Summary
The mechanisms by which cells destabilize and rapidly disassemble filamentous actin networks have remained elusive; however, Coronin, Cofilin, and AIP1 have been implicated in this process. Here, using multi-wavelength single molecule fluorescence imaging, we show that mammalian Cor1B, Cof1, and AIP1 work in concert through a temporally ordered pathway to induce highly efficient severing and disassembly of actin filaments. Cor1B binds to filaments first, and dramatically accelerates the subsequent binding of Cof1, leading to heavily decorated, stabilized filaments. Cof1 in turn recruits AIP1, which rapidly triggers severing and remains bound to the newly-generated barbed ends. New growth at barbed ends generated by severing was blocked specifically in the presence of all three proteins. This activity enabled us to reconstitute and directly visualize single actin filaments being rapidly polymerized by formins at their barbed ends while simultanteously being stochastically severed and capped along their lengths, and disassembled from their pointed ends.
Keywords: actin, single molecule analysis, Cofilin, Coronin, AIP1
Introduction
Actin is a highly abundant cytosolic protein and polymerizes to form dynamic filamentous networks that drive many biological processes, including cell morphogenesis, cell motility, endocytosis, and intracellular transport. Dynamic rearrangements of the actin cytoskeleton are achieved through controlled nucleation and elongation of filaments from a finite pool of ATP-actin monomers. This is counterbalanced by rapid destabilization and disassembly of aged (ADP) filaments, which serves to both replenish the actin monomer pool and sculpt network geometry. We now have a mature understanding of how actin filament arrays are initially formed in cells, revolutionized by the elucidation of the molecular mechanisms underlying actin filament nucleation and elongation1, 2. In contrast, our understanding of how these networks are disassembled remains highly incomplete. The only well understood step in the disassembly process is filament severing. In a wide range of organisms this step is mediated by ADF/Cofilin (hereafter referred to as Cofilin), which binds cooperatively to the sides of ADP-actin filaments and induces structural changes, leading to breaks between the Cofilin-decorated and undecorated regions 3, 4, 5, 6, 7. Severing amplifies the number of filament ends from which subunit dissociation can occur, thus reducing the time to full depolymerization.
Despite these advances in our knowledge of how Cofilin severs filaments, a number of critical questions have remained unanswered. First, Cofilin alone binds surprisingly slowly to ADP-actin filaments (kon =0.013 μM−1s−1), suggesting that additional cellular factors may be required to enhance Cofilin recruitment to F-actin for inducing rapid actin disassembly 8, 9. Second, even though Cofilin is present at concentrations of 5–20 μM in mammalian cells 10, 11, 12, maximal severing by Cofilin in vitro was observed at nanomolar concentrations, and micromolar concentrations of Cofilin perplexingly led to overdecoration and stabilization of filaments 8, 13. Third, it has been unclear how actin polymerization is prevented at the new barbed ends of filaments generated by severing. Cells maintain high levels of ATP-actin monomers, approximately two orders of magnitude above the critical concentration for barbed end assembly, and thus cytosolic conditions strongly favor actin assembly over disassembly 14, 15. As a consequence, severing without simultaneous capping of barbed ends will result in net growth rather than disassembly 8, 10. Together, these observations suggest that additional cellular factors must work in concert with Cofilin to achieve highly efficient severing and disassembly in vivo.
Mounting genetic and biochemical evidence has implicated three proteins (Srv2/CAP, AIP1, and Coronin) in functioning with Cofilin to promote actin disassembly in vivo. Yeast and mammalian homologues of Srv2/CAP facilitate Cofilin-mediated actin disassembly by enhancing filament severing 4–8 fold 16, 17, 18, 19, 20. However, the roles of AIP1 and Coronin are less well understood. AIP1 binds to F-actin and Cofilin, but it has remained controversial whether AIP1 enhances Cofilin-mediated severing and/or caps the barbed ends of filaments after severing 21, 22, 23, 24, 25. The role of Coronin has been even more enigmatic, with ostensibly conflicting genetic and biochemical observations. While genetic data strongly support a role for Coronin in promoting actin turnover, purified yeast and mammalian Coronins both inhibit rather than enhance Cofilin-mediated severing in vitro 26, 27, 28, 29. Thus, the functions and mechanisms of AIP1 and Coronin in regulating actin disassembly have remained poorly understood.
One of the important advances in our understanding of actin disassembly mechanisms came from the recent biochemical work of Brieher and colleagues, who isolated Cofilin, AIP1, and Coronin from cell extracts as factors that together induce the disassembly of Listeria actin tails or purified actin filaments even under assembly-promoting conditions 30, 31. These studies linked together for the first time the functions of AIP1 and Coronin in Cofilin-mediated actin disassembly. Here, we have employed multi-wavelength total internal reflection fluorescence (TIRF) microscopy to directly observe Cofilin, AIP1, and Coronin during actin filament severing and disassembly, and thus define better the roles of each protein and their collective mechanism for inducing rapid actin disassembly. Our study reveals that these three proteins work in a temporally ordered fashion to rapidly disassemble F-actin. This process is initiated by binding of Coronin to filaments, which in turn greatly enhances Cofilin recruitment to filaments sides. Last to arrive is AIP1, which invariably triggers severing. After inducing severing, AIP1 remains bound to the newly generated barbed ends, and together with Cofilin and Coronin blocks new growth, thus enabling filament disassembly even under assembly-promoting conditions.
Results
Rapid actin filament severing by a three-component mixture
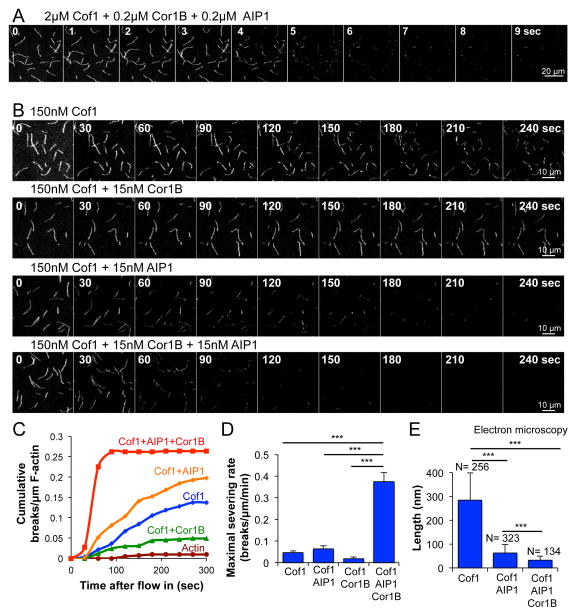
To investigate how mammalian Cofilin-1 (referred to herein as Cof1), AIP1, and Coronin-1B (referred to herein as Cor1B) work together to regulate actin filament disassembly, we first compared their individual and combined effects on severing of surface-tethered Oregon-green (OG) labeled filaments in TIRF assays. Using low micromolar concentrations of Cof1 combined with 10-fold lower concentrations of Cor1B and AIP1 (approximating the ratio found in cells; 29, 30, 32, we observed complete severing and disassembly of filaments (10–15 μm in length) only 5 sec after flow-in (Fig 1A; Supplementary Movie 1). This precluded quantitative measurement of severing rates; therefore we reduced the concentrations of Cof1, AIP1, and Cor1B while maintaining the 10:1:1 ratio (150 nM, 15 nM, and 15 nM, respectively), enabling differences in severing rates to be quantified. Under these conditions (Fig 1B and 1C; Supplementary Movie 2; also see Supplementary Figure 1), Cof1 alone induced an average of 2–3 severing events per filament during the 300 sec observation window. Further addition of AIP1 led to a modest but significant (p < 0.05, one-way ANOVA) increase in severing rate over the 300 sec observation period (1.9 ± 0.2 × 10−4 breaks/μm/sec for Cof1 vs. 3.4 ± 0.45 × 10−4 breaks/μm/sec for Cof1+AIP1, n=3). In contrast, Cor1B strongly and significantly (p < 0.05, one-way ANOVA) inhibited severing by Cof1 (0.5 ± 0.1 × 10−4 breaks/μm/sec, n=3), consistent with its previously reported inhibitory effects in bulk assays 26, 28. In the absence of Cof1, AIP1 and Cor1B each showed no severing activity (Supplementary Figure 1). The most striking effects though were observed in the combined presence of all three proteins (Cof1, AIP1 and Cor1B), where greatly enhanced severing was observed at all time points (Fig 1C), and the maximal severing rate (measured between 30 and 60 sec after flow-in) was 10-fold higher for the three-component mixture compared to Cof1 alone (Fig 1D).
Figure 1. High rates of actin filament severing in the combined presence of mammalian Cof1, Cor1B and AIP1.
