Abstract
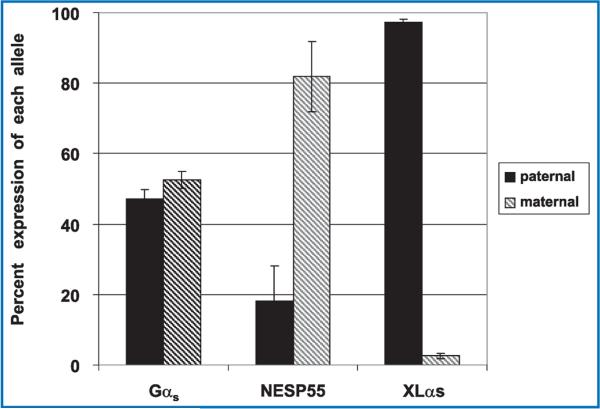
GNAS is a complex gene that through use of alternative first exons encodes signaling proteins Gαs and XLαs plus neurosecretory protein NESP55. Tissue-specific expression of these proteins is regulated through reciprocal genomic imprinting in fully differentiated and developed tissue. Mutations in GNAS account for several human disorders, including McCune–Albright syndrome and Albright hereditary osteodystrophy, and further knowledge of GNAS imprinting may provide insights into variable phenotypes of these disorders. We therefore analyzed expression of Gαs, NESP55, and XLαs prior to tissue differentiation in cell cultures derived from human primordial germ cells. We found that the expression of Gαs was biallelic (maternal allele: 52.6% ± 2.5%; paternal allele: 47.2% ± 2.5%; p = 0.07), whereas NESP55 was expressed preferentially from the maternal allele (maternal allele: 81.9% ± 10%; paternal allele: 18.1% ± 10%; p = 0.002) and XLαs was preferentially expressed from the paternal allele (maternal allele: 2.7% ± 0.3%; paternal allele: 97.3% ± 0.3%;p = 0.007). These results demonstrate that imprinting of NESP55 occurs very early in development, although complete imprinting appears to take place later than 5–11 weeks postfertilization, and that imprinting of XLαs occurs very early postfertilization. By contrast, imprinting of Gαs most likely occurs after 11 weeks postfertilization and after tissue differentiation.
Keywords: imprinting, Albright hereditary osteodystrophy, pseudohypoparathyroidism, GNAS, Gαs, NESP55, XLαs, embryonic germ cells, embryoid body-derived cells
Introduction
Genomic imprinting is an epigenetic process whereby one allele undergoes a partial or total loss of expression.1 Most commonly, imprints are established during gametogenesis. After fertilization, these epigenetic markers are maintained as chromosomes duplicate and separate in the developing organism.1 The associated patterns of imprinting are also often both tissue and developmental-stage specific, and the silencing is only partial.2 Imprinting may cause disease if disrupted and if allelic transcription is either activated or suppressed inappropriately. Abnormal imprinting may be secondary to changes in methylation, histone tail modifications, or by disruptions in chromatin binding.3 In the absence of an imprinting defect, the phenomenon of imprinting can lead to unusual, non-Mendelian patterns of inheritance that introduce parental origin effects on the phenotype.
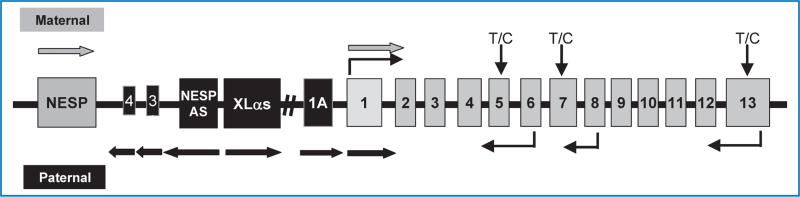
The GNAS locus, located at chromosome 20q13.3,4 exhibits a complex pattern of reciprocal genomic imprinting (Figure 1),5,6 with five alternative promoters that generate multiple sense and antisense transcripts. Transcripts that encode Gαs, the alpha chain of the heterotrimeric guanine nucleotide binding protein that couples hepatahelical receptors to stimulation of adenylyl cyclase, are derived from exons 1–13. Gαs was initially shown to be paternally imprinted in a tissue-specific manner through studies of murine models.7–9 The tissue-specific imprinting was subsequently found to be partial in the renal cortex, thyroid, pituitary, and ovaries from studies of both murine and human tissue.7–13 Upstream of exon 1 are three alternative first exons that each splice onto exons 2–13 to create novel sense transcripts (Figure 1). XLαs encodes a signaling protein that stimulates adenylyl cyclase but lacks a known receptor and has been shown to be maternally imprinted (paternally expressed).14,15 NESP55 encodes a secretory protein and has been shown to be paternally imprinted (maternally expressed).16–19 Exon 1A (also termed as exon A/B) does not encode a known protein but the transcript has been shown to be maternally imprinted (paternally expressed).6,20–22 The mechanism of imprinting of these three alternative transcripts is via differentially methylated regions (DMRs) in their respective promoter regions, whereas tissue-specific imprinting of exon 1 depends upon the methylation status of the DMR for exon 1A.6,16,17,21,23–25 Heterozygous GNAS mutations that alter function or expression of Gαs are associated with several human disorders and the phenotypes are influenced by a parent of origin effect that reflects predominant expression of Gαs from maternal alleles in certain tissues. Loss of function mutations causes an unusual constellation of somatic defects termed as Albright hereditary osteodystrophy (AHO), which includes short stature, subcutaneous