Abstract
Background
Copy number variation on chromosome 15q11.2 (BP1-BP2) causes deletion of CYFIP1, NIPA1, NIPA2 and TUBGCP5; it also affects brain structure and elevates risk for several neurodevelopmental disorders that are associated with dendritic spine abnormalities. In rodents, altered cyfip1 expression changes dendritic spine morphology, motivating analyses of human neuronal cells derived from iPSCs (iPSC-neurons).
Methods
iPSCs were generated from a mother and her offspring, both carrying the 15q11.2 (BP1-BP2) deletion, and a non-deletion control. Gene expression in the deletion region was estimated using quantitative real-time PCR assays. Neural progenitor cells (NPCs) and iPSC-neurons were characterized using immunocytochemistry.
Results
CYFIP1, NIPA1, NIPA2 and TUBGCP5 gene expression was lower in iPSCs, NPCs and iPSC-neurons from the mother and her offspring in relation to control cells. CYFIP1 and PSD95 protein levels were lower in iPSC-neurons derived from the CNV bearing individuals using Western blot analysis. At 10 weeks post-differentiation, iPSC-neurons appeared to show dendritic spines and qualitative analysis suggested that dendritic morphology was altered in 15q11.2 deletion subjects compared with control cells.
Conclusions
The 15q11.2 (BP1-BP2) deletion is associated with reduced expression of four genes in iPSC-derived neuronal cells; it may also be associated altered iPSC-neuron dendritic morphology.
Keywords: intellectual disability, 15q11.2, dendrite, Autism, Copy Number Variation, Dendritic spine, Dendritogenesis, Induce pluripotent stem cells, iPSC, Schizophrenia
INTRODUCTION
The proximal long arm of chromosome 15 (15q11.2-q13) harbors several copy number variants (CNVs) that can increase the risk for common, severe neuropsychiatric disorders (1, 2, 3). The CNVs arise from mispaired low copy number repeats (LCRs) at three breakpoints denoted BP1, BP2 and BP3. A deletion between BP1 and BP2 denoted 15q11.2 (BP1-BP2) elevates risk for intellectual disability (ID), autism spectrum disorders (ASD), schizophrenia (SZ) and seizure disorders, with associated dysmorphic features and neurocognitive developmental delays (4–10). The 15q11.2 (BP1-BP2) deletion is usually inherited (but not imprinted) and has an estimated population frequency of 0.25%. Discrete disabilities in mathematics learning, reading skills and marginally reduced intelligence quotient (IQ) have been observed among individuals with the 15q11.2 (BP1-BP2) deletion, even if the individuals have not been diagnosed with psychiatric disorders (11). Furthermore, comparison of individuals with 15q11.2 (BP1-BP2) deletions/duplications has shown allele-dosage effects in brain regions implicated in psychoses (11). It is noteworthy that the 15q11.2 (BP1-BP2) deletion is distinct from imprinted CNVs arising between either BP1-BP3 or BP2-BP3 regions that cause the Prader-Willi and Angelman syndromes (1, 8).
The BP1-BP2 deletion region encodes four genes (12): (i) Non imprinted in Prader-Willi/Angelman syndrome-1 (NIPA1) mediates Mg++ transport in mouse neuronal tissue (10) (13); (ii) Non imprinted in Prader-Willi/Angelman syndrome-2 (NIPA2) mediates renal Mg++ transport (14); (iii) Cytoplasmic FMRP-interacting Protein-1 (CYFIP1) regulates cytoskeletal dynamics (15); (iv) Tubulin, gamma complex associated protein-5 (TUBGCP5) is required for microtubule nucleation at the centrosome (16). Recently, Pathania and colleagues reported that cyfip1 is enriched at mouse neuronal synapses (17). Rodent knockdown studies indicate that neurons from cyfip1 heterozygous mice show reduced dendritic arborization. Consistent with the role of cyfip1 in dendritic arborization, in vitro cyfip1 overexpression leads to increased dendritic complexity (17, 18). Thus, modulation of CYFIP1 expression levels influences dendritic complexity and spine morphology in mouse neuronal cultures and mouse brain sections. Haploinsufficiency of CYFIP1 could provide a mechanism whereby the 15q11.2 (BP1-BP2) deletion confers risk for neuropsychiatric disorders, as human postmortem studies revealed an important role for dendritic spine structure abnormalities in the pathogenesis of ID, ASD, and SZ (19, 20). However, none of the post-mortem studies, to our knowledge, have taken into account the role of CNVs such as the 15q11.2 (BP1-BP2) deletion that increase risk for these disorders.
Studies using human induced pluripotent stem cells (iPSC) could provide further insight into the neurodevelopmental effects of the 15q11.2 (BP1-BP2) deletion. Recently developed technologies enable the derivation of neuron-like cells from iPSCs generated from human fibroblasts (21, 22). Such human-derived ‘iPSC-neurons’ display many properties characteristic of brain neurons and iPSC-based models can recapitulate key pathologic features of several neuropsychiatric disorders (23–27). Thus, iPSC-neurons may enable us to examine the putative CYFIP1-mediated alteration in dendritic spine architecture among individuals with the 15q11.2 (BP1-BP2) deletion. In this work, we have investigated the expression of CYFIP1 and its flanking genes in the 15q11.2 (BP1-BP2) deletion region in both iPSCs and iPSC-neurons, followed by morphological analysis of dendritic spine development.
