Abstract
Non-typeable Haemophilus influenzae contains an N6-adenine DNA-methyltransferase (ModA) that is subject to phase-variable expression (random ON/OFF switching). Five modA alleles, modA2, modA4, modA5, modA9 and modA10, account for over two-thirds of clinical otitis media isolates surveyed. Here, we use single molecule, real-time (SMRT) methylome analysis to identify the DNA-recognition motifs for all five of these modA alleles. Phase variation of these alleles regulates multiple proteins including vaccine candidates, and key virulence phenotypes such as antibiotic resistance (modA2, modA5, modA10), biofilm formation (modA2) and immunoevasion (modA4). Analyses of a modA2 strain in the chinchilla model of otitis media show a clear selection for ON switching of modA2 in the middle ear. Our results indicate that a biphasic epigenetic switch can control bacterial virulence, immunoevasion and niche adaptation in an animal model system.
Non-typeable Haemophilus influenzae (NTHi) is a significant bacterial pathogen commonly associated with paediatric infection as a predominant cause of otitis media (OM) or middle ear infection. While NTHi contributes to ~40% of all cases of acute OM, it remains the major aetiological agent of chronic OM, recurrent acute OM and OM treatment failure1. It is also a major cause of community-acquired pneumonia and exacerbations of chronic obstructive pulmonary disease. With the rising prevalence of antimicrobial resistance, the success rate in the treatment of NTHi has recently declined. There is no vaccine to prevent infection by NTHi.
Phase variation is the high frequency reversible ON/OFF switching of gene expression, and is a common feature of many virulence determinants expressed by bacterial pathogens2–5. While phase variation is typically associated with genes that encode surface structures, several host-adapted bacterial pathogens, including NTHi, have DNA methyltransferases (mod genes) associated with type III restriction modification systems that are subject to phase-variable expression. Simple tandem DNA repeats have been shown to mediate phase variation of mod genes by rapid reversible loss or gain of a repeat unit leading to frame shifts and ON/OFF switching of expression6–10.
The variable central region of Mod methyltransferases contains the DNA-recognition domain (DRD; see Fig. 1e) that dictates methylation-sequence specificity11–13. In H. influenzae strain Rd, our previous work has demonstrated that random switching of the modA1 gene controls expression of multiple genes via differential methylation of the genome in the modA1 ON and OFF states10. This novel genetic system, termed the phasevarion (phase-variable regulon) regulates gene expression in four other important human pathogens: Neisseria gonorrhoeae, Neisseria meningitidis7, Helicobacter pylori9 and Moraxella catarrhalis14. Differences in the DRD have previously been observed in a genetically diverse collection of H. influenzae strains and these differences define 20 distinct mod alleles (modA1 to modA20)6,15. Different DRDs are proposed to recognize and methylate different target sequences. Our studies with pathogenic Neisseria, which also contains modA, confirmed that different modA alleles methylate different target sequences and thereby regulate different sets of genes, that is, have distinct phasevarions. Conversely, strains that harbour the same modA allele regulate the same phasevarion of genes7,16.
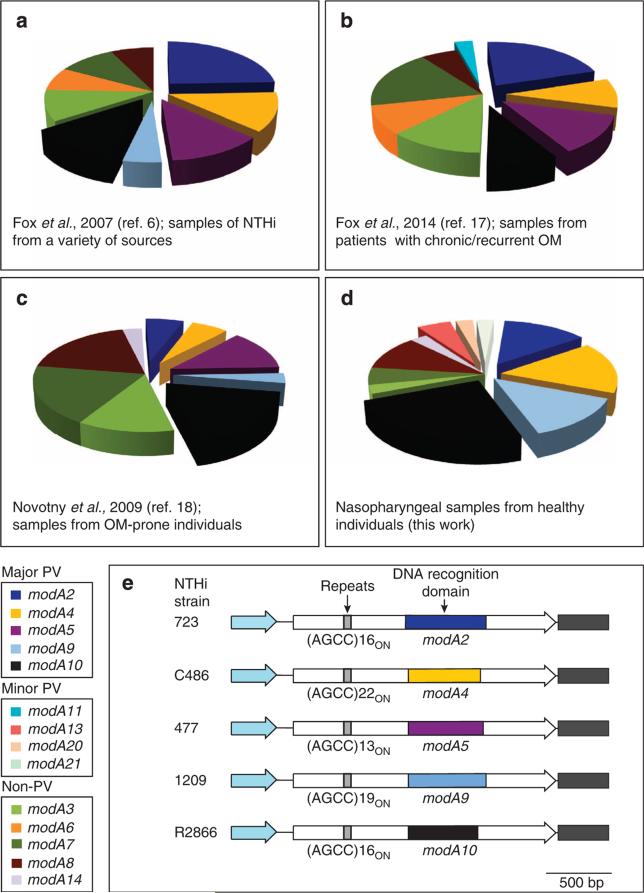
Figure 1. Analysis of NTHi strain collections to ascertain the proportion of isolates containing phase variable modA alleles.
(a–d) Presents the four separate strain collections that were analysed to ascertain the distribution of phase variable modA alleles in NTHi populations; (a) collection containing a broad selection of NTHi isolates from a variety of sources (for example, culture collections, OM-prone individuals)6; (b) samples of paired isolates of NTHi isolated from patients presenting with chronic/recurrent OM17; (c) sample collection of NTHi isolates taken from OM-prone individuals18; (d) NTHi isolates from the nasopharynx of healthy children (collected as part of this study); and (e) the five modA alleles most commonly associated with OM—modA2, modA4, modA5, modA9 and modA10. Each modA gene is represented as a white arrow, with the DRD represented by a coloured box that matches the colour in panels a–d. The black box to the right of each gene represents the 5′ region of the downstream gene (an inactive restriction endonuclease). The blue arrow to the left of each modA gene represents the 3′ end of the gene upstream of each modA (in all the cases, a ribonuclease, rnhB). Details of the strain collections used in c and d are presented in Supplementary Table 1.
In this study, we sought to assess the role that candidate NTHi phasevarions may play in gene expression, immunoevasion and in the pathogenesis of NTHi during experimental OM. Five modA alleles are present in over two-thirds of all isolates of NTHI. We demonstrate that phase variation of all five of these modA alleles controls gene expression differences in all the strains studied. These expression differences include outer-membrane proteins and key vaccine candidates. Biphasic switching of specific modA alleles also influences fitness in opsonophagocytic killing assays and in susceptibility to antibiotics. In an in vivo animal model, we demonstrate, for the first time, direct selection for a particular state of modA expression in the major phase variable modA allele present in NTHi, modA2.
Results
Distribution of modA alleles in NTHi isolates
We conducted a detailed analysis examining the modA allele frequency in a diverse set of NTHi isolates taken from healthy individuals and OM patients (Fig. 1). The modA alleles modA3, modA6, modA7, modA8 and modA14 do not contain tetranucleotide repeats, and thereby do not phase vary in expression. In a previous survey6, we reported that ~70% of NTHi strains contain a phase variable modA allele (n = 41; Fig. 1a). Of all the modA alleles, modA2 was the most prevalent, being found in almost a quarter of all isolates (24%).
We then analysed modA allele distribution in three further strain collections (Fig. 1). The first collection contained paired isolates, isolated from the nasopharynx and the middle ear, from children presenting with chronic and/or recurrent OM (Fig. 1b)17. This collection contained 27 pairs of isolates: 16 pairs were collected from 1982 to 1986 and 11 pairs were collected from 2004 to 2006. Almost two-thirds of strains (59.3%) within this particular collection contain a phase variable modA allele (n = 33), with modA2 most prevalent (22% of all isolates). The second strain collection contained clinical isolates from OM-prone children (Fig. 1c)18. Of these 34 separate isolates (25 recovered from nasopharyngeal swabs and 9 recovered from middle ear effusions), 65% contained a phase variable modA allele. The most common was modA10 (19%). As NTHi is frequently isolated from the upper respiratory tract, we collected samples and analysed modA allele distribution in isolates from the nasopharynges of healthy children (Fig. 1d), with a reported colonization rate of ~80% (ref. 19). In addition, a correlation between NTHi colonization frequency and the incidence of recurrent OM in children has been suggested20. We observed that the predominant modA alleles in healthy nasopharyngeal isolates were modA10 (24%), modA4 (16%), modA2 (14%) and modA9 (14%) (Fig. 1d). These four modA alleles are all phase variable, and alone comprise over two-thirds of all alleles from this collection. One new modA allele was also identified in this study in strain 515 (Supplementary Table 1). We have designated this new allele as modA21.
Taken together, this analysis of four distinct strain collections shows that not only are phase variable modA alleles frequently associated with disease, they are also prevalent in NTHi isolated from healthy individuals. From all the four studies, phase variable modA alleles are present in approximately two-thirds of all the isolates (n = 135), with the modA2 and modA10 alleles, the two most common overall, at 17.2 and 15.4%, respectively. Based on these surveys, we selected the five modA alleles most commonly associated with OM—modA2, 4, 5, 9 and 10—for further analysis in this study (Fig. 1e).
Generation of natural modA ON and OFF strains
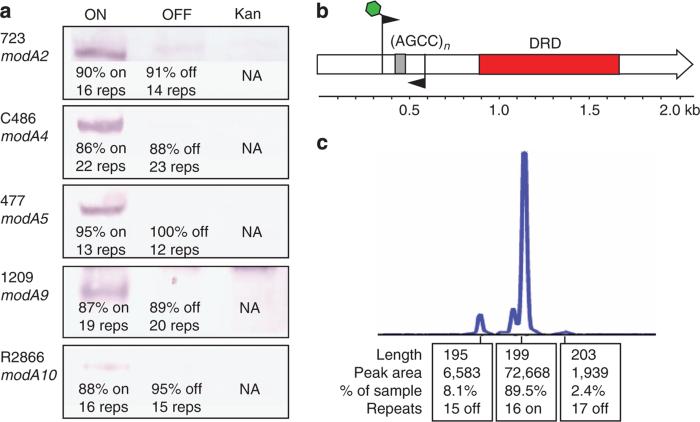
We generated modA natural ON- and OFF-enriched populations (>90% ON or OFF) from the five NTHi strains containing the modA2, 4, 5, 9 and 10 alleles (Fig. 1e). All ON strains contain a number of AGCC repeats that places the modA open reading frame (ORF) in frame, and therefore express a full-length functional protein, as confirmed by western blot using an anti-ModA antibody (Fig. 2a). All the natural ON and OFF strains were continually verified using our well-established FAM-labelled primer PCR screen coupled to fragment length analysis (an example of the methodology and results is given in Fig. 2b,c)6,10. Natural modAOFF strains contain a number of AGCC repeats that place the modA ORF out of frame, leading to a frame shift mutation and premature termination at stop codons in the alternate reading frames (Fig. 2a). Kanamycin knockout modA::kan mutants were also generated in all the five NTHi strains bearing these alleles (Fig. 1e) by disruption of the modA gene via insertion of a kanamycin resistance cassette, as described previously6,10.
