Abstract
The discovery of methods for reprogramming adult somatic cells into induced pluripotent stem cells (iPSCs) has raised the possibility of producing truly personalized treatment options for numerous diseases. Similar to embryonic stem cells (ESCs), iPSCs can give rise to any cell type in the body and they are amenable to genetic correction by homologous recombination. These ESC properties of iPSCs allow for the development of permanent corrective therapies for many currently incurable disorders, including inherited skin diseases, without using embryonic tissues or oocytes. Here, we review recent progress and limitations of iPSC research with a focus on clinical applications of iPSCs, and using iPSCs to model human diseases for drug discovery in the field of dermatology.
INTRODUCTION
Direct reprogramming of somatic cells into induced pluripotent stem cells (iPSCs) through the ectopic expression of reprogramming factors (Takahashi and Yamanaka 2006; Okita et al. 2007; Takahashi et al. 2007; Wernig et al. 2007; Yu et al. 2007) has opened up a new era in research and therapy. Similar to embryonic stem cells (ESCs), iPSCs can be expanded indefinitely and are capable of differentiating into all three germ layers (Takahashi and Yamanaka 2006; Okita et al. 2007; Takahashi et al. 2007; Wernig et al. 2007; Yu et al. 2007). Traditional techniques for the isolation of human ESCs rely on the use of surplus in vitro fertilization embryos (Mitalipova and Palmarini 2006). Therefore, unlike iPSC technology, ESC-based techniques do not allow for the generation of genetically diverse patient-specific cells. Furthermore, the use of ESC-derived cells for therapeutic applications may result in immune rejection, which is not anticipated to be a concern if patient-specific iPSC-derived cells are returned to the same patient. Thus, iPSC technology addresses many obstacles associated with the use of ESCs, including ethical concerns, and allows for the generation of patient-specific pluripotent stem cells, which can be genetically corrected, differentiated into adult lineages, and returned to the same patient as an autograft (Yamanaka 2007; Nishikawa et al. 2008; Yamanaka 2009; Takahashi 2012).
Although iPSCs have tremendous potential for cell-based drug discoveries, cell therapy and disease modeling, extensive analyses are still required to show the safety and reliability of the reprogramming technology. Until recently, progress in this area has been significantly impeded by the lack of efficient protocols for the differentiation of iPSCs into relevant adult lineages/tissues. This was especially apparent in the field of dermatology, which is unfortunate, because the skin may be an ideal tissue to initially apply an iPSC-based therapy. Skin is readily accessible, easy to monitor, and if an adverse event should occur, the affected area could be excised.
Nevertheless, significant advances have recently been achieved in the differentiation of both mouse and human iPSCs into keratinocytes (Bilousova et al. 2011a; Itoh et al. 2011; Bilousova and Roop 2013), melanocytes (Ohta et al. 2011), and fibroblasts (Hewitt et al. 2011); thus, opening the possibility of expanding iPSC technology into the field of dermatology. This article discusses the prospect of using iPSC technology as a tool to study the skin and its pathology and cure genetic skin diseases.
IN SEARCH OF PLURIPOTENCY
The remarkable phenotypic stability and low proliferative capacity of differentiated adult cells limit their applications in personalized regenerative medicine and have triggered an extensive search for sources of pluripotent stem cells suitable for the clinic.
One of the potential sources of pluripotent stem cells is ESCs. In mammals, embryonic development is characterized by a gradual decrease in differentiation potential and an increase in the specialization of cells as they commit to the formation of adult lineages and tissues that constitute the embryo. The developmentally versatile pluripotent ESCs residing in the inner cell mass of the blastocyst (Thomson et al. 1998), exist for a brief period of time during development and eventually differentiate into more specialized multipotent stem cells (Fig. 1). Whereas human pluripotent ESCs still hold great promise in regenerative medicine and drug discoveries, ethical concerns and the possibility of immune rejection of tissues derived from allogeneic ESCs have hindered the therapeutic application of these cells.
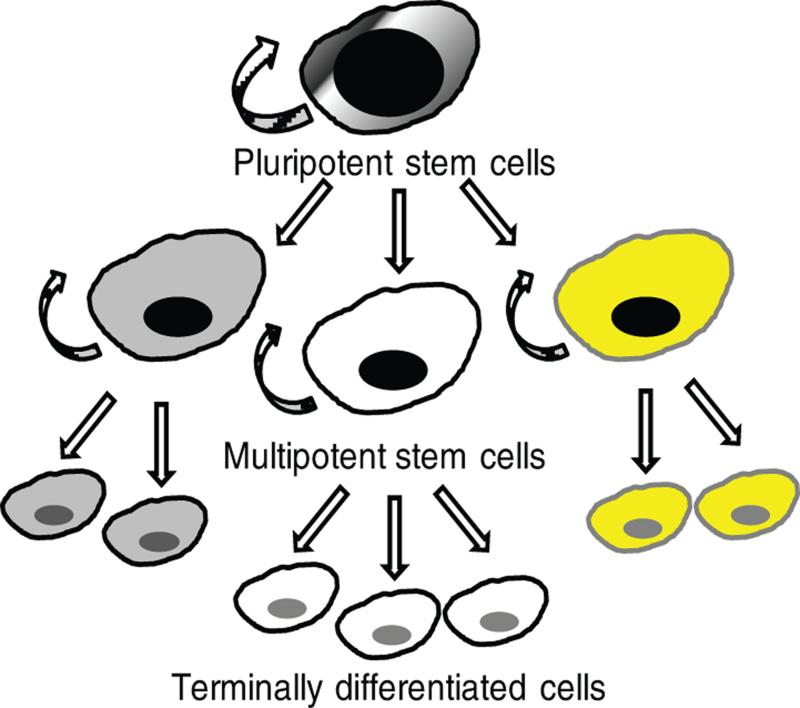
Figure 1. Stem cell hierarchy.
Pluripotent stem cells have the capacity for self-renewal and only exist in an early stage of embryogenesis. They give rise to all types of more specialized multipotent stem cells of the adult organism. Multipotent stem cells also show a self-renewal capacity; however, they are committed to produce only a restricted range of adult somatic cells and terminally differentiated progeny.
Attempts to derive pluripotent stem cells from adult somatic cells were influenced by early nuclear transfer experiments performed in the 1950s using frogs, Rana pipiens (Briggs and King 1952) and Xenopus laevis (Gurdon et al. 1958), as a model system. These early studies documented the feasibility of reprogramming adult frog somatic cell nuclei by the cytoplasm of enucleated unfertilized frog oocytes and generation of cloned frogs. Similar reports of successful nuclear reprogramming, either by transferring somatic cell nuclei into oocytes (Kimura and Yanagimachi 1995; Wakayama et al. 1998) or by fusing somatic cells with pluripotent stem cells (Ambrosi and Rasmussen 2005) were published. However, it was the cloning of Dolly the sheep (Wilmut et al. 1997) that demonstrated the possibility of complete reprogramming of somatic cells from mammals back into the pluripotent state. Following somatic cell nuclear transfer (SCNT), the adult cell’s nucleus is exposed to the cytoplasm of an unfertilized egg and is returned to a pluripotent state from which it can develop an entire animal. Dolly’s birth (Wilmut et al. 1997) and the first report of the derivation of human ESCs shortly thereafter (Thomson et al. 1998) led to wide speculations about the possibility of therapeutic cloning. However, the difficulty of obtaining donated oocytes, which are required to perform SCNT, as well as ethical concerns, limits the applicability of human therapeutic cloning in the clinic.
