Abstract
The vault complex is the largest cellular ribonucleoprotein complex ever characterized and is present across diverse Eukarya. Despite significant information regarding the structure, composition and evolutionary conservation of the vault, little is know about the complex’s actual biological function. To determine if intracellular vaults are morphologically similar to previously studied purified and recombinant vaults, we have used electron cryo-tomography to characterize the vault complexes found in the thin edges of primary human cells growing in tissue culture. Our studies confirm that intracellular vaults are similar in overall size and shape to purified and recombinant vaults previously analyzed. Results from subtomogram averaging indicate that densities within the vault lumen are not ordered, but randomly distributed. We also observe that vaults located in the extreme periphery of the cytoplasm predominately associate with granule-like structures and actin. Our ultrastructure studies augment existing biochemical, structural and genetic information on the vault, and provide important intracellular context for the ongoing efforts to understand the biological function of the native cytoplasmic vault.
Keywords: Vault, Ribonucleoprotein complex, Granules, RNA, Electron cryo-tomography
Introduction
The vault particle is a large (13 MDa; [1]) cellular ribonucleoprotein (RNP) complex, and is the largest RNP complex ever described [2]. Vaults have earned their descriptive name as a result of their symmetric barrel-shaped morphology that is reminiscent of the vaulted ceilings of gothic cathedrals. First observed as a contaminant in preparations of clathrin-coated vesicles purified from rat liver tissue [3], the vault has been subsequently identified in diverse eukaryotes including primitive slime molds, fish, birds and mammals. Curiously, vaults are missing from important model organisms including Caenorhabditis elegans, Drosophila melanogaster, Saccharomyces cerevisiae, and the plant Arabidopsis thaliana [4]. Nevertheless, the wide-ranging presence and evolutionary conservation of vaults across Eukarya suggest that they serve an important and basic biological function [5]. The robust structural conservation of the vault implies that the particle’s unique structure is essential to this role.
Just three proteins comprise the vault complex, with the major vault protein (MVP; 100 kDa) accounting for about 75 % of the complex’s protein content [6]. MVP is an elongated protein that arranges as vertical “staves” to form the thin shell of the vault [7]. Thirty-nine MVPs form the “half-vault” structure with their N-termini defining a plane of twofold symmetry at the particle’s waist. Nine structural repeat domains within the MVP stack to form the “barrel” part of the vault, while a αβ globular domain defines its shoulder region. The distinctive cap structure at the end of the particle is formed by an extended α-helix with a unique C-terminal U-shaped fold [8, 9]. The joining of two half-vault structures encloses approximately 5 × 104 nm3 [2], a volume sufficient to hold 2–3 ribosomes.
The other two minor vault proteins include vault-associated poly(ADP-ribose) polymerase (vPARP; 193 kDa) and telomerase-associated protein 1 (TEP1; 290 kDa). TEP1 and vPARP localize to the particle interior, primarily at the cap and waist regions, respectively [1]. In human cells, there are at least three vault-related non-coding RNAs (vRNA), hvg1, hvg2 and hvg3, enclosed within the vault through their association with TEP1 [1, 10]. However, previous estimates suggest that as little as 20 % of the total vRNA actually associates with the complex, while the other 80 % is free in the cell [11].
Despite the extensive information regarding vault structure, content, prevalence, evolution, and its potential use as a nano-particle platform (reviewed in [12]), the biological function of the native vault complex remains enigmatic. Vaults have been implicated in diverse cellular processes including development, nucleocytoplasmic transport, drug resistance and oxidative stress response (reviewed in [12]). Still, definitive proof of a role for the vault complex in any of these processes is lacking.
There is significant evidence that vaults are part of the host response to infection. MVP−/− mice are viable and appear normal [13]. However, when challenged with Pseudomonas aeruginosa, MVP−/− mice demonstrated reduced levels of bacteria internalization by lung epithelial cells, reduced clearance of the pathogen and increased mortality compared to WT mice. In apparent contrast to this example, MVP was reported to be a virulence factor for the intracellular pathogen Listeria monocytogenes [14]. The L. monocytogenes surface protein InlK binds MVP, and the binding is thought to help the bacterium avoid autophagy and targeting for lysosomal degradation. Vaults also appear to be part of the innate immune signaling response during viral infection. For example, vRNA is up-regulated during Epstein Barr virus infection [15], and MVP-dependent type-I interferon induction has been documented in response to diverse RNA virus infection [16]. More recently, the vPARP protein has been shown to be under positive selection, a hallmark of cellular proteins that are involved in evolutionary “arms races” with viral factors [17]. These data suggest an important role for the vault in the cellular response to infection and implore continued effort to understand its biological function in situ.