Figure 2. Western blots showing ModA presence in modA ON OFF and kanamycin knockout strains and an example of the fragment analysis methodology.
(a) Western blot analysis using ON/OFF/kan cells from all the five modA alleles; anti-modA primary antibody at 1:5,000; AP conjugated anti-rabbit secondary at 1:20,000. A modA band is only present in those cells possessing the ON variant of each strain. The number of repeats (reps) represents the major number of repeats present in that particular isolate, and the percentage ON of that particular strain is noted underneath each relevant blot. Full western blots are presented in Supplementary Fig. 4; (b) an illustration of the methodology used during fragment analysis to ascertain modA ON/OFF state and percentages of cell populations. A 6-carboxyflourescein (FAM)-labelled forward primer (green hexagon) and unlabelled reverse primer was used to generate a PCR product over the repeat tract (AGCCn; grey box) that was analysed using GeneScan technology; and (c) an example trace produced by GeneScan analysis on a representative genomic sample from an NTHi strain containing a phase variable modA gene.
Methylome analysis
Single molecule, real-time (SMRT) sequencing21 was carried out to determine the complete genomes and identify the methylation recognition sequence of these five distinct ModA N6-adenine methyltransferases. DNA isolated from each modAON and modA::kan pair was sequenced and analysed. By comparing the methylomes of the modAON with the modA::kan knockout mutants, we were able to identify the motif methylated by the ModA methyltransferases of ModA4, 5, 9, 10 (Table 1). The ModA2 motif was identified by heterologous expression of ModA2 in Escherichia coli, as described previously22. ModA2, ModA5, ModA9 and ModA10 all have prototypical type III methyltransferase recognition sequences (Table 1), in that they methylate an adenine residue on the N6 position in a 5 base pair (5 bp) non-palindromic sequence23,24. The recognition sequence of ModA4 was different, instead recognizing a four base sequence: 5’-CG(m6A)G-3′ (Table 1). In accordance with REBASE25 naming conventions, we have given the ModA2, ModA4, ModA5, ModA9 and ModA10 methyltransferases the designations M.Hin723I, M.HinC486I, M.Hin477I, M.Hin1209I and M.Hin2866I, respectively. Summary data from the SMRT methylome analysis of the five modA ON/OFF pairs is shown in Supplementary Fig. 1. Complete closed genome sequences for each of the five strains containing these modA alleles were also generated (NTHi strains 723, C486, 477, 1209 and R2866; the R2866 genome sequence had already been deposited in NCBI Genbank; accession number CP002277). Accession codes for the complete annotated genomes of NTHi strains 723, C486, 477 and 1209 are listed in the ‘Accession codes’ section.
Table 1.
Summary of SMRT sequencing and methylome analysis of representative strains containing the five modA alleles under study.
| modAallele | NTHi strain |
Clinical symptoms |
Accession number |
Methylation sequence |
Systematic name |
Number of sites in genome |
Genome size (bp) |
Predicted ORFs |
|---|---|---|---|---|---|---|---|---|
| modA2 | 723 | OM | CP007472 | 5′-CCGA(m6A)-3′ | M.Hin723I | 2,270 | 1,887,620 | 1,868 |
| modA4 | C486 | OM | CP007471 | 5′-CG(m6A)G-3′ | M.HinC486I | 6,203 | 1,846,507 | 1,783 |
| modA5 | 477 | OM | CP007470 | 5′-AC(m6A)GC-3′ | M.Hin477I | 2,548 | 1,846,259 | 1,813 |
| modA9 | 1209 | OM | JMQP01000000 | 5′-CCTG(m6A)-3′ | M.Hin1209I | 2,504 | 1,895,979 | 2,247 |
| modA10 | R2866 | Blood | CP002277* | 5′-CCT(m6A)C-3′ | M.Hin2866I | 1,244 | 1,932,238 | 1,905 |
Strain already annotated and submitted (October 2010) to the EMBL/GenBank/DDBJ databases. A full summary of SMRT sequencing/methylome analysis derived data is presented in Supplementary Fig. 1.
Distribution of ModA sites in NTHi genomes
Bioinformatic analysis of the recognition sequences of ModA2, 4, 5, 9, 10 showed that these sites are widely and evenly distributed in their respective genomes (see Supplementary Fig. 1). ModA4 occurs much more frequently than ModA2, 5, 9 or 10, as is expected for a 4 bp recognition sequence compared with 5 bp recognition sequences. For ModA2, A4, A5 and A9, sites appear to be equally distributed between coding and non-coding regions of the genome. For ModA10, 69.1% of sites are present in non-coding regions, compared with only 30.9% of ModA10 sites in coding regions. Further, there are far fewer ModA10 sites in the R2866 genome than would be expected by chance (1,244 observed/2,714 expected = 45.8%; Supplementary Fig. 1). Other ModA recognition sequences occurred at frequencies close to that predicted by chance: ModA2 (91.2%), ModA4 (79%) ModA5 (104.7%), ModA9 (94%).
modA ON/OFF pairs show outer-membrane protein expression differences
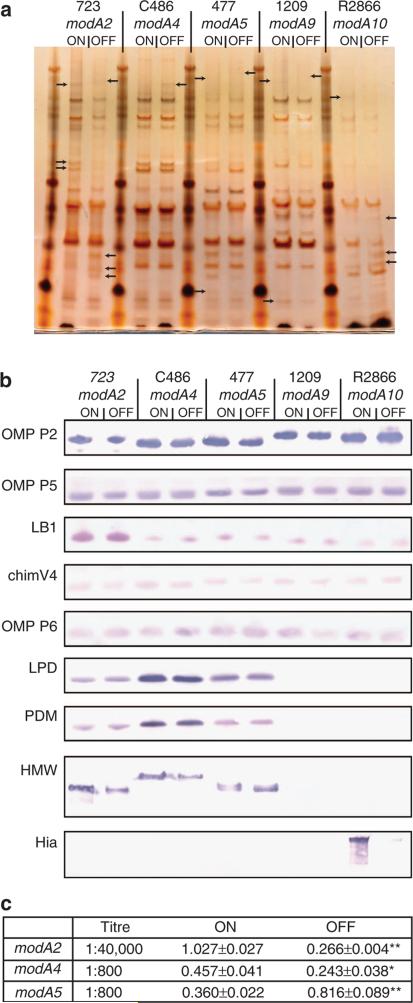
To determine whether modA switching altered the expression of outer-membrane proteins (OMPs), outer-membrane samples were separated using Bis-Tris PAGE gels and silver stained (Fig. 3a) to explore whether any gross protein differences existed within each modA ON/OFF strain pair. Clear differences could be visualized within the OMP profile of each modA ON/OFF pair (noted by arrows in Fig. 3a), indicating that modA phase variation influences the OMP profile, and may therefore influence immunoevasion, pathogenesis and virulence.
Figure 3. Analysis of outer-membrane protein (OMP) preparations.
(a) Silver-stained gel of each modA ON/OFF pair; (b) western blots using antibodies against currently investigated vaccine candidates. Full western blots are presented in Supplementary Fig. 5; (c) ELISA results for HMW in modA2, modA4 and modA5 ON/OFF strain pairs using primary antibody AD6. Values presented are mean values ± s.d. Each experiment was carried out in triplicate. P values were calculated using Student's t-test based on specific titres in the linear range of the response curve (* = <0.01; ** = <0.001). Full ELISA data are presented in Supplementary Data 1.
Vaccine candidate expression in modA ON/OFF strain pairs
OMP preparations from modA ON and OFF strains were studied using western blot with primary antibodies raised against several well-studied vaccine candidates (Fig. 3b): major outer membrane proteins P2 (ref. 26), P5 (refs 27,28) and P6 (refs 29,30); lipoprotein D31 and its deacylated derivative PDM31,32; the high molecular weight (HMW) proteins33,34; and the adhesin Hia35. Three different antisera were used to probe for relative expression of OMP P5 (anti-P5, anti-LB1 and anti-chimV4), as these sera contained antibodies specific to unique epitopes of the protein27,28,36 (Fig. 3b).
We saw an expression difference of OMP P6 in the modA9 ON/OFF pair, with a greater level of OMP P6 seen in modA9ON relative to modA9OFF cells (Fig. 3b). Lipoprotein D showed stable expression independent of modA status in NTHi strains 723 (modA2), C486 (modA4) and 477 (modA5) (Fig. 3b). We could not detect the presence of lipoprotein D in NTHi strains 1209 or R2866, regardless of modA status (Fig. 3b). The analysis of our NTHi strain 1209 SMRT-generated genome revealed that the gene-encoding lipoprotein D (glpQ) is truncated in this strain (1209_00054) to just the carboxy (C)-terminal portion (~65 residues). In strain R2866, a full-length gene is present (R2866_1787), but it appears to not be expressed at a level detectable by western blotting.
Expression of HMW1/2A was greater in OMPs prepared from modA2ON versus modA2OFF; modA4ON versus modA4OFF; and modA5OFF versus modA5ON (Fig. 3b). In these latter two examples, a single protein band was clearly more prevalent in modA4ON and modA5OFF, when compared with their respective partners (Fig. 3b). However, in modA2ON, a higher MW band was more abundant in modA2ON compared with modA2OFF, and a lower MW band was visible only in modA2ON, implying that both HMW1A and HMW2A are present in greater amounts in OMP preparations from NTHi modA2ON when compared with modA2OFF (Fig. 3b). Apparent differences in HMW expression in western blot results were verified using whole-cell enzyme-linked immunosorbent assays (ELISAs) to quantitate the relative expression of HMW1/2A. Statistical significance was calculated using Student's t-test, and significant differences were observed for these three ON/OFF strain pairs (Fig. 3c).
