Abstract
Changes in cell fate and identity are essential for endothelial-to-hematopoietic transition (EHT), an embryonic process that generates the first adult populations of hematopoietic stem cells (HSCs) from hemogenic endothelial cells. Dissecting EHT regulation is a critical step towards production of in vitro derived HSCs. Yet, we do not know how distinct endothelial and hematopoietic fates are parsed during the transition. Here we show that genes required for arterial identity function later to repress hematopoietic fate. Tissue-specific, temporally controlled, genetic loss of arterial genes (Sox17 and Notch1) during EHT results in increased production of hematopoietic cells due to loss of Sox17-mediated repression of hematopoietic transcription factors (Runx1, Gata2). However, the increase in EHT can be abrogated by increased Notch signaling. These findings demonstrate that the endothelial-hematopoietic fate switch is actively repressed in a population of endothelial cells, and that de-repression of these programs augments hematopoietic output.
Introduction
The first hematopoietic stem cells (HSCs) emerge in the embryo from a specialized subset of endothelial cells, collectively termed hemogenic endothelium (HE). The concept of endothelial-derived HSCs has broad clinical implications as it may open new avenues for in vitro blood production. However, the hemogenic capacity of the endothelium is transient and its precise regulation remains unknown. During a narrow developmental time period (approximately embryonic day (E)10–12 in the mouse1,2, and 4–6 weeks in the human3), hemogenic endothelial cells acquire cell morphology and gene expression consistent with hematopoietic identity, in a process called endothelial to hematopoietic transition (EHT)4–6. In the mammalian system, the “hemogenic window” is short lived and typified by groups (or clusters) of rounded cells that are observed within the vascular wall. The hematopoietic cell clusters have been demonstrated to contain both hematopoietic stem and progenitor cells (HSPCs)7,8. Regions known to harbor hemogenic endothelium include the aorta-gonado-mesonephros region (AGM)1,9–12, vitelline and umbilical arteries9,13,14 yolk sac15,16, placenta17,18, and others19,20, but generally encompass arterial vascular beds, as opposed to veins or capillaries21.
Interestingly, regulators of arterial fate including the transcription factor Sox1722 and Notch123, are implicated in hematopoietic emergence from HE, as early loss of either results in hematopoietic defects24,25. Sox17 positively regulates Notch1 for both arterial fate acquisition and hemogenic endothelial specification22,26. How these arterial fate specifiers function in endothelial to hematopoietic conversion, separate from their role in artery-vein specification is unclear.
Here we present data that demonstrates after artery-vein specification, Sox17 actively prevents the transition to hematopoietic fate by repression of key hematopoietic transcription factors, thereby maintaining endothelial identity. The loss of Sox17 promotes hematopoietic conversion, and its dynamic expression imparts a previously unappreciated, but critical step, in endothelial to hematopoietic cell fate transition.
Results
Hematopoietic clusters and endothelial gene expression
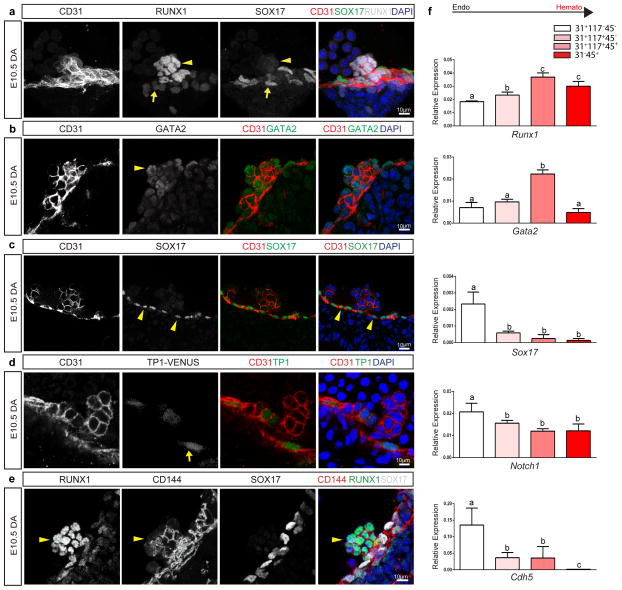
We first evaluated the expression patterns of Sox17, Notch1, Runx1, and Gata2 in the embryonic dorsal aorta (AGM) as all four factors are shown to be required for hematopoietic stem cell emergence. The endothelium of this region can be identified by immunofluorescence of the pan-endothelial cell surface marker PECAM-1 (CD31), and HSPC clusters are easily apparent through their rounded morphology and shared endothelial marker expression (Figure 1a–d). RUNX127 and GATA228, two transcription factors known to be required for HSPC emergence from hemogenic endothelium, are localized to HSPC clusters, as compared to the adjacent endothelium (Figure 1a, b, e). When known regulators of the arterial program including notch signaling23,29 (visualized by the TP1-Venus reporter mouse line30,31) and SOX1722 are evaluated, immunofluorescence is localized to the endothelium and not the HSPC clusters (Figure 1c–e). The appearance of HSPC clusters along the aortic wall is coincident with changes in cell surface marker expression, as cluster cells acquire c-Kit (CD117)7,32,33 and CD4134,35 markers (Supplementary Figure 1a, b), in addition to maintaining endothelial markers CD31 and VE-cadherin (CD144)36, Figure 1a–e. Eventually HSPCs also acquire CD45, a pan hematopoietic surface marker (Supplementary Figure 1c). Sox17 expression is largely undetectable in cluster cells, but rarely can be seen in a perinuclear pattern with co-expression of golgi markers (Supplemental Figure 2d–f), suggesting it no longer functions as a transcription factor in the cluster cell population. As arterial markers can be flow sensitive37–39, we also evaluated the expression patterns of SOX17 and RUNX1 in Mcl2a−/− circulation mutants40 (Supplemental Figure 1g), and found the segregation of SOX17 immunofluorescence to the endothelium and RUNX1 to hematopoietic cell clusters is preserved. The differential expression of surface markers allows for separation of endothelial and hematopoietic populations, as well as HSPC clusters (CD31+CD117+), by fluorescent activated cell sorting (FACS) (Supplementary Figure 1h, i). Transcriptional analyses of sorted populations demonstrate that endothelial subsets (CD31+CD117−CD45−) exhibit lower Runx1 and Gata2 transcript levels when compared to HSPC cluster populations (CD31+CD117+CD45−), or as compared to differentiated hematopoietic cells (CD31−CD45+) (Figure 1f, and Supplementary Table 1). In contrast, genes associated with arterial identity (Sox17 and Notch1) are decreased in HSPC clusters as compared to the endothelium. Sox1726,41 and Notch124,42–45 are known to be important for hemogenic endothelial specification. Thus, the finding that their transcripts and protein levels are actually decreased in HSPC clusters is intriguing. As relatively small populations of primordial germ cells can express CD31 and CD11732,46, we also evaluated populations based on CD41 expression and found the same trend is observed when we identify hemogenic cluster cells with the marker CD4134,35 (Supplementary Figure 1b, i-j). Together the data suggest that endothelial to hematopoietic fate conversion may require down-regulation of critical arterial genes.
Figure 1. Hematopoietic cell clusters down-regulate arterial gene expression.
(a) – (e) Single channels in black and white, scale bars as shown. E10.5 wildtype dorsal aorta (DA).
(a) Hematopoietic cell clusters of the AGM at E10.5. The endothelial layer and attached hematopoietic cell clusters are CD31+ (red). RUNX1 (grey) is notable in cells comprising the hematopoietic cluster (arrowhead). SOX17 (green) expression is localized to the endothelial layer (arrow). DAPI in blue.
(b) GATA2 (green) is notable in the hematopoietic cell cluster (arrowhead). CD31 (red), and DAPI (blue).
(c) SOX17 (green) immunofluorescence is noted in the cell nuclei of the endothelial layer (arrowheads), as compared to the associated cell cluster. CD31 in red, and DAPI in blue.
