Abstract
Histone methyltransferases and demethylases epigenetically regulate gene expression by modifying histone methylation status in numerous cellular processes, including cell differentiation and proliferation. These modifiers also control methylation levels of various non-histone proteins, such as effector proteins that play critical roles in cellular signaling networks. Dysregulated histone methylation modifiers alter expression of oncogenes and tumor suppressor genes and change methylation states of effector proteins, frequently resulting in aberrant cellular signaling cascades and cellular transformation. In this review, we summarize the role of histone methylation modifiers in regulating the following signaling pathways: NF-κB, RAS/RAF/MEK/MAPK, PI3K/Akt, Wnt/β-catenin, p53, and ERα.
Keywords: Histone methylation, histone methyltransferase, histone demethylase, oncogenic signaling, tumor suppressor pathway
Introduction
Chromatin, a complex of eukaryotic DNA and multiple proteins, serves as the cellular information center, receiving and sending various signals during numerous cellular processes. Cellular signaling events are coupled with covalent and non-covalent modifications of chromatin. Chromatin modifications regulate chromatin architecture and gene expression by affecting multiple interactions among DNA, histones, and chromatin-binding proteins. Of the chromatin modifications, histone methylation has emerged as a key epigenetic mark that regulates gene expression. Histone methylation is modulated by histone methyltransferases and demethylases. Notably, these methylation modifiers also modulate methylation states of many non-histone proteins, including key effectors and components in cellular signaling pathways. In cancer, certain histone methylation modifiers are frequently dysregulated, and this dysregulation is linked to aberrations in gene expression and cellular signaling cascades. This review focuses on how cellular signaling pathways are regulated by histone methylation modifiers.
Histone methylation and its modifiers
Histone methylation
Histone methylation occurs predominantly on two highly abundant histone residues, lysine (K) and arginine (R), although methylation can take place on other amino acids, including histidine, aspartic acid, and glutamic acid, and on the carboxyl groups of proteins [1–4]. Unlike acetylation and phosphorylation, methylation marks do not alter the charge of histones but serve as docking sites for specific binding proteins called histone readers [5]. Histone methylation, together with other modifications, also can be recognized by combinatory binding modules in histone readers that ultimately affect chromatin architecture and regulate gene expression [6].
Histone lysine methylation occurs at three different levels: mono-, di-, and tri-methylation. This modification is highly conserved across different species, from unicellular organisms to mammals [7], and is linked to either gene activation or repression depending on the target site. For example, methylation at histone H3 lysine 4 (H3K4), H3K36, and H3K79 is generally related to gene activation, whereas that at H3K9, H3K27, and H4K20 is commonly linked to gene silencing. Apart from histones, this modification also occurs in other cellular proteins. Lysine methylation is associated with multiple cellular processes, including cellular signaling pathways, cell fate determination, terminal differentiation, X inactivation, and the spatiotemporal patterning of Hox genes [8–15].
Like lysine methylation, arginine methylation occurs on both histones and non-histone proteins. It takes place at a guanidino nitrogen of arginine [16–18]. Arginine residues can be methylated mainly in three different ways: ω-NG-monomethyl arginine (MMA); ω-NG, NG-asymmetric dimethyl arginine (ADMA); and ω-NG,N′G-symmetric dimethyl arginine (SDMA). None of these methyl groups, when added to an arginine residue, change its positive charge, but they may affect the protein-protein interaction by eliminating formation of a potential hydrogen bond and changing the bulkiness of arginine side chain [17,19,20]. Arginine methylation regulates a number of different cellular processes, including cellular signaling, transcriptional regulation, RNA metabolism, and DNA damage repair [21].
Histone methylation modifiers
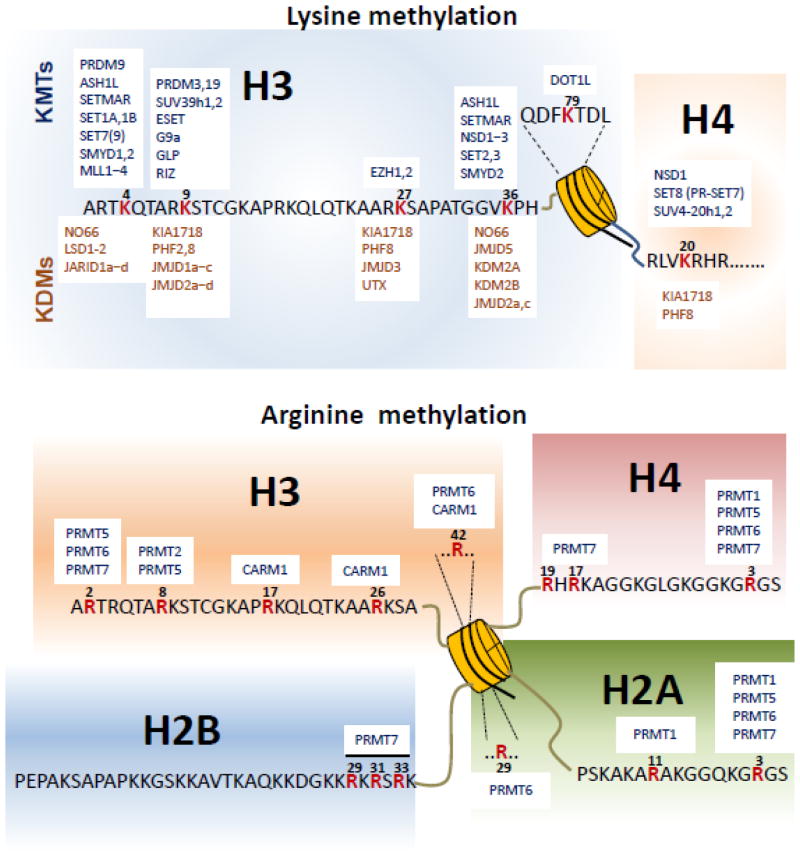
Histone methylation at individual lysine residues is catalyzed by specific lysine methyltransferases (KMTs) and can be removed by specific lysine demethylases (KDMs). SUV39H1 was the first histone KMT identified, and it methylates H3K9 [22]. Since then, numerous KMTs have been identified; they can be divided into two classes on the basis of their conserved catalytic domains. One class contains a highly conserved SET [Su(var)3–9, Enhancer of Zeste, and Trithorax] domain [23]. The other class does not have a SET domain but consists of highly conserved proteins yeast DOT1 (disruptor of telomeric silencing-1; also known as KMT4) and its eukaryotic homologs, such as human and mouse DOT1L (DOT1-Like) [24]. SET-containing KMTs generally methylate lysines within the histone N-terminal tails, whereas DOT1 and DOT1L methylate H3K79 within the histone globular core [25,26] (Figure 1).
Figure 1. Histone methylation and its modifiers.
