Abstract
cAMP-dependent protein kinase (PKA) was the second protein kinase to be discovered and the PKA catalytic (C) subunit serves as a prototype for the large protein kinase superfamily that contains over 500 gene products. The protein kinases regulate much of biology in eukaryotic cells and they are now also a major therapeutic target. Although PKA was discovered nearly 50 years ago and the subsequent discovery of the regulatory subunits that bind cAMP and release the catalytic activity from the holoenzyme followed quickly. Thus in PKA we see the convergence of two major signaling mechanisms - protein phosphorylation and second messenger signaling through cAMP. Crystallography provides a foundation for understanding function, and the structure of the isolated regulatory (R) and C-subunits have been extremely informative. Yet it is the R2C2 holoenzyme that predominates in cells, and one can only appreciate the allosteric features of PKA signaling by seeing the full length protein. The symmetry and the quaternary constraints that one R:C hetero-dimer exerts on the other in the holoenzyme simply are not present in the isolated subunits or even in the R:C hetero-dimer.
Keywords: Cyclic AMP (cAMP), cAMP-dependent protein kinase (PKA), PKA catalytic (C) subunit, PKA Regulatory (R) Subunit, Allostery
Cyclic Nucleotide Binding (CNB) Domain While protein phosphorylation was being discovered as a regulatory mechanism for biological systems through the pioneering studies of Krebs and Fischer in 1959 (Krebs, Graves et al. 1959), the fundamental principles of allostery were being elucidated by Changeux (Monod, Wyman et al. 1965). Independently, Sutherland discovered cAMP as a second messenger for hormone signaling (Rall and Sutherland 1958). The second protein kinase to be discovered in 1968 was cAMP-dependent protein kinase (PKA) (Walsh, Perkins et al. 1968). The discovery that the regulatory (R) subunits of PKA were the major receptors for cAMP (Gill and Garren 1970, Tao, Salas et al. 1970, Brostrom, Corbin et al. 1971) brought together two major regulatory mechanisms, phosphorylation and second messenger signaling, and also introduced the concept of oligomerization and allostery into PKA signaling.
Discovery of PKA holoenzymes and their allosteric regulation
The PKA catalytic (C) subunit, discovered initially as the enzyme responsible for phosphorylating and activating glycogen phosphorylase kinase, was named phosphorylase kinase kinase (Walsh, Perkins et al. 1968). PKA thus originally introduced the concept of cascades in kinase signaling. Only later when its regulatory mechanism was elucidated was it renamed cAMP-dependent protein kinase. PKA was distinct from phosphorylase kinase in several important ways. Phosphorylase kinase is part of a large oligomeric complex that does not dissociate (α4β4,γ4,δ4) whereas the PKA subunits could readily be isolated as free and soluble proteins, which gave PKA a major advantage in terms of biochemical and biophysical characterization. The discovery of the R-subunits defined PKA as an oligomeric protein that contained an R-subunit dimer and two C-subunits (Gill and Garren 1970, Tao, Salas et al. 1970, Brostrom, Corbin et al. 1971). The C-subunit contained the catalytic activity while the R-subunits had high affinity binding sites for cAMP. It was only with the holoenzymes that we came to appreciate that activation of PKA was also highly cooperative with Hill coefficients that were greater than 1. Understanding the molecular mechanism for allosteric activation, however, has taken over four decades, and the mechanistic details are still being elucidated. Understanding PKA allostery emphasizes the importance of biological complexity and oligomerization and also demonstrates why it is essential to reach across scales of time and space and use a range of interdisciplinary techniques.
Our enormous advances in X-ray crystallography also began in the 1950s with the pioneering work of Perutz and Kendrew on myoglobin and hemoglobin (Kendrew, Dickerson et al. 1960, Perutz, Rossmann et al. 1960). The fundamental importance of oligomers for allostery was recognized immediately by Changeux even though at that time hemoglobin was the only oligomeric protein where a structure was available. Describing proteins at atomic level resolution has been a driving force for understanding biological processes ever since. We have made enormous advances in the kinase signaling community beginning with the structure of the PKA C-subunit (Knighton, Zheng et al. 1991), but it is now essential that we understand the large macromolecular signaling complexes. This will require both high and low-resolution data and certainly we will need computational tools to understand the dynamics. Understanding the higher levels of complexity in signaling systems is often challenging not only because of the increased size of the complex but also because of the inherent dynamic properties of signaling proteins in contrast to the stable hemoglobin tetramer and the stable phosphorylase kinase oligomer. The fundamental question for PKA is to understand at atomic level resolution how the binding of a ligand leads to the observed allosteric activation. Structural methods, combined with advanced computational and kinetic studies, make such an approach now feasible. Clearly with PKA one could never fully appreciate the allostery without seeing the oligomers. Visualizing these oligomers and appreciating the fundamental differences between the isoforms, defines for the first time the challenge that we face.
PKA Regulatory Subunits
In the absence of cAMP the dimeric R-subunits bind to and inhibit two C-subunits. Activation is mediated by cAMP binding to the R-subunits, and this process is clearly allosteric based on Hill Coefficients for activation of 1.4–1.8. Initially two classes of R-subunits, referred to as I and II, based on the order in which they eluted from an ion exchange column (Hofmann, Beavo et al. 1975, Rosen, Rubin et al. 1975). Tissue differences were also recognized early on by the work of Rubin who found that there was a distinct form of RII in brain compared to heart (Erlichman, Sarkar et al. 1980). These two RII isoforms were designated as RIIα (heart) and RIIβ (brain). In the intervening decades the sequence, structure, and isoform diversity of the four PKA R-subunits was elucidated; however, the mechanism for allosteric regulation remained obscure. We learned much from structures of the free C- and R-subunits and from truncated R:C heterodimers; however, we did not learn about the fundamental mechanisms for allosteric activation. For this we needed to see the full-length R2C2 holoenzyme structures. Only then did we appreciate the symmetry and beauty of PKA signaling where binding of a small molecule, distal from the R:C interface, unleashes the catalytic activity that is trapped in the R2C2 holoenzyme complex. Here we use RIIβ as a prototype for defining these concepts of allostery since we have high-resolution structures of some of the different states.