(d) Notch pathway activation (green) as measured in the TP-1 Venus mouse line is notable in the endothelial layer (arrow) but less so in the associated hematopoietic cell cluster, CD31 in red. DAPI in blue.
(e) CD144 (red) labels the endothelium and hematopoietic cluster cells (arrowhead), Sox17 in grey, and Runx1 in green.
(f) Embryos at E10.5 were sorted based on cell surface markers to isolate endothelial cells (CD31+CD117−CD45−), hematopoietic cluster cells (CD31+CD117+CD45−), maturing cluster cells and HSPCs (CD31+CD117+CD45+), and mature hematopoietic cells (CD31−CD45+). Bar graphs depict transcript expression (RT-PCR) in each subgroup for Runx1, Gata2, Sox17, Notch1, and Cdh5 (CD144). Differing letters represent significance between groups where a versus b, or b versus c, or a versus c, is significant to a p value < 0.01 or less, n=3 litters, 24 embryos
Sox17 negatively regulates hematopoietic fate
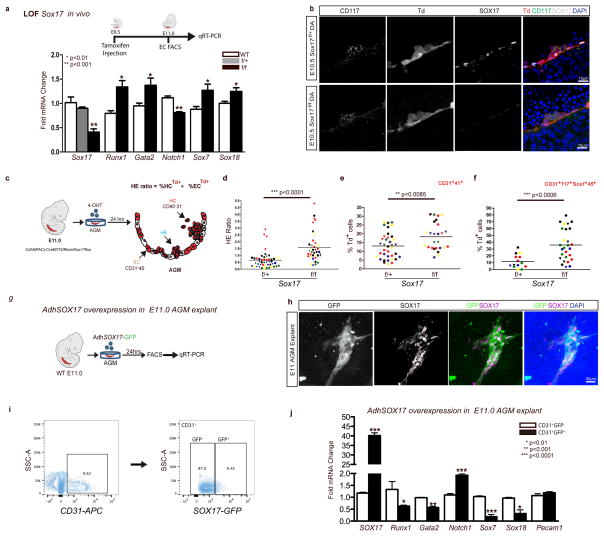
To evaluate the impact of Sox17 on EHT, we undertook both loss and gain-of-function approaches. In vivo endothelial genetic deletion of Sox17 during EHT (induction at E9.5, evaluation at E11, Figure 2a) was evaluated using a endothelial specific Cre recombinase (Cdh5(PAC)-CreERT247) mouse line crossed to a Sox17 floxed line25 with a ROSA26Cre reporter48 (RTom, tdtomato, Td+). The induction strategy is similar to that used in fate tracing studies49 and allows for timing of Sox17 endothelial recombination early in the hemogenic window and during EHT. Transcript analysis of sorted endothelial cells after in vivo induction uncovered a significant increase in Runx1 and Gata2, two hematopoietic transcription factors known to be critical for HSC development during EHT27,50,51 (Figure 2a). Notch1 transcripts are also notably decreased (Figure 2a), in agreement with previous studies that show Sox17 positively regulates the Notch pathway22,26. In addition other members of the SoxF family (Sox7 and Sox18) were increased, possibly due to a compensatory response (Figure 2a). There were no observed differences in endothelial labeling or cell number across homozygous Sox17f/f, heterozygous Sox17f/+, or control animals (Supplementary Figure 2a). Immunohistochemical analysis demonstrates the presence of HSPC clusters in the aorta with a marked decrease of endothelial SOX17 in Sox17f/f mutants (Figure 2b). Also, we did not observe any obvious changes in endothelial morphology as evaluated by scanning electron microscopy (Supplementary Figure 2b).
Figure 2. Endothelial to hematopoietic conversion is increased after Sox17 loss.
(a) Schema and bar graph of qRT-PCR analyses of sorted endothelial cells from E11 embryos after in vivo Sox17 ablation at E9.5. Error bars indicate standard error of the mean (n=3 litters, embryos pooled by genotype). LOF, loss of function.
(b) Immunofluorescence of Sox17 heterozygous and homozygous embryos at E10.5 after in vivo Cre induction (tamoxifen induction at E9.5). Hematopoietic clusters are labeled by CD117 (green), Cre traced endothelial and cluster cells in red (Td+). SOX17 (grey) is absent in homozygous mutant endothelium. DAPI in blue. DA, dorsal aorta. Scale bar denotes 10 μm. Single channels in black and white.
(c) Schematic of AGM explant analysis depicts in vitro Cre lineage tracing and calculation of hemogenic output (HE ratio); the ratio between percent labeled (Td+) hematopoietic cells (CD45+CD31−) to percent labeled (Td+) endothelial cells (CD31+CD45−). 4OHT, 4-hydroxytamoxifen.
(d–f) Each data point represents a separate embryo/AGM explant, littermates are depicted by the same data point color and shape. Bar indicates group mean. P-values calculated on student’s t-test between groups, significance also validated by two-way ANOVA, (Supplementary Table 2).
(d) The HE ratio of Sox17 homozygous (f/f) and heterozygous (f/+) mutant explants. f/+ n=45, f/f n=38, 15 litters.
(e) Percentage of traced Td+ hemogenic endothelial cluster cells, designated as CD31+CD41+. f/+ n=37, f/f n=26, 9 litters.
(f) Percentage of traced (Td+) maturing HSPCs (identified as CD31+CD117+Sca1+CD45+), f/+ n=14, f/f n=27, 7 litters.
(g) Schema depicts overexpression analyses in wildtype AGM explants at E11.0.
(h) Immunofluorescence of E11 AGM explant after human adenoviral SOX17-GFP infection. GFP in green, SOX17 in magenta, and DAPI in blue. Scale bar as indicated.
(i) Cell sorting strategy for endothelial cells (CD31+) after infection to AdhSOX17-GFP (GFP), where GFP+ and GFP− populations were gated.
(j) Bar graph of qRT-PCR analyses of sorted E11 AGM CD31+ cells after AdhSOX17-GFP infection. Error bars indicate standard error of the mean. CD31+GFP− population served as a control, set to one for comparisons of fold change, n=3 litters, embryos pooled, p values as indicated.
(a,j) P-values reflect student’s t-test.
Currently it is not possible to predict which specific endothelial cell within a hemogenic vascular bed will transition to a hematopoietic fate. Also not known is whether endothelial cells comprising the same hemogenic site are all capable of EHT. So whether the actual cell fate conversion is a stochastic event or a predetermined fate change remains to be seen. To circumvent the current obstacles of EHT prediction, we adopted a fate tracing strategy49 that allows measurement of traced hematopoietic cell populations from labeled endothelial precursors within a specific hemogenic vascular site. By inducing endothelial recombination of Sox17 in AGM explants using the Cdh5(PAC)-CreERT2/RTom/Sox17flox transgenic mouse line, the number of EHT derived hematopoietic cells can be quantified through fate mapping (Figure 2c, Supplementary Figure 2c, and Supplementary Table 2). Tamoxifen induction in vitro with the active metabolite 4-hydroxytamoxifen (4-OHT) at E11.0 allows immediate ablation in AGM explants during EHT, and the calculation of a HE ratio which we define as traced hematopoietic cells (HCs) compared to traced endothelial cells (ECs). Using this assay to temporally and conditionally ablate Sox17, we demonstrate that timed loss of endothelial Sox17 promotes conversion to hematopoietic cell fate in situ (Figure 2c–f). Sox17f/f mutants exhibit a significant 3-fold increase in HE ratios indicating increased hematopoietic output, in addition to significantly increased labeled hemogenic cluster populations (CD31+CD41+Td+) and maturing HSPC populations (CD31+CD117+Sca1+CD45+Td+), Figure 2c–f. The observed increase in HE ratios and HSPC number is not due to proliferation effects (as measured by BrdU incorporation, Supplementary Figure 2d), nor is the higher HE ratio due to changes in cell death (Annexin-V staining, Supplementary Figure 2d). We also observe increases in other hematopoietic populations (CD31−CD117+Sca1+CD45+Td+), Supplementary Figure 2e. In addition, when a similar strategy is applied to earlier explants (E9.5) prior to hematopoietic cell cluster emergence, we observe similar trends in the HE ratio (Supplementary Figure 2f). So while Sox17 has been shown to be critical for HE specification prior to EHT, the loss of Sox17 actually promotes hematopoietic fate over endothelial fate during EHT. To further evaluate the role of Sox17 in this process, we undertook gain-of-function studies in wildtype AGM explants using adenoviral-mediated overexpression of human SOX17 (AdhSOX17-GFP), Figure 2g. GFP expression in explants overlapped with SOX17 co-staining (Figure 2h), allowing for cell sorting of AGM endothelial cells (CD31+) that were either successfully infected (GFP+) or not infected (GFP−) by AdhSOX17-GFP, Figure 2i. Transcript analysis of endothelial cells with SOX17 overexpression demonstrates significant increases in Sox17 and Notch1 transcripts with significant reduction in Runx1, Gata2, Sox7 and Sox18 transcripts, Figure 2j. The data altogether suggest that Sox17 negatively regulates hematopoietic fate through repression of Runx1 and Gata2. We also show the known positive regulation of Notch1 by Sox17, and regulation of other SoxF family members, Sox7 and Sox18.