As the genes encoding HMW1A and HMW2A proteins contain variable heptanucleotide repeats in their promoter region that are also reported to influence gene expression4,37, we sequenced across this repeat tract in all the three strain pairs where a difference in HMW expression was evident. This analysis showed that the differences in repeat tract length in each ON/OFF pair were not responsible for the expression changes observed (Supplementary Fig. 2). The literature states that the longer the repeat tract, the lesser the expression level of HMW37. As the repeat tracts of HMW1 and HMW2 are of identical or similar length within each strain pair where differences are seen, it is likely that expression differences are ModA-dependent, and not due to HMW phase variation.
Finally, our analysis of the adhesin Hia revealed that the hia gene was only present in the genome of NTHi strain R2866 (R2866_0725) containing the modA10 allele (Fig. 3b). Hia was expressed at a greater level in modA10ON relative to modA10OFF (Fig. 3b). However, our subsequent analysis showed that this difference was owing to modA10-independent phase variation event in a poly-T tract in the Hia promoter region38.
iTRAQ proteomics analysis of modA ON/OFF strain pairs
To further define the impact of modA ON/OFF status on the expression of existing and potential vaccine candidates, as well as the OMP profile as a whole, preparations of OMPs used in silver staining and western blotting (Fig. 3) were characterized using iTRAQ 1D nanoLC ESI MS/MS. All the data have been deposited to the ProteomeXchange Consortium39 via the PRIDE partner repository, with the data set identifier PXD002210. Proteins with either > 1.5− or <0.65-fold expression differences between ON/OFF pairs when comparing two biological replicates of each OMP preparation are reported (Table 2). Expression changes in OMPs were seen in the modA2, modA4, modA5 and modA10 ON/OFF strain pairs.
Table 2.
iTRAQ quantitative mass spectrometry analysis using OMPs from each ON/OFF pair.
| Gene ID | Gene | Ratio ON:OFF* | Stouffers P-value | Stouffer Adj |
|---|---|---|---|---|
| Increased expression in 723 modA2 ON | ||||
| 723_01555 | N-acetylneuraminate epimerase NanM | 2.36 | 5.40E - 14 | 6.32E - 13 |
| 723_01788 | Adhesin translocation protein HMW2B | 2.05 | 0 | 0 |
| Decreased expression in 723 modA2 ON | ||||
| 723_01435 | Haem/haemopexin utilization protein C HxuC1 | 0.28 | 0 | 0 |
| 723_00620 | Transferrin-binding protein 1 | 0.32 | 4.74E - 08 | 3.45E - 07 |
| 723_01615 | Major ferric iron-binding protein HitA† | 0.37 | 0 | 0 |
| 723_01434 | Haem/haemopexin transporter protein HxuB† | 0.41 | 0 | 0 |
| 723_01596 | Haemin receptor, HemR | 0.42 | 0 | 0 |
| 723_01306 | 15 kDa peptidoglycan-associated lipoprotein OMP P6 | 0.6 | 2.57E - 04 | 1.12E - 03 |
| Increased expression in C486 modA4 ON | ||||
| C486_00892 | OMP P2 | 1.52 | 0 | 0 |
| Increased expression in 477 modA5 ON | ||||
| 477_00572 | OMP P5 | 1.65 | 0 | 0 |
| Increased expression in R2866 modA10 ON | ||||
| R2866_0725 | Adhesin Hiaठ| 11.53 | 0 | 0 |
| R2866_0192 | OMP P6 | 1.85 | 0.046 | 0.17 |
| R2866_1237 | OMP P5 | 1.65 | 7.39E - 03 | 0.043 |
Gene designations from analysis with our SMRT produced genomes for each strain; differences are represented as ON:OFF; statistical significance is measured using Stouffers P value and Stouffers adjusted (Adj) value, as the results are the interpretation and comparison of two independently prepared sets of OMPs from each ON/OFF pair. Schematics of differentially regulated genes with locations of ModA methylation motifs are depicted in Supplementary Fig. 3.
Only samples with an ON:OFF ratio >1.5 or <0.7 were included. Complete iTRAQ data for all the five strain pairs are presented as Supplementary Data 2.
Indicates identification by microarray and iTRAQ.
Identified by western blotting and iTRAQ.
Hia was subsequently shown to be regulated in a modA10-independent manner.
Several OMPs involved in the sequestration of iron and haem were downregulated in modA2ON (Table 2). This included HxuC and HxuB (723_01435 and 723_01434; a haem/haemopexin utilization and haem/haemopexin-binding OMP, respectively), transferrin-binding protein 1 (723_00620), a hemin receptor (HemR, also annotated as HxuC2; 723_01596); and a major ferric iron-binding protein (HitA, 723_01615). This analysis also revealed OMP P6 to be differentially regulated in the modA2 pair (Table 2), a finding not evident from our western blotting analysis (Fig. 3b), but having implications for its use as a suitable vaccine antigen.
HMW2B is an outer membrane-associated protein responsible for the translocation of the adhesins HMW1/2A to the cell surface40. The increase in an accessory protein in modA2ON with specificity for both HMW1/2A proteins would explain the increase in both HMW-A proteins in modA2ON seen in our western blotting with the modA2 strain pair (Fig. 3b). NanM, a sialic acid mutarose, was also present in greater amounts in the outer membrane of modA2ON cells (Table 2).
Phase variation of modA influences susceptibility to antibiotics
Phasevarion-mediated differential susceptibility to antibiotics has been reported in other human-adapted pathogens41, so we sought to investigate whether this was also the case for our five modA ON/OFF pairs in NTHi. Minimum inhibitory concentration (MIC) analysis showed that phase-variable expression of modA2, modA5 and modA10 led to 2-fold changes in susceptibility to ampicillin, erythromycin and gentamicin, respectively (Supplementary Table 2). These findings suggest that gene regulation through DNA methylation is an additional element that may contribute to antibiotic susceptibility.
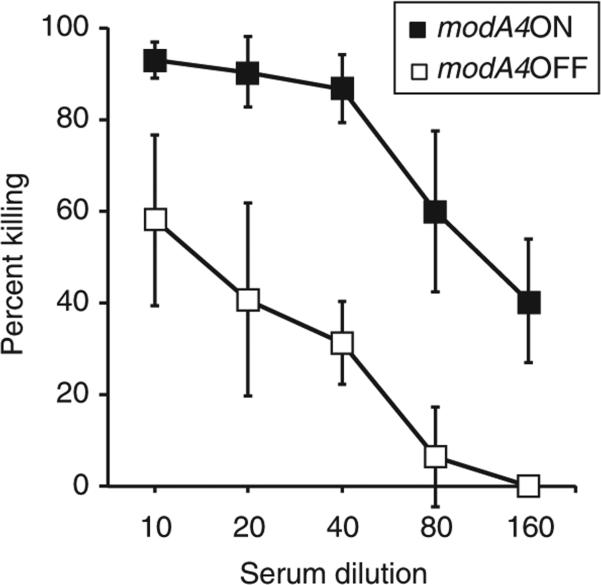
ModA4 mediates evasion of opsonophagocytic killing
Differential expression of HMW protein had been observed in strain C486, containing the modA4 phasevarion. Antiserum was available that would recognize the HMW protein in this strain. We tested our C486 modA4 ON/OFF strain pair in an opsonophagocytic killing assay42. The modA4ON strain was significantly (P<0.05 at all the data points; calculated using Student's t-test) more susceptible to killing at every dilution of antibody tested compared with modA4OFF (Fig. 4). No killing was observed with this strain pair using pre-immune sera from animals used to raise anti-HMW sera, nor was killing observed with antisera raised against the protein Hia (data not shown).
Figure 4. Opsonophagocytic killing curves using strain C486 modA4 ON/OFF strain pair.
Assays were carried out using guinea pig antiserum raised against purified HMW proteins from NTHi strain 12, with NTHi strain C486 harbouring the modA4 ON/OFF pair. Three independent assays were carried out per strain, with error bars representing one s.d. Statistical significance was calculated using Student's t-test, and was <0.05 at all dilutions of antiserum. Individual P values—1:10 = 0.034; 1:20 = 0.0185; 1:40 = 0.0012; 1:80 = 0.009; 1:160 = 0.0065.
Differentially expressed genes in the modA2 phasevarion
Our analysis of samples from a wide variety of sources showed that modA2 is the most prevalent modA allele present in NTHi isolates (Fig. 1), suggesting that modA2 may have an important role in both asymptomatic colonization and disease. iTRAQ proteomic analysis also showed more differences in expression profile analysis between modA2ON and modA2OFF than the other four modA strain pairs. These observations could lead to a greater potential for phenotypic and virulence differences in vivo (Table 2). We therefore selected the modA2 phasevarion for microarray analysis. RNA was isolated and compared from the modA2ON strain and the modA2::kan mutant. We found 36 genes with an expression ratio of 1.4-fold or greater, with 27 genes upregulated in modA2::kan relative to modA2ON and nine genes downregulated, thereby demonstrating modA2 phase variation has a global impact on gene expression (Table 3; full results in Supplementary Table 3). Quantitative real-time PCR specific for several of the differentially expressed genes confirmed the array data (Table 3). This analysis adds further evidence to our observations using iTRAQ quantitative mass spectrometry that there is differential regulation of genes involved in iron acquisition in the modA2 strain pair (Table 2; Table 3). For example both hxuB 723_01434 and hitA 723_1615 are identified as differentially regulated by both iTRAQ and microarray analysis. Microarray analysis revealed several additional iron-regulated genes that were not identified by iTRAQ as they are not OMPs. These include hitB, which encodes a cytoplasmic permease, with both HitA and HitB being essential for the utilization of iron by NTHi43; and hxuA, a secreted protein involved in the binding of haem–haemopexin. Functional HxuA and HxuB proteins are required for virulence in H. influenzae44. Also upregulated in 723 modA2::kan was the yfeABCD operon, which encodes an iron transport system45. This locus has homology to the yfeABCD locus of Yersinia pestis, where it is associated with virulence46. Several genes involved in anaerobic metabolism also showed increased expression in modA2::kan (Table 3). Many enzymes involved in anaerobic respiration appear to play an important role in colonization and virulence in bacterial pathogens47,48.
Table 3.
