Abstract
Although the impact of microRNAs (miRNAs) in development and disease is well established, understanding the function of individual miRNAs remains challenging. Development of competitive inhibitor molecules such as miRNA sponges has allowed the community to address individual miRNA function in vivo. However, the application of these loss-of-function strategies has been limited. Here we offer a comprehensive library of 141 conditional miRNA sponges targeting well-conserved miRNAs in Drosophila. Ubiquitous miRNA sponge delivery and consequent systemic miRNA inhibition uncovers a relatively small number of miRNA families underlying viability and gross morphogenesis, with false discovery rates in the 4–8% range. In contrast, tissue-specific silencing of muscle-enriched miRNAs reveals a surprisingly large number of novel miRNA contributions to the maintenance of adult indirect flight muscle structure and function. A strong correlation between miRNA abundance and physiological relevance is not observed, underscoring the importance of unbiased screens when assessing the contributions of miRNAs to complex biological processes.
The last decade in biomedical sciences has brought renewed appreciation for the ancient world of RNAs and unanticipated dimensions of genome regulation by non-coding RNAs. In particular, microRNAs (miRNAs) have emerged as versatile rheostats of gene expression in development and disease. MiRNAs are ~22 nucleotide endogenous non-coding RNAs that bind to specific miRNA recognition elements (MREs) in target RNAs1, 2. The overt consequence of miRNA activity is post-transcriptional silencing of gene expression primarily via RNA decay or translational inhibition3–6.
Despite rapid progress in understanding the molecular mechanisms underlying miRNA biogenesis and mechanisms of action, the biological functions of most miRNAs remain elusive at an organismal level. Aside from experiments in cell culture7, 8, relatively little comprehensive screening has been performed in vivo to assess the functional complexity of the miRNA landscape9–13. This is partly due to a paucity of genome-wide resources for assessing miRNA loss-of-function (LOF). Null miRNA mutations obtained by targeted approaches will be invaluable for analysis of in vivo function13–17. However, comprehensive analyses of miRNA functions in specific tissues and in the dynamic context of the developing organism will also require precise spatiotemporal and gene dosage control. For this reason, we set out to develop a resource for conditional miRNA LOF that could enable unbiased screens for tissue-specific phenotypes.
The specificity of miRNA target recognition and binding is determined by Watson-Crick base pair complementarity. Recent studies suggest the existence of endogenous competitive inhibition regulatory systems that exploit this mechanism to control endogenous miRNA activity18–24. The same concept inspired the design of artificial competitive inhibitors that offer a powerful experimental approach for miRNA LOF studies. Such miRNA “sponge” and “decoy” technologies were successfully used to define a handful of miRNA functions in multiple species and biological contexts25. Mechanistically, this approach relies on the overexpression of transgenes encoding multiple copies of perfect complementary or “bulged” miRNA target sites. Sponge (SP) transcripts sequester miRNAs, blocking access of target transcripts to endogenous target mRNAs and thus creating a knock-down of miRNA activity that closely resembles hypomorphic or null mutants. When transgenically encoded, SPs can be deployed using binary modular expression systems, providing a versatile tool to study miRNA functions in vivo with spatial and temporal resolution26–32.
RESULTS
A transgenic library of conditional miRNA competitive inhibitors
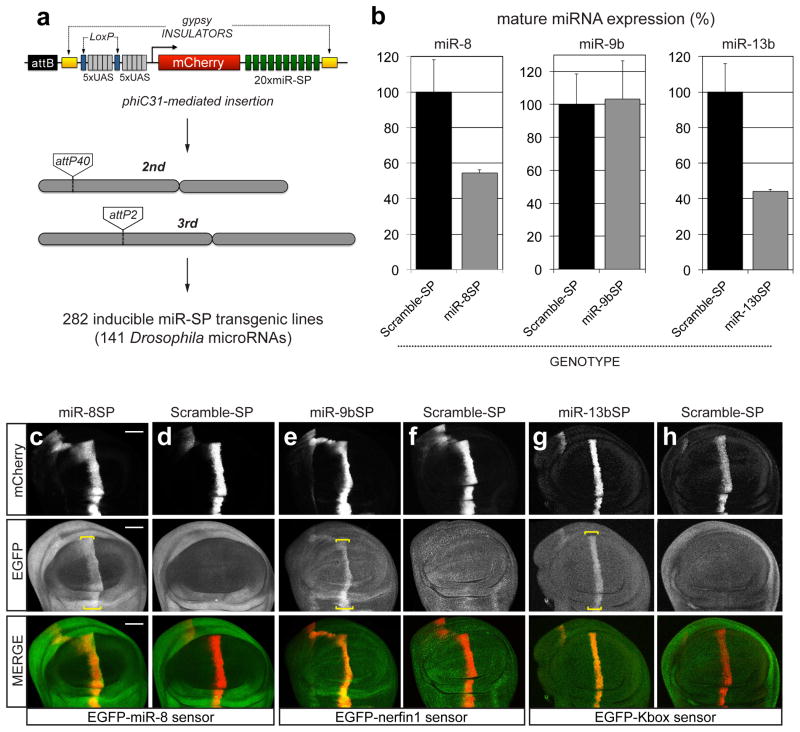
We have previously demonstrated that transgenic SP constructs can faithfully recapitulate known LOF phenotypes for several well-characterized miRNA genes26. Here, we report the first transgenic library of conditional miRNA SPs (miR-SPs), and describe several screens to detect novel miRNA functions required for adult viability, external morphology and flight muscle function in Drosophila. Second generation (GenII) SP constructs were designed and cloned as recently described (Fig. 1a; see Methods)33. A sliding window of 7–8 nucleotides encompassing linker and adjacent SP sequence was scanned to avoid cryptic overlap with existing Drosophila miRNA seed sequences in order to prevent off-target effects (Supplementary Data 1). For the purpose of this study, we focused on a subset of 141 high-confidence miRNAs34, 78 of which display ≥70% sequence similarity between Drosophila and humans35. Using the øC31 site-directed integrase system, we generated 282 transgenic lines carrying one miR-SP transgene on either the 2nd or the 3rd autosome, for each miRNA. Because we observed dose-dependence when comparing expression of single and multiple SP insertions (see below), double transgenic lines were then created for each construct and used throughout this study. Analysis of endogenous miRNA levels following ubiquitous miR-SPGenII expression in larvae (tubulin-Gal4 driver) indicated that the effect of miR-SP expression can vary depending on the miRNA. In some cases we observed no effect on normal miRNA homeostasis (e.g. miR-9b), in other cases a significant decrease in the abundance of mature target miRNAs was apparent (e.g. miR-8 and miR-13b) (Fig. 1b). However, an in vivo miRNA reporter assay in wing imaginal discs revealed that a comparable decrease in miRNA activity is observed in all three cases (Fig. 1c–h).
