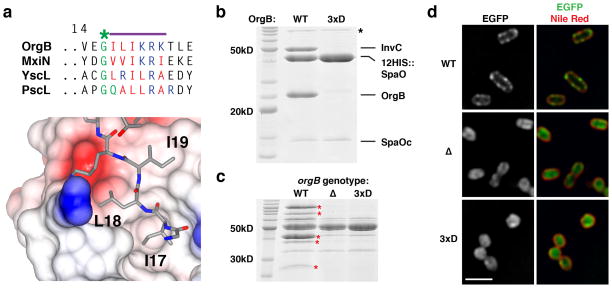
Independent NMR analyses of SpaO(140-297) bound to OrgB(APAR) in solution are consistent with the interface defined in the crystal (Fig. 4). Compared to apo-SpaO, the largest chemical shift deviations (CSD) of backbone amide resonances in SpaO-OrgB map on the crystal structure to residues involved in the interface, which are highly conserved across both the Salmonella/Shigella and Yersinia/Pseudomonas clades (Fig. 4, Supplementary Fig. 7). In the crystal, these residues form the docking site for OrgB residues Ile17, Leu18, and Ile19 (Fig. 5a). The OrgB surface area buried by these three residues (360Å2) accounts for approximately one third of the APAR’s total buried area. Here, the APAR shows noteworthy sequence homology: immediately following a conserved glycine (Gly16, pseudo-lariat apex) is a string of aliphatic and basic amino acids in each homologue (Fig. 5a).
Figure 4. The APAR binding site of SpaO.
(a) The SpaO residues at the APAR interaction site are highly conserved across homologues in other species. Excerpts of the M-COFFEE alignment of SpaO, Shigella flexneri Spa33, Yersinia enterocolica YscQ, and Pseudomonas aeruginosa PscQ are shown with conserved APAR-interacting residues highlighted in red. (b) A surface representation of SpaO with the conserved interfacial residues identified in (a) colored red and the OrgB APAR backbone is yellow. (c) Overlayed 15N-HSQC of apo- (green) and APAR-bound- (violet) SpaO(140-297). The five largest peak shifts are noted. (d) The solution interaction data from (c) are mapped onto the SpaO-OrgB crystal structure. Surface residues are color coded by the size of their weighted CSD in units of standard deviation. Residues not assigned an amide resonance in one of the two data sets are left white. The same view of SpaO is shown with and without OrgB (gray surface).
Figure 5. Structure-guided disruption of the SPOA1,2-APAR interaction.
(a) Clustal Omega alignment of the APAR regions of OrgB, S. flexneri MxiN, Y. enterocolica YscL, and P. aeruginosa PscL. The conserved pseudo-lariat apex glycine is highlighted in green and the subsequent patch of aliphatic (red) and basic (blue) amino acids is highlighted with a purple bar. Beneath, the binding site for OrgB(17-19, gray) is shown as an electrostatic surface. OrgB(1-15) have been removed for clarity. (b) Coomassie stained PAGE of the protein elution from NiNTA resin shows that double hexahistidine-tagged SpaO can co-affinity purify wild-type InvC-OrgB but not InvC-OrgB(I17D,L18D,I19D) when co-expressed in E. coli. 3xD indicates the OrgB(I17D,L18D,I19D) triple mutant. Asterisk denotes nonspecific co-purifying E. coli proteins, likely chaperones. SpaOc indicates the cryptically expressed SPOA2-containing carboxy-terminal fragment. (c) Coomassie stained culture supernatant from wild-type (WT, strain SB1741), orgB deletion(Δ) and orgB(I17D,L18D,I19D) (3xD) S. typhimurium shows loss of injectisome substrate (red asterisks) secretion in the mutants, while flagellar secretion remains intact. (d) Widefield microscopic imaging of fixed S. typhimurium shows exclusive perimembranous localization of SpaO in the wild-type background but cytoplasmic localization in the orgB mutants (scale bar is 2μm, single z-slices shown). Data shown in (b)–(d) are representative of three experiments.
The SPOA1,2-APAR interaction is necessary for T3SS function
To test whether the SPOA1,2-APAR interaction per se is necessary for T3SS function, we constructed an OrgB triple mutant (I17D,L18D,I19D) to disrupt its interaction with SpaO. As predicted, SpaO failed to co-affinity purify OrgB(I17D,L18D,I19D)-InvC when co-expressed in E. coli (Fig. 5b), and the aspartate triple mutation completely abolished T3SS in vivo (Fig. 5c). Fluorescence microscopic analyses of the Yersinia SpaO homologue have shown it to localize in discrete perimembranous punctae11. Might the SPOA1,2-APAR interaction function to localize SpaO to the bacterial inner membrane? In an otherwise wild-type genomic background, an EGFP::3xFLAG::SpaO fusion exhibits punctate, perimembranous localization, consistent with its recruitment to injectisome basal body channels (Fig. 5d). Deletion of orgB disrupts proper SpaO localization, producing a more diffuse, cytoplasmic pattern, and the asparate triple mutation was sufficient to phenocopy the orgB deletion mutant (Fig. 5d). Together, these data suggest that the SpaO(SPOA1,2)-APAR assembly is necessary for the proper localization of SpaO to discrete perimembranous puncta, and that this arrangement is required for T3SS function.