(A) Time points from TIRF microscopy (Supplementary Movie 1) of pre-polymerized OG-labeled actin filaments after flowing in a mixture of the indicated concentrations of Cof1, Cor1B, and AIP1. (B) Time points from TIRF microscopy (Supplementary Movie 2) as above, except flowing in lower concentrations (as indicated) of Cof1, Cor1B, and/or AIP1. (C) Analysis of filament severing kinetics from TIRF assays performed as above. Each data point is the cumulative number of severing events per μm of filament at that time point, averaged for at least 60 filaments pooled from 3 independent trials of at least 20 filaments each. (D) Maximal rates of severing for each condition were determined by averaging the slopes of curves from three independent trials in the time interval from 30–60 sec. Slope measurements at later time points confirmed that these were the maximal rates during the 300 sec observation window. Error bars, SD. (E) Electron microscopy of actin filament severing products. F-actin (2 μM) was incubated with different combinations of 2 μM Cof1, 0.2 μM Cor1B, and 0.2 μM AIP1, then imaged by negative stain electron microscopy (example images in Supplementary Figure 2). Average filament length (± SD) is graphed with the number of filaments analyzed (N) above each bar. Statistical significance and p-values for (D) and (E) were determined by ANOVA and Tukey’s multiple comparison test; ***, p < 0.05.
TIRF microscopy only provides limited information about the size of F-actin severing products, since many of them are below the resolution of light microscopy, and some of the untethered fragments diffuse away. Therefore, we incubated filaments for 10 min with Cof1 alone or the combination of Cof1, AIP1 and Cor1B, and examined the severing products by electron microscopy (Fig 1E; also see Supplementary Figure 2). Filaments incubated with Cof1 alone had an average length of 284 ± 115 nm (n= 256), compared to control filaments of 6184 ± 533 nm (n= 19). Strikingly, filaments incubated with Cof1 and AIP1 had an average length of only 54 ± 18 nm or ≈ 20 actin subunits (n= 323). Filaments incubated with Cof1, AIP1, and Cor1B were even shorter, with an average length of 32 ± 18 nm or ≈ 12 actin subunits (n= 134). Taken together with our TIRF analysis, these data show that while AIP1 alone only modestly increases the initial rate of Cof1-mediated severing (Fig 1C and D), it dramatically reduces the average size of the severing end products after prolonged incubation. In contrast, the contribution of Cor1B appears to be primarily to increase the initial rate of severing by Cof1 and AIP1 (Fig 1C and D), while it only modestly affects the average size of the severing end products (Fig 1E).
Coronin accelerates Cofilin binding to actin filaments
Next, we fluorescently labeled Cof1, AIP1, and Cor1B, so that we could observe each of them in real time during actin filament disassembly by multi-wavelength TIRF microscopy. For Cof1, we reengineered its surface residues to leave only one exposed cysteine at a position where Cy3-maleimide labeling did not interfere with function (Supplementary Figure 3). For AIP1 and Cor1B, SNAP tags were introduced at their C-termini, and the resulting fusion proteins were directly labeled with benzyl guanine-conjugated dyes in the far-red spectrum (see Methods and Supplementary Figure 3). The resulting proteins, Cy3-Cof1, Cor1B-SNAP649, and AIP1-SNAP647, had actin disassembly activities similar to unlabeled counterparts (Supplementary Figure 3b–d).
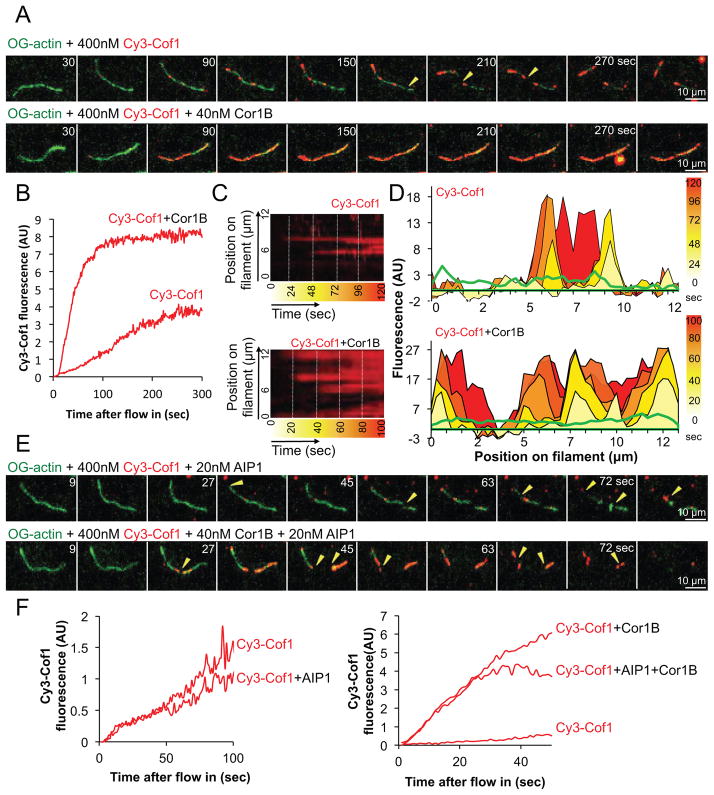
We first examined the kinetics of Cy3-Cof1 binding to OG-actin filaments. Cy3-Cof1 associated slowly with filaments, accumulating in patches that gradually increased in brightness, consistent with a cooperative binding mechanism (Fig 2A and 2B; Supplementary Movie 3). Further, Cy3-Cof1 decoration led to filament severing, with breaks occurring between the decorated and undecorated regions (Fig 2A, yellow arrows). These results using mammalian Cof1 are consistent with those reported for yeast Cof1 7, suggesting that the fundamental mechanism by which Cofilin interacts with and severs filaments is conserved between yeast and mammals.
Figure 2. Cor1B dramatically increases binding of Cy3-Cof1 to actin filaments.
(A) Time points from TIRF movies comparing binding of Cy3-Cof1, in the presence and absence of Cor1B, to preassembled OG-labeled actin filaments. Severing events are indicated by yellow arrow heads. (B) Kinetics of total Cy3-Cof1 fluorescence accumulation on actin filaments in the presence and the absence of Cor1B. The traces are averages from 3 independent trials (analyzing 10–15 filaments each). (C) Kymographs each showing Cy3-Cof1 decoration of a single actin filament in the presence or absence of Cor1B. (D) Spatiotemporal profiles of Cy3-Cof1 distribution along the same filaments as in (C), with time points in sec as color-coded in the heat-bars at the far right. (E) Effect of AIP1 on Cy3-Cof1 binding to actin filaments in the presence of Cor1B. Due to enhanced severing in the presence of AIP1, Cy3-Cof1 binding could only be monitored for 60–90 sec after flow in. Severing events are indicated by yellow arrow heads. (F) Kinetics of Cy3-Cof1 fluorescence accumulation on actin filaments in the presence of AIP1, with or without Cor1B. Traces are averages from 3 independent trials (analyzing 10–15 filaments each).
Addition of unlabeled Cor1B in TIRF reactions led to a dramatic increase in the rate of Cy3-Cof1 binding to actin filaments, and produced heavily decorated, hyper-stabilized filaments (Fig 2A and 2B; Supplementary Movie 3). Kymographs revealed an increase in the Cy3-Cof1 spot density on filaments in the presence of Cor1B relative to that observed in the absence of Cor1B (Fig 2C and D). Each Cy3-Cof1 patch followed a similar pattern, starting as a small dot and gradually increasing in intensity and often merging with other patches. These results show that Cor1B markedly accelerates Cy3-Cof1 binding to filaments. By comparison, AIP1 had little if any effect on the kinetics of Cy3-Cof1 binding to filaments (Fig 2F; Supplementary Movie 3). However, AIP1 had a profound effect on severing in the combined presence of Cor1B and Cy3-Cof1, inducing rapid fragmentation of filaments, including those heavily decorated by Cy3-Cof1 (Fig 2E and F). Quantification of Cy3-Cof1 recruitment to filaments in the presence of AIP1 and Cor1B showed that AIP1 does not alter the kinetics of Cy3-Cof1 recruitment by Cor1B (Fig 2F; Supplementary Movie 3).
Taken together, these observations suggest the following: 1) Cof1 binding to actin filaments is one of the rate-limiting steps in severing, 2) Cof1 binding is accelerated greatly by Cor1B, and 3) the presence of AIP1 strongly enhances severing after Cof1 decoration.