Figure 1.
A transgenic library of conditional miRNA competitive inhibitors.
(a) Second-generation SP elements consist of twenty miRNA binding sites with mismatches at positions 9–12 placed in the 3′UTR of mCherry under the control of 10 tunable Gal4 UAS binding sites. The entire cassette was cloned in an attB vector containing gypsy insulators. phiC31-mediated positional integration was used to generate a library of 282 inducible lines covering 141 high confidence Drosophila miRNAs, at defined landing sites on the 2nd (attP40) and 3rd (attP2) autosomes. (b) Quantification of endogenous miR-8, miR-9b and miR-13b mature miRNA levels using Taqman qPCR in third instar larvae following ubiquitous expression (tubulin-Gal4) of corresponding miR-SP constructs compared to Scramble controls (Student’s t-test: 13b P=0.02; error bars, sem, n =3 biological replicates with 10 animals per sample). (c–h) Targeted expression of miR-8SP (c, Scale bar is 50μm), miR-9bSP (e) and miR-13bSP (g) with ptc-Gal4 in wing imaginal discs ubiquitously expressing tubulinEGFP-miR-8, tubulinEGFP-nerfin1 and tubulinEGFP-Kbox sensors respectively. Tissue-specific up-regulation of sensor levels was observed in cells along the anterior-posterior boundary of the disc. No change was apparent following expression of a Scramble-SP control (d, f, h).
miRNA regulation of adult viability and external morphology
The importance of miRNA-dependent post-transcriptional regulation in animal development and disease is well documented in a large number of case studies. Surprisingly though, a comprehensive in vivo screen of 95 miRNA genes in C. elegans revealed that most individual miRNAs are dispensable or have limited impact on gross organismal development and innate adult behavior9–11. To obtain an initial assessment of miRNA regulatory activities in Drosophila, we screened our attP2 and attP40 double insertion miR-SP library with the ubiquitous tubulin-Gal4 driver and assayed viability and gross morphological defects in eclosing adults. We also included in our screen two SP lines designed and characterized independently (e.g. bantam36 and miR-1). Lines that displayed significant reduction in viability, defined by a stringent cutoff at a value equal or less than one standard deviation of percent viability across the entire collection, were further validated in triplicate (see Methods).
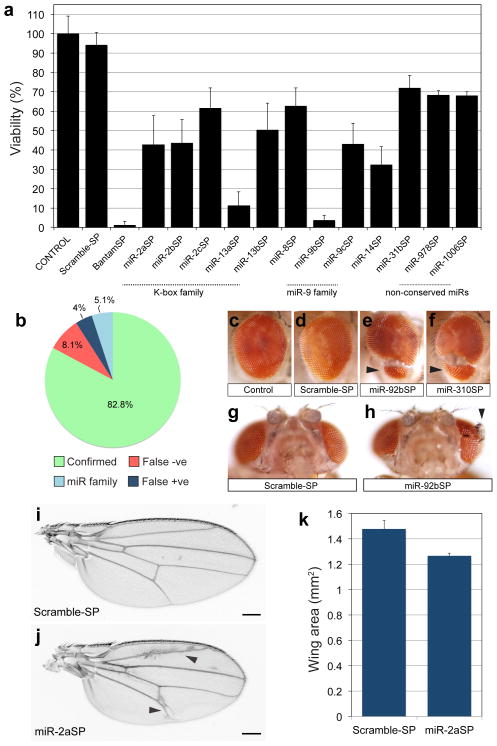
In total, 9% (13/143) of individual miR-SP transgenes rendered a statistically significant viability phenotype, ranging from lethal (0–5% viability) to semilethal (6–50% viability) to subviable (50–70% viability; Fig. 2a). Some lines displayed penetrance below our stringent cut-off that may reflect partial LOF in essential miRNA functions (Supplementary Data 2). In principle, some SPs should be able to inhibit multiple miRNAs in a conserved family. Supporting this argument, several hits in the viability screen belonged to the K-box family (miR-2a, 2b, 2c and miR-13a, 13b) and the miR-9 family (miR-9b, 9c). Previous analysis of K-box miRNA double mutants revealed functional redundancy for lethality37. We tested several of our hits using a complementation assay where the lethal phenotype of a single SP insertion was compared to the same SP carried over a deficiency (Df/+) at the endogenous locus (as described in 26). By this classical criterion, miR-2aSP, miR-2bSP and miR-8SP displayed increased penetrance and thus non-complementing behavior, over Df (Supplmentary Fig. 1a). Interestingly, among miR-9 family members tested (miR-9bSP, miR-9cSP), only miR-9cSP was strongly uncovered by Df (Supplementary Fig. 1a), suggesting some degree of specialization for endogenous miRNA functions within the conserved family.
Figure 2.
Drosophila miRNA phenotypes in viability and external morphology. (a) Viability defects following ubiquitous expression of SP library under the tubulin-Gal4 driver. Data is displayed as average percent viability relative to control; at least six independent replicate batches were analyzed for each genotype (ANOVA, post hoc analysis with Tukey-Kramer multiple comparisons test P≤ 0.01; error bars, s.d). (b) Benchmark comparison of viability phenotypes to Drosophila miRNA null mutants13. “Confirmed” indicates same viability phenotype shared with SP and null. False negative (“False −ve”) indicates miRNAs where null demonstrated viability impaired phenotypes, but miR-SP lines were viable. False positive (“False +ve”) indicates miRNAs where a phenotype was observed with miR-SP but not in the null animal. “miR family” represents miRNAs for which a similar seed sequence is shared and miRSP lines display a viability phenotype that is only confirmed by an individual family member. The denominator for this chart was 99; we excluded all lines for which there was no null available, lines that were not tested by Chen and colleagues13, mutants removing entire clusters of multiple miRs, or mutants for which complementation was inconclusive (Supplementary Data 3). (c-h) Eye morphology defects (arrowheads) following inhibition of miR-92 activity. Genotypes: +/tubulin-Gal4 (c), +/Scramble-SP;tubulin-Gal4/Scramble-SP (d, g), +/miR-92bSP;tubulin-Gal4/miR-92bSP (e, h) +/miR-92bSP;tubulin-Gal4/miR-310SP (f). (i–k) Expression of miR-2aSP results in wing vein patterning abnormalities (j, arrowheads) and an overall reduction in wing blade size (j, Scale bar is 200μm). Average wing size area was quantified in triplicate samples (P=0.004; error bars, sem) from +/Scramble-SP;tubulin-Gal4/Scramble-SP and +/miR-2aSP;tubulin-Gal4/miR-2aSP animals (k).