A divergent SPOA1,2-APAR assembly in the flagellar T3SS
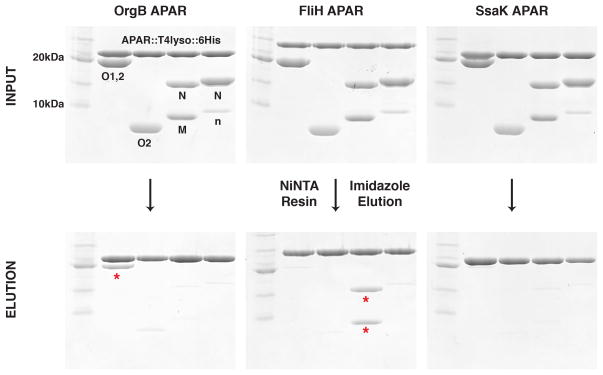
The flagellar C-ring is primarily composed of three proteins: FliM, FliN, and FliG8. The SpaO homologues FliM and FliN are predicted to contain one SPOA domain each, which we designate as SPOA1 and SPOA2, respectively. Paralleling the injectisome, FliN is known to interact with the OrgB homologue FliH14. The evolutionary relationship between injectisomes and flagella creates a practical conundrum: how are homologous T3SS components segregated to their corresponding secretion systems within a common cytoplasmic milieu? To qualitatively assess the subtype specificity of SPOA-APAR interactions, we co-affinity purified a panel of Salmonella SPOA domains with hexahistidine-tagged APAR::lysozyme fusions (Fig. 6). Indeed, the OrgB and FliH APARs robustly co-affinity purify their cognate SPOA1,2 but not that of the other T3SS subtype (Fig. 6, red asterisks). Neither SpaO nor FliM-FliN are pulled down by the APAR from a second pathogenic T3SS found in S. typhimurium (SPI-2 SsaK). Consistent with the observation that the OrgB APAR interacts with surfaces on both SPOA1 and SPOA2, the OrgB and FliH APARs more robustly pull down their cognate SPOA1,2 than homodimeric SPOA2 (Fig. 6).
Figure 6. APARs preferentially interact with their cognate SPOA1,2.
Coomassie stained gels showing the input and imidazole elution for APAR-SPOA co-affinity purification experiments. Red asterisks indicate the cognate SPOA1,2 band(s). Abbreviations: T4lyso: T4 lysozyme; O1,2: SpaO(140-297); O2: SpaO(232-297); M: FliM(245-320); N: FliN (1-137); n: co-purifying amino-terminal FliN degradation product. Data shown are representative of three experiments.
We hypothesized that divergence of the SPOA1,2-APAR assembly architecture contributes to proper component segregation among T3SS subtypes, and sought to structurally characterize the flagellar SPOA-APAR interactions. While complexes of FliM and FliN were stable, they were resistant to crystallization. Interestingly, FliM and FliN can be fused and still support flagellin secretion (Supplementary Fig. 8a) and some swarming motility15. We crystallized the SPOA of FliM (residues 245-334) fused to FliN(5-137), and its structure was solved to 2.56Å (Table 2; Supplementary Fig. 8). Architecturally, the FliM(SPOA1)-FliN(SPOA2) interaction is similar to that of SpaO (Supplementary Fig. 8e, 2.28Å RMSD), with the exception of additional helices present at the carboxy-terminus of each SPOA, as observed in FliN homodimers from Thermotoga maritima7. The similarity of these structures is consistent with the SPOA heterotypic interaction being generalizable across T3SS subtypes.
Table 2.
Data collection and refinement statistics for flagellar structures
| FliM(245-334)::FliN(5-137), SeMet | FliM(245-334)::FliN(5-137) + FliH(1-18)::lysozyme | |
|---|---|---|
| Data collection | ||
| Space group | P 21 21 21 | P 21 21 21 |
| Cell dimensions | ||
| a, b, c (Å) | 75.15, 81.50, 89.96 | 43.21, 76.37, 119.4 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 57.67 - 2.56 (2.67 – 2.56) | 64.33 – 2.30 (2.38 – 2.30) |
| Rmerge | 0.097 (1.215) | 0.070 (0.923) |
| I / σI | 18.5 (2.7) | 20.2 (2.6) |
| CC1/2 | 0.999 (0.814) | 0.999 (0.811) |
| Completeness (%) | 100 (100) | 99.8 (99.8) |
| Redundancy | 13.8 (14.3) | 12.9 (12.9) |
| Refinement | ||
| No. reflections | 18372 | 18174 |
| Rwork / Rfree | 0.2175/0.2593 | 0.1967/0.2620 |
| No. atoms | 2633 | 2739 |
| Protein | 2605 | 2668 |
| Ligand/ion | 5 | 0 |
| Water | 23 | 71 |
| B factors | ||
| Protein | 68.30 | 69.70 |
| Ligand/ion | 73.40 | |
| Water | 64.50 | 65.00 |
| r.m.s. deviations | ||
| Bond lengths (Å) | 0.010 | 0.009 |
| Bond angles (°) | 1.31 | 1.15 |
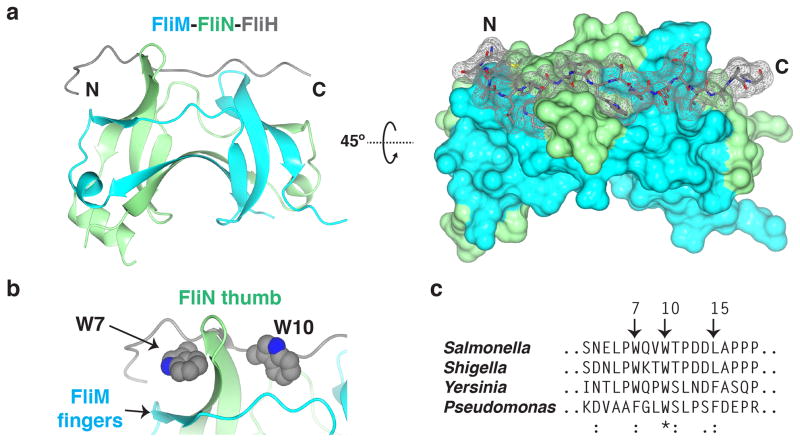
To elucidate the mechanism of FliH-specific assembly with FliM-FliN, we co-crystalized the FliM(SPOA)::FliN fusion with a FliH(1-18)::lysozyme fusion (Fig. 7; Table 2; Supplementary Fig. 9). As with its injectisome counterparts, the FliM-FliN SPOA1,2 did not undergo large conformational changes upon APAR binding (Supplmentary Fig. 9c, 1.11Å RMSD); however, the binding mode for the FliH APAR is radically different. In contrast to the OrgB pseudo-lariat, the FliH APAR adopts a near-linear conformation along the “top” of FliM-FliN (Fig. 7a). As observed in the SpaO-OrgB assembly, the FliH APAR makes extensive contact with both SPOA1 and SPOA2 (Fig. 7a), supporting the observation that the FliH APAR interacts more strongly with the FliM-FliN heterodimer than the FliN homodimer (Fig. 6).