Both cryo-EM single particle analysis and X-ray crystallography have provided high-resolution structural details for purified native and recombinant vaults (reviewed in [18]), but structural information for the complex as it exists within the cell is lacking. Here, we use whole-cell electron cryo-tomography (ECT) of intact cells to image intracellular vaults within their native cytoplasmic micro-environments. We find that intracellular vaults are similar in overall size and shape to purified and recombinant vaults previously analyzed. Results from subtomogram averaging indicate that inside cells, densities within the vault lumen are not ordered, but randomly distributed. We also observe that vaults located in the extreme periphery of the cytoplasm predominately associate with granule-like structures and actin. In contrast to previous studies [19], we did not observe a direct interaction between vaults and microtubules. Our findings provide important intracellular context that may assist in defining the long elusive biological function of the native cytoplasmic vault.
Methods and materials
Electron cryo-tomography of whole mammalian cells
Human umbilical vein endothelia cells (HUVECs, Lonza, Walkersville, MD), COS7 cells (ATCC), HeLa cells or NIH3t3 cells were seeded in six-well plates containing carbon-coated gold EM finder grids (2 μm hole-size and 2 μm hole-spacing; Quantifoil, Jena, Germany) and cultured for 24 h in a humidified incubator maintaining 37 °C and 5 % CO2. After 24 h, EM grids on which cells were growing were removed from the culture plate using forceps and treated with 3 μl of warm media containing 10 nm gold fiducials. Forceps and grid were transferred to the environment chamber of a Vitrobot Mark III (FEI) maintained at 37 °C and 80 % R.H. Excess liquid was manually blotted from the grids on one side before plunging into liquid ethane. Cryo-preserved grids were then imaged in a 300-kV FEI G2 Polara, or FEI Titan Krios transmission electron microscope both equipped with a field emission gun and energy filter (slit width set at 20 eV). Data were collected with either a Gatan Ultracam 4k × 4k lens-coupled charge-coupled device or a K2 Summit direct detector. Tilt-series were collected over a series of angles ranging from −60 to +60 using a step size of 1°; 22,500× magnification (effective pixel size of raw data is 5 Å), a total dose of 150 e/Å2, and a defocus of −6 μm. UCSF Tomo was used to collect tilt-series and 3-D reconstructions were carried out using a weighted back-projection algorithm tracking 10 nm fiducials in IMOD. We estimate the resolution in our reconstructions to be approximately 5.6 nm based on our ability to resolve the lateral spacing of protofilaments [20] comprising the microtubules found within the tomograms (data not shown).
Segmentation of whole-cell cryo-tomogram
The segmentation was performed in the 3dmod software. Individual vaults were segmented by manually tracing each particle’s delimitation in all z slices, followed by meshing the contours to give a continuous surface. The granule bed was segmented by manually tracing the boundary of the bed in each z slice of the tomogram, and meshing the contours afterwards. Because the missing wedge effect obscures the top and bottom of the vaults, they appear flat in the segmentation.
Subtomogram averaging of intracellular vault particles
Twenty intracellular vaults were subtomogram averaged and symmeterized using PEET in IMOD [21].
Results
Intracellular vault structure is conserved among mammalian species
We used ECT to image the thin edge of mammalian cells growing on EM grids (Fig. 1). Because sample thickness is a critical limitation for ECT [22], we used cells whose edges were less than 500 nm thick. Cell lines that we found appropriate for these studies included HUVECs (human, primary; Fig. 1a–c), COS7 (simian, established; Fig. 1d, left panel), NIH3T3 (murine, established; Fig. 1d, center panel), and HeLa (human, established; Fig. 1d, right panel). For each of the cell types, vaults were readily identified in the thin cytoplasmic areas imaged by ECT (Fig. 1a, b) indicating that the vault particles are abundant in the cytoplasmic periphery. We also observed that the vaults imaged in murine, simian and human cells were morphologically very similar (compare Fig. 1b–d), suggesting that vault structure is highly conserved among mammals.
Fig. 1.
