Abstract
Alcohol use disorder (AUD) is a serious health challenge. Despite a large hereditary component to AUDs, few genes have been unambiguously implicated in their etiology. The fruit fly, Drosophila melanogaster, is a powerful model for exploring molecular-genetic mechanisms underlying alcohol-related behaviors and therefore holds great promise for identifying and understanding the function of genes that influence AUD. The use of the Drosophila model for these types of studies depends on the availability of assays that reliably measure behavioral responses to ethanol. This report describes an assay suitable for assessing ethanol sensitivity and rapid tolerance in flies. Ethanol sensitivity measured in this assay is influenced by the volume and concentration of ethanol used, a variety of previously reported genetic manipulations, and also the length of time the flies are housed without food immediately prior to testing. In contrast, ethanol sensitivity measured in this assay is not affected by the vigor of fly handling, sex of the flies, and supplementation of growth medium with antibiotics or live yeast. Three different methods for quantitating ethanol sensitivity are described, all leading to essentially indistinguishable ethanol sensitivity results. The scalable nature of this assay, combined with its overall simplicity to set-up and relatively low expense, make it suitable for small and large scale genetic analysis of ethanol sensitivity and rapid tolerance in Drosophila.
Keywords: ethanol, alcohol, behavior, sensitivity, Drosophila, fruit fly, assay
Introduction
Alcohol use disorder (AUD) is an enormous health problem worldwide (reviewed in 1). Although the mechanisms driving the development of AUD are complex, these disorders have a major genetic component (e.g. 2). The large heritability of AUDs and the conserved behavioral responses to ethanol across many species (reviewed in 3,4) have generated strong interest in using genetic model organisms to investigate the involvement of specific genes in ethanol-related behaviors toward better understanding the molecular basis of AUD. The fruit fly, Drosophila melanogaster, has emerged as a leading model organism for exploring molecular-genetic mechanisms of ethanol-related behaviors (reviewed in 3,4). Studies in flies have highlighted roles for several signaling pathways in behavioral responses to ethanol (reviewed in 5). Intriguingly, some of the genes and pathways that influence behavioral responses to ethanol in flies have also been implicated in rodent ethanol-related behaviors and/or human AUDs (e.g. 6-14). The conservation of mechanisms driving ethanol-related behaviors across species, coupled with the suite of genetic tools available in the Drosophila model system, underscore the utility of the fruit fly model for investigating the genetics of behavioral responses to ethanol.
Sensitivity 15,16 and tolerance (reviewed in 17) to ethanol in humans is linked to the development of AUD. Both of these behavioral responses to ethanol can be modeled in flies via a variety of laboratory assays (reviewed in 3,4). All of the fly assays known to the authors are based on either time-dependent ethanol-induced sedation/incoordination or time-dependent recovery from ethanol sedation.
In a previous article from our group on the genetics of ethanol sensitivity and rapid tolerance in Drosophila, a behavioral assay based on ethanol vapor-induced sedation of flies was used 18. Testing in this assay was initiated by transferring live adult flies without anesthesia to empty food vials, trapping the flies in the vials with a cellulose acetate plug, adding ethanol to the top (i.e. non-fly side) of the cellulose acetate plug, and sealing the vial containing flies, cellulose acetate plug and ethanol with a silicone stopper (see schematic in Figure S3, reference 18). Multiple vials representing different groups of flies were assessed in parallel, increasing throughput of this assay. Vials were given an anonymous code and experimenters were blinded to treatment group to prevent unintended bias in the assessment of sedation. In a standard experiment, flies in vials were tapped gently at 6 min intervals and, after a 30 sec recovery, the number of sedated flies in each vial was counted and converted to percent active flies. Flies absorbed ethanol vapor from the cellulose acetate plug in a time-dependent fashion, causing progressive increases in internal ethanol18 and sedation (c.f. reference 18 and Figure 1A and 1B in this report). Sedation in this assay was operationally defined as flies (i) standing in the absence of walking or (ii) lying on their backs with or without flapping their wings. Here, this ethanol sedation assay is described in detail, further operational optimization relevant to using it is provided, and the assay is used to address the contribution of food supplementation options on fly sedation sensitivity.