Coronin binds rapidly to actin filaments preceding Cofilin
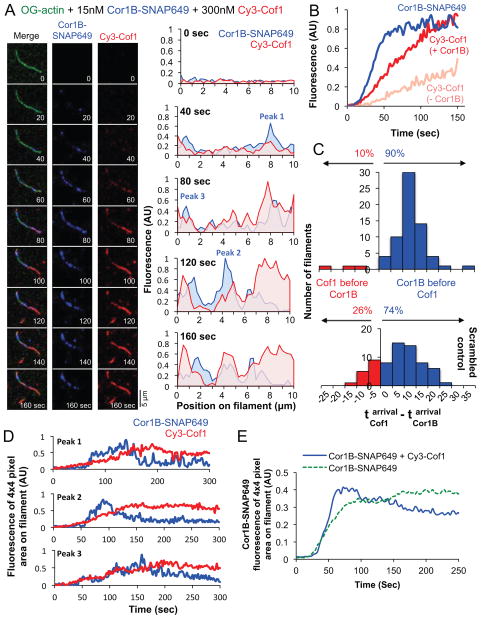
To better understand the spatiotemporal relationship between Cor1B and Cof1 binding to filaments, we next performed three-color TIRF microscopy in order to simultaneously observe binding of Cy3-Cof1 and Cor1B-SNAP649 to OG-actin filaments (Fig 3A; Supplementary Movie 4). Cor1B-SNAP649 appeared on filaments rapidly, even faster than the appearance of Cy3-Cof1 accelerated by the presence of Cor1B-SNAP649 (Fig 3B). Further, at the first observed binding location on a filament, Cor1B-SNAP649 almost always arrived ahead of Cy3-Cof1 (90% of the time; blue bars in upper panel Fig 3C). As shown by a randomized control analysis (lower panel, Fig 3C), this behavior was not merely a coincidence, caused by faster appearance of Cor1B-SNAP649 on filaments. Instead, the data show that there was a tendency for Cy3-Cof1 to bind where Cor1B-SNAP649 was already bound. Further, we often observed that following Cy3-Cof1 recruitment, the Cor1B-SNAP649 signal at these sites would decline, suggesting that Cof1 might be able to displace Cor1B. Analysis of randomly chosen individual Cor1B-SNAP649 spots on filaments, showed that in the absence of Cy3-Cof1, the Cor1B-SNAP649 average fluorescence peaked and remained constant (Fig 3E; Supplementary Figure 4a and c). However, in the presence of Cy3-Cof1, the Cor1B-SNAP649 average fluorescence peaked and then slowly decreased, suggesting that accumulation of Cof1 may displace Cor1B-SNAP649 from these sites (Fig 3D and E; Supplementary Figure 4b). Altogether, these results suggest a preferentially ordered pathway, in which Cor1B typically binds to filaments first and then recruits Cof1 to the same sites, providing a means by which the otherwise slow association of Cof1 with filaments is greatly accelerated.
Figure 3. Simultaneous imaging of Cor1B-SNAP649 and Cy3-Cof1 binding to actin filaments.
(A) Time points from a triple-color TIRF movie (Supplementary Movie 4) showing binding of Cor1B-SNAP649 and Cy3-Cof1 to preassembled OG-labeled actin filaments after flow-in (time 0). Graphs show spatiotemporal profiles of Cy3-Cof1 and Cor1B-SNAP649 distributions along the same filament at different time points. (B) Kinetics of Cor1B-SNAP649 and Cy3-Cof1 fluorescence accumulation on the same set of actin filaments (blue and red traces). Traces are averages from 3 independent trials (analyzing 10–15 filaments each). For comparison, the same analysis was performed for Cy3-Cof1 in the absence of Cor1B (faded red trace). (C) Distribution of time intervals between first appearance of Cor1B-SNAP649 and first appearance of Cy3-Cof1 on actin filaments (top panel). Data are from 3 independent trials (>20 filaments each). A control analysis in which randomly generated values were substituted in place of the observed times of Cor1B-SNAP649 binding yielded a similar distribution (bottom panel) indicating that Cy3-Cof1 had a high tendency to bind where Cor1B-SNAP649 was already bound. (D) Intensity profiles of Cor1B-SNAP649 and Cy3-Cof1 fluorescence of peaks (area of 4×4 pixels) indicated in the profiles in (A). More examples are shown in Supplementary Figure 4b and c. (E) Average Cor1B-SNAP649 fluorescence (derived from profiles as shown in D) for three different regions of 4×4 pixels per filament, for 30 filaments in total (from three independent experiments) over the course of 300 sec in the presence (blue curve) and absence (green curve) of Cof1.
AIP1 is recruited by Cofilin and triggers filament severing
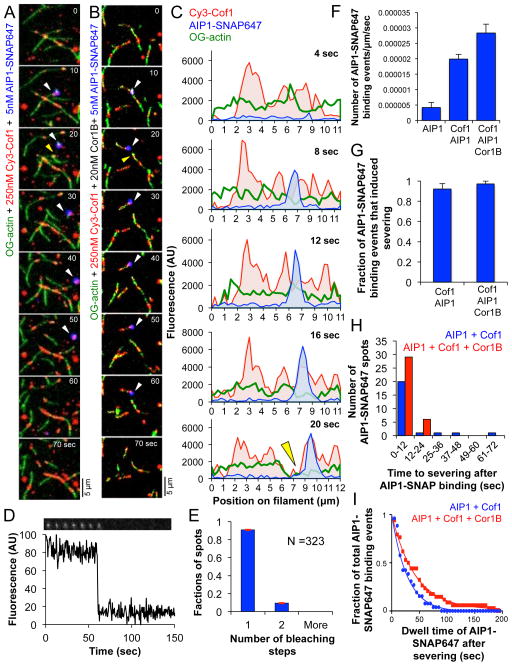
To investigate the spatiotemporal relationship of AIP1 and Cof1 binding to filaments we performed three-color TIRF microscopy experiments as above, except using AIP1-SNAP647, Cy3-Cof1, and OG-actin filaments, with and without unlabeled Cor1B (Fig 4A–C, Supplementary Figure 5, and Supplementary Movie 5). For these experiments, we used a low concentration of AIP1-SNAP647 (5 nM) to reduce background fluorescence in the TIRF images, which was required to enable single molecule observations. We quantified the frequency of AIP1-SNAP647 binding events on filaments with and without Cof1. The AIP1-SNAP647 spots we observed on filaments were likely single molecules, as 90% of surface adsorbed AIP1-SNAP647 spots photobleached in a single step (Fig 4D and E). AIP1-SNAP647 binding to filaments was approximately 4-fold higher in the presence of Cof1, and 6-fold higher in the combined presence of Cof1 and Cor1B (Fig 4F). These results are consistent with previous studies showing that AIP1 alone has low affinity for actin filaments (2–3 μM), but that its association with F-actin is enhanced by Cof122, 33, 34. We also observed that AIP1-SNAP647 binding always occured at sites where Cy3-Cof1 was already bound. Together, these observations suggest that Cof1 plays an important role in recruiting AIP1 to filament sides.
Figure 4. Recruitment of AIP1-SNAP647 to actin filaments induces rapid severing.
(A and B) Time points from TIRF movies (Supplementary Movie 5) showing Cy3-Cof1 and AIP1-SNAP647 binding to preassembled OG-labeled actin filaments after flow-in, both in the absence (A) and presence (B) of Cor1B. White arrowheads indicate appearance of AIP1-SNAP647 fluorescence; yellow arrowheads indicate severing. (C) Cy3-Cof1 and AIP1-SNAP647 spatiotemporal profiles; same filament as in (B). Green curve represents OG-actin fluorescence; yellow arrowhead indicates severing. (D) Stepwise photobleaching of surface-adsorbed AIP1-SNAP647. (E) Distribution of number of steps to complete photobleaching of AIP1-SNAP647 spots (± SE, shown in red, n=323). (F) Frequency of binding of AIP1-SNAP647 (± SEM, n=3) on actin filaments for the indicated conditions. (G) Fraction (± SEM, n=3) of AIP1-SNAP647 binding events that induce actin filament severing. No statistically significant difference was observed by chi-square test (p > 0.05). (H) Distribution of time intervals between appearance of AIP1-SNAP647 on filaments and severing. (I) Fitted curve for the distribution of AIP1-SNAP647 dwell times on actin filaments for the indicated conditions.
In addition, we observed that more than 90% of the observed events in which AIP1-SNAP647 appeared on filaments were rapidly followed by severing in the presence of Cof1, with or without Cor1B (Fig 4G and 4H). The average time interval between binding of AIP1-SNAP647 and filament severing was 9 sec (95% c.i. [5, 17]) in the absence of Cor1B, and this did not change significantly in the presence of Cor1B (6 sec [5, 8]). In most instances (56 of 63 events) AIP1-SNAP647 was also observed to remain bound to one of the new filament ends produced by severing (Fig 4A–C, Supplementary Figure 5, and Supplementary Movie 5). The average dwell time of AIP1-SNAP647 on severed ends was slightly longer in the presence of Cof1 and Cor1B (29.11 sec; 95% c.i. [27.71, 30.66]) compared to Cof1 alone (19.54 sec; 95% c.i. [18.72, 20.45]) (Fig 4I).