While our manuscript was under review, a screen of miRNA deletion mutants for lethal phenotypes was published13, thus allowing a broad benchmark comparison of miR-SPs with independent viability data (summary in Fig. 2b). Of the miRNAs we tested for viability (141 miR-SPGenII strains plus two other constructs; Supplementary Data 2), null alleles exist for 115. Sixteen of these mutants were deemed as not comparable as benchmarks because they either (a) remove multiple clustered miRNA genes, (b) fail to display non-complementation over large Dfs at each locus (ie. not genetically validated), or (c) they were not tested for lethality by Chen and colleagues (Supplementary Data 3). In addition, 27 miR-SPGenII constructs correspond to miRNAs for which no null allele currently exists (Supplementary Data 3). Thus, we compared adult viability phenotypes of null and tubulin-Gal4;UAS-miR-SP for 99 genes (see Fig. 2b and Supplementary Data 3;13). The vast majority of the viability phenotypes in our screen match the published data (82.8%; green in Fig. 2b and Supplementary Data 3). Several miR-SPs did show viability defects that were not observed in corresponding nulls; however, several were members of highly conserved families likely to display functional redundancy as previously observed for K-box miRNAs (light blue in Fig. 2b and Supplementary Data 3)37, thus leaving 4% as conclusive false-positives (miR-14, miR-79, miR-307, and miR-975; dark blue in Fig. 2b and Supplementary Data 3). Finally, some null mutants displayed lethality that was not detected in our tubulin-Gal4;UAS-miR-SP screen, as expected in screens of hypomorphic mutants (e.g. using RNA interference or chemical mutagenesis). Overall, the false-negative rate for viability was 8.1% (red in Fig. 2b and Supplementary Data 3).
We expected that SP activity would be dose-dependent relative to endogenous levels of targeted miRNA, thus allowing us to control the strength of conditional inhibition. To test this, we compared the viability of 1x and 2x SP insertions with tubulin-Gal4 for several of the hits in our screen, including miR-2bSP, miR-8SP, miR-9bSP and miR-9cSP. In each case, the 2x SP gave a more penetrant adult lethal phenotype than 1x SP (Supplementary Fig. 1b). In addition, it is likely that intrinsic differences in miR-SP architecture can influence their efficacy. For example, a previous study using a different lethality assay and SP design reported viability defects following miR-92 competitive inhibition28. However, our individual strains with miR-SPGenII constructs directed against miR-92 family members (miR-92a,b, miR-310/311/312/313) did not display significant lethality, despite the fact that other phenotypes can be detected with SPs directed against this family (see below).
Examining external morphology, we have previously reported that miR-SPs can replicate the deformed adult leg phenotype caused by loss of miR-8 function26, 38. This was confirmed with our GenII miR-8SP lines (Supplementary Fig. 2a–c). GenII SP strains also recapitulated the miR-9-dependent notching of the posterior wing blade margin39–41 (Supplementary Fig. 2d–f). In the adult compound eye, we also observed a novel and highly penetrant morphological phenotype using miR-92bSP, characterized by an apparent invasion of the head cuticle into the retina (Fig. 2c–e) or even more dramatic ectopic outgrowth within the retinal field (Fig. 2g, h). The identical phenotype was observed for miR-310SP, another member of the miR-92 family (Fig. 2f), suggesting some degree of functional redundancy between endogenous members of this miRNA family. In addition, we found that surviving miR-2aSP adults displayed a novel vein patterning defect and a decreased wing size (Fig. 2i, j, k). However, the overall frequency of gross morphological phenotypes was quite low (0.7% to 1.4%, depending on phenotype).
Tissue-specific miRNA function in Drosophila muscle
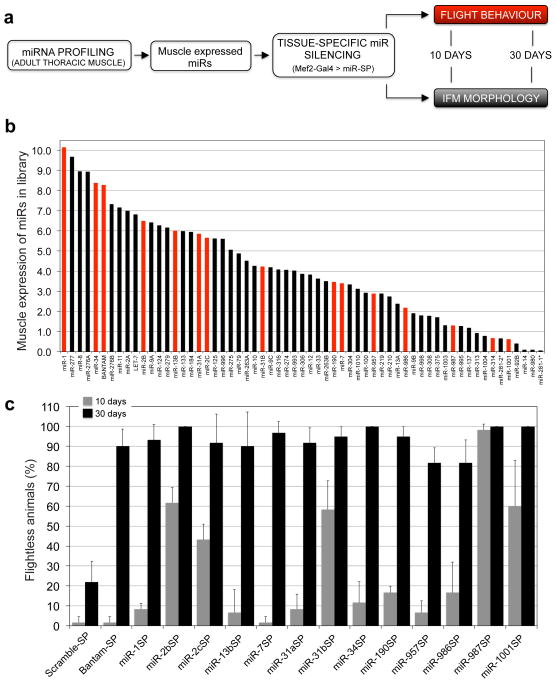
Although a growing body of evidence suggests that miRNAs play vital roles in maintaining the integrity and function of adult tissues42–44, comprehensive interrogation of such phenotypes has been challenging. To realize the potential of our SP library for unbiased discovery of tissue-specific miRNA functions, we next sought to screen for miRNAs that regulate adult muscle morphology, maintenance, and function. We first determined the muscle expression of the miRNAs present in our collection. Total RNA was isolated from dissected adult thoracic muscles, and relative expression levels were determined using a miRNA microarray platform (Fig. 3a). Drosophila miRNA array signals were obtained by fitting a linear model to the log2 transformed probe intensities. This analysis detected 61 miRNAs expressed across a broad range of relative levels in adult thoracic muscles (Fig. 3b; miR-SPGenII strains were available to test 58 of these).
Figure 3.
Tissue-specific in vivo screen for miRNAs regulating muscle function and maintenance. (a) Screen strategy diagram. Muscle-expressed miRNAs were profiled in the adult thoracic muscle tissue, and silenced by driving corresponding miR-SPs with the dMef2-Gal4 driver. Flight behavior and IFM morphology was assessed at 10 day old and 30 day old animals. (b) miRNA microarray profiling of the thoracic muscle tissue. For simplicity, only miRNAs with detectable expression are shown. Red bars denote positive hits in the primary muscle screen. (c) Positive hits from the flight screen. “Flightless phenotype” was defined at a value above twice the s.d. of Scramble-SP controls in 30 day old animals (ANOVA, post hoc analysis with Tukey-Kramer multiple comparisons test P≤ 0.001, error bars, s.d., n=3 replicates of 20 animals). Flight behavior is shown for 10 day old animals in grey bars and for 30 day old animals in black bars.
