Abstract
HTS with microtiter plates has been the major tool used in the pharmaceutical industry to explore chemical diversity space and to identify active compounds and pharmacophores for specific biological targets. However, HTS faces a daunting challenge regarding the fast-growing numbers of drug targets arising from genomic and proteomic research, and large chemical libraries generated from high-throughput synthesis. There is an urgent need to find new ways to profile the activity of large numbers of chemicals against hundreds of biological targets in a fast, low-cost fashion. Chemical microarray can rise to this challenge because it has the capability of identifying and evaluating small molecules as potential therapeutic reagents. During the past few years, chemical microarray technology, with different surface chemistries and activation strategies, has generated many successes in the evaluation of chemical–protein interactions, enzyme activity inhibition, target identification, signal pathway elucidation and cell-based functional analysis. The success of chemical microarray technology will provide unprecedented possibilities and capabilities for parallel functional analysis of tremendous amounts of chemical compounds.
The study of the interaction between chemical compounds and biological targets has dominated modern drug discovery research. Genomic and proteomic studies have indicated the potential existence of ~10,000 druggable targets; however, <500 of these targets have corresponding FDA-approved drugs [1]. Therefore, finding drug candidates by screening large numbers of chemicals against the new targets in a quick and economical fashion has become one of the most challenging tasks for today’s drug discovery process.
Over the past decade, HTS has become a powerful tool for the identification of active compounds and pharmacophores against specific biologicaltargets. At the same time, high-throughput synthesis of small molecules is also widely practiced to generate large numbers of chemicals in a short time period. As a result, different HTS methods have been introduced and widely utilized. To further increase the throughput and reduce the cost of chemicals and targets, the miniaturization of biological assays has been the trend in today’s assay development and laboratory automation. The conventional microtiter-plate-based assays have moved away from the 96-well format to the 384-well system, or even the 1536-well (or more) platform, or microfluidic systems [2]. However, the cost associated with replacing old systems by high-throughput and high-precision automations is substantial.
The success of microarray technology in genomic and proteomic research has given new impetus to miniaturization, providing unprecedented possibilities and capabilities for parallel functional analyses of hundreds or even thousands of bioentities. Microarrays, such as DNA and protein microarrays, have played indispensable roles in genomic and proteomic research [3–7]. By contrast, the development of chemical microarrays, also called chemical compound microarrays or small-molecule microarrays, capable of evaluating a large number of chemical structures against hundreds of biological targets, has been a slow process [8–12]. One of the key problems of developing such chemical microarrays is that compounds with different structures and properties are traditionally screened in a solution phase, and individual reactions need to be isolated in wells. When reactions are reduced to microarray size no existing automatic liquid handling system can process these reactions individually, separately and sequentially if each reaction is still kept in a solution phase. Therefore, chemical microarray technology, which directly links chemicals to the chip surface, was introduced [13–16]. However, compared with DNA and protein microarrays, no general linking technology can be established to immobilize compounds processing different structures and functional groups on the same chip surface. To avoid the potential complicated immobilization process, microarray with dry compounds has been developed [17,18]. This technology can deposit any chemical compound library on the same chip surface in a dry form; however, different compounds possessing different dissolution rates can create problems with reaction uniformity and endpoint data comparison. Recently, chemical microarray technology has also been successfully used for cell-based assays by infusing small molecules with biodegradable polymer [19], but the different physical properties of chemical compounds would also result in different diffusion endpoints. Given that there are >1040 potential low molecular weight chemical compounds available in theory [20], one might imagine that many difficulties could be encountered with the approaches outlined above.
To avoid these problems, a new solution-phase chemical compound microarray has been created [21]. In this format each compound is individually arrayed on a glass surface – with a reaction buffer containing a low concentration of glycerol to prevent evaporation. Therefore, as in conventional well-based screening, the chemical compounds and biological targets are always in reaction solutions, which are activated with biological targets by delivering analytes using aerosol deposition technology [21–23]. With this approach all compounds [21–25] and peptide libraries [26,27] can be microarrayed, and activated by biological targets in the form of pure protein [21–27] or cell lysate [24]. In this review, we will discuss the recent development of all forms of chemical microarrays as well as their applications and limitations in direct drug screening and development.
Chemical microarray with immobilization technology
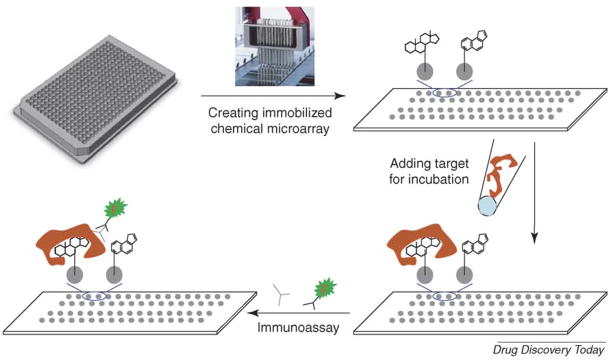
Chemical arrays could have arisen as a byproduct of split-pool techniques, following the development of high-throughput library synthesis in the early 1990s [12]. Lam and co-workers [28] have described this ‘one-bead one-compound’ approach of generating a peptide library on 80 μm beads. Libraries were screened against receptors conjugated to fluorescent dyes or reporter enzymes. This approach eventually merged with the new DNA microarray technology in the late 1990s, creating the first type of chemical microarray with many pioneering studies performed in Schreiber’s laboratory at Harvard [13–16]. This platform, usually called a small-molecule microarray (SMM), uses diverse linking techniques to immobilize chemical compounds with specific moieties covalently coupled to the surface of glass [8,9], and then screens the compounds against chosen biotargets (Figure 1).