ossifications, brachydactyly, and dental abnormalities. When a GNAS mutation occurs on the maternally inherited allele, AHO is associated with resistance to multiple hormones, such as PTH, TSH, GHRH, and gonadotropins, as well as obesity26 and cognitive defects.27 This condition is termed as pseudohypoparathyroidism (PHP) type 1a. In contrast, when the GNAS mutations occur on the paternally inherited allele, only the AHO features are present, a condition termed as pseudopseudohypoparathyroidism (pseudoPHP).26,28–35 Mutations that abrogate normal methylation of the exon 1A DMR on the maternal allele suppress maternal expression of Gαs in hormone-target tissues, leading to an isolated form of PTH resistance termed as PHP type 1b.25 Gain of function mutations in GNAS is also implicated as the basis of human disease. Missense mutations of Arg201 or Gln227 that lead to constitutive activation of Gαs cause isolated endocrine tumors or McCune–Albright syndrome, a disorder characterized by autonomous function and proliferation of endocrine tissues, fibrous dysplasia, and pigmented café au lait skin lesions. The clinical presentation can be affected by the parent of origin of the mutated allele because Gαs is imprinted and expressed specifically by the maternal allele in certain tissues.23–25
Figure 1. The GNAS gene complex located at 20q13.3 consists of 13 exons that encode the signaling protein Gαs.
Upstream of exon 1 are three alternative exons, labeled exon 1A, XLαs, and NESP55. These three alternative first exons are spliced to exons 2–13 to produce unique transcripts. NESP55 is transcribed exclusively from the maternal allele; XLαs and exon 1A are transcribed exclusively from the paternal allele. RT-PCR using an upstream primer specific for Gαs, XLαs, and NESP55 first exons of GNAS and a common downstream primer within exons 6, 8, and 13 of GNAS enabled us to genotype the alleles using a highly variable single nucleotide polymorphism in codon 131 (T/C) of exon 5, codon 185 (T/C) of exon 7, and codon 371 (T/C) of exon 13. The primers are indicated by thin black arrows; direction of transcription are indicated by thick arrows (thick gray arrows indicate transcription from maternal allele; black arrows indicate transcription from paternal allele).
The ontogeny and regulation of GNAS imprinting during development are still incompletely understood. Studies in the mouse have shown that the Nespas-Gnasxl DMR is a gametic imprint,36 which together with the earlier identification of a germ line DMR at Gnas exon 1A,21 predicts that the locus could contain two imprint control regions (ICRs) and is divided between separate domains regulated by independent imprinting mechanisms. Support for this hypothesis derives from imprinting anomalies of human GNAS encountered in the disorder PHP type 1b.6,25 Similar studies that have examined human imprinting as early as 6 to 13 weeks of fetal development in differentiated tissues, specifically heart, spinal cord, muscle, kidney, lung, gut, eye, brain, adrenal, stomach, and ovaries showed expression of Gαs to be biallelic, whereas expression of XLαs and NESP55 was monoallelic.16,17,37 However, these studies were imprecise as the techniques used to assess the parental origin of Gαs transcripts also included the amplification of XLαs and NESP55 transcripts. Recent studies in sheep revealed that GNAS was maternally expressed in the fetus but paternally expressed in the chorioallantois at day 21.38 Studies in human embryonic stem (hES) cells demonstrated imprinting of NESP55 with some expression from the paternal allele.39
In order to further elucidate the timing of imprinting of Gαs and XLαs, as well as confirm early imprinting of NESP55, we examined cell cultures derived from primordial germ cells. Primordial germ cells obtained from the gonadal ridges and attached mesenteries of 5 to 11 week postfertilization male and female embryos were used to derive pluripotent embryonic germ (EG) cell lines.40 EG cells differentiate by forming complex three-dimensional cell aggregates termed as embryoid bodies (EBs), which may contain pluripotent stem cells and cells in various stages of differentiation. The cultured cell lines derived from EBs, embryoid body-derived cells (EBDs), proliferate robustly with a normal karyotype and contain precursors and progenitor cells of various lineages.41 The EG cells have the ability to form all three germ layers and therefore potentially all the organs of the body, and thus are considered a good model for hES cells.