In mammals there are four functionally non-redundant R-subunits (RIα, RIβ, RIIα, and RIIβ) (Brandon, Idzerda et al. 1997), and all have the same general domain organization (Fig. 1). At the N-terminus is a four helix bundle that mediates dimerization as well as binding to PKA scaffold proteins that are referred to as A Kinases Anchoring Proteins or AKAPs. Each AKAP contains a signature amphipathic helix that docks with high affinity onto the surface of the dimerization/docking (D/D) domain, and this mechanism allows PKA to be localized to discrete sites in the cell where it is committed to a specific function such as phosphorylation of the tail of a channel (Wong and Scott 2004, Taylor, Ilouz et al. 2012). The D/D domain is joined by a flexible linker to two tandem cyclic nucleotide binding (CNB) domains at the C-terminus of each R-subunit (CNB-A and CNB-B). Embedded within the flexible linker is an Inhibitor Site (IS) that resembles a PKA substrate and docks into the active site cleft of the C-subunit in the absence of cAMP. The RI subunits have a pseudo-substrate IS (RRXG/AX) where the P-site residue is either Gly or Ala while the RII subunits are substrates as well as inhibitors since the P-site residue is Ser. This is a fundamental difference between the RI and RII subunits that has significant functional consequences (Herberg, Doyle et al. 1999, Martin, Deerinck et al. 2007).
Figure 1. Organization of the PKA R-subunits.
A) The organization of the RIα and RIIβ R-subunits and the sequence of the linker regions for all four isoforms are shown. B) SAXS profiles of the PKA RII-homodimers and holoenzymes show the isoform differences. C) Following binding to the C-subunit, the inhibitor site docks to the active site cleft of the catalytic subunit (indicated by a red box) and the C-linker becomes ordered. The N-linker plays an important role in defining the quaternary structure of each holoenzyme and is ordered differently in RIα, RIβ, and RIIβ following their binding to the C-subunit. This isoform-specific positioning of the N-linker contributes in unique ways to the organization of each tetrameric holoenzyme. Highlighted are the positions of the two symmetry related CNB-A domains in the same holoenzyme. Each CNB-A domain also allosterically regulates the adjacent C-subunit in the holoenzyme.
Although the inhibitor site is the most common way to distinguish RI and RII-subunits, there are also structural differences that can clearly be demonstrated even at low resolution. In this regard small angle X-ray and neutron scattering (SAXS/SANS) are especially informative (Fig. 1). What is most striking about the SAXS data is that each of the four R-dimers and holoenzymes is different (Heller, Vigil et al. 2004, Vigil, Blumenthal et al. 2004, Vigil, Blumenthal et al. 2006). Although one might have predicted that having one tetrameric structure would allow us to model all four, the SAXS data says that this will not be possible. The RIα and RIβ homodimers have a similar relatively compact Y-shaped geometry while the two RIIα and RIIβ homodimers are much more extended and rod-like(Taylor, Ilouz et al. 2012). SAXS also showed that there are very substantial differences between RIIα and RIIβ. When the C-subunits bind to RIIα, for example, the structure remains extended and rod-like, and this was confirmed in recent EM studies of RIIα(Smith, Reichow et al. 2013). In contrast, the RIIβ holoenzyme is very compact and almost globular. Linker swaps between RIIα and RIIβ showed that the linker region is responsible for the remarkably different quaternary structures (Vigil, Blumenthal et al. 2006). The RIIβ holoenzyme has the smallest Dmax of the four holoenzymes. For this reason, the compactness and the globular nature based on SAXS, we focused initially on the RIIβ holoenzyme as a candidate for further structural studies.
The four R-subunits are also functionally non-redundant. Only RIα is embryonically lethal when removed(Amieux and McKnight 2002), and most of the known diseases that are associated with PKA signaling are linked to RIα(Horvath, Bertherat et al. 2010). RIα undergoes nonsense mediated decay (nmd) and the resulting haplo-insufficiency, as well as a number of single point mutations, leads to Carney Complex Disease (CNC) which is associated with numerous cardiac and endocrine disorders. CNC is characterized by unregulated PKA and high basal levels of PKA activity. Another disease associated with RIα is acrodysostosis(Linglart, Menguy et al. 2011). This is an autosomal dominant genetic syndrome, and in these patients the activity of PKA is inhibited. The consequences are more severe and include dwarfism and learning disorders. Genetic knockouts of RIβ, in contrast, lead to learning defects and defects in long-term potentiation(Brandon, Zhuo et al. 1995, Huang, Kandel et al. 1995). A recent study shows that a point mutation in the RIβ D/D domain causes severe neurological defects (Wong, Chiu et al. 2014). RIIβ(−/−) mice have a lean phenotype. They do not become obese with a high fat diet nor do they become insulin resistant(Cummings, Brandon et al. 1996, Schreyer, Cummings et al. 2001). A prevailing challenge is to understand how or whether the structure and/or localization of the specific holoenzymes contribute to their functional diversity.
Intrinsically disordered linkers
It is important to appreciate that PKA in cells exists as an R2C2 holoenzyme due to the D/D domain at the N-terminus. Although the entire linker is completely disordered in the free R-subunits, the portion of the linker that extends from the inhibitory site to the CNBA domain becomes partially ordered following binding of the C-subunit (Fig. 1). The remaining part of the linker, referred to as the N-linker, however, remains mostly disordered as discussed later and is mostly missing from the initial PKA holoenzyme structures. The N-linkers exhibit the highest sequence variability among the four isoforms of the R-subunits (Fig. 1) and are major determinants for the organization of the oligomeric (R2C2) holoenzymes.
Much biological information is embedded within the flexible linkers including phosphorylation sites and PEST sequences that target the R-subunits for proteolytic degradation (Fig. 1). Such flexible or intrinsically disordered regions (IDRs) allow for highly dynamic interactions between the various domains and subunits and are characteristic features of many eukaryotic signaling proteins; however, they introduce major challenges for trying to understand at high resolution how such complexes are regulated, as IDRs typically interfere with crystallization by favoring a more dynamic state. Thus, the tendency is to simply delete these regions to conduct structural studies, even though IDRs are extremely important for biological function and regulation. While elucidating structures of the component parts is a logical way to start, ultimately it is the full-length proteins that reflect the physiological state of PKA in cells, and the essential information for mediating allosteric activation by cAMP can only be seen in the R2C2 holoenzymes.