Differentially expressed genes in the NTHi strain 723 modA2ON microarray.
| Gene | Microarray ID | Description | Ratio | qRT-PCR | B-Stat |
|---|---|---|---|---|---|
| Reduced expression in the H. influenzae strain 723 modA2 mutant | |||||
| 723_01712 | NTHI0007 | Formate dehydrogenase major subunit fdxG | –1.50 | 2.77 | |
| 723_01520 | Hflu203001461 | Alcohol dehydrogenase, class III | –1.53 | 2.42 | |
| 723_01710 | NTHI0010 | Formate dehydrogenase, iron-sulfur subunit fdxH | –1.53 | 1.66 | |
| 723_00429 | Hflu103001790 | L-lactate permease lctP | –1.79 | 3.08 | |
| 723_00946 | Hflu203000185 | DL-methionine transporter ATP-binding subunit metN | –1.96 | –3.56 ±0.741* | 4.80 |
| Increased expression in the H. influenzae strain 723 modA2 mutant | |||||
| 723_01433 | ORF02024 | Haem/haemopexin-binding precursor hxuA | 1.45 | 0.86 | |
| 723_00220 | NTHI1806 | Glycogen synthase glgA | 1.49 | 1.84 | |
| 723_00222 | NTHI1808 | Glycogen debranching enzyme glgX | 1.52 | 1.27 | |
| 723_01160 | HI0534 | Aspartate ammonia-lyase aspA | 1.52 | 2.66 | |
| 723_00775 | ORF01899 | Fumarate reductase subunit D frdD | 1.56 | 0.89 | |
| 723_01524 | HI0181 | Formate transporter focA | 1.57 | 1.14 | |
| 723_01327 | Hflu203001649 | Iron (chelated) ABC transporter yfeB | 1.61 | 1.32 | |
| 723_00221 | NTHI1807 | Glucose-1-phosphate adenylyltransferase glgC | 1.61 | 3.76 | |
| 723_00223 | NTHI1809 | Glycogen branching enzyme glgB | 1.66 | 2.21 | |
| 723_00419 | HI0809 | Phosphoenolpyruvate carboxykinase pckA | 1.69 | 1.00 | |
| 723_01614 | ORF00565 | Iron (III) ABC transporter, permease protein hitB | 1.75 | 4.39 | |
| 723_01328 | Hflu203001648 | Iron chelated ABC transporter permease yfeC | 1.90 | 1.69 | |
| 723_01329 | Hflu203001647 | Iron chelated ABC transporter permease yfeD | 2.08 | 3.98 | |
| 723_00591 | Hflu203000516 | Anaerobic DMSO chain C dmsC | 2.09 | 1.26 | |
| 723_01434 | Hflu103000469 | Haem/haemopexin-binding hxuB† | 2.14 | 1.15 | |
| 723_01326 | Hflu203001650 | Iron chelated ABC transporter yfeA | 2.17 | 2.166 ± 0.667* | 5.83 |
| 723_00589 | Hflu203000518 | Anaerobic DMSO chain A dmsA | 2.22 | 2.14 ± 0.839* | 2.38 |
| 723_00437 | HI1210 | Malate dehydrogenase mdh | 2.28 | 5.50 | |
| 723_00590 | Hflu203000517 | Anaerobic DMSO chain B dmsB | 2.65 | 2.67 | |
| 723_01615 | Hflu203001388 | Iron-utilization hitA† | 4.04 | 2.08 ± 0.397* | 4.72 |
The genes listed are either down- or upregulated in the H. influenzae 723 modA2::kan mutant strain compared with the modA2ON strain. The identity of the gene is indicated with our SMRT-derived genomic annotation (accession number CP007472), the original identifier from the microarray, and a description of each gene. The average ratio presented is the mean of NTHi strain 723 modA2::kan mutant:723 modA2ON from six replicate spots on three independent microarrays, incorporating a dye swap. Only those genes with an expression value >1.4-fold were included in this table.
Gene expression was confirmed by quantitative RT-PCR in NTHi strain 723 modA2ON and 723 modA2OFF strain variants.
Identified by both microarray and iTRAQ. B-stat (B-statistic) represents the log-odds that the gene is differentially expressed. A threshold in the B statistic of 0.0 was adopted as genes with a B score >0 have a >50% probability of being truly differentially expressed. All the microarray data are presented in Supplementary Table 3. Schematics of differentially regulated genes with locations of ModA methylation motifs are depicted in Supplementary Fig 3.
Strain 723 modA2ON is preferentially selected in vivo
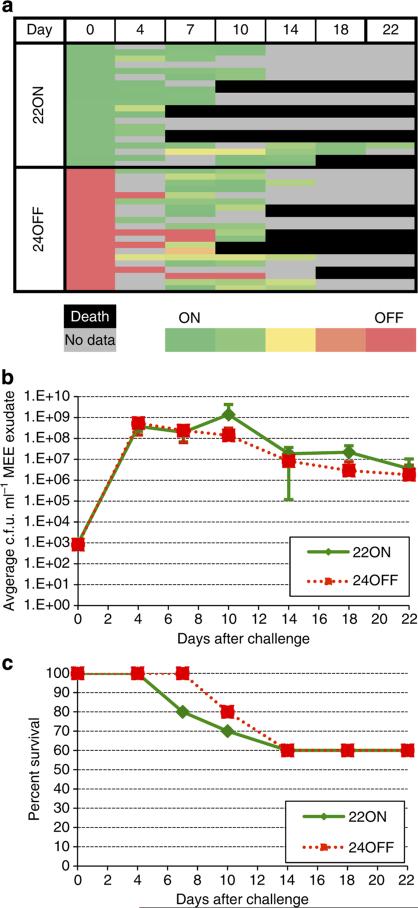
To determine whether the gene expression differences between the modA2 natural ON and OFF states impacted virulence, a variant of NTHi strain 723 selected for its modA2ON status (~90% ON, 22 AGCC repeats; modA2(22)ON) and a variant selected for its modA2OFF status (~90% OFF. 24 AGCC repeats; modA2(24)-OFF) were used as challenge strains in the well-established chinchilla model of OM49. These variants were free to phase-vary during the 22 days of the experiment. In two separate studies, cohorts of five chinchillas each (n = 10 total) were challenged intranasally and transbullarly with NTHi strain 723 modA2(22)ON or strain 723 modA2(24)OFF. Middle ear fluids were collected at regular intervals to assess any differences between these strains in their ability to infect the middle ear. Samples taken directly from the left and right middle ear were immediately snap frozen in liquid nitrogen, and were not subcultured, thereby reflecting the exact ON/OFF status of the bacterial population from the site and time of sampling.
The ON/OFF status of the repeat tracts in both the modA2(22)ON and modA2(24)OFF infected cohorts from the starting inoculum (day 0) and subsequent days 4, 7, 10, 14, 18 and 22 after challenge were verified by fragment analysis (Fig. 5a; full data set presented in Supplementary Table 4). No gross difference in colony-forming units (c.f.u.) number or mortality was observed (Fig. 5b,c). Figure 5a shows a heat map of the modA2ON status of the inoculum, and the bacteria isolated from both middle ears. Animals challenged with the predominantly modA2ON variant (green) remained ON, whereas those animals challenged with the predominantly modA2OFF variant (red) showed a consistent switch from the OFF to the ON state during the course of the experimental OM study (Fig. 5a). This observation suggests that selection for modA2ON over modA2OFF occurred during overt infection of the middle ear.
Figure 5. Results from chinchilla middle ear samples.
(a) Heat map showing fragment analysis results from animals challenged with either NTHi 723 modA2(22)ON or modA2(24)OFF. The time of sampling is indicated at the top of the figure by days after initial infection, for example, day 0 = initial inoculum, day 4 = 4 days after initial infection and so on. All fragment analysis data are presented in Supplementary Table 4; (b) middle ear exudate (MEE) bacterial CFU mean counts from animals infected with modA2(22)ON and modA2(24)OFF. Error bars represent s.d.; and (c) survival of animals infected with modA2(22)ON and modA2(24)OFF.
modA2ON forms more robust biofilms in vitro than modA2OFF
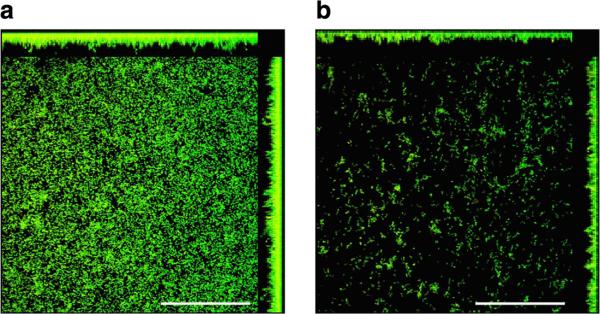
Given the selection for modA2ON in the chinchilla middle ear and the importance of biofilm formation in chronicity and recurrence of OM, we assayed these ON and OFF variants for relative ability to form a biofilm in vitro. Chamberslides were inoculated with either NTHI strain 723 modA2(22)ON or modA2(24)OFF and allowed to form biofilms for 24 h before being stained with a viable bacterial stain and subjected to confocal imaging. As shown in Fig. 6, biofilms formed by the modA2ON variant were notably more robust than those formed by modA2OFF.
Figure 6. Biofilm formation using modA2ON and OFF strains.
(a) Shows biofilm formation using modA2(22)ON. (b) Shows biofilm formation using modA2(24)OFF after 24 h growth on chambered coverglass and visualized using confocal microscopy following LIVE/DEAD BacLight staining. Scale bar shown represents 50 μm.
Discussion
Phenotypic analysis of all modA ON/OFF pairs showed that all phasevarions influence the protein profile of the outer membrane, with gross phenotypic changes evident with the five strain pairs on silver-stained gels (Fig. 3). Our in-depth analysis of multiple current vaccine candidates50 through western blotting, and iTRAQ quantitative mass-spectrometry of OMPs, showed that modA phase variation influenced the expression of several of these proteins, with some current vaccine candidates50 differentially expressed in multiple phasevarions (HMW1/2A, OMP P6). Most compelling in this analysis was the expression and phenotypic differences related to HMW1/2A. The HMW protein is a well-characterized adhesin expressed by NTHi, and has been investigated as a potential vaccine candidate33,34. However, our findings suggest that differential expression is occurring in three separate modA phasevarions. This influence appears to be direct (modA4 and modA5 directly affect HMW-A expression) or indirect (modA2 ON cells show higher expression of the accessory protein HMW2B). Although HMW1/2A is known to be phase variable itself, through heptanucleotide repeats in its promoter region37, sequencing of HMW in all the three strain pairs show that phase-variable expression of HMW1/2A itself is likely not responsible for the large differences in expression demonstrated by western blot and ELISA in the modA2, modA4 and modA5 ON/OFF strain pairs. The difference in HMW expression mediated by the modA4 phasevarion is sufficient to result in marked differences in the rate of HMW-specific opsonophagocytic killing (Fig. 4). This would likely result in selection for a modA4OFF subpopulation that express lower amounts of HMW. HMW has adhesion functions51 after selection has been relaxed; therefore, selection for adherent cells may counter select for the subpopulation of modA4OFF.