The success in using a cell from an adult animal to generate another healthy, fertile adult has shown that unfertilized eggs and ESCs contain a set of factors that can confer pluripotency to somatic cells. Encouraged by these findings, Takahashi and Yamanaka (2006) analyzed 24 genes that were specifically expressed in ESCs as candidates for factors that induce pluripotency in murine somatic cells. They identified a combination of four factors that when ectopically expressed was sufficient to revert differentiated mouse somatic cells into embryonic-like cells that were similar to ESCs (Takahashi and Yamanaka 2006). The factors were Oct-4 (also known as POU5F1), sex determining region Y (SRY)-box2 (Sox-2), Kruppel-like factor 4 (Klf-4) and c-Myc (Takahashi and Yamanaka 2006). The iPSCs resulting from this manipulation functioned in a manner undistinguished from mouse ESCs: they were capable of forming multiple cell types in vitro and in vivo, they expressed markers associated with pluripotent cells, and perhaps most importantly, they could be used to make fertile mice (Takahashi and Yamanaka 2006; Okita et al. 2007). A similar combination of factors (Takahashi et al. 2007; Wernig et al. 2007) as well as an alternative combination of Oct-4, Sox-2, Nanog and Lin-28 (Yu et al. 2007) were later shown to be sufficient to reprogram human somatic cells into the pluripotent state (Fig. 2); thus, launching a new era in regenerative medicine and tissue engineering.
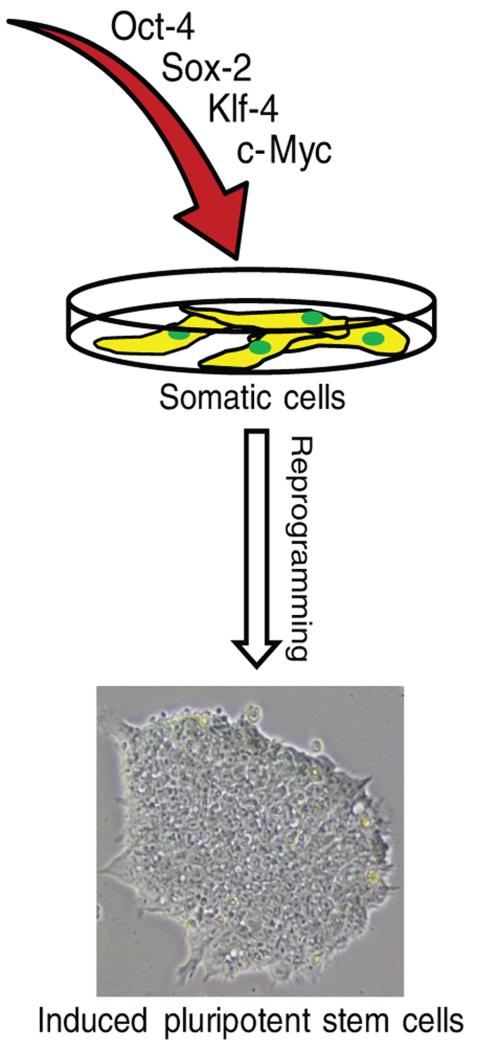
Figure 2. Reprogramming human somatic cells with four transcription factors.
Both mouse and human somatic cells can be reprogrammed into embryonic stem cell-like iPSCs as a result of exogenous expression of four transcription factors: Oct-4, Sox-2, Klf-4 and c-Myc. An alternative combination of Oct-4, Sox-2, Nanog and Lin-28 can also be used to achieve reprogramming of human somatic cells. The resulting iPSCs resemble ESCs at the phenotypic, functional, and molecular levels.
THE GENERATION OF iPSCS
Since the development of the first protocols for the generation of mouse iPSCs, followed by the generation of human iPSCs, a variety of reprogramming protocols have been published, and various combinations of reprogramming factors in the form of transcription factors or microRNAs (miRNAs) have been used. The initial protocols for the generation of both mouse and human iPSCs relied on the use of integrating retro- and lentiviral vectors to deliver the exogenous reprogramming factors into somatic cells (Takahashi and Yamanaka 2006; Takahashi et al. 2007; Yu et al. 2007; Wernig et al. 2008; Sommer et al. 2009; Sommer et al. 2012). However, the observation that the expression of reprogramming factors was only essential in the initial steps of reprogramming and silencing of exogenous reprogramming factors must occur for the successful directed differentiation of iPSCs (Takahashi and Yamanaka 2006; Okita et al. 2007) has opened the possibility for developing genome integration-free approaches without the risk for permanent genetic modifications caused by insertional mutagenesis. A number of genome integration-free techniques for the generation of iPSCs have subsequently been published with a focus on accelerating the transition of iPSC-based technology into the clinic.
Somatic cells have been successfully reprogrammed with expression plasmids encoding the reprogramming factors (Okita et al. 2008) and adenoviruses (Stadtfeld et al. 2008; Zhou and Freed 2009). However, these approaches are not considered clinically safe due to a potential risk of genomic integrations and mutagenesis. Similarly, the use of PiggyBac transposons (Yusa et al. 2009) may be associated with point mutations and chromosomal rearrangements. Recently, iPSCs were generated with a set of miRNAs alone (Anokye-Danso et al. 2011). However, the method for the delivery of reprogramming miRNAs also relies on the use of integrating lentiviral vectors (Anokye-Danso et al. 2011), and the only report of reprogramming with direct transfections of mature miRNAs showed an extremely low efficiency (Miyoshi et al. 2011). More clinically relevant approaches require a DNA-free delivery of reprogramming factors to avoid the potential integration of the exogenous DNA into the genome. Examples of DNA-free reprogramming approaches are shown in Fig. 3.
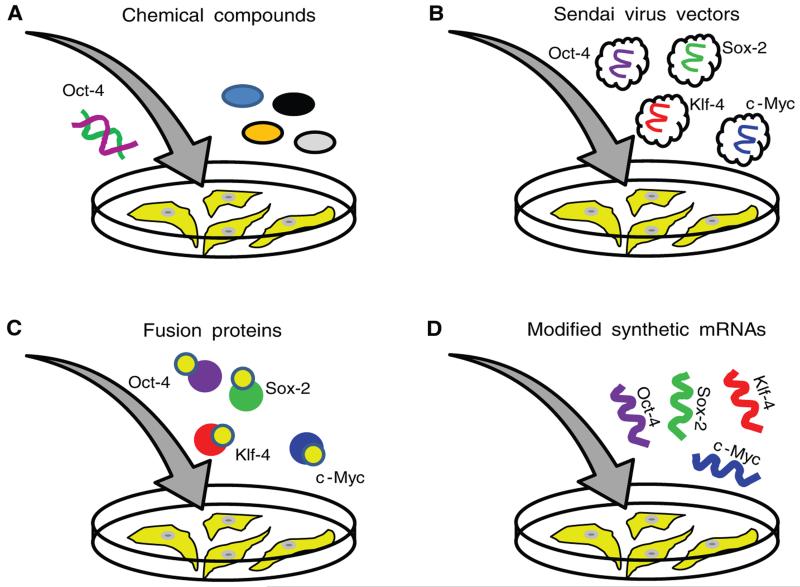
Figure 3. Clinically relevant DNA-free approaches to reprogram human somatic cells into iPSCs.
(A) Chemical compounds that substitute for the expression of ectopic reprogramming factors have been identified and may be used for the reprogramming of human somatic cells. However, exogenous expression of Oct-4 is still required for this method to produce iPSCs. (B) RNA-containing Sendai viruses can be used to deliver reprogramming factors into somatic cells. The Sendai viral genome is not able to integrate into the host genomic DNA, and viral vectors are slowly diluted during the robust cell division of iPSCs. (C) Reprogramming can be achieved with purified recombinant reprogramming proteins fused to either HIV-TAT or polyarginine peptides. Both peptides are cell-penetrating peptides and can deliver large cargo proteins into cells. (D) The direct transfection of modified synthetic messenger RNAs (mRNAs) encoding the reprogramming factors can also be used to reprogram somatic cells.