MATERIALS AND METHODS
Clinical recruitment and initial screening
The participants were selected from an earlier genetic research study in which individuals were assessed using the Diagnostic Interview for Genetics Studies (DIGS) and provided venous blood samples for genomic DNA analysis (28). The participants (N=791) were screened for deletions in the 15q11.2 region using the RNase P Copy Number Reference qPCR Assay, with a VIC-labeled TAMRA probe (Life Technologies) and TaqMan Copy Number Assays probes for NIPA2 and TUBGCP5 (Assay IDs: Hs01842079_cn and Hs00956290_cn for NIPA2; Hs0128273_cn and Hs02106285_cn for TUBGCP5). All qPCR reactions were run in triplicate on an ABI 7900HT instrument (Applied Biosystems) and thermal cycling conditions were 95 °C for 10 min followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min, as per manufacturer's instructions. Five CEPH DNA samples were analyzed in each PCR plate for each assay as reference controls. CopyCaller Software 2.0 was used to perform relative quantitation analysis of genomic DNA targets using the real-time PCR data.
iPSC generation and quality control
Two participants with 15q11.2 deletions were identified for further analysis based on the initial qPCR screening of genomic DNA (Id numbers: 9000 for the proband and 9001 for the proband’s mother). Skin biopsies were obtained from both participants as well as a control individual without the deletion as previously described (29, 30). Fibroblasts were reprogrammed to produce iPSC lines using Sendai virus transfection at the NIMH funded Rutgers University Cell and DNA Repository (RUCDR) (31).
Array Comparative Genomic Hybridization (CGH)
Array-CGH was carried out using a PerkinElmer array platform. Genomic DNA samples from iPSCs and a reference control were first digested using BglIII enzyme. Adaptors were ligated with the fragmented DNA followed by PCR amplification of the fragmented DNA. Following purification, samples were labeled with Cy3 and Cy5 labelled dCTP. Labeled samples were combined with Cot-1 DNA and hybridized into the chip at 42 °C for 14–16 h. After hybridization, chips were washed and analyzed using the PerkinElmer Cytogenomics software at Magee-Women’s Hospital, Pittsburgh in a CLIA certified laboratory.
Neuronal Differentiation
iPSCs were cultured with neuronal precursor (NP) selection medium, followed by NP expansion medium containing fibroblast growth factor 2 (FGF-2) for generation of neural stem cells as previously described elsewhere (30). After 5–7 days in culture, neural rosettes were identified, manually dissected and plated into low attachment plates where embryoid body like structures – denoted as neurospheres – emerged. On plating neurospheres into matrigel coated plates, neural progenitor cells (NPCs) were collected manually for monolayer culture. NPCs were then cultured in neurobasal medium containing B27 supplement and brain-derived neurotropic factor (BDNF, 10 ng/ml) for neuronal differentiation.
Quantitative PCR analysis of genes in deletion region
Cellular RNA was isolated using Qiagen total RNA isolation kit. A total of 1 µg RNA was used to synthesize cDNA with the Superscript III First Strand Synthesis kit (Invitrogen). Quantitative RT-PCR was then performed using commercially available Taqman probes (Life Technologies Inc.). Analysis was performed using the ΔΔCt method and data were normalized to housekeeping gene β-actin (32).
Immunocytochemistry
Immunofluorescence staining was performed as previously described (33). Primary antibodies used were as follows: mouse anti-human nestin monoclonal antibody (1:200; R&D Systems), rabbit polyclonal anti-SOX1 (1:200; Abcam), rabbit polyclonal anti-musashi (1:200; Abcam), mouse anti-β tubulin III monoclonal (1:50; R&D System), mouse monoclonal anti-MAP2 (1:200; Millipore), chicken polyclonal anti-MAP2 (1:5000; PA1-10005, Thermo Fisher Scientific Pierce), rabbit polyclonal anti-VGLUT1 (1:200; Synaptic Systems), rabbit anti-NMDAR1 monoclonal (1:400; Abcam), rabbit polyclonal calbindin antibody (1:200; Abcam), mouse monoclonal anti-CART (Abcam, 1:100 dilution), mouse monoclonal anti-CYFIP1 (1:100; Abcam), mouse monoclonal anti-GFP (1:3000; MAB3580, Millipore), mouse monoclonal anti-PSD-95 (?) and rabbit polyclonal anti-PSD95 (?). Secondary antibodies were Alexa Fluor 488 goat anti-rabbit (1:200; Life Technologies), Alexa Fluor 488 goat anti-mouse (1:200; Life Technologies), Alexa Fluor 488 donkey anti-rabbit (1:1000; A21206, Life Technologies), Cy3 donkey anti-chicken (1:1000; 703-165-155, Jackson ImmunoResearch), and 647 donkey anti-mouse (1:1000; A31571, Life Technologies). Nuclear staining utilized bisBenzimide Hoechst 33342 trihydrochloride (1:1000; Life Technologies) or Hoechst 33342 (1:3000; B2261, Sigma). Briefly, samples were fixed in 4% PFA, rinsed in PBS, incubated with blocking solution, incubated with primary antibodies overnight at 4 °C, washed in PBS, incubated for 1–2 h with secondary antibodies, rinsed in PBS, counterstained with Hoechst 33342 and mounted using gelvatol medium.
Morphological features of dendrites
After two and half months in neurobasal medium, neurons were used for transfection experiments. Transfection was carried out with pEGFP plasmid using lipofectamine 2000 following the manufacturer protocol (Life Technologies; EGFP is the mutant type of GFP where Phe-64-->Leu, Ser-65-->Thr mutations are introduced to increase the sensitivity of the reporter protein and to improve resistance to photobleaching). At 72 h post transfection, neurons were fixed for 10 min in 4% PFA and mounted onto coverslips for imaging. Fluorescent images were acquired with constant power and pinhole aperture on a Nikon A1 confocal microscope. The analysis was carried out at 60x magnification with sequential acquisition setting at 2048×2048 pixels resolution. Each image was a Z-series projection of ~10–12 images each, averaged two times and taken at 0.2 µm depth intervals. Neurites (dendrites) and dendritic spines were reconstructed and subjected to Sholl analysis using Imaris software (Bitplane, v. 7.4). Dendritic spines were manually identified on 100–200 µM dendrites and automatically analyzed. The classification was automated. Statistical analyses utilized SPSS software.
The studies were approved by the Institutional Review Board (IRB) and the Institutional Biosafety Committee (IBC) at the University of Pittsburgh.