In this study, iTRAQ proteomic analysis of the OMP profile, and detailed gene expression studies of NTHi strain 723, containing modA2, the most prevalent modA allele found in all the clinical isolates (Fig. 1), showed that modA2 has a major effect on gene expression and phenotype. Together with animal studies showing consistent selection for cells that have switched from modA2OFF to ON in our chinchilla model system of OM, this indicates that switching of the modA2 phasevarion plays an important role in niche adaptation to the middle ear. This could be due, in part, to increased HMW expression, despite this making the cells more susceptible to an adaptive immune response. Here, the balance between adherence to host cells, a key step in colonization, and expression of this immunogenic surface protein appears to have been achieved: a modA2ON strain (increased in HMW1/2A expression) may be more adherent when compared with modA2OFF (decreased HMW1/2A). Perhaps the advantage of increased adhesion early in infection through increased HMW1/2A expression gives a selective advantage, with a gradual decrease in expression of HMW1/2A occurring over prolonged periods of host colonization, thus reducing the potential for an adaptive immune response to HMW1/2A52.
Expression of iron and haem-acquisition-associated genes have previously been shown to increase in NTHi recovered from middle ear effusions in patients with OM53. Thus, strains lacking a functional ModA2 protein (modA2OFF and modA2::kan) appear to be primed to cope well in the iron-restricted host environment through increased production of a number of iron-acquisition factors. However, this may, in fact, be counterproductive: high iron concentrations are known to increase oxidative stress through the Fenton reaction54, and may, in fact, decrease the fitness of modA2OFF strains in some host environments. Several genes involved in anaerobic metabolism are upregulated in modA2::kan (Table 3). Dimethylsulfoxide reductase is a virulence factor in the Pasteurellaceae55; and phosphoenolpyruvate carboxykinase, which feeds phosphoenolpyruvate into gluconeogenesis to generate a pool of glucose phosphate, is key to the pathogenesis of Staphylococcus aureus56. Several gene-encoding enzymes involved in glycogen synthesis are also upregulated—glycogen is important for biofilm formation in Salmonella enteritidis57 and E. coli58. However, many of these genes were shown to be key when studying mutants in these systems: an all or nothing approach. The subtle changes resulting from modA2 phase variation (for example, phosphoenolpyruvate carboxykinase is only ~1.7-fold upregulated in modA2::kan; enzymes involved in glycogen synthesis are all ~1.5-fold upregulated in modA2::kan) appear to give no competitive advantage to modA2OFF compared with modA2ON when infecting the middle ear. Indeed, the changes in modA2ON are, in fact, preferentially selected. Whether this is due to the increased adhesion afforded by increased HMW-A proteins in modA2ON, the potential for increased oxidative stress through increased iron accumulation in modA2OFF leading to decreased fitness, some result of metabolic flux due to differences in the levels of respiratory and associated enzymes, or a combination of all these factors, remains to be elucidated. Moreover, as the NTHi 723 modA2ON strain was able to form more robust biofilms in vitro, this may also contribute to selection for the ON variant in the middle ear niche in vivo. Thus, selection against modA2OFF, and selection for modA2ON, may combine in vivo.
Recent work17,59,60 supports a key role for phase variation of individual factors in niche adaptation, independent of the established role of phase variation in immunoevasion. In this study, we have focused on the biphasic epigenetic switch that results in pleiotropic differential regulation of a phase-variable regulon of genes—the phasevarion. Previous studies have suggested a role for phasevarions in virulence6,7,9,10,16,41, but the impact of this novel biphasic epigenetic switch was untested in vivo. In this study, we observed for the first time a consistent selection for individuals that switched from modA2OFF to ON within the middle ear niche in the chinchilla model of OM. This in vivo data reveals a clear fitness advantage for modA2ON in this niche. Furthermore, we have shown that five phase-variable modA alleles predominate in clinical OM isolates and healthy carriers, suggesting a link between NTHi phasevarions and the potential to transmit and cause disease. All candidate phasevarions examined were shown to regulate multiple genes, including potential vaccine candidates. Defining the stable immunological target that NTHi represents requires a full analysis of the impact of phasevarions on NTHi gene expression, and future vaccine candidates will need to be assessed to confirm that their expression is not influenced by the epigenetic changes that result from phasevarion ON/OFF switching. Our recent studies describing a distinct, six-phase epigenetic switch in the major Gram-positive pathogen Streptococcus pneumoniae61 indicates that bacterial epigenetics is a key emerging field in bacterial pathogenesis and a new challenge to vaccine development for these important human pathogens.
Methods
Bacterial strains and cultures
NTHi strains 723, 477 and 1209 were received from the Finnish Otitis Media study group62. NTHi strain C486 was isolated from a child with otitis media63. Strain R2866 was isolated from a child with sepsis64. NTHi were routinely cultured in BHI broth (Oxoid) supplemented (sBHI) with hemin (1% v/v) and NAD (2 μg ml−1) or sBHI agar (as broth but with 1% w/v bacteriological agar; Oxoid). Liquid cultures were grown aerobically at 37 °C with shaking at 90 r.p.m. Plates were grown at 37 °C supplemented with 5% (v/v) CO2. E. coli DH5α (Coli Genetic Stock Centre, Yale University, USA) and BL21(DE3) (Merck Millipore) strains were grown at 37 °C in Luria-Bertani (LB) broth supplemented with ampicillin (100 μg ml−1) or kanamycin (50 μg ml−1) as required.
Molecular biology
All restriction endonucleases were purchased from New England Biolabs. Primers were purchased from Sigma-Aldrich and are detailed in Supplementary Table 5. PCR was carried out as recommended by the manufacturer's instructions (Promega; EMD Millipore, USA). Sequencing was carried out using Big Dye 3.1 (Perkin Elmer) and PCR products purified using the Qiagen PCR purification kit according to the manufacturer's instructions. Samples were sequenced by the Griffith University DNA sequencing facility (GUDSF), Brisbane, Australia, or the Australian Equine Genetics Research Facility, University of Queensland, Brisbane. Primers used for sequencing the HMW promoter region (HMW1-F; HMW2-F; HMW3-F and HMW-R; Supplementary Table 5) were based on those used previously37. The modA DRD region was analysed as previously described6. Briefly, PCR products encompassing the DRD were amplified using primers Him6 and Him11 (Supplementary Table 5), sequenced and compared with modA allele reference sequences described6,15. Natural ON and OFF strains were isolated by fragment length analysis of the modA repeat tract of multiple single colonies using the fluorescently labelled forward primer Him1F and the reverse primer Him3 (Supplementary Table 5)6, and fragments were analysed by AEGRC or GUDSF. Strains containing >90% ON or OFF were considered to be natural ON or OFF, respectively, and were used in subsequent studies.
PCR products generated for cloning into the pET51 Ek/LIC cloning vector (EMD Millipore) were prepared using KOD Hot-start DNA polymerase (EMD Millipore) using gene-specific primers for modA2 or siaB, (Supplementary Table 5) according to the manufacturer's instructions. Overexpression of ModA2 and SiaB was carried out using E. coli BL21 cells, which were induced by the addition of IPTG to a final concentration of 0.5 mM for 2 h at 37 °C with shaking at 120 r.p.m.
RNA extraction
Triplicate cultures of NTHi strains 723 modA2ON and 723 modA2::kan were grown to exponential phase (optical density at 600 nm = 0.3 to 0.4) in sBHI broth before RNA extraction. Growth rates of strain pairs used to make RNA for microarray comparison were equivalent, ensuring that the samples taken were in the same growth phase. Culture media for RNA preps were free of antibiotics. Approximately 100 μg of total RNA was prepared from each sample using the RNeasy Midi Kit according to the manufacturer's instructions (Qiagen). The triplicate samples were pooled and the integrity and concentration of RNA was determined via micro-fluidic analysis on a bio-analyser (Agilent Technologies).
Microarray analysis
All the microarray analyses were performed on H. influenzae genome arrays (TIGR; http://pfgrc.tigr.org/). Each microarray consists of 4,454 70-mer oligonucleotides representing ORFs from H. influenzae strains Rd KW20, 86-028NP, R2846 and R2866. Methods and analysis were as previously described65. Briefly, 5 μg of each total RNA sample was labelled using random hexamers and direct incorporation of fluorescently Cy3- or Cy5-labelled nucleotides as described66. Hybridizations were performed in triplicate and incorporated a dye swap to account for dye bias. After hybridization, arrays were washed and scanned on an Agilent G2565BA microaray scanner. Images of the hybridizations were analysed using Imagene 5.5 (BioDiscovery) and the mean foreground, mean background and spot/signal quality determined. Primary data were imported into an in-house installation of BASE (http://kidney.scgap.org/base). After print-tip intensity-independent Lowess normalization, differential expression was defined using a robust statistical method rather than simple fold change. All the genes were ranked using the B-statistic method where both fold change and variance of signals in replicates are used to determine the likelihood that genes are truly differentially expressed.
Quantitative real-time PCR
Oligonucleotides (Supplementary Table 5) were designed using Primer Express 1.0 software (ABI Prism; PE Biosystems). Real-time PCR reactions were performed in triplicate using RNA isolated from 723 modA2ON and 723 modA2OFF. cDNA was synthesized using NEB Protoscript II and random hexamers (Invitrogen; 50 ng μl−1) according to the manufacturer's instructions. Reverse transcriptase reactions lacking Protoscript II were performed as a negative control. All the real-time PCR reactions were performed in a 25-μl mixture containing a 1 in 5 dilution of the cDNA preparation (5 μl), 10×SYBR Green buffer (PE Applied Biosystems) and 2 μM of each primer. 16S RNA was used as the standard control in each quantitative PCR. Amplification and detection of specific products were performed with the ABI Prism 7700 sequence-detection system (PE Applied Biosystems) with the following cycle profile: 95 °C for 10 min, followed by 45 cycles of 95 °C for 15 s and 60 °C for 1 min. The data were analysed with ABI prism 7700 (version 1.7) analysis software. Relative gene expression between 723 modA2OFF and 723 modA2ON was determined using the 2–ΔΔCT relative quantification method.