Sox17 represses Runx1 and Gata2
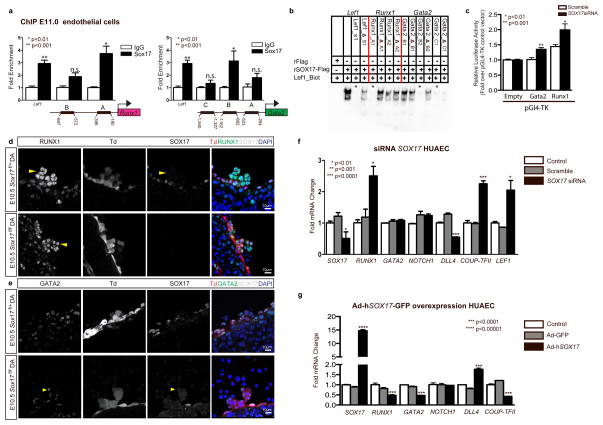
To determine whether the observed changes in Runx1 and Gata2 were due to regulation by SOX17, chromatin immunoprecipitation (ChIP) was carried out in sorted endothelial cells at E11 (Figure 3a), as well as in human umbilical arterial endothelial cell lines, HUAECs (Supplementary Figure 3a, and Supplementary Table 3). Two predicted SOX17 binding sites upstream of Runx1 and Gata2 5′UTRs showed significant enrichment (Figure 3a). To demonstrate whether SOX17 was capable of direct DNA binding of specific sequences in vitro, electrophoretic mobility shift assays (EMSA) were conducted for sites with high species homology between human and mouse (Supplementary Figure 3b, and Supplementary Table 4). Specific areas within ChIP enriched regions were capable of out-competing the known SOX17 regulatory site in the Lef1 promoter52 (Figure 3b). We further analyzed the regulation of Runx1 and Gata2 using luciferase reporter assays, which demonstrate de-repression of both Runx1 and Gata2 activity after Sox17 siRNA knockdown (Figure 3c). In vivo loss of Sox17 demonstrates intact hematopoietic clusters with normal localization of RUNX1 and GATA2 expression (Figure 3d, e). To investigate how Sox17 may regulate Runx1 and Gata2 in mature endothelium in the human system, we conducted in vitro gain and loss-of-function experiments. SOX17 siRNA inhibition of human umbilical arterial cell lines resulted in significantly elevated RUNX1 transcripts, at similar levels to the control LEF152, a SOX17 repressive target (Figure 3f). In addition, genes important in arterial and venous identity are altered with decreased arterial gene transcripts (DLL4)53,54 and elevated transcript levels of COUP-TFII, an important determinant of venous fate21 (Figure 3f). In contrast, when SOX17 is overexpressed after adenoviral infection, RUNX1 and GATA2 transcript levels are significantly decreased (Figure 3g). SOX17 overexpression also altered levels of DLL4 (increased) and COUP-TFII (decreased) (Figure 3g). The data suggest a novel role of Sox17 as a repressor of hematopoietic fate, while confirming Sox17 as a pro-arterial fate regulator.
Figure 3. SOX17 directly binds Runx1 and Gata2 for repression of hematopoietic fate.
(a) SOX17 chromatin immunoprecipitation (ChIP) qRT-PCR of E11.0 sorted endothelial cells. Letters denote regions with SOX17 binding site consensus sequences upstream of Runx1 and Gata2 promoters, and Lef1 as a positive control. Error bars indicate standard error of the mean (SEM). IgG control set to one for comparisons of fold change, n=3 litters, embryos pooled, p values as indicated.
(b) Electrophoretic mobility shift assay (EMSA) of putative SOX17 binding sites within ChIP sequences designated by letters in (a). Each lane represents biotin-labeled duplexed oligonucleotides containing the Lef1 promoter SOX17 binding site (Lef1_Biot). Addition of rSox17-Flag produces a specific shift, indicating protein-DNA complex (lane 2), which is competed away by unlabeled Lef1 (Lef1_s1), while mutant probe does not compete (Lef1_Δ_s1). Similar designations are used for putative binding sites (and mutants) in Runx1 and Gata2 sequences. Asterisks denote competitive binding.
(c) Bar graph depicts luciferase activity of Gata2 and Runx1 promoters after Sox17 siRNA versus control (scramble). P-values as indicated. Error bars represent SEM.
(d) Immunofluorescence of hematopoietic cell clusters (arrowhead) in E10.5 dorsal aorta (DA) of Sox17f/f and Sox17f/+ mutants (after tamoxifen mediated Cre induction at E9.5). Traced cells labeled in red (Td+), SOX17 in grey, and RUNX1 in green. DAPI in blue. Scale bar 10μm. Single channels in black and white.
(e) GATA2 (green) and SOX17 (grey) immunofluorescence of hematopoietic cell clusters in E10.5 dorsal aorta (DA) of Sox17f/+ and Sox17f/f (arrowhead) mutants (Cre induction at E9.5). Traced cells in red (Td+), SOX17 in grey, and RUNX1 in green. DAPI in blue. Scale bar denotes 10μm. Single channels in black and white.
(f) SOX17 siRNA knockdown in HUAECs and qRT-PCR analysis. Control represents treatment with lipofectin alone, SOX17 siRNA compared to scrambled (n=3 experiments, error bars indicate SEM). P-values as indicated.
(g) Adenoviral-mediated overexpression of hSOX17 in HUAECs and qRT-PCR analyses, p values calculated with respect to Adeno-GFP infected cells, control represents uninfected cells (n=3 experiments, error bars indicate SEM).
