Abstract
Follicular helper T cells (TFH) are essential for germinal centers and most long term humoral immunity. Differentiation of TFH cells depends on the transcriptional repressor, Bcl6. Bcl6 mediates gene repression via the recruitment of co-repressors. Currently, it is unknown how Bcl6 recruits corepressors to regulate gene expression of TFH cells. Here we demonstrate, using a mutant form of Bcl6 with two BTB (bric-a-brac, tramtrack, broad-complex) mutations that abrogate corepressor binding, that the Bcl6 BTB domain is required for proper differentiation of TFH and GC TFH cells in vivo. Importantly, we also observe a significant defect in germinal center B cell development. These results are consistent in multiple contexts, including a novel LCMV NP-specific TCR transgenic mouse model. Taken together, these data suggest that the Bcl6 BTB domain is a key mediator of the differentiation of TFH cells.
INTRODUCTION
B cell CLL/lymphoma 6 (Bcl6) is a transcriptional repressor that is essential for the differentiation of T follicular helper cells (TFH) and germinal center B cells. TFH cells are CD4 T cells specialized in providing help for B cells (1). The absence of TFH cells results in the loss of germinal centers (GC) and, consequently, abrogated memory B cell, plasma cell, and neutralizing antibody responses. Thus, TFH cells have critical roles in protective immune responses against pathogens, as well as deleterious roles in numerous autoimmune diseases (1, 2).
Bcl6 consists of a bric-a-brac, tramtrack, broad-complex (BTB/POZ) domain, a middle domain (also known as RDII), and a zinc finger domain consisting of six krueppel-like zinc fingers (1). BTB domains are evolutionarily conserved protein interaction domains that are widely present in transcription factors (3, 4). The BTB domain forms the interface of the obligate homodimer and the corepressors BCOR, SMRT, and NCOR bind at the cleft formed by this interface (5-8). While Bcl6 is required for TFH differentiation (9-12), the contributions of its functional domains in CD4+ T cells are not well understood. In this study, we sought to examine to role of the Bcl6 BTB domain in TFH differentiation and function.
MATERIALS AND METHODS
Mice and vectors
C57BL/6J (B6) and CreCD4 mice were purchased from the Jackson Laboratory. Bcl6fl/fl (13), CD45.1 congenic and Smarta TCR transgenic (‘SM’, specific for LCMV GP (glycoprotein) 66-77 on I-Ab) (14) mice were on a full B6 background and were bred at LJI. Bcl6BTBMUT mice, engineered to express the Bcl6 BTBmut from the endogenous Bcl6 locus, were generously provided by Dr. Ari Melnick (15), which were crossed to homozygosity at LJI for use in all experiments. NIP TCR transgenic mice were generated as described below and in Supplementary Figure 1. TCR hybridomas were generated (Janice White and Philippa Marrack, unpublished data) and TCR sequences were cloned and sequenced using cDNA isolated from LCMV-reactive clones. TCR sequences were expressed in 58 αβ− T cell hybridomas and tested for reactivity against LCMV-infected dendritic cells. The TCRαβ pair showing the strongest reactivity (Vα1-Jα8 and Vβ6-Dβ1-Jβ2.3 rearrangements) was chosen and cloned into genomic TCR expression cassette vectors. Linearized DNA fragments were injected into fertilized C57BL/6 eggs at the UCSD transgenic mouse facility. Pups were genotyped (Fig. S1). A single α/β TCR transgenic founder mouse (“NIP”) was selected and crossed to B6.SJL mice to generate CD45.1+ NIP mice. All animal experiments were conducted in accordance with approved animal protocols. The green fluorescent protein (GFP)-expressing retroviral (RV) expression vector pMIG was used. BTB domain mutant Bcl6 RV (BTBmut) was generated by inducing two point mutations in the protein interaction domain that do not affect dimerization (16). RV particles were produced as previously described (9).
Infections and immunizations
Lymphocytic choriomeningitis virus (LCMV) Armstrong stocks were prepared and quantified as previously described (9). Infections were performed by intraperitoneal injection of 0.5-2 × 105 plaque-forming units of LCMV Armstrong per mouse. GP61-keyhole limpet hemocyanin (KLH) was prepared in alum and injected as described previously (17). A total of 20 μg GP61-KLH was resuspended in alum for bilateral footpad injections.
