Abstract
Intravital microscopy is a uniquely powerful tool, providing the ability to characterize cell and organ physiology in the natural context of the intact, living animal. With the recent development of high-resolution microscopy techniques such as confocal and multiphoton microscopy, intravital microscopy can now characterize structures at subcellular resolution and capture events at sub-second temporal resolution. However, realizing the potential for high resolution requires remarkable stability in the tissue. Whereas the rigid structure of the skull facilitates high-resolution imaging of the brain, organs of the viscera are free to move with respiration and heartbeat, requiring additional apparatus for immobilization. In our experience, these methods are variably effective, so that many studies are compromised by residual motion artifacts. Here we demonstrate the use of IMART, a software tool for removing motion artifacts from intravital microscopy images collected in time series or in three dimensions.
Keywords: intravital microscopy, fluorescence microscopy, multiphoton microscopy, image processing, image registration, motion artifact, in vivo imaging, image analysis
Introduction
Although intravital microscopy has been conducted for more than 200 years,1 its scope has significantly expanded with the development of confocal and multiphoton microscopy, whose optical sectioning provides biomedical researchers with the capability to conduct in vivo studies at cellular and subcellular resolution. However, achieving subcellular resolution in intravital microscopy requires that tissues be free of even the slightest motion. For studies of the brain, tissue stability is provided by the rigid constraints of the skull. However, for most parts of the body, tissue motion is not similarly constrained. As a result, high-resolution in vivo imaging of most organs and tissues requires methods to reduce, if not eliminate, tissue motion resulting from respiration and heartbeat.
A variety of systems have been developed to eliminate tissue motion in intravital microscopy. The most widely used system is the dorsal skin-fold chamber,2 which clamps a flap of skin tissue into a rigid metal frame, supporting a variety of studies of angiogenesis,3 microvascular function,4 tumor biology, 5 and bio-material interactions.6 Tumor development has also been imaged using surgically inserted imaging windows—for example a mammary gland imaging window—which is immobilized by rigid attachment to a specially designed stage-top box.7,8 The Weigert laboratory developed a holder system that immobilizes the sub-mandibular salivary glands of rodents sufficiently to support resolution of the dynamics of individual endosomes and secretory vesicles in vivo.9-11
Studies of visceral organs, which are free to move beneath an imaging window, require additional steps to immobilize the organ without compromising its function. High-resolution intravital microscopy of the rodent kidney can be accomplished by placing the kidney in a specially designed “kidney cup,”12,13 although satisfactory stability can also be provided by the pressure of the animal's own weight, when the externalized kidney is imaged from below on an inverted microscope.14 Several groups have achieved high-resolution imaging of the rodent liver, after immobilization by direct adhesion to a coverglass window.15-19 High-resolution imaging of the lung has been accomplished by synchronizing image acquisition with respiration and adhering the lung to a coverslip via an adhesive20 or via vacuum.21-23 High-resolution imaging of the beating heart can be accomplished by synchronizing image collection with signals from an electrocar-diogram.24 Stabilization of the intestine for intravital microscopy is simplified by the fact that a small loop can be externalized from the body,25,26 thus isolating the tissue from body motions. Recently, several general solutions have been developed to stabilize abdominal organs for intravital microscopy, including a “suctioning stabilizer,”27 a “microstage device,”28 and a unique window whose frame is bonded to the target tissue.29
Each of these techniques can provide effective immobilization of tissue with sub-micron precision. However, in our experience, the effectiveness of organ immobilization varies according to the morphology of the animal, resulting in residual motion artifacts in many imaging data sets. Here we demonstrate the use of novel image registration software that we have developed to address the residual motion-induced artifacts characteristic of images collected by intravital microscopy. Equally capable of correcting motion artifacts in images collected in time series or in three dimensions, this software can be used to restore intravital microscopy data to a quality that supports quantitative analysis at the highest resolution. We also describe how interested researchers can obtain, install and use this software.
Results
IMART: intravital microscopy artifact reduction tool
As mentioned above, methods of organ stabilization are frequently very effective in eliminating tissue motion. Successful stabilization of the kidney with a kidney cup is depicted in Figure 1, which shows the results of multiphoton microscopy of the kidney of a living rat, following intravenous injection of Hoechst 33342 (labeling nuclei blue), 3000 MW TexasRed dextran (red, internalized into endosomes of proximal tubule cells), and 500000 MW fluorescein dextran (green, in intertubular capillaries). In the time series of 120 images collected over two minutes (Video S1), the only discernible motion is that of the blood flowing in the vasculature. This stability is demonstrated graphically in Figure 1B, which shows a volume rendering of this data, with the sequential images of the time series arrayed vertically.
Figure 1.
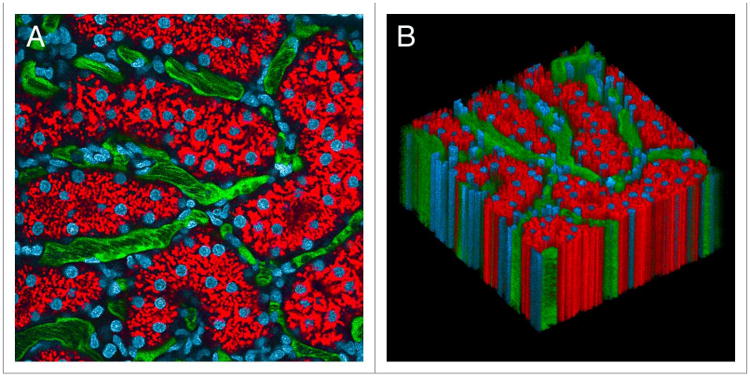
Intravital microscopy of the rat kidney mounted in a kidney cup. (A) First of 120 images collected from the kidney of a living rat, following intravenous injection of hoechst 33342 (labels nuclei blue), 3000 MW texasred dextran (red, internalized into endosomes of proximal tubule cells) and 500 000 MW fluorescein dextran (green, in intertubular capillaries). (B) XYt volume rendering of the time series, with sequential images arrayed vertically in the volume. Image volume is 200 microns across. the time series and volume rendering are presented in Video S1.