Figure 7. Structure of the SPOA1,2-APAR interaction in the flagella.
(a) Ribbon diagram (left) and surface representation (right) of the FliM-FliN-FliH structure. T4 lysozyme has been omitted. N and C indicate the amino- and carboxy-termini of the FliH APAR, respectively. (b) A zoomed view of the FliH aromatic clamp, with the side-chain atoms of FliH W7 and W10 represented as spheres. (c) Excerpted M-COFFEE alignment of FliH with its homologues from S. flexneri, Y. enterocolica, and P. aeruginosa. Highly conserved residues of interest are noted (S. typhimurium numbering).
The FliH-FliM-FliN assembly is characterized by the burial of several highly conserved hydrophobic FliH side-chains. Two tryptophan side-chains form an aromatic clamp, which binds hydrophobic pockets on opposite faces of the FliN thumb (Fig. 7b). These residues are critical for flagellar function14 and are highly conserved (Fig. 7c). Similarly, the bulky side-chain of FliH Leu15 fills a hydrophobic pocket on the thumb of FliM (Supplementary Fig. 10). The binding interfaces for these three residues are formed by both FliM and FliN and are highly conserved across species (Supplementary Fig. 10). This structure presents a conserved model for FliH-FliM-FliN interaction, which is distinct from that of SpaO-OrgB.
DISCUSSION
We present here a series of structures that yield critical mechanistic insights into T3SS sorting platform assembly across multiple species and secretion subtypes. The existence of heterotypic SPOA interactions provides a structural explanation for the observed 1:~3 stoichiometry of SPOA1 to SPOA2 in SpaO homologues5. While two of these SPOA2 domains could be accounted for by a homodimer interacting with full length SpaO, the conformation of the third SPOA2 (located in the full length protein) was unclear. Previous reports had proposed the existence of an alternate autostabilizing conformation for the third SPOA25. We show here that the third SPOA2 can be stabilized by a SPOA1-SPOA2 interaction.
Similar to SpaO and its injectisome homologues, the ratio of FliM to FliN in situ is estimated to be 1:38. In the context of our FliM-FliN structure, this suggests a model for FliM-FliN interaction similar to that of SpaO. FliM(SPOA1) would engage FliN(SPOA2) in a heterotypic SPOA-SPOA interaction, and additional homodimeric FliN would interact with FliM-FliN in an as of yet undetermined fashion (analogous to the SpaO SPOA2 homodimer interaction with full length SpaO). However, reports of FliN tetramerization and FliM:FliN ratios between 1:3 and 1:4 suggests that more complicated higher order structures may be utilized by the flagellar apparatus7. It should also be noted that while previous investigations of the flagellar T3SS have focused on the interaction between FliH and FliN specifically14, our structures and biochemical data show that the FliH APAR more strongly interacts with the FliM-FliN complex than with FliN alone, suggesting that the FliM-FliN complex is the physiologically relevant binding partner for FliH.
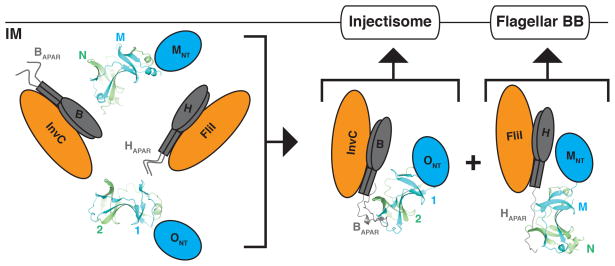
Our structures suggest a partial model for the subtype-specific assembly of the T3SS sorting platforms: the heterotypic interaction between SPOA domains within a given T3SS subtype functions as an adaptor for the ATPase and its regulator through interaction with the APAR peptide (Fig. 8). However, a number of questions remain regarding the higher order architecture of the sorting platform in situ. We hypothesize that the puncta formed by SpaO in vivo represent the high-molecular weight sorting platforms described by Galán and colleagues4. Armitage and colleagues have quantified the stoichiometry and dynamics of these puncta in Yersinia, showing them to possess ~22 copies of SpaO-homologue per punctum and to be in dynamic exchange with the cytoplasm11. In contrast, Liu and colleagues’ recent tomographic reconstruction of Shigella injectisomes revealed the presence of only six SpaO homologue-dependent pods of density beneath the injectisome, and their localization was OrgB homologue-independent10. Taken together with our findings, these results suggest that there may be two subpopulations of SpaO in vivo: one stably associated with the injectisome, and a second dynamic population in exchange with the cytoplasm, requiring the SPOA1,2-APAR interaction to form high molecular weight, perimembranous sorting platforms. Recent analyses of FliI ATPase dynamics by Minamino and colleagues suggest a similar two population model, which they hypothesize functions to deliver secretion substrates to the assembling flagella16.