In all EBD cell lines that we examined, the expression of Gαs was biallelic whereas expression of NESP55 and XLαs was preferentially from the maternal or paternal allele, respectively. These results strongly imply that the tissue-specific imprinting of Gαs occurs after 11 weeks postfertilization and after tissue differentiation; the imprinting of XLαs occurs very early postfertilization. In addition, these data are consistent with the prior findings that imprinting of NESP55 in ES cell lines occurs very early in development, although complete imprinting takes place later than 5–11 weeks postfertilization.39
Materials and Methods
EBD cell cultures (LVEE, SLRC, BBEP, SCEC, EDEC, CDEP, and EUEE) were cultured and maintained under conditions as previously described.40 The karyotypes of LVEE were 46XX and of SLRC were 46XY. RNA was isolated using an RNA isolation kit with modified cultured cell protocol (Gentra Systems, Minneapolis, MN, USA). First strand cDNA was synthesized as previously described with appropriate controls.10 NESP55, XLαs, and Gαs PCR products were amplified using specific forward primers corresponding to nucleotide sequences in the first exon for each and a common reverse primer corresponding to nucleotide sequences in exon 6, 8, and 13 (Table 1). To further amplify PCR products from NESP55 and XLαs, nested PCR was performed after DNA was purified from the initial PCR product (Qiagen, Valencia, CA, USA) (Table 1). After PCR, 30 μL samples were electrophoresed in 1.5% agarose gels, stained with ethidium bromide, and appropriate sized bands were isolated. Reactions without reverse transcriptase and without RNA produced no bands. DNA from the PCR products was extracted using QIAquick gel extraction kit (Qiagen). DNA fragments were sequenced and analyzed by phosphoimager quantitation, calculating for percent expression of uniallelic bands minus background as previously described.10 Data are expressed as mean ± SEM. Statistical significance between groups was determined using unpaired t test with differences considered significant at p < 0.05.
Table 1.
Primers and PCR conditions.
| Transcript and primers | PCR conditions | |||
|---|---|---|---|---|
| Specific first exon forward primers | ||||
| NESP55 (Accession no. NMJH6592) | ||||
| Round 1 | 5′-GAAGAGTCGAAGGAGCCCAAG-3′ | 95°C × 6 minutes; 95°C × 30 seconds, 57°C × 30 seconds, 72°C | ||
| Round 2 | 5′-GTCACTAATGGAGGACGCCGT-3′ | × 1 minute 45 seconds × 35 cycles; 72°C × 10 minutes | ||
| XLαs (Accession no. NM_001077490) | ||||
| Round 1 | 5′-GGATGCCTCCGCTGGTTTCAGCATCG-3′ | 95°C × 6 minutes; 95°C × 30 seconds, 57°C × 30 seconds, 72°C | ||
| Round 2 | 5′-CTTTCTCGTGCAAGCCTTC-3′ | × 1 minute 45 seconds × 35 cycles; 72°C × 10 minutes | ||
| Gas (Accession no. NM_000516) | ||||
| Round 1 | 5′-ATGGGCTGCCTCGGGAACAGT-3′ | 95°C × 3 minutes; 94°C × 30 seconds, 66°C × 2 minutes, × 39 cycles; 68°C × 5 minutes | ||
| Common downstream reverse primers | Product size (bp) | |||
| Exon 6 | NESP55 | XLαs | Gαs | |
| Round 1 | 5′-TTCGTAGCAGGCACGCACTCC-3′ | 482 | 616 | 451 |
| Round 2 | 5′-GCCTTGGCATGCTCATAGA-3′ | 329 | 517 | 416 |
| Exon 8 | ||||
| Round 1 | 5′-GTCCACCTGGAACTTGGTCTC-3′ | 635 | 769 | 604 |
| Round 2 | 5′-CCAGAAGTCAGGACACGGCAG-3′ | 489 | 677 | 576 |
| Exon 13 | ||||
| Round 1 | 5′-AGAGCAGCTCGTACTGACGAAG-3′ | 1,172 | 1,306 | 1,141 |
| Round 2 | 5′-GCGCTGAATGATGTCACGGC-3′ | 1,027 | 1,215 | 1,115 |
Results and Discussion
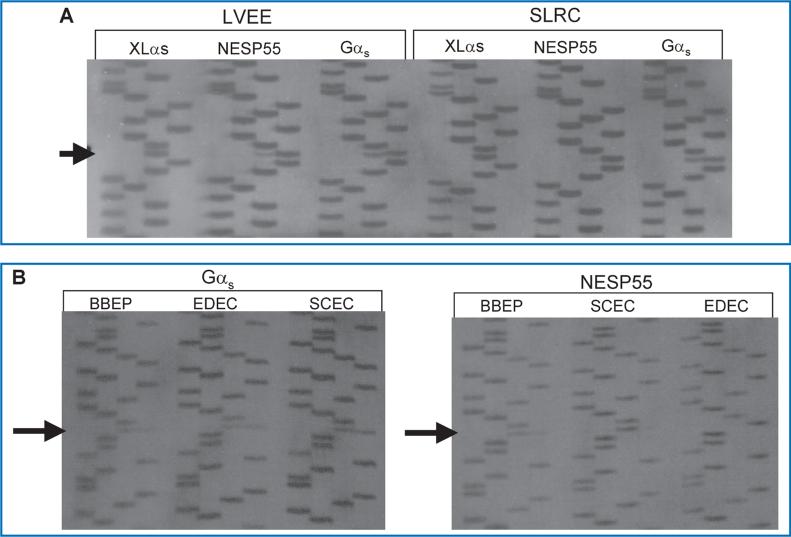
Nearly 100 genes in the human and mouse genomes are imprinted and are therefore monoallelically expressed in a tissue and developmental stage-specific manner (http://www.mgu.har.mrc.ac.uk/research/genomic_imprinting/maps.html).42 Abnormal imprinting of many of these genes results in either anomalous activation or suppression of transcription that leads to a variety of clinical consequences, including abnormal fetal growth, disturbances in neurocognitive development, and neoplasia.43,44 In the present study, we assessed allelic expression of GNAS transcripts in seven different cell cultures (LVEE, SLRC, BBEP, SCEC, EDEC, CDEP, and EUEE) using single nucleotide polymorphisms (SNPs) within exons 5, 7, and 13 of GNAS to distinguish between parent of origin of transcripts.45–47 Some cell lines were also examined under culture conditions to ensure reproducibility regardless of external factors (LVEE and LVEC). Five of the seven cell lines were informative as there was heterozygosity for at least one of the three GNAS