Cyclic Nucleotide Binding (CNB) Domains: mediators of the allosteric transitions
The CNB domain has been conserved throughout biology from bacteria to man as the major receptor for cAMP(Berman, Ten Eyck et al. 2005), and nature has coupled this CNB domain to many signaling nodes(Kannan, Taylor et al. 2007). In prokaryotes there are many catabolite gene activator proteins that regulate gene transcription while in eukaryotes the major CNB domains are coupled to ion channels, guanine nucleotide exchange factors (EPAC) and kinases (PKA and PKG). Here we focus on PKA where the cAMP binding R-subunits are encoded by genes that are distinct from the C-subunit in contrast to cGMP-dependent protein kinase (PKG) where the CNB domains are fused directly to the kinase domain as a single gene product(Francis and Corbin 1999). The sequences of the PKA CNB domains are compared in Fig. 2. Each CNB domain has a helical subdomain that flanks an 8-stranded β sandwich. Located between β strands 6 and 7 is a short loop containing a small helix. This is the signature motif of all CNB domains and is referred to as the phosphate binding cassette (PBC). A highly conserved Arg in the PBC of every cAMP binding CNB domain binds to the phosphate of cAMP while a conserved Glu the beginning of the PBC binds to the ribose OH (Fig. 2). The β sandwich is flanked by two helical motifs. The N3A motif at the N-terminus precedes β1, while the B/C helix follows β8. The three helical domains toggle between two different states, the cAMP bound state (B state or active state) and the holoenzyme state (H state or inactive state), and the correlated motions of these motifs are summarized in general in Fig. 2(Kornev, Taylor et al. 2008). After summarizing the dynamic and allosteric features of the CNB domains, we will describe the two states – one (Active) which is captured in the cAMP bound R-subunit and the other (Inactive) which is captured in the R:C complex.
Figure 2. CNB Domains of R-subunits.
A) Sequence alignments of the R-subunits C-linker and CNB domains. Helices are colored in red and strands in blue. B) The CNB-A domain of RIα is used as a prototype for this highly conserved structural domain. C-subunit bound Inactive (H conformation) and cAMP bound Active (B conformation) (PDB ID 1CX4) conformations are shown. In the absence of cAMP (left) the B/C helix moves “out”, while the N3A motif moves “in”. Upon cAMP binding (right) the PBC moves towards the cAMP phosphate moving the B/C-helix “in” and causing the N3A motif to move “out”. The middle panel shows how the cyclic nucleotide is bound to the Phosphate Binding Cassette, which is the signature morif for this domain.
The dynamic CNB domains function as regulators of PKA activity(Kornev, Taylor et al. 2008). The correlated motions of the three helical motifs in the CNB domain propagate signals that are initiated by the binding and release of cAMP and are sensed by both the flanking CNB domains and in the holoenzyme by the C-subunit even though cAMP per se does not directly contact the C-subunit. The dynamic features of the CNB domain are best described by extensive studies of the CNB-A of RIα(Das, Abu-Abed et al. 2006, Akimoto, Selvaratnam et al. 2013), which serves as a prototypical CNB domain (Fig. 2). Although we have not yet done extensive NMR work with the RIIβ subunits, NMR studies of RIα suggest that the two states (Active and Inactive) are highly populated even in the absence of cAMP and the C-subunit and that the energy barrier between the two states is low(Akimoto, Selvaratnam et al. 2013). This supports a conformational selection model for allostery where the equilibrium between the two states is altered as a consequence of ligand binding or C-subunit binding. These NMR predictions are also reflected in recent Markov models of the RIα CNB-A domain (Malmstrom, Kornev et al. 2015) and for activation of the RIα holoenzyme (Boras, Kornev et al. 2014).
In PKA there are two contiguous CNB domains, and in the presence of cAMP these two CNB domains interact with each other to form a network of allosteric contacts. There are thus two protein interfaces that are sensed by the CNB-A domain. First is the interface with the CNB-B domain. Second is the interface with the C-subunit in the holoenzyme. The cAMP bound conformation represents the Active conformation, and this state is captured in many different isoforms (Su, Dostmann et al. 1995, Diller, Madhusudan et al. 2001, Rinaldi, Wu et al. 2010) (Fig. 3A,B). Although the folds for each cAMP-bound CNB domain can be easily superimposed, the orientation of the two domains relative to each other is actually quite distinct for each R-subunit. In this active conformation, the B/C helix of the CNB-A domain moves “in” towards the cAMP bound PBC while the N3A motif of CNB-A moves “out”. These same motions are seen in the CNB-B domain. In addition to the internal motions of each CNB domain, there is a hydrophobic residue that caps the adenine ring of cAMP, and this residue typically comes from outside the CNB domain. In all PKA isoforms the CNB-B domain provides the hydrophobic capping residue for cAMP that is bound to the CNB-A domain although the location of the capping residue is isoform specific (Berman, Ten Eyck et al. 2005). In RIα the N3A motif of the CNB-B domain, which is contiguous with the B/C helix of CNB-A, caps the cAMP in CNB-A thereby creating a major interface between CNB-A and CNB-B. In RIIβ the capping residue also comes from the CNB-B domain but in this case it is located in the B Helix. The interface between the CNB-A and CNB-B domains is thus very different in RIα and RIIβ (Fig. 3). The recent structure solution of bcy1, the yeast homolog of the PKA R-subunit, shows yet another variation on this theme (Rinaldi, Wu et al. 2010) and demonstrates why it is essential to solve each structure. Having a structure of the CNB domains of RIα is not sufficient to model the interfaces between the two CNB domains in RIIβ, and bcy1.
Figure 3. Flexibility of the Cyclic Nucleotide Binding Domains in the PKA Regulatory Subunits.