Flow cytometry
Flow cytometry was done with monoclonal antibodies against SLAM (CD150, Biolegend) and CD4, CD8, CD44, CD62L, CD25, B220, Fas, and GL7 (eBioscience). Stains were done for 30 minutes at 4 °C in PBS supplemented with 0.5% bovine serum albumin and 0.1% sodium azide, unless specified otherwise. CXCR5 staining was done as described (9, 18). Intracellular staining for Bcl6 was performed with an Alexa 647-conjugated monoclonal antibody to Bcl6 (clone K112-91) (BD Pharmingen) using the FoxP3 ICS kit buffers and protocol (eBioscience).
Statistical Analysis
Statistical tests were performed using Prism 5.0 (GraphPad). P values were calculated by two-tailed unpaired Student’s t tests with a 95% confidence interval. Error bars depict the standard error of the mean. *, p < 0.05. **, p < 0.01, ***, p < 0.001.
RESULTS AND DISCUSSION
The Bcl6 BTB domain regulates TFH differentiation
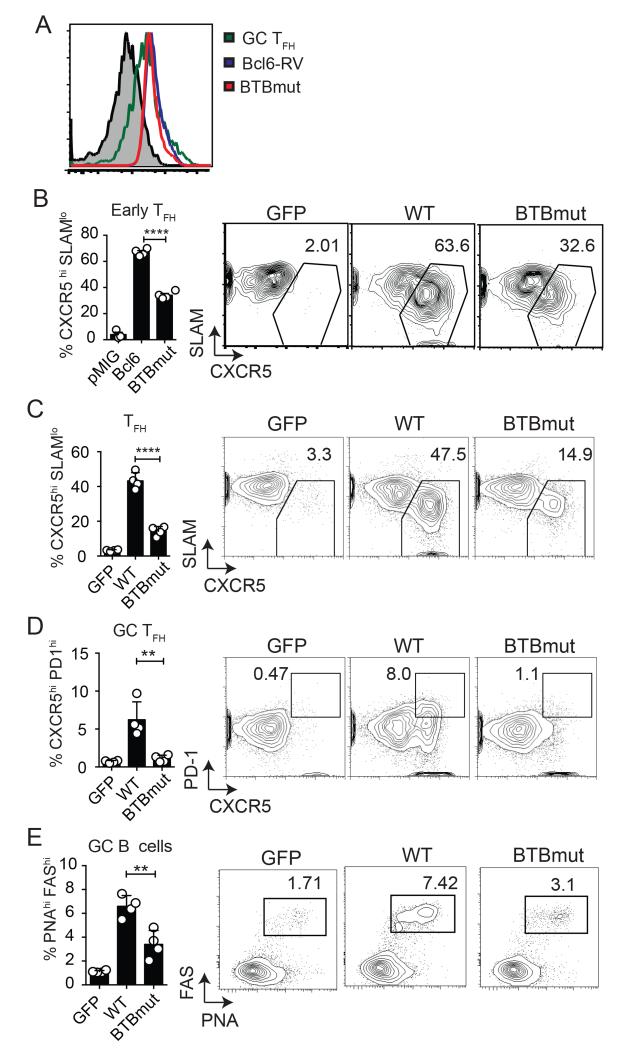
Given that the Bcl6 BTB domain is known to interact with corepressors (such as BCOR) in B cells that are also expressed in CD4 T cells, we sought to determine the role of the Bcl6 BTB domain in TFH differentiation. To this end, we generated N21K and H116A mutations in the Bcl6 BTB domain. These point mutations together have been shown to prevent co-repressor binding to the BTB domain without affecting the ability of Bcl6 to dimerize (16). Similar levels of Bcl6 protein expression were observed in CD4 T cells transduced with an RV expressing either WT-Bcl6 or the mutant form (BTBmut), and the amount of Bcl6 expression was comparable to that of GC TFH cells (Fig. 1A). To examine the role of the Bcl6 BTB domain in vivo, we determined if expression of the Bcl6 BTBmut protein in Bcl6-deficient cells could rescue Tfh development in response to acute LCMV infection. LCMV glycoprotein-specific Bcl6fl/fl CreCD4 Smarta (SM) TCR transgenic CD45.1 cells were transduced with Bcl6 WT, BTBmut, or an empty GFP vector (GFP) and transferred to Bcl6fl/fl CreCD4 hosts. We first examined early TFH development 3 days following an acute LCMV infection. Cells transduced with the empty vector (GFP+) did not differentiate into TFH cells, confirming the requirement of Bcl6 for TFH differentiation. Ectopic expression of WT Bcl6 was sufficient to rescue TFH development (Fig. 1B). We observed a defect in early TFH differentiation in BTBmut+ SM cells compared with Bcl6-WT+ SM (p < 0.0001, Fig. 1B). We then examined TFH and GC B cell development at 7 days following acute LCMV infection. Again, there was a significant impairment in TFH differentiation in BTBmut+ cells (p < 0.0001, Fig. 1C) compared with WT. This was coupled with an even larger reduction in GC TFH cells (p = 0.006, Fig. 1D). GC TFH cells are the more polarized TFH cells that are present in the germinal centers and are identifiable as CXCR5hiPD1hiPSGL1loBcl6hi cells (1). In parallel we observed a significant defect in GC B cell development (p = 0.005, Fig. 1E. Thus, Bcl6 BTB domain functions appeared to be necessary for the majority of TFH differentiation and B cell help.