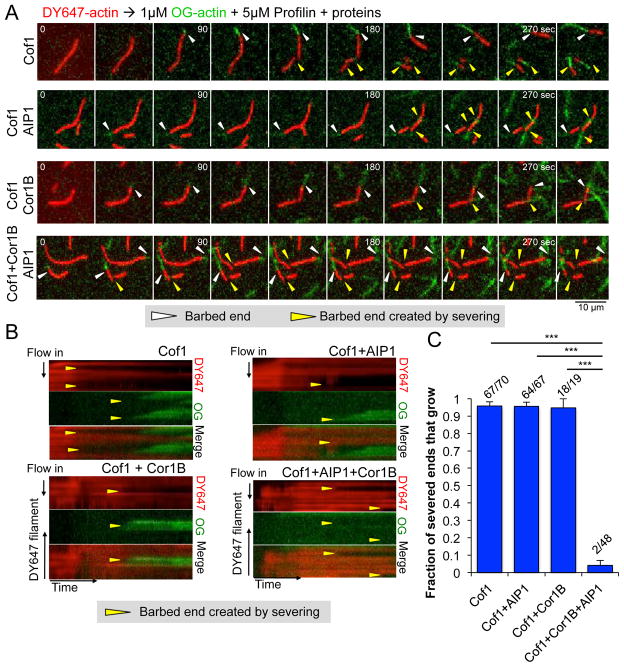
Capping at severed ends requires the three-component mixture
As mentioned earlier (see Introduction), cytosolic conditions strongly favor actin assembly over disassembly because of the high concentration of ATP-actin monomers. For this reason, cells require a mechanism for efficient capping or blocking of new barbed ends generated by severing; otherwise, severing will promote increased growth rather than disassembly. To address whether Cof1, Cor1B, and/or AIP1 can block actin polymerization at newly generated barbed ends after severing, we developed a two-color actin assay, in which we first polymerized and tethered DY647-labeled actin filaments, then replaced the solution by flow-in with a mixture of OG-actin monomers, profilin, and different concentrations of Cof1, Cor1B, and/or AIP1 (Fig 5; Supplementary Movie 6). Under each condition, the original barbed end (white arrows, Fig 5A) continued to polymerize, indicated by the appearance of new (green) polymer. In the presence of Cof1, Cof1 and AIP1, or Cof1 and Cor1B, polymerization was also observed at nearly all of the barbed ends generated by severing (yellow arrows in Fig 5A and 5B; Supplementary Movie 6). In striking contrast, polymerization was rarely observed at barbed ends generated by severing in the presence of Cof1, Cor1B, and AIP1 (Fig 5A–C; Supplementary Movie 6). These observations show that efficient obstruction of new growth at barbed ends generated by severing requires the combination of all three proteins, Cof1, Cor1B, and AIP1. Further, they show that these proteins do not block polymerization at the original, growing barbed end, and thus their ability to inhibit barbed end growth may be closely coupled to severing.
Figure 5. Growth at barbed ends generated by severing is blocked specifically by the combination of Cor1B, Cof1, and AIP1.
(A) Time points from TIRF movies showing the polymerization of OG-actin (green) from the existing and newly generated barbed ends of preassembled, tethered DY647-actin filaments (red) after severing. After DY647-actin filaments were preassembled, the indicated proteins were flowed in, and both severing and new barbed end polymerization were monitored for 300 sec. White arrow heads designate the original barbed ends; yellow arrow heads indicate new barbed ends generated by severing. (B) Kymographs showing DY647- and OG-actin fluorescence for the same filaments, severed by the indicated combination of proteins. (C) Fraction (± SEM, n=3) of newly generated barbed ends that grow within 300 sec after flow-in. The fraction of filaments that grew after severing is shown above each bar. Statistical significance and p-value were determined by a chi-square test; ***, p < 0.05.
Reconstitution of rapidly treadmilling actin filaments
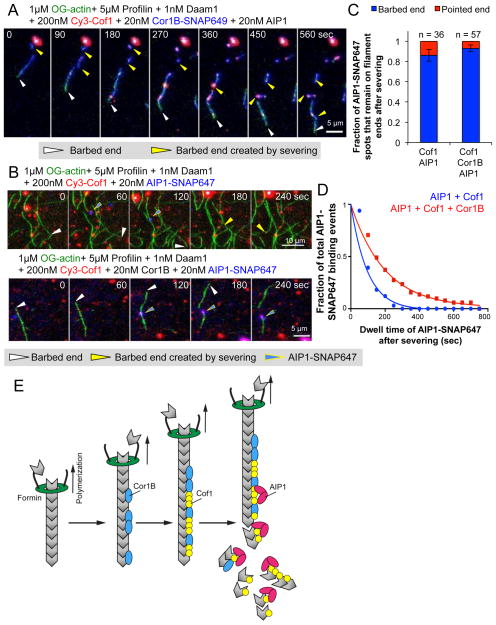
Finally, we asked whether the Cor1B-Cof1-AIP1 disassembly system could be used to reconstitute filament treadmilling under assembly-promoting conditions (as found in vivo), in which filaments are being actively polymerized by formins at their barbed ends while at the same time undergoing stochastic and coupled severing/capping along their lengths, producing new pointed ends that disassemble. For these experiments, we used multi-wavelength TIRF microscopy in reactions containing OG-actin, profilin, formin (Daam1 construct consisting of its FH1, FH2, and C-terminal tail domains35), Cy3-Cof1, Cor1B-SNAP649, and AIP1. Filaments were observed to polymerize at their barbed ends at the ~4-fold accelerated rate expected for this formin in the presence of profilin 38, while being severed along their length (Fig 6A; Supplementary Movie 7). Cor1B-SNAP649 was observed to bind rapidly to the newly polymerized regions of the filaments, leaving only a short undecorated region behind the growing barbed end (white arrows, Fig 6A). Cy3-Cof1 binding followed behind Cor1B-SNAP649 binding, in agreement with our kinetic analysis on preassembled filaments (Fig 3B). Severing events correlated with sites of Cy3-Cof1 decoration (yellow arrows, Fig 6A), and the fragments released by severing did not polymerize but rather disassembled (presumably from their pointed ends), despite the assembly-promoting conditions.
Figure 6. Reconstitution of rapid actin filament treadmilling.
(A and B) Time points from TIRF movies showing barbed end growth stimulated by formins, concurrent with stochastic severing and capping by Cof1, AIP1, and Cor1B, leading to filament disassembly. Fluorescently labeled proteins are color-coded. White arrowheads indicate the formin-capped barbed end of the filament; yellow arrowheads indicate new barbed ends generated by severing; and yellow-outlined blue arrow heads indicate binding of AIP1-SNAP647 to filaments. (C) Fraction of AIP1-SNAP647 spots that remain on the barbed versus the pointed end after severing. Statistical significance and p-value were determined by a chi-square test (not significant, p > 0.05). (D) Fitted curve for the distribution of dwell times for AIP1-SNAP647 spots at newly generated barbed ends after severing in the presence and absence of Cor1B. (E) Model of temporally ordered disassembly by Cor1B, Cof1 and AIP1. Fast-growing filaments (bound at their barbed ends by formins) are decorated first by Cor1B. Cor1B then enhances Cof1 recruitment, leading to Cof1-stabilized filaments. Once Cof1 is present, AIP1 binds to the filaments and induces fast severing. New barbed ends generated by severing in the presence of Cor1B, Cof1, and AIP1 fail to grow, leading to net disassembly of these F-actin fragments.
We also took advantage of this system to analyze the association of AIP1-SNAP647 with filament ends after severing, because the growing barbed ends could be unambiguously identified (Fig 6B; Supplementary Movie 8). This confirmed that AIP1-SNAP647 was recruited to sites of Cy3-Cof1 decoration on filaments, and that binding of AIP1-SNAP647 almost invariably led to severing (33/34 events for AIP1 and Cof1; 54/56 events for AIP1, Cof1, and Cor1B). Further, AIP1-SNAP647 remained bound primarily to the barbed ends of the severed filaments, both in the presence and absence of Cor1B (Fig 5C). However, Cor1B substantially increased the average dwell time of AIP1-SNAP647 on filament barbed ends after severing, from 66.94 sec for Cof1 and AIP1 (95% c.i. [61.41, 73.56]) to 137.4 sec for Cof1, AIP1 and Cor1B (95% c.i. [128.3, 147.9]) (Fig 6B and 6D). These results help explain why all three proteins are required for efficient obstruction of growth at barbed ends generated by severing (Fig 5), although they do not rule out additional mechanistic contributions from Cor1B. Together, these experiments using actively growing filaments validate the key aspects of the Cor1B-Cof1-AIP1 mechanism observed above using preassembled filaments, and demonstrate that filament treadmilling can be reconstituted in vitro under assembly-promoting conditions.