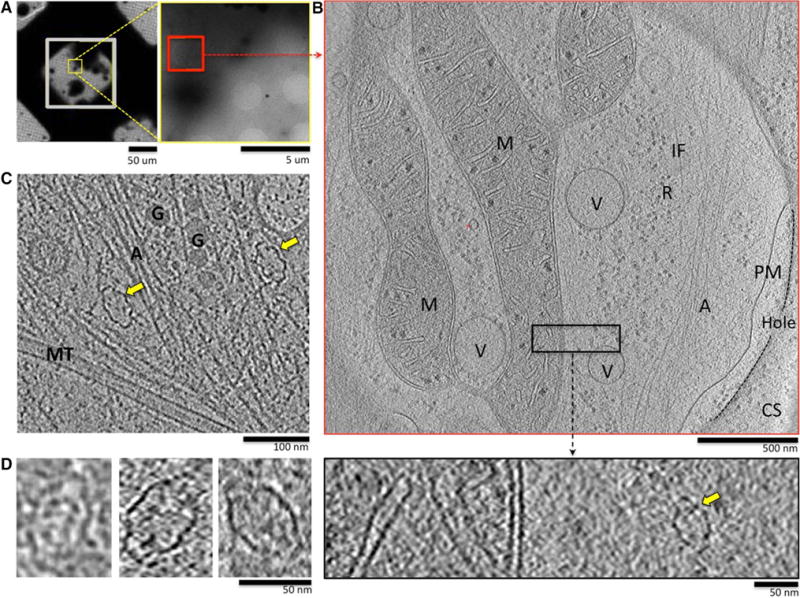
Vaults are readily observed by ECT in the thin cytoplasmic edges of cultured mammalian cells including HUVECs (a–c), Cos7 (d, left panel), NIH3T3 (d, middle panel) and HeLa (d, right panel), yellow arrows identify vaults in larger fields of view. a Left panel low magnification view of HUVECs growing on the carbon support of a gold EM grid; right panel higher magnification view of the yellow inset from left. b Top panel 14-nm-thick tomographic slice from a reconstruction of a tilt-series collected over the red inset in the right panel of a; the edge of the plasma membrane extends over a hole in the carbon support (CS); bottom panel close-up view of the black inset from tomogram in top panel. The tomographic slice has been adjusted to reveal the vault particle present (yellow arrow). A actin, G granule, IF intermediate filament, M mitochondria, MT microtubule, PM plasma membrane, R ribosomes, V vesicle, CS carbon support
Intracellular vault morphology is similar to that of purified vaults
While each of the four cell types above had recognizable vault complexes in their cytoplasmic periphery, vaults were more readily imaged in the very thin cytoplasmic periphery of HUVECs. For this reason, our subsequent analysis of intracellular vault architecture is based on vaults that were imaged in HUVECs. To determine if intracellular vaults have the same overall dimensions as purified vaults, we measured 40 vault particles (Fig. 2a). The height and width mean values were 65.94 ± 1.5 and 33.37 ± 0.99 nm, respectively, and agreed very well with the previously published vault measurements [9, 23].
Fig. 2.
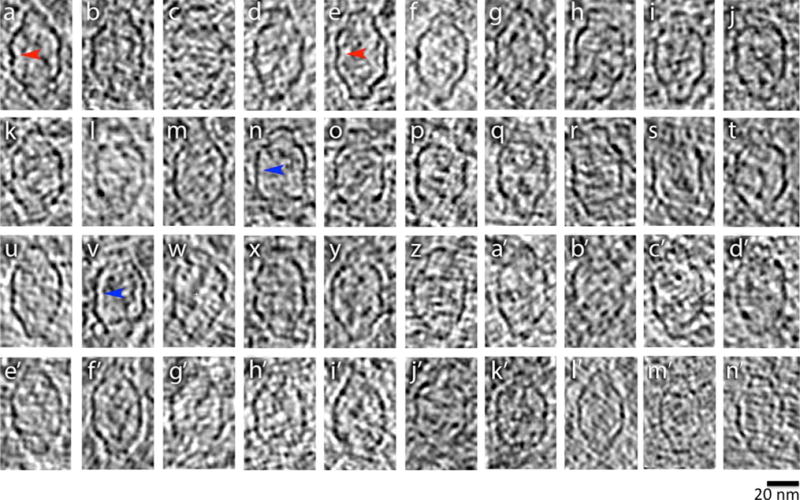
Montage of intracellular vaults. Red arrows identify indentations at individual particle midsections and blue arrows identify vault barrels vault midsections that are relatively straight
Consistent with structural features reported for purified vaults [23], the intracellular vaults also showed an indentation at the waist of the particle. For some vaults the waist indentation was very pronounced (Fig. 2, panels a, e, red arrows), but not for others (Fig. 2, panels n, v, blue arrows). Our observations suggest that within the cell, the vault structure is not entirely rigid, but is able to adopt a range of conformations without significantly altering its overall dimensions, a characteristic that may be related to vault cellular function.