FIGURE 1. Small-molecule microarray.
Chemical compounds synthesized with the same linking functional group are arrayed and covalently immobilized on the surface of microarray chips using a standard microarrayer. Biological targets in cell lysates or in purified forms are added to the chips, this is followed by several washing steps to eliminate non-specific and weak binding. The compounds that bind to the target with high affinity are then identified by immunoassays (the chemical structures shown in all figures are for illustration purposes only).
The essential requirements for this form of chemical microarray are specific reactive groups on both the microarray surface and library. For example, MacBeath et al. [13] immobilized compounds containing a thio group to the surface of maleimide-derivatized glass. Hergenrother et al. [14] generated an alcohol-reactive glass surface for immobilizing compounds containing hydroxyl functionalities. Winssinger et al. [29] have covalently linked small molecules with a peptide nucleic acid tag, and then arrayed them on an oligonucleotide surface. With this approach, Winssinger et al. [30] have screened a small-molecule microarray against proteases in crude cell lysates, and identified caspase inhibitors by detecting caspase activation. Kohn et al. [31] have coupled azide-functionalized molecules on a phosphane-derivatized microarray and Dillmore et al. [32] have developed photochemistries for linking chemicals.
Kuruvilla and co-workers have published an interesting study using this approach [15]. They have synthesized 3780 structurally complex 1,3-dioxane small molecules using ‘one-bead one-stock-solution’ technology, and microarrayed them on glass slides. This SMM was then screened with fluorescently (Cy5) labeled yeast prion-like protein Ure2p, a repressor of transcription factors for Nil1p and Gln3. One out of eight hits from the primary binding screening had activity in a secondary cell-based screening assay, with a dissociation constant (Kd) value of 18.1 μM. After SAR analysis, a small group of derivatives was synthesized, based on the lead structure, and a second-generation compound emerged with better solubility and an improved Kd of 7.5 μM. Interestingly, this compound selectively activated only the Nil1p-dependent glucose-sensitive signaling pathway, but not the Gln3-dependent nitrogen nutrient-responsive signaling pathway. This research effectively utilized DNA microarray and chemical microarray technologies and demonstrated that chemical microarray technology can be a powerful tool for selecting chemical ligands with high binding activities to perturb bioactivities.
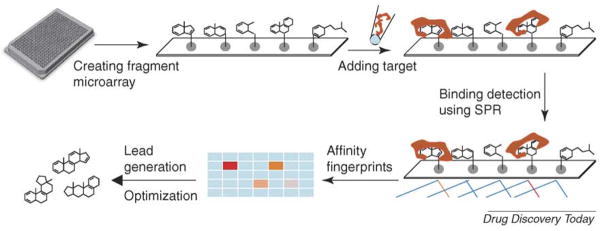
However, immobilizing a large diversified chemical library has many limitations. As a result, scientists from Graffinity Pharmaceuticals have created a fragment chemical microarray system that immobilizes a few thousand drug fragments or pharmacophores (http://www.graffinity.com/t_elements.php#). In this unique platform, the low-complexity drug-like compounds are synthesized and immobilized on microarrays with a specially designed self-assembled monolayer (SAM), and a thin gold layer provides the base for this SAM and surface plasmon resonance (SPR) detection. The microarray chips are then treated with the biological target without labeling. A Plasmon Imager®, based on the phenomenon of SPR, is then used to record the mass change generated by soluble proteins bound on immobilized chemical surfaces. A wavelength shift in the SPR setting, corresponding to the amount of proteins bound to the chemical surface, helps to create the protein–ligand affinity fingerprints. With this approach, drug-like fragments with high binding affinity towards protein targets were quickly identified [33] (Figure 2). Compared with more-traditional small-molecule screening, binding affinity is generally weaker in this system (Kd values in μM range) but the percentage of fragments capable of binding to particular proteins is higher. The quality of the lead compounds will also depend heavily on how the initial screening data are interpreted. Therefore, Graffinity has created a whole process that is termed RAISE® (rapid array informed structure evolution), which includes SAR analysis for identification and development of drug candidates from their screening database (Figure 2). This label-free affinity binding assay can detect weak protein–ligand interactions and reduce labor and reagent costs by simplifying the initial screening work. However, because of the use of chemical fragments, this technology cannot utilize the tremendous values of existing chemical libraries. There is also no clear answer as to how many fragments screened will be enough, therefore this approach can potentially lose leads because of biased microarrays, created by lacking certain chemical fragments.
FIGURE 2. Fragment chemical microarray.
Thousands of chemical fragments are synthesized and immobilized on microarrays, then coated with a thin layer of gold. After treating the chips with target proteins, a surface plasmon resonance (SPR)-based Plasmon Imager® is used to record the mass change when soluble proteins bind immobilized chemicals. Wavelength shifts in SPR corresponding to the amount of protein binding to the chemical surface, creating protein–ligand affinity fingerprints. After SAR analysis, the first generation of drug-like compounds and analogs are synthesized and tested, and lead candidates will then be further optimized.