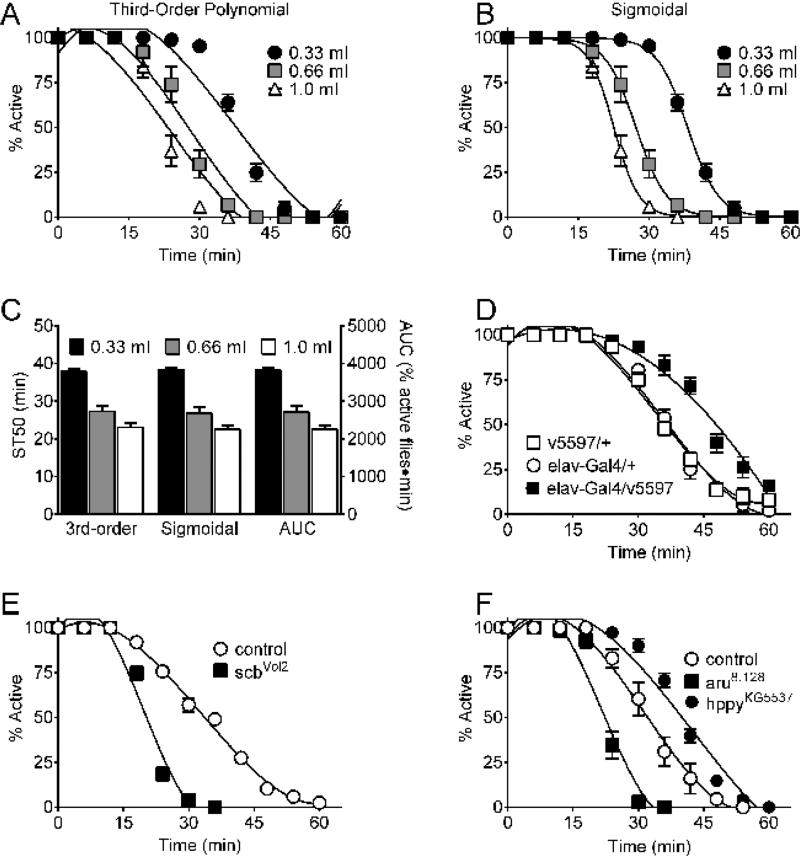
Figure 1. (A-C) Sedation assay data analysis options.
Control w[A] females (w1118 isogenic, Bloomington Indiana Drosophila Stock Center stock #5905) were exposed to vapor from the indicated volumes of 85% (vol/vol) ethanol. (A) Sedation time-course data fit with third-order polynomials. (B) Sedation time-course data fit with sigmoidal curves. (C) Ethanol sedation sensitivity quantitated as (left Y-axis) ST50 values interpolated from third-order polynomials and sigmoidal curves or (right Y-axis) area under the curve (AUC, % active flies x time). Ethanol volume, but not analysis method, impacted ethanol sensitivity (two-way ANOVA; effect of volume, p <0.0001; effect of analysis method, n.s.; n = 8/group; AUC data transformed 1/100 to account for the difference in magnitude of the values). (D-F) Representative time-course data from. (D) Expression of Cnx14D RNAi in the nervous system (elav-Gal4/v5597) blunted ethanol sensitivity relative to controls (v5597/+ and elav-Gal4/ +). (E and F) Mutation of scb (scbVol2) and aru (aru8.128) enhanced ethanol sedation sensitivity and mutation of hppy (hppyKG5537) blunted ethanol sedation sensitivity compared to controls. Data in E-F were previously reported as ST50 values 18.
Protocol
1. Day Before Assay
Collect flies into fresh food vials in groups of 11 (single sex) under brief (1-5 min) CO2.
Allow flies to recover overnight in food vials in an environmentally controlled space (typically 25 °C, 60% relative humidity, 12 hr light/dark cycle).
Prepare ethanol solution(s) by diluting pure (100%) ethanol in purified (≥18 MΩ) water to final concentration(s) appropriate for the planned experiment. Allow solution(s) to return to room temperature overnight.
Note: Dilution of ethanol is exothermic.
2. Day of Assay
For each vial of flies to be tested, prepare (i) a clean, empty food vial; i.e. testing vial, (ii) a new cellulose acetate plug, (iii) a silicone stopper and (iv) 1 ml of ethanol solution (see Table 3).
Prepare testing room by adjusting temperature to 20-25 °C and relative humidity to 55-65%.
Have another worker assign a unique code to each group of vials and record the code for later. Place coded vials with flies in testing room to acclimate for a few min.
Label empty testing vials to match codes on fly vials from 2.3.
Construct a hard copy testing log by entering the codes into columns (one column/vial) in a spreadsheet similar to Table 1.
Using the testing log as a guide, arrange coded food vials with flies and empty testing vials into matching arrays in the testing room. Note: A reasonable maximum number of vials to test is 24 (i.e. 6 sets of 4 vials each).
Transfer flies from food vials into matched/labeled empty testing vials and immediately insert cellulose acetate plugs into testing vials until cellulose acetate plugs are 2 cm below the vial tops.
Hereafter, handle each row of four vials as a set at staggered one min intervals.
For the time 0 assessment, grasp each vial individually with thumb and forefinger, tap gently on the table three times to knock flies to the bottom of the vial, wait 30 sec and then count the number of flies that are immobile/dead. Record the number of immobile/dead flies for each vial at time 0 min in the hardcopy testing log.
Start timer counting up continuously at time 0 and immediately begin adding 1 ml of ethanol to cellulose acetate plugs in the vials for the first row/set of 4 vials. Add 1 ml of ethanol to the cellulose acetate plugs in the vials at 5 sec intervals in the order they will be tested. Add ethanol to the cellulose acetate plugs in a circular motion so that the ethanol is absorbed evenly throughout the cellulose acetate plugs. When ethanol has been added to all 4 testing vials in the set, insert a silicone plug in each vial to seal it.
At times 1, 2, 3, 4 and 5 min, add 1 ml of ethanol to the second, third, fourth, fifth and sixth sets of 4 vials, respectively. Continue inserting silicone stoppers after adding ethanol to each set of 4 vials.