Figure 8. Subtype-specific assembly of the T3SS sorting platform by SPOA1,2-APAR interactions.
Schematic illustration of the proposed role for the SPOA1,2-APAR assembly in organizing and localizing the T3SS sorting platforms in a subtype-specific fashion. IM indicates the inner membrane; ONT, the SpaO amino-terminal domain(s); 1 and 2, SpaO SPOA1 and SPOA2; B, OrgB; H, FliH; MNT, the FliM amino-terminal domains; M and N, the SPOA domains of FliM and FliN; Injectisome, the membrane integral components of the pathogenic T3SS; Flagellar BB, the flagellar basal body and associated integral membrane components.
What might be the mechanism for sorting platform targeting, and how might this factor into T3SS machine function? Perhaps APAR binding to the SPOA1,2 scaffold induces conformational changes in OrgB, InvC, and/or the amino-terminus of SpaO that facilitates interaction with the membrane-integral components of the T3SS. Alternatively, the SPOA1,2-APAR assembly might function simply by inducing proximity between sorting platform components. Intriguingly, the FliH APAR region has previously been shown to interact with the membrane integral export gate protein FlhA17, suggesting that OrgB/FliH may function as a hub, bridging the ATPase, export gate, and SpaO/FliM-FliN. How the sorting platform component OrgA – which lacks a clear flagellar homologue – factors into complex assembly remains to be determined. Similarly, the implications of sorting platform assembly for substrate recruitment, dechaperoning, and secretion remain unclear. Our structures will support the precise interrogation of sorting platform interactions in the biomechanics of secretion, and the necessity of the SPOA1,2-APAR interaction makes it a novel target for the design of anti-virulence compounds.
METHODS
Bioinformatics
Sequence alignments were performed using Clustal Omega18 or M-COFFEE19. Secondary structure and disorder predictions were performed using the PSIPRED server20.
Molecular Biology
Polymerase chain reaction (PCR) was performed using OneTaq (New England Biolabs), Phusion (New England Biolabs), or PfuTurbo (Agilent) as per manufacturer guidelines with oligonucleotides purchased from Integrated DNA Technologies. All mutations or gene fusions were created by overlap extension PCR. Gene sequences from S. typhimurium were PCR amplified from the T3SS-competent strains SB300 (wild-type; gift of J. Galán) or SB1741 (3xFLAG::SpaO, silent SpaO L79CTG to L79CTA variant; gift of J. Galán)4. The T4 lysozyme (C54T, C97A) sequence was obtained from Addgene plasmid 18111. An additional mutation (D20N) in T4 lysozyme was made to decrease toxicity in E. coli21, and the terminal three residues were mutated to alanines to decrease conformational entropy. Standard molecular biology protocols were followed to clone sequences of interest into modified pCDFduet or pETduet vectors for expression in E. coli or pBAD for expression in S. typhimurium. Restriction enzymes and Quick Ligase (New England Biolabs) were used as per manufacturer specifications. Salmonella genomic mutants were produced using homologous recombination from SacB-expressing suicide plasmids22. All SpaO and OrgB mutants were prepared on the SB1741 background. FliM and FliN mutants were prepared on the SB300 background.
Protein Expression and Purification
Constructs were transformed into BL21(DE3)Gold E. coli for heterologous expression and protein expression induced mostly as described23. Specifically, bacteria were grown to an OD600 of 0.5–0.6 at 37°C in LB medium, the cultures were cooled to 18°C, induced with 250μM IPTG, and grown overnight at 18°C. Selenomethionine (SeMet) substituted protein was produced in the methionine auxotrophic E. coli B834(DE3) grown in methionine-free media supplemented with SeMet23. Uniformly labeled 15N/13C- or 2H/15N/13C- protein samples were produced by overexpression in isotopically enriched minimal media. Deuterium oxide, 15N-ammonium chloride, and 13C-glucose were obtained from Cambridge Isotope Labs.
After induction overnight at 18°C, cells were harvested by centrifugation and resuspended in lysis buffer (200mM NaCl, 20mM Tris.Cl pH=8.0, 5%v/v glycerol, 3mM imidazole.Cl pH=8.0, 5mM MgCl2, 5mM 2-mercaptoethanol, 1mM PMSF, and 0.1mg/mL DNaseI). Cells were lysed by 1–2 passes through a mechanical homogenizer (Avestin C5) at 4°C.
Proteins were purified from E. coli cell lysates under native or denaturing conditions (as indicated for each downstream application below) and affinity purified on NiNTA resin (Qiagen). For purification under native conditions, all steps were performed at 4°C. Lysate was clarified by centrifugation for 30min at 30,000g and loaded onto NiNTA resin by gravity flow. The column was washed with 5–10 volumes of wash buffer (200mM NaCl, 20mM Tris.Cl pH=8.0, 5%v/v glycerol, 30mM imidazole.Cl pH=8.0) and then eluted in elution buffer (200mM NaCl, 20mM Tris.Cl pH=8.0, 5%v/v glycerol, 360mM imidazole.Cl pH=8.0). The elution was supplemented with 1mM EDTA and dialyzed overnight against 200mM NaCl, 20mM Tris.Cl pH=8.0, 1mM dithiothreitol. Affinity tags were removed by cleavage with HRV 3C protease.