Discussion
In this study, we have used multi-wavelength TIRF microscopy to directly visualize the integrated actions of Coronin, Cofilin, and AIP1 during the process of actin filament severing and disassembly. Our results reveal that Coronin, Cofilin, and AIP1 arrive sequentially on filaments (the majority of the time in that order) and that each protein plays a key role in recruiting the next one and makes a distinct contribution to inducing highly rapid filament disassembly. These findings resolve several long-standing dilemmas about the functions and mechanisms of Coronin, Cofilin, and AIP1, and provide new insights into how cells induce rapid destabilization and disassembly of actin filaments under assembly-promoting conditions. They also highlight the importance of studying the activities of groups of proteins, rather than individual ones, to define their roles and mechanisms in biological processes.
The mechanism we have defined may be used by cells to induce the highly rapid disassembly of filaments in locations such as the leading edge of cells and sites of endocytosis, where filament turnover is observed to occur in 5–20 seconds 36, 37. Indeed, we observed complete disassembly only a few seconds after we exposed preassembled ~10 micron long filaments to a mixture of Cor1B, Cof1, and AIP1 at their estimated cellular ratios and close to their estimated cellular concentrations. The highly rapid actin filament disassembly using near cellular concentrations of Cor1B, Cof1 and AIP1 also demonstrates that the three proteins acting together overcome many of the limitations reported for Cofilin alone (see Introduction), including delayed severing due to slow binding of Cofilin to filaments, and sub-optimal severing at high (micromolar) concentrations compared to that seen at low (nanomolar) concentrations of Cofilin 8, 9. By lowering the concentrations of Cor1B, Cof1, and AIP1, while maintaining their cellular ratio, we were able to quantify the contributions of each protein to the three-part mechanism, and to directly observe each protein acting on filaments during disassembly. In our system, Cof1, Cor1B and AIP1 did not cause filaments to disassemble in bursts, as described by Kueh and colleagues 31; however, there are differences in the experimental designs which could account for the different results (see Methods).
Our experiments using labeled Coronin and Cofilin expose critical aspects of their spatiotemporal relationship, and resolve several long-standing dilemmas. First, they show that the slow binding of Cofilin to filaments is accelerated dramatically by Coronin, demonstrating that this slow step in severing can be enhanced by co-factors, and may be regulated in vivo. In addition, we observed that Cofilin is recruited to sites on filaments where Coronin is already bound. Precisely how this is achieved is not yet clear, but may involve Coronin altering the conformation of F-actin and/or providing additional contacts for Cofilin on filaments. Indeed, recent cryo-EM studies show that Cofilin and Coronin occupy closely juxtaposed sites on ADP-actin filaments and may be in direct contact 4, 38. Coronin could have additional effects on the nucleotide state of F-actin, e.g., in promoting Pi release, but our observation that Coronin dramatically recruits Cofilin to preassembled filaments, which should be composed entirely of ADP-actin subunits, argues that Coronin most likely recruits Cofilin to ADP-actin filaments by providing additional contacts and/or inducing subtle changes in F-actin conformation that favor Cofilin binding.
A second issue resolved by our data is the seemingly paradoxical observation that, despite multiple lines of genetic evidence suggesting that Coronin promotes actin disassembly in vivo, alone Coronin inhibits rather than enhances Cofilin-mediated severing in vitro 26, 28. We observed that Coronin accelerates Cofilin recruitment to filaments, both in the presence or absence of AIP1, but that without AIP1 this leads to over-decoration and hyper-stabilization of filaments by Cofilin. However, in the added presence of AIP1 it leads to strongly enhanced severing and disassembly. Thus, Coronin biochemically stimulates Cofilin-mediated actin disassembly specifically in the presence of AIP1. We also made the observation that AIP1 (without Coronin) only marginally improves the severing efficiency of Cofilin. This suggests that in addition to recruiting Cofilin to filaments, Coronin binding to filaments greatly enhances severing by AIP1 and Cof1. Moreover, after severing, Coronin increased the dwell time of AIP1 on barbed ends generated by severing, and was critical for blocking growth at those ends. Thus, Coronin makes multiple mechanistic contributions to promoting actin filament disassembly, and each of these roles is highly integrated with the functions of Cofilin and AIP1.
Our data also shed important light on the long-standing debate over the AIP1 mechanism in actin disassembly. A role for AIP1 in capping barbed ends of severed filaments has been supported by genetic interactions between AIP1 and capping protein 39, 40, 41, live-imaging analysis of GFP-AIP1 speckles at the leading edge 42, and specific biochemical observations 21, 25, 39. However, a different set of biochemical studies reported enhanced Cofilin-mediated severing in the presence of AIP1, but saw no clear evidence of barbed end capping 12, 23, 43. Our observations resolve these discrepancies. We found that low concentrations of AIP1 (10–15 nM) were insufficient to cap the newly generated barbed ends. On the other hand, in the presence of Cor1B (10–15 nM), 10–15 nM AIP1 strongly enhance severing by Cofilin, but also capped the severed ends. Thus, our results show that low concentrations of AIP1 and Cor1B can together promote actin disassembly by both mechanisms, i.e., enhancing severing and capping/blocking the new barbed ends generated by severing. This may explain why some previous studies (which did not include Coronin, and used different protocols and protein concentrations) did not observe barbed end capping. Using direct imaging of labeled molecules on actin filaments, we also observed that Cofilin recruited AIP1 to filament sides, and upon binding invariably induced severing. AIP1 remained attached to the barbed end after severing, and the presence of Cor1B substantially increased the AIP1 dwell time on the severed end. Taken together, these data demonstrate unequivocally that AIP1 both enhances Cofilin severing and caps barbed ends generated by severing, but the efficiency of both effects of AIP1 are modulated by Coronin.
Since the strongest severing activity that we observed occurred in the combined presence of Coronin, Cofilin, and AIP1, a future challenge will be to understand the structural basis for how these three proteins simultaneously interact with filaments. As mentioned above, Coronin and Cofilin appear to bind adjacent sites on filaments, so it will be important next to determine how their combined presence affects F-actin conformation, nucleotide state, and mechanical properties. In addition, it has yet to be determined precisely how AIP1 binding to a Cofilin-decorated region of a filament triggers severing. This may involve AIP1 displacing one or more Cof1 molecules from a patch to generate a local discontinuity that destabilizes filaments 44, or AIP1 displacing one of Cofilin’s two binding interactions with F-actin 45. Finally, it will be important to study if and how Cofilin, Coronin and AIP1 work alongside other actin disassembly factors, e.g., Srv2/CAP, GMF, and possibly Twinfilin 16, 18, 46, 47. GMF binds with high affinity to Arp2/3 complex and stimulates filament debranching. Srv2/CAP binds to filaments, independently of Cofilin, and enhances severing not by accelerating Cofilin recruitment but instead by reducing the time from Cofilin binding to severing 16, 17, 18. These mechanistic effects are distinct from, and possibly complementary to those we have defined here for Coronin and AIP1: Coronin accelerates the recruitment of Cofilin; AIP1 is recruited to filaments by Cofilin and greatly enhances severing; and all three (Cofilin, AIP1 and Coronin) together cap severed barbed ends. Thus, each of these conserved proteins may have distinct capabilities in promoting actin filament disassembly, providing cells with a diverse tool kit with which to locally tune and shape actin networks, tailoring their architectures and dynamics for a wide range of functions.
Methods
Plasmids
A plasmid for expressing mouse Coronin-1B (Cor1B) with a C-terminal 8His tag in mammalian cells (vector pTT5SH8Q2) was kindly provided by Dr. Jim Bear (University of North Carolina). A plasmid carrying mouse AIP1 was kindly provided by Dr. Naoki Watanabe (Tohoku University), and the insert was cloned into the same mamalian expression vector as Cor1B. Plasmids for expressing Cor1B-SNAP and AIP1-SNAP were generated by cloning a SNAP-tag at the C-terminus of each protein in the same expression vectors as above. The plasmid for expressing human Cof1 in E. coli was generously provided by Dr. David Kovar (University of Chicago). For fluorescent labeling of human Cof1, a mutagenesis strategy was followed, related to that used to label yeast Cof1 16, in which the following substitutions were introduced by site-directed mutagenesis: C139A, C147A, C39S, and T63C. All constructs were verified by DNA sequencing.