SNPs. Two cell lines (LVEE and SLRC) were heterozygous for the exon 5 SNP (Figure 2A), three cell lines (BBEP, EDEC, and SCEC) were heterozygous for the exon 7 SNP (Figure 2B), and three cell lines (BBEP, EDEC, and SCEC) were heterozygous for the exon 13 SNP (data not shown). Gαs transcripts were derived equally from the maternal (52.6% ± 2.5% of total, range: 48.7% to 55.2%) and paternal (47.2% ± 2.6% of total, range: 44.8% to 51.3%) alleles (p = 0.07). By contrast, for all five cell cultures, NESP55 was preferentially expressed from the maternal allele (81.9% ± 10% of total, range: 69.6% to 95.3%), with only a minor contribution derived from the paternal allele (18.1% ± 10%, range: 4.7% to 30.4%) (p = 0.002) (Figure 3). We were only able to amplify XLαs through the exon 5 polymorphism, and thus were limited to two samples for analysis, which showed 97.3% ± 0.3% (range: 96.8% to 97.8%) expression from the paternal allele and 2.7% ± 0.3% (range: 2.2% to 3.2%) expression from the maternal allele (p = 0.007).
Figure 2. (A) Sequence analysis of the product of RT-PCR of XLαs, NESP55, and Gαs exon 5 RNA derived from the two EBD cell cultures (LVEE and SLRC) that were heterozygous for the polymorphism in codon 131 (T/C).
(Polymorphism indicated by arrow; order of base pairs: GATC) (B) Sequence analysis of the product of RT-PCR of Gαs and NESP55 exon 7 RNA derived from three EBD cell cultures (BBEP, EDEC, SCEC) that were heterozygous for the polymorphism in codon 185 (T/C). (Poly morphism indicated by arrows; order of base pairs: GATC).
Figure 3. Relative expression of Gαs, NESP55, and XLαs alleles in EBD cell cultures.
Quantification of band intensity of the polymorphic alleles was performed with the Phosphorimager system (Bio-Rad, Hercules, CA, USA) and corrected for variability of sample loading between lanes. In all five cell cultures heterozygous for the exon 5, 7, or 13 polymorphism, the expression of Gαs was biallelic (maternal allele = 52.6% ± 2.5%; paternal allele = 47.2% ± 2.5%; p = 0.07), whereas NESP55 was expressed preferentially from the maternal allele (maternal allele = 81.9% ± 10%; paternal allele = 18.1% ± 10%; p = 0.002) and XLαs was preferentially expressed from the paternal allele (maternal allele = 2.7% ± 0.3%; paternal allele = 97.3% ± 0.3%; p = 0.007). Error bars indicate SEM.
In the cell lines examined, expression of the imprinted NESP55 allele was greater (18%) than the imprinted XLαs allele (3%). These results are consistent with the report by Rugg-Gunn et al., in which the expression of NESP55 in human ES cells was generally monoallelic but did have random low expression of the imprinted paternal allele (approximately 20% in 2 of 5 cells examined).48 These results are in contrast to similar analyses performed in adult tissues, in which expression of both NESP55 and XLαs appears to be uniallelic.8–13 One explanation for this discrepancy is that imprinting of GNAS, as for some other genes,49 is dynamic during development. Alternatively, imprinting of GNAS does not become fully established until later in tissue differentiation.
Previous studies of specific imprinted genes have shown various patterns regarding the timing of imprinting and have shown discrepancies of imprinting between species. For example, in mice some imprinted genes, including Snrpn and H19, are initially monoallelically expressed postfertilization, then become demethylated and biallelically expressed later in gestation.50–53 However, studies in humans have found that the imprinting of Snrpn may only become established in early postzygotic development.54 Further analyses of H19 in mice have shown that the methylation of the promoter-proximal region of H19 was erased during preimplantation development and reestablished by midgestation, but the methylation of the distal region was preserved throughout embryogenesis.55 By contrast, Rugg-Gunn found that the H19 promoter was differentially methylated in hESCs.48 Our current results extend these interspecies discrepancies to GNAS. In mice, the DMR of Nesp is not established until postimplantation development after the blastocyst stage, and the DMR of Gnasxl is partially methylated in both sperm and oocytes, indicating either that methylation of the maternal allele is not erased in the germ cells or that methylation occurs de novo in these cells.56 By contrast, we found that imprinting of NESP55 is not yet complete whereas XLαs imprinting is essentially complete prior to tissue differentiation.