Intracellular vaults have disordered internal densities
Similar to purified vaults, intracellular vaults are mostly hollow protein cages with discreet internal densities (Fig. 3a, green arrows). To determine if the internal densities are positioned consistently within the intracellular vault lumen, we averaged the 3-dimensional structures of 20 vaults shown in Fig. 2. The averaged vault volume (Fig. 3b) agreed well with the single particle reconstruction of vaults purified from rat tissue [23] and the high-resolution structure determined by X-ray crystallography ([9]; shown in Fig. 3c). The averaged volume also demonstrated that the internal densities are neither ordered, nor consistently localized to a region within the vault lumen.
Fig. 3.
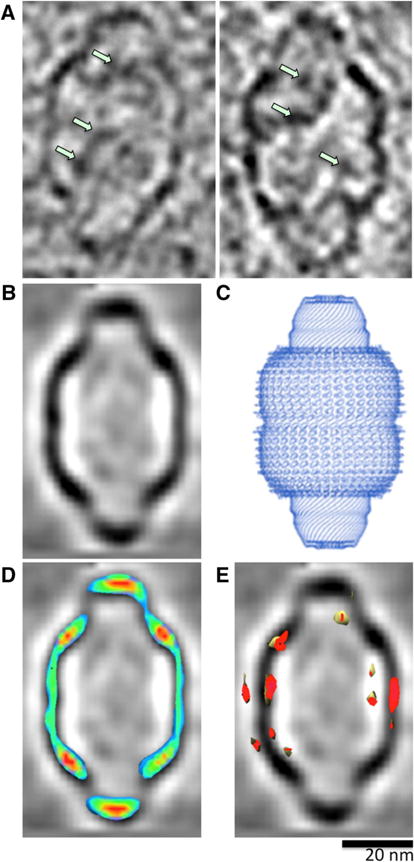
Intracellular vault ultrastructure. a Intracellular vaults demonstrate discreet internal densities (green arrows). b Subtomogram average of intracellular vault particles. c Crystal structure of purified rat liver vaults [9]. d Overlay of density heat map with averaged volume; areas of highest electron density are red and low-density areas are green. e Overlay of variance map showing the areas of highest variability (red) localized to the particle’s waist region
The averaged vault volume showed stronger densities at the shoulders and at the caps of the vaults (Fig. 3d, red). As expected, a density variance analysis of the same averaged volume localized the greatest structural variability to the waist region (Fig. 3e).
Association of vaults with intracellular structures
To investigate the intracellular context in which the vaults were found, we analyzed 15 different tomograms taken at the thin edge of HUVEC cells and identified a total of 155 vaults. We then sought to identify the intracellular structures in the vicinity of each vault. Of the 155 vaults, 97 were either embedded in, or in close proximity to beds of granules (Fig. 4a). The granules were irregular in size varying from 30 to 50 nm in diameter (Fig. 4b), and were often concentrated within “granule-beds” of varying size and shape. The “granule-beds” typically excluded ribosomes from their interior, and were at times associated with microtubules (Fig. 5b, and movie). Eighteen vaults were found in close proximity to ribosomes and 11 were associated with actin, while a total of 16 vaults co-localized with both ribosomes and actin. Only 13 of the vaults analyzed were isolated from other recognizable structures (Fig. 4c).
Fig. 4.
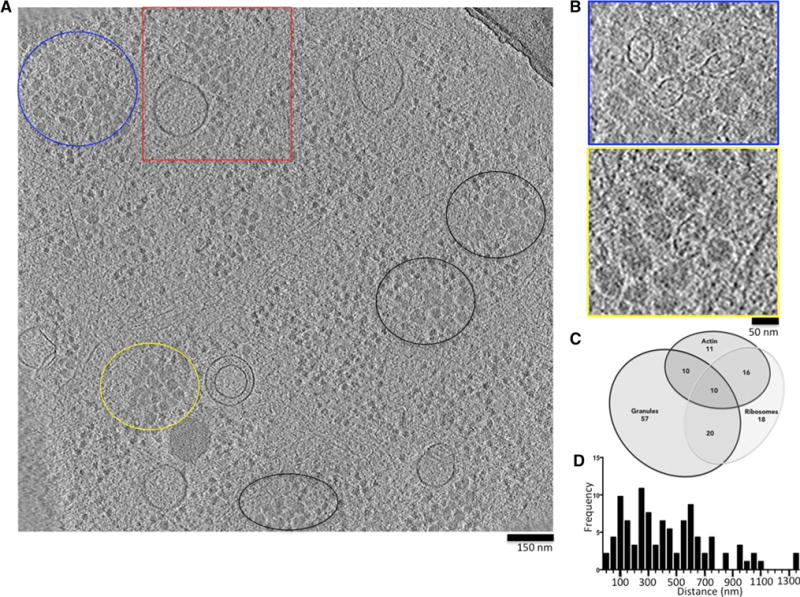
Vaults located in the periphery of the cytoplasm preferentially associate with specific intracellular features. a Single tomographic sliced through the edge of a HUVEC with granule beds circled. b Blue top panel and yellow bottom panel are close-up views of granule beds in a enclosed by blue and yellow circles, respectively. c 3-set proportional Venn diagram of the distribution of 155 vaults in relation to their surrounding elements. d Frequency distribution of the distance to the nearest microtubule of 92 vaults
Fig. 5.