We have found that the kidney may be imaged with minimal motion artifacts even without a kidney cup by collecting images from below on an inverted microscope stand, a condition in which the weight of the animal effectively immobilizes the kidney against the imaging cover glass. However, the effectiveness of either approach varies according to the morphology of each rat, resulting in unforeseen motion artifacts in some studies. An example is presented in Figure 2. Figure 2A shows the first of a series of 45 images collected from a rat following intravenous injection of Hoechst 33342 and TexasRed-labeled albumin. The motion artifact is apparent in Figure 2B, which shows an “XT” image, an image in which sequential images collected along the white line shown in Figure 2A are arrayed vertically. However, the motion artifact is better appreciated by viewing the animation of the time series, shown in Video S2. This video demonstrates the unique nature of the motion artifact encountered in intravital microscopy; unlike the rigid translation of the field that results from stage drift, tissue motion in intravital microscopy can result in substantial warping of each individual image. This intra-scene distortion reflects the fact that the field is disturbed several times during the collection of a single image, reflecting the relatively slow rate of image capture relative to the high respiratory and heart rate of laboratory rodents.
Figure 2.
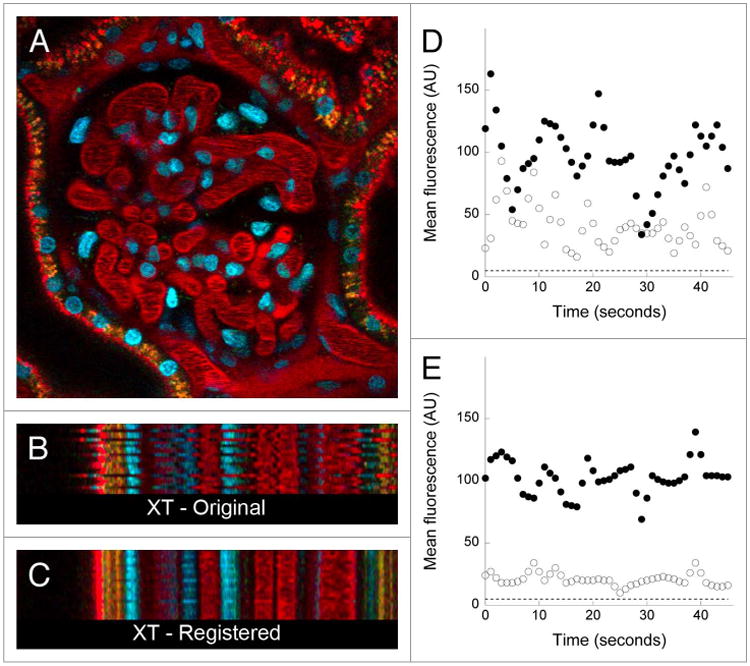
Digital correction of motion artifacts in a time series of images collected from the kidney of a living rat. (A) First of 45 images collected from the kidney of a living rat, following intravenous injection of hoechst 33342 (labels nuclei blue) and texasred-labeled albumin. (B) Xt projection of the region identified by the horizontal line in Panel A, in which sequential linescans are arrayed vertically. (C) Xt projection of the same region shown in B, but after digital correction of the time series. image field is 160 microns across. the time series of the original and corrected image are presented in Video S2. (D and E) Quantification of mean fluorescence in a ten-pixel region located in the lumen of a glomerular capillary (closed circles), or in a region located 4 pixels (1.6 microns) away in the adjacent Bowman's space (open circles) before (D) or after (E), digital correction. dashed line—background signal level.
There are a number of software solutions available for correcting motion artifacts in microscopy data, many provided as plugins for ImageJ. One of these plugins, Intravital_Microscopy_Toolbox,30 can be used to automatically identify and eliminate distorted frames from a set of images collected in time series. Most motion-correction solutions are designed to correct for the effects of simple translation, such as those occurring in studies of cells grown in culture. Although there are a few ImageJ plugins designed to correct for the kinds of non-rigid distortions characteristic of intravital microscopy images, we have found none that provide an efficient, effective solution for motion artifacts in sequences of intravital microscopy images. In order to provide researchers with a simple tool that can be used to correct motion artifacts in sequences of images collected in time series or in three dimensions, we developed IMART (Intravital Microscopy Artifact Reduction Tool), which corrects motion artifacts through a combination of rigid and non-rigid image registration techniques.
Image registration is a process in which two or more images acquired at different times, from different sensors with different resolutions or dimensions, or from different viewpoints/perspectives are matched to one another.31 This matching is accomplished via a process in which all of the images in the data set are transformed and aligned into a shared coordinate system. The process is often described as finding an explicit function that performs a mapping of a target image onto a source image.32 Our approach is based upon the idea that two sequentially collected images are, for the majority of the field, identical to one another. Thus, the quality of the registration of two images can be measured in terms of their overall similarity after correction. Rigid registration is first performed to correct global translations throughout the image sequence. This process performs an optimization to determine the corresponding horizontal and vertical displacement values that maximize the similarity between two sequential images. This similarity may be defined using one of many metrics, with a commonly used metric being the sum of squared differences between the pixel intensity values of the moving and reference images. This pairwise optimization is performed for all images in the data set.
Rigid registration is then followed by a non-rigid registration step, whose focus is on correcting small-scale, nonlinear/ non-rigid distortions. Non-rigid registration is accomplished by deformation of an underlying grid of control points. Initially evenly spaced, this grid is iteratively deformed, according to a cost function consisting of two terms with competing goals. The first goal is to maximize the similarity and alignment between the reference image and the image being deformed, using similarity metrics similar to those used for rigid registration. The second goal is to smooth and regularize the deformation to create a realistic transformation. Finding a procedure that effectively and efficiently balances the priorities of similarity and smoothness is the major challenge of non-rigid image registration. Details of the operation of the algorithm, how various similarity metrics are defined, and how these control points are mathematically optimized can be found in a description of a previous version of the software.33
Use of IMART for correcting motion artifacts in time series intravital microscopy data
The results of applying this software to the data shown in Figure 2 are shown in the right panel of Video S2, and in the XT image shown in Figure 2C. Both figures show that the software has effectively eliminated horizontal motion in the field, so that the positions of the nuclei, capillaries and even the endosomes (punctate red and yellow structures) are stable over the time series. Correction of the motion artifacts has also salvaged the utility of this data for quantifying glomerular permeability. The graphs on the right side of Figure 2 show the results of quantifying the mean fluorescence in a region located in the lumen of a glomerular capillary (filled circles), or in a region 4 pixels (1.6 microns) away in the Bowman's space (open circles). Quantitations of the original data (Fig. 2D) vary wildly over time, reflecting the motion of the capillary wall across both regions over the time series. These variations are much reduced after image registration (Fig. 2E), providing more reliable measures of local glomerular permeability. Residual variation in the graph of the registered data (also seen in Video S2 and Fig. 2C) is likely to reflect axial motion of the tissue, whose effects cannot be corrected using post-collection approaches.