Figure 1. The Bcl6 BTB domain is necessary for proper TFH cell differentiation.
(A) Similar levels of Bcl6 expression in Bcl6-RV, BTBmut-RV and GC TFH. Bcl6 MFI of Bcl6fl/fl CreCD4 CD4 T cells transduced with either BTBmut-RV or Bcl6-RV compared to ex vivo GC TFH isolated from mice at 7 days following LCMV infection. (B-E) Bcl6fl/fl CreCD4 SM cells were retrovirally transduced with empty GFP vector, Bcl6 WT, or BTBmut, then transferred to Bcl6fl/fl CreCD4 mice and analyzed at day 3 or 7 following an acute LCMV infection. (B) Early Tfh differentiation (CXCR5hi SLAMlo) by transduced Bcl6fl/fl CreCD4 SM cells at day 3. (C) Tfh differentiation (CXCR5hi SLAMlo) by transduced Bcl6fl/fl CreCD4 SM cells at day 7. (D) GC TFH (CXCR5hi PD1hi) by transduced Bcl6fl/fl CreCD4 SM cells at day 7. (E) GC B cells (PNAhi Fashi) at day 7. At least 3 mice were used for each condition. Experiments were repeated at least 3 times. * p < 0.05, ** p < 0.01, *** p < 0.001.
Impaired TFH differentiation by germline Bcl6BTBMUT CD4 T cells
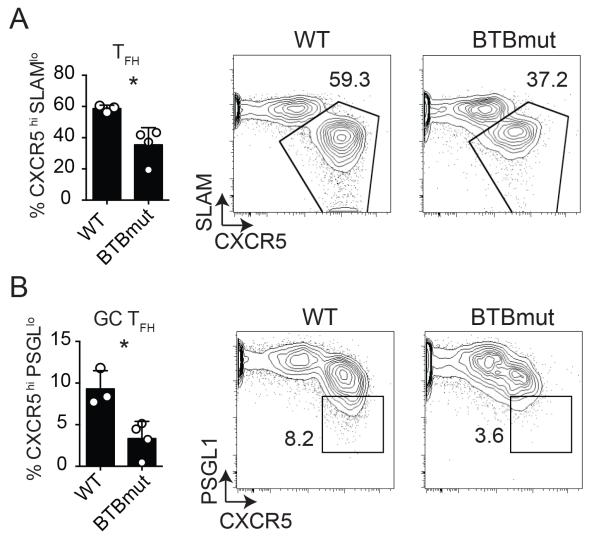
These results are different from a recent report using mixed bone marrow chimeric mice with Bcl6BTBMUT and Tcrb−/− donor cells, in which no difference in total Tfh frequencies were observed upon immunization with sheep red blood cells (SRBC), though antigen-specific cells were not directly assessed in that study (19). Therefore, in light of those data, it was incumbent upon us to explore the role of the Bcl6 BTB domain in TFH differentiation in multiple contexts, directly examining antigen-specific Tfh cells in each case. Thus, we obtained the germline “knock-in” Bcl6BTBMUT mice for further studies. CD4 T cells were isolated from Bcl6BTBMUT SM CD45.1+ or WT SM CD45.1+ mice and transferred to Bcl6fl/fl CreCD4 hosts. Antigen-specific TFH and GC TFH differentiation was examined at 7 days following acute LCMV infection. We observed a significant defect in both TFH (p = 0.017, Fig. 2A) and GC TFH differentiation (p = 0.014, Fig. 2B), confirming the requirement for the Bcl6 BTB domain in the generation of TFH cells.