RESULTS
Identifying individuals with the 15q11.2 (BP1-BP2) deletion
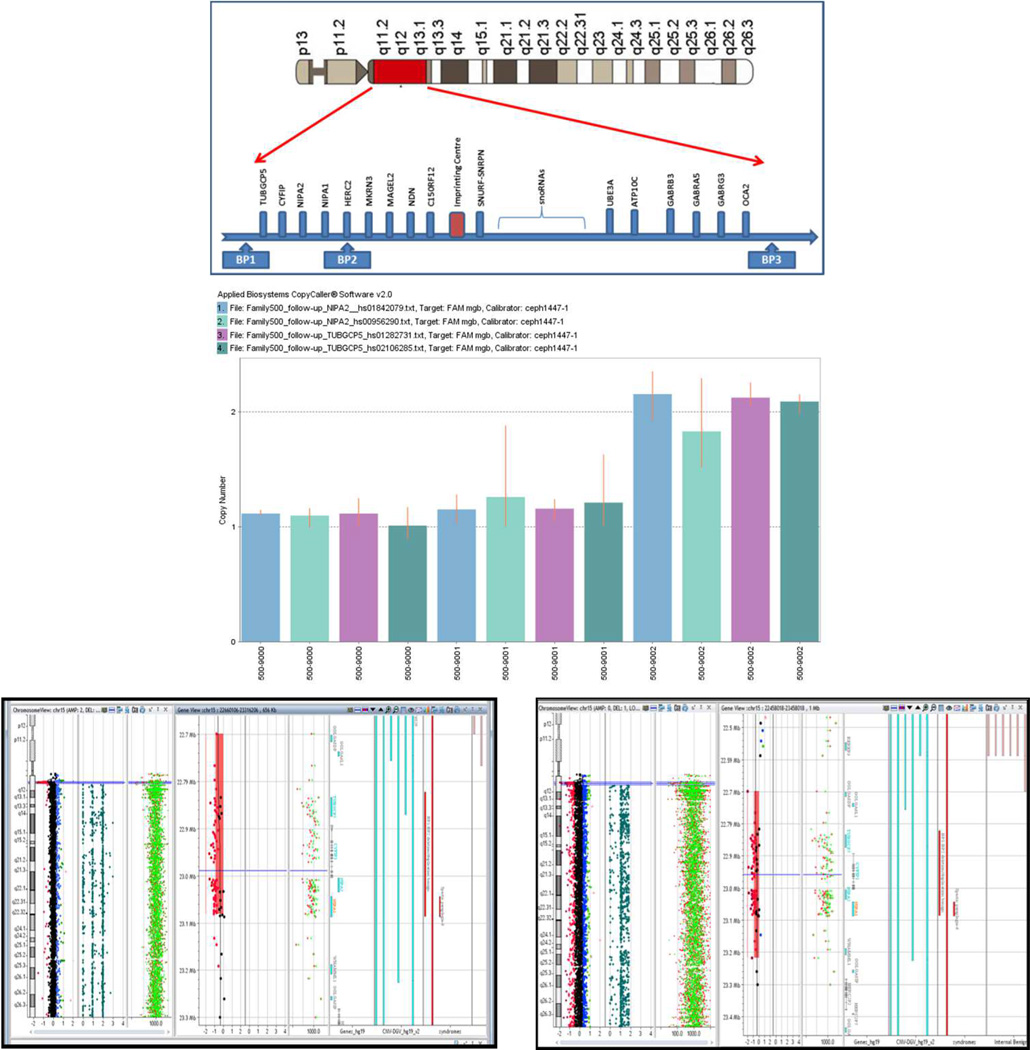
Using custom qPCR assays, an individual with schizoaffective disorder (DSM IV criteria, Id 500–9000) and the proband’s mother (Id 500–9001) were identified as carriers of the 15q11.2 (BP1-BP2) deletion, in contrast to the proband’s sibling (Id 500–9002, Figure 1, middle panel). The ~382kb deletion was confirmed using array comparative genomic hybridization (array CGH) in genomic DNA from iPSCs derived from the same probands and the proband’s mother (Fig 1, bottom panel).
Figure 1. Analysis of the 15q11.2 (BP1-BP2) deletion region.
Top panel: schematic diagram of the CNVs in the 15q proximal arm. Middle panel: qPCR assays using genomic DNA from blood samples drawn from individuals in a nuclear family: Id 500–9000 (proband), 500–9001 (mother) and 500–9002 (sibling without the deletion). Bottom panel: Array-CGH studies indicated the deletion of 15q11.2 region in genomic DNA from iPSCs; left: 500–9000 (proband), right: 500–9001 (mother).