In the example shown in Figure 2, the tissue was grossly immobilized, leaving only minor distortions in the field resulting from respiration and perhaps heartbeat. However, in some cases, animal motion can actually shift the entire field of view—an example of which is shown in Figure 3, which shows the results of a study in which images were collected from a living rat injected with Hoechst 33342, 500000 MW TexasRed-dextran and 3000 MW fluorescein dextran. In this study, the tissue slowly translated upwards and then later rapidly downwards over the course of the image series (left panel of Video S3). This translation is also apparent in the YT and XT sections shown in Figure 3B and C, respectively, and in the XYT volume rendering shown in Panel D. Interestingly, the rigid registration process was capable of correcting for the slow translation, but only partially successful at correcting for the second, more rapid translation (middle images of Fig. 3B and C; middle panel of Video S2). However, this translation, as well as the nonlinear warping of the field, was eliminated in the second stage of the registration process, where the non-rigid algorithm is applied (bottom of Fig. 3B and C, Fig. 3D, right panel of Video S2).
Figure 3.
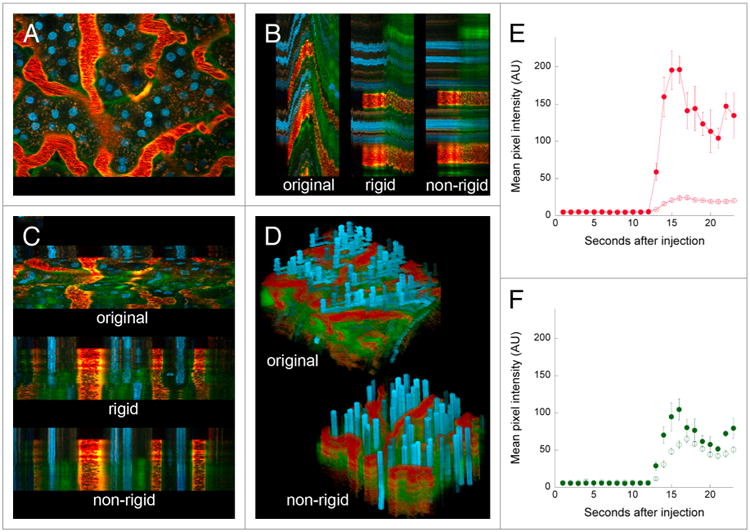
Digital correction of rigid and non-rigid motion artifacts in a time series of images collected from the kidney of a living rat. (A) First of 68 images collected from the kidney of a living rat, following intravenous injection of hoechst 33342, 500 000 MW texasred-dextran and 3000 MW fluorescein dextran. (B) Yt projection of the region identified by the vertical line in Panel A, before and after rigid and non-rigid registration. (C) Xt projection of the region identified by the horizontal line in Panel A, before and after rigid and non-rigid registration. (D) Volume rendering of XYt volume before and after non-rigid registration. image field is 195 microns across. The time series of the original and corrected image are presented in Video S3. (E) Graphs of mean intensity (± SE) of the 500 000 MW red dextran in 5 regions of the interstitium (open circles) or 5 regions in the lumen of adjacent capillaries (closed circles). (F) Graphs of mean intensity (± se) of the 3000 MW green dextran in 5 regions of the interstitium (open circles) or 5 regions in the lumen of adjacent capillaries (closed circles).
This particular study was conducted to evaluate microvascular permeability, as measured by the relative leakage of large and small molecular-weight dextrans from the intertubular capillaries into the surrounding interstitium. The original data were essentially useless for this purpose; the field translations precluded following fluorescence changes in any particular region. However, the vascular leakage of the small molecular-weight green dextran is easily discernible in the registered image series, appearing outside capillaries almost immediately upon per-fusion, well before it appears in the filtrate in tubular lumens (appearing as green borders to the yellow capillaries in the XT and YT images). Quantitative analysis of the registered image series was easily accomplished, demonstrating that the intertubular capillaries were minimally permeable to the 500000 MW dextran (Fig. 3E), but highly permeable to the 3000 MW dextran (Fig. 3F)
As mentioned above, the image registration strategy is based upon the idea that sequential frames of a time series will be very similar to one another in the absence of motion artifacts. For the data shown in Figures 1 and 2, this condition clearly applies—the distribution of fluorescence over nuclei and endosomes, and the overall outlines of the vasculature should be essentially identical in sequential frames. Real changes in the distribution of the fluorescence in the vasculature due to blood flow occur at a rate high enough to appear random, thus having little to no effect on the control grid. Similarly, in studies of white cell migration in the liver, we find that the software is capable of correcting for motion artifacts without distraction from the small population of motile cells migrating around an otherwise rigid field. However, in many cases, an investigator will need to characterize a time series over the course of a large perturbation. The data shown in Figure 3, which features a sudden infusion of green and red dextran in the microvasculature, with the green dextran subsequently appearing in the interstitium and later in the tubular lumens is one such example. For this image series, the registration software was clearly able to distinguish and correct motion-induced perturbations in the distribution of fluorescence without responding to the real changes occurring in the field. However, the data shown in Figure 4 apparently presented a greater challenge to the software.
Figure 4.
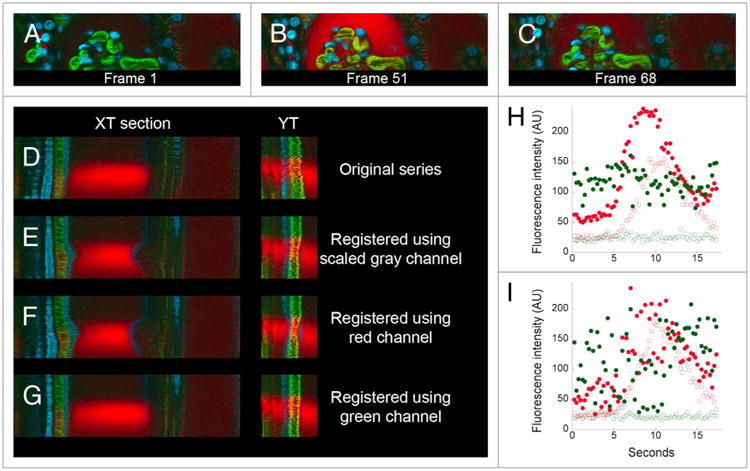
Use of different color channels for digital correction of motion artifacts in a time series of images collected from the kidney of a living rat. (A) First of 68 images collected from the kidney of a living rat, following intravenous injection of 500,000 MW fluorescein dextran, 3000 MW TexasRed dextran and Hoechst 33342. (B and C) 51st and 68th frame after injection of a second injection of 3000 MW TexasRed dextran. (D–G) XT and YT sections from regions identified with white lines in Panel C. (D) Original data. (E) After registration based upon scaled gray channel. (F) After registration based upon green channel. (G) After registration based upon red channel. Image fields are 160 microns wide. (H and I) Quantification of the green and red signals in a region in the capillary (closed circles) or in a region 4 pixels (1.6 microns) away, in the Bowman's space (open circles). (H) After registration based upon the green channel. (I) original data.