Figure 2. Impaired TFH differentiation by Bcl6BTBMUT CD4 T cells.
Bcl6BTBMUT SM or WT SM cells were transferred to Bcl6fl/fl CreCD4 mice and analyzed 7 days following acute LCMV infection. (A) Tfh (CXCR5hi SLAMlo) Bcl6BTBMUT SM cells. (B) GC TFH (CXCR5hi PSGL1lo) Bcl6BTBMUT SM cells. At least 3 mice were used for each condition. Experiments were repeated at least 3 times. * p < 0.05, ** p < 0.01, *** p < 0.001.
BTB dependent TFH differentiation in NIP-transgenic Bcl6BTBMUT CD4 T cells
One possibility is that this observed phenotype may be selective to the SM CD4 T cell response, though SM CD4 T cells have been very informative in numerous contexts (9, 14, 17, 20). To this end, we developed a new LCMV-specific TCR transgenic mouse. The LCMV nucleoprotein (NP) is the major target of the antibody response during LMCV infection, and the currently used GP-specific SM CD4 T cells do not allow for the study of the immunodominant antibody response because GP-specific CD4 T cells do not provide help to NP-specific B cells (14). Epitope mapping defined a specific peptide (NP 311-325) of the NP protein efficiently recognized by CD4 T cells during LCMV infection (21, 22). For this reason, NP 311-325 was chosen as a target to generate a novel TCR transgenic mouse model (Fig. S1). NIP-transgenic CD4 T cells transferred into B6 mice exhibited robust expansion and differentiated into TFH and GC TFH cells following LCMV infection (Fig. S1B-E). NIP CD4 T cells transferred into B6 hosts were able to induce robust B cell responses, including the generation of GC B cells (Fig. S1F), plasma cells (Fig. S1G), and LCMV-specific antibodies (Fig. S1H).
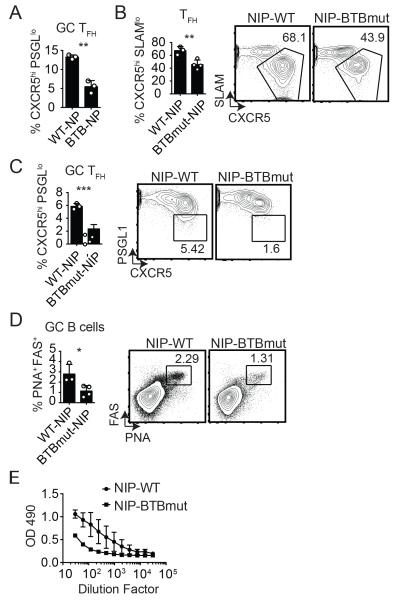
We examined the role of the Bcl6 BTB domain in the context of a retrogenic mouse model where bone marrow (BM) from Bcl6BTBMUT or B6 (WT) mice was transduced with an NP-specific TCRα and TCRβ retroviral construct (NIP-RV) and the transduced bone marrow was transferred into irradiated WT recipients (23). WT or Bcl6BTBMUT NP-specific naive CD4 T cells were isolated from these mice after reconstitution and transferred to Bcl6fl/fl CreCD4 hosts. The NIP+ WT and NIP+ Bcl6BTBMUT CD4 T cells expanded robustly in response to an acute LCMV infection. At 7 days following LCMV infection, we observed a significant defect in TFH differentiation (Fig. S2) and GC TFH differentiation (Fig. 3A, Fig. S2) for Bcl6BTBMUT NIP-RV+ compared to WT NIP-RV+ mice (p = 0.01 and p = 0.0014). We then made use of germline NIP TCR transgenic mice crossed with Bcl6BTBMUT mice. Bcl6BTBMUT NIP or WT NIP CD4 T cells were transferred into Bcl6fl/fl CreCD4 recipients followed by LCMV infection. We observed severe defects in TFH (p = 0.0088, Fig. 3B) and GC TFH (p = 0.0006, Fig. 3C) differentiation in Bcl6BTBMUT NIP CD4 T cells. The consequence of the substantial loss of GC Tfh was a significant loss of GC B cell development (Fig. 3D) and reduced anti-LCMV IgG response (Fig. 3E). Taken together, these data demonstrate that the Bcl6 BTB domain is important for TFH differentiation and function in response to at least two different antigens.