This study is unique in that expression of Gαs, NESP55, and XLαs is evaluated individually and prior to tissue differentiation in a unique model, EBD cell cultures. Thus, we overcame the limitations of a previous study of Gαs expression in fetal tissues that was confounded by use of primers that amplified all transcripts from the GNAS locus.37 Moreover, EBD cell cultures represent a homogenous population of cells and avoid artifacts introduced using tissues that contain heterogenous populations of cells that may have different patterns of imprinting. EBD cell cultures have been shown to express simultaneously a wide array of mRNA and protein markers that are normally associated with distinct developmental lineages. In addition, these cells are able to differentiate in vitro into derivatives of the three embryonic germ layers, and thus meet the definition of pluripotent stem cells. Although EBD cells are not identical to hES cells, it is important to note that they appear to have comparable epigenetic stability based on analyses showing similar imprinting patterns for IGF2.57
Our study has several limitations. First, we were unable to assess expression of exon 1A alleles. Biallelic expression of Gαs in EBD cell cultures could indicate transient methylation of the paternal exon 1A DMR, or more likely, lack of expression of a putative repressor protein that has been proposed to silence transcription of Gαs from the paternal allele in imprinted tissues.23–25 Second, we did not directly assess methylation status of the DMRs corresponding to NESP, XLαs, and exon 1A ICRs. Finally, because EBD cell cultures contain precursors of various cell lineages, our studies do not necessarily reflect in vivo timing of tissue-specific imprinting of the various gene products nor do they address what occurs in vivo in somatic tissues. Nevertheless, our results further support the hypothesis that Gαs is biallelically expressed in early embryonic development and becomes imprinted after 11 weeks postfertilization and after tissue differentiation. In addition, we have confirmed early imprinting in NESP55 in these human embryonic germ cell cultures as found in ES cell lines by others,39 although complete imprinting takes place later than 5–11 weeks postfertilization. Finally, we have demonstrated that XLαs is imprinted as early as 5–11 weeks postfertilization in human embryonic germ cell cultures. Knowledge regarding the timing of GNAS imprinting may provide further clues regarding the causes of the variable phenotypes associated with mutations in this gene.
Acknowledgments
This work was supported by the National Institutes of Health T-35 HD007446 to the American Pediatric Society/Society for Pediatric Research Student Research Program and a generous donation from the Bosworth Family.
Footnotes
Conflict of Interest
J.L.C., J.A., S.H., M.A.L., and E.L.G-L. have nothing to declare. M.J.S. and Johns Hopkins University School of Medicine have stock and stock options in Geron.
References
- 1.Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2(1):21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 2.Morison IM, Ramsay JP, Spencer HG. A census of mammalian imprinting. Trends Genet. 2005;21(8):457–465. doi: 10.1016/j.tig.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 3.Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet. 2005;14(Spec No 1):R47–R58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- 4.Bastepe M, Juppner H. Identification and characterization of two new, highly polymorphic loci adjacent to GNAS1 on chromosome 20q13.3. Mol Cell Probes. 2000;14(4):261–264. doi: 10.1006/mcpr.2000.0308. [DOI] [PubMed] [Google Scholar]
- 5.Plagge A, Kelsey G. Imprinting the Gnas locus. Cytogenet Genome Res. 2006;113(1–4):178–187. doi: 10.1159/000090830. [DOI] [PubMed] [Google Scholar]
- 6.Bastepe M, Juppner H. GNAS locus and pseudohypoparathyroidism. Hormone Res. 2005;63(2):65–74. doi: 10.1159/000083895. [DOI] [PubMed] [Google Scholar]
- 7.Yu S, Yu D, Lee E, Eckhaus M, Lee R, Corria Z, Accili D, Westphal H, Weinstein LS. Variable and tissue-specific hormone resistance in heterotrimeric Gs protein alpha-subunit (Gsalpha) knockout mice is due to tissue-specific imprinting of the gsalpha gene. Proc Natl Acad Sci USA. 1998;95(15):8715–8720. doi: 10.1073/pnas.95.15.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Germain-Lee EL, Schwindinger W, Crane JL, Zewdu R, Zweifel LS, Wand G, Huso DL, Saji M, Ringel MD, Levine MA. A mouse model of albright hereditary osteodystrophy generated by targeted disruption of exon 1 of the Gnas gene. Endocrinology. 2005;146(11):4697–4709. doi: 10.1210/en.2005-0681. [DOI] [PubMed] [Google Scholar]