For the study shown in Figure 4, a series of images of a small region of the rat kidney, a cross-section of the glomerular capsule, was collected at the rate of four frames per second in order to capture the dynamics of filtration of 3000 MW TexasRed dextran. Prior to collection of this time series, the rat had been injected with 500 000 MW fluorescein dextran (which is not filtered, thus labeling the lumen of capillaries), an earlier dose of 3000 MW TexasRed dextran (which, having been previously filtered, labeled endosomes of the proximal tubule and the lumen of distal tubules) and the nuclear label Hoechst 33342 (Fig. 4A). Within five seconds of injection into the tail vein, the bolus of 3000 MW TexasRed dextran arrived at the glomerulus, where it was rapidly filtered into the Bowman's capsule, from which it cleared into the renal tubule (Fig. 4B and C). The collected time series (shown in the top panel of Video S4) shows motion artifacts that, while relatively modest, prevent characterization of the kinetics of dextran filtration, which is measured by the rate of appearance of the dextran in the Bowman's space immediately adjacent to a capillary.
Interestingly, our initial attempts to correct this motion artifact were only partially successful. While the registration clearly removed the wobbling motion of the original series, it introduced a new motion—a contraction of the Bowman's capsule (compare first and second row in Video S4, and XT and YT sections in Fig. 4D and E). This artifact appears to be based upon the dramatic change in the overall image that occurs when the bolus of red dextran arrives at the glomerulus. The registration software operates on grayscale images, but provides the capability to perform registration and correction of color images using either one of the color channels or a composite channel based upon the scaled combination of the color channels. The “scaled gray” channel, which contains the most information and typically gives the best results, was used in this first attempt. A similar, and perhaps worse, distortion of the Bowman's capsule was obtained when we repeated the correction using the red channel alone (third row of Video S4; Fig. 4F). However, when we eliminated the contribution of the red channel by basing the correction on images from the green channel, the contraction of the Bowman's space was eliminated (bottom row in Video S4; Fig. 4G). Unlike the example shown in Figure 3, the registration software required the removal of the red channel information from the data set in order to distinguish motion-induced perturbations from the actual changes occurring in the field.
Digital correction of the motion artifact in this image series made it possible to accurately measure the kinetics of filtration of the red dextran. Figure 4H shows graphs of the levels of red and green fluorescence in the capillaries (closed circles) and in the Bowman's space (open circles). This graph demonstrates that the red dextran is immediately filtered into the Bowman's space, whereas the green dextran is retained in the vasculature. This conclusion would be difficult to draw from a similar analysis of the original data (Fig. 4I).
The problem of axial motion in time series data
In nearly all of the examples shown above, a residual motion artifact is apparent even after registration. Manifest as a rapid blinking of particular structures, this artifact reflects axial motion of the tissue, so that certain structures appear and disappear over the course of the time series. The effects of axial motion are masked somewhat by the fact that axial resolution is approximately 3-fold worse than lateral resolution in optical microscopy. Nonetheless, this kind of motion presents a serious challenge for studies involving time series measurements of small structures and cannot be corrected using post-collection approaches. An example is shown in Video S5 (left panel), which shows a time series of images collected from the liver of a living rat, following intravenous injection of sodium fluorescein, an organic anion that is rapidly transported from the blood into the bile by hepatocytes. The narrow diameter of the bile canaliculi is such that axial motions as small as a micron can result in the rapid appearance and disappearance of canaliculi in sequential images collected from a single plane. Thus, even modest amounts of motion confound our ability to quantify canalicular secretion of fluorescein, the objective of this particular study.
In order to address this problem, we developed an approach in which we collect short image stacks (typically encompassing six microns of depth) at each time point, and then generate a time series of the maximum projection of each volume. While this approach reduces the temporal resolution of the study, it ensures that canaliculi located near the center of the vertical stack are continuously captured through the time series (see right panel of Video S5). In addition to improving the continuity of the time series images of canaliculi, this approach also increases the number of canaliculi collected at each time point (compare Fig. 5A and B). The effect of this procedure on quantitation is shown in Figures 5C and D, which show the mean fluorescence quantified in regions of interest located over four canaliculi, as measured either from a series of single images (Fig. 5C) or from a series of volume projections (Fig. 5D). Whereas the variation of fluorescence measurements obtained from a single plane is such that it would be essentially impossible to quantify the rate of canalicular secretion, the rates are easily obtained from an analysis of the volume projections, which additionally demonstrate impressive reproducibility between cells.
Figure 5.