Expression of genes in the 15q11.2 (BP1-BP2) deletion region
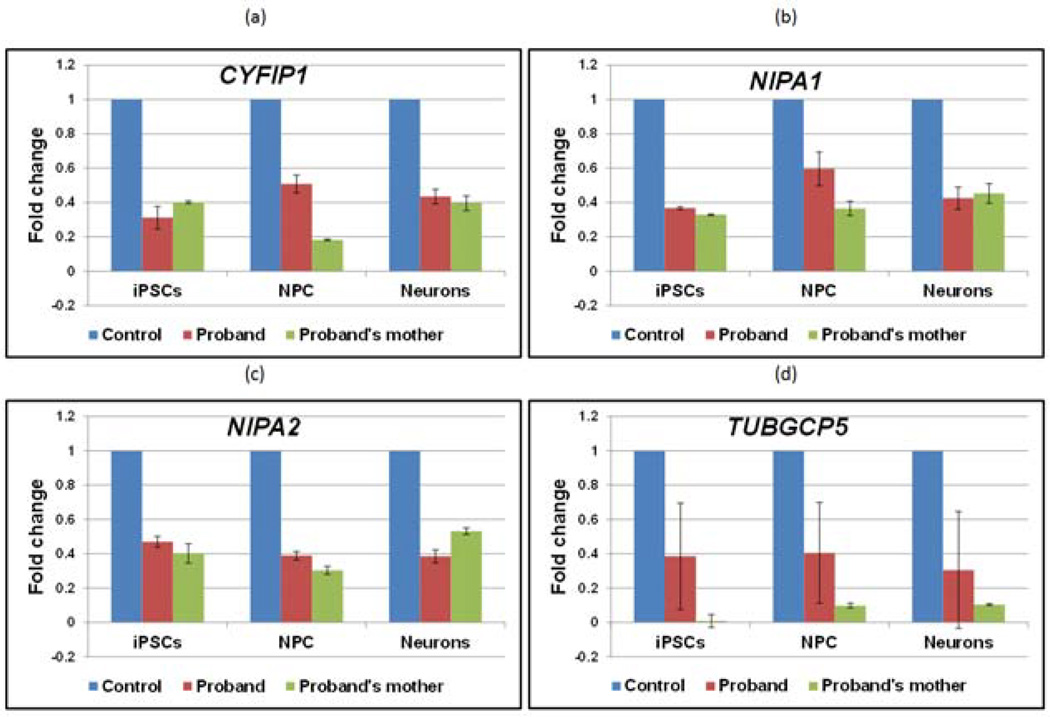
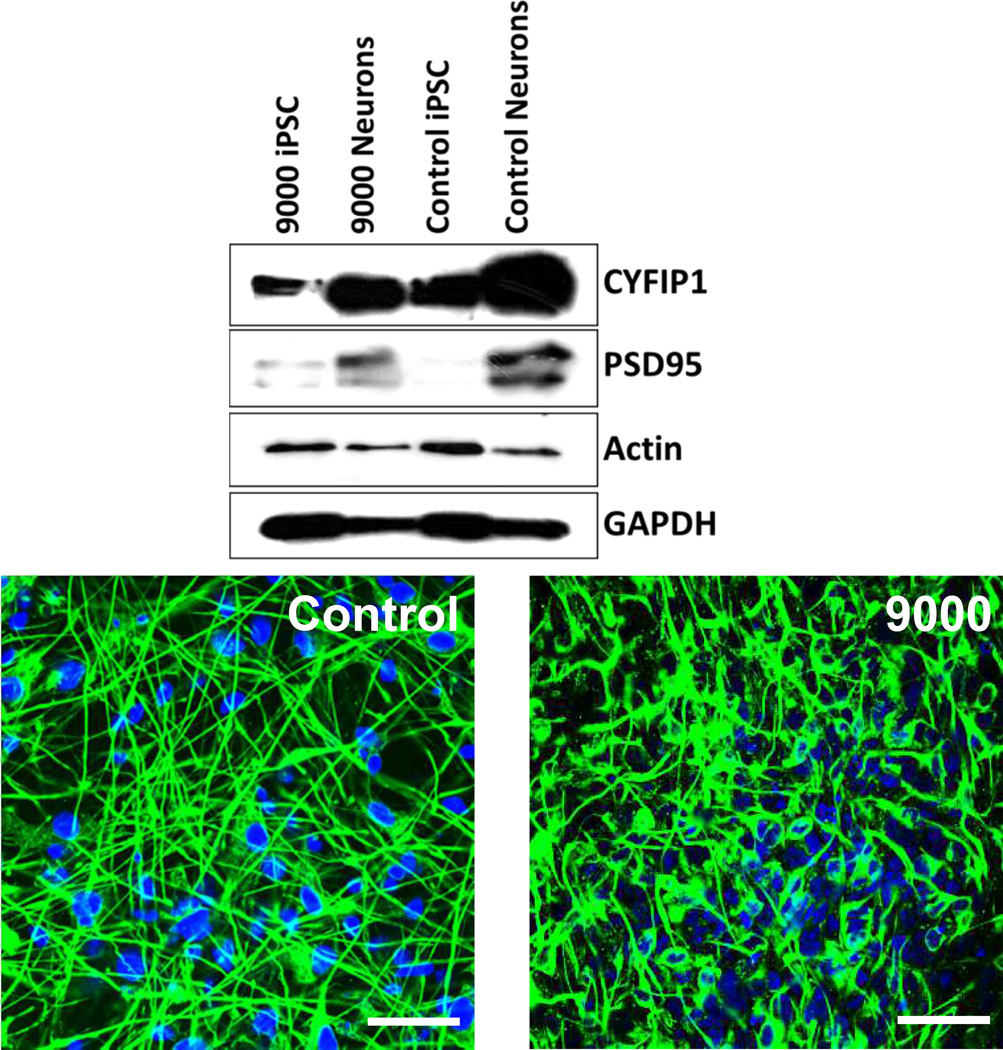
Quantitative real time PCR analysis indicated that the expression of CYFIP1, NIPA1, NIPA2 and TUBGCP5 encoded in the deletion region were reduced by approximately 50% levels in iPSCs, NPCs and iPSC-neurons bearing the 15q11.2 deletion compared to control (without carrying the deletion) (Figure 2). Further, Western blot and immunocytochemical analysis indicated that CYFIP1 and PSD95 protein levels were reduced in the proband’s cells (9000) compared with cells without the BP1-BP2 deletion (Figure 3).
Figure 2. Expression of genes in 15q11.2 (BP1-BP2) deletion region.
Levels of expression of each gene were normalized to β-actin levels; deletion lines (red/green): fold changes for each gene relative to control values (blue).
Figure 3. CYFIP1 and PSD95 protein levels in iPSC-neurons.
Top panel: Western blot analysis of CYFIP1 in iPSCs and neurons from the proband (9000) and a control. Bottom panel: Immunostaining of neurons with CYFIP1 antibody from the control and proband. Scale bar = 40 µm.