To disrupt the activity of these candidate miRNAs selectively in the muscle tissue from embryonic stages through adulthood, 2x miR-SP constructs were expressed using a dMef2-Gal4 driver. Flight behavior and indirect flight muscle (IFM) morphology were assessed in adult progeny at 10 days and 30 days post eclosion (Fig. 3a). Analysis of 30 day-old animals revealed that 14 miR-SP lines rendered a penetrant “flightless” phenotype (black bars in Fig. 3c). These included miR-SPs targeting bantam, miR-1, the K-box family (miR-2b, 2c, and 13b displayed strong phenotypes; miR-2a and 13a were flight impaired but fell below our stringent cutoff; Supplementary Data 4), miR-7, the miR-31 family, miR-34, miR-190, miR-957, miR-986, miR-987 and miR-1001. All but one of these lines (miR-987SP) appeared normal or displayed mildly impaired flight behavior at 10 days (grey bars in Fig. 3c). However, when we then assayed miR-987SP adults at 4 days post-eclosion, we found normal flight behavior relative to control (1.7 +/− 2.9% non-fliers in miR-987SP compared to 0% in Scramble-SP). Therefore, all behavioral phenotypes recovered in our muscle screen displayed a progressive, age-dependent loss of flight. These miRNA genes were also evenly distributed across the range of expression levels (Fig. 3b red bars), showing little correlation with endogenous miRNA abundance.
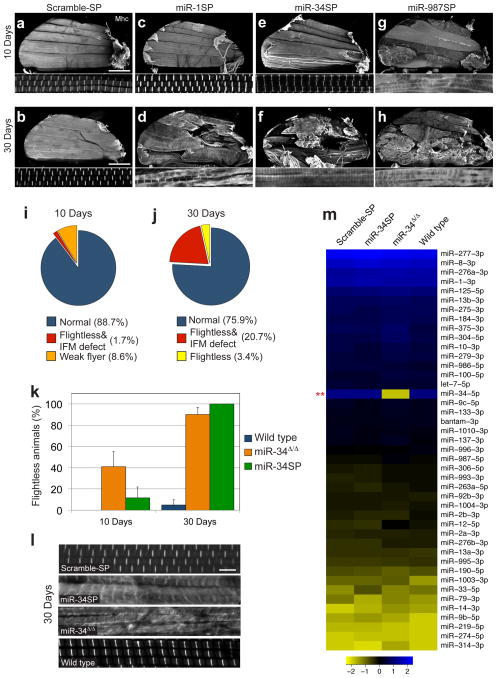
Adult flight behavior is primarily dependent on the activity of the IFMs. To assess the impact of miRNA inhibition on muscle morphology, IFM myofibril structure was examined in sagittal bisections of the thorax stained for F-actin and Myosin heavy chain (Mhc). At 30 days, 12 of the 14 flightless SP lines showed dramatic defects in IFM muscle integrity and sarcomere organization (Fig. 4a–h; Supplementary Fig. 3), with a relatively broad range of penetrance (Supplementary Figure 4). Only miR-7SP and miR-13bSP showed no detectable IFM abnormalities at this level of resolution (yellow wedge in Fig. 4j). Notably, miR-987SP animals displayed a detectable defect in gross IFM or myofibril morphology at 10 days (red wedge in Fig. 4i). Thus, despite strong dMef2-Gal4-dependent expression starting in the mesoderm at embryonic stage 7, our SP screen detected many age-dependent IFM phenotypes, but no obvious defects in muscle development.
Figure 4.
Twelve miRNAs are required to maintain flight muscle structure.
58 lines were assayed for flight at 10 and 30 days post-eclosion, and all lines that displayed significant flight deficits were then assayed for IFM morphology (a–h) Sagittal bisections of the adult thorax stained for actin and Myosin heavy chain (Mhc) shown at low (top panel) and high magnification (bottom panel). Normal IFM and sarcomere morphology in 10 days and 30 days old Scramble-SP controls (a, b, Scale bar is 200μm), late onset IFM phenotype following miR-1SP expression (c, d), or miR-34SP expression (e, f) and early onset IFM defects in miR-987SP animals (g, h). A summary of the lines that display flight and IFM phenotypes at 10 days post-eclosion (i) is shown for comparison to the 30 day results shown in (j); red represents all SP lines that display both flight and IFM defects, whereas orange and yellow represent animals with no detectable IFM morphology defect that were flight impaired or flightless, respectively. (k–l) Comparison of miR-34SP and miR-34Δ/Δ null mutants. Null mutant adults (orange bars) display a stronger flightless phenotype at 10 days but are comparable to miR-34SP (green bars) at 30 days (k) error bars, sem, n=3 replicates of 20 animals. IFM sarcomere morphology and Mhc distribution and pattern are comparable in miR-34SP and miR-34Δ/Δ null mutants at 30 days (displaying 15.7% penetrance [n=19], compared to 25% in miR-34SP) (l, Scale bar is 5μm). (m) NanoString nCounter profiling of adult thoracic muscle. All miRNAs expressed above background values are represented. Only the levels of mature miR-34-5p were substantially reduced in the null mutant. Statistical significance was established in this case by comparing the expression values of miR-34Δ/Δ to the wild type control using the NanoStringNorm package in R (t-test, **P<0.003). For all other genotypes, statistical significance was established by comparing the miR-SP values, against miR-34Δ/Δ, Scramble-SP and wild type controls. No other endogenous miRNA levels change significantly in miR-34SP or miR-34Δ/Δ animals compared to Scramble-SP or wild type controls (see Methods).
Among the candidate miRNAs identified in our screen, miR-1 is considered an “archetypal” muscle miRNA whose sequence and expression pattern appears to be evolutionarily conserved from flies to mammals. In Drosophila, miR-1 null mutations display paralysis, severe disruption of somatic muscle tissues and early larval death, preventing analysis of function during adult life45–47. Examination of 30 day-old escapers expressing miR-1SP uncovered a highly penetrant flightless phenotype and severe degeneration of IFM muscle fibers (Fig. 3c and Fig. 4d). These results highlight the capacity of miRNA SPs to complement studies where complete LOF renders early developmental lethality.