For purification under denaturing conditions, guanidinium chloride was added to the lysate to a final concentration of 6M. The post extraction lystate was clarified by centrifugation at 30,000g for 15min at 4°C and loaded onto NiNTA resin in batch at 25°C. Still at 25°C, the resin was washed with denaturing wash buffer (8M urea, 500mM NaCl, 20mM Tris.Cl pH=8.0, 30mM imidazole.Cl pH=8.0) and eluted in denaturing elution buffer (8M urea, 200mM NaCl, 20mM Tris.Cl pH=8.0, 360mM imidazole.Cl pH=8.0). The elution was supplemented with 5mM EDTA and 5mM DTT and protein refolded by dialysis against 200mM NaCl, 20mM Tris.Cl pH=8.0, 1mM DTT (3–4 changes, dialysis time of 24h total, 4°C). For T4 lysozyme fusions, Hepes.Na pH=7.0 was substituted for Tris.Cl pH=8.0. Insoluble material was removed by centrifugation or filtration and affinity tags were removed by cleavage with HRV 3C protease.
Affinity purified proteins were further purified by ion exchange chromatography using an AKTA FPLC and the following columns (GE Healthcare): T4 lysozyme fusions were purified by cation exchange on a SourceS column; all other constructs were purified by anion exchange on a SourceQ column. For cation exchange chromatography, proteins were loaded in batch in 10mM Hepes.Na pH=7.0, 50–100mM NaCl and eluted by a NaCl gradient (from 0 to 1000mM) in the same buffer. For anion exchange, proteins were loaded in batch in 20mM Tris.Cl pH=8.0, 50–100mM NaCl and eluted by a NaCl gradient (from 0 to 1000mM) in the same buffer.
Prior to crystallography, ion exchange purified proteins were further purified by gel filtration chromatography on a Superdex 75 column (GE Healthcare) in final buffer (200mM NaCl, 20mM Tris.Cl pH=8.0, 2mM DTT) and concentrated using centrifugal concentrators (Amicon). To form the SpaO-OrgB::lysozyme complex for crystallization, cation exchange purified OrgB(1-30)::T4 lysozyme was mixed with an excess of anion exchange purified SpaO(145-213) + SpaO (232-297) and allowed to incubate overnight at 4°C. The SpaO-OrgB::lysozyme complex was then purified by gel filtration chromatography. To form the FliM::FliN-FliH::lysozyme complex for crystallization, anion exchange purified FliM(245-334)::FliN(5-137) was mixed with an excess of cation exchange purified FliH(1-18)::T4 lysozyme and allowed to incubate overnight at 4°C. The FliM::FliN-FliH::lysozyme complex was then purified by gel filtration chromatography.
Crystallization
All proteins were crystallized by hanging drop vapor diffusion with 1:1 and 2:1 ratios of protein (in final buffer) to precipitant at 25°C (except where noted). For crystallization, SpaO(232-297) and FliM(245-334)::FliN(5-137) were purified under native conditions; SpaO(145-213) and SpaO (232-297) were purified under denaturing conditions and co-refolded; the T4 lysozyme fusions were purified under denaturing conditions, refolded, and mixed with their cognate SPOA1,2 as described above. The protein concentrations, crystallization buffers, and cryoprotection conditions for each protein or complex are as follows:
SpaO(232-297) was concentrated to 8mg per mL and crystallized with 35% PEG400, 200mM calcium acetate, 100mM sodium acetate pH=5.0. Crystals were cryoprotected in the mother liquor. Microseeding was employed to enhance crystal uniformity and diffraction. Briefly, crystals to be seeded were harvested in precipitant solution and vortexed in a microfuge tube with a small stir bar for ~60 seconds. The slurry of microseeds was serially dilluted (5–10-fold steps) in precipitant solution and 5 selected microseed-precipitant mixtures were mixed with fresh protein as in a normal hanging drop experiment.
SpaO(145-213) + SpaO (232-297) was concentrated to 12mg per mL and crystallized with 25% PEG400, 10% isopropanol, 100mM sodium citrate pH=5.6 at 4°C. Microseeding (as above) was employed to enhance crystal uniformity and diffraction. Crystals were cryoprotected in mother liquor with the PEG400 concentration raised to 37.5%.
SpaO(145-213) + SpaO (232-297) + OrgB(1-30)::T4 lysozyme was concentrated to 18.5mg per mL and crystallized with 25% PEG3350, 200mM ammonium formate, 100mM sodium acetate pH=5.0. Microseeding (as above) was employed to enhance crystal uniformity and diffraction. Crystals were cryoprotected in 30% PEG3350, 10% glycerol, 200mM ammonium acetate, 100mM sodium acetate pH=5.0.
SpaO(145-213, SeMet) + SpaO (232-297, SeMet) + OrgB(1-30)::T4 lysozyme (native) was concentrated to 18mg per mL, supplemented with 50mM maltose, and crystallized with 25% PEG3350, 200mM ammonium formate, 100mM sodium acetate pH=5.0. Microseeding (as above) was employed to enhance crystal uniformity and diffraction. Crystals were cryoprotected in 25% PEG3350, 10% ethylene glycol, 200mM ammonium formate, 100mM sodium acetate pH=5.0, 50mM maltose.
FliM(245-334)::FliN(5-137) was concentrated to 7.5mg per mL and crystallized with 2.2M NaCl, 100mM imidazole.Cl pH=8.0. Crystals were cryoprotected with 2M NaCl, 100mM imidazole.Cl pH=8.0, 30% glycerol.