Characteristics of differentiated iPSC-neurons
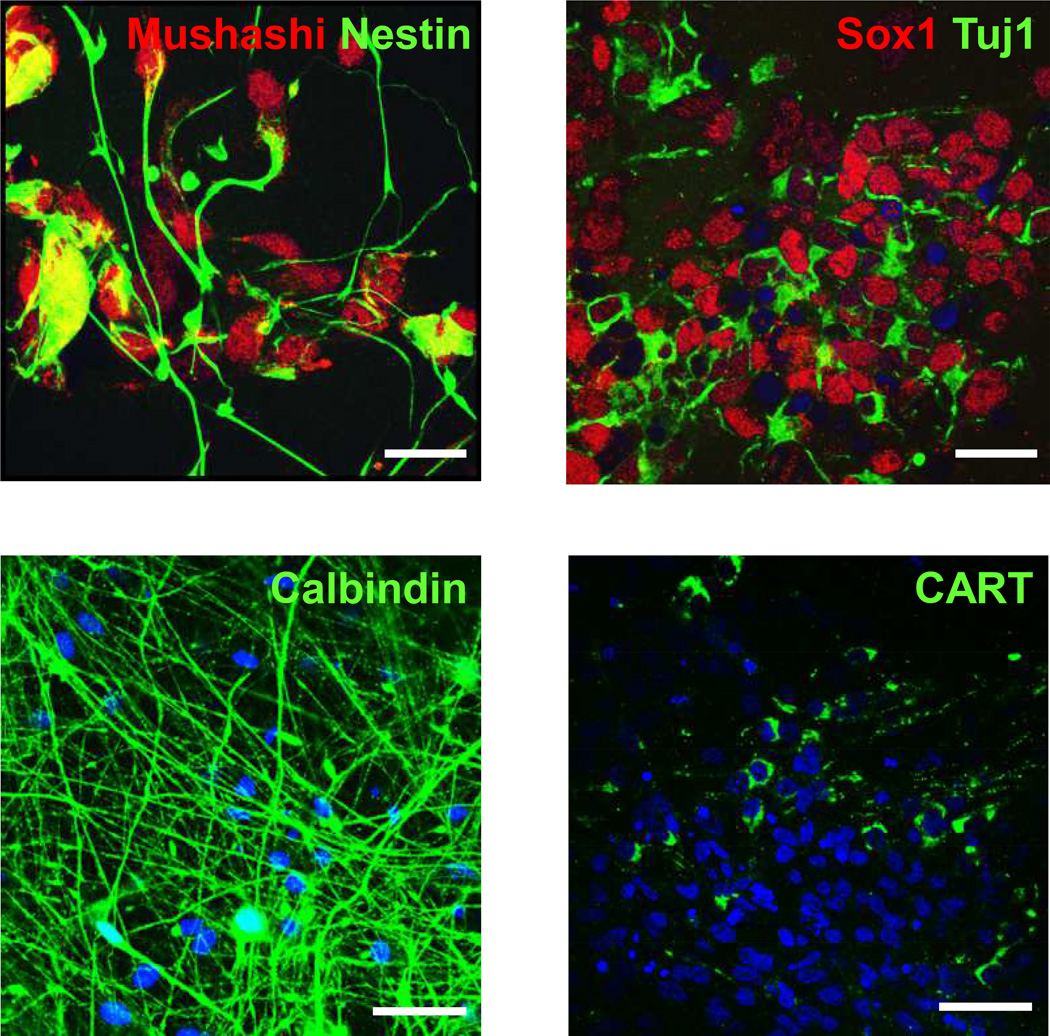
Initially, iPSCs were differentiated into neural progenitor cells (NPC) in monolayer culture, as indicated by immunoreactivity for Mushashi, Nestin, Sox1 and Tuj1 (Figure 4, top panel). In addition, the iPSC-derived neurons expressed Calbindin and cocaine- and amphetamine-regulated transcript (CART) (Figure 4, bottom panel).
Figure 4. Characterization of neural progenitor and neuron-like cells derived from iPSCs.
Cells were derived from an individual without the 15q11.2 BP1-BP2 deletion Top panel: neural progenitor cells (NPCs). Left: Mushashi (Red) & Nestin (Green); right: Sox1 (Red) & Tuj1 (Green). Scale bar = 20 µm.
Bottom panel: Neurons. Left: Calbindin, Right CART. Scale bar = 40 µm.
Dendritic spine morphology
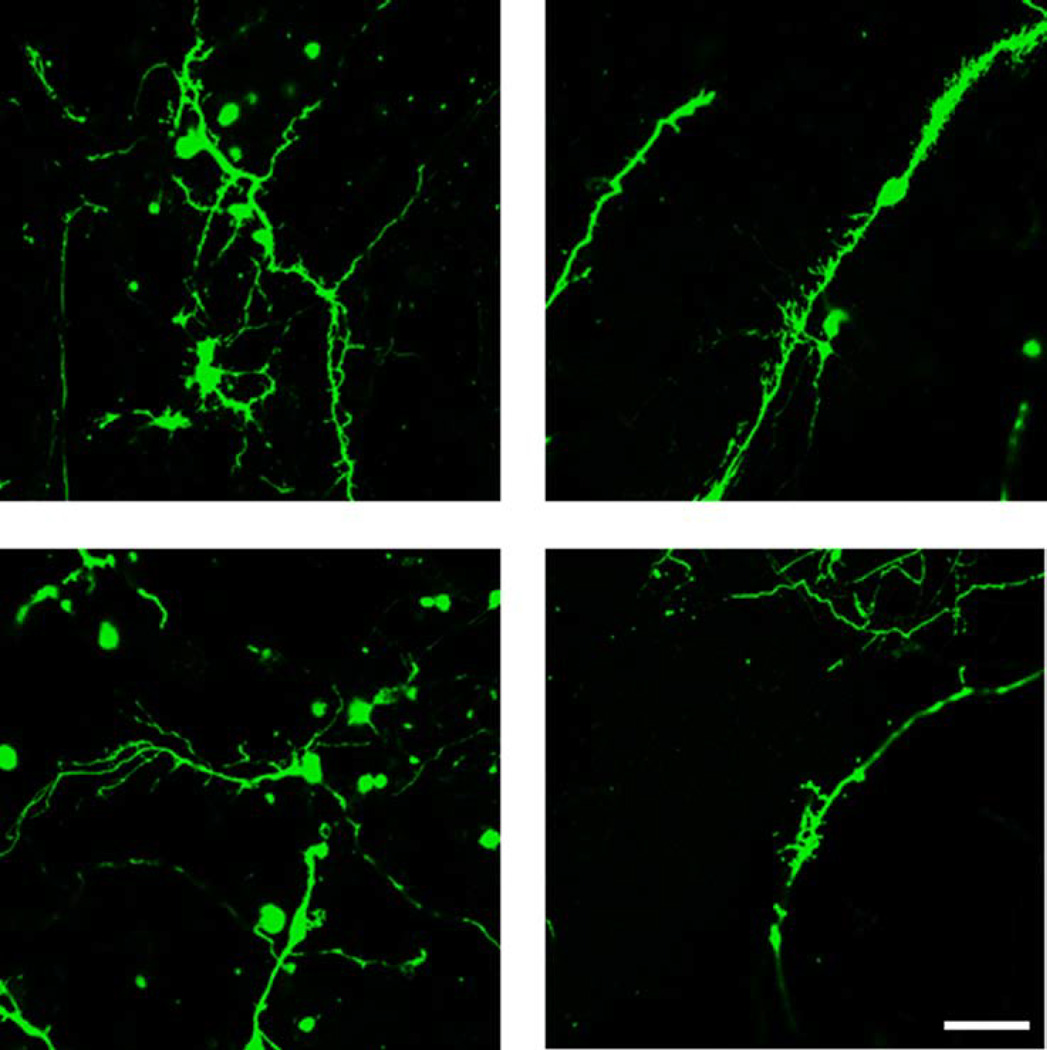
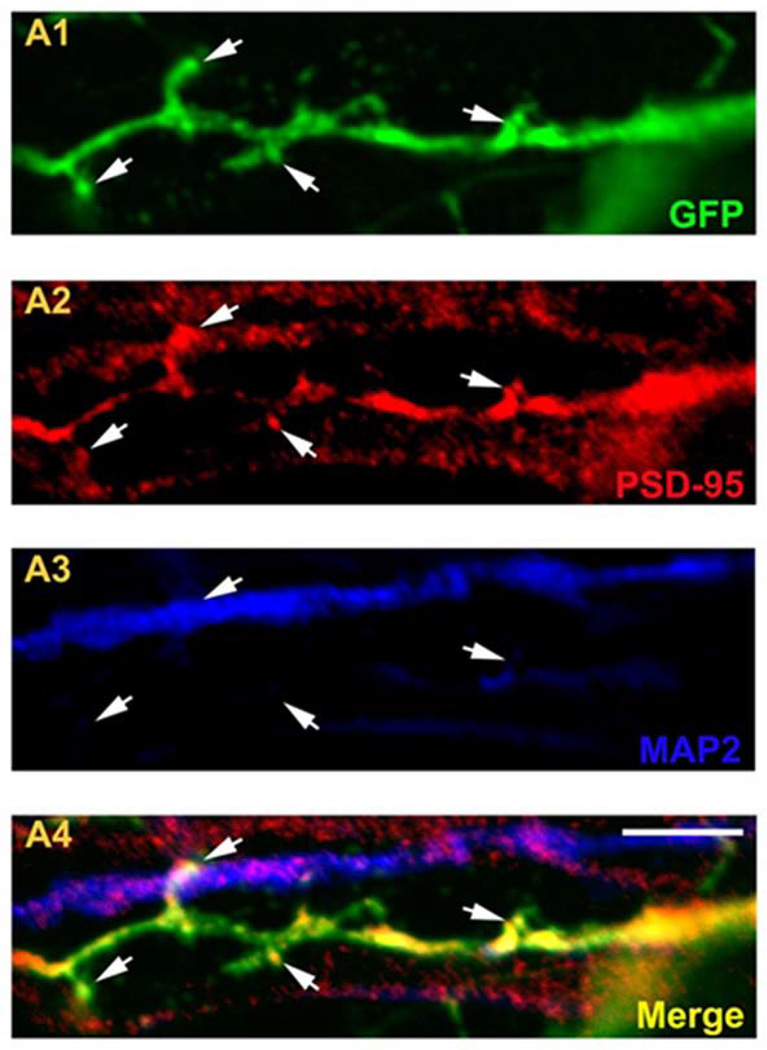
As previously reported (34), iPSC- were relatively immature at 10 weeks in culture (Figure 5). Qualitative analysis suggested that iPSC-neurons bearing the 15q11.2(BP1-BP2) deletion had altered dendritic morphology (Figure 5). Dendritic spines were detectable in the iPSC-neurons, indicated by co-localization of PSD-95 staining in EGFP labeled neurites (Figure 6).
Figure 5. Dendritic spines in iPSC- neurons.
Confocal images of iPSC-neurons transfected with EGFP. Top panel: control; Bottom panel: mother with deletion (500–9001). Scale bar = 20 µm.
Figure 6. Dendritic spines in iPSC derived neurons.
Immunostaining of dendrites from a control iPSC derived neuron. A1: GFP; A2: PSD-95; A3: MAP2; A4: Merge. Scale bar = 20 µm.