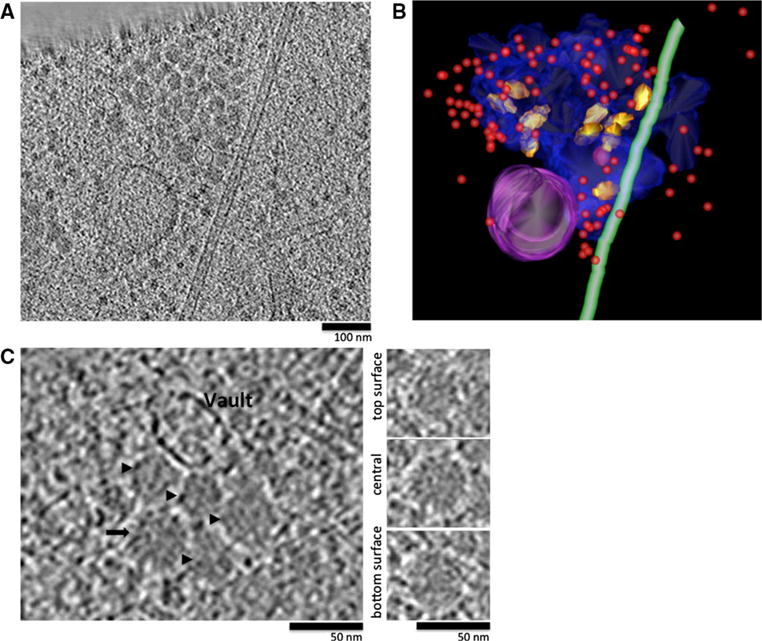
a Tomographic slice (40 nm in thickness) of granule bed in Fig. 4a, red boxed area. b Segmentation of the same region in a showing the granule bed delimitation (transparent blue), a microtubule (green), a vesicle (purple), vaults (gold), a round structure (pink) and ribosomes (red spheres). Rotational view of segmentation can be found in Supplemental Movie 1. c Left panel close-up view of granules (black arrowheads and arrow) and a vault; right panels series of tomographic slices through granule identified in left panel with black arrow
Contrary to what is described in the literature, we did not find consistent evidence for the direct association of vaults with microtubules. Of the 15 tomograms analyzed, 7 contained microtubules. We plotted the distance of each one of the 92 vaults in those 7 tomograms to the nearest microtubule in a frequency distribution graph (Fig. 4d), and found only one vault in close proximity to a microtubule (22 nm). Although no direct association was seen between vaults and microtubules, we note that they are both found in the same cellular microenvironment, the thin edge of the cytoplasm. Also, because ECT can image only thin edges of cells, we cannot rule out a direct association between vaults and microtubules in thicker regions of the cell that are closer to the nucleus.
To better depict the interaction of vaults with granule beds, a 3-D segmentation of one of the granule beds from Fig. 4a (red boxed area) is shown in Fig. 5 and movie 1.
Potentially open vaults
In a very few cases it appeared as if vaults were opening up at their waist and caps to interact with the closely associated granules (Fig. 6a–c).
Fig. 6.

Variable vault morphologies. Examples of vaults opening up to the granules at the waist region (a, b) and the cap (c)
Discussion
Prior to this paper, all reported vault structures were from purified vaults analyzed apart from their natural microenvironment; this is the first study to structurally characterize the vault complex as it exists within intact cells. Consistent with fluorescence microscopy studies that showed localization of the MVP protein to the cytoplasm (reviewed in [12]), we have demonstrated that assembled vaults are abundant in the cytoplasmic edges of diverse mammalian cells, and can be readily imaged by ECT. It is therefore remarkable that traditional thin-section EM techniques have not previously detected the distinct structural signature of the vault within cells or tissue. This is most likely explained by the loss of a subset of intracellular structures during the fixation, dehydration, and staining processes concomitant to the preparation of samples for traditional EM imaging [12, 24]. In contrast, ECT is a direct imaging technique that does not require a contrasting agent, and can be applied to samples that are cryo-preserved and thus analyzed in a frozen, hydrated and near-native state (reviewed in [22]).