Histone lysine methyltransferases (KMTs) and lysine demethylases (KDMs) for six major lysine methylation sites in histones are aligned for their cognate sites (top panel). Arginine methylation sites and their corresponding protein arginine methyltransferases (PRMTs) are also depicted (bottom panel).
The protein arginine N-methyltransferases (PRMTs) catalyze addition of the methyl groups to the arginine residues. PRMTs are classified as type I, type II, type III, or type IV enzymes. Types I, II, and III catalyze methylation on the terminal (i.e., ω) guanidino nitrogen atom. Although types I and II both generate an MMA intermediate, type I PRMTs (PRMT1, 2, 3, 6, and 8 and PRMT4, also known as co-activator–associated arginine methyltransferase 1 [CARM1]) further modify this intermediate to ADMA, whereas type II PRMTs (PRMT5 and 9) catalyze the generation of SDMA [20,27,28]. PRMT7 appears to exhibit type III enzymatic activity to catalyze the formation of MMA. However, PRMT7 was also reported to generate SDMA in vitro and in cells, although this activity may be indirect [29–32]. The type IV RMT2 catalyzes monomethylation of the internal (i.e., δ) guanidino nitrogen atom. Most of the PRMTs are known to methylate glycine- and arginine-rich (GAR) motifs in their substrates [33]. In contrast, PRMT4 methylates arginine residues in proline-, glycine-, and methionine-rich (PGM) motifs [34]. Interestingly, PRMT5 can symmetrically dimethylate arginine residues in both GAR and PGM motifs [35]. Like KMTs, PRMTs methylate both histones (Figure 1) and several non-histone proteins [20,36,37].
Histone methylation was once considered a stable and static modification. However, it has been shown that the lysine-specific demethylase 1 (LSD1; also known as KDM1A) removes methyl groups from H3K4me1/2 by utilizing FAD as a co-factor [38]. LSD1 requires Co-REST to demethylate H3K4me1/2 on nucleosomal substrates [39]. Interestingly, it was reported that LSD1 in the presence of androgen receptor may demethylate H3K9me1/2 [40]. Later, JHDM1A, a Jumonji C (JmjC) domain–containing protein, was identified as a demethylase that removes methyl groups from H3K36me1/2 [41,42]. Since then, numerous JmjC-domain-containing histone lysine demethylases, including trimethylated lysine demethylases, have been identified (Figure 1) [40,42–44]. This family of demethylases requires Fe (II) and α-ketoglutarate as cofactors and exhibits a high specificity for target lysine residues. Interestingly, some demethylases demethylate di- and monomethylated but not trimethylated lysines, whereas others preferentially erase methyl groups from tri- and dimethylated lysines or monomethylated lysines [45]. In contrast to lysine demethylases, it remains still unclear whether there is a bona fide arginine demethylase. JMJD6 was reported to have arginine demethylation activity on H4R3 and H3R2 [46,47]. However, JMJD6 was also shown to be rather a hydroxylase that adds a hydroxyl group at the 5-C of a lysine side chain of the splicing factor U2AF65 [48].
It has been shown that histone methylation modifiers control methylation states in non-histone substrates to regulate their activities, as described later in this review. Notably, these non-histone substrates include key components of multiple cellular signaling pathways (e.g., nuclear factor–kappa B [NF-κB], epidermal growth factor receptor [EGFR], RAF1, mitogen-activated protein kinase (MAPK) kinase kinase 2 [MAP3K2], p53, and estrogen receptor [ER],) (Table 1). Aberrant methylation of histones and these non-histone proteins has been linked to various human cancers [49,50].
Table 1.
Non-histone targets of histone methylation modifiers
| Signaling pathway | Methylation modifiers | Substrate | Residue(s) | Phenotypic changes/functions | Refs |
|---|---|---|---|---|---|
| NF-κB signaling | NSD1 | p65 | K218, K221 | Activates NF-κB signaling | [57] |
| KDM2A | p65 | K218, K221 | Inhibits NF-κB signaling | [57] | |
| SET9 | p65 | K37, K314, K315 | K37 activates NF-κB signaling; K314 and K315 reduce p65 stability and promote degradation |
[59] [64] |
|
| SETD6 | p65 | K310 | Inhibits p65-driven transcription | [65] | |
| PRMT5 | p65 | R30 | Activates NF-κB signaling | [66] | |
| MAPK signaling | SMYD3 | MAP3K2 | K260 | Activates MAP3K2 and thereby activates ERK1/2 | [81] |
| PRMT5 | EGFR | R1175 | Inhibits EGFR signaling | [76] | |
| PRMT5 | RAF1 | R563 | Enhances RAF1 degradation and inhibits ERK1/2 | [77] | |
| PI3K/Akt signaling | PRMT1 | FOXO | R248, R250 | Enhances FOXO degradation and apoptosis | [95] |
| PRMT1 | BAD | R94, R96 | Promotes mitochondrial localization and apoptosis | [96] | |
| Wnt signaling | PRMT1 | Axin | R378 | Negatively regulates Wnt signaling | [115] |
| EZH2 | PAF, β-catenin | independent of its catalytic activity | Activates Wnt signaling | [109, 110] | |
| ER signaling | PRMT4 | p300 | R2142 | Disrupts interaction between GRIP1 and p300 | [163] |
| SRC-3 | Inhibits ERα-mediated transcription | [155] | |||
| SMYD2 | ERα | K266 | Inhibits ERα activity | [150] | |
| PRMT1 | ERα | R260 | Activates ER signaling | [97] | |
| RIP140 | R240, R650, R948 | Activates ER signaling | [157] | ||
| PGC-1α | R665, R667, R669 | Enhances co-activation | [158] | ||
| p53 pathway | SET7/SET9 | p53 | K372 | Positive effect on stability, apoptosis | [62] |
| SMYD2 | p53 | K370 | Reduces DNA binding ability, p53-mediated apoptosis | [127] | |
| LSD1 | p53 | K370me2 | Inhibits 53BP1 binding and p53 function | [128] | |
| SET8 | p53 | K382 | Reduces transcriptional activity, apoptosis | [131] | |
| G9a/GLP | p53 | K373 | Inhibits p53 activity and its dependent apoptosis | [132] |
Regulation of signaling pathways by histone methylation modifiers
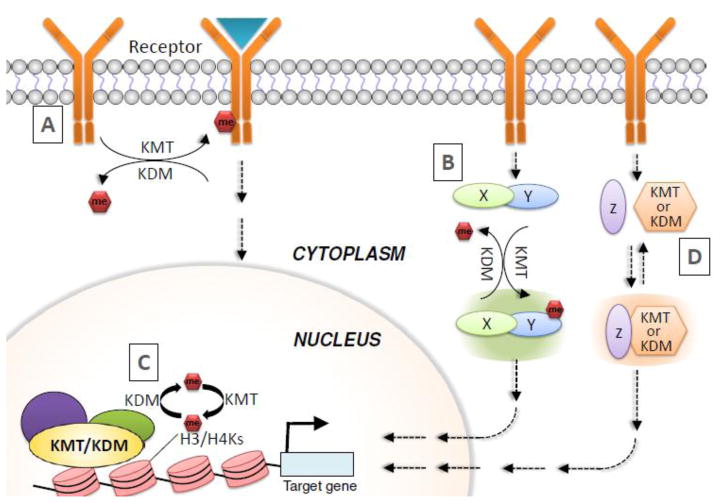
Generally, methylation modifiers can regulate various signaling pathways by I) directly methylating and demethylating their components, including receptors and downstream effectors; II) transcriptionally modulating expression of their components; and III) modulating the activities of their components via physical interaction (Figure 2).