Figure 3. Bcl6BTBMUT LCMV-specific NIP transgenic CD4 T cells are defective in TFH cell differentiation and function.
CD4 T cells from Bcl6BTBMUT NIP-TCR+ retrogenic mice were transferred to Bcl6fl/fl CreCD4 mice and analyzed at day 7 following acute LCMV infection. (A) GC Tfh cells (CXCR5hi PSGL1lo) among Bcl6BTBMUT NIP-TCR+ cells. (B-E) Bcl6BTBMUT NIP or WT NIP cells were transferred to Bcl6fl/fl CreCD4 mice and analyzed following acute LCMV infection. (B) Tfh (CXCR5hi SLAMlo) Bcl6BTBMUT NIP cells. (C) GC TFH (CXCR5hi PSGL1lo) Bcl6BTBMUT NIP cells. (D) GC B cells (PNAhi Fashi). (E) LCMV-specific IgG. At least 3 mice were used for each condition. Experiments in B-D were repeated at least 3 times. * p < 0.05, ** p < 0.01, *** p < 0.001.
The Bcl6 BTB domain regulates TFH differentiation and GC B cell development following protein immunization
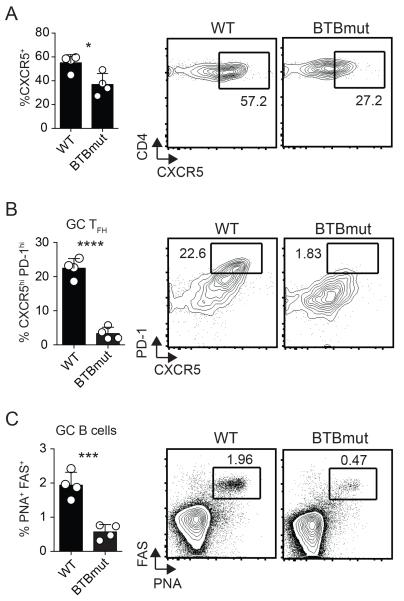
To examine the role of the Bcl6 BTB domain in another experimental context we utilized a protein immunization model, keyhole limpet hemocyanin (KLH) conjugated with gp61-80 peptide in alum (17). We transferred Bcl6BTBMUT SM or WT SM CD4 T cells into Bcl6fl/fl CreCD4 hosts followed by immunization with GP61-KLH in alum. At 10 days following immunization, lymph nodes were harvested and examined for antigen-specific TFH differentiation and function. We observed a significant reduction in CXCR5 expression by Bcl6BTBMUT CD4 T cells (p = 0.0195, Fig. 4A) and a severe defect in GC TFH differentiation (p < 0.0001, Fig. 4B). Bcl6BTBMUT CD4 T cells were functionally deficient as they were unable to promote GC B cell responses (p = 0.0008, Fig. 4C). We also observed defects in TFH and GC TFH differentiation in Bcl6fl/fl CreCD4 mice receiving BTBmut-RV+ SM CD4 T cells as compared with Bcl6-WT-RV+ SM CD4 T cells at 10 days following immunization (data not shown). Thus, the Bcl6 BTB domain is needed for optimal TFH differentiation and function in both the context of protein immunizations and an acute viral infection.
Figure 4. Defective TFH differentiation and function in the absence of a function Bcl6 BTB domain following protein immunization.
Bcl6BTBMUT SM or WT SM cells were transferred to Bcl6fl/fl CreCD4 mice and analyzed following GP61-KLH (alum) immunization, day 10. (A) Tfh (CXCR5hi SLAMlo) Bcl6BTBMUT SM cells. (B) GC TFH (CXCR5hi PD1hi) Bcl6BTBMUT SM cells. (C) GC B cells (PNAhi Fashi). At least 3 mice were used for each condition. Experiments were repeated at least 3 times. * p < 0.05, ** p < 0.01, *** p < 0.001.