One potentially safe approach to generate iPSCs is to use small chemical compounds to substitute for the expression of reprogramming factors. Many small molecules that can functionally replace exogenous reprogramming factors and enhance the efficiency of reprogramming have been identified (Zhang et al. 2012). However, the requirement for the exogenous expression of Oct-4 in the human reprogramming protocol that utilizes a cocktail of defined small molecules limits the applicability of this approach in the clinic (Zhu et al. 2010; Zhang et al. 2012). Nonetheless, a recent study by Hou et al (2013) found a set of small-molecule compounds that can reprogram mouse cells into iPSCs without the expression of any exogenous reprogramming factors, suggesting that a complete combination of chemical compounds suitable for the reprogramming of human somatic cells may be identified in the future.
Another possibility for exogenous DNA-free reprogramming utilizes a nonintegrating RNA-containing Sendai virus (Fusaki et al. 2009; Ban et al. 2011). Sendai virus is a negative-strand RNA virus that belongs to the Paramyxoviridae family (Nakanishi and Otsu 2012). It replicates in the cytoplasm of infected cells, does not go through a DNA phase that can integrate into the host genome, and has been successfully used to introduce foreign genes in a wide spectrum of host cells (Nakanishi and Otsu 2012). Sendai viral vectors are currently being applied in gene therapy studies for cystic fibrosis and vaccines (Nakanishi and Otsu 2012) and have been shown to induce reprogramming with high efficiency (Fusaki et al. 2009; Ban et al. 2011). Although Sendai viral vectors are slowly diluted during the robust cell division of iPSCs, more studies are required to confirm the clinical safety of reprogramming with this approach.
Several reports indicate that iPSCs can also be generated without the use of any nucleic acids by fusing reprogramming factors with cell penetrating peptides (Kim et al. 2009; Zhou et al. 2009). The identification of a group of proteins with an enhanced ability to cross the plasma membrane in a receptor-independent manner has led to the discovery of a class of protein domains with cell-penetrating properties. The fusion of these domains with heterologous proteins is sufficient to allow their rapid transduction into different cells in a concentration-dependent manner. The most commonly used cell penetrating peptides are the HIV-1 transcriptional activator TAT protein and polyarginine (Frankel and Pabo 1988; Green and Loewenstein 1988). The fusion of reprogramming factors with TAT or polyarginine protein transduction domains has been shown to be sufficient for reprogramming somatic cells (Kim et al. 2009; Zhou et al. 2009). Although being potentially the safest approach, the low efficiency of iPSC generation with cell-penetrating proteins (0.001%), and technical difficulties associated with protein purification, prevent an easy transition of this methodology into the clinic. A more promising approach for the transgene-free generation of iPSCs may be the use of synthetic modified mRNAs encoding the reprogramming factors. This approach has been shown to reprogram a variety of cell lines with the efficiency that surpasses that of the other integration-free protocols (Warren et al. 2010; Warren et al. 2012). The only disadvantage of mRNA-based reprogramming, however, is that mRNA molecules must be delivered into the cells every day until the reprogramming is complete.
DONOR CELL TYPES FOR REPROGRAMMING
Unlike ESCs, which can be derived only from the developing embryo, iPSCs have been generated from multiple easily accessible cell types, such as dermal fibroblasts, keratinocytes, melanocytes, peripheral blood CD 34+ cells, etc.
Fibroblasts are the most commonly used cell type for reprogramming due to their accessibility and easy culture conditions. As a result, the reprogramming process has been extensively studied in these cells and has been shown to follow an organized sequence of events, which begins with the down-regulation of somatic gene expression (Polo et al. 2012). The first step of reprogramming in fibroblasts requires a mesenchymal-to-epithelial transition (MET) (Esteban et al. 2012), which is followed by the activation of the early pluripotency markers SSEA-1 and alkaline phosphatase before pluripotency genes Nanog and Oct-4 become involved and provide an independence from exogenous factor expression (Polo et al. 2012). The requirement for MET as a first step in reprogramming of fibroblasts may partly explain why cells of epithelial origin, such as keratinocytes, are easier to reprogram (Aasen et al. 2008; Gadue and Cotsarelis 2008). Although the exact mechanism of reprogramming remains unknown, the removal of epigenetic modifications, which restrict expression of pluripotency genes in somatic cells, seems to play a predominant role in the process (Polo et al. 2012; Watanabe et al. 2013).
Somatic cells exhibit different reprogramming capacity depending on their origin, rate of proliferation and gene expression profiles; and selecting the appropriate donor line for reprogramming may be critical not only to achieve the highest efficiency and quality of reprogramming, but also to attain maximum differentiation capacities of the resulting iPSCs. For instance, epigenetic profiling of iPSCs revealed that the reprogrammed cells retain epigenetic marks of the cell type of origin (Kim et al. 2010; Kim et al. 2011), although these marks disappear upon continued passaging (Polo et al. 2010). This short-term genetic memory may provide an advantage in achieving the higher differentiation efficiency of early-passage iPSCs into a target adult lineage/tissue, which might be particularly relevant to iPSCs derived from keratinocytes or melanocytes. Not only can keratinocytes and melanocytes be reprogrammed with a higher efficiency than fibroblasts (Aasen et al. 2008; Utikal et al. 2009), the residual epigenetic memory in keratinocyte- and melanocyte-derived iPSCs may allow for easier differentiation back into the corresponding original cell type for cell replacement. One drawback in using skin cells such as keratinocytes and melanocytes for reprogramming is the potential accumulation of DNA mutations due to continuous exposure of the skin to UV light. However, this problem can be avoided if biopsies are taken from nonsun exposed sites such as armpits.
APPLICATION OF iPSCS IN DERMATOLOGY
Essentially all of the most devastating forms of inherited skin diseases such as the epidermolysis bullosa (EB) subtypes and congenital ichthyoses (Khavari 1997; DiGiovanna and Robinson-Bostom 2003; Smith 2003) are caused by known monogenic defects, which, in theory, should enable their correction at the genetic level. The correction achieved in iPSCs derived from a patient with a skin disease may be followed by the differentiation of these corrected iPSCs into autologous skin cells for transplantation. In addition to genetic skin disorders, iPSC technology may potentially be applicable to wound healing, depigmentation disorders, and cosmetic dermatology.
The successful development of iPSC-based therapies for inherited skin disorders depends on four important steps (Fig.4). First, cells need to be isolated from a patient’s skin biopsy. Second, these cells have to be reprogrammed into iPSCs. Third, the genetic defects in generated iPSCs need to be corrected by safe approaches, preferentially through homologous recombination (HR). Fourth, these genetically corrected patient-specific pluripotent cells have to be differentiated into the cell type relevant to their disease, followed by transplantation onto the same patient as an autograft. Uncorrected iPSCs may also be utilized as a source of disease-relevant patient-specific cells for in vitro disease modeling and in vivo xenograft modeling, which would offer platforms for yielding new insights into disease mechanisms and drug discovery.