Figure 2. Overview of the molecular mechanisms underlying regulation of signaling pathways by methylation modifiers.
Methylation modifiers can modulate signaling pathways by I) methylating and demethylating receptor kinases (A), effectors (e.g., kinases) (B), and activators/repressors of effectors (B); II) transcriptionally regulating expression of components in signaling pathways (C); and III) controlling the activities of signaling components via physical interaction (D). KDM, lysine demethylases; KMT, lysine methyltransferases; Me, methylation.
Histone methylation modifiers and NF-κB signaling
NF-κB signaling plays an important role in regulating multiple biological processes, including immune response, cell proliferation, and animal development. The NF-κB family of transcription factors comprises five members: p65 (RelA), RelB, c-Rel, p105/p50, and p100/p52. The precursor subunits p105 and p100 undergo proteolytic processing to become p50 and p52, respectively. Of the five members, the p50-p65 heterodimer is the key contributor to canonical NF-κB signaling. The p50-p65 heterodimer is inactive in a heterotrimeric complex consisting of p50, p65, and IκB in the cytoplasm (Figure 3). In response to a wide variety of cellular stimuli, IκB is phosphorylated by the IκB kinase complex (IKK) and is removed by ubiquitination-mediated degradation. Then, the p50–p65 heterodimer is released to be translocated to the nucleus. The p50-p65 heterodimer binds to the promoters of its target genes and induces gene expression [51–53]. Constitutive activation of NF-κB signaling is linked to numerous pathological states, including tumorigenesis and inflammation [54,55]. NF-κB signaling is controlled by multiple post-translational modifications [56]. Interestingly, NF-κB activities are modified by methylation and demethylation, as described in the subsequent paragraphs.
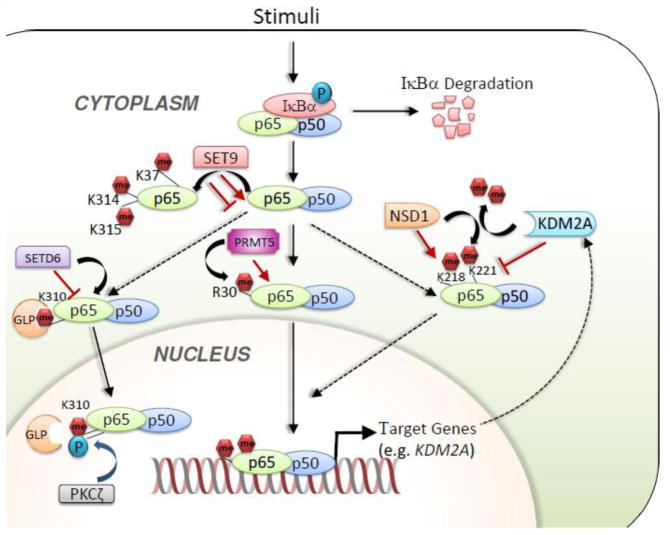
Figure 3. Regulation of NF-κB signaling pathways by methylation modifiers.
Components of NF-κB, including p65, can be regulated by multiple methyltransferases and demethylases.
Lysine methylation of NF-κB
Using genetic and biochemical approaches, Lu et al. showed that K218 and K221 of p65 can be methylated by the H3K36 methyltransferase NSD1 and demethylated by the H3K36me1/2 demethylase KDM2A (also known as FBXL11 and JHDM1A). NSD1 activates NF-κB activity, whereas KDM2A reduces it. They showed that the proliferation of HT29 colon cancer cells was promoted by NSD1-mediated methylation of p65 at K218/K221 but was antagonized by KDM2A-catalyzed demethylation of the same sites. Interestingly, NF-κB also increased expression of the KDM2A gene to form a negative feedback regulatory loop [57] (Figure 3). Subsequently, Zhang et al. documented that the plant homeodomain finger protein 20 (PHF20) promotes NF-κB transcriptional activity by interacting with methylated p65 at K218 and K221 [58]. Specifically, the interaction between PHF20 and methylated p65 blocks the association between p65 and the serine-threonine protein phosphatase 2A (PP2A) and thereby maintains the active, phosphorylated status of p65.
Ea and Baltimore showed that the SET domain-containing protein 9 (SET9) methylates p65 at K37 upon activation of NF-κB by tumor necrosis factor alpha (TNFα) [59]. It should be noted that SET9 was initially shown to be a H3K4 mono-methyltransferase [60,61] but later was reported to be unable to methylate nucleosomal H3K4 [62]. This p65 methylation facilitates the binding of p65 to the promoters of several NF-κB–regulated genes, such as IKBA, interferon gamma–induced protein 10, and TNFA, during TNFα stimulation. In line with this, expression of these genes was significantly reduced in p65−/− mouse embryonic fibroblast cells expressing the K37Q mutant as compared to those that expressed wild type p65 [59]. Lu et al. indicated that p65 methylation at K37 regulates genes distinct from those regulated by NSD1-mediated methylation of p65 at K218/K221 [63]. Seemingly contradictory to these studies was the report by Yang et al. that SET9-mediated monomethylation of p65 at the K314 and K315 residues brings about proteosomal degradation of p65, leading to decreased NF-κB activity in response to TNF-α stimulation [64] (Figure 3).
Levy et al. demonstrated that the SET domain–containing protein 6 (SETD6) can monomethylate p65 on K310 [65]. SETD6-mediated methylation of p65 inhibits p65-driven transcriptional programs, including inflammatory responses in primary immune cells. Mechanistically, SETD6-catalyzed methylation of p65 is recognized by the ankyrin repeat of the histone methyltransferase GLP (G9a-like protein), which modulates p65 target genes to be in a repressed chromatin state through H3K9 methylation [65]. Interestingly, phosphorylation of p65 at Ser311 by protein kinase C-ζ inhibits the association of GLP with p65 K310me1 and de-represses p65 target genes [65] (Figure 3).
Arginine methylation of NF-κB
NF-κB also undergoes arginine methylation. Wei et al. showed that R30 of p65 can be dimethylated by PRMT5, leading to activation of NF-κB signaling [66]. It was also shown that the expression of most NF-κB–inducible genes (~85%) that are downregulated by the p65-R30A mutant is also reduced by PRMT5 loss, suggesting that PRMT5-mediated R30 methylation of p65 is critical for NF-κB activity (Figure 3). Because PRMT5 expression is frequently elevated in many types of cancer, it is possible that PRMT5 overexpression promotes tumorigenic events by enhancing NF-κB signaling [67].