Immobilized chemical microarrays, such as the SMM and fragment chemical microarray technology, are particularly useful in probing binding partners or identifying targets. Microarrays containing thousands of compounds or pharmacophores, at the microliter level, can be screened in parallel with multiple biotargets. However, the major disadvantage of this platform is that it requires specific chemistry for coupling, limiting the use of existing chemical libraries. In addition, factors such as the length and flexibility of linkers, compound binding orientation, spatial hindrance and microarray surface properties also affect target binding. Nevertheless, in combination with proteomic and genomic research, lead compounds perturbing a biological signaling pathway can be rapidly identified with this technology [15,33].
Dry chemical microarray
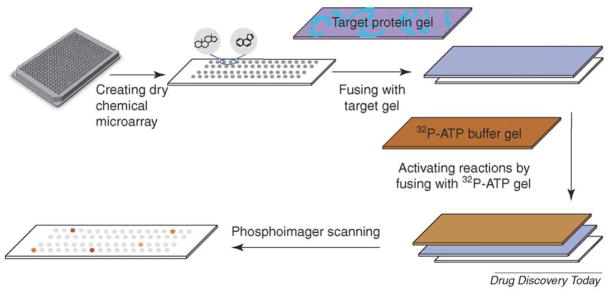
To process any chemical library and avoid the limitation of immobilization, scientists from Abbott Laboratories generated a new form of chemical microarray by putting compounds dissolved in dimethyl sulfoxide (DMSO) on polystyrene sheets, and then drying them. These chemical microarrays were then processed by diffusion toward targets immobilized in agarose gels (Figure 3); this process is called microarrayed compound screening (μARCS) [17,18].
FIGURE 3. Dry chemical microarray.
Chemical compounds are arrayed and dried on polystyrene sheets that have the same footprint as a 384-well plate. An agarose gel embedded with enzyme and substrate is applied to the surface of the array. After a short incubation, a second gel containing radioactive ATP is applied to initiate the biological reaction. The final reactions are detected using a standard phosphorimager.
David and colleagues [17] first reported this technology to identify inhibitors for HIV integrase (using agarose matrices to contain and introduce various reagents involved in the assay). In this study, a library of ~250,000 compounds from different structural classes was screened and novel inhibitors from different classes identified. Using similar approaches, Abbott scientists have applied the μARCS technology to different enzymes. For example, Gopalakrishnan and co-workers [18] used the same technology to screen for caspase inhibitors. First, 8640 discrete compounds were spotted and dried onto a polystyrene sheet the same size as a conventional microtiter plate. Then, an agarose gel containing caspase-3 was laid on top of the compounds, followed by a second agarose gel with peptide substrate placed on top of the enzyme gel – starting the reaction. Groebe et al. [34] and Freiberg et al. [35] have reported other protease- and kinase-based screening approaches – using radiometric detection methods for kinases (Figure 3). One interesting application of this technology was to use it for identifying agonists of G-protein-coupled receptors [36]. HEK-293 cells coexpressing human dopamine D(4.4) receptor and chimeric G(αqo5) protein, preloaded with the Ca2+ indicator Fluo-4 acetoxymethyl ester, were cast into a 1% agarose gel, then placed above the compound sheet. Agonists will increase the cytosolic Ca2+ level, which will result in enhanced fluorescence upon binding to the intracellular Fluo-4.
Discovery Partners International (DPI) has licensed the mARCS technology from Abbott for commercialization, and further improved the whole process from microarray spotting and agarose gel casting to imaging and data analysis [37]. At DPI, scientists arrayed a total of 4608 chemical compounds (20 nl), in duplicate on a standard array that had a footprint of a microtiter plate, using the modified Gilson Constellation™ system with syringe-solenoid dispensing technology. With this method, the compounds are dried immediately after spotting and can be stored in a gas-tight inert atmosphere for future assays. Although the platform has been tested for many diversified targets, including enzymes and receptors [37], one of the major problems with this approach is that the rate and capability of resolubilization and diffusion of different classes of dry compounds will complicate the dynamic range of arrayed compounds versus bioavailable compounds. To address this question, Cheng et al. [38] have characterized the diffusion of 59 compounds with diverse properties by using liquid chromatography–mass spectrometry (LC–MS) to determine how much compound was left on the polystyrene sheet after incubation with the agarose gel for defined periods of time. As expected, the study found that compounds of lower ClogP showed a higher rate of transfer to agarose gels, whereas other physical properties, such as molecular weight, size, acidity and H-bonding properties, have less effect on diffusion rate. The study demonstrated that the majority of the 59 compounds had >20% transfer after a 10 min incubation with agarose gels, providing sufficient amounts of compounds for screening purposes. The study also revealed that certain compounds might have transfer problems, thus yielding insufficient concentrations of compound for interaction with biotargets. This is a particular problem for in vitro screening campaigns in which fast enzyme reactions, weak inhibitors, weak receptor binders, hydrophobic compounds and complex natural products might be involved. Differential transfer rates in HTS will detect inhibitors at different kinetic time points, resulting in incomparable inhibition profiles. This would be a major challenge when IC50 profiling a group of compounds against the same target, under the same reaction conditions.