Our results demonstrate the importance of the Bcl6 BTB repressor domain in CD4 T cells in the differentiation of TFH cells and in the ability of TFH cells to provide help to B cells. A previous study did not observe a TFH defect in Bcl6BTBMUT CD4 T cells (19), possibly because antigen-specific TFH were not directly analyzed, or possibly due to a confounding negative feedback Bcl6 autoregulatory mechanism involving cullin3, whereby cullin3 is most likely recruited via the Bcl6 BTB domain (24). Alternatively, the BTB domain of Bcl6 has more prominent roles for Bcl6 activity in some in vivo contexts than others. Bcl6 functions in TFH cells via binding to thousands of genes, demonstrating that Bcl6 is deeply integrated into TFH cell biology (25). In this study, TFH defects in the absence of BTB domain function were consistent when assessing antigen-specific TFH cells, using two different antigen specificities, as well as in the context of both acute viral infection and protein immunization. The defect in TFH differentiation and function that was observed is likely due to the inability of Bcl6 to recruit corepressors such as SMRT, NCOR, and BCOR and thus repress target genes. Indeed, it has been found that BCOR-deficient CD4 T cells are defective for GC TFH differentiation and B cell help functions (26). Thus, BCOR may be the main co-repressor interacting with the Bcl6 BTB domain in TFH cells. SMRT, NCOR, and BCOR are all expressed in CD4 T cells, and further investigation of the comparative roles of each of these Bcl6 BTB domain interacting transcription factors is needed. This report highlights the importance of the repressive ability of the Bcl6 BTB domain in TFH cells for successful germinal center reactions and robust antibody responses.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Marc Jenkins, Jessica Yang, Vivian Bardwell, and Noah Tubo for sharing unpublished data. We thank Ari Melnick for sharing the Bcl6BTBMUT mice. We thank Janice White and Philippa Marrack for generating T cell hybridomas for LCMV NP 311-325.
GRANT SUPPORT
This work was supported by NIH NIAID grants R01 AI063107 and U19 AI109976 (to S.C.).
REFERENCES
- 1.Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. 2014;41:529–542. doi: 10.1016/j.immuni.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ueno H, Banchereau J, Vinuesa CG. Pathophysiology of T follicular helper cells in humans and mice. Nat. Immunol. 2015;16:142–152. doi: 10.1038/ni.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardwell VJ, Treisman R. The POZ domain: a conserved protein-protein interaction motif. Genes & Development. 1994;8:1664–1677. doi: 10.1101/gad.8.14.1664. [DOI] [PubMed] [Google Scholar]
- 4.Beaulieu AM, Sant’Angelo DB. The BTB-ZF Family of Transcription Factors: Key Regulators of Lineage Commitment and Effector Function Development in the Immune System. J. Immunol. 2011;187:2841–2847. doi: 10.4049/jimmunol.1004006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huynh KD, Bardwell VJ. The BCL-6 POZ domain and other POZ domains interact with the co-repressors N-CoR and SMRT. Oncogene. 1998;17:2473–2484. doi: 10.1038/sj.onc.1202197. [DOI] [PubMed] [Google Scholar]
- 6.Huynh KD, Fischle W, Verdin E, Bardwell VJ. BCoR, a novel corepressor involved in BCL-6 repression. Genes & Development. 2000;14:1810–1823. [PMC free article] [PubMed] [Google Scholar]
- 7.Dhordain P, Albagli O, Lin RJ, Ansieau S, Quief S, Leutz A, Kerckaert JP, Evans RM, Leprince D. Corepressor SMRT binds the BTB/POZ repressing domain of the LAZ3/BCL6 oncoprotein. Proc. Natl. Acad. Sci. USA. 1997;94:10762–10767. doi: 10.1073/pnas.94.20.10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghetu AF, Corcoran CM, Cerchietti L, Bardwell VJ, Melnick A, Prive GG. Structure of a BCOR corepressor peptide in complex with the BCL6 BTB domain dimer. Mol. Cell. 2008;29:384–391. doi: 10.1016/j.molcel.2007.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, Dent AL, Craft J, Crotty S. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kroenke MA, Eto D, Locci M, Cho M, Davidson T, Haddad EK, Crotty S. Bcl6 and Maf cooperate to instruct human follicular helper CD4 T cell differentiation. J. Immunol. 2012;188:3734–3744. doi: 10.4049/jimmunol.1103246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, Wang Y-H, Dong C. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu D, Rao S, Tsai LM, Lee SK, He Y, Sutcliffe EL, Srivastava M, Linterman M, Zheng L, Simpson N, Ellyard JI, Parish IA, Ma CS, Li Q-J, Parish CR, Mackay CR, Vinuesa CG. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 13.Kaji T, Ishige A, Hikida M, Taka J, Hijikata A, Kubo M, Nagashima T, Takahashi Y, Kurosaki T, Okada M, Ohara O, Rajewsky K, Takemori T. Distinct cellular pathways select germline-encoded and somatically mutated antibodies into immunological memory. J. Exp. Med. 2012;209:2079–2097. doi: 10.1084/jem.20120127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oxenius A, Bachmann MF, Zinkernagel RM, Hengartner H. Virus-specific MHC-class II-restricted TCR-transgenic mice: effects on humoral and cellular immune responses after viral infection. Eur. J. Immunol. 1998;28:390–400. doi: 10.1002/(SICI)1521-4141(199801)28:01<390::AID-IMMU390>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 15.Huang C, Gonzalez DG, Cote CM, Jiang Y, Hatzi K, Teater M, Dai K, Hla T, Haberman AM, Melnick A. The BCL6 RD2 Domain Governs Commitment of Activated B Cells to Form Germinal Centers. Cell Rep. 2014;8:1497–1508. doi: 10.1016/j.celrep.2014.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahmad KF, Melnick A, Lax S, Bouchard D, Liu J, Kiang C-L, Mayer S, Takahashi S, Licht JD, Prive GG. Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain. Mol. Cell. 2003;12:1551–1564. doi: 10.1016/s1097-2765(03)00454-4. [DOI] [PubMed] [Google Scholar]
- 17.Choi YS, Yang JA, Yusuf I, Johnston RJ, Greenbaum J, Peters B, Crotty S. Bcl6 expressing follicular helper CD4 T cells are fate committed early and have the capacity to form memory. J. Immunol. 2013;190:4014–4026. doi: 10.4049/jimmunol.1202963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi YS, Kageyama R, Eto D, Escobar TC, Johnston RJ, Monticelli L, Lao C, Crotty S. ICOS Receptor Instructs T Follicular Helper Cell versus Effector Cell Differentiation via Induction of the Transcriptional Repressor Bcl6. Immunity. 2011;34:932–946. doi: 10.1016/j.immuni.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang C, Hatzi K, Melnick A. Lineage-specific functions of Bcl-6 in immunity and inflammation are mediated by distinct biochemical mechanisms. Nat. Immunol. 2013;14:380–388. doi: 10.1038/ni.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harker JA, Lewis GM, Mack L, Zuniga EI. Late interleukin-6 escalates T follicular helper cell responses and controls a chronic viral infection. Science. 2011;334:825–829. doi: 10.1126/science.1208421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dow C, Oseroff C, Peters B, Nance-Sotelo C, Sidney J, Buchmeier M, Sette A, Mothé BR. Lymphocytic choriomeningitis virus infection yields overlapping CD4+ and CD8+ T-cell responses. Journal of Virology. 2008;82:11734–11741. doi: 10.1128/JVI.00435-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Homann D, Lewicki H, Brooks D, Eberlein J, Mallet-Designé V, Teyton L, Oldstone MBA. Mapping and restriction of a dominant viral CD4+ T cell core epitope by both MHC class I and MHC class II. Virology. 2007;363:113–123. doi: 10.1016/j.virol.2006.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holst J, Szymczak-Workman AL, Vignali KM, Burton AR, Workman CJ, Vignali DAA. Generation of T-cell receptor retrogenic mice. Nat. Protoc. 2006;1:406–417. doi: 10.1038/nprot.2006.61. [DOI] [PubMed] [Google Scholar]
- 24.Mathew R, Mao A-P, Chiang AH, Bertozzi-Villa C, Bunker JJ, Scanlon ST, McDonald BD, Constantinides MG, Hollister K, Singer JD, Dent AL, Dinner AR, Bendelac A. A negative feedback loop mediated by the Bcl6-cullin 3 complex limits Tfh cell differentiation. J. Exp. Med. 2014;211:1137–1151. doi: 10.1084/jem.20132267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hatzi K, Nance JP, Kroenke MA, Bothwell M, Haddad EK, Melnick A, Crotty S. BCL6 orchestrates Tfh cell differentiation via multiple distinct mechanisms. The Journal of Experimental Medicine. 2015 doi: 10.1084/jem.20141380. doi:10.1084/jem.20141380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang JA, Tubo NJ, Gearhart MD, Bardwell VJ, Jenkins MK. BCL6 interacting corepressor contributes to germinal center T follicular helper cell formation and B cell helper function. 2015 doi: 10.4049/jimmunol.1500201. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.