Physical interaction between histone methyltransferases and NF-κB
The H3K36 methyltransferase NSD2 (also known as MMSET or WHSC1) acts as a coactivator of NF-κB [68]. NSD2 was overexpressed in prostate cancer and was recruited to the promoters of NF-κB target genes, including IL6, IL8, VEGFA, CCND1, BCL2, and BIRC5, in castration-resistant prostate cancer cells. NSD2 then activated NF-κB target genes by increasing H3K36me2 and H3K36me3 levels. In addition, NSD2 interacts with the transcriptional co-activator and acetyltransferase p300 and facilitates cytokine-induced recruitment of NF-κB and p300 to the promoters of NF-κB target genes, resulting in increased levels of p300-catalyzed histone acetylation for gene activation [68].
The H3K27 methyltransferase EZH2 generally plays a critical role in epigenetic gene silencing. However, EZH2 functions differently in ER-negative basal-like breast cancer cells in which it physically interacts with p65 and RelB and constitutively activates NF-κB target genes [69]. This function of EZH2 is not dependent on its catalytic activity. It should be noted that in ER-positive lumina-like breast cancer cells, EZH2 downregulates expression of NF-κB target genes by interacting with ER and depositing H3K27 methylation [69]. Thus, EZH2 may have a dichotomous function in regulating NF-κB signaling in breast cancer cells. The histone H3K9 methyltransferase G9a (also known as EHMT2) was reported to interact with RelB and to induce RelB-mediated gene silencing [70]. These studies suggest that NF-κB interacts with different methylation modifiers to have a dual and context-dependent function in regulating expression of its target genes.
Histone methylation modifiers and RAS/RAF/MEK/MAPK signaling
The MAPKs regulate diverse cellular processes, including cell proliferation, cell migration, cellular differentiation, and survival, in response to extracellular signals. The best-studied MAPK family of proteins includes extracellular signal-regulated kinases 1 and 2 (ERK1/2), c-Jun amino-terminal kinases 1 to 3 (JNK1 to 3), p38 (α, β, γ, and δ), and ERK5. Aberrant regulation of these MAPKs is associated with various pathological conditions, including cancer. It has been well documented that the activities of MAPKs are modulated by phosphorylation states that are controlled by multiple kinases and phosphatases [71–75]. Recent studies have shown that the activities of MAPKs are also regulated by arginine and lysine methylation, as described in this section.
Arginine methylation of EGFR and RAF1
PRMT5 regulates MAPK signaling by methylating the upstream activators of MAPKs, such as EGFR and RAF1. PRMT5 monomethylates EGFR receptor at R1175 to increase trans- autophosphorylation of EGFR at Tyr (Y) 1173 [76], which in turn provides a docking site for the SH2 domain of the phosphatase SHP1 (also known as PTPN6) (Figure 4). The recruitment of SHP1 downregulates EGFR-ERK signaling. Blocking of R1175 methylation amplifies EGFR signaling and concomitantly increases proliferation, migration, and invasion of breast epithelial cells. Although the mode of action of R1175 methylation is not very well understood, it is possible that R1175 methylation increases the interaction between EGFR and the Y1173 kinase or inhibits the recognition of EGFR by the Y1173 phosphatase [76]. PRMT5 also regulates MAPK signaling by methylating the serine/threonine-protein kinase RAF1 (also known as CRAF) in melanoma cells [77]. PRMT5 enhances RAF1 degradation by methylating RAF1 at R563, reducing the activities of the downstream kinases, such as MEK1/2 and ERK1/2. RNAi-mediated or pharmacological inhibition of PRMT5 activity increases the amplitude and duration of RAS-dependent ERK phosphorylation in response to growth factors. These studies suggest that PRMT5 can have a tumor-suppressive property by downregulating EGFR and RAF1 signaling. However, multiple lines of evidence also indicate that PRMT5 may have an oncogenic function in several tumor types, including leukemia and breast cancer, by activating AKT signaling [78] and repressing tumor suppressor proteins [79].
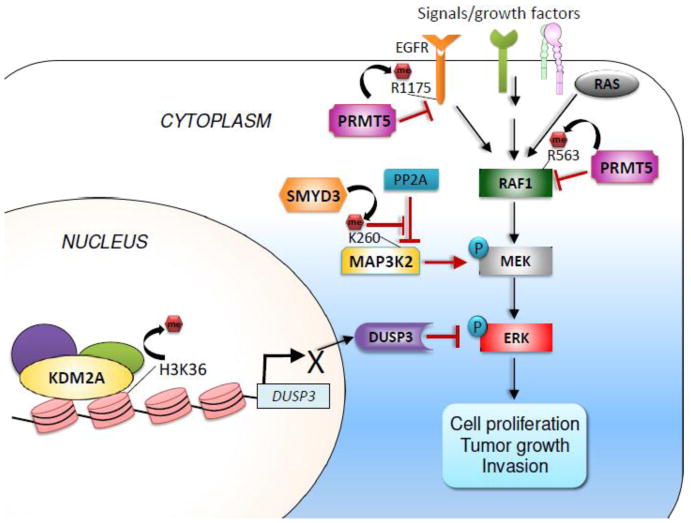
Figure 4. Methylation modifiers and MAPK signaling pathways.
The lysine demethylase KDM2A activates ERK signaling by repressing expression of the ERK phosphatase DUSP3, while the lysine methyltransferase SMYD3 activates MAPK signaling through methylation of MAP3K2. PRMT5 methylates EGFR and RAF1 to downregulate MAPK signaling.
SMYD3-mediated lysine methylation of MAP3K2
SET and MYND domain containing 3 (SMYD3) preferentially catalyzes H4K5 monomethylation, and to a lesser extent dimethylation and trimethylation at H4K5 [80]. Mazur et al. reported that SMYD3 is predominantly localized in cytoplasm and methylates MAP3K2 at K260 [81] (Figure 4). This methylation is dependent on SMYD3’s catalytic activity and also correlates with ERK1/2 phosphorylation levels. Mechanistically, SMYD3-mediated K260 methylation of MAP3K2 inhibits its binding to the PP2A complex, a negative regulator of the MAPK pathway [81]. High SMYD3 expression also correlates with progression of pancreatic and lung cancer [81]. Consistent with this, pancreas-specific loss of Smyd3 prevented inflammation-induced neoplastic lesions and formation of metaplastic ducts in pancreas-conditional K-RASG12D and p53-null mice [81]. Similarly, lung-specific loss of Smyd3 in lung-conditional K-RASG12D significantly inhibited K-RASG12D–driven lung adenocarcinoma formation. The tumor-promoting function of Smyd3 in K-RASG12D–induced tumorigenesis is dependent on its catalytic activity [81]. This study highlights an important role for SMYD3-mediated methylation of MAP3K2 in enhancing oncogenic K-RAS signaling.