Creating new solution-phase chemical microarray technology for HTS
The previously described chemical microarray technologies are all solid-phase formats; because chemical compounds are arrayed on a well-less and wall-less flat surface there is no liquid handler activating each compound individually as in traditional microtiter-plate-based reactions. To perform microarray reactions comparable with reactions done in microtiter plates, solution-phase chemical microarrays have to be developed. One of the first groups to try to combine peptide microarray technologies with conventional liquid handling methods to transfer kinase reactions to microarrays was that of Houseman and colleagues [39]. They first coated glass-surface microarrays with a peptide substrate for kinases, and then prepared a kinase reaction solution, in conventional 384-well plates, that included the chemical compound, the kinase and ATP. The kinase reaction was initiated by transferring the kinase reaction solution from the 384-well plate to the substrate-coated microarrays. This study demonstrated that the phosphorylation reaction, and its inhibition by chemical compounds, could be detected in this array format. However, this approach is not an ideal solution because the procedure requires premixing reagents in wells (on plates) and then transferring them to a substrate chip, which, of course, demands more labor and time in liquid handling than conventional systems.
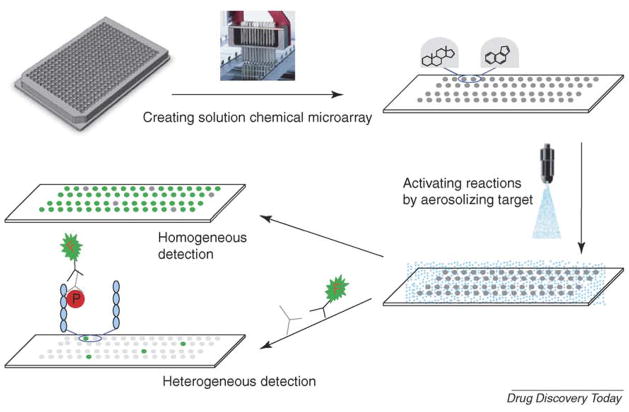
In an attempt to develop a true solution-phase chemical microarray that can perform the same assays as conventional microtiter plates, Diamond’s group from the University of Pennsylvania (USA) created a solution-phase chemical microarray using glycerol as an anti-evaporating reagent that is mixed with chemical compounds to be arrayed on the microarray surface. To activate this solution-phase chemical microarray they adapted an aerosol deposition technology that converts biological targets into a fine mist, which can cover the top of the whole microarray simultaneously [21]. Exploiting this technology as a new tool for drug screening, Reaction Biology has licensed and developed solution-phase chemical microarray technology into a commercial product, the DiscoveryDot™ chemical microarray drug screening platform (Figure 4) [22,23]. DiscoveryDot™ is based on the principle of microarraying chemical libraries on a glass slide (1 inch × 3 inches) in formats with >6000 compounds per microarray –in a nonvolatile glycerol-based format compatible with −80 °C storage. Each compound forms an individual well-less reaction center in a total volume of 1 nl. The biochemical reactions are then started by using aerosol deposition technology, and the reaction products are detected with a laser scanner or imager (Figure 4). One of the key elements of DiscoveryDot™ is that the chemical compounds and targets are in solution throughout the whole process; therefore, all existing chemical libraries can be screened by this platform, making this technology the only universal chemical microarray to date. Because this solution-phase platform mimics the conventional large volume well-based system, the environment and mechanism of biochemical reactions on this microarray are very similar to the well-based reactions – but with higher efficiency for profiling multiple biotargets. For example, we have microarrayed the library of pharmacologically active compounds (LOPAC) on polylysine-coated slides, poly-(Glu-Tyr) peptide-coated slides, and dephosphorylated myelin basic protein (MBP)-coated slides, in parallel. The polylysine slides were then used for protease screening; the poly-(Glu-Tyr) peptide-coated slides were used for screening against Src, a tyrosine kinase, and the MBP-coated slides were used for screening against p38α, a serine–threonine protein kinase. Each round of screening for proteases and for Src has yielded many inhibitors (unpublished), but p38α screening has only yielded one particularly strong inhibitor, 4-(4-fluorophenyl)-2-(4-hydroxyphenyl)-5-(4-pyridyl)-1H-imidazole (SB202190). When the same reaction was performed in a 384-well plate format the same compound was identified with similar potency [40].
FIGURE 4. Solution-phase chemical microarray.
Existing chemical compounds in assay buffer containing dimethyl sulfoxide (DMSO) and 10% glycerol are arrayed on the surface of the microarray. The compounds are always in solution without any chemical linking to the microarray. For homogeneous reactions, the biological target and substrate are added into each reaction dot by aerosol deposition technology, and, in the case of kinases, the reactions are initiated by spraying on ATP. The reaction products are detected by a laser scanner or imager. For heterogeneous ELISA-based reactions, such as kinase assays using whole proteins as substrates, the substrate is immobilized on the microarray surface first, before compounds are microarrayed. The kinase is then sprayed on to the array and the reactions initiated by spraying on ATP. The reactions are then detected by conventional ELISA protocols.