SMRT sequencing and methylome analysis
Genomic DNA from each natural modAON and kanamycin knockout pair was prepared using the Qiagen genomic DNA midi kit according to the manufacturer's instructions. SMRT and methylome analysis was carried out as done previously21,22. Briefly, genomic DNA was sheared to an average length of ~ 10 kb using g-TUBEs (Covaris, Woburn, MA, USA) and SMRTbell template-sequencing libraries were prepared using sheared DNA. DNA was end repaired, then ligated to hairpin adaptors. Incompletely formed SMRTbell templates were degraded with a combination of Exonuclease III (New England Biolabs; Ipswich, MA, USA) and Exonuclease VII (USB; Cleveland, OH, USA). Primer was annealed and samples were sequenced on the PacBio RS II (Menlo Park, CA, USA) using standard protocols for long insert libraries. Plasmid midipreps from E. coli cells expressing NTHi 723 ModA2 and a negative control expressing a non-methylase (SiaB), were prepared using the Qiagen plasmid midi kit according to the manufacturer's instructions, and analysed as above.
Preparation of OMPs from NTHi
NTHi modA ON/OFF pairs were grown in sBHI broth (50 ml) at 37 °C overnight with shaking at 100 r.p.m. The cells were pelleted at 4,500 r.p.m. for 15 min at 4 °C, resuspended in 4 ml 10 mM HEPES-NaOH pH7.5 and OMPs were prepared as detailed previously67. Briefly, cells were lysed by sonication, and debris pelleted as above. Sarcosyl was added to the clarified supernatant to a final concentration of 1% and incubated at 25 °C for 30 min. Supernatants were then centrifuged at 110,000g for 90 min. Pellets were resuspended in 10 mM HEPES-NaOH pH7.5, sarcosyl added to a final concentration of 1%, and incubation and centrifugation steps repeated twice more. Final pellets containing the OMP-enriched fraction were resuspended in 100 μl of 10 mM HEPES-NaOH pH7.5 and the protein concentration quantified using the BCA protein assay kit according to the manufacturer's instructions (Thermo Scientific).
Western blot analysis
Each of the OMP preparations (5 μg) were run on the Novex Bis-Tris pre-cast gel system with MOPS running buffer according to the manufacturer's instructions (Life Technologies). Ammoniacal silver staining was carried out to visualize proteins. Western blotting was carried out using nitro-cellulose membranes (Bio-Rad) and standard protocols68. All mouse (AD6 anti-HMW1/2A34; 1F4 anti-Hia35) and rabbit (LB1 anti-OMP P5 (ref. 28); chimV4 anti-OMP P5; anti-OMP P5) primary antibodies were used at a dilution of 1:2,500; all chinchilla (anti-OMP P2; anti-LPD27; anti-PDM27; anti-OMP P5/P6) primary antibodies at a dilution of 1:250. Anti-mouse-AP and anti-rabbit-AP secondary antibodies were used at a dilution of 1:5,000 (Sigma-Aldrich); protein A-AP secondary antibody was used at a dilution of 1:500 (Sigma-Aldrich) in blots where chinchilla primary antibodies were used. Blots were developed using SigmaFAST NBT/BCIP tablets according to the manufacturer's instructions (Sigma-Aldrich). All the primary antibodies were raised by the authors laboratories, with specific references for those described previously.
ELISA assay
ELISA assays were carried out using standard protocols68 in 96-well Maxisorb plates (NUNC; Thermo Scientific). Cells were diluted to a 0D600 of 0.2 (2 × 108 c.f.u. ml−1), with 50 μl added per well. All the strains were assayed in triplicate. Primary antibody AD6 against HMW34 was used at a starting concentration of 1:200 (NTHi strains C486 and 477) or 1:10,000 (NTHi strain 723) and serially diluted two-fold in 1 × PBS pH 7.9. Secondary antibody (goat anti-mouse HRP conjugate; Sigma-Aldrich) was used at a concentration of 1:10,000. Antibody was detected using TMB single-substrate solution as recommended by the manufacturer (Sigma-Aldrich). The data were plotted as antibody dilution (x axis) versus absorbance at 450 nm (y axis), and data from specific titres in the linear range of the response curve picked for statistical analysis.
iTRAQ analysis
iTRAQ 1D nanoLC ESI MS/MS was carried out by the Australian Proteome Analysis Facility (APAF), Macquarie University, Sydney, Australia. Approximately 10 μg of OMP preparation from duplicate samples of each modA ON/OFF pair was supplied for the analysis. Samples were buffer exchanged into 0.25 M TEAB and 0.05% SDS and quantified samples were reduced with TCEP, alkylated with MMTS and digested with trypsin. Digested samples were labelled, passed through an SDS removal column (Thermo Fisher) and dried. Labelled samples were resuspended in 50 μl of loading/desalting solution (0.1% formic acid and 2% acetonitrile 97.9% water). Sample (20 μl) was injected onto a peptide trap (Michrome peptide Captrap) for pre-concentration and desalted with 0.1% formic acid, 2% acetonitrile. Peptides were eluted from the column using a linear solvent gradient, with steps, from mobile phase A: mobile phase B (98:2) to mobile phase A: mobile phase B (65:35) where mobile phase A is 0.1% formic acid and mobile phase B is 90% ACN/0.1% formic acid at 600 nl min−1 over a 100-min period. After peptide elution, the column was cleaned with 95% buffer B for 15 min and then equilibrated with buffer A for 25 min before next sample injection. The reverse phase nanoLC eluent was subject to positive ion nanoflow electrospray analysis in an information dependent acquisition mode (IDA). In IDA mode, a TOFMS survey scan was acquired (m/z 400–1,500, 0.25 s), with the 10 most intense multiply charged ions (counts > 150) in the survey scan sequentially subjected to MS/MS analysis. MS/MS spectra were accumulated for 200 ms in the mass range m/z 100–1,500 with the total cycle time of 2.3 s. The experimental nanoLC ESI ms/ms data were submitted to ProteinPilot V4.2b (AB Sciex) for data processing.
MIC assay
The MIC was measured by broth microdilution in triplicate experiments as described previously69 using mid-log phase NTHi cells grown aerobically in sBHI. Briefly, 50 μl of each culture was added to 96-well plates in which antibiotics had been serially diluted, and plates grown at 37 °C with 5% CO2 for 24 h. The MIC (mg l−1) was determined as the last dilution at which turbidity was observed following overnight growth. All the assays were performed in triplicate.
Opsonophagocytic killing assays
The growth conditions of the bacteria, the growth and differentiation of the HL-60 cells, (ATCC CCL-240) and the opsonophagocytic assay itself were performed as described previously33,42. The opsonophagocytic assay was performed in 5-ml capped polystyrene tubes (Sarstedt, Newton, NC). The complement source was human serum collected from a single healthy adult that was adsorbed to remove serum IgG by passing aliquots repeatedly over a protein G affinity column at 4 °C. Antibody (guinea pig anti-strain 12 HMW1/HMW2 antiserum42) was serially diluted, and ~ 5 × 103 c.f.u. mid-log phase bacterial cells were added to each dilution, and tubes incubated at 37 °C with 5% CO2 for 15 min. Following this, complement was added, followed by immediate addition of differentiated HL-60 cells. Tubes were incubated at 37 °C with 200 r.p.m. horizontal shaking. At the end of the 90-min incubation period, c.f.u. were calculated by plating 10 μl of each culture and incubating overnight at 37 °C with 5% CO2. The per cent killing at each serum dilution was calculated by determining the ratio of the bacterial colony count at each dilution to that of the complement control using the modA4 ON/OFF pair (NTHi strain C486). Pre-immune sera and anti-Hia sera were used as negative controls.
Biofilm formation on chambered coverglass slides
Formation of NTHI biofilms was performed in eight-well-chambered coverglass slides (Thermo Scientific, Waltham, MA) as described previously70. Briefly, mid-log phase cultures of NTHi strain 723 modA2ON and modA2OFF grown in sBHI were diluted with fresh pre-warmed media and used to inoculate 4 × 104 c.f.u. in 200 μl total volume per well. Slides were incubated at 37 °C with 5% atmospheric CO2 and the growth medium was replaced with fresh medium after 16 h. Twenty-four hours after seeding, biofilms were stained with LIVE/DEAD BacLight stain (Life Technologies) and fixed overnight in fixative (1.6% paraformaldehyde, 2.5% glutaraldehyde, 4% acetic acid in 0.1 M phosphate buffer, pH 7.4). Fixative was replaced with saline before imaging on a Zeiss 510 Meta-laser scanning confocal microscope; images were rendered with Zeiss Zen software.
Chinchilla model of NTHi-induced OM
Adult chinchillas (Chinchilla lanigera) were supplied by Rauscher's Chinchilla Ranch, LaRue, OH. Chinchillas were not sex-differentiated, and were classed as adult if weighing between 500–700 grams. Chinchillas were allowed to acclimate in the vivarium for 7–10 days before beginning the study. In two separate studies, cohorts of five chinchillas each were established, then challenged intranasally and transbullarly with either: (1) NTHi strain 723 modA2(22)ON or (2) NTHi strain 723 modA2(24)OFF at a challenge dose of ~ 1 × 108 c.f.u. intranasally and 750 c.f.u. transbullarly. A total of 10 animals (20 ears) were challenged with each of these variants of NTHI strain 723. The animals were then monitored daily via video otoscopy for 22 days. Delivered doses were confirmed by dilution plate counts of inocula on chocolate agar. On days 2, 4, 7, 10, 14, 18 and 22 after challenge epitympanic taps (removal of a small aliquot of fluid from the middle ear space) were performed as described previously. On collection of samples, 10 μl were placed in a sterile Eppendorf tube on ice. This aliquot was used for evaluation of the colony-forming unit of NTHi per ml sample. The remainder of the collected samples was immediately centrifuged at 4 °C for 3 min, the supernatant was carefully removed and the pellet was snap frozen in liquid nitrogen and stored at − 80 °C. On day 22 (unless removed from the study early owing to morbidity), anaesthetized chinchillas were euthanized and the bullae were dissected from the skull. The left bulla from each chinchilla was aseptically opened and imaged. The middle ear mucosa, including any adherent mucosal biofilm was removed, homogenized in sterile PBS and snap frozen.
With regard to animal treatment and handling, all protocols were approved by the Nationwide Children's Hospital Animal Care and Use Committee, in accordance with the US Department of Health and Human Services Guide for the Care and Use of Laboratory Animals.