Transcriptional regulation of MAPK signaling by histone methylation modifiers
Histone methylation modifiers transcriptionally regulate MAPK signaling. In particular, we recently showed that KDM2A acted as a critical regulator of ERK signaling in lung cancer cells [82]. KDM2A was frequently upregulated in tumor samples from lung cancer patients, and high KDM2A levels were associated with poor prognosis [82]. Thus, KDM2A may be a prognostic marker for lung cancer and a therapeutic target. KDM2A overexpression repressed expression of the dual-specificity phosphatase-3 (DUSP3) gene by demethylating H3K36me2 at the DUSP3 promoter (Figure 4). Because DUSP3 preferentially dephosphorylates ERK1/2 in lung cancer cells, KDM2A-mediated repression of DUSP3 amplifies ERK1/2 signaling to increase cell proliferation and invasion [82]. Consistent with this, KDM2A depletion drastically inhibited tumorigenicity and invasion of lung cancer cells in mouse xenograft models. The tumor-promoting function of KDM2A is dependent largely on its enzymatic activity [82]. Interestingly, Chen et al. showed that the H3K9me1/2 demethylase JMJD1A (also called JHDM2A) positively regulates expression of Spry2, a key negative regulator of ERK1/2 in human bronchial epithelial BEAS-2B cells [83]. Both hypoxia and nickel (an environmental carcinogen) inhibit JMJD1A’s enzymatic activity, resulting in decreased expression of Spry2 and increased ERK1/2 signaling [84].
Interestingly, EZH2 epigenetically represses expression of the DAB2IP gene, which encodes a RAS GTPase-activating protein in prostate cancer cells. Because DAB2IP suppresses RAS and NF-κB through distinct domains, EZH2-mediated repression of DAB2IP expression activates RAS and NF-κB signaling to promote tumor growth and metastasis [85]. In addition, EZH2 interacts with phosphorylated p38 and enhances the p38 signaling pathway in breast cancer cells to promote their migration and metastasis [86].
Histone methylation modifiers and PI3K/AKT signaling
Like the RAS/RAF/MEK/MAPK pathway, the phosphatidylinositol 3-kinase (PI3K)/the serine-threonine kinase AKT (also called protein kinase B) /mammalian target of rapamycin (mTOR) pathway is a major signaling pathway that regulates cell proliferation, growth, and survival. It has been shown extensively that this pathway is modulated by several post-translational modifications, including phosphorylation [87–90], ubiquitination [91,92], and sumoylation [93,94]. Arginine and lysine methylation have emerged as an important type of modification that regulates upstream and downstream factors of AKT signaling, as described in this section.
Regulation of PI3K/AKT signaling pathway by PRMT1
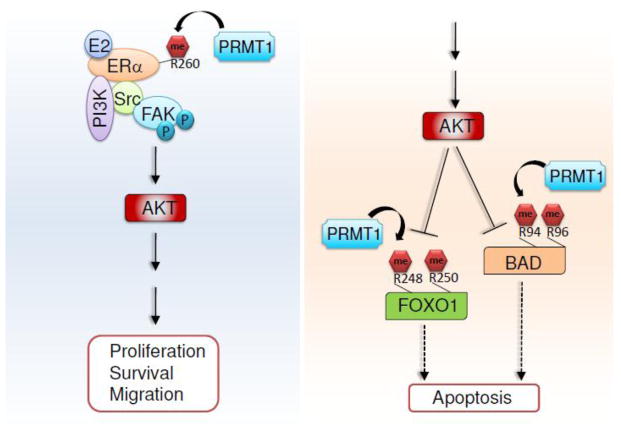
PRMT1 has been shown to regulate the PI3K/AKT signaling pathway through arginine methylation. Downstream of PI3K-AKT signaling, PRMT1 methylates Forkhead box O (FOXO) at conserved R248 and R250 residues within a consensus motif (i.e., R-X-R-X-X-S/T) for AKT-mediated phosphorylation [95] (Figure 5). This methylation blocks AKT-mediated Ser253 phosphorylation of FOXO1, which leads to cytoplasmic localization and ubiquitin-mediated proteosomal degradation of FOXO1. Loss of PRMT1 or its enzymatic activity leads to a decrease in oxidative stress–induced apoptosis dependent on AKT-mediated Ser253 phosphorylation of FOXO1 [95]. In addition, PRMT1 specifically methylates BCL-2 antagonist of cell death (BAD) at the R94 and R96 residues, which reside in an AKT phosphorylation motif (Figure 5). PRMT1-catalyzed methylation of BAD impedes AKT-mediated phosphorylation of BAD at Ser99, blocking the interaction of BAD with phosphoserine binding 14-3-3 proteins. Thus, PRMT1 induces mitochondrial localization of BAD, thereby promoting apoptosis [96].
Figure 5. Regulation of AKT signaling pathways by PRMT1.
PRMT1 enhances AKT signaling by methylating ERα in response to estrogen. In contrast, PRMT1-mediated methylation of FOXO1 and BAD interferes with AKT-mediated inhibition of apoptosis.
In contrast, PRMT1 appears to facilitate AKT activation in response to estrogen treatment. PRMT1 methylates ERα at the R260 residue within the DNA-binding domain of ERα [97]. R260 methylation of ERα takes place in the cytoplasm of normal and malignant epithelial breast cells. ERα-R260 is hypermethylated in a subset of breast cancers. This methylation event induces the interaction of ERα with the Src kinase and the p85 subunit of PI3K (a heterodimer composed of a p110 catalytic subunit and a p85 regulatory subunit) (Figure 5). The focal adhesion kinase (FAK), a Src substrate, is also recruited to ERα/PI3K/Src via Src. Such multi-protein interaction induces AKT activation to promote cell proliferation, survival, and migration [97].
Regulation of PI3K/AKT signaling pathway by EZH2
Gonzales et al. showed that EZH2 overexpression enhances PI3K/AKT signaling through activation of AKT isoform 1 in breast cancer cells [98]. Interestingly, EZH2 interacts with AKT-1. EZH2-induced phenotypes, such as BRCA1 nuclear export, aneuploidy, and mitotic defects, are dependent on AKT-1 activation. Consistent with these findings, high EZH2 protein levels were associated with increased levels of phospho-AKT-1 (Ser473) and decreased nuclear levels of phospho-BRCA1 (Ser1423) in invasive breast cancer samples [98].
Histone methylation modifiers and Wnt/β-catenin signaling
Wnt/β-catenin signaling is critical for normal development and tissue homeostasis [99,100], and its aberrant regulation is linked to tumorigenesis [101]. Notably, β-catenin is stabilized in response to Wnt activation and binds to the DNA-binding proteins T-cell factor (TCF) and lymphoid enhancing factor-1 (LEF-1) to activate β-catenin target genes [102,103]. The Wnt/β-catenin signaling pathway is also controlled by histone methylation modifiers, as summarized in Figure 6.