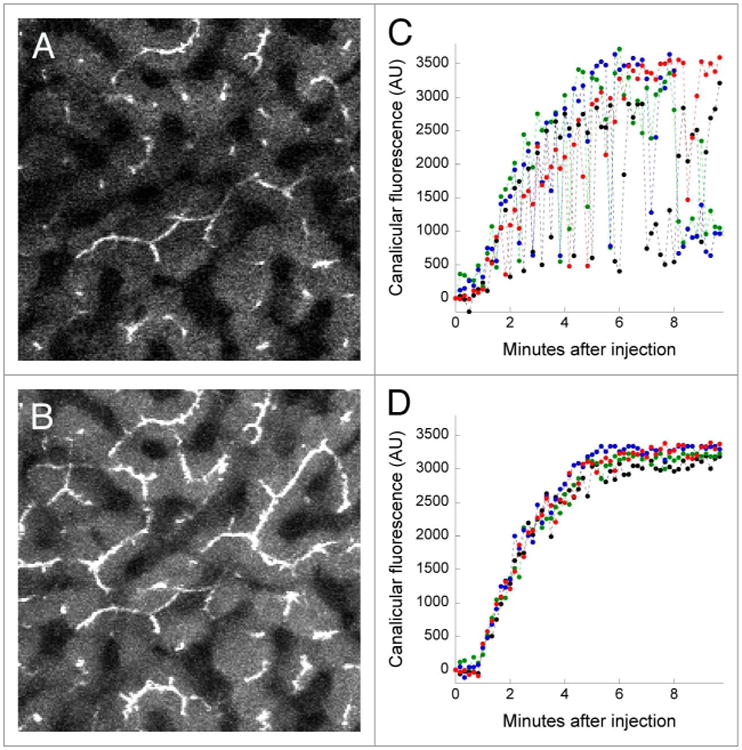
Accommodating axial motion by collection of image stacks. (A) A single multi-photon fluorescence microscopy image collected from the liver of a living rat five minutes after intravenous injection of sodium fluorescein. (B) A maximum-intensity projection of a six-plane image volume collected at the same time point as that shown in Panel A. image field is 383 microns across. (C) Graph of the mean fluorescence quantified in regions of interest (10 pixel lines) located over 4 canaliculi, measured in a series of images collected from a single focal plane. (D) As in Panel C, but measured from a series of maximum-intensity projection images of volumes collected over time. lateral motion of this time series was minimal and thus did not require IMART correction.
Use of IMART for correcting motion artifacts in three-dimensional intravital microscopy data
The optical sectioning ability of confocal and multiphoton microscopy provides the capability for three-dimensional volumetric imaging of living animals. However, sample motion occurring during collection of the sequential focal planes of the volume can result in a distorted volume that may be difficult to interpret. We have found that when image volumes are collected in such a way that sequential focal planes overlap with one another and share some structural information, this overlap can be used to identify and correct motion artifacts. The working principle for correcting motion artifacts in images collected from sequential planes is thus similar to that for correcting motion artifacts in images collected in time series—the goal of the algorithm is to reduce the dissimilarity between sequential images.
An example of tissue motion occurring during collection of a 3D volume of the kidney of a living rat is shown in Figure 6 and Video S6A. A slow shift in the tissue results in a progressive vertical translation of the volume (left panel in Video S6A). Since the major component of the motion is a simple translation, rigid registration removes most of the motion artifact (middle panel in Video S6A). Subsequent non-rigid registration removes residual nonlinear warping in the field (right panel in Video S6A). The net effect of image registration is improvement in the quality of the resulting 3D image volume such that the structure of the glomerular capillaries, obscured in a projection of the original data (Fig. 6A) is much clearer after registration (Fig. 6B). The improvement is also demonstrated in the volume renderings of the original and registered volumes (left and right panels, respectively, in Video S6B).
Figure 6.
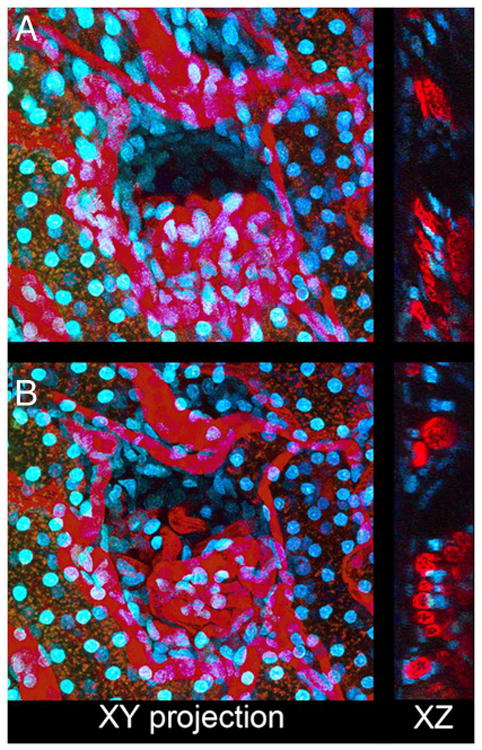
Correction of motion artifacts in three-dimensional image volumes. (A) XY maximum projection of a series of 50 images collected 0.6 microns apart from the kidney of a living rat following intravenous injection of Hoechst 33342 and 500 000 MW TexasRed-dextran. Right—single XZ section of the same volume. (B) As in Panel A, but following rigid and non-rigid registration. Image field is 212 microns across.
Demonstration of the digital correction of nonlinear motion artifacts in a three-dimensional volume is shown in Figure 7. Figure 7A shows a single image from a three-dimensional image volume collected from the kidney of a living rat following injection of 110000 MW fluorescein dextran. The characteristic rippling effect of a high-frequency motion in the tissue is apparent when the sequential images are viewed in sequence (left panel of Video S7). However, this subtle motion artifact is essentially eliminated by non-rigid registration (right panel of Video S7). The effect of image registration on resolution is more clearly demonstrated in Figure 7B, which shows XZ sections of volumes constructed from the original data (top), following rigid registration (middle), and following non-rigid registration (bottom). Individual lysosomes (small, punctate yellow structures) that are indistinct in the original volume are clearly defined after non-rigid registration.
Figure 7.
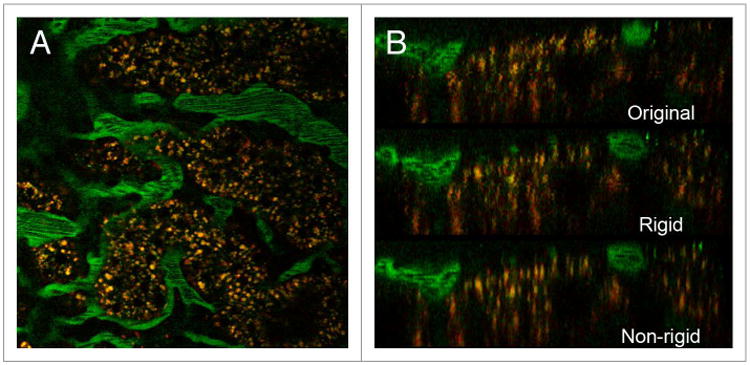
Correction of nonlinear motion artifacts in three-dimensional image volumes. (A) single image from a three-dimensional image volume collected from the kidney of a living rat after intravenous injection of 110 000 MW fluorescein dextran. (B) XZ sections of volumes reconstructed from original data (top), following rigid registration (middle) and following non-rigid registration (bottom). Field is 197 microns across.