Fragment analysis to determine modA2 ON/OFF status in chinchilla samples
Fluid samples were taken from the left and right middle ears on days 0, 2, 4, 7, 10, 14, 18 and 22 from each cohort of 10 chinchillas that had been challenged with either strain 723 modA2(22)ON or strain 723 modA2(24)OFF. The ratio of modA2ON and modA2OFF from the starting inoculum (day 0) compared with days 4, 7, 10, 14, 18 and 22 was verified via fragment analysis for each of the chinchillas. Samples were thawed briefly on ice, with 1 μl serving as the template in a 25 μl GoTaq PCR reaction (Promega) using primers Him1F and Him3 (Supplementary Table 5) as described previously6. Samples were run using GeneScan fluorescent-PCR fragment-length analysis by GUDSF, and analysed using Peak Scanner software (Applied Biosystems).
Supplementary Material
Acknowledgements
Work was supported by NHMRC (Australia) Project Grant 1034401 to M.P.J. and L.O.B., NHMRC Program Grant 565526 to A.G.M. and M.P.J., NHMRC Program Grant to M.P.J. 1071659, Grant NIH/NIDCD (USA) R01DC003915 to L.O.B., and grant NIH/NIAID (USA) R01AI 81887 to S.J.B. iTRAQ analysis was undertaken at APAF, the infrastructure provided by the Australian Government through the National Collaborative Research Infrastructure Strategy (NCRIS).
Footnotes
Author contributions
L.O.B. and M.P.J. conceived the project; J.M.A., L.O.B. and M.P.J. designed the experiments; J.M.A., Y.N.S., K.L.F., M.B., T.A.C., J.A.J., K.L.B., F.E.-C.J., P.M.P. and S.J.B. performed the experiments; J.M.A. and Y.N.S. analysed the data; A.G.M., S.J.B. S.M.G., J.K. and A.L.S. provided advice; J.M.A., L.O.B. and M.P.J. wrote the paper.
Accession codes. The SMRT methylome data for strains 723, C486, 477, 1209 and R2866 have been deposited in REBASE. Annotated genomic sequences for strains 723, C486, 477 and 1209 have been deposited in NCBI GenBank with accession codes CP007472 (strain 723), CP007471 (strain C486), CP007470 (strain 477) and JMQP01000000 (strain 1209). iTRAQ proteomic data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD002210. The microarray data have been deposited in the NCBI Gene Expression Omnibus database with accession codes GSE69831.
Supplementary Information accompanies this paper at http://www.nature.com/naturecommunications
Competing financial interests: T.A.C., M.B. and J.K. are full-time employees of Pacific Biosciences, a company commercializing the use of single molecule, real-time (SMRT) sequencing. The remaining authors declare no competing financial interests.
How to cite this article: Atack, J. M. et al. A biphasic epigenetic switch controls immunoevasion, virulence and niche adaptation in non-typeable Haemophilus influenzae. Nat. Commun. 6:7828 doi: 10.1038/ncomms8828 (2015).
References
- 1.Haggard M. Otitis media: prospects for prevention. Vaccine. 2008;26(Suppl 7):G20–G24. doi: 10.1016/j.vaccine.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 2.Bayliss CD. Determinants of phase variation rate and the fitness implications of differing rates for bacterial pathogens and commensals. FEMS Microbiol. Rev. 2009;33:504–520. doi: 10.1111/j.1574-6976.2009.00162.x. [DOI] [PubMed] [Google Scholar]
- 3.Moxon R, Bayliss C, Hood D. Bacterial contingency loci: the role of simple sequence DNA repeats in bacterial adaptation. Annu. Rev. Genet. 2006;40:307–333. doi: 10.1146/annurev.genet.40.110405.090442. [DOI] [PubMed] [Google Scholar]
- 4.Power PM, et al. Simple sequence repeats in Haemophilus influenzae. Infect. Genet. Evol. 2009;9:216–228. doi: 10.1016/j.meegid.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saunders NJ, et al. Repeat-associated phase variable genes in the complete genome sequence of Neisseria meningitidis strain MC58. Mol. Microbiol. 2000;37:207–215. doi: 10.1046/j.1365-2958.2000.02000.x. [DOI] [PubMed] [Google Scholar]
- 6.Fox KL, et al. Haemophilus influenzae phasevarions have evolved from type III DNA restriction systems into epigenetic regulators of gene expression. Nucleic Acids Res. 2007;35:5242–5252. doi: 10.1093/nar/gkm571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Srikhanta YN, et al. Phasevarions mediate random switching of gene expression in pathogenic Neisseria. PLoS Pathog. 2009;5:e1000400. doi: 10.1371/journal.ppat.1000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Srikhanta YN, Fox KL, Jennings MP. The phasevarion: phase variation of type III DNA methyltransferases controls coordinated switching in multiple genes. Nat. Rev. Microbiol. 2010;8:196–206. doi: 10.1038/nrmicro2283. [DOI] [PubMed] [Google Scholar]
- 9.Srikhanta YN, et al. Phasevarion mediated epigenetic gene regulation in Helicobacter pylori. PLoS ONE. 2011;6:e27569. doi: 10.1371/journal.pone.0027569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Srikhanta YN, Maguire TL, Stacey KJ, Grimmond SM, Jennings MP. The phasevarion: A genetic system controlling coordinated, random switching of expression of multiple genes. Proc. Natl Acad. Sci. USA. 2005;102:5547–5551. doi: 10.1073/pnas.0501169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Humbelin M, et al. Type III DNA restriction and modification systems EcoP1 and EcoP15. Nucleotide sequence of the EcoP1 operon, the EcoP15 mod gene and some EcoP1 mod mutants. J. Mol. Biol. 1988;200:23–29. doi: 10.1016/0022-2836(88)90330-0. [DOI] [PubMed] [Google Scholar]
- 12.Saha S, Ahmad I, Reddy YV, Krishnamurthy V, Rao DN. Functional analysis of conserved motifs in type III restriction-modification enzymes. Biol. Chem. 1998;379:511–517. doi: 10.1515/bchm.1998.379.4-5.511. [DOI] [PubMed] [Google Scholar]
- 13.Timinskas A, Butkus V, Janulaitis A. Sequence motifs characteristic for DNA [cytosine-N4] and DNA [adenine-N6] methyltransferases. Classification of all DNA methyltransferases. Gene. 1995;157:3–11. doi: 10.1016/0378-1119(94)00783-o. [DOI] [PubMed] [Google Scholar]
- 14.Blakeway LV, et al. ModM DNA methyltransferase methylome analysis reveals a potential role for Moraxella catarrhalis phasevarions in otitis media. FASEB J. 2014;28:5197–5207. doi: 10.1096/fj.14-256578. [DOI] [PubMed] [Google Scholar]
- 15.Gawthorne JA, Beatson SA, Srikhanta YN, Fox KL, Jennings MP. Origin of the diversity in DNA recognition domains in phasevarion associated modA genes of pathogenic Neisseria and Haemophilus influenzae. PLoS ONE. 2012;7:e32337. doi: 10.1371/journal.pone.0032337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seib KL, et al. Specificity of the ModA11, ModA12 and ModD1 epigenetic regulator N6-adenine DNA methyltransferases of Neisseria meningitidis. Nucleic Acids Res. 2015;43:4150–4162. doi: 10.1093/nar/gkv219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fox KL, et al. Selection for phase variation of LOS biosynthetic genes frequently occurs in progression of non-typeable Haemophilus influenzae infection from the nasopharynx to the middle ear of human patients. PLoS ONE. 2014;9:e90505. doi: 10.1371/journal.pone.0090505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Novotny LA, et al. Epitope mapping immunodominant regions of the PilA protein of nontypeable Haemophilus influenzae (NTHI) to facilitate the design of two novel chimeric vaccine candidates. Vaccine. 2009;28:279–289. doi: 10.1016/j.vaccine.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harabuchi Y, et al. Nasopharyngeal colonization with nontypeable Haemophilus influenzae and recurrent otitis media. Tonawanda/Williamsville Pediatrics. J. Infect. Dis. 1994;170:862–866. doi: 10.1093/infdis/170.4.862. [DOI] [PubMed] [Google Scholar]
- 20.Faden H, et al. Relationship between nasopharyngeal colonization and the development of otitis media in children. Tonawanda/Williamsville Pediatrics. J. Infect. Dis. 1997;175:1440–1445. doi: 10.1086/516477. [DOI] [PubMed] [Google Scholar]
- 21.Murray IA, et al. The methylomes of six bacteria. Nucleic Acids Res. 2012;40:11450–11462. doi: 10.1093/nar/gks891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clark TA, et al. Characterization of DNA methyltransferase specificities using single-molecule, real-time DNA sequencing. Nucleic Acids Res. 2012;40:e29. doi: 10.1093/nar/gkr1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyer HW. DNA restriction and modification mechanisms in bacteria. Ann. Rev. Microbiol. 1971;25:153–176. doi: 10.1146/annurev.mi.25.100171.001101. [DOI] [PubMed] [Google Scholar]
- 24.Hadi SM, Bachi B, Iida S, Bickle TA. DNA restriction--modification enzymes of phage P1 and plasmid p15B. Subunit functions and structural homologies. J. Mol. Biol. 1983;165:19–34. doi: 10.1016/s0022-2836(83)80240-x. [DOI] [PubMed] [Google Scholar]
- 25.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE--a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2010;38:D234–D236. doi: 10.1093/nar/gkp874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy TF, Bartos LC. Human bactericidal antibody response to outer membrane protein P2 of nontypeable Haemophilus influenzae. Infect. Immun. 1988;56:2673–2679. doi: 10.1128/iai.56.10.2673-2679.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bakaletz LO, et al. Protection against development of otitis media induced by nontypeable Haemophilus influenzae by both active and passive immunization in a chinchilla model of virus-bacterium superinfection. Infect. Immun. 1999;67:2746–2762. doi: 10.1128/iai.67.6.2746-2762.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bakaletz LO, Leake ER, Billy JM, Kaumaya PT. Relative immunogenicity and efficacy of two synthetic chimeric peptides of fimbrin as vaccinogens against nasopharyngeal colonization by nontypeable Haemophilus influenzae in the chinchilla. Vaccine. 1997;15:955–961. doi: 10.1016/s0264-410x(96)00298-8. [DOI] [PubMed] [Google Scholar]