At time 6 min, test the first set of 4 vials by grasping each vial with thumb and forefinger, tapping gently on the table three times to knock flies to the bottom of the vial, waiting 30 sec and then counting and recording the total number of flies that are sedated. Score flies as sedated if they (i) stand on the floor of the vial but do not walk or (ii) lie on their backs with or without flapping their wings.
Handle each vial within the set at 5 sec intervals using the schedule in Table 2.
At times 7, 8, 9, 10 and 11 min, test the second, third, fourth, fifth and sixth sets of vials, respectively, as done for the first set.
At time 12 min, test the first set of 4 vials again as described in 2.12 and continue testing the second, third, fourth, fifth and sixth sets of vials at 13, 14, 15, 16 and 17 min, respectively.
Continue testing flies as described in 2.12 until all flies are sedated.
Enter the total number of flies in each vial in the hard copy testing log. Censor immobile/dead flies at time 0 from the total number of flies.
Calculate the percent active flies at each assessment time-point and plot data as % active flies (y-axis) vs. time (x-axis). Quantitate ethanol sedation by interpolating Sedation Time 50 values (ST50, time to 50% sedation) from third-order polynomial or sigmoidal curve fits or calculating area under the curve.
Compile data from decoded vials and perform statistical analyses (e.g. one-way ANOVA with Bonferroni multiple comparison test) as appropriate for the experimental design.
Table 3.
Materials.
| Name of Material/ Equipment | Company | Catalog Number | Comments/Description |
|---|---|---|---|
| food vials | VWR | 89092-772 | narrow |
| Flugs | Genesee/flystuff.com | 49-102 | narrow |
| silicone stopper | Fisher Scientific | 09-704-1l | #4 |
| ethanol | Pharmaco-Aaper | 111000200 | 200 proof |
Table 1.
Typical testing log. See protocol step 2.5.
| vial → | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
| code → | ||||||||
| total # → | ||||||||
| 0 min | ||||||||
| 6 min | ||||||||
| 12 min | ||||||||
| 18 min | ||||||||
| 24 min | ||||||||
| 30 min | ||||||||
| 36 min | ||||||||
| 42 min | ||||||||
| 48 min | ||||||||
| 54 min | ||||||||
| 60 min |
Table 2.
Typical tapping and testing schedule. See protocol step 2.13.
| Vial | Tap | Assess |
|---|---|---|
| 1 | 6 min 0 sec | 6 min 30 sec |
| 2 | 6 min 5 sec | 6 min 35 sec |
| 3 | 6 min 10 sec | 6 min 40 sec |
| 4 | 6 min 15 sec | 6 min 45 sec |
Representative Results
The raw data from this ethanol sedation assay are the numbers of flies that are sedated as a function of ethanol vapor exposure time. Raw data are converted to the percent active flies as a function of time (primary data, Figures 1A, B, D-F). Sensitivity to ethanol sedation from the primary data can be quantitated as Sedation Time 50 (ST50), the time required for 50% of flies to become sedated or aea under the curve (AUC), via interpolation from curve fits. Previously reported18 third-order polynomial curves fit the primary data well as expected (average R2 = 0.934, n = 24, Figure 1A), although sigmoidal curves fit the primary data somewhat better (average R2 = 0.997, n = 24; Figure 1B). As an alternative to determining ST50 values from curve fits, ethanol sedation sensitivity from the primary data can also be quantitated as area under the curve (AUC, % active flies x time) (Figure 1C). The results from these three methods of quantitation correlate extremely well with each other (Pearson r2 ≥0.998, n = 24) and are essentially indistinguishable (Figure 1C), although the units are necessarily different for ST50 (min) and AUC (% active flies x time). Details regarding all calculations, spreadsheets used, etc. are available from the corresponding author. Note that higher and lower values of ST50 (or AUC) indicate blunted and enhanced sensitivity to ethanol sedation, respectively. For consistency with the previous report18, ST50 values derived from third-order polynomial curve fits were used throughout the remainder of this study.Importantly, the assay detects the effects of RNAi against Cnx14D (Figure 1D) and mutations in scb (Figure 1E), aru and hppy (Figure 1F), Clic and NPFR1 (data not shown) on ethanol sensitivity as reported in 18 and the articles originally describing the effects of these genetic manipulations 7,19-22.
During routine use of this assay, tapping of vials with cellulose acetate plugs wetted with 2 ml of ethanol caused the cellulose acetate plugs to travel toward the bottoms of some vials in a few individual experiments (not shown), thereby potentially changing the flies’ free space and the concentration of ethanol vapor. Additionally, approximately 0.25 ml of ethanol reached the bottom (i.e. fly side) of the cellulose acetate plugs when they were wetted with 2 ml of ethanol (Figure 2A), raising the possibility that flies in these assays were exposed to ethanol liquid in addition to ethanol vapor. Decreasing the volume of ethanol in the cellulose acetate plug from 2 ml to 1 ml eliminated the downward migration of the cellulose acetate plugs during the assay even after vigorous tapping (not shown) and also eliminated measureable amounts of ethanol solution on the bottoms of cellulose acetate plugs (Figure 2A). Sedation of flies was sensitive to the volume of ethanol used even with volumes less than 1 ml (Figure 1A-C). Using 1 ml of ethanol therefore helps maintain a constant open space for flies during the assay and ensures that liquid ethanol is not being ingested. Flies are therefore presumably exposed to ethanol solely as a vapor in this assay. One ml of 85% ethanol (i.e. vapor from 1 ml of 85% (vol/vol) ethanol) and 1-5 day old w[A] flies were used in all subsequent studies reported here unless indicated otherwise.