Intersecting roles of Sox17, Runx1, and the Notch pathway
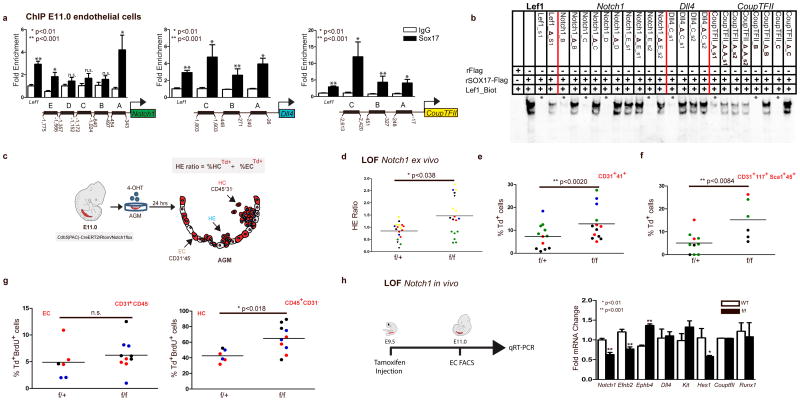
As Sox17 was previously shown to promote arterial identity upstream of the Notch pathway22, we evaluated SOX17 regulation of notch pathway members in our system. SOX17 ChIP demonstrates enriched occupancy upstream of the Notch1 5prime;UTR, and of the Notch ligand Dll4 (Figure 4a). In addition, we also observe occupancy upstream of CoupTFII, which has not been previously described (Figure 4a). Similar enrichment of these sites was observed in HUAECs (Supplementary Figure 4a). We further validated direct binding of SOX17 within the enriched ChIP sites via EMSA, and demonstrated multiple SOX17 binding sites are capable of outcompeting Lef1 controls (Figure 4b, and Supplementary Figure 4b, c). To understand whether Notch1, a putative downstream target of SOX17, also plays a repressive role in EHT, we evaluated Notch1 loss-of-function. Similar to Sox17, loss of Notch1 in AGM explants increased the HE ratio, as well as populations of HSPCs (Figure 4c–f, Supplementary Figure 4d, and Supplementary Table 5). We also observed increased HE ratios after AGM explants were exposed the γ-secretase inhibitor DAPT (Supplementary Figure 4e). However, when BrdU incorporation was evaluated in Notch1 mutant explants, significantly higher levels of incorporation occurred in the hematopoietic compartment (Figure 4g), suggesting the observed changes may be due to hematopoietic cell proliferation, and not due to an increase in EHT. Annexin-V levels were not notably changed (Supplementary Figure 4f, g). In vivo loss of Notch1 (induction at E9.5), demonstrates expected changes in arterial and venous identity genes23 EfnB2 and EphB4 within sorted endothelial cells (Figure 4h). No changes in Runx1 transcripts were noted, while expectedly Hes1 transcripts were decreased (Figure 4h). There were no observed differences in endothelial labeling or cell number across homozygous Notch1f/f, heterozygous Notch1f/+, or control animals (Supplementary Figure 4h). Interestingly, we also noted expected changes in endothelial morphology55 (Supplementary Figure 4i).
Figure 4. The role of the Notch pathway in endothelial to hematopoietic fate decisions.
(a) SOX17 (ChIP) qRT-PCR of E11.0 sorted endothelial cells. Letters denote regions upstream of Notch1, Dll4, and CoupTFII promoters, and Lef1 as a positive control. Error bars indicate standard error of the mean. IgG control set to one for comparisons of fold change, n=3 litters, embryos pooled, p values as indicated.
(b) EMSA of putative SOX17 binding sites within ChIP sequences (designated by letters in (a)). Each lane represents biotin-labeled duplexed oligonucleotides spanning the Lef1 promoter SOX17 binding site (Lef1_Biot). Addition of rSox17-Flag produces a specific shift, indicating protein-DNA complex (lane 2), which is competed away by unlabeled Lef1 (Lef1_s1), while mutant probe does not compete (Lef1_Δ_s1). Similar designations are used for putative binding sites (and mutants) in Notch1, Dll4, and CoupTFII sequences. Asterisks denote competition.
(c) Schematic of AGM explant analysis depicts in vitro Cre lineage tracing and calculation of hemogenic output (HE ratio); the ratio between percent labeled (Td+) hematopoietic cells (CD45+CD31−) to percent labeled (Td+) endothelial cells (CD31+CD45−).
(d–g) Each data point represents a separate embryo/AGM explant, littermates are depicted by the same data point color and shape. Bar indicates group mean. P-values calculated on student’s t-test between groups, significance also validated by two-way ANOVA, Supplementary Table 5)
(d) The HE ratio of Notch1 homozygous (f/f) and heterozygous (f/+) mutant explants. f/+ n=18, f/f n=21, 6 litters. LOF, loss of function.
(e) Percentage of traced Td+ hemogenic endothelial cluster cells, designated as CD31+CD41+. f/+ n=12, f/f n=13, 4 litters.
(f) Percentage of traced (Td+) maturing HSPCs (identified as CD31+CD117+Sca1+CD45+) f/+ n=10, f/f n=6, 3 litters.
(g) BrdU+ cells measured after 2 hour incubation with BrdU in traced ECs (left) and traced HCs (right) demonstrates a significant increase in HC proliferation, f/+ n=6, f/f n=10, 3 litters.
(h) Schema and bar graph of qRT-PCR analyses of sorted endothelial cells from E11 embryos after in vivo Notch1 ablation at E9.5. Error bars indicate standard error of the mean (n=3 litters, embryos pooled by genotype). LOF, loss of function.
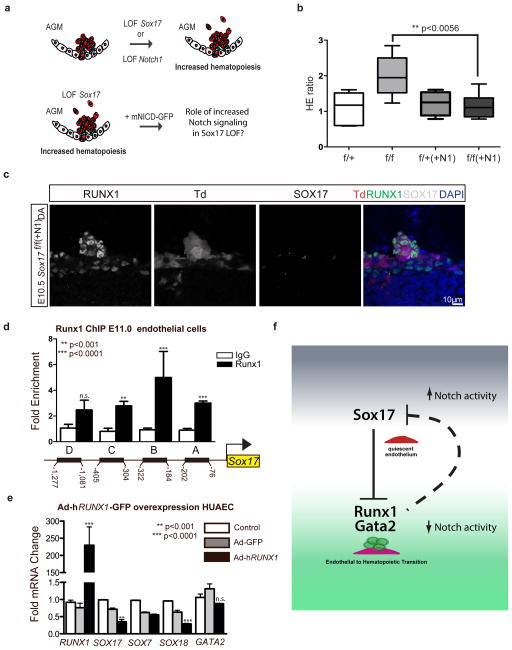
To understand the role of Notch1 signaling in the context of Sox17 loss, we bred R26RNotch1IC-nEGFP lines56 (+mNICD-GFP) that overexpress the Notch1 intracellular domain (NICD) upon Cre activation into our temporal endothelial-specific Sox17 loss-of-function models (Figure 5a). Increased Notch activation in E11.0 AGM explants was capable of abrogating the observed EHT increase in Sox17 mutants (Figure 5b) with normal appearing HSPC clusters in vivo after induction of Sox17 loss and NICD overexpression at E9.5 (Figure 5c). Thus, the conversion to hematopoietic fate in hemogenic endothelium requires loss of arterial identity programs in addition to de-repression of hematopoietic genes by SOX17. While our data has shown the regulation of Runx1 by SOX17, previous reports suggest RUNX1 may directly bind and repress Sox1757. To evaluate whether there may be bi-directional regulation in the endothelium, we performed RUNX1 ChIP of conserved sites upstream of Sox17 transcriptional start sites and found multiple areas of enrichment (Figure 5d and Supplementary Figure 4j, k). Adenoviral overexpression of RUNX1 (AdhRUNX1-GFP) in HUAECs demonstrates decreased SOX17 and SOX18 transcripts (Figure 5e). Overall, the data presents a complex regulatory network for the maintenance of endothelial cell fate and the conversion to a hematopoietic fate (Figure 5f). Once hematopoietic fate is achieved, both Sox17 and Notch1 have known roles in hematopoietic cell survival25 and lineage differentiation58, which is also evident in our hematopoietic colony assay evaluation of mutant hematopoietic cells (Supplementary Figure 5).
Figure 5. Parsing endothelial and hematopoietic fates during EHT.
(a) Schematic depicting Sox17 or Notch1 loss of function (LOF) and strategy for evaluating Notch overexpression (mNICD-GFP) in Sox17 mutants. NICD, Notch1 intracellular domain.