DiscoveryDot™ technology has been used in general proteomic research and for drug discovery. Gosalia and Diamond [21] published this technology first, showing its potential power to multiplex enzymatic reactions with microarrays containing different peptide substrates for a variety of proteases. They identified small molecules that were capable of inhibiting caspases. Similarly, Ma et al. [24] have also identified specific small molecules that inhibited different caspase isoforms. The caspase peptide-substrates microarray could also be activated by cell lysate that had been pretreated with apoptosis inducer [24]. This specially designed peptide microarray can be used for identifying compounds capable of modulating apoptosis pathways.
Recently, using a 722-member peptide library microarray of fluorogenic protease substrates with the general structure Ac-Ala-X-X-(Arg/Lys)-coumarin, Gosalia et al. [25] profiled human blood serine proteases (human thrombin, factor Xa, plasmin and urokinase plasminogen activator). The results provided a complete map of protease specificity for all of the substrates tested, and identified cooperative interactions between substrate subsites. Gosalia et al. [26] have further profiled these peptides with 13 serine proteases (activated protein C, plasma kallikrein, factor VIIa, factor IXaβ, factor XIa, factor αXIIa, activated complement C1s, C1r and D, tryptase, trypsin, subtilisin Carlsberg, and cathepsin G) and 11 papain-like cysteine proteases (cathepsins B, H, K, L, S and V, rhodesain, papain, chymopapain, ficin, and stem bro-melain). This study was the first to report the substrate specificity of rhodesain, a papain-like cysteine protease expressed in Trypanasoma brucei rhodesiense which is a parasitic protozoan responsible for causing sleeping sickness.
However, the greatest potential of this technology is for drug screening, primary for hit identification and confirmation and secondary for lead optimization. Ma et al. [22] have reported the commercial side of the technology by revealing the details of aerosol technology and its application in creating homogeneous-based screening formats for enzymes such as caspases, matrix metalloproteinases, thrombin, factor Xa, histone deacetylases and protein kinase A. Because of the nature of homogeneous reaction systems, time-dependent and dose-dependent reactions can also be performed on microarrays, delivering kinetic parameters such as Km, Vmax, and kcat. In fact, the authors demonstrated that these microarray-generated parameters were quantitatively similar to those derived from well-based assays. For example, both technologies yielded similar IC50 values for caspase-6 when the same reaction conditions were used. Furthermore, taking advantage of miniaturization pays off in larger scale HTS: the total cost of microarray-based reactions could be reduced over 20-fold compared with standard low volume 384-well-based assays [22].
This type of homogeneous-based HTS with proteases has recently reached a milestone with the discovery of a cathepsin L inhibitor, MDL28170, from such a screening campaign. This compound could potentially be used in the treatment of severe acute respiratory syndrome (SARS), because it has been proposed that SARS infection is a three-step process that involves receptor-binding and induced conformational changes in the S glycoprotein, followed by proteolysis by cathepsin L and subsequent activation of membrane fusion in the endosomal compartment. Inhibition of cathepsin L with MDL28170 was shown to block the entry of SARS coronavirus (SARS-CoV) into host cells [27]. This study opened a new target class for drug development that is therapeutically valuable to fight against the SARS virus.
Horiuchi et al. [23] recently published the application of DiscoveryDot™ in kinase primary screening and profiling with a heterogeneous format, utilizing traditional ELISA for the reporter. In this format, the kinase substrate (either peptide or native protein) was immobilized on the surface of the chip before the chemical library was arrayed on top of this. Then, the biochemical reactions were activated by aerosol deposition of the enzyme. The product of phosphorylated peptide or protein was then detected by an antibody that recognizes the phosphorylated products. For example, using biotinylated poly-(Glu-Tyr) peptide as a substrate, the p60c-src assay on microarray produced a signal-to-noise ratio of 36.3 and a Z′ factor of 0.63 for HTS, in addition to accurate enzyme kinetic parameters ( ) and IC50 values for staurosporine (210 nM) and PP2 (326 nM) at 10 μM ATP. Similarly, using native MEK protein as a substrate, the nanoliter reactions of b-Raf on microarray were inhibited by GW5074 at an expected IC50 of 9 nM. Compound selectivity profiling against multiple targets, b-Raf (V599E), kinase insert domain receptor (KDR), Met, Flt-3 (D835Y), Lyn, epidermal growth factor receptor (EGFR), platelet-derived growth factor receptor β (PDGFRβ) and Tie2, was also carried out.
DiscoveryDot™ has been commercialized mainly for biochemical-based assays, ELISA-based detection for protein–protein interaction and antibody library screening. Binding-based assays for receptor–ligand interactions (with fluorescent polarization as the detection method) are also under development.