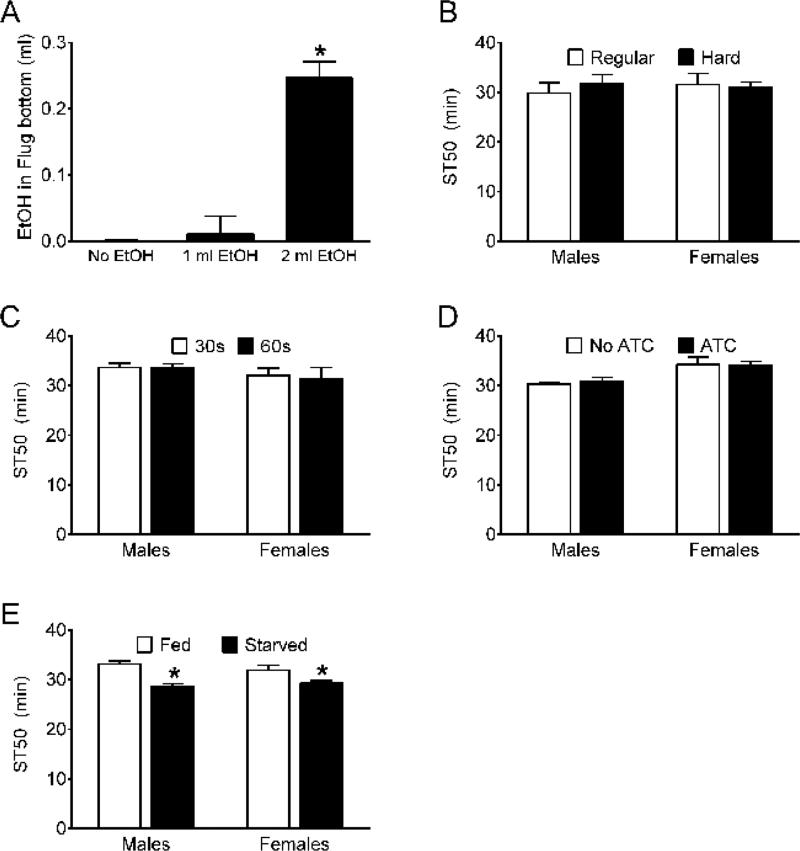
Figure 2. Assessment of operational parameters in the sedation assay.
(A) Amount of ethanol in the bottom 2 mm of cellulose acetate plugs. The volume of ethanol added to the tops of cellulose acetate plugs had a significant effect on the volume of ethanol that moved into the bottom 2 mm of cellulose acetate plugs during a mock 60 min experiment (one-way ANOVA, p <0.0001; *Bonferroni multiple comparison test, 2 ml vs 0 and 1 ml, p <0.05 n = 4). Ethanol was quantitated as a change in mass of the cellulose acetate plugs. (B) Neither vigor of tapping the vials during testing (Regular, Hard) nor sex of the flies tested affected ST50s (two-way ANOVA; effect of tapping, n.s.; effect of sex, n.s.; n = 12). (C) Neither recovery time after tapping nor sex had significant effects on ST50s (two-way ANOVA; effect of recovery time, n.s.; effect of sex, n.s.; n = 6). (D) Inclusion of antibiotics (ATC; ampicillin, tetracycline and chloramphenicol) in the growth medium had no overall effect on ST50s, but there was an effect of sex on ST50 (two-way ANOVA; effect of ATC, n.s.; effect of sex, p = 0.001; interaction, n.s.; n = 6). (E) Effect of starvation on ethanol sedation sensitivity. ST50s were significantly lower in flies deprived of food and water for 6 hr (Starved) compared to normally fed (Fed) flies; ST50s in males and females were indistinguishable overall (two-way ANOVA; effect of starvation, p <0.0001; effect of sex, n.s.; n = 12; *Bonferroni multiple comparison tests, effect of starvation in males and females, p <0.05).
In experiments with 24 vials, flies are tapped gently at 6 min intervals to allow the experimenter to assess (i.e. cycle through) all vials on a reasonable, prescribed schedule. ST50s from studies with control flies using fewer vials and 5 min intervals were comparable to ST50s from studies with 6 min intervals (data not shown). Flies are assessed for sedation 30 sec after tapping to allow the experimenter time to tap a set of four vials in sequence prior to determining the sedation status of the flies in each vial. To determine if differences in tapping vigor or recovery time after tapping impact ethanol sedation sensitivity, ST50 values in male and female control flies were measured while being tapped normally (Regular) or very sharply (Hard) and after a standard 30 sec or longer 60 sec recovery. Tapping vigor (Figure 2B) and recovery time (Figure 2C) had no measurable effect on ST50 values in control males or females.