Unlike miR-1, miR-34 had not previously been analyzed in Drosophila muscle despite mounting evidence implicating this conserved miRNA in muscle function (Supplementary Table 1). Thus, we sought to confirm this function for miR-34 by examining a null mutation42. Indeed, homozygous null animals (miR-34Δ/Δ) display age-dependent deficits in flight behavior that are slightly more severe than miR-34SP at 10 days but reach comparable levels at 30 days (Fig. 4k). Moreover, IFM morphology comparisons confirm that miR-34Δ/Δ nulls display the same abnormal morphology and distribution of Mhc characteristic of dMef2-Gal4;miR-34SP animals at comparable penetrance (Fig. 4l). Because the miR-34 muscle phenotype was qualitatively similar to many of the other hits in our flight screen, we wanted to confirm that miR-34Δ/Δ and miR-34SP did not cause altered expression of other muscle-expressed miRNAs required for muscle maintenance. Thus, we used sensitive NanoString nCounter profiling to monitor miRNAs levels in the adult thorax (see Methods). Aside from the loss of miR-34-5p in null mutants, no other miRNAs were significantly changed compared to controls (Fig. 4m, Supplementary Data 5), suggesting that miR-34 acts independently of other conserved miRNAs in this context.
DISCUSSION
In this study, we describe a transgenic Drosophila resource for conditional competitive inhibition for 141 high-confidence miRNAs, as a versatile toolkit for discovery and tissue-specific analysis of miRNA functions in vivo. This resource is highly complementary to collections of miRNA gene deletions that offer chronic, complete and systemic LOF48, 13. Like the chemical mutagenesis and RNAi methods typically used to detect novel loci in genome-wide functional screens49, miR-SPs typically produce partial LOF; however, this feature combined with the spatial-temporal specificity conferred by the huge arsenal of Gal4 drivers (e.g. http://flystocks.bio.indiana.edu/) empowers the miR-SP approach with many advantages for analysis of post-embryonic and cell or tissue-specific functions.
Overall, the occurrence of significant adult viability and external morphology defects following ubiquitous miRNA inhibition in Drosophila appears to be comparable to the frequency of phenotypes resulting from systemic loss of miRNA function in C. elegans10. The relatively low frequency of external morphology defects (3.5% overall; n=5/143; Fig. 2 and Supplementary Fig. 2) and the low false discovery rates observed in our tubulin-Gal4 screens (Fig. 2b), suggest that transgenic SPs are largely free from significant off-target effects. Interestingly, our novel tissue-specific screen identified a much greater percentage of miRNAs required for the form and function of adult flight muscle (24%; n=14/58; Fig. 4h). Our analysis suggests that disruption of miR-34 and 11 other miRNAs can induce a progressive disruption of IFM structure and function, thus uncovering a substantial regulatory landscape for muscle maintenance and/or homeostasis.
Recent studies suggest that vertebrate orthologs for several of the conserved miRNAs required for muscle maintenance in our screen (miR-1, 7, 31, 34 and the K-box ortholog miR-23) are associated with muscle physiology in vertebrate species (Supplementary Table 1). However, to our knowledge, only miR-1 and miR-34 have been implicated by LOF in vertebrate cardiac and/or skeletal muscle function43, 50. Interestingly, loss of Drosophila miR-34 has been reported to induce late-onset brain degeneration42, raising the intriguing possibility of a general tissue maintenance theme. Of course, future study is needed to distinguish between events that may trigger active degenerative processes versus those that disrupt ongoing replenishment of protein networks in muscle. It may also be interesting to test these muscle-maintenance miRNAs for degenerative phenotypes in other tissues. Although future comparisons with null mutations will be required to validate many of these novel loci, the fact that most of these miRNAs were not previously known to support muscle maintenance highlights the potential of the miR-SP library for tissue-specific screening. In conclusion, the library of transgenic SPs reported here represents a valuable resource for unbiased and conditional LOF screens in the intact organism.
METHODS
Genetics and miR-SP library generation
Drosophila stocks
The following Gal4 drivers were obtained from the Bloomington Stock center and crossed with miR-SP lines to drive ubiquitous, wing disc, and mesodermal expression: tubulin-Gal4, patched (ptc)-Gal4, and dMef2-Gal4, respectively. The transgenic lines containing 3′UTR sensors for miR-8, nerfin, and k-box miRNAs, were previously described51–53. miR-1SP construct was generated in the Han lab by introducing ten repetitive miRNA complementary sequences (GGTACGTTTAGCGTAAGTTAT synthesized by GenScript) separated by four-nucleotide linkers CGCG into the pUAST vector. Bantam-SP was a generous gift from Steve Cohen.
Conditional miR-SP collection
miR-SP constructs were designed with a silencing cassette of 20 repetitive miRNA complementary sequences separated by variable four-nucleotide linker sequences, and assembled as previously described26. To avoid off-target effects the combined miRNA and linker sequences were checked against every mature miRNA sequence in the Drosophila genome. The entire cassette was then cloned into the 3′UTR of mCherry between NotI and XbaI in a modified pWALIUM10-moe vector (54, http://www.flyrnai.org/TRiP-HOME.html) carrying the white+ selectable marker and flanking insulator sequences (as described in33). To obtain miR-SPs with relatively equal expression and avoid epigenetic positional effects, transgenic flies were generated using phiC31 site-specific genomic integration in specific landing sites on the 2nd (attP40) and 3rd (attP2) Drosophila autosomes (Genetic Services Inc.). Both attP2 and attP40 insertion site stocks are viable as homozygotes and have been characterized by the Perrimon lab as controls for genetic analysis of muscle aging and viability phenotypes that run out to 56 days55; attP40 insertions are carried as heterozygotes in all SP screens carried out. Insertion of Scramble-SP sequence at the attP2 and attP40 sites acted as control. The sequences of all designed miR-SP constructs are listed in Supplementary Data 1.
Mature miRNA quantification
Total-RNA was isolated according to the miRVana miRNA kit protocol without enrichment for miRNAs (Invitrogen) from ubiquitously expressing miR-SP or Scramble wandering 3rd instar larvae with intestines removed. Real-time qPCR was performed using a standard TaqMan MicroRNA assay kit protocol on an Applied Biosystems 7900HT Sequence Detection System (Applied Biosystems). Reaction volumes, cycles and analysis were performed as described56 with the exception that expression values are expressed relative to S2 rRNA expression.