Our ECT analyses of intracellular vaults confirmed that purification does not significantly alter vault structure since intracellular vaults displayed the same unique symmetrical dome-shaped morphology as purified vaults. Subtomogram averaging of the intracellular vaults produced an averaged structure with high electron density localized at the caps. This result is consistent with the localization of vRNA to the vault caps where it may function to stabilize their structure, thus resulting in an increased averaged density for the caps compared to the barrel or shoulder regions [10, 25].
A variance analysis of the same averaged volume confirmed that the barrel midsection was the most variable region of the complex, and a subset of intracellular vaults also demonstrated the waist indentation characteristic of purified vaults (Fig. 2, panels a, e). These observations underscore the role of the midsection in mediating vault dynamics, and are not surprising since the twofold axis at the vault waist region delineates the half-vault structures that comprise the full particle [8, 26]. Previous work suggested that within cells half-vaults could exchange with each other by separating at the midsection [27], but we did not see structures consistent with this type of complex dynamic. This, however, does not exclude the possibility that such dynamics are prevalent in parts of the cell that we were not able to image by ECT.
In our analysis of individual intracellular vaults, we consistently observed densities within the lumen of the particles. Based on density difference maps showing that the minor vault proteins localized to the interior of purified particles [1], these densities could represent vPARP and TEP1. Because the averaged volume of the vaults demonstrated no regular internal densities, we conclude, however, that the contents are not ordered within the intracellular vault lumen, but distributed randomly.
Perhaps most importantly, our ECT studies described the cellular ultrastructure with which native vaults associate. The overwhelming majority of vaults imaged by ECT were embedded in “granule beds”. Vaults did not appear to adopt a preferred orientation within the granule beds, but a few vaults appeared to interact with granules at their caps, or by opening up at their waist region (Fig. 6), observations consistent with previous reports [8, 26]. Given the previously reported localization of vPARP to the waist region of the vault [1], it is possible that the granules are enzymatically targeted by vault-associated vPARP. Alternatively, vPARP can polyadenylate MVP directly [28], and by doing so may regulate interactions between vaults and granules.
Although we are not able to conclusively say what the granules are, we infer that they are related to the biological function of the vault due to their strong co-localization with vault particles. Earlier work using fixed and negatively stained vaults purified from Dictyostelium discoideum [29] identified structures that are similar to the granules we have observed in association with vaults in situ. The authors of this paper postulated that the granule-like structures they observed might be morphological variants of the vault. However, the nature of the granule-like particles and their relationship to vaults were not confirmed. While it is possible that the intracellular granules we describe here are structural variants of the vault, we think it is more likely that granules are distinct cellular structures that associate with, and are related to, the biological function of the vault.
Recent work by the Rome lab [30] has provided an elegant model for vault assembly as a process that is tightly coupled to MVP expression from polysomes. Because we did not directly observe structures consistent with this proposed model (i.e., polyribosomes next to partial vaults), the granule beds may not be sites of coupled MVP translation and vault assembly. This conclusion is also supported by the observation that ribosomes are largely excluded from the interior of the granule beds. Intracellular granules have been proposed to be key elements of the mesoscale organization of the cell by mediating the concentration of soluble proteins through liquid phase transitions (reviewed in [31, 32]). This model of cellular organization is independent of membrane boundaries, and instead relies on the formation of “droplets”, or localized concentrations of proteins to produce distinct functional regions of cytoplasm. Confirming the identity of the granules with which the vaults associate may shed light on not only the cellular role of the vault, but also the role of granules in cellular organization.
Earlier studies using fluorescence microscopy of GFP-tagged MVP, or immuno-fluorescence strategies [19] showed that vaults interacted with polymerized microtubules within cells. Interestingly, we did not observe direct interactions between vaults and microtubules in the cytoplasmic periphery of mammalian cells. Indeed, more than 80 % of vaults were located more that 100 nm away from the nearest microtubule (Fig. 4d). One possible explanation for not observing this interaction by ECT is that vaults may not preferentially interact with microtubules in the thin areas of the cell where we are able to image. Vaults may traffic along microtubules from the interior of the cell to its extreme edges, but once in the periphery they may preferentially interact with other cellular factors, for example granules. Another possible explanation is that in the cell, the interaction of vaults with granule beds may mediate trafficking of vaults along microtubules instead of a direct interaction between vaults and microtubules. The unique association of vaults with intracellular granules may be key to understanding the complex’s cellular function.
Supplementary Material
Acknowledgments
We thank Zhiheng Yu and M. Jason de la Cruz of the Howard Hughes Medical Institute CryoEM Shared Resource at Janelia Farm for assistance with data collection. This work was supported by NIH Grant 2P50GM082545 (to GJJ).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00018-015-1898-y) contains supplementary material, which is available to authorized users.