Antibiotics can be used to suppress bacterial growth in fly food medium. To determine if supplementation of the food medium with antibiotics affects ethanol sedation sensitivity, ST50 values were measured in flies reared for one generation in the presence or absence of three antibiotics (ampicillin, 100 μg/ml; tetracycline, 20 μg/ml; chloramphenicol, 125 μg/ml). Rearing flies on these three antibiotics had no effect on ethanol sedation sensitivity (Figure 2D).
Flies are transferred to empty food vials—and are therefore deprived of food and water for ~5 min—immediately prior to being exposed to ethanol vapor in the sedation assay. To determine if the amount of time flies are kept in empty food vials before initiating the assay impacts ethanol sedation sensitivity, flies were starved for 6 hr (at least 70-fold of normal) and then ST50 values were measured. Starvation for 6 hr significantly decreased ST50s in both male and female control flies (Figure 2E). Although the effect of starvation on ST50 was relatively small (9-14% in these experiments), the amount of time flies spend in empty vials prior to being exposed to ethanol vapor in the assay should be held uniform across groups within an experiment and also between experiments.
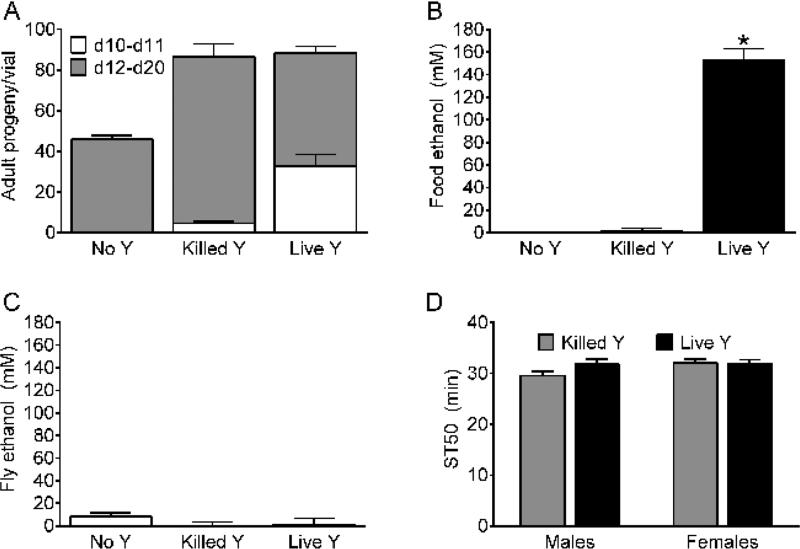
Laboratory fly food can be supplemented with live yeast (e.g. 18,20,23,24) to promote production of progeny. For example, food vials supplemented with live yeast (Saccharomyces cerevisiae) produced adults earlier than those with heat-killed yeast or no live yeast (compare d10-d11, white bars, Figure 3A), although the total number of progeny produced over 20 days was indistinguishable in vials supplemented with live and heat-killed yeast (Figure 3A). To address the possibility that live yeast produce meaningful amounts of ethanol on fly food, the effects of live yeast on (i) ethanol in the food medium, (ii) ethanol in flies, and (iii) ethanol sedation sensitivity in flies were measured. Food medium supplemented with live yeast contained substantial amounts of ethanol compared to food supplemented with heat-killed or no yeast (Figure 3B). Nevertheless, control flies grown on medium supplemented with live, killed or no yeast had indistinguishably low concentrations of internal ethanol (Figure 3C). Furthermore, ethanol sedation sensitivity was indistinguishable in control flies grown on medium supplemented with heat-killed or live yeast (Figure 3D).
Figure 3. Effect of live yeast on growth, ethanol content and sedation sensitivity.
(A) Adult progeny from vials containing food medium supplemented with no yeast (No Y), heat killed yeast (Killed Y) or live yeast (Live Y). White bars, adult progeny emerging during days 10-11; grey bars, days 12-20. Overall, yeast treatment had a significant effect on progeny production (one-way ANOVAs; all (10-20) days, p <0.0001; days 10-11, p <0.0001; days 12-20, p = 0.0002; n = 5). Total number of progeny produced during all (10-20) days was greater from vials with Killed Y and Live Y than No Y (Bonferroni multiple comparison test, p<0.05). During days 10-11, vials with Live Y produced more progeny than Killed Y and vials with Killed Y produced more progeny than No Y (white bars, Bonferroni multiple comparison tests, p<0.05). During days 12-20, vials with Killed Y produced more progeny than vials with No Y or Live Y (grey bars, Bonferroni multiple comparison test, p <0.05). (B) Ethanol in the food medium was significantly higher in vials supplemented with Live Y compared to vials with No Y and Killed Y (one-way ANOVA, p <0.0001, n = 5; Bonferroni multiple comparison test, p <0.05). (C) Internal ethanol in w[A] female flies was not significantly affected by supplementation with yeast (one-way ANOVA, n.s., n = 5-10). Ethanol content in B and C determined as described18. (D) ST50 values were not significantly affected by the supplementation of the growth medium with yeast in male or female w[A] (two-way ANOVA; effect of yeast, n.s.; effect of sex, n.s.; n = 12).