FliM(245-334)::FliN(5-137) + FliH(1-18)::T4 lysozyme was concentrated to 17mg per mL and crystallized with 11% PEG400, 100mM sodium potassium phosphate pH=6.5. Crystals were cryoprotected with 40% PEG400, 200mM sodium potassium phosphate pH=6.5.
Structure Determination
Data were collected at the National Synchrotron Light Source (Brookhaven National Laboratory) beamline X29A at a temperature of −173°C using the following x-ray wavelengths: 0.979Å for SeMet crystals, 1.075Å for native crystals. Diffraction data sets were indexed and integrated in iMOSFLM24 and scaled and reduced with AIMLESS25. Data sets were truncated at I/σI>2.0, and all sets were determined to have a CC1/2>0.7 in the outermost resolution shell26 (Tables 1,2).
The PHENIX program suite27 was used to solve the crystallographic phase problem. SpaO(232-297), SpaO(145-213) + SpaO (232-297), and FliM(245-334)::FliN(5-137) were solved by SeMet single wavelength anomalous diffraction in Autosol. The SPOA1,2-APAR::lysozyme structures were solved by molecular replacement in Phaser-MR using the experimentally phased cognate SPOA1,2 structure and T4 lysozyme (PDB 2LZM). Structures were built in Phenix (Autobuild) with additional manual model building performed in Coot28.
Structures were refined and validated in Phenix (Tables 1,2). SpaO(145-213) + SpaO (232-297) + OrgB(1-30)::T4 lysozyme crystals exhibited twinning and were refined in Phenix using the twin law l,-k,h. Ramachandran statistics for all models are as follows: SpaO(232-297, SeMet): 98% favored, 0% outliers; SpaO(145-213) + SpaO (232-297): 89% favored, 3% outliers; SpaO(145-213) + SpaO (232-297) + OrgB(1-30)::T4 lysozyme: 94% favored, 0.8% outliers; SpaO(145-213, SeMet) + SpaO (232-297, SeMet) + OrgB(1-30)::T4 lysozyme: 89% favored, 1.8% outliers; FliM(245-334)::FliN(5-137, SeMet): 92% favored, 0.9% outliers; FliM(245-334)::FliN(5-137) + FliH(1-18)::T4 lysozyme: 94% favored, 0.9% outliers.
ANODE29 was used to perform post-hoc analysis of anomalous scatters in SpaO(145-213, SeMet) + SpaO (232-297, SeMet) + OrgB(1-30)::T4 lysozyme crystals, providing additional empirical support for the SpaO-OrgB model coordinates (Supplementary Fig. 6b). Except where indicated, all representations of models and maps for figures were produced in QtMG30.
NMR Spectroscopy
The NMR sample of refolded SpaO(140-297) consisted of 0.3mM U-2H/15N/13C labeled protein in 10mM citrate buffer at pH 5.6 with 90%H2O/10%D2O (v/v), 100mM NaCl, and 1mM dithiothreitol. For comparison of the apo and APAR-bound forms, 15N/13C-labeled SpaO(140-297) was co-refolded with an excess of unlabeled thioredoxin::OrgB(2-30). The thioredoxin solublization tag was cleaved off by overnight incubation with HRV 3C protease. Protease and affinity tags were removed on NiNTA resin and the SpaO-OrgB complex was separated from the majority of free thioredoxin by Superdex75 gel filtration chromatography. The final concentration of the protein complex was 0.2mM in 10mM citrate buffer at pH 5.6 supplemented with 10% v/v deuterium oxide, 100mM NaCl, and 2mM dithiothreitol.
The NMR data were collected on Bruker 600, 800, and 900 MHz AVANCE spectrometers equipped with TCI/TXI CryoProbes™ at 20°C for the apo-SpaO and 30°C for the APAR-bound forms. For resonance assignments of apo-SpaO, transverse relaxation optimized (TROSY) triple resonance31 experiments including trHNCO, trHN(CA)CO, trHNCA, trHN(CO)CA, trHNCACB and trHN(CO)CACB were acquired at 600 and 900 MHz. A 15N-NOESY-HSQC spectrum with 100ms mixing time was also acquired at 900 MHz. To assign APAR-bound SpaO, a suite of conventional backbone experiments32 were acquired at 600 and 800 MHz.
The data were processed in Topspin 2.1 spectra and analyzed using the Autolink module in CARA 1.533. In both apo-SpaO and its complex with APAR, we were able to successfully assign >95% of the backbone resonances. The heteronuclear chemical shifts were analyzed using the TALOS+34 database to predict the secondary structure of the protein. The weighted CSD were calculated from amide proton (H) and nitrogen chemical shifts (15N) using the following equation: CSD=√((ΔδH)2+((Δδ15N)/5)2)
Co-affinity Purification Assay
For co-affinity purification of the SpaO-OrgB-InvC complex (Figs. 3,5), the proteins indicated were co-expressed and purified under native conditions as described above. For the SPOA-APAR::lysozyme pulldown experiment (Fig. 6), the indicated SPOA-containing proteins were Ni-affinity purified under native conditions, their affinity tags removed by overnight incubation with HRV protease 3C, and they were further purified by anion exchange chromatography (as above). APAR::lysozyme fusions were separately purified under denaturing conditions and were subjected to cation exchange chromatography after refolding (as above). 1mg of hexahistidine-tagged APAR::lysozyme fusion protein was mixed with 2mg of the indicated SPOA-containing protein in 0.2M NaCl, 20mM Tris.Cl pH=8.0 (final volume 4mL) and incubated on ice for 2h. The mixture was twice passed over 2mL of NiNTA resin, washed with 8mL wash buffer (200mM NaCl, 20mM Tris.Cl pH=8.0, 5%v/v glycerol, 30mM imidazole.Cl pH=8.0) and then eluted in 3.5mL elution buffer (200mM NaCl, 20mM Tris.Cl pH=8.0, 5%v/v glycerol, 360mM imidazole.Cl pH=8.0).