DISCUSSION
iPSCs, NPCs and neurons derived from both individuals with the inherited 15q11.2 (BP1-BP2) deletion showed reduced expression of all four genes encoded in the deleted region; reduced CYFIP1 protein levels were also observed in deletion bearing iPSC-neurons. The iPSC-neurons from the control and the deletion bearing individuals were relatively premature. Our preliminary observations, which need to be confirmed using unbiased quantitative analyses, are an important step towards understanding the changes in dendritic spine number, shape and plasticity that have been reported in the pathogenesis of intellectual disability, Fragile X syndrome, ASD and SZ (19, 35, 36). Earlier rodent studies indicate that CYFIP1 overexpression or haploinsufficiency increase immature spine number, suggesting an important role for CYFIP1 in dendritic spine morphology (17). Yoon and colleagues recently reported altered differentiation patterns in neuronal and glial lineages of human iPSCs bearing the 15q11.2 (BP1-BP2) deletion and attributed the abnormalities to hemizygous expression of CYFIP1 (37). Whether Yoon’s observations can be related to altered dendritic spine density needs to be addressed through future studies. If the 15q11.2 (BP1-BP2) deletion leads to haploinsufficiency of CYFIP1 and reduced expression of CYFIP1 induces changes in dendritic spine architecture in the human brain, it may provide a plausible explanation for the risk for ID, ASD and SZ conferred by the 15q11.2 (BP1-BP2) deletion.