Discussion
Straightforward assays that reproducibly quantitate meaningful phenotypes are of great value for the analysis of behavior. The work described here addresses several practical aspects of an assay for measuring ethanol sedation sensitivity and rapid tolerance in Drosophila. Although not a focus of this work, behavioral analyses are facilitated by maintaining the environment and genetic background constant for test subjects within a study. Furthermore, comparisons should typically be made between groups of flies reared and tested side-by-side. To this end, all flies within individual experiments in this work had the same genetic background and were reared side by side under uniform environmental conditions (25 °C, 60% relative humidity, 12 hr light/dark cycle) with standard food medium (10% sucrose, 3.3% cornmeal, 2% yeast and 1% agar) except for those experiments in which the food medium was explicitly manipulated.
The ethanol sedation assay described here requires very few materials (plastic vials, cellulose acetate plugs, silicone stoppers and ethanol), all of which are inexpensive and readily available from commercial sources (Table 3). This ethanol sedation assay is similar to and to a large extent based on previously reported methods (e.g. 6,20,25-27, comprehensively reviewed in 3,28). Although all of these assays have utility for measuring ethanol sedation sensitivity, advantages of the assay described here include (i) flies are not exposed to liquid ethanol during the assay, (ii) the reduced possibility that flies can become entangled in cotton or other material in the testing vial during the assay, (iii) the ability to test many groups of flies in parallel, and (iv) three options for the objective quantification of ethanol sensitivity from primary data sets. Together, these advantages largely eliminate the possibility that flies drink ethanol, that ethanol could wet the external surfaces of flies thereby inhibiting their overall locomotor abilities, and that performance of flies in the assay could be confounded by difficulties associated with entangling material in the testing environment. Additionally, these advantages increase the overall throughput and reproducibility of the assay. Furthermore, ethanol sensitivity measured in this assay is not influenced by expression of endogenous white or the transgenic marker mini-white18, making it suitable for studies with transgenes and genetic backgrounds commonly used in Drosophila genetic analyses.
Several key parameters should be controlled to obtain reliable results from ethanol sedation assays. In addition to controlling the growth environment and genetic background of strains being tested (see above), the volume and concentration of ethanol are of course extremely important to the quantitation of ethanol sensitivity. Note that the dilution of ethanol in water is exothermic. Consequently, diluted ethanol solutions should be allowed to equilibrate to room temperature prior to the initiation of sedation assays. An additional parameter that should be made uniform across the groups being tested is the length of time that flies are housed in empty food vials prior to starting the sedation assay. Two additional parameters should also be controlled. The number of flies per vial should ideally be (a) the same in all vials and groups, (b) a number that can be easily counted quickly, and (c) a number large enough to allow for relatively smooth sedation time-courses. Eleven flies/ vial works well in our laboratory and is a suggested starting point. A final parameter to consider is the age of the flies used in sedation assays. Although no reproducible effects of age on ethanol sedation sensitivity in 1-10 day old flies has been found (data not shown), the use of young age-matched animals seems justified given the large literature on age-related behavioral changes in flies 29,30.
Other parameters do not seem as critical in sedation assays, at least when testing control flies using the ranges of parameters described here. Sex, strength of tapping, recovery time from tapping, and supplementation of fly food with antibiotics (ampicillin, tetracycline and chloramphenicol) or live yeast do not alter ethanol sensitivity measured as described here. Similarly, quantitating ethanol sensitivity by interpolation from third-order polynomial curves, interpolation from sigmoidal curves, or determination of AUC from the primary sedation time-course data leads to essentially indistinguishable interpretations. Although no effect of sex was consistently observed in the studies described here, effects of sex on ethanol sensitivity in several Drosophila genetic backgrounds have been reported 31. Thus, it is possible that sex effects were simply masked by genetic variance in the w[A] background used here. Alternatively, it is possible that measuring sex effects on behavioral responses in Drosophila require as yet unidentified assay or growth conditions. In any case, the recommendation is to make all parameters as uniform as possible in sedation assays, including the sex of any groups being explicitly compared.
Given that live yeast produce sizeable amounts of ethanol in fly food, it was somewhat surprising that supplementation of food with live yeast did not increase internal ethanol in flies. One possible explanation is that the ethanol is not produced uniformly across the surface of the food and flies might therefore selectively ingest food with no or relatively low concentrations of ethanol. Additional studies will be required to address this and other possible explanations for this finding.
A large number of modifications to the assay described here are possible depending on the goals of the experiment being performed. For example, the number of vials tested in an experiment, the number of flies tested in each vial, the concentration and volume of ethanol used, the duration of ethanol exposure, the time interval between sedation assessments, the age and sex of the flies tested, and the criteria for sedation can all be modified to suit the needs of an individual laboratory. The recommendation is to start with one or two groups of four vials when initially learning the assay and then scale up to using the number of vials that provides the throughput required for the project. Should unexpectedly short or long ST50s be observed, it would be good practice to ensure that test flies were subjected to short (1-5 min) anesthesia times, were allowed to recover from anesthesia overnight, were less than 10 days old, were not physically damaged during handling, were not wetted by ethanol solution, and did not have locomotor impairments.