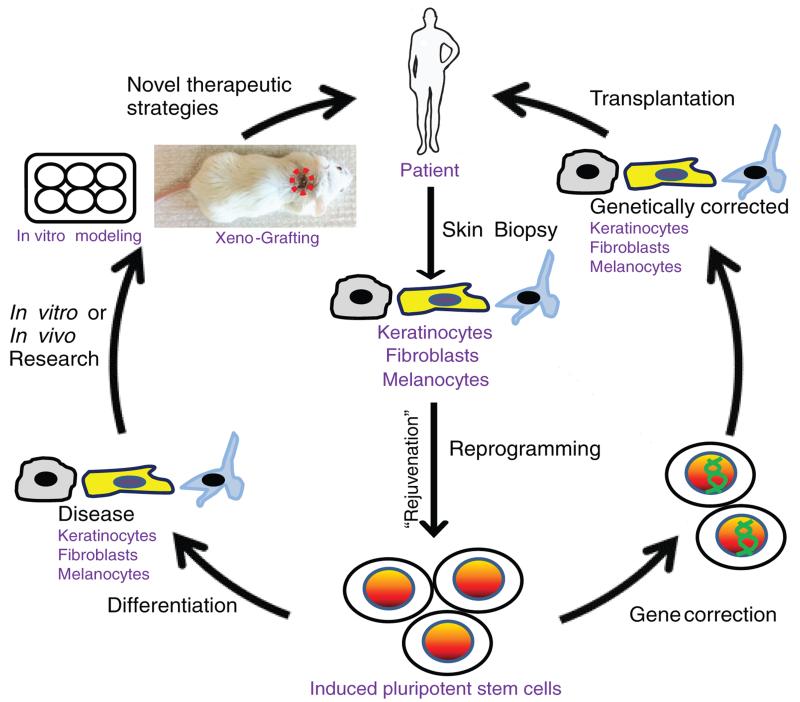
Figure 4. Dermatological applications of iPSCs.
The generation of iPSCs allows for the development of stem cell replacement strategies to treat genetic skin disorders and modeling these diseases with the goal to producing novel therapeutic options for patients. Initially, cells need to be isolated from a skin biopsy of a patient with a genetic skin disease. Suitable cell types for reprogramming are keratinocytes, melanocytes and fibroblasts. During reprogramming, cells acquire ESC properties and undergo “rejuvenation”, as can be determined by the elongation of telomeres and the restoration of the mitochondrial function. The resulting iPSCs can then be genetically corrected by HR. The corrected iPSC clones can be differentiated into relevant cell types for transplantation and returned back to the patient as an autograft. Alternatively, uncorrected iPSCs can be differentiated into cell types relevant to the disease of interest for modeling the disease in organotypic cultures or for recapitulating the disease phenotype in vivo by transplanting onto immunodeficient mice as a xenograft.
Reprogramming Cells from Patients with Inherited Skin Diseases
The generation of iPSC lines from patients with genetic disorders provides an unlimited supply of cells for studies and an opportunity to repair gene defects in vitro, as long as iPSCs can be differentiated into relevant cell types. To date, human iPSCs have been generated for several genetic skin disorders, including type VII collagen (Col7)-deficient Recessive Dystrophic EB (RDEB) (Itoh et al. 2011; Tolar et al. 2011), LAMB3 gene-deficient Junctional EB (JEB) (Tolar et al. 2013), p63 mutant Ectrodactyly, Ectodermal dysplasia, and Cleft lip/palate (EEC) syndrome (Shalom-Feuerstein et al. 2013), Epidermolytic Hyperkeratosis (EHK) with a dominant N188S keratin 1 mutation (Bilousova et al. 2011b), and EB simplex (EBS) with a dominant R125C keratin 14 mutation (Bilousova et al. 2012). In addition, iPSCs have been obtained from patients with Dyskeratosis Congenita (DC), a disease of telomere maintenance that manifests itself in multiple organ failures, cancer, and age-associated skin phenotypes such as dyskeratotic nails, delayed wound healing, and hair loss (Agarwal et al. 2010; Agarwal and Daley 2011; Batista et al. 2011).
Attempts have also been made to correct the defects in iPSCs generated from skin disorders. In the study by Tolar et al.(2013), a LAMB3 encoding lentivirus was used to correct keratinocytes derived from JEB-iPSCs (Tolar et al. 2013). In another study by Tolar et al (2011), transient gene correction of COL7A1 gene-deficient RDEB-iPSCs was achieved via the transfection with an expression plasmid encoding the wild-type human COL7 gene (Tolar et al. 2011). The “corrected” RDEB-iPSCs were differentiated into structures resembling skin and cells of a hematopoietic lineage through teratoma formation (Tolar et al. 2011). Wild-type, congenic bone marrow residing mesenchymal cells have been previously shown to engraft into the skin, regenerate injured epithelia and restore Col7 production in blistered areas of RDEB patients (Tolar et al. 2009) and in mouse models (Fujita et al. 2010; Tamai et al. 2011). Thus, corrected iPSCs may provide an unlimited source of autologous cells of both epidermal and mesenchymal lineages for the treatment of RDEB and potentially other skin blistering diseases.
Differentiation of iPSCs into Skin Cells
The ability to differentiate iPSCs into relevant cell types is an important prerequisite for the successful development of iPSC-based treatment strategies. Keratinocytes, melanocytes and fibroblasts have been recently obtained from iPSCs. Mouse and human iPSCs have been differentiated into keratinocytes by sequential applications of retinoic acid and bone morphogenetic protein-4 (BMP4) (Bilousova et al. 2011a; Itoh et al. 2011; Bilousova and Roop 2013). Mouse iPSC-derived keratinocytes were shown to form an epidermis and skin appendages when grafted with mouse fibroblasts onto athymic nude mice (Bilousova et al. 2011a), whereas human iPSC-derived keratinocytes were able to establish a functional organotypic skin in culture (Itoh et al. 2011). Keratinocytes were also generated from RDEB-iPSCs (Itoh et al. 2011), JEB-iPSCs (Tolar et al. 2013), EEC-iPSCs (Shalom-Feuerstein et al. 2013), and EBS-iPSCs (Bilousova et al. 2012) and were shown to recapitulate the phenotypes of corresponding diseases when grown in vitro. Melanocytes can be derived from iPSCs by supplementing the culture with Wnt3a, Stem Cell factor, and Endothelin-3 (Ohta et al. 2011), and the treatment of iPSC cultures with EGF and BMP4 can produce fibroblasts (Hewitt et al. 2011). Although protocols for the differentiation of iPSCs into the main skin cell types now exist, rigorous studies have not been performed to determine whether these iPSC-derived cells are identical to their normal skin equivalents.
Customized Gene Correction Therapy
Current technologies for gene transfer into somatic skin cells suffer from many limitations. One of the biggest obstacles is the availability of safe vectors that are able to produce long lasting therapeutic effects with low risk of insertional mutagenesis. Plasmid- and adenoviral vector-based approaches for gene delivery are inefficient in inducing a long-term corrective effect in keratinocytes and are associated with biosafety concerns (Khavari 2000; Ortiz-Urda et al. 2002). Thus, a genetic correction therapy via HR may be the only permanent cure for inherited skin diseases, especially for those involving recessive loss-of-function mutations, as occurs in JEB and RDEB. Because the epidermis is continuously renewed by adult stem cells residing in the proliferative basal layer, any permanent genetic correction must target this stem cell population. However, to date, no one has reported the successful use of HR technology to correct a defective gene in human keratinocyte stem cells. The only successful clinical trial for permanent gene correction of a blistering disease, in this case, JEB, was performed by Mavilio et al (2006) and utilized a retroviral vector to restore expression of laminin 322. That trial was placed on hold because of safety concerns with the use of retroviral vectors (De Luca et al. 2009).