Obtaining and using IMART
The IMART registration software is derived from an earlier version of the software described in33 and demonstrated in.22 Directions for obtaining a complete version of the software and a user's guide may be found at http://www.medicine.iupui.edu/icbm/software/. As of the time of writing, IMART is available only for the Windows operating system. Written using the MATLAB programming environment, IMART uses MATLAB libraries and thus requires either prior installation of MATLAB or installation of the MATLAB Compiler Runtime (also provided as part of the download). Although there are no specific hardware requirements, it is recommended that systems be equipped with at least 8 GB of memory.
IMART is highly flexible, providing the user with the ability to specify registration parameters, including optimization method, image similarity metric, image interpolation method, grid spacing, smoothness penalty coefficient, and optimization step size. By default, the program first conducts a rigid registration, which is then followed by non-rigid registration of the results of the rigid registration. Since the algorithm used to correct linear translation is much less computationally intensive than the algorithm required to correct for non-rigid distortions, we provide the user with the option of conducting only rigid registration, which provides faster output for data sets with simple translational shifts.
For multi-channel data, the program allows the user to specify whether registration is to be conducted on one particular channel, or upon a “scaled gray” combination of all channels. The user is also able to specify whether registration is based upon a “moving” reference frame, in which each frame is registered to the previous frame in the series, or upon registration to a single reference frame. Guidance for program settings and the effects of different registration parameters is provided in the user's guide.
Discussion
Intravital microscopy is a powerful technique that provides biomedical researchers with a unique window into physiology and cell biology in the most relevant biological context—within the intact, living animal. However, intravital microscopy is far from routine, requiring the development of specialized techniques that ensure the health of the animal, while still providing access to the tissue of interest. For visceral organs, an additional challenge is that the tissue must function normally, but also be immobilized to sub-micron precision. A variety of devices have been developed to immobilize and present various structures and organs for intravital microscopy. However, these devices are not fool-proof, so that residual motion artifacts that complicate or prevent analysis are frequently encountered. These experimental failures are intolerable; in addition to sacrificing a laboratory animal for no purpose, they result in the loss of data from crucial time points, confounding the careful design of studies. Here we present a software solution to correct for the effects of residual motion artifacts, providing a tool that can effectively improve and even salvage intravital microscopy data.
A variety of software solutions have been developed to address the problem of motion artifacts in microscopy, but few address the unique and complex motion artifacts encountered in intravital microscopy. To the degree that the effects of motion result in the distortion of a small number of images collected in time series, one solution is to simply eliminate the distorted frames from the time series. This approach has been incorporated into the “Intravital_Microscopy_Toolbox” plugin for ImageJ.30 Existing approaches to correct distorted images are based upon the general strategy of minimizing differences between a target frame and a reference frame. For time series that feature structures that can be effectively segmented, an effective approach has been developed based upon registration of skeletonized shapes derived from the structure.34 A more common approach is to minimize differences in pixel intensity between the target image and a template reference image.35,36
All of these techniques are based upon a correction scheme in which a target frame is compared with a single reference frame that is selected from the time series,34 obtained under conditions that lack motion artifacts,35 or derived from the entire time series.36 While generally effective, this approach is poorly suited to image sequences that feature wholesale changes in the field. For studies such as those shown in Figures 3–5, dramatic changes in the field make it impossible to register the time series to a single reference frame. IMART's successful correction of the motion artifacts in these examples is based upon a correction scheme in which each target frame is compared with the preceding frame in the sequence. This approach makes IMART capable of correcting for the effects of motion on images collected in time series, but also on images collected from sequential focal planes to construct three-dimensional image volumes. In principle, this implies that IMART could be extended to correct motion artifacts in four-dimensional data, that is, three-dimensional image data collected over time. However, correction of both lateral and axial motion artifacts in these kinds of studies is significantly more complex, and is complicated by the fact that the temporal relationship between sequential planes of a three-dimensional volume (collected within a second of each another) is completely different than that between sequential images of a particular focal plane (whose collection may be separated by several seconds). We are currently exploring methods to address these additional complexities, and plan to incorporate the capability to correct motion artifacts in four-dimensional data in an upcoming version of IMART.
Methods
Intravital microscopy
All animal studies were conducted in compliance with the Institutional Animal Care and Use Committee guidelines of Indiana University. The methods used for intravital microscopy of the kidney are as described in12 and the methods of intravital microscopy of the liver are as described in.15 Imaging was conducted using either Olympus Fluoview 1000 (Olympus, Inc.) or BioRad MRC1024 (Carl Zeiss, Inc.) confocal/multiphoton microscope systems at the Indiana Center for Biological Microscopy.
IMART development
The non-rigid registration methods employed by IMART are derived from a method described in.37 This method focused on the pairwise non-rigid registration of two magnetic resonance images. This was accomplished through the use of B-splines, or basis splines. The registration of a point in an image when using B-splines is only determined by the area immediately surrounding the nearest control points, resulting in very localized deformations. We have extended this method to address the specific challenges associated with intravital microscopy, including the registration of not only of a pair of images but of a series of many images (both in time series and in three dimensions), appropriate selection of a reference image, appropriate selection of similarity metrics, and the incorporation of pixel intensity data presented in multi-channel image sets. The use of B-splines for non-rigid registration allows for easy visualization of the deformation, and also gives way to a method for evaluating motion artifact reduction—despite the lack of ground-truth data—using tools borrowed from the image and video compression community.38
Image analysis and presentation
Quantitative image analysis was conducted using Metamorph image processing software (Molecular Devices). Quantitative analysis was conducted on raw image data, but images presented in figures were contrast-enhanced (adjusting intensity minimum, maximum, and gamma). In some cases, images were smoothed using a Gaussian filter. In color images, the visibility of Hoechst-labeled nuclei was enhanced by selectively adjusting the hue, saturation and lightness of the blue channel. In all cases, images to be compared were processed identically and in such a way that the processing preserved the visibility of both the dim and bright structures of the original image. Images were processed, assembled into figures and annotated using Adobe Photoshop (Adobe). Graphics were produced and summary statistics obtained using Kaleidagraph (Synergy Software).
Volume rendering and video production
Volume rendering was conducted using Voxx software,39 and videos produced using either Voxx or Metamorph, and compressed using TMPGEnc 2.5 (Pegasys, Inc.).