The assay measures ethanol sedation sensitivity only and therefore is not designed or suited for assessing other behavioral responses to ethanol. Like all known ethanol behavioral paradigms in flies, this assay requires flies to have largely normal locomotor abilities and therefore genotypes with impaired locomotion should not be tested. Another limitation of the assay is that the concentration of ethanol vapor in the vial is presumably rising continuously as the drug volatilizes from the cellulose acetate plug. Although the flies’ internal ethanol rises progressively with time in all fly ethanol behavioral assays, it seems possible that delivering a fixed concentration of ethanol vapor to flies could improve the consistency of results from this assay. A method for delivering a fixed concentration of ethanol vapor has not yet been identified that would allow the assay to be used at the scale described here.
The ethanol sedation assay described here is well-suited for genetic analysis of ethanol sensitivity and rapid tolerance 18. Growth environment, genetic background, the concentration and volume of ethanol, the amount of time flies spend in empty food vials, and the number and age of flies used should all be controlled. Assuming these parameters are adequately controlled, the sedation assay should be suitable for both reverse and forward genetic approaches that investigate the molecular basis for behavioral responses to ethanol in Drosophila.
Acknowledgements
These studies were supported by grants from the National Institutes of Health, National Institute for Alcoholism and Alcohol Abuse to M.G. (P20AA017828, R01AA020634, P50 AA022537). The authors thank Jill Bettinger for helpful discussions and Jacqueline DeLoyht for technical assistance.
Footnotes
Citation: Sandhu, S., Kollah, A., Chan, R., Lewellyn, L., Grotewiel, M. An Inexpensive, Scalable Behavioral Assay for Measuring Ethanol Sedation Sensitivity and Rapid Tolerance in Drosophila. J. Vis. Exp. (), e52676, doi:10.3791/52676 (2015).
Video Link
The video component of this article can be found at http://www.jove.com/video/52676/
Disclosures
The authors have nothing to disclose.
References
- 1.Rehm J, et al. Global burden of disease and injury and economic cost attributable to alcohol use and alcohol-use disorders. Lancet. 2009;373:2223–2233. doi: 10.1016/S0140-6736(09)60746-7. doi:10.1016/S0140-6736(09)60746-7. [DOI] [PubMed] [Google Scholar]
- 2.Prescott CA, Kendler KS. Genetic and environmental contributions to alcohol abuse and dependence in a population-based sample of male twins. The American journal of psychiatry. 1999;156:34–40. doi: 10.1176/ajp.156.1.34. [DOI] [PubMed] [Google Scholar]
- 3.Devineni AV, Heberlein U. The evolution of Drosophila melanogaster as a model for alcohol research. Annual review of neuroscience. 2013;36:121–138. doi: 10.1146/annurev-neuro-062012-170256. doi:10.1146/annurev-neuro-062012-170256. [DOI] [PubMed] [Google Scholar]
- 4.Scholz H, Mustard JA. Invertebrate Models of Alcoholism. Current topics in behavioral neurosciences. 2011;13:433–457. doi: 10.1007/7854_2011_128. doi:doi: 10.1007/7854_2011_128. [DOI] [PubMed] [Google Scholar]
- 5.Rodan AR, Rothenfluh A. In: Functional Plasticity and Genetic Variation: Insights into the Neurobiology of Alcoholism. Reilly MT, Lovinger DM, editors. Vol. 91. Elsevier; 2010. [DOI] [PubMed] [Google Scholar]
- 6.Schumann G, et al. Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7119–7124. doi: 10.1073/pnas.1017288108. doi:10.1073/pnas.1017288108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corl AB, et al. Happyhour, a Ste20 family kinase, implicates EGFR signaling in ethanol-induced behaviors. Cell. 2009;137:949–960. doi: 10.1016/j.cell.2009.03.020. doi: 10.1016/j.cell.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 8.Moore MS, et al. Ethanol intoxication in Drosophila: Genetic and pharmacological evidence for regulation by the cAMP signaling pathway. Cell. 1998;93:997–1007. doi: 10.1016/s0092-8674(00)81205-2. [DOI] [PubMed] [Google Scholar]
- 9.Scholz H, Franz M, Heberlein U. The hangover gene defines a stress pathway required for ethanol tolerance development. Nature. 2005;436:845–847. doi: 10.1038/nature03864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riley BP, et al. Alcohol dependence is associated with the ZNF699 gene, a human locus related to Drosophila hangover, in the Irish affected sib pair study of alcohol dependence (IASPSAD) sample. Molecular psychiatry. 2006;11:1025–1031. doi: 10.1038/sj.mp.4001891. [DOI] [PubMed] [Google Scholar]