A) Different cAMP bound R subunits. Although each CNB domain is highly conserved, the two domains of RIIβ (Diller, Madhusudan et al. 2001)(PDB ID 1CX4) and RIα (Su, Dostmann et al. 1995) (PDB ID 1RGS) are oriented in distinctly different ways. To emphasize this, the two structures have their CNBA domains oriented the same position. B) The R:C conformation of an RIIβ subunit complexed with a C subunit in the holoenzyme complex. C) The conformational changes of the RIIβ in cAMP bound (active) and holoenzyme (inactive) states. The B/C-helix in CNBA is kinked in the cAMP-bound state and extended into a single long helix in the holoenzyme. In the presence of cAMP RIIβ has a compact configuration with two CNB domains packed against each other. In the cAMP free form RIIβ unfolds and forms an extensive interface with the C-subunit of PKA (Zhang, Smith-Nguyen et al. 2012).
The essence of the conformational switch between the active and inactive conformations is captured in the structure of the R:C heterodimer. The major conformational change is seen in the A domain where the B/C helix is extended and moves “out” away from the PBC. Folded on top of the extended B/C helix is the C-terminal portion of the linker that is now locked into place in the holoenzyme by the docking of the Inhibitor Site to the active site cleft of the C-subunit (Fig. 3C).
R:C Heterodimers
While the free R- and C-subunits allowed us to understand how cAMP binds to the R-subunits and how the C-subunit functions as a catalyst, they did not tell us how the C-subunit was inhibited by the R-subunit nor did they explain how catalytic activity is unleashed by cAMP. This required structures of R:C heterodimers that were engineered with monomeric forms of the R-subunits that included the inhibitor site. These structures allowed us to appreciate for the first time the inherent conformational flexibility of the CNB domain and the domain reorganization of the two CNB domains. In these R:C heterodimers we also see how part of the linker is organized(Kim, Xuong et al. 2005, Kim, Cheng et al. 2007, Wu, Brown et al. 2007). As predicted the Inhibitor Site, which is disordered in free R-subunits, is locked into the active site cleft of the catalytic subunit so that it sterically blocks binding of other substrates. The C-linker region is wrapped around the C-Lobe of the C-subunit where is covers the B/C Helix of CNB-A. The two CNB domains are then docked onto the C-Lobe of the C-subunit. The heterodimers revealed, in particular, the flexibility of the CNB domains as they extend across the C-Lobe. Binding of cAMP to the CNB-A domain thus stabilizes a new protein:protein interface between CNB-A and the C-subunit and alters the previous interface between the CNB-A and CNB-B domains. This extended conformation seen in the inhibited R:C heterodimers is very similar for all four isoforms, which is in striking contrast to the active conformations that are seen in the dissociated cAMP-bound R-subunits.
Full-length R2C2 Holoenzyme
Although the R:C heterodimer structures contain all of the information for high affinity binding of the C-subunit, the Hill Coefficients for activation of the R:C heterodimers is still close to 1.0. These structures thus do not capture the allostery that is embedded within the full-length holoenzyme where the Hill coefficients are 1.4–1.8 (Table 1). The structure of the R2C2 RIIβ holoenzyme allowed us for the first time to appreciate not only the symmetry of the holoenzyme but also the extensive cross talk between the two heterodimers and the importance of this cross talk for mediating allosteric activation (Zhang, Smith-Nguyen et al. 2012). The domain organization of the RIIβ holoenzyme as well as the detailed interactions between the two RC heterodimers (R:C and R′:C′) are shown in Fig. 4. Although the dimerization domain is present in the RIIβ construct, we do not see density for the D/D domain, most likely because it is flexible. The oligomeric constraints of one heterodimer on the other defines this holoenzyme complex clearly as a dimer of dimers.
Table 1.
Isoform-specific allosteric activations of PKAa
| Complex | Ka | Hill Coefficient |
|---|---|---|
| RIα2:C2 | 101nM | 1.7 |
| RIβ2:C2 | 29nM | 1.4 |
| RIIα2:C2 | 137nM | 1.5 |
| RIIβ2:C2 | 584nM | 1.8 |
| RIIβ(108–402):C | 65nM | 0.82 |
| RIIβ(1–280)2:C2 | 60nM | 1.2 |
| RIIβ(R230K)2:C2 | 12.9μM | 0.99 |
| RIIβ(R359K)2:C2 | 490nM | 0.62 |
| RIIα(90–400):C | 40nM | 0.81 |
The Ka and Hill Coefficient numbers are from (Zawadzki and Taylor 2004, Ilouz, Bubis et al. 2012, Zhang, Smith-Nguyen et al. 2012)
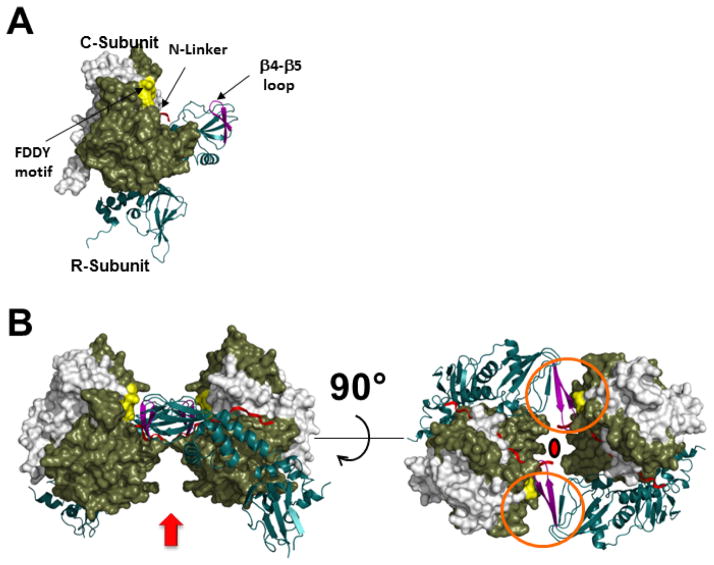
Figure 4. Structure of the R2C2 RIIβ Holoenzyme.