There are several approaches to induce HR that can be coupled with the generation of iPSCs to achieve genetic repair of inherited skin diseases. Enhanced HR in iPSCs could be achieved using chimeric molecules composed of a nuclease domain and separate, customized DNA-recognition domains. These chimeric molecules can introduce a double-strand break (DSBs) in a specific DNA sequence. The presence of exogenous donor DNA carrying the correct gene sequence and the homology to the sequence flanking the DSB triggers HR and replacement of the defective gene with the corrected one (Carlson et al. 2012). Zinc-finger nucleases (ZFNs) (Kim et al. 1996; Bibikova et al. 2003; Porteus and Baltimore 2003) and transcription activator-like effector nucleases (TALENs) (Boch et al. 2009; Moscou and Bogdanove 2009; Christian et al. 2010) are the main representatives of chimeric endonucleases capable of introducing specific DSBs into genomic DNA and are widely used as tools for research and gene therapy. In addition to ZFNs and TALENs, a novel nuclease-based technique has recently been adapted to achieve genomic editing in human iPSCs. This technique uses the type II prokaryotic CRISPR (clustered regularly interspaced short palindromic repeats)/CRISPR-associated (Cas) systems, which when directed by short RNAs can induce precise cleavage at endogenous genomic loci with high efficiency (Jinek et al. 2013; Mali et al. 2013). ZFN- , TALEN- and CRISPR/Cas -based gene modification strategies have been successfully used on human and mouse ESCs and iPSCs (Hockemeyer et al. 2009; Hockemeyer et al. 2011; Mali et al. 2013). In addition, a recent study by Osborn et al (2013) shows the potential for using chimeric endonucleases in personalized genome-editing strategies for the treatment of inherited skin disorders (Osborn et al. 2013). Using TALENs, the authors achieved a specific heterozygous correction of COL7A1 gene mutation in primary fibroblasts isolated from a RDEB patient. These corrected fibroblasts were then used to generate iPSCs suitable for the differentiation into corrected autologous keratinocytes (Osborn et al. 2013). The proof-of-principal research by Osborn et al (2013) provides an important set of data that warrants further studies on the applicability of nuclease-based genome-editing approaches for the correction of inherited skin diseases.
The Potential of Cellular Reprogramming for Tissue Rejuvenation
The generation of iPSCs coupled with gene targeting may solve many obstacles that are associated with gene correction in somatic cells. Unlike somatic cells, iPSCs can be expanded indefinitely, allowing for the easier selection and expansion of corrected clones. In addition, the process of reprogramming leads to the “rejuvenation” of fully competent cells, as shown by the down-regulation of senescent pathways, elongation of telomeres, and restoration of the mitochondrial function (Marion et al. 2009; Suhr et al. 2009; Prigione et al. 2010; Suhr et al. 2010; Lapasset et al. 2011). Reports indicate that iPSCs can be derived from very old patients (Dimos et al. 2008; Lapasset et al. 2011), and these iPSCs can be differentiated into fibroblasts that show a “rejuvenated” phenotype with respect to telomere length and the mitochondrial function (Lapasset et al. 2011). Reprogramming also restores telomere length in cells derived from patients with autosomal dominant DC, caused by heterozygous mutations in the catalytic protein component of telomerase TERT (Agarwal et al. 2010), promising novel therapeutic benefits for these patients. Thus, reprogramming may hold a key toward tissue rejuvenation and provide insights into mechanisms underlying aging and longevity. Although it is now clear that cells from older patients can be reprogrammed, more studies will be needed to better understand how age impacts iPSC quality and how the “rejuvenating” properties of reprogramming can be employed for reversing skin aging and improving outcomes of stem-cell based therapies for skin disorders. As a particular example, the generation of iPSCs from a patient with a genetic skin disorder may potentially provide a source of “rejuvenated” adult skin stem cells that are most likely exhausted as a result of unsuccessful attempts to repair blistered tissues. The exhaustion of keratinocyte stem cells was especially apparent in the reported case of using genetically corrected keratinocyte stem cells for the treatment of a patient with JEB (Mavilio et al. 2006). Also, the “rejuvenating” properties of reprogramming may potentially be applicable for the treatment of chronic wounds in the elderly, providing autologous, young-like skin cells for transplantation.
Revertant Mosaicism as Natural Gene Therapy
Although ZFNs, TALENs and CRISPR/Cas systems can dramatically increase the efficiency of genetic correction in iPSCs, extensive genetic analysis will be required to eliminate the concern of off-target events that may occur if iPSCs are genetically corrected with these approaches. Naturally occurring somatic reversions may offer an alternative source of genetically corrected cells suitable for transplantation.
“Natural” somatic reversions of inherited mutations are observed in many human genetic diseases, such as Fanconi anemia (Kalb et al. 2007), Bloom syndrome (Ellis et al. 2001), Wiskott-Aldrich syndrome (Stewart et al. 2007), as well as inherited skin disorders (reviewed in (Lai-Cheong et al. 2011; Pasmooij et al. 2012)). There are a number of reports of revertant mosaicism in patients with inherited skin disorders including RDEB (Jonkman et al. 1997), JEB (Pasmooij et al. 2007), Epidermolysis Bullosa Simplex (Schuilenga-Hut et al. 2002), and ichthyosis with confetti (Choate et al. 2010). Revertant mosaicism manifests as patches of normal skin in patients with genetic skin disorders. These patches represent a clonal outgrowth of cells with acquired secondary mutations that negate the effects of the primary mutation and reverse the phenotype of the disease. Several mechanisms are responsible for the second “correcting” gene event in revertant cells and include gene conversion, second-site mutation, intragenic crossovers, etc (Lai-Cheong et al. 2011; Pasmooij et al. 2012). Thus, the generation of iPSCs from spontaneously revertant skin cells in diseases such as RDEB or JEB may provide an unlimited supply of naturally corrected cells for a cell replacement therapy.
iPSCs for Disease Modeling
Although skin cells can be easily isolated from a patient’s biopsy, expanded and used for modeling inherited skin disorders in organotypic cultures, their short lifespan limits their applicability for in vitro studies. Immortalization of somatic cell lines relies on the constitutive expression of oncogenes, which may dramatically influence the phenotype of cell types of interest. The iPSC technology provides a renewable source of patient-specific cells, which, upon subsequent differentiation into relevant cell types, can be used to generate patient-specific organotypic or in vivo xenograft models (Fig.4). The skin cells derived from disease-specific iPSCs maintain their phenotype in 3D cultures, as shown by the differentiation of RDEB-iPSCs into keratinocytes (Itoh et al. 2011), and may be invaluable for research on many aspects of skin biology and inherited skin diseases. The ability to genetically manipulate iPSCs will also allow the modeling of skin diseases by targeted mutagenesis of the relevant genes without the use of ESCs. This may be of particular importance for modeling rare inherited skin disorders such as RDEB, EBS, and JEB. A low incidence of these diseases worldwide limits the accrual of data for research and impedes progress in developing therapeutic strategies for these skin conditions.
CHALLENGES IN iPSC RESEARCH
Although considerable progress has been made in deriving iPSCs from patients, and differentiating them into tissues of interest, the use of iPSCs as a platform for studying diseases and the development of therapies is just emerging. There are several challenges that must be addressed before the transition of iPSCs into the clinic. They include the safety of methodologies for the generation, genetic correction, and differentiation of iPSCs, and the high cost associated with the repair of genetic defects in patient-derived iPSCs and subsequent transplantation of corrected iPSC-derived cells back into the patient.
The first major challenge is in the reprogramming method itself. Although genome integration-free approaches are widely used for the generation of iPSCs, the safety and reliability of these methods are currently under investigation.
Second, iPSCs, like ESCs, are predisposed to teratoma formation when they are not completely differentiated into somatic cells. Although efficient differentiation protocols have been developed, currently available cell purification technologies may not completely separate out the differentiated cells from undifferentiated iPSCs. This may pose a risk of transplanting undifferentiated or partially differentiated iPSCs into the patient. In addition, several recent genomic and epigenetic analyses have suggested that genomic abnormalities such as the accumulation of mutations and aberrant DNA methylation of certain single bases occur in iPSCs, either by reprogramming itself or by subsequent culture conditions (Mayshar et al. 2010; Gore et al. 2011). However, the observed genomic abnormalities and accumulation of mutations may be caused by the method used for the generation of iPSCs, rather than reprogramming itself. Indeed, a recent study indicates that iPSCs generated by nonintegrating approaches show a significantly lower number of somatic mutations than the cells generated with integrating approaches (Cheng et al. 2012). Therefore, more extensive and thorough genomic and epigenetic studies must be performed before applying iPSCs in the clinic.