Protein purification and fluorescent labeling
Rabbit skeletal muscle actin (RMA) was purified as described 48. In brief, RMA was purified first by generating an acetone powder from ground muscle tissue, which was stored in aliquots at 80°C. Aliquots of acetone powder were then pulverized using a coffee grinder, resuspended in G-buffer, and cleared by low speed centrifugation. The actin was polymerized overnight and then pelleted. The pellet was disrupted by douncing, dialyzed against G-buffer for 2–3 d, and then gel filtered on a 16/60 S200 column (GE Healthcare). Column fractions were stored at 4°C. Actin was labeled on Cys374 with either Oregon Green (OG) maleimide or DY647 maleimide (Dyomics, Jena, Germany), as described in Kuhn and Pollard49. Briefly, monomeric actin reconstituted from an actin pellet was dialyzed against two changes of G-buffer without DTT for 1 h each. After clarification at 500 × g for 5 min, actin was polymerized by mixing an equal volume of cold 2× label buffer (2× = 50 mM imidazole, pH 7.5, 0.2 M KCl, 4 mM MgCl2, 6 mM NaN3, 0.6 mM ATP). After 5 min polymerized actin was diluted to 1 mg/ml with cold 1× label buffer, then a 10-fold molar excess of OG or DY647 maleimide was added dropwise to the actin while stirring, and the solution was stirred gently overnight. Labeled actin was clarified at 500 × g for 5 min and centrifuged at 105× g for 2 h to pellet actin filaments. The pellet was resuspended in G-buffer by douncing, dialyzed for 2 days against two changes of G-buffer and gel filtered on a 16/60 S200 column (GE Healthcare). Peak fractions were combined and stored at −20°C. Labeling efficiency of OG-actin was measured by absorbance at 290 and 491 nm, and extinction coefficient E491=77,800 M−1cm−1. Labeling efficiency of DY647-actin was measured by absorbance at 290 and 653 nm, and extinction coefficient E653=250,000 M−1cm−1. The absorption at 290 nm was corrected for background fluorescence from the dye (correction factor 0.016991 for OG-actin and 0.024 for DY647-actin). The formin Daam1 (6his-FH1-FH2-C) was inducibly-expressed in yeast, and purified by sequential Ni2+-NTA and gel filtration chromatography steps35. Cor1B and AIP1 were expressed and purified from transfected HEK293T cells (ATCC). Cells were grown on plates at 37°C under a humidified atmosphere containing 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM), supplemented with 10% (v/v) heat-inactivated foetal bovine serum, glucose (4.5 g/l), penicillin (100 units/ml) and streptomycin (100 μg/ml). Cells at 30–40% confluence were transiently transfected using 25 kDa linear polyethylenimine (Polysciences, Warrington, PA). 72 h post-transfection, cells were harvested in PBS, pelleted by centrifugation at 1,000 × g for 5 min, and lysed by repeated freeze-thawing in 20 mM Tris/HCl pH 7.5, 150 mM NaCl, 1% (v/v) Triton X-100 and a standard cocktail of protease inhibitors (Roche, Germany). After a 30 min incubation on ice, cell lysates were cleared by centrifugation at 20,000 × g at 4°C using an eppendorf tabletop centrifuge and incubated with Ni2+-NTA beads (Qiagen, Valencia, CA) for 90 min at 4°C in the presence of 10 mM imidazole. After washing with Buffer A (20 mM Tris pH 7.5, 300 mM NaCl, 50 mM imidazole and 1 mM DTT), proteins were eluted in Buffer A supplemented with 250 mM imidazole, concentrated, and purified further on a Superose 6 gel filtration column (GE Healthcare Biosciences, Pittsburgh, PA) equilibrated in Buffer B (20 mM Tris pH 8.0, 50 mM KCl and 1 mM DTT). For fluorescent labeling of SNAP-tagged proteins, the fusion proteins were bound to Ni2+-NTA beads, washed extensively in PBS with 1 mM DTT and incubated with a 5-fold excess of benzylguanine or benzylchloropyrimidine SNAP-Surface® 649 (New England Biolabs, Ipswich, MA) for 2 h at room temperature. Next, beads were washed extensively in PBS with 1 mM DTT and eluted in PBS with 250 mM imidazole. To remove free dye, proteins were exchanged into Buffer B on PD-10 columns (GE Healthcare Biosciences). Cor1B-SNAP was first labeled with SNAP-Surface® Alexa Fluor® 647 (New England Biolabs, Ipswich, MA), but this preparation showed severe non-specific binding to slides in TIRF microscopy assays. This issue was overcome by labeling of Cor1B with SNAP-Surface® 649. A labeling efficiency of 30% was obtained for Cor1B-SNAP649 and AIP1-SNAP647. Labeling efficiencies were determined spectrophotometrically using the absorbance at 650 nm and an extinction coefficient of 250,000 M−1 cm−1 for SNAP- SNAP-Surface® Alexa Fluor® 647 or Surface® 649, combined with absorbance at 280 nm and an estimated extinction coefficient of 82,850 M−1 cm−1 for Cor1B-SNAP or 118,720 M−1 cm−1 for AIP1-SNAP. The absorption at 280 nm was corrected for background fluorescence from the dye (correction factor 0.024). Human Cof1 was expressed in BL21 (DE3) E. coli by growing cells at 37°C in TB medium to log phase, then inducing expression with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at 18°C for 16 h. Cells were harvested by centrifugation and stored at −80°C, then lysed by sonication in 20 mM Tris pH 8.0, 50 mM NaCl, 1 mM DTT and protease inhibitors. Lysates were cleared by centrifugation at 30,000 × g for 20 min in a Fiberlite F13-14X50CY rotor (Thermo Scientific, Rockport, Illinois), and applied to a 5 ml HiTrap HP Q column (GE Healthcare Biosciences). The flow-through containing Cof1 was collected and dialyzed into 20 mM Hepes pH 6.8, 25 mM NaCl and 1 mM DTT. Next, the protein was applied to a 5 ml HiTrap SP FF column (GE Healthcare Biosciences) and eluted with a linear gradient of NaCl (25 to 500 mM). Fractions containing Cof1 were concentrated and dialyzed into Buffer B, aliquoted, snap-frozen in liquid N2, and stored at −80°C until use. Dye-labeled Cof1 was purified similarly except that the protein was eluted from the SP FF column with PBS, and then incubated with a 10-fold excess of Cy3-maleimide (GE Healthcare Biosciences) for 2 h at room temperature in the presence of 0.3 mM TCEP. Excess dye was removed by passing the protein over a PD-10 column equilibrated in Buffer B. Final labeling efficiency was 30%. Labeling efficiency was determined as described above, using absorbance at 550 nm and an extinction coefficient of 150,000 M−1 cm−1 for Cy3, combined with absorbance at 280 nm and an estimated extinction coefficient of 14,440 M−1 cm−1 for Cof1.
Total internal reflection fluorescence (TIRF) microscopy
For all experiments, coverslips were first cleaned by sonication in detergent for 60 min, followed by successive sonications in 1M KOH and 1M HCl for 20 min each, then sonication in ethanol for at least 60 min. Coverslips were then washed extensively with H2O, dried in an N2-stream, layered with 200 μl of 80% ethanol pH 2.0, 2 mg/ml PEG-silane and 2 μg/ml biotin-PEG-silane (Laysan Bio Inc., Arab, AL), and incubated for 16 h at 70°C. Flow cells were assembled by rinsing PEG-coated coverslips extensively with H2O, then attaching them to a flow chamber (Ibidi, Martinsried, Germany) using double-sided tape (2.5 cm × 2 mm × 120 μm) and epoxy resin. For all TIRF experiments in this study except for those shown in Figure 6, the actin filaments were tethered. To accomplish this, flow cells were incubated for 3 min with HBSA (HEK buffer with 1% BSA), followed by 30 s incubation with 0.1 mg/ml Streptavidin in HEK buffer. Flow cells were washed HBSA and equilibrated with TIRF buffer (10 mM imidazole pH 7.4, 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 0.2 mM ATP, 10 mM DTT, 15 mM glucose, 20 μg/ml catalase, 100 μg/ml glucose oxidase, and 0.5% methylcellulose (4000 cP)). Reactions were initiated by rapidly diluting actin monomers (1 μM final, 10% OG-labeled, 0.5 % biotinylated) into TIRF buffer, followed by transferring that mixture into a flow chamber. After filaments had polymerized to lengths of ~10–15 μm, the reaction mixture was replaced with TIRF buffer containing the indicated proteins. For assays using two differentially labeled actin preparations, filaments were first polymerized as above using 1 μM actin (10% DY647-labeled, 0.5 % biotinylated), then the reaction mixture was replaced by flow-in with TIRF buffer containing 1 μM actin (10% OG-labeled, 0.5% biotinylated) and the indicated proteins. To induce a manageable severing rate that allows one to follow filament growth at severed ends without over-crowding the field with filaments, different concentrations of Cof1, Cor1B, and AIP1 were used in each condition. Concentrations were as follows: 250 nM Cof1 alone, 250 nM Cof1 + 25 nM Cor1B, 100 nM Cof1 + 10 nM AIP1, 75 nM Cof1 + 7.5 nM AIP1 + 7.5 nM Cor1B. For experiments in Figure 6, non-attached filaments (no biotin-actin) were polymerized in TIRF buffer containing 2% dextran instead of 0.5% methylcellulose as the crowding agent, which minimized filament diffusion. Single- and multi-wavelength time-lapse TIRF imaging was performed using a Nikon-Ti200 inverted microscope equipped with a 150 mW Ar-Laser (Mellot Griot, Carlsbad, CA), a TIRF-objective with a N.A. of 1.49 (Nikon Instruments Inc., New York, NY), and an EMCCD camera (Andor Ixon, Belfast, Northern Ireland). During measurements, optimal focus was maintained by the perfect focus system (Nikon Instruments Inc., New York, NY). For single-molecule photobleaching of AIP1-SNAP647, flow cells were assembled using uncoated, acid-washed coverslips and washed with HBSA. Next, 0.4 nM AIP1-SNAP647 was flowed in (in TIRF buffer without glucose oxidase and catalase), and after a 5 min incubation the adsorbed spots were exposed to continuous illumination at high laser power, and images were acquired every 0.5 sec.