References
- 1.Mikyas Y, Makabi M, Raval-Fernandes S, Harrington L, Kickhoefer VA, Rome LH, Stewart PL. Cryoelectron microscopy imaging of recombinant and tissue derived vaults: localization of the MVP N termini and VPARP. J Mol Biol. 2004;344(1):91–105. doi: 10.1016/j.jmb.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 2.Kedersha NL, Heuser JE, Chugani DC, Rome LH. Vaults. III. Vault ribonucleoprotein particles open into flower-like structures with octagonal symmetry. J Cell Biol. 1991;112(2):225–235. doi: 10.1083/jcb.112.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kedersha NL, Rome LH. Isolation and characterization of a novel ribonucleoprotein particle: large structures contain a single species of small RNA. J Cell Biol. 1986;103(3):699–709. doi: 10.1083/jcb.103.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kickhoefer VA, Vasu SK, Rome LH. Vaults are the answer, what is the question? Trends Cell Biol. 1996;6(5):174–178. doi: 10.1016/0962-8924(96)10014-3. [DOI] [PubMed] [Google Scholar]
- 5.Kedersha NL, Miquel MC, Bittner D, Rome LH. Vaults. II. Ribonucleoprotein structures are highly conserved among higher and lower eukaryotes. J Cell Biol. 1990;110(4):895–901. doi: 10.1083/jcb.110.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stephen AG, Raval-Fernandes S, Huynh T, Torres M, Kickhoefer VA, Rome LH. Assembly of vault-like particles in insect cells expressing only the major vault protein. J Biol Chem. 2001;276(26):23217–23220. doi: 10.1074/jbc.C100226200. [DOI] [PubMed] [Google Scholar]
- 7.Anderson DH, Kickhoefer VA, Sievers SA, Rome LH, Eisenberg D. Draft crystal structure of the vault shell at 9-A resolution. PLoS Biol. 2007;5(11):e318. doi: 10.1371/journal.pbio.0050318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casanas A, Querol-Audi J, Guerra P, Pous J, Tanaka H, Tsukihara T, Verdaguer N, Fita I. New features of vault architecture and dynamics revealed by novel refinement using the deformable elastic network approach. Acta Crystallogr D Biol Crystallogr. 2013;69(Pt 6):1054–1061. doi: 10.1107/S0907444913004472. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka H, Kato K, Yamashita E, Sumizawa T, Zhou Y, Yao M, Iwasaki K, Yoshimura M, Tsukihara T. The structure of rat liver vault at 3.5 angstrom resolution. Science (New York, NY) 2009;323(5912):384–388. doi: 10.1126/science.1164975. [DOI] [PubMed] [Google Scholar]
- 10.Kong LB, Siva AC, Kickhoefer VA, Rome LH, Stewart PL. RNA location and modeling of a WD40 repeat domain within the vault. RNA (New York, NY) 2000;6(6):890–900. doi: 10.1017/s1355838200000157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kickhoefer VA, Rajavel KS, Scheffer GL, Dalton WS, Scheper RJ, Rome LH. Vaults are up-regulated in multidrug-resistant cancer cell lines. J Biol Chem. 1998;273(15):8971–8974. doi: 10.1074/jbc.273.15.8971. [DOI] [PubMed] [Google Scholar]
- 12.Berger W, Steiner E, Grusch M, Elbling L, Micksche M. Vaults and the major vault protein: novel roles in signal pathway regulation and immunity. Cell Mol Life Sci CMLS. 2008;66(1):43–61. doi: 10.1007/s00018-008-8364-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mossink MH, van Zon A, Fränzel-Luiten E, Schoester M, Kickhoefer VA, Scheffer GL, Scheper RJ, Sonneveld P, Wiemer EAC. Disruption of the murine major vault protein (MVP/LRP) gene does not induce hypersensitivity to cytostatics. Cancer Res. 2002;62(24):7298–7304. [PubMed] [Google Scholar]
- 14.Dortet L, Mostowy S, Samba-Louaka A, Louaka AS, Gouin E, Nahori M-A, Wiemer EAC, Dussurget O, Cossart P. Recruitment of the major vault protein by InlK: a Listeria monocytogenes strategy to avoid autophagy. PLoS Pathog. 2011;7(8):e1002168. doi: 10.1371/journal.ppat.1002168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mrazek J, Kreutmayer SB, Grasser FA, Polacek N, Huttenhofer A. Subtractive hybridization identifies novel differentially expressed ncRNA species in EBV-infected human B cells. Nucleic Acids Res. 2007;35(10):e73. doi: 10.1093/nar/gkm244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu S, Hao Q, Peng N, Yue X, Wang Y, Chen Y, Wu J, Zhu Y. Major vault protein: a virus-induced host factor against viral replication through the induction of type-I interferon. Hepatology. 2012;56(1):57–66. doi: 10.1002/hep.25642. [DOI] [PubMed] [Google Scholar]