Immunostaining of imaginal discs
Larvae were dissected in ice cold PBS. Discs were fixed in 4% PFA at room temperature for 20 min., washed in PBS, and PBST (0.01% Triton-X100), blocked with 5% normal goat serum in PBST and incubated with primary antibody anti-GFP (Molecular Probes A6455, 1:500), overnight at 4°C washed 3 times in PBST and then incubated with secondary Alexa fluor 488 goat anti-rabbit IgG (Molecular Probes A-11008, 1:2,000) for 3 hours at room temperature. Discs were then washed, mounted in SlowFade Gold antifade reagent (Invitrogen) and imaged with a 20x objective on a Nikon A1R confocal.
Ubiquitous miR-SP expression and analysis
Crosses to examine lethality were carried out at 27°C with 12 males carrying the miR-SP on the second and third chromosome and 25 tubulin-Gal4/TM3 virgins allowed to mate for 1 day in a vial, then transferred to a bottle on the second day and finally transferred to a second bottle on Day 4. After 2 days, the adult flies in the final bottle were discarded. Eclosed animals were collected every day up to 6 days after first eclosion, and promptly counted. Flies were scored for and against mCherry expression, or against the TM3 balancer. To account for subtle contribution(s) of TM3 to viability phenotypes, balanced drivers were crossed to Canton-S to establish a correction factor. Raw data is shown in Supplementary Data 2. miR-SP lines displaying lethality in the above assay were crossed again in vials with 6 miR-SP males and 10 tubulin-Gal4 virgins. Each genotype was set up in triplicate. Crosses were flipped every two days into a new vial for 5 days after which eclosed animals were collected and counted as described above. All lines were screened as double insert (2x) stocks to increase phenotypic penetrance because comparisons between 1x for several miR-SPs showed consistent dose-dependence (Supplementary Fig. 1b).
Gross morphology of ubiquitously miR-SP expressing animals was examined with specific attention to retina (size/shape/pattern/pigmentation/bristle), wing (size/shape/veination/bristle and hair pattern), leg (length/shape/segment morphology/bristle pattern), and body (size/shape/bristle pattern). For the analysis of leg and wing morphology, cuticle preparations were prepared by dehydrating in a series of ethanol dilutions. Muscle was then cleared in xylene and tissues mounted in Cytoseal-60 (Cole-Parmer). Wings and legs were imaged with Nikon Digital Sight DS-Fi1 color camera on a Nikon 80i upright microscope at 4x. A 2×2 montage was taken and stitched together using NIS-Elements software. Area was calculated from stitched images using Fiji image processing package (http://fiji.sc/Fiji).
Muscle specific miR-SP expression and analysis
Crosses to examine adult flight behavior were carried out as described above (lethality assay) with the exception that dMef2-Gal4 virgins were used to drive muscle specific expression of the miR-SP. Eclosed animals were collected every day up to 6 days after first eclosion, and then aged for 10 or 30 days. Animals were flipped to new food vials every other day to maintain integrity of the collection. Flight assay was carried out at ambient temperature in a dark room in an illuminated arena with the following dimensions: H=65 cm, W=64 cm, D=51 cm. Flies were sorted into 3 groups of 20 for each gender a minimum of 1 hr prior to the assay. After 1 hour recovery from brief anesthesia, we found that no wild type control flies (Canton-S; dMef2-Gal4 raised at 27°C to elevate Gal4 activity) hit the target area at 10 days of age. A small number of control animals fly poorly, ending up in the outer ring, when the flies are reared and aged to 30 days at 27°C. This result consistent with observations following 24 hr of recovery from CO2. All tests were completed at the same time of day. Animals were flipped into a vial with no food directly preceding the assay. Flies were dropped through a funnel from a height of 74 cm centered above three concentric circles (diameter: 7cm inner circle, 15cm middle circle, and 21 cm outer circle) and the number of animals in each circle was scored from an image of the arena taken immediately after landing. Flies falling within the first two circles were counted as “non-fliers” (raw data in Supplementary Data 4). A threshold for “non-fliers” was set at two standard deviations of Scramble-SP, followed by ANOVA analysis and Tukey-Kramer multiple comparisons test to assess significance for each SP line above threshold.
IFM morphology was assessed only in flightless animals and controls. Wings, legs and abdomen removed from adult flies and the thorax muscles were soaked in relaxing solution (20mM phosphate buffer, pH 7.0; 5 mM MgCl; 5 mM EGTA) for 5 min. Thoraces were then moved to 4% paraformaldehyde (PFA) in relaxing solution for 10 min then transferred to a 5% agarose gel plate covered with PBT (PBS + 0.2% Triton X-100). Thoraces were then bisected sagitally with a scalpel blade (Fine Science Tools) and blocked in relaxing solution + 3% HIGS for 20 min. before fixation in 4% PFA in PBT for 10 min. Immunohistochemistry was carried out on the hemi-thoraces with mouse α-MHC (1:50 in PBT, 57) followed with incubation in goat α-mouse 568 (1:200 in PBT, Invitrogen A-11031) and Alexa Fluor® phalloidin 488 (1:500 in PBT, Invitrogen A12379). Samples were mounted in SlowFade Gold antifade reagent (Invitrogen). Hemi-thoraces were imaged using a Nikon Ti-E and A1R confocal with 10x and 100x objectives using NIS-Elements acquisition software. A minimum of n=4 hemi-thoraces were imaged for each genotype to account for variable expressivity (Supplementary Fig. 4); samples with evidence of tissue damage due to improper dissection were excluded prior to analysis of the results. Max-Intensity projections were obtained using the NIS-Elements analysis software.
Profiling miRNA muscle expression
Muscle tissue was dissected from the thorax of adult flies and isolated using the standard Trizol (Invitrogen) protocol followed by RNeasy Plus kit (Qiagen) clean up with DNAse treatment. RNA was labeled with Cy5 following Agilent standard protocol. Agilent microarrays covering 152 Drosophila miRNAs were designed as previously described58, with miRNA probes of varied lengths to equalize melting temperatures to 55°C. MiRNA expression data was analyzed using the AgiMicroRNA Bioconductor Package version 2.0.1 (59). The software is implemented in the open-source statistical scripting language R and is integrated into the Bioconductor project (http://www.bioconductor.org) under the GPL license. For data pre-processing, a target file was generated to assign each scanned data file to the appropriate experimental group. Scanned data from the Agilent Feature Extraction image analysis software was imported into an R object that stores the relevant probe and raw intensity data information needed for the pre-processing. Raw array data was normalized using quantile normalization and we obtained the miRNA gene signal by fitting a linear model to the log2 transformed probe intensities. This model produced an estimate of the miRNA gene signal corrected for probe effects. To evaluate differences in the individual gene expression between experimental groups, the absolute value of the difference in total expression was computed for each of the miRNAs sampled on the array.