A) The structure of the RIIβ:C heterodimer is shown. The RIIβ is shown as a ribbon while the C-subunit is shown as a space filing model with the N-terminal residues (14–121 are in white and the C-terminal residues (12–350) in tan. Two motifs and the N-linker of RIIβ-subunit (red) are exposed to solvent in the R heterodimers, but have important roles in the assembly of the holoenzyme. The β4–β5 loop (purple) in RIIβ is located within the CNB-A domain of the R-subunits. The Phe-Asp-Asp-Tyr (FDDY) motif (yellow) resides in the C-tail of the C-subunit and is an integral part of the ATP-binding site. B) The quaternary structure of the R2C2 RIIβ holoenzyme is shown. The possible position of the D/D domain is shown by an arrow. The linkers are shown in red. Rotation allows one to appreciate the twofold symmetry that is found in the holoenzyme and also how the two motifs, the β4–β5 loops and the FDDY motifs, enclosed in red circles, contribute to the assembly of the holoenyzme. The twofold axis of symmetry is indicated by a red dot.
The allosteric properties of the RIIβ holoenzyme are distinct from the other three PKA isoforms (Zhang, Smith-Nguyen et al. 2012). The R2C2 holoenzyme, for example, has a very high Ka for activation by cAMP (584 nM) in contrast to the heterodimer (Ka = 65nM) (Table 1). The Ka for the RIIβ tetrameric holoenzyme is also significantly higher than the Ka for other holoenzymes (101nM in RIα, 29nM in RIβ, and 137nM in RIIα tetrameric holoenzymes). The Hill Coefficient for the holoenzyme is also 1.8 in contrast to 0.82 for the heterodimer. The increased Ka for the RIIβ tetrameric holoenzyme can be explained by the extensive interfaces between the two heterodimers where the PBC docking site for cAMP in CNB-A is juxtapositioned directly against the ATP binding site in N-Lobe of the C-subunit in the opposite heterodimer (Fig. 4). One also needs to recruit the cAMP capping residue from ~40Å away to form a cAMP-bound R-dimer (Fig. 3). Hence the RIIβ holoenzyme is more resistant to cAMP activation. The considerable crosstalk between the two CNB domains and the between the two heterodimers is also reflected by several mutations. When the CNB-B domain is deleted entirely, for example, the Ka for activation is 60 nM and the Hill coefficient is reduced to 1.2 indicating that there is still significant allosteric cross talk between the two CNB-A domains(Zhang, Smith-Nguyen et al. 2012). When the cAMP binding site in the CNB-A domain is destroyed by mutating Arg230 to Lys, the Ka for activation is very high, 13μM, confirming that cAMP binding to CNB-A is essential for activation. However, when the same mutation is introduced into the CNB-B domain, the enzyme still has a similar Ka for activation (490nM) but all cooperativity is lost. The molecular basis for this complex allostery could simply not be appreciated in the absence of the holoenzyme structure.
The major interface between the two heterodimers in the holoenzyme is between the N-Lobe of the C-subunit and the CNB-A domain of the R′ subunit (Fig. 4). Two motifs form this interface. One is the FDDY motif in the C-terminal tail of the C-subunit (Kannan, Haste et al. 2007, Romano, Kannan et al. 2009) and the other is the β4-β5 loop in the R′ subunit. In the RIIα:C heterodimer this portion of the C-tail in was disordered, and the temperature factors for the entire N-Lobe of the C-subunit were very high (Fig. 5A) In contrast in the R2C2 holoenzyme, these two motifs are now engaged by the opposite R:C heterodimer, and the temperature factors are very low (Fig. 5C). This quaternary constraint was not seen in the R:C heterodimer. The consequence of the β4-β5 loop of the R′-subunit being buttressed up against the C-Tail of the C-subunit is that the C-tail is now pushed into a closed conformation even though there is no nucleotide bound. The C-tail, and in particular the FDDY motif, is typically disordered in the nucleotide free C-subunit (Bastidas, Wu et al. 2014) (Fig. 5B), and it was also highly disordered in the RIIα:C heterodimer(Wu, Brown et al. 2007). Here it is ordered independent of the nucleotide. The β4-β5 loop from the RIIβ ′-subunit engages most of the ATP binding machinery of the N-lobe in the opposite C-subunit and assembles it into a closed conformation. Based on previous structures of the ligand-free C-subunit, ATP is required to bring these elements together (Fig. 5B). The apo C-subunit has an open conformation, and the FDDY motif in the C-tail is disordered. In the R2C2 holoenzyme, the β4-β5 loop of the R′ heterodimer accomplishes the same thing. Instead of assembling the ATP binding pocket by ATP, the β4-β5 loop pushes the C-Tail into a closed conformation. The holoenzyme gave a stunning view of how allostery might be achieved by the cooperative interaction of the R and C domains in the tetrameric oligomer. cAMP bound to one domain not only influences the interaction of that domain with the C-linker and in turn the active site of its own C-subunit but at the same time through its β4-β5 loop is contacting the C-Tail of the C′ subunit. The cAMP does not directly interact with the inhibitor site of its own C-subunit nor does it directly contact the C-tail of the C-subunit yet binding of this small molecule to the CNB-A domain has a major effect on disrupting the protein interfaces. One could never appreciate these interactions from a heterodimer that contains only one R and one C-subunit (Fig. 5D).
Figure 5. The conformation and B-factor analysis of the C-subunit in RII:C heterodimer and the RIIβ2:C2 holoenzyme structures.
A) In RIIα:C (Wu, Brown et al. 2007) (PDB ID: 2QVS) heterodimer, the C subunit is dynamic with high B factors and has the active site open in the absence of ATP. B) The dynamic nature of the C-tail. The intermediate state (PDB ID:4NTT), colored red, adopts a conformation that does not conform to the open state colored olive (PDB ID: 4NTS), or the closed state colored gray (PDB ID: 1ATP). The residues Phe327 and Tyr330 are displayed in surface representation which are important for ATP binding, colored black. C) In RIIβ2:C2 (PDB ID: 3TNP) holoenzyme, the C subunit is more stable with relatively low B factors and has the active site closed even in the absence of ATP. D) Allosteric interactions of the two RIIβ subunits in the holoenzyme.