- 9.Chen M, Haluzik M, Wolf NJ, Lorenzo J, Dietz KR, Reitman ML, Weinstein LS. Increased insulin sensitivity in paternal Gnas knockout mice is associated with increased lipid clearance. Endocrinology. 2004;145(9):4094–4102. doi: 10.1210/en.2004-0038. [DOI] [PubMed] [Google Scholar]
- 10.Germain-Lee EL, Ding CL, Deng Z, Crane JL, Saji M, Ringel MD, Levine MA. Paternal imprinting of Galpha(s) in the human thyroid as the basis of TSH resistance in pseudohypoparathyroidism type 1a. Biochem Biophys Res Commun. 2002;296(1):67–72. doi: 10.1016/s0006-291x(02)00833-1. [DOI] [PubMed] [Google Scholar]
- 11.Hayward BE, Barlier A, Korbonits M, Grossman AB, Jacquet P, Enjalbert A, Bonthron DT. Imprinting of the G(s)alpha gene GNAS1 in the pathogenesis of acromegaly. J Clin Invest. 2001;107(6):R31–R36. doi: 10.1172/JCI11887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J, Erlichman B, Weinstein LS. The stimulatory G protein alpha-subunit Gs alpha is imprinted in human thyroid glands: implications for thyroid function in pseudohypoparathyroidism types 1A and 1B. J Clin Endocrinol Metab. 2003;88(9):4336–4341. doi: 10.1210/jc.2003-030393. [DOI] [PubMed] [Google Scholar]
- 13.Mantovani G, Ballare E, Giammona E, Beck-Peccoz P, Spada A. The Gαs gene: predominant maternal origin of transcription in human thyroid gland and gonads. J Clin Endocrinol Metab. 2002;87(10):4736–4740. doi: 10.1210/jc.2002-020183. [DOI] [PubMed] [Google Scholar]
- 14.Klemke M, Pasolli HA, Kehlenbach RH, Offermanns S, Schultz G, Huttner WB. Characterization of the extra-large G protein alpha -subunit XLalpha s. II. Signal Transduction Properties. J Biol Chem. 2000;275(43):33633–33640. doi: 10.1074/jbc.M006594200. [DOI] [PubMed] [Google Scholar]
- 15.Pasolli HA, Klemke M, Kehlenbach RH, Wang Y, Huttner WB. Characterization of the extra-large G protein alpha-subunit XLαs. I. Tissue distribution and subcellular localization. J Biol Chem. 2000;275(43):33622–33632. doi: 10.1074/jbc.M001335200. [DOI] [PubMed] [Google Scholar]
- 16.Hayward BE, Moran V, Strain L, Bonthron DT. Bidirectional imprinting of a single gene: GNAS1 encodes maternally, paternally, and biallelically derived proteins. Proc Natl Acad Sci USA. 1998;95(26):15475–15480. doi: 10.1073/pnas.95.26.15475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayward BE, Kamiya M, Strain L, Moran V, Campbell R, Hayashizaki Y, Bonthron DT. The human GNAS1 gene is imprinted and encodes distinct paternally and biallelically expressed G proteins. Proc Natl Acad Sci USA. 1998;95(17):10038–10043. doi: 10.1073/pnas.95.17.10038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ischia R, Lovisetti-Scamihorn P, Hogue-Angeletti R, Wolkersdorfer M, Winkler H, Fischer-Colbrie R. Molecular cloning and characterization of NESP55, a novel chromogranin-like precursor of a peptide with 5-HT1B receptor antagonist activity. J Biol Chem. 1997;272(17):11657–11662. doi: 10.1074/jbc.272.17.11657. [DOI] [PubMed] [Google Scholar]
- 19.Leitner B, Lovisetti-Scamihorn P, Heilmann J, Striessnig J, Blakely RD, Eiden LE, Winkler H. Subcellular localization of chromogranins, calcium channels, amine carriers, and proteins of the exocytotic machinery in bovine splenic nerve. J Neurochem. 1999;72(3):1110–1116. doi: 10.1046/j.1471-4159.1999.0721110.x. [DOI] [PubMed] [Google Scholar]
- 20.Jan de Beur SM, Ding DL, Germain-Lee EL, Cho J, Maret A, Levine MA. Discordance between genetic and epigenetic defects in pseudohypoparathyroidism type 1b revealed by inconsistent loss of maternal imprinting of GNAS1. Am J Hum Genet. 2003;73:314–322. doi: 10.1086/377136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J, Litman D, Rosenberg MJ, Yu S, Biesecker LG, Weinstein LS. A GNAS1 imprinting defect in pseudohypoparathyroidism type IB. J Clin Invest. 2000;106(9):1167–1174. doi: 10.1172/JCI10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swaroop A, Agarwal N, Gruen JR, Bick D, Weissman SM. Differential expression of novel Gsαsignal transduction protein cDNA species. Nucleic Acids Res. 1991;17:4725–4729. doi: 10.1093/nar/19.17.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, Chen M, Deng C, Bourc‘his D, Nealon JG, Erlichman B, Bestor TH, Weinstein LS. Identification of the control region for tissue-specific imprinting of the stimulatory G protein alpha-subunit. Proc Natl Acad Sci USA. 2005;102(15):5513–5518. doi: 10.1073/pnas.0408262102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juppner H, Bastepe M. Different mutations within or upstream of the GNAS locus cause distinct forms of pseudohypoparathyroidism. J Pediatr Endocrinol Metab. 2006;19(Suppl 2):641–646. doi: 10.1515/jpem.2006.19.s2.641. [DOI] [PubMed] [Google Scholar]