Although using a patient’s own cells to generate iPSC-derived cells is thought to eliminate the concern with immune rejection, one report indicates that iPSCs may be immunogenic (Zhao et al. 2011). This study assessed the immunogenicity of iPSC-derived teratomas formed by subcutaneous injection of undifferentiated mouse iPSCs. Given that teratomas are a type of tumor, their rejection by the recipient is more likely an indication of tumor immunity and may not be relevant to immune responses triggered by somatic cell transplantation. Indeed, a recent publication indicates that terminally differentiated cells derived from mouse iPSCs do not trigger an immune response in syngeneic recipients (Araki et al. 2013), suggesting that iPSC-derived cells might be well tolerated by the immune system. Nevertheless, further studies will need to be performed to rule out any possibilities of an iPSC-mediated immune response in patients. Another aspect of immune rejection with iPSC-based therapy is related to gene correction. This may be of a particular importance for the genetic correction of skin diseases with homozygous null mutations of relevant genes such as RDEB. Reintroduction of a protein unfamiliar to the host may trigger an immune response and eventual rejection of corrected iPSC-derived cells. Prescreening for patients with compound heterozygous mutations or the expression of truncated, nonfunctional forms of the protein of interest may partially solve the issue of immune rejection on transplantation of corrected cells.
CONCLUSION
Because of its easy accessibility, the skin is an attractive organ to test novel concepts of regenerative medicine, and iPSC-based therapeutic strategies for the treatment of inherited skin diseases are a great example. The potential of iPSCs for generating platforms to better understand disease mechanisms, drug screening and ultimately for iPSC-based therapeutic approaches is enormous and offers the possibility for truly personalized medicine. iPSCs would not only eliminate the need for generating ESCs from fertilized human embryos, but also avoid the complication of immune responses inevitable during the transplantation of allogeneic cells. Although still in its youth, the iPSC field is gaining momentum and it holds great promise for changing the way we treat patients with inherited skin diseases and reversing age-associated conditions that affect the skin. It is no surprise that 2012 Nobel Prize in Medicine was co-awarded to Sir John Gurdon for his pioneering discovery that the cytoplasm of frog oocytes contained factors that could reprogram the nucleus of adult somatic cells to generate viable frogs, and to Shinya Yamanaka for discovering the actual identity of these reprogramming factors, which opened the door for using iPSC-based technology for the treatment of a variety of human diseases including skin diseases.
ACKNOWLEDGMENTS
We are grateful for funding support from the National Institutes of Health (R01AR059947 and P30 AR057212), the US Department of Defense (PR110793), the Foundation for Ichthyosis & Related Skin Types (F.I.R.S.T.) and Dystrophic Epidermolysis Bullosa Research Association (DEBRA) International. We apologize to authors whose work was not cited due to space constrains.
REFERENCES
- Aasen T, Raya A, Barrero MJ, Garreta E, Consiglio A, Gonzalez F, Vassena R, Bilic J, Pekarik V, Tiscornia G, et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nature biotechnology. 2008;26:1276–1284. doi: 10.1038/nbt.1503. [DOI] [PubMed] [Google Scholar]
- Agarwal S, Daley GQ. Telomere dynamics in dyskeratosis congenita: the long and the short of iPS. Cell research. 2011;21:1157–1160. doi: 10.1038/cr.2011.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal S, Loh YH, McLoughlin EM, Huang J, Park IH, Miller JD, Huo H, Okuka M, Dos Reis RM, Loewer S, et al. Telomere elongation in induced pluripotent stem cells from dyskeratosis congenita patients. Nature. 2010;464:292–296. doi: 10.1038/nature08792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosi DJ, Rasmussen TP. Reprogramming mediated by stem cell fusion. Journal of cellular and molecular medicine. 2005;9:320–330. doi: 10.1111/j.1582-4934.2005.tb00358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, Zhang Y, Yang W, Gruber PJ, Epstein JA, et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell stem cell. 2011;8:376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki R, Uda M, Hoki Y, Sunayama M, Nakamura M, Ando S, Sugiura M, Ideno H, Shimada A, Nifuji A, et al. Negligible immunogenicity of terminally differentiated cells derived from induced pluripotent or embryonic stem cells. Nature. 2013;494:100–104. doi: 10.1038/nature11807. [DOI] [PubMed] [Google Scholar]
- Ban H, Nishishita N, Fusaki N, Tabata T, Saeki K, Shikamura M, Takada N, Inoue M, Hasegawa M, Kawamata S, et al. Efficient generation of transgene-free human induced pluripotent stem cells (iPSCs) by temperature-sensitive Sendai virus vectors. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:14234–14239. doi: 10.1073/pnas.1103509108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista LF, Pech MF, Zhong FL, Nguyen HN, Xie KT, Zaug AJ, Crary SM, Choi J, Sebastiano V, Cherry A, et al. Telomere shortening and loss of self-renewal in dyskeratosis congenita induced pluripotent stem cells. Nature. 2011;474:399–402. doi: 10.1038/nature10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300:764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- Bilousova G, Chen J, Roop DR. Differentiation of mouse induced pluripotent stem cells into a multipotent keratinocyte lineage. The Journal of investigative dermatology. 2011a;131:857–864. doi: 10.1038/jid.2010.364. [DOI] [PubMed] [Google Scholar]
- Bilousova G, Chen J, Roop DR. Exploring the therapeutic potential of induced pluripotent stem cells for the treatment of epidermolytic hyperkeratosis and epidermolysis bullosa simplex. The Journal of investigative dermatology. 2011b;131:s70. doi: 10.1038/jid.2010.364. abstract. [DOI] [PubMed] [Google Scholar]
- Bilousova G, Chen J, Yasuda M, Roop DR. Developing a stem cell-based therapy for epidermolysis bullosa simplex by promoting an allele-specific knockout of mutant keratin 14 in induced pluripotent stem cells. The Journal of investigative dermatology. 2012;132:s74. abstract. [Google Scholar]
- Bilousova G, Roop DR. Generation of functional multipotent keratinocytes from mouse induced pluripotent stem cells. Methods Mol Biol. 2013;961:337–350. doi: 10.1007/978-1-62703-227-8_22. [DOI] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- Briggs R, King TJ. Transplantation of Living Nuclei From Blastula Cells into Enucleated Frogs’ Eggs. Proceedings of the National Academy of Sciences of the United States of America. 1952;38:455–463. doi: 10.1073/pnas.38.5.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson DF, Fahrenkrug SC, Hackett PB. Targeting DNA With Fingers and TALENs. Molecular therapy Nucleic acids. 2012;1:e3. doi: 10.1038/mtna.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Hansen NF, Zhao L, Du Y, Zou C, Donovan FX, Chou BK, Zhou G, Li S, Dowey SN, et al. Low incidence of DNA sequence variation in human induced pluripotent stem cells generated by nonintegrating plasmid expression. Cell stem cell. 2012;10:337–344. doi: 10.1016/j.stem.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choate KA, Lu Y, Zhou J, Choi M, Elias PM, Farhi A, Nelson-Williams C, Crumrine D, Williams ML, Nopper AJ, et al. Mitotic recombination in patients with ichthyosis causes reversion of dominant mutations in KRT10. Science. 2010;330:94–97. doi: 10.1126/science.1192280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca M, Pellegrini G, Mavilio F. Gene therapy of inherited skin adhesion disorders: a critical overview. The British journal of dermatology. 2009;161:19–24. doi: 10.1111/j.1365-2133.2009.09243.x. [DOI] [PubMed] [Google Scholar]