The initial holoenzyme was crystallized in the absence of ATP; however, seeing that the ATP binding chamber was perfectly assembled we asked whether we could add ATP to the crystals and trap it in the preformed site. Surprisingly we did not trap ATP but instead trapped both ADP and the phosphorylated R-subunit (Fig. 6). In other words, we were able to carry out a full catalytic cycle in this crystal. The temperature factors were not high for the phosphorylated inhibitor site suggesting that product release is not significantly influenced by phosphorylation of the inhibitor site Ser. This is the first time that both products have been trapped in the crystal lattice of a protein kinase. The structure of the ADP bound holoenzyme shows clearly how these two nucleotides, ADP and cAMP, compete for the two subunits of the holoenzyme Fig. 6. The essence of the allosteric regulation of the RIIβ holoenzyme is embedded within this interface. While we do not have a structure of the cAMP bound holoenzyme, we know that the RR/AA mutant, which cannot bind to the active site, still forms a compact holoenzyme that is no longer inhibited (Wang, Scott et al. 1991). Our challenge is to trap these different conformational states of the holoenzyme even at low resolution.
Figure 6. Allosteric interactions in the RIIβ holoenzyme allow for single turnover phosphorylation of RIIβ.
A) Left: The active site of the C-subunit in the RIIβ2:C2 tetrameric holoenzyme is fully closed in the absence of ATP. Right: By diffusing MgATP into the apo-crystals, the reaction products (ADP, (P)-RIIβ, and 2 Mg2+ ions are trapped. B) The binding site for cAMP to CNB-A′ in R′C′ heterodimer (left arrow) is juxta-positioned against the ATP binding site in the N-Lobe of the C-subunit in the symmetry related RC dimer. The trajectory for binding of cAMP to the PBC is indicated by a red arrow.
Future challenges
While we were able to learn much from high resolution structures of the PKA RIIβ- and C-subunits, these structures did not explain how the activity of PKA was inhibited by the R-subunit nor did they explain cAMP-mediated activation. They also did not shed light on the allosteric mechanism nor did the R:C heterdimers. It was only the full length R2C2 holoenzyme that allowed us to at least begin to understand symmetry and allostery although many questions remain unanswered. We still do not understand, for example, how the flexible linkers communicate with the folded D/D domains and the folded CNB domains and the C-subunit. Most importantly, we do not understand the dynamic intermediate states that result from cAMP binding to the holoenzyme. Without this we will never have a full mechanistic understanding of how the allosteric activation of PKA is achieved.
Acknowledgments
We acknowledge members of the Taylor laboratory over many decades for the interdisciplinary work, both structural and biochemical, that has allowed us to achieve an understanding of these proteins. We especially acknowledge earlier discussions and encouragement from Professor Shmuel Shaltiel from the Weizmann Institute who never let us forget the importance of the holoenzyme after we solved the first crystal structures of the C-subunit. We are also enormously grateful for discussions and encouragement and guidance from Professor Jean Pierre Changeux whose visits to UCSD over the past seven years have helped us to understand and appreciate and, hopefully in the end, to visualize the mechanistic details of the allosteric activation of PKA. Obviously we are all still learning and challenges remain!
Funding: This study was funded by NIH grants GM34921 and GM19301 to SST and from the Howard Hughes Medical Institute to SST.
Footnotes
Conflict of interest: None.
Human & animal studies: This article does not contain any studies with human or animal subjects performed by the author, with the exception of those carried out on electric fish.
Informed consent: Informed consent was obtained from all individual participants included in the study.
References
- Akimoto M, Selvaratnam R, McNicholl ET, Verma G, Taylor SS, Melacini G. Signaling through dynamic linkers as revealed by PKA. Proc Natl Acad Sci U S A. 2013;110(35):14231–14236. doi: 10.1073/pnas.1312644110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amieux PS, McKnight GS. The essential role of RI alpha in the maintenance of regulated PKA activity. Ann N Y Acad Sci. 2002;968:75–95. doi: 10.1111/j.1749-6632.2002.tb04328.x. [DOI] [PubMed] [Google Scholar]
- Bastidas AC, Wu J, Taylor SS. Molecular features of product release for the PKA catalytic cycle. Biochemistry. 2014 doi: 10.1021/bi500684c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Ten Eyck LF, Goodsell DS, Haste NM, Kornev A, Taylor SS. The cAMP binding domain: An ancient signaling module. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(1):45–50. doi: 10.1073/pnas.0408579102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boras BW, Kornev A, Taylor SS, McCulloch AD. Using Markov state models to develop a mechanistic understanding of protein kinase A regulatory subunit RIalpha activation in response to cAMP binding. J Biol Chem. 2014;289(43):30040–30051. doi: 10.1074/jbc.M114.568907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandon EP, Idzerda RL, McKnight GS. PKA isoforms, neural pathways, and behaviour: making the connection. Curr Opin Neurobiol. 1997;7(3):397–403. doi: 10.1016/s0959-4388(97)80069-4. [DOI] [PubMed] [Google Scholar]