New chemical microarray for cell-based assays
The chemical microarray formats mentioned above have mainly been used for screening chemical-binding proteins and potential drugs using biochemical assays [13–18,21–27], but cell-based assays are potentially capable of being developed. For example, Ziauddin and Sabatini [41] have developed a reverse-transfection microarray by using gelatin to immobilize cDNA vectors on regular glass-surface chips (upon which cells can be grown) to study gene function. Using a similar approach, Stockwell and Sabatini’s groups [19] have recently published the first chemical microarray format for cell-based high-content screening. In this study, biodegradable polylactide-co-glycolide (PLGA®) copolymer was used to impregnate the microarray surface with 200 μm diameter discs for each compound, and cells were then seeded on top of these compounds that were slowly diffused out to affect proximal cells. PLGA®, which has been used for delivering protein drugs, enabled the controlled release of proteins and small molecules through a combination of drug diffusion and polymer erosion. This chemical microarray technology has the potential for high-throughput cell-based assays, but must overcome a few obstacles before becoming useful as a broad platform. For example, finding one polymer for imbedding and releasing possibly millions of chemical compounds (with different physical properties) is a challenge. In addition, the release and stability of chemicals will affect the assays and comparison studies. Furthermore, the authors have noticed that chips printed >72 h before use are not as active as freshly printed chips. Data analysis is another challenge – because the concentration of compounds in cells decreases from the center of the array tothe skirt. As a result, the effects of the compounds are also decreased at the outer edges, increasing the burden of data analysis in addition to the already complex format for microtiter-plate-based screening.
Conclusions
The new development of chemical microarray technology, capable of analyzing hundreds of thousands of chemicals with multiple biological targets in parallel, will further advance the HTS with its miniaturized reaction and parallel screening systems. Their applications, from target identification to the discovery of lead compounds, have made it a promising platform, useful in many drug discovery processes to reduce cost and shorten cycle time, while increasing productivity.
Another advantage is that these technologies are easily accessed or evaluated, scientists from academia and industry with access to a DNA and protein microarray facility could perform some of the assays mentioned in this review, with certain modification, based on instrument availability. In addition, the markets for arrayers, assay detectors and data analysis are readily available (Table 1). However, for one to take advantage of these chemical microarrays, a correct platform has to be selected. The summary in Table 2 can be used as a general guide for biochemical assays. For example, the immobilized microarray platform is preferred for fishing biological targets from a mixture of biofluid. Either dry forms or gel-pad forms can be used for almost all chemical and enzyme reactions, and cell-based assays are possible by using the slow-release property of polymers used for embedding the chemicals; however, the difference in rates of compound dissolution, diffusion and stability makes it difficult to observe uniform reactions. The solution-phase chemical microarray, which mimics the conventional well-based assays, can be used for homogeneous and heterogeneous assays with virtually all chemical compounds, enzymes and protein–protein interactions. However, the assay formats are limited at present by the availability of suitable detectors. This will continue to limit assay choice until detectors are developed with the necessary resolution and sensitivity to recognize the μm reaction centers.
TABLE 1.
Sample instruments for assembling chemical microarray reactions
| Process | Device | Technology | Manufacture | Specificity |
|---|---|---|---|---|
| Arrayer or spotter | QArray microarrayer | Solid pin | http://www.genetix.com | Contact nanoliter pin printing, higher coefficient of variation (CV), humidity control, fast, great for making multiple chips with large chemical library. |
| NanoPrint™ microarrayer | Solid pin | http://www.arrayit.com | Contact nanoliter pin printing, humidity control, automatic plate handler, higher CV, fast, great for making multiple chips with large chemical library. | |
| Manual arrayer and pin tool | Solid pin | http://www.vp-scientific.com | Contact nanoliter pin printing, higher CV, great for research. | |
| Gilson Constellation | Solenoid dispensing | http://www.gilson.com | Non-contact printing, slow, 5–500 nl, low CV. Similar machines are available in most pharmaceutical laboratories. | |
| ArrayJet | Inkjet printing | http://www.arrayjet.co.uk | Non-contact printing, μl range, low CV, good for small chemical library printing. | |
| Piezorray | Piezo printing | http://las.perkinelmer.com | Non-contact printing, slow, pl to nl range, low CV. | |
| Echo 550 | Acoustic | http://www.labcyte.com | With potential for nl range spotting. | |
| ATS-100 | Acoustic | http://www.edcbiosystems.com | With potential for nl range spotting. | |
| Activator | Sprayer | Aerosol | http://www.reactionbiology.com | Fast activation, low reagent consumption. |
| Gel caster | SDS-page gel apparatus | http://www.bio-rad.com | Available in all biology laboratories. | |
| Imager and data analysis | NovaRay® | Charge-coupled device(CCD) camera imager | http://www.alphainnotech.com | Fluorescence intensity, read microarray and plate, 4 μm resolution. |
| ViewLux™ | CCD camera imager | http://www.perkinelmer.com | Fluorescence polarization, fluorescence intensity, time-resolved fluorescence, luminescence and absorbance assays. Low resolution for high-density microarray. | |
| GenePix® | Laser scanner | http://www.moleculardevices.com | Fluorescence intensity, four lasers, 5–100 μm resolution, slide autoloader. | |
| LS Reloaded™ | Laser scanner | http://www.tecan.com | Read microarray and plate, fluorescence intensity, four colors, 4–40 μm resolution, adjustable angle of laser beam for dual color scanning. | |
| Typhoon™ | Laser scanner | http://www4.amershambiosciences.com | Applicable for fluorescence, luminescence and isotopic imaging, four colors, 10–100 μm resolution. | |
| IsoCyte™ | Laser scanner | http://www.blueshiftbiotech.com | A plate reader with four channel intensity, two channel anisotropy, but with potential of fluorescence intensity and polarization detection for microarray. |
TABLE 2.