Supplementary Material
Acknowledgments
This work was supported by a George M. O'Brien Award from the National Institutes of Health NIH/NIDDK P50 DK 61594. Microscopy was conducted at the Indiana Center for Biological Microscopy, which is supported by a grant (INGEN) from the Lilly endowment. Images were kindly provided by Jennifer Ryan, Clifford Babbey, Seokmin Hong, Ruben Sandoval, George Rhodes and Amy Zollman.
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
References
- 1.Pittet MJ, Weissleder R. Intravital imaging. Cell. 2011;147:983–91. doi: 10.1016/j.cell.2011.11.004. http://dx.doi.org/10.1016/j.cell.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lehr HA, Leunig M, Menger MD, Nolte D, Messmer K. Dorsal skinfold chamber technique for intravital microscopy in nude mice. Am J Pathol. 1993;143:1055–62. [PMC free article] [PubMed] [Google Scholar]
- 3.Sckell A, Leunig M. The dorsal skinfold chamber: studying angiogenesis by intravital microscopy. Methods Mol Biol. 2009;467:305–17. doi: 10.1007/978-1-59745-241-0_19. http://dx.doi.org/10.1007/978-1-59745-241-0_19. [DOI] [PubMed] [Google Scholar]
- 4.Menger MD, Laschke MW, Amon M, Schramm R, Thorlacius H, Rucker M, Vollmar B. Experimental models to study microcirculatory dysfunction in muscle ischemia-reperfusion and osteomyocutaneous flap transfer. Langenbeck's archives of surgery / Deutsche Gesellschaft fur Chirurgie. 2003;388:281–90. doi: 10.1007/s00423-003-0426-y. [DOI] [PubMed] [Google Scholar]
- 5.Baron VT, Welsh J, Abedinpour P, Borgström P. Intravital microscopy in the mouse dorsal chamber model for the study of solid tumors. Am J Cancer Res. 2011;1:674–86. [PMC free article] [PubMed] [Google Scholar]
- 6.Laschke MW, Vollmar B, Menger MD. The dorsal skinfold chamber: window into the dynamic interaction of biomaterials with their surrounding host tissue. Eur Cell Mater. 2011;22:147–64. doi: 10.22203/ecm.v022a12. discussion 164-7. [DOI] [PubMed] [Google Scholar]
- 7.Gligorijevic B, Kedrin D, Segall JE, Condeelis J, van Rheenen J. Dendra2 photoswitching through the Mammary Imaging Window. J Vis Exp. 2009;28:1278. doi: 10.3791/1278. http://dx.doi.org/10.3791/1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kedrin D, Gligorijevic B, Wyckoff J, Verkhusha VV, Condeelis J, Segall JE, van Rheenen J. Intravital imaging of metastatic behavior through a mammary imaging window. Nat Methods. 2008;5:1019–21. doi: 10.1038/nmeth.1269. http://dx.doi.org/10.1038/nmeth.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masedunskas A, Sramkova M, Parente L, Sales KU, Amornphimoltham P, Bugge TH, Weigert R. Role for the actomyosin complex in regulated exocytosis revealed by intravital microscopy. Proc Natl Acad Sci U S A. 2011;108:13552–7. doi: 10.1073/pnas.1016778108. http://dx.doi.org/10.1073/pnas.1016778108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masedunskas A, Sramkova M, Parente L, Weigert R. Intravital microscopy to image membrane trafficking in live rats. Methods Mol Biol. 2013;931:153–67. doi: 10.1007/978-1-62703-056-4_9. http://dx.doi.org/10.1007/978-1-62703-056-4_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masedunskas A, Weigert R. Intravital two-photon microscopy for studying the uptake and trafficking of fluorescently conjugated molecules in live rodents. Traffic. 2008;9:1801–10. doi: 10.1111/j.1600-0854.2008.00798.x. http://dx.doi.org/10.1111/j.1600-0854.2008.00798.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunn KW, Sutton TA, Sandoval RM. Live-animal imaging of renal function by multiphoton microscopy. Curr Protoc Cytom. 2007;Chapter 12 doi: 10.1002/0471142956.cy1209s41. Unit12 9. [DOI] [PubMed] [Google Scholar]
- 13.Tanner GA, Gretz N, Shao Y, Evan AP, Steinhausen M. Organic anion secretion in polycystic kidney disease. J Am Soc Nephrol. 1997;8:1222–31. doi: 10.1681/ASN.V881222. [DOI] [PubMed] [Google Scholar]
- 14.Dunn KW, Sandoval RM, Kelly KJ, Dagher PC, Tanner GA, Atkinson SJ, Bacallao RL, Molitoris BA. Functional studies of the kidney of living animals using multicolor two-photon microscopy. Am J Physiol Cell Physiol. 2002;283:C905–16. doi: 10.1152/ajpcell.00159.2002. http://dx.doi.org/10.1152/ajpcell.00159.2002. [DOI] [PubMed] [Google Scholar]
- 15.Babbey CM, Ryan JC, Gill EM, Ghabril MS, Burch CR, Paulman A, Dunn KW. Quantitative intravital microscopy of hepatic transport. IntraVital. 2012;1:44–53. http://dx.doi.org/10.4161/intv.21296. [Google Scholar]
- 16.Liu Y, Chen HC, Yang SM, Sun TL, Lo W, Chiou LL, Huang GT, Dong CY, Lee HS. Visualization of hepatobiliary excretory function by intravital multiphoton microscopy. J Biomed Opt. 2007;12:014014. doi: 10.1117/1.2710237. http://dx.doi.org/10.1117/1.2710237. [DOI] [PubMed] [Google Scholar]
- 17.Abboud G, Kaplowitz N. Drug-induced liver injury. Drug safety: an international journal of medical toxicology and drug experience. 2007;30:277–94. doi: 10.2165/00002018-200730040-00001. [DOI] [PubMed] [Google Scholar]
- 18.Li FC, Liu Y, Huang GT, Chiou LL, Liang JH, Sun TL, Dong CY, Lee HS. In vivo dynamic metabolic imaging of obstructive cholestasis in mice. Am J Physiol Gastrointest Liver Physiol. 2009;296:G1091–7. doi: 10.1152/ajpgi.90681.2008. http://dx.doi.org/10.1152/ajpgi.90681.2008. [DOI] [PubMed] [Google Scholar]
- 19.Recknagel P, Claus RA, Neugebauer U, Bauer M, Gonnert FA. In vivo imaging of hepatic excretory function in the rat by fluorescence microscopy. Journal of biophotonics. 2012;5:571–81. doi: 10.1002/jbio.201100118. [DOI] [PubMed] [Google Scholar]