- 11.Morozova TV, et al. Alcohol sensitivity in Drosophila: translational potential of systems genetics. Genetics. 2009;183:733–745. 731SI–712SI. doi: 10.1534/genetics.109.107490. doi:10.1534/genetics.109.107490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ogueta M, Cibik O, Eltrop R, Schneider A, Scholz H. The influence of Adh function on ethanol preference and tolerance in adult Drosophila melanogaster. Chemical senses. 2010;35:813–822. doi: 10.1093/chemse/bjq084. doi:10.1093/chemse/bjq084. [DOI] [PubMed] [Google Scholar]
- 13.Han S, et al. Integrating GWASs and human protein interaction networks identifies a gene subnetwork underlying alcohol dependence. American journal of human genetics. 2013;93:1027–1034. doi: 10.1016/j.ajhg.2013.10.021. doi:10.1016/j.ajhg.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lind PA, et al. A genomewide association study of nicotine and alcohol dependence in Australian and Dutch populations. Twin Res Hum Genet. 2010;13:10–29. doi: 10.1375/twin.13.1.10. doi:10.1375/twin.13.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schuckit MA. Low level of response to alcohol as a predictor of future alcoholism. The American journal of psychiatry. 1994;151:184–189. doi: 10.1176/ajp.151.2.184. [DOI] [PubMed] [Google Scholar]
- 16.Schuckit MA, Smith TL. An 8-year follow-up of 450 sons of alcoholic and control subjects. Archives of general psychiatry. 1996;53:202–210. doi: 10.1001/archpsyc.1996.01830030020005. [DOI] [PubMed] [Google Scholar]
- 17.Tabakoff B, Cornell N, Hoffman PL. Alcohol tolerance. Ann Emerg Med. 1986;15:1005–1012. doi: 10.1016/s0196-0644(86)80119-6. [DOI] [PubMed] [Google Scholar]
- 18.Chan RF, et al. Contrasting Influences of Drosophila white/mini-white on Ethanol Sensitivity in Two Different Behavioral Assays. Alcohol Clin Exp Res. 2014;38:1582–1593. doi: 10.1111/acer.12421. doi:10.1111/acer.12421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eddison M, et al. arouser reveals a role for synapse number in the regulation of ethanol sensitivity. Neuron. 2011;70:979–990. doi: 10.1016/j.neuron.2011.03.030. [DOI] [PubMed] [Google Scholar]
- 20.Wen T, Parrish CA, Xu D, Wu Q, Shen P. Drosophila neuropeptide F and its receptor, NPFR1, define a signaling pathway that acutely modulates alcohol sensitivity. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:2141–2146. doi: 10.1073/pnas.0406814102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhandari P, Kendler KS, Bettinger JC, Davies AG, Grotewiel M. An assay for evoked locomotor behavior in Drosophila reveals a role for integrins in ethanol sensitivity and rapid ethanol tolerance. Alcohol Clin Exp Res. 2009;33:1794–1805. doi: 10.1111/j.1530-0277.2009.01018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhandari P, et al. Chloride intracellular channels modulate acute ethanol behaviors in Drosophila, Caenorhabditis elegans and mice. Genes, brain, and behavior. 2012;11:387–397. doi: 10.1111/j.1601-183X.2012.00765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gargano JW, Martin I, Bhandari P, Grotewiel MS. Rapid iterative negative geotaxis (RING): a new method for assessing age-related locomotor decline in Drosophila. Experimental gerontology. 2005;40:386–395. doi: 10.1016/j.exger.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 24.Chen J, Wang Y, Zhang Y, Shen P. Mutations in Bacchus reveal a tyramine-dependent nuclear regulator for acute ethanol sensitivity in Drosophila. Neuropharmacology. 2013;67:25–31. doi: 10.1016/j.neuropharm.2012.10.013. doi:10.1016/j.neuropharm.2012. [DOI] [PubMed] [Google Scholar]
- 25.Lasek AW, Giorgetti F, Berger KH, Tayor S, Heberlein U. Lmo genes regulate behavioral responses to ethanol in Drosophila melanogaster and the mouse. Alcohol Clin Exp Res. 2011;35:1600–1606. doi: 10.1111/j.1530-0277.2011.01506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maples T, Rothenfluh A. A simple way to measure ethanol sensitivity in flies. J Vis Exp. 2011:e2541. doi: 10.3791/2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rothenfluh A, et al. Distinct behavioral responses to ethanol are regulated by alternate RhoGAP18B isoforms. Cell. 2006;127:199–211. doi: 10.1016/j.cell.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 28.Rothenfluh A, Troutwine BR, Ghezzi A, Atkinson NS. In: Neurobiology of Alcohol Dependence. Noronha A, editor. Vol. 23. Elsevier; 2014. pp. 467–495. [Google Scholar]
- 29.Jones MA, Grotewiel M. Drosophila as a model for age-related impairment in locomotor and other behaviors. Experimental gerontology. 2010;46:320–325. doi: 10.1016/j.exger.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin I, Grotewiel MS. Oxidative damage and age-related functional declines. Mechanisms of ageing and development. 2006;127:411–423. doi: 10.1016/j.mad.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 31.Devineni AV, Heberlein U. Acute ethanol responses in Drosophila are sexually dimorphic. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:21087–21092. doi: 10.1073/pnas.1218850110. [DOI] [PMC free article] [PubMed] [Google Scholar]