As noted in the Discussion, in our experiments Cof1, Cor1B and AIP1 did not cause filaments to disassemble in bursts, as reported by Kueh and colleagues 31. However, there are two major differences in the experimental design between the studies. First, there are differences in the filament attachment mode. In most of our reactions, we incorporated a low percentage (0.5%) of biotin-actin into filaments and attached them to the surface using streptavidin. In other experiments (e.g., our treadmilling assays in Fig 6) we used untethered filaments. In both cases, we did not observe a burst of disassembly at filament ends. In constrast, Kueh and colleagues tethered filaments using filamin, raising the possibility that the bursting is promoted by this mode of attachment, and indeed Kueh et al. observed less bursting when filaments were tethered instead with N-ethylmaleimide-inactivated myosin. Second, the two studies used different actin-labeling strategies. Kueh and colleagues assembled filaments with a relatively high percentage of labeled actin (30%). Further, they used actin monomers that were labeled heterogenously on exposed lysines; thus, each monomer could have multiple dye molecules attached at different locations. In contrast, we used a substantially lower percentage of labeled actin (10% Oregon green-actin), in which the dye molecules uniformly labeled on Cys374, so that there was only one dye molecule per actin. This strategy limits formation of photo-induced actin dimers, which is reported to cause ‘pausing’ in actin filament depolymerization 50.
TIRF data analysis
All TIRF data was analyzed using ImageJ software (NIH, Bethesda, MD). Before each analysis, the background was subtracted using the standard background subtraction tool (rolling ball radius 50 pixels). Severing rates were calculated by measuring the initial lengths of filaments before flow-in, and counting severing events observed during the next 300 sec after flow-in of the indicated protein combinations. For binding of Cy3-Cofilin and Cor1B-SNAP, the OG-actin filaments were first traced (at 488 nm) over time using the Plot Z-axis profile tool, and then the trace was saved as a ROI and used to determine the corresponding fluorescence profiles in other channels. After correction for bleed through between the Cy3 and SNAP649 channels, fluorescence in all channels was normalized to the OG actin signal. Fluorescent traces at specific time points were obtained in a similar manner, except that they were additionally normalized to the highest signal detected in the indicated channel. The same approach was followed to monitor the change in Cor1B-SNAP649 fluorescence for three different regions of 4×4 pixels per filament, as shown in Fig 3D and E, and Fig S4B and C. The appearance of Cor1B-SNAP649 and Cy3-Cof1 were scored by eye and confirmed by obtaining the fluorescent traces in both channels at the time of detection as described above. Subsequently, filaments were observed until the first sight of a molecule in the other channel. For kinetic analysis, measured time intervals (Fig 4H) were fit to an exponential model using maximum likelihood methods, and fit parameter confidence intervals were estimated by bootstrapping 51. Other statistical analyses and curve fittings (Fig 4I and 6D) were performed with Prism 5.0.
Electron microscopy
Skeletal muscle Ca-ATP-G actin (24 μM) was polymerized by addition of inorganic salts (2 mM MgCl2, 50 mM KCl) for 1 h at 25°C. Then F-actin was diluted to 2 μM in F-buffer (50mM KCl, 2mM MgCl2, 0.2 mM EGTA, 1 mM DTT, 5 mM Tris, pH 8) and incubated for 10 min at 25°C with one or more of the following proteins: 2 μM Cofilin, 0.2 μM AIP1, and/or 0.2 μM Cor1B. Samples were diluted another two-fold in F-buffer and droplets (5–7 μl) were applied for 20–30 sec to glow discharged formvar-carbon coated 200 mesh copper grids, then the grids were blotted to remove excess solution, negatively stained with 1% (w/v) uranyl acetate for 1 min, blotted again, and allowed to air-dry. Images were recorded on a CCD camera using a FEI Morgani 268 transmission electron microscope at an acceleration voltage of 80 kV and magnifications of 5,600, 8,900 and 18,000. For measurement of filament lengths, pictures at magnification 5,600 or 8,900 were used, and micrographs from adjacent areas (7–15 images) on the grid were combined into one picture using Adobe Photoshop. The contour traces of each filament were measured using the ruler tool in Adobe Photoshop.
Supplementary Material
Acknowledgments
We are indebted to Dennis Breitsprecher for making the initial observations that inspired this work, and to Dr. Chen Xu for assistance with the Morgani electron microscope. This work was supported by NIH grants to J.G. and B.G (GM098143) and B.G. (GM063691). In addition, we acknowledge support from the Brandeis University NSF MRSEC, DMR-1420382.
Footnotes
Author contributions
S.J. performed all of the TIRF experiments and most of the data analysis; A.C. performed the EM; C.Y and J.G. performed some of the data analysis; S.J. and B.G. designed the experiments and wrote the manuscript. J.G. edited the manuscript.
Competing financial interests statement
The authors declare no competing financial interests.
References
- 1.Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nature reviews Molecular cell biology. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chesarone MA, Goode BL. Actin nucleation and elongation factors: mechanisms and interplay. Current opinion in cell biology. 2009;21:28–37. doi: 10.1016/j.ceb.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De La Cruz EM. How cofilin severs an actin filament. Biophysical reviews. 2009;1:51–59. doi: 10.1007/s12551-009-0008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galkin VE, et al. Remodeling of actin filaments by ADF/cofilin proteins. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:20568–20572. doi: 10.1073/pnas.1110109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayakawa K, Sakakibara S, Sokabe M, Tatsumi H. Single-molecule imaging and kinetic analysis of cooperative cofilin-actin filament interactions. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:9810–9815. doi: 10.1073/pnas.1321451111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGough A, Pope B, Chiu W, Weeds A. Cofilin changes the twist of F-actin: implications for actin filament dynamics and cellular function. The Journal of cell biology. 1997;138:771–781. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suarez C, et al. Cofilin tunes the nucleotide state of actin filaments and severs at bare and decorated segment boundaries. Curr Biol. 2011;21:862–868. doi: 10.1016/j.cub.2011.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andrianantoandro E, Pollard TD. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Molecular cell. 2006;24:13–23. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 9.Cao W, Goodarzi JP, De La Cruz EM. Energetics and kinetics of cooperative cofilin-actin filament interactions. Journal of molecular biology. 2006;361:257–267. doi: 10.1016/j.jmb.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh M, Song X, Mouneimne G, Sidani M, Lawrence DS, Condeelis JS. Cofilin promotes actin polymerization and defines the direction of cell motility. Science. 2004;304:743–746. doi: 10.1126/science.1094561. [DOI] [PubMed] [Google Scholar]
- 11.Hotulainen P, Paunola E, Vartiainen MK, Lappalainen P. Actin-depolymerizing factor and cofilin-1 play overlapping roles in promoting rapid F-actin depolymerization in mammalian nonmuscle cells. Molecular biology of the cell. 2005;16:649–664. doi: 10.1091/mbc.E04-07-0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nadkarni AV, Brieher WM. Aip1 destabilizes cofilin-saturated actin filaments by severing and accelerating monomer dissociation from ends. Curr Biol. 2014;24:2749–2757. doi: 10.1016/j.cub.2014.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dedova IV, Nikolaeva OP, Mikhailova VV, dos Remedios CG, Levitsky DI. Two opposite effects of cofilin on the thermal unfolding of F-actin: a differential scanning calorimetric study. Biophysical chemistry. 2004;110:119–128. doi: 10.1016/j.bpc.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 14.Kiuchi T, Ohashi K, Kurita S, Mizuno K. Cofilin promotes stimulus-induced lamellipodium formation by generating an abundant supply of actin monomers. The Journal of cell biology. 2007;177:465–476. doi: 10.1083/jcb.200610005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vitriol EA, Wise AL, Berginski ME, Bamburg JR, Zheng JQ. Instantaneous inactivation of cofilin reveals its function of F-actin disassembly in lamellipodia. Molecular biology of the cell. 2013;24:2238–2247. doi: 10.1091/mbc.E13-03-0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaudhry F, Breitsprecher D, Little K, Sharov G, Sokolova O, Goode BL. Srv2/cyclase-associated protein forms hexameric shurikens that directly catalyze actin filament severing by cofilin. Molecular biology of the cell. 2013;24:31–41. doi: 10.1091/mbc.E12-08-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jansen S, Collins A, Golden L, Sokolova O, Goode BL. Structure and Mechanism of Mouse Cyclase-associated Protein (CAP1) in Regulating Actin Dynamics. The Journal of biological chemistry. 