Nanostring nCounter miRNA profiling
All crossed were carried out at 27°C. Thoraxes from 1–2 days old adult females of relevant genotypes (dMef2-Gal4>miR34b; dMef2-Gal4>Scramble-SP; miR-34Δ/Δ (a gift from Nancy Bonini) and Iso white-1,2,3) were dissected (n ≥ 4) in PBS in biological duplicates. Total RNA was extracted using the miRNeasy kit (Qiagen). Purified RNA was concentrated using Amicon Ultra-0.6 Centrifugal Filters (Millipore). For each sample, ~100 ng total RNA was loaded into the nCounter Drosophila miRNA Assay (Nanostring) and processed according to the manufacturer protocols. Briefly, miRNAs were ligated, hybridized to reporter probes at 65°C for 12h and prepared on the nCounter Prep Station before being digitally counted at 555 FOV on the nCounter Digital Analyzer. The raw data counts were analyzed using the NanoStringNorm R package60. The data was normalized using the geometric mean of the six positive controls and then it was background corrected by subtracting the mean and two standard deviations of the six negative controls. Finally, the data were normalized for sample/RNA content using the geometric mean of three housekeeping genes. Normalized miRNA expression levels were log2 transformed and analyzed using a t-test to identify differentially expressed miRNA between samples. Heatmaps were generated using the gplots R package with the log2 transformed and normalized values of the experiment. Subsequently, for each condition the mean of the two replicates was taken and the data was centered and scaled by subtracting for each condition the mean values and dividing it by the STDEV.
Supplementary Material
Acknowledgments
We thank Ms. Alicia Taylor and Ms. Heather Faherty (Harvard Medical School) for excellent technical support, and Emmanouela Repapi (Computational Biology Research Group, University of Oxford) for help with the analysis of Nanostring profiling data. We thank Drs. Leslie Griffith and Ronald Davis for critical feedback on our manuscript, and we thank Dr. Stephen Cohen and colleagues for personal communication of results prior to publication and sharing several mutant strains. We thank Dr. Jennifer Waters and Lauren Alvarenga in the Nikon Imaging Center at Harvard Medical School for microscopy assistance. We thank the Bloomington Drosophila Stock Center (NIH P40OD018537) for stocks used in this study as well as acceptance of the miRSP library for distribution. Work in the T.F. laboratory is supported by WIMM Strategic Award, MRC #G0902418. Work in the Z.H. laboratory was supported by grants from the NIH (R01HL090801, R01DK098410). Work in the E.C.L. laboratory was supported by the Burroughs Wellcome Fund [1004721], the Starr Cancer Consortium [I3-A139] and the NIH [R01-GM083300]. Work in the D.W. laboratory was supported by Simons Foundation SFARI award 240253, Nancy Lurie Marks Family Foundation and NIH grant 1R01MH090611-01A1. Work in the N.P. laboratory was supported by NIH/NIGMS R01-GM084947 and the Howard Hughes Medical Institute. Work in the D.V.V. laboratory and all publication costs were supported by NIH/NINDS R01-NS069695.
Footnotes
Author Contributions
T.A.F, E.M.M, N.P. and D.VV conceived and designed the study. T.A.F, E.M.M, R.B, J.Y, and A.B, performed most of the experiments. B.R.S carried out the Nanostring profiling assays. Y.Z and M.S-L performed the miRNA microarray profiling of muscle tissues. M.B wrote the miR-SP design algorithm. F.B and E.C.L generated wing disc sensor assay data. T.DL and D.W carried out the bioinformatics analysis pertaining to miRNA muscle profiling. Z.H provided unpublished miR-SP reagents. T.A.F, E.M.M and D.VV wrote the manuscript and generated the figures and N.P. edited them.
Competing Financial Interests
There are no competing financial interests to disclose.
References
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nature reviews. Genetics. 2008;9:831–842. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336:233–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meijer HA, et al. Translational repression and eIF4A2 activity are critical for microRNA-mediated gene regulation. Science. 2013;340:82–85. doi: 10.1126/science.1231197. [DOI] [PubMed] [Google Scholar]
- 7.Mullokandov G, et al. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nature methods. 2012;9:840–846. doi: 10.1038/nmeth.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Polesskaya A, et al. Genome-wide exploration of miRNA function in mammalian muscle cell differentiation. PloS one. 2013;8:e71927. doi: 10.1371/journal.pone.0071927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miska EA, et al. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS genetics. 2007;3:e215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alvarez-Saavedra E, Horvitz HR. Many families of C. elegans microRNAs are not essential for development or viability. Current biology : CB. 2010;20:367–373. doi: 10.1016/j.cub.2009.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brenner JL, Jasiewicz KL, Fahley AF, Kemp BJ, Abbott AL. Loss of individual microRNAs causes mutant phenotypes in sensitized genetic backgrounds in C. elegans. Current biology : CB. 2010;20:1321–1325. doi: 10.1016/j.cub.2010.05.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park CY, et al. A resource for the conditional ablation of microRNAs in the mouse. Cell reports. 2012;1:385–391. doi: 10.1016/j.celrep.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen YW, et al. Systematic Study of Drosophila MicroRNA Functions Using a Collection of Targeted Knockout Mutations. Developmental cell. 2014;31:784–800. doi: 10.1016/j.devcel.2014.11.029. [DOI] [PubMed] [Google Scholar]
- 14.Chen YW, Weng R, Cohen SM. Protocols for use of homologous recombination gene targeting to produce microRNA mutants in Drosophila. Methods in molecular biology. 2011;732:99–120. doi: 10.1007/978-1-61779-083-6_8. [DOI] [PubMed] [Google Scholar]
- 15.Xiao A, et al. Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic acids research. 2013;41:e141. doi: 10.1093/nar/gkt464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kondo S, Ueda R. Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics. 2013;195:715–721. doi: 10.1534/genetics.113.156737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, et al. Inheritable and precise large genomic deletions of non-coding RNA genes in zebrafish using TALENs. PloS one. 2013;8:e76387. doi: 10.1371/journal.pone.0076387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cesana M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karreth FA, et al. In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell. 2011;147:382–395. doi: 10.1016/j.cell.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tay Y, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–357. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen TB, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 22.Memczak S, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 23.Denzler R, Agarwal V, Stefano J, Bartel DP, Stoffel M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Molecular cell. 2014;54:766–776. doi: 10.1016/j.molcel.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ebert MS, Sharp PA. MicroRNA sponges: progress and possibilities. Rna. 2010;16:2043–2050. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loya CM, Lu CS, Van Vactor D, Fulga TA. Transgenic microRNA inhibition with spatiotemporal specificity in intact organisms. Nature methods. 2009;6:897–903. doi: 10.1038/nmeth.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoon WH, Meinhardt H, Montell DJ. miRNA-mediated feedback inhibition of JAK/STAT morphogen signalling establishes a cell fate threshold. Nature cell biology. 2011;13:1062–1069. doi: 10.1038/ncb2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Z, Liang S, Zhao Y, Han Z. miR-92b regulates Mef2 levels through a negative-feedback circuit during Drosophila muscle development. Development. 2012;139:3543–3552. doi: 10.1242/dev.082719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herranz H, Hong X, Cohen SM. Mutual repression by bantam miRNA and Capicua links the EGFR/MAPK and Hippo pathways in growth control. Current biology : CB. 2012;22:651–657. doi: 10.1016/j.cub.2012.02.050. [DOI] [PubMed] [Google Scholar]