Several other lines of work are necessary to enable more firm conclusions. Analysis of additional cell lines, particularly of individuals from other families and with additional neurodevelopmental disorders, are required to see whether the results are replicable and to investigate whether the dendritic spine abnormalities are influenced by the individual’s genomic background. PSD-95 labeling showed a punctate staining in iPSC-neurons from a control individual, similar to observed in postmortem human studies (38). In addition, consistent with other iPSC studies, PSD-95 appeared to be more diffuse in neurites which may represent relative immaturity in the iPSC-neurons (34). To test this possibility, further agnostic, unbiased analyses of dendritic architecture are needed in iPSC-neurons differentiated for longer periods. Moreover, electrophysiological studies in the deletion bearing iPSC-neurons could evaluate the functional impact of the observed morphological abnormalities in dendritic spines.
In conclusion, our human iPSC based cellular model indicates hemizygous reduction in the expression of four deleted genes during different stages of neuronal development. Qualitative analysis of iPSC-neurons suggests altered dendritic morphology is associated with the 15q11.2 (BP1-BP2) deletion. Future studies of additional individuals with and without the 15q11.2 (BP1-BP2) deletion are necessary to fully evaluate the functional effects of the deletion.
Acknowledgements
This work was supported by the following grants to VN: NIH grants MH63480, MH093246 and 07R-1712 from the Stanley Medical Research Institute. We also thank Indo-US Science & Technology Forum (IUSSTF) for providing a fellowship to DKD.
Footnotes
Disclosure Statement
There are no actual or potential conflicts of interest, including any financial, personal or other relationships with people or organizations during the development of the work submitted.
REFERENCES
- 1.Cafferkey M, Ahn JW, Flinter F, Ogilvie C. Phenotypic features in patients with 15q11.2(BP1-BP2) deletion: further delineation of an emerging syndrome. Am J Med Genet A. 2014;164A(8):1916–1922. doi: 10.1002/ajmg.a.36554. [DOI] [PubMed] [Google Scholar]
- 2.Cook EH, Jr, Lindgren V, Leventhal BL, Courchesne R, Lincoln A, Shulman C, et al. Autism or atypical autism in maternally but not paternally derived proximal 15q duplication. Am J Hum Genet. 1997;60(4):928–934. [PMC free article] [PubMed] [Google Scholar]
- 3.Cox DM, Butler MG. The 15q11.2 BP1-BP2 Microdeletion Syndrome: A Review. Int J Mol Sci. 2015;16(2):4068–4082. doi: 10.3390/ijms16024068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burnside RD, Pasion R, Mikhail FM, Carroll AJ, Robin NH, Youngs EL, et al. Microdeletion/microduplication of proximal 15q11.2 between BP1 and BP2: a susceptibility region for neurological dysfunction including developmental and language delay. Hum Genet. 2011;130(4):517–528. doi: 10.1007/s00439-011-0970-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43(9):838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Kovel CG, Trucks H, Helbig I, Mefford HC, Baker C, Leu C, et al. Recurrent microdeletions at 15q11.2 and 16p13.11 predispose to idiopathic generalized epilepsies. Brain. 2010;133(Pt 1):23–32. doi: 10.1093/brain/awp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doornbos M, Sikkema-Raddatz B, Ruijvenkamp CA, Dijkhuizen T, Bijlsma EK, Gijsbers AC, et al. Nine patients with a microdeletion 15q11.2 between breakpoints 1 and 2 of the Prader-Willi critical region, possibly associated with behavioural disturbances. Eur J Med Genet. 2009;52(2-3):108–115. doi: 10.1016/j.ejmg.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 8.Murthy SK, Nygren AO, El Shakankiry HM, Schouten JP, Al Khayat AI, Ridha A, et al. Detection of a novel familial deletion of four genes between BP1 and BP2 of the Prader-Willi/Angelman syndrome critical region by oligo-array CGH in a child with neurological disorder and speech impairment. Cytogenet Genome Res. 2007;116(1-2):135–140. doi: 10.1159/000097433. [DOI] [PubMed] [Google Scholar]
- 9.Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455(7210):232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Zwaag B, Staal WG, Hochstenbach R, Poot M, Spierenburg HA, de Jonge MV, et al. A cosegregating microduplication of chromosome 15q11.2 pinpoints two risk genes for autism spectrum disorder. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(4):960–966. doi: 10.1002/ajmg.b.31055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stefansson H, Meyer-Lindenberg A, Steinberg S, Magnusdottir B, Morgen K, Arnarsdottir S, et al. CNVs conferring risk of autism or schizophrenia affect cognition in controls. Nature. 2014;505(7483):361–366. doi: 10.1038/nature12818. [DOI] [PubMed] [Google Scholar]
- 12.Chai JH, Locke DP, Greally JM, Knoll JH, Ohta T, Dunai J, et al. Identification of four highly conserved genes between breakpoint hotspots BP1 and BP2 of the Prader-Willi/Angelman syndromes deletion region that have undergone evolutionary transposition mediated by flanking duplicons. Am J Hum Genet. 2003;73(4):898–925. doi: 10.1086/378816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rainier S, Chai JH, Tokarz D, Nicholls RD, Fink JK. NIPA1 gene mutations cause autosomal dominant hereditary spastic paraplegia (SPG6) Am J Hum Genet. 2003;73(4):967–971. doi: 10.1086/378817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goytain A, Hines RM, El-Husseini A, Quamme GA. NIPA1(SPG6), the basis for autosomal dominant form of hereditary spastic paraplegia, encodes a functional Mg2+ transporter. J Biol Chem. 2007;282(11):8060–8068. doi: 10.1074/jbc.M610314200. [DOI] [PubMed] [Google Scholar]