In vitro Secretion Assay
S. typhimurium of the indicated genotype were grown for 6h at 37°C in LB medium with NaCl supplemented to a final concentration of 0.3M. Cells were pelleted by centrifugation at 3400g for 0.5–1h and the supernatants were 0.22μm filtered. Secreted proteins were precipitated from the filtered supernatants with 15% trichloroacetic acid overnight at 4°C. The precipitate was pelleted by centrifugation at 3400g for 1h at 4°C, resuspended in ice-cold acetone and transferred to a microfuge tube. After 0.25h on ice, the precipitate was harvested by centrifugation at 16,000g for 0.75h at 4°C and resuspended in 0.2M Tris.Cl pH=8.0, 0.2M NaCl to neutralize any residual acid before the addition of sodium dodecyl sulfate polyacrylamide gel electrophoresis (PAGE) loading buffer. For plasmid complementation analysis, S. typhimurium were electroporated with SpaO sequences cloned into the pBAD vector and expression was induced with 0.01% arabinose for the entire duration of the experiment.
Fluorescence Microscopy
S. typhimurium were grown as for the in vitro secretion assay. Cells were harvested by centrifugation, washed 3 times in PBS, and fixed overnight with 4% formaldehyde in PBS at 4°C. Cells were again washed 3 times in PBS, counterstained with DAPI (Sigma Aldrich) and 10mM Nile Red (Sigma-Aldrich), and immobilized on poly-L-lysine (Sigma Aldrich) coated coverslips. Covers were mounted in Prolong Diamond (Life Technologies) and sealed with nail polish. Slides were imaged on a DeltaVision Image Restoration Microscope with a 100× objective (Applied Precision). Images were deconvoluted in Softworx (Applied Precision) and processed identically in ImageJ (NIH) and Photoshop (Adobe).
Supplementary Material
Acknowledgments
The authors would like to thank Deena Oren and Alison North (Rockefeller University), and Wuxian Shi (Brookhaven National Laboratory) for technical assistance and discussions. We are grateful to Matthew Lefebre (Yale University) and F. Nina Papavasiliou (Rockefeller University) for discussions. Jorge Galán (Yale University) graciously donated plasmids and S. typhimurium strains. This work was funded in part by NIH grants to C.E.S. (AI52182 and AI093704) and research funds from the Rockefeller University. R.Q.N. was supported by the Hearst Foundation and by a Medical Scientist Training Program grant from the National Institute of General Medical Sciences of the National Institutes of Health under award number T32GM07739 to the Weill Cornell/Rockefeller/Sloan-Kettering Tri-Institutional MD-PhD Program. X-ray diffraction data for this study were measured at beamline X29A of the National Synchrotron Light Source. Financial support comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy, and from the National Institute of General Medical Sciences (P41GM103473) and the National Institute of Biomedical Imaging (P30-EB-00998). Preliminary x-ray diffraction experiments performed on the Rigaku/MSC microMax 007HF in the Rockefeller University Structural Biology Resource Center were made possible by Grant Number 1S10RR022321-01 from the National Center for Research Resources of the NIH. The NMR data was collected at the New York Structural Biology Center, made possible by a grant from NYSTAR and ORIP/NIH facility improvement grant CO6RR015495. The 900 MHz NMR spectrometers were purchased with funds from NIH grant P41GM066354, the Keck Foundation, New York State Assembly, and U.S. Dept. of Defense. The content of this study is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Author Contributions: R.Q.N. conceived of the project, designed and performed experiments, analyzed data, and wrote and revised the paper. S.B. designed and performed experiments, analyzed data, and revised the paper. M.L. performed experiments and revised the paper. C.E.S. conceived of the project, analyzed data, and wrote and revised the paper.
Conflict of interest statement: The authors declare that they have no competing financial interests.
Accession numbers: Atomic coordinates have been deposited in the Protein Data Bank under the following deposition codes: 4YX1, SpaO(232-297, SeMet); 4YX5, SpaO(145-213) + SpaO (232-297); 4YX7, SpaO(145-213) + SpaO (232-297) + OrgB(1-30)::T4 lysozyme; 4YXA, SpaO(145-213, SeMet) + SpaO (232-297, SeMet) + OrgB(1-30)::T4 lysozyme (native); 4YXB, FliM(245-334)::FliN(5-137) SeMet; 4YXC, FliM(245-334)::FliN(5-137) + FliH(1-18)::T4 lysozyme. NMR chemical shifts are deposited in the BMRB under ID 26543 (apo-SpaO) and 26546 (APAR-bound SpaO).