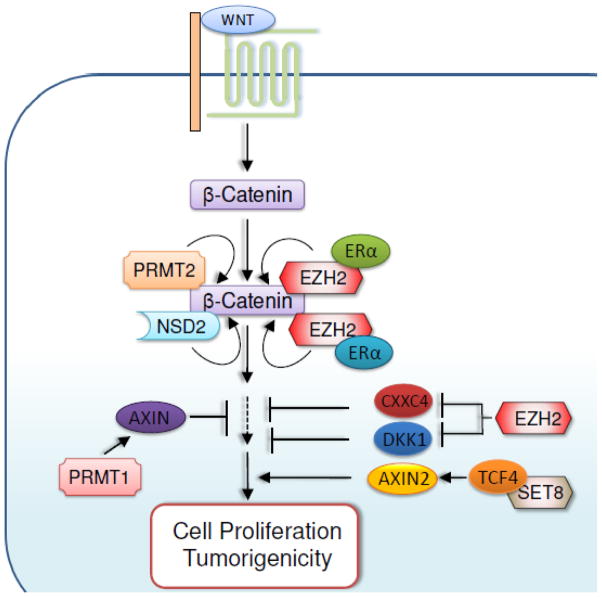
Figure 6. Regulation of Wnt/β-catenin signaling pathways by methylation modifiers.
Wnt/β-catenin signaling can be enhanced by the methyltransferases PRMT2, EZH2, SET8, and NSD2, but can be inhibited by PRMT1.
Regulation of Wnt/β-catenin signaling pathway by EZH2
EZH2 enhances RAF1-ERK-β-catenin signaling in breast tumor initiating cells (BTICs) [104]. In BTICs, EZH2 overexpression induces RAF1 gene amplification by downregulating expression of the DNA damage repair protein RAD51. RAF1 amplification enhances ERK-β-catenin signaling to increase survival and expansion of BTICs [104]. ERK phosphorylates and primes glycogen synthase kinase 3β (GSK-3β) for its subsequent inactivation [105]. Inactivation of GSK-3β stabilizes functional β-catenin, because GSK-3β–catalyzed phosphorylation of β-catenin promotes β-catenin degradation.
EZH2 also activates Wnt/β-catenin signaling by silencing expression of Wnt pathway antagonists. In hepatocellular carcinoma, for instance, ectopic overexpression of EZH2 promoted proliferation of immortalized hepatocytes by concomitantly reducing expression of several Wnt inhibitors, including AXIN2, NKD1, PPP2R2B, PRICKLE1, and SFRP5 [106]. In gastric cancer cells, EZH2 activated Wnt/β-catenin-signaling by epigenetically repressing the Wnt signaling antagonist CXXC4, thereby promoting tumorigenic phenotypes [107]. In addition, EZH2-mediated transcriptional repression of the Wnt signaling antagonist DKK1 contributes to increased tumorigenicity of lung cancer cells that were exposed to tobacco smoke condensate. [108].
Interestingly, EZH2 acts as a transcriptional activator for β-catenin target genes independent of its methyltransferase activity linked to gene repression. EZH2 directly binds to β-catenin and ERα and subsequently enhances transactivation of MYC and CCND1 genes by ERα and β-catenin. This gene activation by EZH2 leads to cell cycle progression in breast cancer cells [109]. Recently, it was also shown that PCNA-associated factor (PAF) is associated with the β-catenin transcriptional complex and upregulates β-catenin target genes by recruiting EZH2 to their promoters [110].
Regulation of Wnt/β-catenin signaling by other histone methylation modifiers
Dot1L was shown to activate senseless, a Wnt target gene, by methylating H3K79 at its promoter in Drosophila [111]. Interestingly, Dot1L-mediated H3K79 methylation of the senseless promoter requires the monoubiquitination of H2B by the Rad6/Bre1 complex [111]. The SET domain containing lysine methyltransferase-8 (SET8; also known as SETD8, KMT5A) also acts as a mediator of Wnt signaling [112]. Specifically, SET8 directly associates with LEF1/TCF4, and this interaction is controlled by Wnt3a. Thus, SET8 is recruited to Wnt-activatable genes, such as AXIN2, and positively regulates them, possibly by monomethylating H4K20 [112]. NSD2 has been found to be overexpressed in several cancer types and to interact with some Wnt-signaling regulators, including β-catenin. NSD2 positively regulates expression of CCND1, a target gene of the β-catenin/Tcf-4 complex, via H3K36 trimethylation [113].
It has been shown that PRMT2 is required for Wnt/β-catenin–dependent establishment of the dorsal developmental program in Xenopus. Specifically, PRMT2 is recruited by β-catenin to β-catenin target genes and may establish poised chromatin structure by generating asymmetrically dimethylated H3R8 at their promoters [114]. PRMT1 negatively regulates β-catenin–dependent transcription by stabilizing Axin, an inhibitor of Wnt signaling. PRMT1 physically interacts with and methylates Axin at R378. This PRMT1-mediated methylation stabilizes Axin [115].
Histone methylation modifiers and the p53 pathway
p53 is a well-studied tumor suppressor that is mutated in approximately 50% of human cancers; it is a transcription factor that regulates the cell cycle, apoptosis, and DNA repair in response to a variety of genotoxic stresses. It has been well documented that the activity and stability of p53 are modulated by multiple types of post-translational modifications [116–118], including phosphorylation, acetylation, ubiquitination, and sumoylation [119–121]. Because p53 methylation has already been reviewed elsewhere [122–125], we will only briefly summarize a few selected studies regarding p53 methylation.
p53 methylation was first documented by the Reinberg group, who showed that SET9 (alias SET7) monomethylates p53 at K372 [62]. This methylation increases the stability of nuclear p53, resulting in both enhanced expression of the p53 target gene p21 and increased levels of p53-mediated apoptosis [62]. p53-K372 monomethylation is important for subsequent acetylation of p53 that is also linked to p21 gene activation [126].
In contrast to p53-K372 methylation, SMYD2-catalyzed K370 monomethylation (K370me1) of p53 represses gene-activating function of p53 by reducing chromatin-associated p53, suggesting that K370 monomethylation is a repressive mark for p53 activity [127]. Consistent with this, RNAi-mediated depletion of SMYD2 significantly induces p53 target gene expression and enhances p53-mediated apoptosis [127]. SMYD2-mediated methylation of K370 is also inhibited by SET9-directed K372 methylation. Interestingly, p53-K370 methylation can be reversed. Berger and colleagues also reported that K370me2 of p53 is the preferred substrate of LSD1, although LSD1 demethylates both K370me1 and K370me2 [128]. In contrast to K370me1 of p53, K370me2 of p53 enhances p53 transcriptional activity by interacting with 53BP1, a p53 co-activator that plays an important role in DNA damage response [129,130]. Thus, LSD1-mediated demethylation of K370me2 inhibits p53 function by abolishing the interaction of K370me2 with 53BP1 [128].