Chemical microarray platform comparisons
| Small-molecule microarray | Fragment chemical microarray | Dry chemical microarray | Solution-phase chemical microarray | ||
|---|---|---|---|---|---|
| Chemical library | Specific synthesis | Specific synthesis | Any library | Any library | |
| Assays | Protein binder | Immunoassay detection | Label-free surface plasmon resonance(SPR) detection | ELISA, isotopic | ELISA, isotopic, time-resolved fluorescence |
| Protein–protein interrupter | Difficult to perform | Difficult to perform | Difficult to perform | ELISA, isotopic, fluorescence labeling, time-resolved fluorescence | |
| Enzyme inhibitor | Difficult to perform | Difficult to perform | Easy to use; low density, but the resolubilization of compounds might affect reaction kinetics. | Easy to use, high-density array, homogeneous and heterogeneous assays, time dependent and endpoint assays. | |
| Data analysis | DNA microarray software compatible | Specifically designed | Spot finder compatible | DNA microarray software compatible |
Acknowledgments
This work was funded in part by multiple small-business innovation research (SBIR) grants from the National Cancer Institute (NCI) and the National Institute of Dental and Craniofacial Research (NIDCR) of NIH, and a RO1 grant from NIH Roadmap initiative to Dr Ma. The authors wish to thank Mr Yuan Wang for his skillful work and Dr Scott Diamond for his scientific input.
References
- 1.Drews J. Drug discovery, a historical perspective. Science. 2000;287:1960–1964. doi: 10.1126/science.287.5460.1960. [DOI] [PubMed] [Google Scholar]
- 2.Sundberg SA. High-throughput and ultra-high-throughput screening: solution- and cell-based approaches. Curr Opin Biotechnol. 2000;11:47–53. doi: 10.1016/s0958-1669(99)00051-8. [DOI] [PubMed] [Google Scholar]
- 3.Zhu H, et al. Proteomics. Annu Rev Biochem. 2003;72:783–812. doi: 10.1146/annurev.biochem.72.121801.161511. [DOI] [PubMed] [Google Scholar]
- 4.Stoll D, et al. Protein microarrays, applications and future challenges. Curr Opin Drug Discov Devel. 2005;8:239–252. [PubMed] [Google Scholar]
- 5.Poetz O, et al. Protein microarrays, catching the proteome. Mech Ageing Dev. 2005;126:161–170. doi: 10.1016/j.mad.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 6.Smith MG, et al. Global analysis of protein function using protein microarrays. Mech Ageing Dev. 2005;126:171–175. doi: 10.1016/j.mad.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 7.Templin MF, et al. Protein microarray technology. Drug Discov Today. 2002;7:815–822. doi: 10.1016/s1359-6446(00)01910-2. [DOI] [PubMed] [Google Scholar]
- 8.Lam KS, Renil M. From combinatorial chemistry to chemical microarray. Curr Opin Chem Biol. 2002;6:353–358. doi: 10.1016/s1367-5931(02)00326-5. [DOI] [PubMed] [Google Scholar]
- 9.Uttamchandani M, et al. Small molecule microarrays, recent advances and applications. Curr Opin Chem Biol. 2005;9:4–13. doi: 10.1016/j.cbpa.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 10.Stockwell BR, et al. Exploring biology with small organic molecules. Nature. 2004;432:846–854. doi: 10.1038/nature03196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walsh DP, Chang YT. Recent advances in small molecule microarrays, applications and technology. Comb Chem High Throughput Screen. 2004;7:557–564. doi: 10.2174/1386207043328427. [DOI] [PubMed] [Google Scholar]
- 12.He XG, et al. Ligand discovery using small molecule microarrays. J Pharmacol Exp Ther. 2005;313:1–7. doi: 10.1124/jpet.104.076943. [DOI] [PubMed] [Google Scholar]
- 13.MacBeath G, et al. Printing small molecules as microarrays and detecting protein-ligand interactions en masse. J Am Chem Soc. 1999;121:7967–7968. [Google Scholar]
- 14.Hergenrother PJ, et al. Small-molecule microarrays: covalent attachment and screening of alcohol-containing small molecules on glass slides. J Am Chem Soc. 2000;122:7849–7850. [Google Scholar]
- 15.Kuruvilla FG, et al. Dissecting glucose signalling with diversity-oriented synthesis and small-molecule microarrays. Nature. 2002;416:653–657. doi: 10.1038/416653a. [DOI] [PubMed] [Google Scholar]
- 16.Vetter D. Chemical microarrays, fragment diversity, label-free imaging by plasmon resonance–a chemical genomics approach. J Cell Biochem Suppl. 2002;39:79–84. doi: 10.1002/jcb.10408. [DOI] [PubMed] [Google Scholar]
- 17.David CA, et al. Microarray compound screening (microARCS) to identify inhibitors of HIV integrase. J Biomol Screen. 2002;7:259–266. doi: 10.1177/108705710200700309. [DOI] [PubMed] [Google Scholar]
- 18.Gopalakrishnan SM, et al. Application of Micro Arrayed Compound Screening (microARCS) to identify inhibitors of caspase-3. J Biomol Screen. 2002;7:317–323. doi: 10.1177/108705710200700403. [DOI] [PubMed] [Google Scholar]