2014;289:30732–30742. doi: 10.1074/jbc.M114.601765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Normoyle KP, Brieher WM. Cyclase-associated protein (CAP) acts directly on F-actin to accelerate cofilin-mediated actin severing across the range of physiological pH. The Journal of biological chemistry. 2012;287:35722–35732. doi: 10.1074/jbc.M112.396051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ono S. The role of cyclase-associated protein in regulating actin filament dynamics - more than a monomer-sequestration factor. Journal of cell science. 2013;126:3249–3258. doi: 10.1242/jcs.128231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quintero-Monzon O, et al. Reconstitution and dissection of the 600-kDa Srv2/CAP complex: roles for oligomerization and cofilin-actin binding in driving actin turnover. The Journal of biological chemistry. 2009;284:10923–10934. doi: 10.1074/jbc.M808760200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okada K, Blanchoin L, Abe H, Chen H, Pollard TD, Bamburg JR. Xenopus actin-interacting protein 1 (XAip1) enhances cofilin fragmentation of filaments by capping filament ends. The Journal of biological chemistry. 2002;277:43011–43016. doi: 10.1074/jbc.M203111200. [DOI] [PubMed] [Google Scholar]
- 22.Okada K, Obinata T, Abe H. XAIP1: a Xenopus homologue of yeast actin interacting protein 1 (AIP1), which induces disassembly of actin filaments cooperatively with ADF/cofilin family proteins. Journal of cell science. 1999;112 (Pt 10):1553–1565. doi: 10.1242/jcs.112.10.1553. [DOI] [PubMed] [Google Scholar]
- 23.Ono S, Mohri K, Ono K. Microscopic evidence that actin-interacting protein 1 actively disassembles actin-depolymerizing factor/Cofilin-bound actin filaments. The Journal of biological chemistry. 2004;279:14207–14212. doi: 10.1074/jbc.M313418200. [DOI] [PubMed] [Google Scholar]
- 24.Rodal AA, Tetreault JW, Lappalainen P, Drubin DG, Amberg DC. Aip1p interacts with cofilin to disassemble actin filaments. The Journal of cell biology. 1999;145:1251–1264. doi: 10.1083/jcb.145.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi M, et al. Oryza sativa actin-interacting protein 1 is required for rice growth by promoting actin turnover. The Plant journal: for cell and molecular biology. 2013;73:747–760. doi: 10.1111/tpj.12065. [DOI] [PubMed] [Google Scholar]
- 26.Cai L, Makhov AM, Bear JE. F-actin binding is essential for coronin 1B function in vivo. Journal of cell science. 2007;120:1779–1790. doi: 10.1242/jcs.007641. [DOI] [PubMed] [Google Scholar]
- 27.Cai L, Marshall TW, Uetrecht AC, Schafer DA, Bear JE. Coronin 1B coordinates Arp2/3 complex and cofilin activities at the leading edge. Cell. 2007;128:915–929. doi: 10.1016/j.cell.2007.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gandhi M, Achard V, Blanchoin L, Goode BL. Coronin switches roles in actin disassembly depending on the nucleotide state of actin. Molecular cell. 2009;34:364–374. doi: 10.1016/j.molcel.2009.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goode BL, et al. Coronin promotes the rapid assembly and cross-linking of actin filaments and may link the actin and microtubule cytoskeletons in yeast. The Journal of cell biology. 1999;144:83–98. doi: 10.1083/jcb.144.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brieher WM, Kueh HY, Ballif BA, Mitchison TJ. Rapid actin monomer-insensitive depolymerization of Listeria actin comet tails by cofilin, coronin, and Aip1. The Journal of cell biology. 2006;175:315–324. doi: 10.1083/jcb.200603149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kueh HY, Charras GT, Mitchison TJ, Brieher WM. Actin disassembly by cofilin, coronin, and Aip1 occurs in bursts and is inhibited by barbed-end cappers. The Journal of cell biology. 2008;182:341–353. doi: 10.1083/jcb.200801027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin MC, Galletta BJ, Sept D, Cooper JA. Overlapping and distinct functions for cofilin, coronin and Aip1 in actin dynamics in vivo. Journal of cell science. 2010;123:1329–1342. doi: 10.1242/jcs.065698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohri K, Vorobiev S, Fedorov AA, Almo SC, Ono S. Identification of functional residues on Caenorhabditis elegans actin-interacting protein 1 (UNC-78) for disassembly of actin depolymerizing factor/cofilin-bound actin filaments. The Journal of biological chemistry. 2004;279:31697–31707. doi: 10.1074/jbc.M403351200. [DOI] [PubMed] [Google Scholar]
- 34.Okada K, Ravi H, Smith EM, Goode BL. Aip1 and cofilin promote rapid turnover of yeast actin patches and cables: a coordinated mechanism for severing and capping filaments. Molecular biology of the cell. 2006;17:2855–2868. doi: 10.1091/mbc.E06-02-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jaiswal R, Breitsprecher D, Collins A, Correa IR, Jr, Xu MQ, Goode BL. The formin Daam1 and fascin directly collaborate to promote filopodia formation. Curr Biol. 2013;23:1373–1379. doi: 10.1016/j.cub.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaksonen M, Sun Y, Drubin DG. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- 37.Watanabe N, Mitchison TJ. Single-molecule speckle analysis of actin filament turnover in lamellipodia. Science. 2002;295:1083–1086. doi: 10.1126/science.1067470. [DOI] [PubMed] [Google Scholar]
- 38.Ge P, Durer ZA, Kudryashov D, Zhou ZH, Reisler E. Cryo-EM reveals different coronin binding modes for ADP- and ADP-BeFx actin filaments. Nature structural & molecular biology. 2014;21:1075–1081. doi: 10.1038/nsmb.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Balcer HI, et al. Coordinated regulation of actin filament turnover by a high-molecular-weight Srv2/CAP complex, cofilin, profilin, and Aip1. Curr Biol. 2003;13:2159–2169. doi: 10.1016/j.cub.2003.11.051. [DOI] [PubMed] [Google Scholar]
- 40.Berro J, Pollard TD. Synergies between Aip1p and capping protein subunits (Acp1p and Acp2p) in clathrin-mediated endocytosis and cell polarization in fission yeast. Molecular biology of the cell. 2014;25:3515–3527. doi: 10.1091/mbc.E13-01-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Michelot A, Grassart A, Okreglak V, Costanzo M, Boone C, Drubin DG. Actin filament elongation in Arp2/3-derived networks is controlled by three distinct mechanisms. Developmental cell. 2013;24:182–195. doi: 10.1016/j.devcel.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsuji T, Miyoshi T, Higashida C, Narumiya S, Watanabe N. An order of magnitude faster AIP1-associated actin disruption than nucleation by the Arp2/3 complex in lamellipodia. PloS one. 2009;4:e4921. doi: 10.1371/journal.pone.0004921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Q, Courtemanche N, Pollard TD. Aip1 promotes actin filament severing by cofilin and regulates the constriction of the cytokinetic contractile ring. The Journal of biological chemistry. 2014 doi: 10.1074/jbc.M114.612978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elam WA, Kang H, De La Cruz EM. Competitive displacement of cofilin can promote actin filament severing. Biochemical and biophysical research communications. 2013;438:728–731. doi: 10.1016/j.bbrc.2013.07.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aggeli D, et al. Coordination of the filament stabilizing versus destabilizing activities of cofilin through its secondary binding site on actin. Cytoskeleton. 2014;71:361–379. doi: 10.1002/cm.21178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gandhi M, et al. GMF is a cofilin homolog that binds Arp2/3 complex to stimulate filament debranching and inhibit actin nucleation. Curr Biol. 2010;20:861–867. doi: 10.1016/j.cub.2010.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moseley JB, Okada K, Balcer HI, Kovar DR, Pollard TD, Goode BL. Twinfilin is an actin-filament-severing protein and promotes rapid turnover of actin structures in vivo. Journal of cell science. 2006;119:1547–1557. doi: 10.1242/jcs.02860. [DOI] [PubMed] [Google Scholar]
- 48.Graziano BR, Jonasson EM, Pullen JG, Gould CJ, Goode BL. Ligand-induced activation of a formin-NPF pair leads to collaborative actin nucleation. The Journal of cell biology. 2013;201:595–611. doi: 10.1083/jcb.201212059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuhn JR, Pollard TD. Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophysical journal. 2005;88:1387–1402. doi: 10.1529/biophysj.104.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Niedermayer T, et al. Intermittent depolymerization of actin filaments is caused by photo-induced dimerization of actin protomers. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:10769–10774. doi: 10.1073/pnas.1121381109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith BA, Gelles J, Goode BL. Single-molecule studies of actin assembly and disassembly factors. Methods in enzymology. 2014;540:95–117. doi: 10.1016/B978-0-12-397924-7.00006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.