Shi et al. showed that SET8 monomethylates p53 at K382 to inhibit its transcriptional activity and to consequently decrease expression of its target genes [131]. The H3K9 methyltransferases G9a and GLP were shown to dimethylate p53 at K373 [132]. These methyltransferases were suggested to inhibit p53 function, because knockdown of G9a and GLP increases apoptosis [132].
Histone methylation modifiers and estrogen receptor signaling
ER signaling contributes to normal cell growth, development, and tumorigenesis [133–137]. ERα is a ligand-dependent transcription factor that regulates gene expression [138–140]. In response to estrogen, ER’s transcriptional activity is modulated by numerous coactivators and corepressors [141–143], including histone-modifying enzymes [144].
Regulation of ER signaling by lysine methyltransferases and demethylases
Dreijerink et al. showed that menin, a tumor suppressor and an important component of MLL1 and MLL2 H3K4 methyltransferase complexes, functions as a transcriptional co-activator of ERα [145]. MLL1–4 are associated with the estrogen-induced activation of HOXC13 [146]. In addition, the H3K4 methyltransferase MLL4 (also known as MLL2, KMT2D and ALR) was shown to directly interact with ERα and to cooperate with ERα for transcriptional activation of ERα target genes. Consistent with this, MLL4 knockdown inhibited proliferation of ERα-positive MCF-7 cells [147]. Interestingly, MLL4 depletion was also shown to impede proliferation and invasion of ERα-negative cells [148]. Together, MLL1–4 each may play a role in ERα-mediated transcriptional activation.
Kim et al. demonstrated that SMYD3 acts as a co-activator of ERα and potentiates ERα-mediated activation of ERα target genes [149]. Recently, Zhang et al. reported that SMYD2 directly methylates ERα at K266. This methylation Inhibits acetylation of ERα at K266/268 to decrease ERα’s transactivation activity and may be antagonized by LSD1-mediated demethylation [150]. LSD1 is recruited to most ERα target genes and is required for ERα-dependent gene activation [151], although LSD1 is also linked to gene repression. It was reported, similarly, that ERα-mediated chromatin looping requires LSD1-mediated demethylation of the repressive H3K9me2 [152].
Regulation of ER signaling by arginine methyltransferases
Arginine methyltransferases play an important role in ER signaling by methylating arginine residues in both histones and non-histone proteins [141]. Metivier et al. showed that activation of ERα by estrogen induces the oscillatory recruitment of ERα coactivators, including PRMT4 and PRMT1, on its target genes [153]. PRMT4 is required for estrogen-induced cell cycle progression of MCF-7 breast cancer cells [154]. It dimethylates H3R17 at the E2F1 promoter in an ERα-dependent manner, leading to increased expression of the cell cycle transcriptional factor E2F1 [154]. PRMT4-mediated methylation of steroid receptor coactivator 3 (SRC-3) dissociates the interaction between PRMT4 and SRC-3, resulting in decrease of ERα-mediated transcription [155]. Thus, it was proposed that PRMT4 has a dual-function coactivator that activates transcription by histone arginine methylation but terminates ER signaling by SRC-3 methylation [155].
PRMT1 facilitates ERα-induced transcriptional activation by asymmetrically dimethylating H4R3 at ERα target genes [156]. As mentioned above, PRMT1 methylates ERα at R260 within ERα’s DNA-binding domain, inducing the association of ERα with PI3K and Src. This association leads to AKT phosphorylation at S473 and subsequent cell cycle progression [97]. In addition to ERα, PRMT1 also methylates ERα cofactors and modulates their activities. For example, PRMT1 methylates receptor-interacting protein 140 (RIP140), a ligand-dependent corepressor for ERα and other nuclear receptors, at the R240, R650, and R948 residues. PRMT1-mediated methylation of RIP140 inhibits the repressive activity of RIP140 [157]. PRMT1 methylates peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α), a transcriptional coactivator for ERα and other nuclear receptors, at R665, R667, and R669. PRMT1-catalyzed methylation of PGC-1α enhances PGC-1α’s coactivator activity [158].
Future perspectives
Recent systematic sequencing studies of cancer genomes have revealed that multiple histone methylation modifiers are frequently mutated and amplified in diverse cancer types. For instance, the H3K27 demethylase UTX and MLL4 undergo somatic mutations in several tumor types (e.g., renal cancer and medulloblastoma) and thus may act as tumor suppressor genes in such tumors. Indeed, mouse genetic experiments showed that UTX may act as a tumor suppressor in acute lymphoid leukemia (ALL) [159]. EZH2 is often amplified and overexpressed in multiple malignancies, such as prostate and breast cancer, in which EZH2 may be an oncogene. It should be noted that EZH2 may have a tumor suppressive role in the myeloid malignancies and ALL. In fact, EZH2 often undergoes loss-of-function mutations in these malignancies (reviewed in [160]), and its loss in hemopoietic tissues in mice showed the high frequency of spontaneous T-cell ALL [161]. Taken together, these studies highlight the importance of histone methylation modifiers in properly regulating signaling pathways and maintaining cellular homeostasis.
As summarized herein, dysregulated histone methylation modifiers may drive or promote tumorigenesis and metastasis by altering transcriptional programs and cellular signaling pathways. Importantly, aberrantly expressed modifiers are in principle targetable, because they have intrinsic enzymatic activities. In fact, many small molecule inhibitors against specific histone methylation modifiers have been reported. Some specific inhibitors, including inhibitors against LSD1 (ORY-1001 and GSK2879552), DOT1L (EPZ-5676), and EZH2 (E7438, GSK2816126, and CPI-1205), have entered clinical phase 1 trials [162]. Successful results of these trials may provide new avenues for therapeutic interventions. We believe that an exciting era has opened in chromatin and epigenetics research.
Acknowledgments
We apologize for not reviewing numerous pertinent articles because of space limitations. We are thankful to Kathryn Hale for manuscript editing. The work of this laboratory is supported by grants to M.G.L. from the NIH (R01 GM095659 and R01 CA157919), the Center for Cancer Epigenetics at The University of Texas MD Anderson Cancer Center, and Cancer Prevention and Research Institute of Texas (RP110183) and by a fellowship to H.A. from the Odyssey Program at MD Anderson Cancer Center.