(b) HE ratios of E11 AGM explants in Sox17 mutants with and without Notch overexpression (+N1). Center-lines represent median values, box represents 25th–75th percentiles, bars represent minimum and maximum values. f/+(−N1) n=7, f/+(+N1) n=3, f/f (−N1) n=5, f/f (+N1) n=8, 3 litters. P-values calculated on student’s t-test between groups, significance also validated by two-way ANOVA, (Supplementary Table 2)
(c) Immunofluorescence of a representative hematopoietic cluster in a E10.5 Sox17f/f(+N1) AGM after in vivo induction of Cre and NICD at E9.5. SOX17 in grey, traced ECs (Td+) in red and RUNX1+ in green. DAPI in blue. Scale bar denotes 10μm.
(d) RUNX1 chromatin immunoprecipitation (ChIP) PCR of E11.0 sorted endothelial cells. Letters denote evaluated regions containing RUNX1 binding site consensus sequences upstream of the Sox17 promoter. Error bars indicate standard error of the mean. IgG control set to one for comparisons of fold change, n=3 litters, embryos pooled, p values as indicated.
(e) Adenoviral-mediated overexpression of hRUNX1 in HUAECs and qRT-PCR analyses, p values calculated with respect to Adeno-GFP infected cells, control represents uninfected cells (n=3 experiments, error bars indicate SEM).
(f) Schematic depicting the cell fate switch from endothelial to hematopoietic fate, and the governing regulatory pathways of EHT. Sox17 inhibition of Runx1 and Gata2 maintains endothelial fate. Loss of Sox17 inhibition in the context of decreased Notch activity promotes hematopoietic fate conversion.
Discussion
An important obstacle in recapitulating hemogenic endothelium in culture for in vitro blood production is identification of possible activators and silencers of the hemogenic program. Here we demonstrate important altering requirements for Sox17 and Notch1, which highlights the refinements needed for translational models recapitulating EHT. Previous studies have identified Runx127, Gata228, Notch124,59, and Sox1725,26 as critical for endothelial to hematopoietic transition. However, dissecting the contributions of these pathways to vascular development versus the process of hematopoietic emergence from the endothelium has not been previously reported. Notch1, and more recently Sox17 have important roles in arterial specification22,43,60. As the major vessels that harbor hemogenic endothelium are arterial sites9,13, it may be that arterial identity is a prerequisite to hemogenic endothelial activity. However, hemogenic activity also occurs in yolk sac and placental vascular beds that are not overtly arterial9,16,18. In addition, recent evidence in human ESC cultures suggest that while hemogenic endothelial cells incorporate into arterial vascular walls, they have differential surface marker expression profiles than arterial cells61. There is also evidence that arterial identity can be uncoupled from hemogenic capacity62,63. So it may be that hemogenic endothelial specification requires the same pathways mobilized in the acquisition of arterial identity, but not arterial identity per se43. However, for the direct transition to hematopoietic fate, the expression levels of arterial/hemogenic specifiers need to be reduced. The complex temporal requirements, elucidated here, explains previous data where continued or overexpression of Sox17 was noted to prevent hematopoietic conversion in culture26,64. In addition, the reciprocal repression of Sox17 by RUNX1 introduces another unique aspect of fate determination where once endothelial Sox17 levels decrease, Runx1 levels can rapidly rise during the fate switch, and together they function as a classical bistable system; similar to those described in mesodermal progenitors65. Lastly, the data also demonstrate that the EHT program can be manipulated for increased hematopoietic output, suggesting that hemogenic endothelial cell number may not be a fixed entity. If EHT is not restricted to a fixed number of endothelial cells within a hemogenic vascular compartment, but instead occurs as a more global transient stochastic process of developing endothelium, it allows for the possibility of endothelial expansion for hematopoietic stem cell production.
Methods
Animal care and use
Animal protocols were conducted in accordance with University of California at San Francisco and Baylor College of Medicine Laboratory Animal Research Committee guidelines. Cdh5(PAC)-CreERT2 (Tg(Cdh5-cre/ERT2)1Rha) mice47, Notch1tm2Rko and Sox17tm2Sjm floxed lines25,66, and R26RNotch1IC-nEGFP (Gt(ROSA)26Sortm1(Notch1)Dam) lines56 were crossed to R26RTd Cre reporter lines (Gt(ROSA)26Sortm14(CAG-tdTomato)Hze)48. TP1-Venus (Tg(Rbp4*-Venus)#Okn) mice30,31 were generously provided by RIKEN BioResource Center. Myosin light chain 2 alpha (Mlc2a −/−) mutant lines were provided by Mary Dickinson (Baylor College of Medicine)40,67. Pregnancies were dated by presence of a vaginal plug (day 0.5 of gestation). Genomic DNA from adult tail tips or conceptus yolk sacs was genotyped using MyTaq Extract PCR Kit (Bioline, BIO21127). Genotype PCR was performed using primers listed in Supplementary Table 6.
Immunofluorescence and confocal microscopy
E10.5 to E11.5 embryos (in vivo induction with maternal tamoxifen injection at E9.5) were fixed in 2% paraformaldehyde solution overnight and frozen in Tissue-Tek OCT Compound (Sakura Finetek, 4583). 20–30 μm cryosections were obtained (Thermo Scientific Micron, HM550). Slides were dried for 1hr at room temperature, washed with PBST (0.5% Tween or Triton-X100) and incubated in blocking buffer (PBST, 1% BSA, 5% donkey serum) for 1hr. Primary antibodies (for full list of antibodies please see Supplementary Table 7) were incubated at 4°C overnight or room temperature for 6hrs in blocking buffer. Slides were washed with PBST and incubated with the secondary antibody for 2 hrs, washed, stained with 2ug/ul DAPI and mounted in Vectashield (H-1400) or Vectamount (Vector Laboratories, H-5000). Images were captured on a Leica SPE Confocal Microscope and compiled using ImageJ and Imaris 7.6 (Bitplane; Belfast, UK) software.
Flow cytometric analyses and cell sorting
Whole embryos or AGMs underwent mechanical dissociation by pipetting to single cell suspension in Hank’s Balanced Salt Solution with 2% fetal bovine serum, 1% penicillin/streptomycin and 10mM HEPES, pH 7.268 and stained for 30min at 4°C with agitation. Single cell suspensions were sorted in a BD FACS Aria III. Flow cytometric analyses were performed on a FACS Verse or FACS Aria III with FACSDiva 8.0 software (BD Biosciences) and data analyzed using FlowJo v10.0.7 (Tree Star). Gating strategy in Supplementary Figures 1h,i, and 2c, see Supplementary Table 7 for a list of antibodies.
Real Time RT-PCR expression analysis
For in vivo transcriptional characterization of the induced endothelium, lineage traced CD31-APC+,CD41-FITC−,CD45-FITC− DAPI-excluded cells were sorted (for full list of antibodies please see Supplementary Table 7) into MCDB-131 complete medium and RNA was immediately extracted using RNeasy Plus Micro Kit (Qiagen, 74034). 50–300ng of RNA was reverse transcribed using Superscript III Reverse Transcriptase (Life Technologies, 18080044) according to manufacturer’s instructions and cDNA was quantified with Fast SYBR Green Master Mix (Life Technologies, 4385612) in a CFX384 Touch Real-Time PCR Detection System (Bio-Rad). Fluorescence was interpreted relative to GAPDH housekeeping gene expression and quantified using the ΔCt method to obtain relative expression or the ΔΔ Ct method for fold change values, as indicated. A full list of oligonucleotide sequences is listed in Supplementary Table 1.
AGM explant culture and in vivo induction
AGMs from Cdh5(PAC)-CreERT2/R26RTd/Sox17 and Notch1 floxed embryos were dissected and cultured for 24hrs on 40μm filters at an air liquid interface in 10μM 4-hydroxytamoxifen 4-OHT (Sigma H7904) in myelocult medium (Stem Cell Technologies) supplemented with 10−6M hydrocortisone (Stem Cell Technologies)49, at E11 (and E9.5 for Sox17 mutants). In vivo induction was achieved by intraperitoneal injection of 0.8 mg of tamoxifen of pregnant dams at E9.5. Tamoxifen powder (MP Biomedical, 156738) was dissolved in a sunflower seed oil/ethanol (10:1) mixture at 10 mg/ml49. DAPT γ-secretase inhibitor (Sigma, D5942) was prepared in DMSO and added directly to explant culture medium at final concentrations of 25μM, 50μM, 100μM, or 200μM. For overexpression studies, AGMs were incubated with 8 x107 adenoviral particles per milliliter at 37C with agitation for 1 hr prior to explant culture49. Adeno-CMV-hSox17-GFP (AdhSox17-GFP) was produced by Vector Biolabs (ADV-224019, Ref Seq: BC140307).