- 30.Li W, et al. MicroRNA-276a functions in ellipsoid body and mushroom body neurons for naive and conditioned olfactory avoidance in Drosophila. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33:5821–5833. doi: 10.1523/JNEUROSCI.4004-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loya CM, McNeill EM, Bao H, Zhang B, Van Vactor D. miR-8 controls synapse structure by repression of the actin regulator enabled. Development. 2014;141:1864–1874. doi: 10.1242/dev.105791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu CS, et al. MicroRNA-8 promotes robust motor axon targeting by coordinate regulation of cell adhesion molecules during synapse development. Philosophical transactions of the Royal Society of London. Series B, Biological sciences. 2014;369 doi: 10.1098/rstb.2013.0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bejarano F, et al. A genome-wide transgenic resource for conditional expression of Drosophila microRNAs. Development. 2012;139:2821–2831. doi: 10.1242/dev.079939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic acids research. 2011;39:D152–157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ibanez-Ventoso C, Vora M, Driscoll M. Sequence relationships among C. elegans, D. melanogaster and human microRNAs highlight the extensive conservation of microRNAs in biology. PloS one. 2008;3:e2818. doi: 10.1371/journal.pone.0002818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Becam I, Rafel N, Hong X, Cohen SM, Milan M. Notch-mediated repression of bantam miRNA contributes to boundary formation in the Drosophila wing. Development. 2011;138:3781–3789. doi: 10.1242/dev.064774. [DOI] [PubMed] [Google Scholar]
- 37.Ge W, et al. Overlapping functions of microRNAs in control of apoptosis during Drosophila embryogenesis. Cell death and differentiation. 2012;19:839–846. doi: 10.1038/cdd.2011.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karres JS, Hilgers V, Carrera I, Treisman J, Cohen SM. The conserved microRNA miR-8 tunes atrophin levels to prevent neurodegeneration in Drosophila. Cell. 2007;131:136–145. doi: 10.1016/j.cell.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 39.Li Y, Wang F, Lee JA, Gao FB. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes & development. 2006;20:2793–2805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biryukova I, Asmar J, Abdesselem H, Heitzler P. Drosophila mir-9a regulates wing development via fine-tuning expression of the LIM only factor, dLMO. Developmental biology. 2009;327:487–496. doi: 10.1016/j.ydbio.2008.12.036. [DOI] [PubMed] [Google Scholar]
- 41.Bejarano F, Smibert P, Lai EC. miR-9a prevents apoptosis during wing development by repressing Drosophila LIM-only. Developmental biology. 2010;338:63–73. doi: 10.1016/j.ydbio.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu N, et al. The microRNA miR-34 modulates ageing and neurodegeneration in Drosophila. Nature. 2012;482:519–523. doi: 10.1038/nature10810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boon RA, et al. MicroRNA-34a regulates cardiac ageing and function. Nature. 2013;495:107–110. doi: 10.1038/nature11919. [DOI] [PubMed] [Google Scholar]
- 44.Sun K, Lai EC. Adult-specific functions of animal microRNAs. Nature reviews. Genetics. 2013;14:535–548. doi: 10.1038/nrg3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kwon C, Han Z, Olson EN, Srivastava D. MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:18986–18991. doi: 10.1073/pnas.0509535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sokol NS, Ambros V. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes & development. 2005;19:2343–2354. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nguyen HT, Frasch M. MicroRNAs in muscle differentiation: lessons from Drosophila and beyond. Current opinion in genetics & development. 2006;16:533–539. doi: 10.1016/j.gde.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 48.Weng R, Chen YW, Bushati N, Cliffe A, Cohen SM. Recombinase-mediated cassette exchange provides a versatile platform for gene targeting: knockout of miR-31b. Genetics. 2009;183:399–402. doi: 10.1534/genetics.109.105213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.St Johnston D. Using mutants, knockdowns, and transgenesis to investigate gene function in Drosophila. Wiley interdisciplinary reviews. Developmental biology. 2013;2:587–613. doi: 10.1002/wdev.101. [DOI] [PubMed] [Google Scholar]
- 50.King IN, et al. A genome-wide screen reveals a role for microRNA-1 in modulating cardiac cell polarity. Developmental cell. 2011;20:497–510. doi: 10.1016/j.devcel.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 52.Lai EC, Tam B, Rubin GM. Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes & development. 2005;19:1067–1080. doi: 10.1101/gad.1291905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shcherbata HR, et al. Stage-specific differences in the requirements for germline stem cell maintenance in the Drosophila ovary. Cell stem cell. 2007;1:698–709. doi: 10.1016/j.stem.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ni JQ, et al. A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics. 2009;182:1089–1100. doi: 10.1534/genetics.109.103630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Demontis F, Patel VK, Swindell WR, Perrimon N. Intertissue control of the nucleolus via a myokine-dependent longevity pathway. Cell reports. 2014;7:1481–1494. doi: 10.1016/j.celrep.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen C, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic acids research. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saide JD, et al. Characterization of components of Z-bands in the fibrillar flight muscle of Drosophila melanogaster. The Journal of cell biology. 1989;109:2157–2167. doi: 10.1083/jcb.109.5.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang H, Ach RA, Curry B. Direct and sensitive miRNA profiling from low-input total RNA. Rna. 2007;13:151–159. doi: 10.1261/rna.234507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lopez-Romero P. Pre-processing and differential expression analysis of Agilent microRNA arrays using the AgiMicroRna Bioconductor library. BMC genomics. 2011;12:64. doi: 10.1186/1471-2164-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Waggott D, et al. NanoStringNorm: an extensible R package for the pre-processing of NanoString mRNA and miRNA data. Bioinformatics. 2012;28:1546–1548. doi: 10.1093/bioinformatics/bts188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.