References
- 1.Galan JE, Lara-Tejero M, Marlovits TC, Wagner S. Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annu Rev Microbiol. 2014;68:415–38. doi: 10.1146/annurev-micro-092412-155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldridge P, Hughes KT. Regulation of flagellar assembly. Curr Opin Microbiol. 2002;5:160–5. doi: 10.1016/s1369-5274(02)00302-8. [DOI] [PubMed] [Google Scholar]
- 3.Kawamoto A, et al. Common and distinct structural features of Salmonella injectisome and flagellar basal body. Sci Rep. 2013;3:3369. doi: 10.1038/srep03369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lara-Tejero M, Kato J, Wagner S, Liu X, Galan JE. A sorting platform determines the order of protein secretion in bacterial type III systems. Science. 2011;331:1188–91. doi: 10.1126/science.1201476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bzymek KP, Hamaoka BY, Ghosh P. Two translation products of Yersinia yscQ assemble to form a complex essential to type III secretion. Biochemistry. 2012;51:1669–77. doi: 10.1021/bi201792p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fadouloglou VE, et al. Structure of HrcQB-C, a conserved component of the bacterial type III secretion systems. Proc Natl Acad Sci U S A. 2004;101:70–5. doi: 10.1073/pnas.0304579101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown PN, Mathews MA, Joss LA, Hill CP, Blair DF. Crystal structure of the flagellar rotor protein FliN from Thermotoga maritima. J Bacteriol. 2005;187:2890–902. doi: 10.1128/JB.187.8.2890-2902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao R, Pathak N, Jaffe H, Reese TS, Khan S. FliN is a major structural protein of the C-ring in the Salmonella typhimurium flagellar basal body. J Mol Biol. 1996;261:195–208. doi: 10.1006/jmbi.1996.0452. [DOI] [PubMed] [Google Scholar]
- 9.Morita-Ishihara T, et al. Shigella Spa33 is an essential C-ring component of type III secretion machinery. J Biol Chem. 2006;281:599–607. doi: 10.1074/jbc.M509644200. [DOI] [PubMed] [Google Scholar]
- 10.Hu B, et al. Visualization of the type III secretion sorting platform of Shigella flexneri. Proc Natl Acad Sci U S A. 2015;112:1047–52. doi: 10.1073/pnas.1411610112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diepold A, Kudryashev M, Delalez NJ, Berry RM, Armitage JP. Composition, formation, and regulation of the cytosolic c-ring, a dynamic component of the type III secretion injectisome. PLoS Biol. 2015 doi: 10.1371/journal.pbio.1002039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bergeron JR, et al. The modular structure of the inner-membrane ring component PrgK facilitates assembly of the type III secretion system basal body. Structure. 2015;23:161–72. doi: 10.1016/j.str.2014.10.021. [DOI] [PubMed] [Google Scholar]
- 13.Mizusaki H, Takaya A, Yamamoto T, Aizawa S. Signal pathway in salt-activated expression of the Salmonella pathogenicity island 1 type III secretion system in Salmonella enterica serovar Typhimurium. J Bacteriol. 2008;190:4624–31. doi: 10.1128/JB.01957-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minamino T, et al. Roles of the extreme N-terminal region of FliH for efficient localization of the FliH-FliI complex to the bacterial flagellar type III export apparatus. Mol Microbiol. 2009;74:1471–83. doi: 10.1111/j.1365-2958.2009.06946.x. [DOI] [PubMed] [Google Scholar]
- 15.Kihara M, Francis NR, DeRosier DJ, Macnab RM. Analysis of a FliM-FliN flagellar switch fusion mutant of Salmonella typhimurium. J Bacteriol. 1996;178:4582–9. doi: 10.1128/jb.178.15.4582-4589.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bai F, et al. Assembly dynamics and the roles of FliI ATPase of the bacterial flagellar export apparatus. Sci Rep. 2014;4 doi: 10.1038/srep06528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hara N, Morimoto YV, Kawamoto A, Namba K, Minamino T. Interaction of the extreme N-terminal region of FliH with FlhA is required for efficient bacterial flagellar protein export. J Bacteriol. 2012;194:5353–60. doi: 10.1128/JB.01028-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wallace IM, O’Sullivan O, Higgins DG, Notredame C. M-Coffee: combining multiple sequence alignment methods with T-Coffee. Nucleic Acids Res. 2006;34:1692–9. doi: 10.1093/nar/gkl091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 2013;41:W349–57. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shoichet BK, Baase WA, Kuroki R, Matthews BW. A relationship between protein stability and protein function. Proc Natl Acad Sci U S A. 1995;92:452–6. doi: 10.1073/pnas.92.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaniga K, Bossio JC, Galan JE. The Salmonella typhimurium invasion genes invF and invG encode homologues of the AraC and PulD family of proteins. Mol Microbiol. 1994;13:555–68. doi: 10.1111/j.1365-2958.1994.tb00450.x. [DOI] [PubMed] [Google Scholar]
- 23.Bhaskaran SS, Stebbins CE. Structure of the catalytic domain of the Salmonella virulence factor SseI. Acta Crystallogr D Biol Crystallogr. 2012;68:1613–21. doi: 10.1107/S0907444912039042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67:271–81. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69:1204–14. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–3. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adams PD, et al. The Phenix software for automated determination of macromolecular structures. Methods. 2011;55:94–106. doi: 10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thorn A, Sheldrick GM. ANODE: anomalous and heavy-atom density calculation. J Appl Crystallogr. 2011;44:1285–1287. doi: 10.1107/S0021889811041768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McNicholas S, Potterton E, Wilson KS, Noble ME. Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr D Biol Crystallogr. 2011;67:386–94. doi: 10.1107/S0907444911007281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salzmann M, Pervushin K, Wider G, Senn H, Wuthrich K. TROSY in triple-resonance experiments: new perspectives for sequential NMR assignment of large proteins. Proc Natl Acad Sci U S A. 1998;95:13585–90. doi: 10.1073/pnas.95.23.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Progress in Nuclear Magnetic Resonance Spectroscopy. 1999;34:93–158. [Google Scholar]
- 33.Keller RLJ. The Computer Aided Resonance Assignment Tutorial. CANTINA Verlag; Goldau, Switzerland: 2004. [Google Scholar]
- 34.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–23. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.