- Brandon EP, Zhuo M, Huang YY, Qi M, Gerhold KA, Burton KA, Kandel ER, McKnight GS, Idzerda RL. Hippocampal long-term depression and depotentiation are defective in mice carrying a targeted disruption of the gene encoding the RI beta subunit of cAMP-dependent protein kinase. Proc Natl Acad Sci U S A. 1995;92(19):8851–8855. doi: 10.1073/pnas.92.19.8851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brostrom CO, Corbin JD, King CA, Krebs EG. Interaction of the subunits of adenosine 3′:5′-cyclic monophosphate-dependent protein kinase of muscle. Proc Natl Acad Sci U S A. 1971;68(10):2444–2447. doi: 10.1073/pnas.68.10.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings DE, Brandon EP, Planas JV, Motamed K, Idzerda RL, McKnight GS. Genetically lean mice result from targeted disruption of the RII beta subunit of protein kinase A. Nature. 1996;382(6592):622–626. doi: 10.1038/382622a0. [DOI] [PubMed] [Google Scholar]
- Das R, Abu-Abed M, Melacini G. Mapping allostery through equilibrium perturbation NMR spectroscopy. J Am Chem Soc. 2006;128(26):8406–8407. doi: 10.1021/ja060046d. [DOI] [PubMed] [Google Scholar]
- Diller TC, Madhusudan, Xuong NH, Taylor SS. Molecular basis for regulatory subunit diversity in cAMP-dependent protein kinase: crystal structure of the type II beta regulatory subunit. Structure. 2001;9(1):73–82. doi: 10.1016/s0969-2126(00)00556-6. [DOI] [PubMed] [Google Scholar]
- Erlichman J, Sarkar D, Fleischer N, Rubin CS. Identification of two subclasses of type II cAMP-dependent protein kinases. Neural-specific and non-neural protein kinases. J Biol Chem. 1980;255(17):8179–8184. [PubMed] [Google Scholar]
- Francis SH, Corbin JD. Cyclic nucleotide-dependent protein kinases: intracellular receptors for cAMP and cGMP action. Crit Rev Clin Lab Sci. 1999;36(4):275–328. doi: 10.1080/10408369991239213. [DOI] [PubMed] [Google Scholar]
- Gill GN, Garren LD. A cyclic-3′,5′-adenosine monophosphate dependent protein kinase from the adrenal cortex: comparison with a cyclic AMP binding protein. Biochem Biophys Res Commun. 1970;39(3):335–343. doi: 10.1016/0006-291x(70)90581-4. [DOI] [PubMed] [Google Scholar]
- Heller WT, Vigil D, Brown S, Blumenthal DK, Taylor SS, Trewhella J. C subunits binding to the protein kinase A RI alpha dimer induce a large conformational change. J Biol Chem. 2004;279(18):19084–19090. doi: 10.1074/jbc.M313405200. [DOI] [PubMed] [Google Scholar]
- Herberg FW, Doyle ML, Cox S, Taylor SS. Dissection of the nucleotide and metal-phosphate binding sites in cAMP-dependent protein kinase. Biochemistry. 1999;38(19):6352–6360. doi: 10.1021/bi982672w. [DOI] [PubMed] [Google Scholar]
- Hofmann F, Beavo JA, Bechtel PJ, Krebs EG. Comparison of adenosine 3′:5′-monophosphate-dependent protein kinases from rabbit skeletal and bovine heart muscle. J Biol Chem. 1975;250(19):7795–7801. [PubMed] [Google Scholar]
- Horvath A, Bertherat J, Groussin L, Guillaud-Bataille M, Tsang K, Cazabat L, Libe R, Remmers E, Rene-Corail F, Faucz FR, Clauser E, Calender A, Bertagna X, Carney JA, Stratakis CA. Mutations and polymorphisms in the gene encoding regulatory subunit type 1-alpha of protein kinase A (PRKAR1A): an update. Hum Mutat. 2010;31(4):369–379. doi: 10.1002/humu.21178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YY, Kandel ER, Varshavsky L, Brandon EP, Qi M, Idzerda RL, McKnight GS, Bourtchouladze R. A genetic test of the effects of mutations in PKA on mossy fiber LTP and its relation to spatial and contextual learning. Cell. 1995;83(7):1211–1222. doi: 10.1016/0092-8674(95)90146-9. [DOI] [PubMed] [Google Scholar]
- Ilouz R, Bubis J, Wu J, Yim YY, Deal MS, Kornev AP, Ma Y, Blumenthal DK, Taylor SS. Localization and quaternary structure of the PKA RIbeta holoenzyme. Proc Natl Acad Sci U S A. 2012;109(31):12443–12448. doi: 10.1073/pnas.1209538109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan N, Haste N, Taylor SS, Neuwald AF. The hallmark of AGC kinase functional divergence is its C-terminal tail, a cis-acting regulatory module. Proc Natl Acad Sci U S A. 2007;104(4):1272–1277. doi: 10.1073/pnas.0610251104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan N, Taylor SS, Zhai Y, Venter JC, Manning G. Structural and functional diversity of the microbial kinome. PLoS Biol. 2007;5(3):e17. doi: 10.1371/journal.pbio.0050017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrew JC, Dickerson RE, Strandberg BE, Hart RG, Davies DR, Phillips DC, Shore VC. Structure of myoglobin: A three-dimensional Fourier synthesis at 2 A. resolution. Nature. 1960;185(4711):422–427. doi: 10.1038/185422a0. [DOI] [PubMed] [Google Scholar]
- Kim C, Cheng CY, Saldanha SA, Taylor SS. PKA-I holoenzyme structure reveals a mechanism for cAMP-dependent activation. Cell. 2007;130(6):1032–1043. doi: 10.1016/j.cell.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Kim C, Xuong NH, Taylor SS. Crystal structure of a complex between the catalytic and regulatory (RIalpha) subunits of PKA. Science. 2005;307(5710):690–696. doi: 10.1126/science.1104607. [DOI] [PubMed] [Google Scholar]
- Knighton DR, Zheng JH, Ten Eyck LF, Ashford VA, Xuong NH, Taylor SS, Sowadski JM. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253(5018):407–414. doi: 10.1126/science.1862342. [DOI] [PubMed] [Google Scholar]
- Kornev AP, Taylor SS, Ten Eyck LF. A generalized allosteric mechanism for cis-regulated cyclic nucleotide binding domains. PLoS Comput Biol. 2008;4(4):e1000056. doi: 10.1371/journal.pcbi.1000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs EG, Graves DJ, Fischer EH. Factors affecting the activity of muscle phosphorylase B kinase. Journal of Biological Chemistry. 1959;234(11):2867–2873. [PubMed] [Google Scholar]