- 25.Bastepe M, Pincus JE, Sugimoto T, Tojo K, Kanatani M, Azuma Y, Kruse K, Rosenbloom AL, Koshiyama H, Jüppner H. Positional dissociation between the genetic mutation responsible for pseudohypoparathyroidism type Ib and the associated methylation defect at exon A/B: evidence for a long-range regulatory element within the imprinted GNAS1 locus. Hum Mol Genet. 2001;10(12):1231–1241. doi: 10.1093/hmg/10.12.1231. [DOI] [PubMed] [Google Scholar]
- 26.Long DN, McGuire S, Levine MA, Weinstein LS, Germain-Lee EL. Body mass index differences in pseudohypoparathyroidism type 1a versus pseudopseudohypoparathyroidism may implicate paternal imprinting of Gα(s) in the development of human obesity. J Clin Endocrinol Metab. 2007;92(3):1073–1079. doi: 10.1210/jc.2006-1497. [DOI] [PubMed] [Google Scholar]
- 27.Mouallem M, Shaharabany M, Weintrob N, Shalitin S, Nagelberg N, Shapira H, Zadik Z, Farfel Z. Cognitive impairment is prevalent in pseudohypoparathyroidism type Ia, but not in pseudopseudohypoparathyroidism: possible cerebral imprinting of Gsα. Clin Endocrinol (Oxf) 2008;68(2):233–239. doi: 10.1111/j.1365-2265.2007.03025.x. [DOI] [PubMed] [Google Scholar]
- 28.Albright F, Burnett CH, Smith PH, Parson W. Pseudo-hypoparathyroidism—an example of “Seabright-Bantam Syndrome”—report of three cases. Endocrinology. 1942;30(6):922–932. [Google Scholar]
- 29.Albright F, Forbes AP, Henneman PH. Pseudopseudohypoparathyroidism. Trans Assoc Am Physicians. 1952;65:337–350. [PubMed] [Google Scholar]
- 30.Levine MA. Pseudohypoparathyroidism. Principles of Bone Biology. San Diego: Academic Press. 2002:1137–1163. [Google Scholar]
- 31.Weinstein LS, Yu S, Warner DR, Liu J. Endocrine manifestations of stimulatory G protein alpha-subunit mutations and the role of genomic imprinting. Endocr Rev. 2001;22(5):675–705. doi: 10.1210/edrv.22.5.0439. [DOI] [PubMed] [Google Scholar]
- 32.Wilson LC, Trembath RC. Albright's hereditary osteodystrophy. [Review]. J Med Genet. 1994;31:779–784. doi: 10.1136/jmg.31.10.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Germain-Lee EL, Groman J, Crane JL, Jan de Beur SM, Levine MA. Growth hormone deficiency in pseudohypoparathyroidism type 1a: another manifestation of multihormone resistance. J Clin Endocrinol Metab. 2003;88(9):4059–4069. doi: 10.1210/jc.2003-030028. [DOI] [PubMed] [Google Scholar]
- 34.Mantovani G, Maghnie M, Weber G, De Menis E, Brunelli V, Cappa M, Loli P, Beck-Peccoz P, Spada A. Growth hormone-releasing hormone resistance in pseudohypoparathyroidism type ia: new evidence for imprinting of the Gs alpha gene. J Clin Endocrinol Metab. 2003;88(9):4070–4074. doi: 10.1210/jc.2002-022028. [DOI] [PubMed] [Google Scholar]
- 35.Plagge A, Kelsey G, Germain-Lee EL. Physiological functions of the imprinted Gnas locus and its protein variants Galpha(s) and XLαs in human and mouse. J Endocrinol. 2008;196(2):193–214. doi: 10.1677/JOE-07-0544. [DOI] [PubMed] [Google Scholar]
- 36.Coombes C, Arnaud P, Gordon E, Dean W, Coar EA, Williamson CM, Feil R, Peters J, Kelsey G. Epigenetic properties and identification of an imprint mark in the Nesp-Gnasxl domain of the mouse Gnas imprinted locus. Mol Cell Biol. 2003;23(16):5475–5488. doi: 10.1128/MCB.23.16.5475-5488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campbell R, Gosden CM, Bonthron DT. Parental origin of transcription from the human GNAS1 gene. J Med Genet. 1994;31:607–614. doi: 10.1136/jmg.31.8.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thurston A, Taylor J, Gardner J, Sinclair KD, Young LE. Monoallelic expression of nine imprinted genes in the sheep embryo occurs after the blastocyst stage. Reproduction. 2008;135(1):29–40. doi: 10.1530/REP-07-0211. [DOI] [PubMed] [Google Scholar]
- 39.Rugg-Gunn PJ, Ferguson-Smith AC, Pedersen RA. Human embryonic stem cells as a model for studying epigenetic regulation during early development. Cell Cycle. 2005;4(10):1323–1326. doi: 10.4161/cc.4.10.2076. [DOI] [PubMed] [Google Scholar]
- 40.Shamblott MJ, Axelman J, Wang S, Bugg EM, Littlefi eld JW, Donovan PJ, Blumenthal PD, Huggins GR, Gearhart JD. Derivation of pluripotent stem cells from cultured human primordial germ cells. Proc Natl Acad Sci USA. 1998;95(23):13726–13731. doi: 10.1073/pnas.95.23.13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shamblott MJ, Axelman J, Littlefi eld JW, Blumenthal PD, Huggins GR, Cui Y, Cheng L, Gearhart JD. Human embryonic germ cell derivatives express a broad range of developmentally distinct markers and proliferate extensively in vitro. Proc Natl Acad Sci USA. 2001;98(1):113–118. doi: 10.1073/pnas.021537998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beechey CV, Cattanach BM, Blake A, Peters J. MRC mammalian genetics unit, Harwell, Oxfordshire. [November 5, 2008];Mouse imprinting data and references. 2005 Available at: http://www.mgu.har.mrc.ac.uk/research/genomic_imprinting/.