- 15.Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134(6):1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 16.Murphy SM, Preble AM, Patel UK, O'Connell KL, Dias DP, Moritz M, et al. GCP5 and GCP6: two new members of the human gamma-tubulin complex. Mol Biol Cell. 2001;12(11):3340–3352. doi: 10.1091/mbc.12.11.3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pathania M, Davenport EC, Muir J, Sheehan DF, Lopez-Domenech G, Kittler JT. The autism and schizophrenia associated gene CYFIP1 is critical for the maintenance of dendritic complexity and the stabilization of mature spines. Transl Psychiatry. 2014;4:e374. doi: 10.1038/tp.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oguro-Ando A, Rosensweig C, Herman E, Nishimura Y, Werling D, Bill BR, et al. Increased CYFIP1 dosage alters cellular and dendritic morphology and dysregulates mTOR. Mol Psychiatry. 2014 doi: 10.1038/mp.2014.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, Woolfrey KM. Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci. 2011;14(3):285–293. doi: 10.1038/nn.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glausier JR, Lewis DA. Dendritic spine pathology in schizophrenia. Neuroscience. 2013;251:90–107. doi: 10.1016/j.neuroscience.2012.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inoue H, Nagata N, Kurokawa H, Yamanaka S. iPS cells: a game changer for future medicine. EMBO J. 2014;33(5):409–417. doi: 10.1002/embj.201387098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagata S, Toyoda M, Yamaguchi S, Hirano K, Makino H, Nishino K, et al. Efficient reprogramming of human and muse primary extra-embryonic cells to pluripotent stem cells. Genes Cells. 2009;14(12):1395–1404. doi: 10.1111/j.1365-2443.2009.01356.x. [DOI] [PubMed] [Google Scholar]
- 23.Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473(7346):221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Giorgio FP, Boulting GL, Bobrowicz S, Eggan KC. Human embryonic stem cell-derived motor neurons are sensitive to the toxic effect of glial cells carrying an ALS-causing mutation. Cell Stem Cell. 2008;3(6):637–648. doi: 10.1016/j.stem.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 25.Israel MA, Yuan SH, Bardy C, Reyna SM, Mu Y, Herrera C, et al. Probing sporadic and familial Alzheimer's disease using induced pluripotent stem cells. Nature. 2012;482(7384):216–220. doi: 10.1038/nature10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koch P, Breuer P, Peitz M, Jungverdorben J, Kesavan J, Poppe D, et al. Excitation-induced ataxin-3 aggregation in neurons from patients with Machado-Joseph disease. Nature. 2011;480(7378):543–546. doi: 10.1038/nature10671. [DOI] [PubMed] [Google Scholar]
- 27.Nguyen HN, Byers B, Cord B, Shcheglovitov A, Byrne J, Gujar P, et al. LRRK2 mutant iPSC-derived DA neurons demonstrate increased susceptibility to oxidative stress. Cell Stem Cell. 2011;8(3):267–280. doi: 10.1016/j.stem.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Talkowski ME, Kirov G, Bamne M, Georgieva L, Torres G, Mansour H, et al. A network of dopaminergic gene variations implicated as risk factors for schizophrenia. Hum Mol Genet. 2008;17(5):747–758. doi: 10.1093/hmg/ddm347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.D'Aiuto L, Di Maio R, Heath B, Raimondi G, Milosevic J, Watson AM, et al. Human induced pluripotent stem cell-derived models to investigate human cytomegalovirus infection in neural cells. PLoS One. 2012;7(11):e49700. doi: 10.1371/journal.pone.0049700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D'Aiuto L, Prasad KM, Upton CH, Viggiano L, Milosevic J, Raimondi G, et al. Persistent Infection by HSV-1 Is Associated With Changes in Functional Architecture of iPSC-Derived Neurons and Brain Activation Patterns Underlying Working Memory Performance. Schizophr Bull. 2015;41(1):123–132. doi: 10.1093/schbul/sbu032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ban H, Nishishita N, Fusaki N, Tabata T, Saeki K, Shikamura M, et al. Efficient generation of transgene-free human induced pluripotent stem cells (iPSCs) by temperature-sensitive Sendai virus vectors. Proc Natl Acad Sci U S A. 2011;108(34):14234–14239. doi: 10.1073/pnas.1103509108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cawthon RM. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002;30(10):e47. doi: 10.1093/nar/30.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.D'Aiuto L, Robison CS, Gigante M, Nwanegbo E, Shaffer B, Sukhwani M, et al. Human IL-12 p40 as a reporter gene for high-throughput screening of engineered mouse embryonic stem cells. BMC Biotechnol. 2008;8:52. doi: 10.1186/1472-6750-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verpelli C, Carlessi L, Bechi G, Fusar Poli E, Orellana D, Heise C, et al. Comparative neuronal differentiation of self-renewing neural progenitor cell lines obtained from human induced pluripotent stem cells. Front Cell Neurosci. 2013;7:175. doi: 10.3389/fncel.2013.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Bartolomeis A, Iasevoli F, Tomasetti C, Buonaguro EF. MicroRNAs in Schizophrenia: Implications for Synaptic Plasticity and Dopamine-Glutamate Interaction at the Postsynaptic Density. New Avenues for Antipsychotic Treatment Under a Theranostic Perspective. Mol Neurobiol. 2014 doi: 10.1007/s12035-014-8962-8. [DOI] [PubMed] [Google Scholar]
- 36.Ebrahimi S, Okabe S. Structural dynamics of dendritic spines: molecular composition, geometry and functional regulation. Biochim Biophys Acta. 2014;1838(10):2391–2398. doi: 10.1016/j.bbamem.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 37.Yoon KJ, Nguyen HN, Ursini G, Zhang F, Kim NS, Wen Z, et al. Modeling a genetic risk for schizophrenia in iPSCs and mice reveals neural stem cell deficits associated with adherens junctions and polarity. Cell Stem Cell. 2014;15(1):79–91. doi: 10.1016/j.stem.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deo AJ, Cahill ME, Li S, Goldszer I, Henteleff R, Vanleeuwen JE, et al. Increased expression of Kalirin-9 in the auditory cortex of schizophrenia subjects: its role in dendritic pathology. Neurobiol Dis. 2012;45(2):796–803. doi: 10.1016/j.nbd.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]