BrdU
AGM explants were incubated for 2hrs with BrdU (10μM), disaggregated, and stained for extracellular markers CD45-percp and CD31-APC for 30min. Cells were then fixed and permeabilized with BD Cytofix/Cytoperm™ (BD Biosciences, 554714) according to manufacturer instructions. Cell pellet was washed and incubated in DNase I (300 μg/mL) for 1hr at 37°C, stained with DAPI and anti-BrdU conjugated with FITC for 30min, and analyzed by flow cytometry.
Annexin-V
AGM explants were disaggregated, washed in PBS and resuspended in buffer (10 mM HEPES, 0.9% NaCl, 2.5 mM CaCl2, 0.1% BSA) containing FITC-conjugated Annexin-V (BioLegend, 640906). Cells were incubated at room temperature in the dark for 15 min followed by the addition of buffer containing DAPI, and analyzed by flow cytometry.
siRNA
Primary human umbilical arterial endothelial cells (HUAEC) (VEC Technologies) were cultured in MCDB-131 Complete medium (VEC Technologies). Sox17 Silencer Select siRNA (Ambion, s34626-8), scramble negative control siRNA (non-targeted sequences), versus water (control) was administered using Lipofectin (Invitrogen, 18292011), and RNA was extracted 48hrs later using the RNeasy Mini Kit (Qiagen, 74104). Real Time RT PCR was conducted as described above. All cell culture experiments were done between passages 4–6. Supplementary Table 1 lists oligonucleotide sequences of Real Time RT PCR primers.
Recombinant adenovirus
Recombinant adenoviral particles were produced by Vector Biolabs (Philadelphia, PA). Human SOX17 adenovirus (Ad-hSOX17-GFP) contains Sox17 cDNA (GenBank RefSeq ID BC140307) and enhanced green fluorescent protein (eGFP) driven by CMV promoters. Human RUNX1 adenovirus (Ad-hRUNX1-GFP) contains eGFP-2A preceding RUNX1 cDNA (RefSeq ID BC136381) driven by a single CMV promoter. Ad-GFP control adenovirus (cat# 1060) contains CMV driving eGFP only. 1–3 x 102 viral particles per cell were used to infect subconfluent HUAECs 36 hrs before RNA extraction. All cell culture experiments were done between passages 4–7.
Chromatin Immunoprecipitation (ChIP)
Briefly, HUAEC or E11 CD31-APC+ cells were cross-linked with 1% formaldehyde, quenched with 0.125M glycine and re-suspended in lysis buffer (50 mM HEPES-KOH pH7.5, 140mM NaCl, 1mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% triton X-100 in ddH20) containing protease inhibitors. The chromatin solution was sonicated, and the supernatant diluted 10-fold. An aliquot of total diluted lysate was used for input gDNA control. Primary antibody or IgG control was incubated with Pierce Protein A/G Magnetic Beads (Thermo Scientific, 88803) at 4°C overnight to preclear the sample. Sox17 antibody (R&D Systems, AF1924) was used to ChIP in both sorted ECs and HUAEC samples, while Runx1 antibody (Cell Signaling, D4A6) was used to perform ChIP in HUAECs. The magnetic bead coated by the antibody was washed (PBS, 0.1% Triton X-100) then incubated with the precleared sample at 4°C overnight. The precipitates were washed, and the chromatin complexes eluted. After reversal of cross-linking (65°C for 4 hours), the DNA was purified using QIAquick PCR purification kit (Qiagen, 28104) and 100pg was used as a template in each qPCR reaction for quantitative analysis. Oligonucleotides used in PCR for quantitative ChIP are listed in Supplementary Table 3. Antibody dilutions in Supplementary Table 7.
Non-radioactive electrophoretic mobility shift assay
Recombinant SOX17-Flag and Flag alone (pcDNA3 vector (Promega)) were expressed in 293T cells. Plasmids were transfected using Lipofectamine 2000 Transfection Reagent (Life Technologies, 11668019) 36hrs before cells were lysed in RIPA buffer containing protease inhibitors. Recombinant protein was immunoprecipitated from lysate overnight at 4°C with Anti-FLAG M2 magnetic beads (Sigma, M8823) and the recombinant protein eluted with excess FLAG peptide. 5–7ul of the first eluate was used in a binding reaction along with 0.3pMol of complementary annealed 3′Biotin-labeled oligonucleotides (Integrated DNA Technologies), 300-fold excess competitor probes, 0.02U Poly(dG-dC) (Sigma, P9389), and binding buffer (100mM HEPES pH 8.0, 50mM KCl, 500 μM DTT, 50 μM EDTA, 1mM MgCl2, and 5% glycerol by volume)69. DNA-protein complexes were resolved on 7% native polyacrylamide gel, transferred to neutrally charged nylon membrane, incubated with Streptavidin-POD (Roche, 11089153001) and imaged by chemiluminescence. See Supplementary Table 4 for probe sequences.
Luciferase reporter assay
Putative regulatory sequences (700–850bp) including Sox17 ChIP-enriched regions and EMSA-competent SOX17 binding sites were synthesized and cloned (Integrated DNA Technologies) based upon UCSC genome browser murine sequences (see Supplementary Methods for fragment sequences). The fragments were amplified by PCR (Phusion, New England Biolabs) with appropriate linkers. The pGL4-TK vector (pGL4.54, Promega), containing the gene encoding Firefly luciferase driven by a TK minimal promoter, was digested using kpnI restriction enzyme (NEB) and mung bean nuclease (NEB) followed by ligation using Gibson Assembly mastermix (NEB) and confirmatory sequencing. 30,000 C166 murine yolk sac endothelial cells (ATCC, CRL-2581) were reverse cotransfected with 400ng of reporter vector along with 10ng of a Renilla luciferase transfection control plasmid (pRL, Promega) and 30pMol of a Sox17-targeted or non-targeted “scramble” siRNA pool (ON-TARGETplus siRNA SMARTpool, GE Dharmacon) using Lipofectamine 3000 (Life Technologies) according to manufacturer’s recommendations. After 48 hours of culture, cells were lysed and luciferase activity assessed using the Dual-Luciferase Reporter Assay System reagents (Promega) in a GloMax 96 Microplate Luminometer with dual injectors. In technical triplicate, relative luciferase activity was calculated by dividing Firefly readings by Renilla readings for each well and then normalized according to baseline values for each treatment condition after transfection of pGL4-TK without a fragment added.
Statistical Analyses
Student’s t-test, one-way and two-way ANOVA analyses were performed as indicated in all experiments where n≥3 unless otherwise noted. Mean and standard error were calculated and graphed using GraphPad Prism 6 software. All statistical measurements are listed in Supplementary Tables 2 and 5.
Supplementary Material
Acknowledgments
We would like to thank Juan Carlos Zúñiga-Pflücker PhD of Sunnybrook Health Sciences Centre in Toronto for the OP9-Dl1 cell lines, Yomiko Saga PhD of National Genetics Institutes of Japan and RIKEN BRC for use of the TP1-Venus (ICR) mouse line, Courtney Griffin PhD of the University of Oklahoma Health Sciences Center for the C166 cell line and advisement on luciferase protocols, Mary Dickinson of the Baylor College of Medicine for the Mlc2a mutant mouse line. Joanna Tober PhD of the Speck lab in the University of Pennsylvania for her help and advice on methycellulose colony excision genotyping, Ralph Adams PhD of Max Planck Institute for use of his Cdh5(PAC)-CreERT2 line. The work was supported by the American Heart Association Scientist Development Grant (JDW), Burroughs Wellcome Fund Career Award for Medical Scientists (ACZ), and the NIH Office of the Director’s Innovator Award program (ACZ).