- 20.Kreisel D, Nava RG, Li W, Zinselmeyer BH, Wang B, Lai J, Pless R, Gelman AE, Krupnick AS, Miller MJ. In vivo two-photon imaging reveals monocyte-dependent neutrophil extravasation during pulmonary inflammation. Proc Natl Acad Sci U S A. 2010;107:18073–8. doi: 10.1073/pnas.1008737107. http://dx.doi.org/10.1073/pnas.1008737107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Looney MR, Thornton EE, Sen D, Lamm WJ, Glenny RW, Krummel MF. Stabilized imaging of immune surveillance in the mouse lung. Nat Methods. 2011;8:91–6. doi: 10.1038/nmeth.1543. http://dx.doi.org/10.1038/nmeth.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Presson RG, Jr, Brown MB, Fisher AJ, Sandoval RM, Dunn KW, Lorenz KS, Delp EJ, Salama P, Molitoris BA, Petrache I. Two-photon imaging within the murine thorax without respiratory and cardiac motion artifact. Am J Pathol. 2011;179:75–82. doi: 10.1016/j.ajpath.2011.03.048. http://dx.doi.org/10.1016/j.ajpath.2011.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tabuchi A, Mertens M, Kuppe H, Pries AR, Kuebler WM. Intravital microscopy of the murine pulmonary microcirculation. J Appl Physiol (1985) 2008;104:338–46. doi: 10.1152/japplphysiol.00348.2007. http://dx.doi.org/10.1152/japplphysiol.00348.2007. [DOI] [PubMed] [Google Scholar]
- 24.Vinegoni C, Lee S, Feruglio PF, Marzola P, Nahrendorf M, Weissleder R. Sequential average segmented microscopy for high signal-to-noise ratio motion-artifact-free in vivo heart imaging. Biomed Opt Express. 2013;4:2095–106. doi: 10.1364/BOE.4.002095. http://dx.doi.org/10.1364/BOE.4.002095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu C, Shen Y, Littman DR, Dustin ML, Velázquez P. Visualization of mucosal homeostasis via single-and multiphoton intravital fluorescence microscopy. J Leukoc Biol. 2012;92:413–9. doi: 10.1189/jlb.0711344. http://dx.doi.org/10.1189/jlb.0711344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marchiando AM, Shen L, Graham WV, Weber CR, Schwarz BT, Austin JR, 2nd, Raleigh DR, Guan Y, Watson AJ, Montrose MH, et al. Caveolin-1-dependent occludin endocytosis is required for TNF-induced tight junction regulation in vivo. J Cell Biol. 2010;189:111–26. doi: 10.1083/jcb.200902153. http://dx.doi.org/10.1083/jcb.200902153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vinegoni C, Lee S, Gorbatov R, Weissleder R. Motion compensation using a suctioning stabilizer for intravital microscopy. Intravital. 2012;1:115–21. doi: 10.4161/intv.23017. http://dx.doi.org/10.4161/intv.23017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao L, Kobayakawa S, Yoshiki A, Abe K. High resolution intravital imaging of subcellular structures of mouse abdominal organs using a microstage device. PLoS One. 2012;7:e33876. doi: 10.1371/journal.pone.0033876. http://dx.doi.org/10.1371/journal.pone.0033876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ritsma L, Steller EJ, Ellenbroek SI, Kranenburg O, Borel Rinkes IH, van Rheenen J. Surgical implantation of an abdominal imaging window for intravital microscopy. Nat Protoc. 2013;8:583–94. doi: 10.1038/nprot.2013.026. http://dx.doi.org/10.1038/nprot.2013.026. [DOI] [PubMed] [Google Scholar]
- 30.Soulet D, Paré A, Coste J, Lacroix S. Automated filtering of intrinsic movement artifacts during two-photon intravital microscopy. PLoS One. 2013;8:e53942. doi: 10.1371/journal.pone.0053942. http://dx.doi.org/10.1371/journal.pone.0053942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown LG. A Survey of Image Registration Techniques. Comput Surv. 1992;24:325–76. http://dx.doi.org/10.1145/146370.146374. [Google Scholar]
- 32.Zitova B, Flusser J. Image registration methods: a survey. Image Vis Comput. 2003;21:977–1000. http://dx.doi.org/10.1016/S0262-8856(03)00137-9. [Google Scholar]
- 33.Lorenz KS, Salama P, Dunn KW, Delp EJ. Digital correction of motion artefacts in microscopy image sequences collected from living animals using rigid and nonrigid registration. J Microsc. 2012;245:148–60. doi: 10.1111/j.1365-2818.2011.03557.x. http://dx.doi.org/10.1111/j.1365-2818.2011.03557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar AN, Short KW, Piston DW. A motion correction framework for time series sequences in microscopy images. Microscopy and microanalysis: the official journal of Microscopy Society of America, Microbeam Analysis Society. Microscopical Society of Canada. 2013;19:433–50. doi: 10.1017/S1431927612014250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dombeck DA, Khabbaz AN, Collman F, Adelman TL, Tank DW. Imaging large-scale neural activity with cellular resolution in awake, mobile mice. Neuron. 2007;56:43–57. doi: 10.1016/j.neuron.2007.08.003. http://dx.doi.org/10.1016/j.neuron.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Greenberg DS, Kerr JN. Automated correction of fast motion artifacts for two-photon imaging of awake animals. J Neurosci Methods. 2009;176:1–15. doi: 10.1016/j.jneumeth.2008.08.020. http://dx.doi.org/10.1016/j.jneumeth.2008.08.020. [DOI] [PubMed] [Google Scholar]
- 37.Rueckert D, Sonoda LI, Hayes C, Hill DL, Leach MO, Hawkes DJ. Nonrigid registration using free-form deformations: application to breast MR images. IEEE Trans Med Imaging. 1999;18:712–21. doi: 10.1109/42.796284. http://dx.doi.org/10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- 38.Lorenz KS, Salama P, Dunn KW, Delp EJ. Non-rigid registration of multiphoton microscopy images using B-splines. Proceedings of the SPIE. 2011;7962 [Google Scholar]
- 39.Clendenon JL, Phillips CL, Sandoval RM, Fang S, Dunn KW. Voxx: a PC-based, near real-time volume rendering system for biological microscopy. Am J Physiol Cell Physiol. 2002;282:C213–8. doi: 10.1152/ajpcell.2002.282.1.C213. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