- Linglart A, Menguy C, Couvineau A, Auzan C, Gunes Y, Cancel M, Motte E, Pinto G, Chanson P, Bougneres P, Clauser E, Silve C. Recurrent PRKAR1A mutation in acrodysostosis with hormone resistance. N Engl J Med. 2011;364(23):2218–2226. doi: 10.1056/NEJMoa1012717. [DOI] [PubMed] [Google Scholar]
- Malmstrom R, Kornev AP, Taylor SS, Amaro R. Allostery through conformational selection: cAMP regulation of the cyclic-nucleotide binding domain in PKA. 2015. (Submitted) [Google Scholar]
- Martin BR, Deerinck TJ, Ellisman MH, Taylor SS, Tsien RY. Isoform-specific PKA dynamics revealed by dye-triggered aggregation and DAKAP1alpha-mediated localization in living cells. Chem Biol. 2007;14(9):1031–1042. doi: 10.1016/j.chembiol.2007.07.017. [DOI] [PubMed] [Google Scholar]
- Monod J, Wyman J, Changeux JP. On the Nature of Allosteric Transitions: A Plausible Model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- Perutz MF, Rossmann MG, Cullis AF, Muirhead H, Will G, North AC. Structure of haemoglobin: a three-dimensional Fourier synthesis at 5.5-A. resolution, obtained by X-ray analysis. Nature. 1960;185(4711):416–422. doi: 10.1038/185416a0. [DOI] [PubMed] [Google Scholar]
- Rall TW, Sutherland EW. Formation of a cyclic adenine ribonucleotide by tissue particles. J Biol Chem. 1958;232(2):1065–1076. [PubMed] [Google Scholar]
- Rinaldi J, Wu J, Yang J, Ralston CY, Sankaran B, Moreno S, Taylor SS. Structure of yeast regulatory subunit: a glimpse into the evolution of PKA signaling. Structure. 2010;18 (11):1471–1482. doi: 10.1016/j.str.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano RA, Kannan N, Kornev AP, Allison CJ, Taylor SS. A chimeric mechanism for polyvalent trans-phosphorylation of PKA by PDK1. Protein Sci. 2009;18(7):1486–1497. doi: 10.1002/pro.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen OM, Rubin CS, Erlighman J. Properties of the cyclid AMP-dependent protein kinase from bovine and porcine heart. Adv Enzyme Regul. 1975;13:173–185. doi: 10.1016/0065-2571(75)90014-x. [DOI] [PubMed] [Google Scholar]
- Schreyer SA, Cummings DE, McKnight GS, LeBoeuf RC. Mutation of the RIIbeta subunit of protein kinase A prevents diet-induced insulin resistance and dyslipidemia in mice. Diabetes. 2001;50(11):2555–2562. doi: 10.2337/diabetes.50.11.2555. [DOI] [PubMed] [Google Scholar]
- Smith FD, Reichow SL, Esseltine JL, Shi D, Langeberg LK, Scott JD, Gonen T. Intrinsic disorder within an AKAP-protein kinase A complex guides local substrate phosphorylation. Elife. 2013;2:e01319. doi: 10.7554/eLife.01319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Dostmann WR, Herberg FW, Durick K, Xuong NH, Ten Eyck L, Taylor SS, Varughese KI. Regulatory subunit of protein kinase A: structure of deletion mutant with cAMP binding domains. Science. 1995;269(5225):807–813. doi: 10.1126/science.7638597. [DOI] [PubMed] [Google Scholar]
- Tao M, Salas ML, Lipmann F. Mechanism of activation by adenosine 3′:5′-cyclic monophosphate of a protein phosphokinase from rabbit reticulocytes. Proc Natl Acad Sci U S A. 1970;67 (1):408–414. doi: 10.1073/pnas.67.1.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SS, Ilouz R, Zhang P, Kornev AP. Assembly of allosteric macromolecular switches: lessons from PKA. Nature Reviews Molecular Cell Biology. 2012;13(10):646–658. doi: 10.1038/nrm3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigil D, Blumenthal DK, Heller WT, Brown S, Canaves JM, Taylor SS, Trewhella J. Conformational differences among solution structures of the type Ialpha, IIalpha and IIbeta protein kinase A regulatory subunit homodimers: role of the linker regions. J Mol Biol. 2004;337(5):1183–1194. doi: 10.1016/j.jmb.2004.02.028. [DOI] [PubMed] [Google Scholar]
- Vigil D, Blumenthal DK, Taylor SS, Trewhella J. Solution scattering reveals large differences in the global structures of type II protein kinase A isoforms. J Mol Biol. 2006;357(3):880–889. doi: 10.1016/j.jmb.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Walsh DA, Perkins JP, Krebs EG. An Adenosine 3′,5′-Monophosphate-Dependant Protein Kinase from Rabbit Skeletal Muscle. Journal of Biological Chemistry. 1968;243(13):3763-&. [PubMed] [Google Scholar]
- Wang YH, Scott JD, McKnight GS, Krebs EG. A constitutively active holoenzyme form of the cAMP-dependent protein kinase. Proc Natl Acad Sci U S A. 1991;88(6):2446–2450. doi: 10.1073/pnas.88.6.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong TH, Chiu WZ, Breedveld GJ, Li KW, Verkerk AJ, Hondius D, Hukema RK, Seelaar H, Frick P, Severijnen LA, Lammers GJ, Lebbink JH, van Duinen SG, Kamphorst W, Rozemuller AJ, Bakker EB, Neumann M, Willemsen R, Bonifati V, Smit AB, van Swieten J. PRKAR1B mutation associated with a new neurodegenerative disorder with unique pathology. Brain. 2014;137(Pt 5):1361–1373. doi: 10.1093/brain/awu067. [DOI] [PubMed] [Google Scholar]
- Wong W, Scott JD. AKAP signalling complexes: focal points in space and time. Nat Rev Mol Cell Biol. 2004;5(12):959–970. doi: 10.1038/nrm1527. [DOI] [PubMed] [Google Scholar]
- Wu J, Brown SH, von Daake S, Taylor SS. PKA type IIalpha holoenzyme reveals a combinatorial strategy for isoform diversity. Science. 2007;318(5848):274–279. doi: 10.1126/science.1146447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawadzki KM, Taylor SS. cAMP-dependent protein kinase regulatory subunit type IIbeta: active site mutations define an isoform-specific network for allosteric signaling by cAMP. J Biol Chem. 2004;279(8):7029–7036. doi: 10.1074/jbc.M310804200. [DOI] [PubMed] [Google Scholar]
- Zhang P, Smith-Nguyen EV, Keshwani MM, Deal MS, Kornev AP, Taylor SS. Structure and Allostery of the PKA RIIbeta Tetrameric Holoenzyme. Science. 2012;335(6069):712–716. doi: 10.1126/science.1213979. [DOI] [PMC free article] [PubMed] [Google Scholar]