Footnotes
Competing financial interests: The authors declare no competing financial interests.
Authors Contributions: CL conducted the majority of the experiments, with support from JH, as well as data analysis, all under the guidance of AZ. JH with supervision by CL and AZ optimized most of the protocols and technical approaches for colony assays, cell culture studies, EMSA, and luciferase assays; with cloning strategy and troubleshooting support from CS, reagents and EMSA troubleshooting help from HP, MD, and JW. FB provided technical expertise with immunofluorescence and scanning electron microscopy. JZ initiated pilot experiments with Notch loss of function and also provided immunofluorescence expertise, while KC generated critical reagents for pilot Sox17 viral experiments. AR and JW obtained and evaluated circulation deficient mutant lines. HY provided technical mouse support and the preliminary analysis of Sox17 and Notch1 cell culture expression. JW and MD helped with reagents and advice. CL and AZ designed the project. CL, JH, and AZ and wrote the manuscript. All authors critically read and contributed to the manuscript.
References
- 1.Medvinsky A, Dzierzak E. Definitive hematopoiesis is autonomously initiated by the AGM region. Cell. 1996;86:897–906. doi: 10.1016/s0092-8674(00)80165-8. [DOI] [PubMed] [Google Scholar]
- 2.Taylor E, Taoudi S, Medvinsky A. Hematopoietic stem cell activity in the aorta-gonad-mesonephros region enhances after mid-day 11 of mouse development. Int J Dev Biol. 2010;54:1055–1060. doi: 10.1387/ijdb.103152et. [DOI] [PubMed] [Google Scholar]
- 3.Tavian M, Péault B. Embryonic development of the human hematopoietic system. Int J Dev Biol. 2005;49:243–250. doi: 10.1387/ijdb.041957mt. [DOI] [PubMed] [Google Scholar]
- 4.Bertrand JY, et al. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature. 2010;464:108–111. doi: 10.1038/nature08738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kissa K, Herbomel P. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature. 2010;464:112–115. doi: 10.1038/nature08761. [DOI] [PubMed] [Google Scholar]
- 6.Eilken HM, Nishikawa SI, Schroeder T. Continuous single-cell imaging of blood generation from haemogenic endothelium. Nature. 2009;457:896–900. doi: 10.1038/nature07760. [DOI] [PubMed] [Google Scholar]
- 7.Boisset JC, et al. Progressive maturation towards hematopoietic stem cells in the mouse embryo aorta. Blood. 2014 doi: 10.1182/blood-2014-07-588954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen MJ, et al. Erythroid/Myeloid Progenitors and Hematopoietic Stem Cells Originate from Distinct Populations of Endothelial Cells. Cell Stem cell. 2011;9:541–552. doi: 10.1016/j.stem.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Bruijn MF, Speck NA, Peeters MC, Dzierzak E. Definitive hematopoietic stem cells first develop within the major arterial regions of the mouse embryo. EMBO J. 2000;19:2465–2474. doi: 10.1093/emboj/19.11.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.North T, et al. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development. 1999;126:2563–2575. doi: 10.1242/dev.126.11.2563. [DOI] [PubMed] [Google Scholar]
- 11.de Bruijn MFTR, et al. Hematopoietic stem cells localize to the endothelial cell layer in the midgestation mouse aorta. Immunity. 2002;16:673–683. doi: 10.1016/s1074-7613(02)00313-8. [DOI] [PubMed] [Google Scholar]
- 12.Jaffredo T, Gautier R, Eichmann A, DIETERLEN-LIEVRE F. Intraaortic hemopoietic cells are derived from endothelial cells during ontogeny. Development. 1998;125:4575–4583. doi: 10.1242/dev.125.22.4575. [DOI] [PubMed] [Google Scholar]
- 13.Gordon-Keylock S, Sobiesiak M, Rybtsov S, Moore K, Medvinsky A. Mouse extraembryonic arterial vessels harbor precursors capable of maturing into definitive HSCs. Blood. 2013;122:2338–2345. doi: 10.1182/blood-2012-12-470971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Bruijn MF, et al. CFU-S(11) activity does not localize solely with the aorta in the aorta-gonad-mesonephros region. Blood. 2000;96:2902–2904. [PubMed] [Google Scholar]
- 15.Yoder MC, Hiatt K, Mukherjee P. In vivo repopulating hematopoietic stem cells are present in the murine yolk sac at day 9.0 postcoitus. Proc Natl Acad Sci USA. 1997;94:6776–6780. doi: 10.1073/pnas.94.13.6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lux CT, et al. All primitive and definitive hematopoietic progenitor cells emerging before E10 in the mouse embryo are products of the yolk sac. Blood. 2008;111:3435–3438. doi: 10.1182/blood-2007-08-107086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ottersbach K, Dzierzak E. The murine placenta contains hematopoietic stem cells within the vascular labyrinth region. Dev Cell. 2005;8:377–387. doi: 10.1016/j.devcel.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 18.Rhodes KE, et al. The emergence of hematopoietic stem cells is initiated in the placental vasculature in the absence of circulation. Cell Stem Cell. 2008;2:252–263. doi: 10.1016/j.stem.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z, et al. Mouse Embryonic Head as a Site for Hematopoietic Stem Cell Development. Stem Cell. 2012;11:663–675. doi: 10.1016/j.stem.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 20.Nakano H, et al. Haemogenic endocardium contributes to transient definitive haematopoiesis. Nature Communications. 1AD;4:1564–10. doi: 10.1038/ncomms2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.You LR, et al. Suppression of Notch signalling by the COUP-TFII transcription factor regulates vein identity. Nature. 2005;435:98–104. doi: 10.1038/nature03511. [DOI] [PubMed] [Google Scholar]
- 22.Corada M, et al. Sox17 is indispensable for acquisition and maintenance of arterial identity. Nature Communications. 2013;4:2609. doi: 10.1038/ncomms3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawson ND, et al. Notch signaling is required for arterial-venous differentiation during embryonic vascular development. Development. 2001;128:3675–3683. doi: 10.1242/dev.128.19.3675. [DOI] [PubMed] [Google Scholar]
- 24.Kumano K, et al. Notch1 but not Notch2 is essential for generating hematopoietic stem cells from endothelial cells. Immunity. 2003;18:699–711. doi: 10.1016/s1074-7613(03)00117-1. [DOI] [PubMed] [Google Scholar]
- 25.Kim I, Saunders TL, Morrison SJ. Sox17 Dependence Distinguishes the Transcriptional Regulation of Fetal from Adult Hematopoietic Stem Cells. Cell. 2007;130:470–483. doi: 10.1016/j.cell.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clarke RL, et al. The expression of Sox17 identifies and regulates haemogenic endothelium. Nat Cell Biol. 2013 doi: 10.1038/ncb2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen MJ, Yokomizo T, Zeigler BM, Dzierzak E, Speck NA. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature. 2009;457:887–891. doi: 10.1038/nature07619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Pater E, et al. Gata2 is required for HSC generation and survival. Journal of Experimental Medicine. 2013;464:116. doi: 10.1084/jem.20130751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krebs LT, et al. Notch signaling is essential for vascular morphogenesis in mice. Genes & Development. 2000;14:1343–1352. [PMC free article] [PubMed] [Google Scholar]
- 30.Kohyama J, et al. Visualization of spatiotemporal activation of Notch signaling: live monitoring and significance in neural development. Developmental Biology. 2005;286:311–325. doi: 10.1016/j.ydbio.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 31.Sasaki N, Kiso M, Kitagawa M, Saga Y. The repression of Notch signaling occurs via the destabilization of mastermind-like 1 by Mesp2 and is essential for somitogenesis. Development. 2011;138:55–64. doi: 10.1242/dev.055533. [DOI] [PubMed] [Google Scholar]