- 29.Berenson CS, Murphy TF, Wrona CT, Sethi S. Outer membrane protein P6 of nontypeable Haemophilus influenzae is a potent and selective inducer of human macrophage proinflammatory cytokines. Infect. Immun. 2005;73:2728–2735. doi: 10.1128/IAI.73.5.2728-2735.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy TF, et al. Identification of a 16,600-dalton outer membrane protein on nontypeable Haemophilus influenzae as a target for human serum bactericidal antibody. J. Clin. Invest. 1986;78:1020–1027. doi: 10.1172/JCI112656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akkoyunlu M, Janson H, Ruan M, Forsgren A. Biological activity of serum antibodies to a nonacylated form of lipoprotein D of Haemophilus influenzae. Infect. Immun. 1996;64:4586–4592. doi: 10.1128/iai.64.11.4586-4592.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akkoyunlu M, Melhus A, Capiau C, van Opstal O, Forsgren A. The acylated form of protein D of Haemophilus influenzae is more immunogenic than the nonacylated form and elicits an adjuvant effect when it is used as a carrier conjugated to polyribosyl ribitol phosphate. Infect. Immun. 1997;65:5010–5016. doi: 10.1128/iai.65.12.5010-5016.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winter LE, Barenkamp SJ. Human antibodies specific for the high-molecular-weight adhesion proteins of nontypeable Haemophilus influenzae mediate opsonophagocytic activity. Infect. Immun. 2003;71:6884–6891. doi: 10.1128/IAI.71.12.6884-6891.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winter LE, Barenkamp SJ. Antibodies specific for the high-molecular-weight adhesion proteins of nontypeable Haemophilus influenzae are opsonophagocytic for both homologous and heterologous strains. Clin. Vaccine Immunol. 2006;13:1333–1342. doi: 10.1128/CVI.00221-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winter LE, Barenkamp SJ. Antibodies specific for the Hia adhesion proteins of nontypeable Haemophilus influenzae mediate opsonophagocytic activity. Clin. Vaccine Immunol. 2009;16:1040–1046. doi: 10.1128/CVI.00090-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Novotny LA, Clements JD, Bakaletz LO. Kinetic analysis and evaluation of the mechanisms involved in the resolution of experimental nontypeable Haemophilus influenzae-induced otitis media after transcutaneous immunization. Vaccine. 2012;31:3417–3426. doi: 10.1016/j.vaccine.2012.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dawid S, Barenkamp SJ, St. Geme JW. Variation in expression of the Haemophilus influenzae HMW adhesins: A prokaryotic system reminiscent of eukaryotes. Proc. Natl Acad. Sci. USA. 1999;96:1077–1082. doi: 10.1073/pnas.96.3.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Atack JM, et al. Selection and counter-selection of Hia expression reveals a key role for phase-variable expression of this adhesin in infection caused by non-typeable Haemophilus influenzae. J. Infect. Dis. 2015 doi: 10.1093/infdis/jiv103. doi:10.1093/infdis/jiv103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vizcaino JA, et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 2014;32:223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.St Geme JW, Grass S. Secretion of the Haemophilus influenzae HMW1 and HMW2 adhesins involves a periplasmic intermediate and requires the HMWB and HMWC proteins. Mol. Microbiol. 1998;27:617–630. doi: 10.1046/j.1365-2958.1998.00711.x. [DOI] [PubMed] [Google Scholar]
- 41.Jen FE, Seib KL, Jennings MP. Phasevarions mediate epigenetic regulation of antimicrobial susceptibility in Neisseria meningitidis. Antimicrob. Agents Chemother. 2014;58:4219–4221. doi: 10.1128/AAC.00004-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winter LE, Barenkamp SJ. Antibodies to the HMW1/HMW2 and Hia adhesins of nontypeable Haemophilus influenzae mediate broad-based opsonophagocytic killing of homologous and heterologous strains. Clin. Vaccine Immunol. 2014;21:613–621. doi: 10.1128/CVI.00772-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanders JD, Cope LD, Hansen EJ. Identification of a locus involved in the utilization of iron by Haemophilus influenzae. Infect. Immun. 1994;62:4515–4525. doi: 10.1128/iai.62.10.4515-4525.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seale TW, et al. Complex role of hemoglobin and hemoglobin-haptoglobin binding proteins in Haemophilus influenzae virulence in the infant rat model of invasive infection. Infect. Immun. 2006;74:6213–6225. doi: 10.1128/IAI.00744-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harkness RE, Chong P, Klein MH. Identification of two iron-repressed periplasmic proteins in Haemophilus influenzae. J. Bacteriol. 1992;174:2425–2430. doi: 10.1128/jb.174.8.2425-2430.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bearden SW, Perry RD. The Yfe system of Yersinia pestis transports iron and manganese and is required for full virulence of plague. Mol. Microbiol. 1999;32:403–414. doi: 10.1046/j.1365-2958.1999.01360.x. [DOI] [PubMed] [Google Scholar]
- 47.Atack JM, et al. Characterization of an ntrX mutant of Neisseria gonorrhoeae reveals a response regulator that controls expression of respiratory enzymes in oxidase-positive proteobacteria. J. Bacteriol. 2013;195:2632–2641. doi: 10.1128/JB.02062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Su S, Hassett DJ. Anaerobic Pseudomonas aeruginosa and other obligately anaerobic bacterial biofilms growing in the thick airway mucus of chronically infected cystic fibrosis patients: an emerging paradigm or ‘Old Hat’? Expert Opin. Ther. Targets. 2012;16:859–873. doi: 10.1517/14728222.2012.708025. [DOI] [PubMed] [Google Scholar]
- 49.Bakaletz LO. Chinchilla as a robust, reproducible and polymicrobial model of otitis media and its prevention. Expert Rev. Vaccines. 2009;8:1063–1082. doi: 10.1586/erv.09.63. [DOI] [PubMed] [Google Scholar]
- 50.Pelton SI, et al. Panel 6: Vaccines. Otolaryngol. Head Neck Surg. 2013;148:E90–E101. doi: 10.1177/0194599812466535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barenkamp SJ, St Geme JW. Genes encoding high-molecular-weight adhesion proteins of nontypeable Haemophilus influenzae are part of gene clusters. Infect. Immun. 1994;62:3320–3328. doi: 10.1128/iai.62.8.3320-3328.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cholon DM, et al. Serial Isolates of persistent Haemophilus influenzae in patients with chronic obstructive pulmonary disease express diminishing quantities of the HMW1 and HMW2 adhesins. Infect. Immun. 2008;76:4463–4468. doi: 10.1128/IAI.00499-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whitby PW, Sim KE, Morton DJ, Patel JA, Stull TL. Transcription of genes encoding iron and heme acquisition proteins of Haemophilus influenzae during acute otitis media. Infect. Immun. 1997;65:4696–4700. doi: 10.1128/iai.65.11.4696-4700.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Halliwell B, Gutteridge JM. Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem. J. 1984;219:1–14. doi: 10.1042/bj2190001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baltes N, Hennig-Pauka I, Jacobsen I, Gruber AD, Gerlach GF. Identification of dimethyl sulfoxide reductase in Actinobacillus pleuropneumoniae and its role in infection. Infect. Immun. 2003;71:6784–6792. doi: 10.1128/IAI.71.12.6784-6792.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chaffin DO, Taylor D, Skerrett SJ, Rubens CE. Changes in the Staphylococcus aureus transcriptome during early adaptation to the lung. PLoS ONE. 2012;7:e41329. doi: 10.1371/journal.pone.0041329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bonafonte MA, et al. The relationship between glycogen synthesis, biofilm formation and virulence in Salmonella enteritidis. FEMS Microbiol. Lett. 2000;191:31–36. doi: 10.1111/j.1574-6968.2000.tb09315.x. [DOI] [PubMed] [Google Scholar]
- 58.Jackson DW, et al. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J. Bacteriol. 2002;184:290–301. doi: 10.1128/JB.184.1.290-301.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Poole J, et al. Analysis of nontypeable Haemophilus influenzae phase variable genes during experimental human nasopharyngeal colonization. J. Infect. Dis. 2013;208:720–727. doi: 10.1093/infdis/jit240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsao D, Nelson KL, Kim D, Smith AL. Infant rat infection modifies phenotypic properties of an invasive nontypeable Haemophilus influenzae. Microbes Infect. 2012;14:509–516. doi: 10.1016/j.micinf.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Manso AS, et al. A random six-phase switch regulates pneumococcal virulence via global epigenetic changes. Nat. Commun. 2014;5:5055. doi: 10.1038/ncomms6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meats E, et al. Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J. Clin. Microbiol. 2003;41:1623–1636. doi: 10.1128/JCM.41.4.1623-1636.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cody AJ, et al. High rates of recombination in otitis media isolates of non-typeable Haemophilus influenzae. Infect. Genet. Evol. 2003;3:57–66. doi: 10.1016/s1567-1348(02)00152-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nizet V, Colina KF, Almquist JR, Rubens CE, Smith AL. A virulent nonencapsulated Haemophilus influenzae. J. Infect. Dis. 1996;173:180–186. doi: 10.1093/infdis/173.1.180. [DOI] [PubMed] [Google Scholar]
- 65.Seib KL, et al. Characterization of the OxyR regulon of Neisseria gonorrhoeae. Mol. Microbiol. 2007;63:54–68. doi: 10.1111/j.1365-2958.2006.05478.x. [DOI] [PubMed] [Google Scholar]
- 66.Grimmond S, et al. Sexually dimorphic expression of protease nexin-1 and vanin-1 in the developing mouse gonad prior to overt differentiation suggests a role in mammalian sexual development. Hum. Mol. Genet. 2000;9:1553–1560. doi: 10.1093/hmg/9.10.1553. [DOI] [PubMed] [Google Scholar]
- 67.Murphy TF, Dudas KC, Mylotte JM, Apicella MA. A subtyping system for nontypable Haemophilus influenzae based on outer-membrane proteins. J. Infect. Dis. 1983;147:838–846. doi: 10.1093/infdis/147.5.838. [DOI] [PubMed] [Google Scholar]
- 68.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A laboratory Manual. 2nd edn Cold Spring Harbour Laboratory Press; 1989. [Google Scholar]
- 69.Peak IR, Jennings CD, Jen FE, Jennings MP. Role of Neisseria meningitidis PorA and PorB expression in antimicrobial susceptibility. Antimicrob. Agents Chemother. 2014;58:614–616. doi: 10.1128/AAC.02506-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jurcisek JA, Dickson AC, Bruggeman ME, Bakaletz LO. In vitro biofilm formation in an 8-well chamber slide. J. Vis. Exp. 2011:2481. doi: 10.3791/2481. doi:10.3791/2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.