- DiGiovanna JJ, Robinson-Bostom L. Ichthyosis: etiology, diagnosis, and management. American journal of clinical dermatology. 2003;4:81–95. doi: 10.2165/00128071-200304020-00002. [DOI] [PubMed] [Google Scholar]
- Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W, Croft GF, Saphier G, Leibel R, Goland R, et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- Ellis NA, Ciocci S, German J. Back mutation can produce phenotype reversion in Bloom syndrome somatic cells. Human genetics. 2001;108:167–173. doi: 10.1007/s004390000447. [DOI] [PubMed] [Google Scholar]
- Esteban MA, Bao X, Zhuang Q, Zhou T, Qin B, Pei D. The mesenchymal-to-epithelial transition in somatic cell reprogramming. Current opinion in genetics & development. 2012;22:423–428. doi: 10.1016/j.gde.2012.09.004. [DOI] [PubMed] [Google Scholar]
- Frankel AD, Pabo CO. Cellular uptake of the tat protein from human immunodeficiency virus. Cell. 1988;55:1189–1193. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- Fujita Y, Abe R, Inokuma D, Sasaki M, Hoshina D, Natsuga K, Nishie W, McMillan JR, Nakamura H, Shimizu T, et al. Bone marrow transplantation restores epidermal basement membrane protein expression and rescues epidermolysis bullosa model mice. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14345–14350. doi: 10.1073/pnas.1000044107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proceedings of the Japan Academy Series B, Physical and biological sciences. 2009;85:348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadue P, Cotsarelis G. Epidermal cells rev up reprogramming. Nature biotechnology. 2008;26:1243–1244. doi: 10.1038/nbt1108-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore A, Li Z, Fung HL, Young JE, Agarwal S, Antosiewicz-Bourget J, Canto I, Giorgetti A, Israel MA, Kiskinis E, et al. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M, Loewenstein PM. Autonomous functional domains of chemically synthesized human immunodeficiency virus tat trans-activator protein. Cell. 1988;55:1179–1188. doi: 10.1016/0092-8674(88)90262-0. [DOI] [PubMed] [Google Scholar]
- Gurdon JB, Elsdale TR, Fischberg M. Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature. 1958;182:64–65. doi: 10.1038/182064a0. [DOI] [PubMed] [Google Scholar]
- Hewitt KJ, Shamis Y, Hayman RB, Margvelashvili M, Dong S, Carlson MW, Garlick JA. Epigenetic and phenotypic profile of fibroblasts derived from induced pluripotent stem cells. PloS one. 2011;6:e17128. doi: 10.1371/journal.pone.0017128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nature biotechnology. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nature biotechnology. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M, Kiuru M, Cairo MS, Christiano AM. Generation of keratinocytes from normal and recessive dystrophic epidermolysis bullosa-induced pluripotent stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:8797–8802. doi: 10.1073/pnas.1100332108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkman MF, Scheffer H, Stulp R, Pas HH, Nijenhuis M, Heeres K, Owaribe K, Pulkkinen L, Uitto J. Revertant mosaicism in epidermolysis bullosa caused by mitotic gene conversion. Cell. 1997;88:543–551. doi: 10.1016/s0092-8674(00)81894-2. [DOI] [PubMed] [Google Scholar]
- Kalb R, Neveling K, Hoehn H, Schneider H, Linka Y, Batish SD, Hunt C, Berwick M, Callen E, Surralles J, et al. Hypomorphic mutations in the gene encoding a key Fanconi anemia protein, FANCD2, sustain a significant group of FA-D2 patients with severe phenotype. American journal of human genetics. 2007;80:895–910. doi: 10.1086/517616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khavari PA. Therapeutic gene delivery to the skin. Molecular medicine today. 1997;3:533–538. doi: 10.1016/S1357-4310(97)01143-X. [DOI] [PubMed] [Google Scholar]
- Khavari PA. Genetic correction of inherited epidermal disorders. Human gene therapy. 2000;11:2277–2282. doi: 10.1089/104303400750035816. [DOI] [PubMed] [Google Scholar]
- Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, Ko S, Yang E, Cha KY, Lanza R, et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell stem cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Zhao R, Doi A, Ng K, Unternaehrer J, Cahan P, Huo H, Loh YH, Aryee MJ, Lensch MW, et al. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nature biotechnology. 2011;29:1117–1119. doi: 10.1038/nbt.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Yanagimachi R. Intracytoplasmic sperm injection in the mouse. Biology of reproduction. 1995;52:709–720. doi: 10.1095/biolreprod52.4.709. [DOI] [PubMed] [Google Scholar]
- Lai-Cheong JE, McGrath JA, Uitto J. Revertant mosaicism in skin: natural gene therapy. Trends in molecular medicine. 2011;17:140–148. doi: 10.1016/j.molmed.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapasset L, Milhavet O, Prieur A, Besnard E, Babled A, Ait-Hamou N, Leschik J, Pellestor F, Ramirez JM, De Vos J, et al. Rejuvenating senescent and centenarian human cells by reprogramming through the pluripotent state. Genes Dev. 2011;25:2248–2253. doi: 10.1101/gad.173922.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion RM, Strati K, Li H, Tejera A, Schoeftner S, Ortega S, Serrano M, Blasco MA. Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell stem cell. 2009;4:141–154. doi: 10.1016/j.stem.2008.12.010. [DOI] [PubMed] [Google Scholar]
- Mavilio F, Pellegrini G, Ferrari S, Di Nunzio F, Di Iorio E, Recchia A, Maruggi G, Ferrari G, Provasi E, Bonini C, et al. Correction of junctional epidermolysis bullosa by transplantation of genetically modified epidermal stem cells. Nature medicine. 2006;12:1397–1402. doi: 10.1038/nm1504. [DOI] [PubMed] [Google Scholar]
- Mayshar Y, Ben-David U, Lavon N, Biancotti JC, Yakir B, Clark AT, Plath K, Lowry WE, Benvenisty N. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell stem cell. 2010;7:521–531. doi: 10.1016/j.stem.2010.07.017. [DOI] [PubMed] [Google Scholar]
- Mitalipova M, Palmarini G. Isolation and characterization of human embryonic stem cells. Methods Mol Biol. 2006;331:55–76. doi: 10.1385/1-59745-046-4:55. [DOI] [PubMed] [Google Scholar]
- Miyoshi N, Ishii H, Nagano H, Haraguchi N, Dewi DL, Kano Y, Nishikawa S, Tanemura M, Mimori K, Tanaka F, et al. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell stem cell. 2011;8:633–638. doi: 10.1016/j.stem.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- Nakanishi M, Otsu M. Development of Sendai virus vectors and their potential applications in gene therapy and regenerative medicine. Current gene therapy. 2012;12:410–416. doi: 10.2174/156652312802762518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa S, Goldstein RA, Nierras CR. The promise of human induced pluripotent stem cells for research and therapy. Nature reviews Molecular cell biology. 2008;9:725–729. doi: 10.1038/nrm2466. [DOI] [PubMed] [Google Scholar]
- Ohta S, Imaizumi Y, Okada Y, Akamatsu W, Kuwahara R, Ohyama M, Amagai M, Matsuzaki Y, Yamanaka S, Okano H, et al. Generation of human melanocytes from induced pluripotent stem cells. PloS one. 2011;6:e16182. doi: 10.1371/journal.pone.0016182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- Ortiz-Urda S, Thyagarajan B, Keene DR, Lin Q, Fang M, Calos MP, Khavari PA. Stable nonviral genetic correction of inherited human skin disease. Nature medicine. 2002;8:1166–1170. doi: 10.1038/nm766. [DOI] [PubMed] [Google Scholar]
- Osborn MJ, Starker CG, McElroy AN, Webber BR, Riddle MJ, Xia L, DeFeo AP, Gabriel R, Schmidt M, von Kalle C, et al. TALEN-based gene correction for epidermolysis bullosa. Molecular therapy : the journal of the American Society of Gene Therapy. 2013;21:1151–1159. doi: 10.1038/mt.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasmooij AM, Jonkman MF, Uitto J. Revertant mosaicism in heritable skin diseases: mechanisms of natural gene therapy. Discovery medicine. 2012;14:167–179. [PubMed] [Google Scholar]