- 32.Yokomizo T, Dzierzak E. Three-dimensional cartography of hematopoietic clusters in the vasculature of whole mouse embryos. Development. 2010;137:3651–3661. doi: 10.1242/dev.051094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tober J, Yzaguirre AD, Piwarzyk E, Speck NA. Distinct temporal requirements for Runx1 in hematopoietic progenitors and stem cells. Development. 2013 doi: 10.1242/dev.094961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robin C, Ottersbach K, Boisset JC, Oziemlak A, Dzierzak E. CD41 is developmentally regulated and differentially expressed on mouse hematopoietic stem cells. Blood. 2011;117:5088–5091. doi: 10.1182/blood-2011-01-329516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boisset JC, Clapes T, Van Der Linden R, Dzierzak E, Robin C. Integrin IIb (CD41) plays a role in the maintenance of hematopoietic stem cell activity in the mouse embryonic aorta. Biology Open. 2013;2:525–532. doi: 10.1242/bio.20133715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taoudi S, et al. Progressive divergence of definitive haematopoietic stem cells from the endothelial compartment does not depend on contact with the foetal liver. Development. 2005;132:4179–4191. doi: 10.1242/dev.01974. [DOI] [PubMed] [Google Scholar]
- 37.Obi S, et al. Fluid shear stress induces arterial differentiation of endothelial progenitor cells. J Appl Physiol. 2009;106:203–211. doi: 10.1152/japplphysiol.00197.2008. [DOI] [PubMed] [Google Scholar]
- 38.Jahnsen ED, et al. Notch1 is pan-endothelial at the onset of flow and regulated by flow. PLoS ONE. 2015;10:e0122622. doi: 10.1371/journal.pone.0122622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chong DC, Koo Y, Xu K, Fu S, Cleaver O. Stepwise arteriovenous fate acquisition during mammalian vasculogenesis. Dev Dyn. 2011;240:2153–2165. doi: 10.1002/dvdy.22706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang C, et al. Embryonic atrial function is essential for mouse embryogenesis, cardiac morphogenesis and angiogenesis. Development. 2003;130:6111–6119. doi: 10.1242/dev.00831. [DOI] [PubMed] [Google Scholar]
- 41.Nakajima-Takagi Y, et al. Role of SOX17 in hematopoietic development from human embryonic stem cells. Blood. 2013;121:447–458. doi: 10.1182/blood-2012-05-431403. [DOI] [PubMed] [Google Scholar]
- 42.Yoon MJ, et al. Mind bomb-1 is essential for intraembryonic hematopoiesis in the aortic endothelium and the subaortic patches. Molecular and Cellular Biology. 2008;28:4794–4804. doi: 10.1128/MCB.00436-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marcelo KL, et al. Hemogenic Endothelial Cell Specification Requires c-Kit, Notch Signaling, and p27-Mediated Cell-Cycle Control. DEVCEL. 2013;27:504–515. doi: 10.1016/j.devcel.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim AD, et al. Discrete Notch signaling requirements in the specification of hematopoietic stem cells. EMBO J. 2014 doi: 10.15252/embj.201488784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richard C, et al. Endothelio-Mesenchymal Interaction Controls runx1 Expression and Modulates the notch Pathway to Initiate Aortic Hematopoiesis. DEVCEL. 2013;24:600–611. doi: 10.1016/j.devcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wakayama T, Hamada K, Yamamoto M, Suda T, Iseki S. The expression of platelet endothelial cell adhesion molecule-1 in mouse primordial germ cells during their migration and early gonadal formation. Histochem Cell Biol. 2003;119:355–362. doi: 10.1007/s00418-003-0528-1. [DOI] [PubMed] [Google Scholar]
- 47.Wang Y, et al. Ephrin-B2 controls VEGF-induced angiogenesis and lymphangiogenesis. Nature. 2010;465:483–486. doi: 10.1038/nature09002. [DOI] [PubMed] [Google Scholar]
- 48.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2009;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zovein AC, et al. Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell. 2008;3:625–636. doi: 10.1016/j.stem.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Pater E, et al. Gata2 is required for HSC generation and survival. Journal of Experimental Medicine. 2013;210:2843–2850. doi: 10.1084/jem.20130751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.North TE, et al. Runx1 expression marks long-term repopulating hematopoietic stem cells in the midgestation mouse embryo. Immunity. 2002;16:661–672. doi: 10.1016/s1074-7613(02)00296-0. [DOI] [PubMed] [Google Scholar]
- 52.Liu X, et al. Sox17 modulates Wnt3A/ -catenin-mediated transcriptional activation of the Lef-1 promoter. AJP: Lung Cellular and Molecular Physiology. 2010;299:L694–L710. doi: 10.1152/ajplung.00140.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shutter JR, et al. Dll4, a novel Notch ligand expressed in arterial endothelium. Genes & Development. 2000;14:1313–1318. [PMC free article] [PubMed] [Google Scholar]
- 54.Duarte A, et al. Dosage-sensitive requirement for mouse Dll4 in artery development. Genes & Development. 2004;18:2474–2478. doi: 10.1101/gad.1239004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Limbourg FP, et al. Essential role of endothelial Notch1 in angiogenesis. Circulation. 2005;111:1826–1832. doi: 10.1161/01.CIR.0000160870.93058.DD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proc Natl Acad Sci USA. 2003;100:14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lichtinger M, et al. RUNX1 reshapes the epigenetic landscape at the onset of haematopoiesis. EMBO J. 2012:1–16. doi: 10.1038/emboj.2012.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Radtke F, et al. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10:547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- 59.Burns CE, Traver D, Mayhall E, Shepard JL, Zon LI. Hematopoietic stem cell fate is established by the Notch-Runx pathway. Genes & Development. 2005;19:2331–2342. doi: 10.1101/gad.1337005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lawson ND, et al. Notch signaling is required for arterial-venous differentiation during embryonic vascular development. Development. 2001;128:3675–3683. doi: 10.1242/dev.128.19.3675. [DOI] [PubMed] [Google Scholar]
- 61.Ditadi A, et al. Human definitive haemogenic endothelium and arterial vascular endothelium represent distinct lineages. Nat Cell Biol. 2015;17:580–591. doi: 10.1038/ncb3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Burns CE, et al. A genetic screen in zebrafish defines a hierarchical network of pathways required for hematopoietic stem cell emergence. Blood. 2009;113:5776–5782. doi: 10.1182/blood-2008-12-193607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robert-Moreno À, et al. Impaired embryonic haematopoiesis yet normal arterial development in the absence of the Notch ligand Jagged1. EMBO J. 2008;27:1886–1895. doi: 10.1038/emboj.2008.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nobuhisa I, et al. Sox17-mediated maintenance of fetal intra-aortic hematopoietic cell clusters. Mol Cell Biol. 2014;34:1976–1990. doi: 10.1128/MCB.01485-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lagha M, et al. Pax3:Foxc2 Reciprocal Repression in the Somite Modulates Muscular versus Vascular Cell Fate Choice in Multipotent Progenitors. DEVCEL. 2009;17:892–899. doi: 10.1016/j.devcel.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 66.Radtke F, et al. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10:547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- 67.Lucitti JL, et al. Vascular remodeling of the mouse yolk sac requires hemodynamic force. Development. 2007;134:3317–3326. doi: 10.1242/dev.02883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zovein AC, et al. Beta1 integrin establishes endothelial cell polarity and arteriolar lumen formation via a Par3-dependent mechanism. Dev Cell. 2010;18:39–51. doi: 10.1016/j.devcel.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Donohoe ME, Zhang LF, Xu N, Shi Y, Lee JT. Identification of a Ctcf cofactor, Yy1, for the X chromosome binary switch. Molecular Cell. 2007;25:43–56. doi: 10.1016/j.molcel.2006.11.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.