- 43.Glaser RL, Ramsay JP, Morison IM. The imprinted gene and parent-of-origin effect database now includes parental origin of de novo mutations. Nucleic Acids Res. 2006;34(Database issue):D29–D31. doi: 10.1093/nar/gkj101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feinberg AP. An epigenetic approach to cancer etiology. Cancer J. 2007;13(1):70–74. doi: 10.1097/PPO.0b013e31803c6e3b. [DOI] [PubMed] [Google Scholar]
- 45.Miric A, Vechio JD, Levine MA. Heterogeneous mutations in the gene encoding the alpha subunit of the stimulatory G protein of adenylyl cyclase in Albright hereditary osteodystrophy. J Clin Endocrinol Metab. 1993;76:1560–1568. doi: 10.1210/jcem.76.6.8388883. [DOI] [PubMed] [Google Scholar]
- 46.Aldred MA, Trembath RC. Activating and inactivating mutations in the human GNAS1 gene. Hum Mutat. 2000;16(3):183–189. doi: 10.1002/1098-1004(200009)16:3<183::AID-HUMU1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 47.Waltman C, Levine MA, Schwindinger WF, Wand GS. Polymorphism of the gene encoding the alpha subunit of the stimulatory G-protein of adenylyl cyclase (GNAS1). Hum Genet. 1994;93:477–478. doi: 10.1007/BF00201682. [DOI] [PubMed] [Google Scholar]
- 48.Rugg-Gunn PJ, Ferguson-Smith AC, Pedersen RA. Epigenetic status of human embryonic stem cells. Nat Genet. 2005;37(6):585–587. doi: 10.1038/ng1556. [DOI] [PubMed] [Google Scholar]
- 49.Trasler JM. Gamete imprinting: setting epigenetic patterns for the next generation. Reprod Fertil Dev. 2006;18(1–2):63–69. doi: 10.1071/rd05118. [DOI] [PubMed] [Google Scholar]
- 50.Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002;117(1–2):15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- 51.Szabo PE, Hubner K, Scholer H, Mann JR. Allele-specific expression of imprinted genes in mouse migratory primordial germ cells. Mech Dev. 2002;115(1–2):157–160. doi: 10.1016/s0925-4773(02)00087-4. [DOI] [PubMed] [Google Scholar]
- 52.Li B, Kadura I, Fu DJ, Watson DE. Genotyping with TaqMAMA. Genomics. 2004;83(2):311–320. doi: 10.1016/j.ygeno.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 53.Shemer R, Birger Y, Riggs AD, Razin A. Structure of the imprinted mouse Snrpn gene and establishment of its parental-specific methylation pattern. Proc Natl Acad Sci USA. 1997;94(19):10267–10272. doi: 10.1073/pnas.94.19.10267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.El Maarri O, Buiting K, Peery EG, Kroisel PM, Balaban B, Wagner K, Urman B, Heyd J, Lich C, Brannan CI, Walter J, Horsthemke B. Maternal methylation imprints on human chromosome 15 are established during or after fertilization. Nat Genet. 2001;27(3):341–344. doi: 10.1038/85927. [DOI] [PubMed] [Google Scholar]
- 55.Tremblay KD, Duran KL, Bartolomei MS. A 5′ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol Cell Biol. 1997;17(8):4322–4329. doi: 10.1128/mcb.17.8.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu J, Yu S, Litman D, Chen W, Weinstein LS. Identification of a methylation imprint mark within the mouse Gnas locus. Mol Cell Biol. 2000;20(16):5808–5817. doi: 10.1128/mcb.20.16.5808-5817.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Onyango P, Jiang S, Uejima H, Shamblott MJ, Gearhart JD, Cui H, Feinberg AP. Monoallelic expression and methylation of imprinted genes in human and mouse embryonic germ cell lineages. Proc Natl Acad Sci USA. 2002;99(16):10599–10604. doi: 10.1073/pnas.152327599. [DOI] [PMC free article] [PubMed] [Google Scholar]