- Pasmooij AM, Pas HH, Bolling MC, Jonkman MF. Revertant mosaicism in junctional epidermolysis bullosa due to multiple correcting second-site mutations in LAMB3. The Journal of clinical investigation. 2007;117:1240–1248. doi: 10.1172/JCI30465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, Borkent M, Apostolou E, Alaei S, Cloutier J, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, Apostolou E, Stadtfeld M, Li Y, Shioda T, et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nature biotechnology. 2010;28:848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- Prigione A, Fauler B, Lurz R, Lehrach H, Adjaye J. The senescence-related mitochondrial/oxidative stress pathway is repressed in human induced pluripotent stem cells. Stem Cells. 2010;28:721–733. doi: 10.1002/stem.404. [DOI] [PubMed] [Google Scholar]
- Schuilenga-Hut PH, Scheffer H, Pas HH, Nijenhuis M, Buys CH, Jonkman MF. Partial revertant mosaicism of keratin 14 in a patient with recessive epidermolysis bullosa simplex. The Journal of investigative dermatology. 2002;118:626–630. doi: 10.1046/j.1523-1747.2002.01715.x. [DOI] [PubMed] [Google Scholar]
- Shalom-Feuerstein R, Serror L, Aberdam E, Muller FJ, van Bokhoven H, Wiman KG, Zhou H, Aberdam D, Petit I. Impaired epithelial differentiation of induced pluripotent stem cells from ectodermal dysplasia-related patients is rescued by the small compound APR-246/PRIMA-1MET. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:2152–2156. doi: 10.1073/pnas.1201753109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith F. The molecular genetics of keratin disorders. American journal of clinical dermatology. 2003;4:347–364. doi: 10.2165/00128071-200304050-00005. [DOI] [PubMed] [Google Scholar]
- Sommer AG, Rozelle SS, Sullivan S, Mills JA, Park SM, Smith BW, Iyer AM, French DL, Kotton DN, Gadue P, et al. Generation of human induced pluripotent stem cells from peripheral blood using the STEMCCA lentiviral vector. Journal of visualized experiments : JoVE. 2012 doi: 10.3791/4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer CA, Stadtfeld M, Murphy GJ, Hochedlinger K, Kotton DN, Mostoslavsky G. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 2009;27:543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science. 2008;322:945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart DM, Candotti F, Nelson DL. The phenomenon of spontaneous genetic reversions in the Wiskott-Aldrich syndrome: a report of the workshop of the ESID Genetics Working Party at the XIIth Meeting of the European Society for Immunodeficiencies (ESID). Budapest, Hungary October 4-7, 2006. Journal of clinical immunology. 2007;27:634–639. doi: 10.1007/s10875-007-9121-z. [DOI] [PubMed] [Google Scholar]
- Suhr ST, Chang EA, Rodriguez RM, Wang K, Ross PJ, Beyhan Z, Murthy S, Cibelli JB. Telomere dynamics in human cells reprogrammed to pluripotency. PloS one. 2009;4:e8124. doi: 10.1371/journal.pone.0008124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhr ST, Chang EA, Tjong J, Alcasid N, Perkins GA, Goissis MD, Ellisman MH, Perez GI, Cibelli JB. Mitochondrial rejuvenation after induced pluripotency. PloS one. 2010;5:e14095. doi: 10.1371/journal.pone.0014095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K. Cellular reprogramming--lowering gravity on Waddington’s epigenetic landscape. Journal of cell science. 2012;125:2553–2560. doi: 10.1242/jcs.084822. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tamai K, Yamazaki T, Chino T, Ishii M, Otsuru S, Kikuchi Y, Iinuma S, Saga K, Nimura K, Shimbo T, et al. PDGFRalpha-positive cells in bone marrow are mobilized by high mobility group box 1 (HMGB1) to regenerate injured epithelia. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:6609–6614. doi: 10.1073/pnas.1016753108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Tolar J, Ishida-Yamamoto A, Riddle M, McElmurry RT, Osborn M, Xia L, Lund T, Slattery C, Uitto J, Christiano AM, et al. Amelioration of epidermolysis bullosa by transfer of wild-type bone marrow cells. Blood. 2009;113:1167–1174. doi: 10.1182/blood-2008-06-161299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolar J, Xia L, Lees CJ, Riddle M, McElroy A, Keene DR, Lund TC, Osborn MJ, Marinkovich MP, Blazar BR, et al. Keratinocytes from induced pluripotent stem cells in junctional epidermolysis bullosa. The Journal of investigative dermatology. 2013;133:562–565. doi: 10.1038/jid.2012.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolar J, Xia L, Riddle MJ, Lees CJ, Eide CR, McElmurry RT, Titeux M, Osborn MJ, Lund TC, Hovnanian A, et al. Induced pluripotent stem cells from individuals with recessive dystrophic epidermolysis bullosa. The Journal of investigative dermatology. 2011;131:848–856. doi: 10.1038/jid.2010.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utikal J, Maherali N, Kulalert W, Hochedlinger K. Sox2 is dispensable for the reprogramming of melanocytes and melanoma cells into induced pluripotent stem cells. Journal of cell science. 2009;122:3502–3510. doi: 10.1242/jcs.054783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakayama T, Perry AC, Zuccotti M, Johnson KR, Yanagimachi R. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature. 1998;394:369–374. doi: 10.1038/28615. [DOI] [PubMed] [Google Scholar]
- Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell stem cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L, Ni Y, Wang J, Guo X. Feeder-free derivation of human induced pluripotent stem cells with messenger RNA. Scientific reports. 2012;2:657. doi: 10.1038/srep00657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe A, Yamada Y, Yamanaka S. Epigenetic regulation in pluripotent stem cells: a key to breaking the epigenetic barrier. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2013;368:20120292. doi: 10.1098/rstb.2012.0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M, Lengner CJ, Hanna J, Lodato MA, Steine E, Foreman R, Staerk J, Markoulaki S, Jaenisch R. A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nature biotechnology. 2008;26:916–924. doi: 10.1038/nbt1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- Yamanaka S. Strategies and new developments in the generation of patient-specific pluripotent stem cells. Cell stem cell. 2007;1:39–49. doi: 10.1016/j.stem.2007.05.012. [DOI] [PubMed] [Google Scholar]
- Yamanaka S. A fresh look at iPS cells. Cell. 2009;137:13–17. doi: 10.1016/j.cell.2009.03.034. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Yusa K, Rad R, Takeda J, Bradley A. Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nature methods. 2009;6:363–369. doi: 10.1038/nmeth.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li W, Laurent T, Ding S. Small molecules, big roles - the chemical manipulation of stem cell fate and somatic cell reprogramming. Journal of cell science. 2012;125:5609–5620. doi: 10.1242/jcs.096032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Zhang ZN, Rong Z, Xu Y. Immunogenicity of induced pluripotent stem cells. Nature. 2011;474:212–215. doi: 10.1038/nature10135. [DOI] [PubMed] [Google Scholar]
- Zhou H, Wu S, Joo JY, Zhu S, Han DW, Lin T, Trauger S, Bien G, Yao S, Zhu Y, et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell stem cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Freed CR. Adenoviral gene delivery can reprogram human fibroblasts to induced pluripotent stem cells. Stem Cells. 2009;27:2667–2674. doi: 10.1002/stem.201. [DOI] [PubMed] [Google Scholar]
- Zhu S, Li W, Zhou H, Wei W, Ambasudhan R, Lin T, Kim J, Zhang K, Ding S. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell stem cell. 2010;7:651–655. doi: 10.1016/j.stem.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]