- 17.Daugherty MD, Young JM, Kerns JA, Malik HS. Rapid evolution of PARP genes suggests a broad role for ADP-ribosylation in host-virus conflicts. PLoS Genet. 2014;10(5):e1004403. doi: 10.1371/journal.pgen.1004403.s018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanaka H, Tsukihara T. Structural studies of large nucleoprotein particles, vaults. Proc Jpn Acad Ser B Phys Biol Sci. 2012;88(8):416–433. doi: 10.2183/pjab.88.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eichenmüller B, Kedersha N, Solovyeva E, Everley P, Lang J, Himes RH, Suprenant KA. Vaults bind directly to microtubules via their caps and not their barrels. Cell Motil Cytoskelet. 2003;56(4):225–236. doi: 10.1002/cm.10147. [DOI] [PubMed] [Google Scholar]
- 20.Ando D, Mattson MK, Xu J, Gopinathan A. Cooperative protofilament switching emerges from inter-motor interference in multiple-motor transport. Sci Rep. 2014;4:7255. doi: 10.1038/srep07255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicastro D, Schwartz C, Pierson J, Gaudette R, Porter ME, McIntosh JR. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 2006;313(5789):944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- 22.Gan L, Jensen GJ. Electron tomography of cells. Q Rev Biophys. 2012;45(1):27–56. doi: 10.1017/S0033583511000102. [DOI] [PubMed] [Google Scholar]
- 23.Kong LB, Siva AC, Rome LH, Stewart PL. Structure of the vault, a ubiquitous cellular component. Structure. 1999;7(4):371–379. doi: 10.1016/s0969-2126(99)80050-1. [DOI] [PubMed] [Google Scholar]
- 24.Pilhofer M, Ladinsky MS, McDowall AW, Petroni G, Jensen GJ. Microtubules in bacteria: ancient tubulins build a five-protofilament homolog of the eukaryotic cytoskeleton. PLoS Biol. 2011;9(12):e1001213. doi: 10.1371/journal.pbio.1001213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kickhoefer VA, Liu Y, Kong LB, Snow BE, Stewart PL, Harrington L, Rome LH. The Telomerase/vault-associated protein TEP1 is required for vault RNA stability and its association with the vault particle. J Cell Biol. 2001;152(1):157–164. doi: 10.1083/jcb.152.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Querol-Audi J, Casanas A, Uson I, Luque D, Caston JR, Fita I, Verdaguer N. The mechanism of vault opening from the high resolution structure of the N-terminal repeats of MVP. EMBO J. 2009;28(21):3450–3457. doi: 10.1038/emboj.2009.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang J, Kickhoefer VA, Ng BC, Gopal A, Bentolila LA, John S, Tolbert SH, Rome LH. Vaults are dynamically unconstrained cytoplasmic nanoparticles capable of half vault exchange. ACS Nano. 2010;4(12):7229–7240. doi: 10.1021/nn102051r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kickhoefer VA, Siva AC, Kedersha NL, Inman EM, Ruland C, Streuli M, Rome LH. The 193-kD vault protein, VPARP, is a novel poly(ADP-ribose) polymerase. J Cell Biol. 1999;146(5):917–928. doi: 10.1083/jcb.146.5.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vasu SK, Rome LH. Dictyostelium vaults: disruption of the major proteins reveals growth and morphological defects and uncovers a new associated protein. J Biol Chem. 1995;270(28):16588–16594. doi: 10.1074/jbc.270.28.16588. [DOI] [PubMed] [Google Scholar]
- 30.Mrazek J, Toso D, Ryazantsev S, Zhang X, Zhou ZH, Fernandez BC, Kickhoefer VA, Rome LH. Polyribosomes are molecular 3D nanoprinters that orchestrate the assembly of vault particles. ACS Nano. 2014 doi: 10.1021/nn504778h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weber SC, Brangwynne CP. Getting RNA and protein in phase. Cell. 2012;149(6):1188–1191. doi: 10.1016/j.cell.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 32.Hyman AA, Brangwynne CP. Beyond stereospecificity: liquids and mesoscale organization of cytoplasm. Dev Cell. 2011;21(1):14–16. doi: 10.1016/j.devcel.2011.06.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