- 19.Bailey SN, et al. Microarrays of small molecules embedded in biodegradable polymers for use in mammalian cell-based screens. Proc Natl Acad Sci U S A. 2004;101:16144–16149. doi: 10.1073/pnas.0404425101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gribbon P, Sewing A. High-throughput drug discovery, what can we expect from HTS? Drug Discov Today. 2005;10:17–22. doi: 10.1016/S1359-6446(04)03275-1. [DOI] [PubMed] [Google Scholar]
- 21.Gosalia DN, Diamond SL. Printing chemical libraries on microarrays for fluid phase nanoliter reactions. Proc Natl Acad Sci U S A. 2003;100:8721–8726. doi: 10.1073/pnas.1530261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma H, et al. Nanoliter homogeneous ultra high throughput screening microarray for lead discoveries and IC50 profiling. Assay Drug Dev Technol. 2005;3:177–187. doi: 10.1089/adt.2005.3.177. [DOI] [PubMed] [Google Scholar]
- 23.Horiuchi KY, et al. Microarrays for the functional analysis of the chemical-kinase interactome. J Biomol Screen. 2006;11:48–56. doi: 10.1177/1087057105282097. [DOI] [PubMed] [Google Scholar]
- 24.Ma H, et al. Frontiers in Biochip Technology. Springer; New York: 2006. A homogeneous microarray for enzymatic functional assays; pp. 3–18. [Google Scholar]
- 25.Gosalia DN, et al. Profiling serine protease substrate specificity with solution phase fluorogenic peptide microarrays. Proteomics. 2005;5:1292–1298. doi: 10.1002/pmic.200401011. [DOI] [PubMed] [Google Scholar]
- 26.Gosalia DN, et al. High throughput substrate specificity profiling of serine and cysteine proteases using solution-phase fluorogenic peptide microarrays. Mol Cell Proteomics. 2005;4:626–636. doi: 10.1074/mcp.M500004-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.Simmons G, et al. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc Natl Acad Sci U S A. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lam KS, et al. A new type of synthetic peptide library for identifying ligand-binding activity. Nature. 1991;354:82–84. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- 29.Winssinger N, et al. From split-pool libraries to spatially addressable microarrays and its application to functional proteomic profiling. Angew Chem Int Ed. 2001;40:3152–3155. doi: 10.1002/1521-3773(20010903)40:17<3152::AID-ANIE3152>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 30.Winssinger N, et al. Profiling protein function with small molecule microarrays. Proc Natl Acad Sci U S A. 2002;99:11139–11144. doi: 10.1073/pnas.172286899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kohn M, et al. Staudinger ligation: a new immobilization strategy for the preparation of small-moleculearrays. Angew Chem Int Ed Engl. 2003;42:5830–5834. doi: 10.1002/anie.200352877. [DOI] [PubMed] [Google Scholar]
- 32.Dillmore WS, et al. A photochemical method for patterning the immobilization of ligands and cells to self-assembled monolayers. Langmuir. 2004;20:7223–7231. doi: 10.1021/la049826v. [DOI] [PubMed] [Google Scholar]
- 33.Dickopf S, et al. Custom chemical microarray production and affinity fingerprinting for the S1 pocket of factor VIIa. Anal Biochem. 2004;335:50–57. doi: 10.1016/j.ab.2004.08.033. [DOI] [PubMed] [Google Scholar]
- 34.Groebe DR, et al. Putting thought to paper, a microARCS protease screen. J Biomol Screen. 2003;8:668–675. doi: 10.1177/1087057103258587. [DOI] [PubMed] [Google Scholar]
- 35.Freiberg G, et al. Utilization of microarrayed compound screening (microARCS) to identify inhibitors of p56lck tyrosine kinase. J Biomol Screen. 2004;9:12–21. doi: 10.1177/1087057103259667. [DOI] [PubMed] [Google Scholar]
- 36.Gopalakrishnan SM, et al. A cell-based microarrayed compound screening format for identifying agonists of G-protein-coupled receptors. Anal Biochem. 2003;321:192–201. doi: 10.1016/s0003-2697(03)00425-1. [DOI] [PubMed] [Google Scholar]
- 37.Hoever M, Zbinden P. The evolution of microarrayed compound screening. Drug Discov Today. 2004;9:358–365. doi: 10.1016/S1359-6446(04)03037-5. [DOI] [PubMed] [Google Scholar]
- 38.Cheng X, et al. Compound transfer efficiency from polystyrene surfaces, application to microarrayed compound screening. J Biomol Screen. 2005;10:293–303. doi: 10.1177/1087057104272498. [DOI] [PubMed] [Google Scholar]
- 39.Houseman BT, et al. Peptide chips for the quantitative evaluation of protein kinase activity. Nat Biotechnol. 2002;20:270–274. doi: 10.1038/nbt0302-270. [DOI] [PubMed] [Google Scholar]
- 40.Horiuchi KY, et al. Biochemical microarrays for studying chemical biology interaction: DiscoveryDot™ technology. Chem Biol Drug Des. 2006;67:87–88. doi: 10.1111/j.1747-0285.2005.00326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ziauddin J, Sabatini DM. Microarrays of cells expressing defined cDNAs. Nature. 2001;411:107–110. doi: 10.1038/35075114. [DOI] [PubMed] [Google Scholar]