References
- 1.Grillo MA, Colombatto S. S-adenosylmethionine and protein methylation. Amino Acids. 2005;28(4):357–362. doi: 10.1007/s00726-005-0197-6. [DOI] [PubMed] [Google Scholar]
- 2.Aletta JM, Cimato TR, Ettinger MJ. Protein methylation: a signal event in post-translational modification. Trends Biochem Sci. 1998;23(3):89–91. doi: 10.1016/s0968-0004(98)01185-2. [DOI] [PubMed] [Google Scholar]
- 3.Stallcup MR. Role of protein methylation in chromatin remodeling and transcriptional regulation. Oncogene. 2001;20(24):3014–3020. doi: 10.1038/sj.onc.1204325. [DOI] [PubMed] [Google Scholar]
- 4.Sprung R, Chen Y, Zhang K, Cheng D, Zhang T, Peng J, Zhao Y. Identification and validation of eukaryotic aspartate and glutamate methylation in proteins. J Proteome Res. 2008;7(3):1001–1006. doi: 10.1021/pr0705338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ng SS, Yue WW, Oppermann U, Klose RJ. Dynamic protein methylation in chromatin biology. Cell Mol Life Sci. 2009;66(3):407–422. doi: 10.1007/s00018-008-8303-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 7.Garcia BA, Hake SB, Diaz RL, Kauer M, Morris SA, Recht J, Shabanowitz J, Mishra N, Strahl BD, Allis CD, Hunt DF. Organismal differences in post-translational modifications in histones H3 and H4. J Biol Chem. 2007;282(10):7641–7655. doi: 10.1074/jbc.M607900200. [DOI] [PubMed] [Google Scholar]
- 8.Cavalli G. Chromatin and epigenetics in development: blending cellular memory with cell fate plasticity. Development. 2006;133(11):2089–2094. doi: 10.1242/dev.02402. [DOI] [PubMed] [Google Scholar]
- 9.Minard ME, Jain AK, Barton MC. Analysis of epigenetic alterations to chromatin during development. Genesis. 2009;47(8):559–572. doi: 10.1002/dvg.20534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Briggs SD, Xiao T, Sun ZW, Caldwell JA, Shabanowitz J, Hunt DF, Allis CD, Strahl BD. Gene silencing: trans-histone regulatory pathway in chromatin. Nature. 2002;418(6897):498. doi: 10.1038/nature00970. [DOI] [PubMed] [Google Scholar]
- 11.Frederiks F, Tzouros M, Oudgenoeg G, van Welsem T, Fornerod M, Krijgsveld J, van Leeuwen F. Nonprocessive methylation by Dot1 leads to functional redundancy of histone H3K79 methylation states. Nat Struct Mol Biol. 2008;15(6):550–557. doi: 10.1038/nsmb.1432. [DOI] [PubMed] [Google Scholar]
- 12.Margueron R, Trojer P, Reinberg D. The key to development: interpreting the histone code? Curr Opin Genet Dev. 2005;15(2):163–176. doi: 10.1016/j.gde.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 13.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, Zeitlinger J, Lewitter F, Gifford DK, Young RA. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122(4):517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 14.Kouzarides T. Histone methylation in transcriptional control. Curr Opin Genet Dev. 2002;12(2):198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- 15.Peterson CL, Laniel MA. Histones and histone modifications. Curr Biol. 2004;14(14):R546–551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 16.Xu C, Henry PA, Setya A, Henry MF. In vivo analysis of nucleolar proteins modified by the yeast arginine methyltransferase Hmt1/Rmt1p. RNA. 2003;9(6):746–759. doi: 10.1261/rna.5020803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Y, Bedford MT. Protein arginine methyltransferases and cancer. Nat Rev Cancer. 2013;13(1):37–50. doi: 10.1038/nrc3409. [DOI] [PubMed] [Google Scholar]
- 18.Bedford MT. Arginine methylation at a glance. J Cell Sci. 2007;120(Pt 24):4243–4246. doi: 10.1242/jcs.019885. [DOI] [PubMed] [Google Scholar]
- 19.Tripsianes K, Madl T, Machyna M, Fessas D, Englbrecht C, Fischer U, Neugebauer KM, Sattler M. Structural basis for dimethylarginine recognition by the Tudor domains of human SMN and SPF30 proteins. Nat Struct Mol Biol. 2011;18(12):1414–1420. doi: 10.1038/nsmb.2185. [DOI] [PubMed] [Google Scholar]
- 20.Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33(1):1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Mol Cell. 2005;18(3):263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 22.Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406(6796):593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 23.Jenuwein T, Laible G, Dorn R, Reuter G. SET domain proteins modulate chromatin domains in eu- and heterochromatin. Cell Mol Life Sci. 1998;54(1):80–93. doi: 10.1007/s000180050127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singer MS, Kahana A, Wolf AJ, Meisinger LL, Peterson SE, Goggin C, Mahowald M, Gottschling DE. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics. 1998;150(2):613–632. doi: 10.1093/genetics/150.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dillon SC, Zhang X, Trievel RC, Cheng X. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 2005;6(8):227. doi: 10.1186/gb-2005-6-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, Zhang Y. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol. 2002;12(12):1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 27.Gary JD, Clarke S. RNA and protein interactions modulated by protein arginine methylation. Prog Nucleic Acid Res Mol Biol. 1998;61:65–131. doi: 10.1016/s0079-6603(08)60825-9. [DOI] [PubMed] [Google Scholar]
- 28.Yang Y, Hadjikyriacou A, Xia Z, Gayatri S, Kim D, Zurita-Lopez C, Kelly R, Guo A, Li W, Clarke SG, Bedford MT. PRMT9 is a type II methyltransferase that methylates the splicing factor SAP145. Nat Commun. 2015;6:6428. doi: 10.1038/ncomms7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dhar SS, Lee SH, Kan PY, Voigt P, Ma L, Shi X, Reinberg D, Lee MG. Trans-tail regulation of MLL4-catalyzed H3K4 methylation by H4R3 symmetric dimethylation is mediated by a tandem PHD of MLL4. Genes Dev. 2012;26(24):2749–2762. doi: 10.1101/gad.203356.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JH, Cook JR, Yang ZH, Mirochnitchenko O, Gunderson SI, Felix AM, Herth N, Hoffmann R, Pestka S. PRMT7, a new protein arginine methyltransferase that synthesizes symmetric dimethylarginine. J Biol Chem. 2005;280(5):3656–3664. doi: 10.1074/jbc.M405295200. [DOI] [PubMed] [Google Scholar]
- 31.Karkhanis V, Wang L, Tae S, Hu YJ, Imbalzano AN, Sif S. Protein arginine methyltransferase 7 regulates cellular response to DNA damage by methylating promoter histones H2A and H4 of the polymerase delta catalytic subunit gene, POLD1. J Biol Chem. 2012;287(35):29801–29814. doi: 10.1074/jbc.M112.378281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jelinic P, Stehle JC, Shaw P. The testis-specific factor CTCFL cooperates with the protein methyltransferase PRMT7 in H19 imprinting control region methylation. PLoS Biol. 2006;4(11):e355. doi: 10.1371/journal.pbio.0040355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boffa LC, Karn J, Vidali G, Allfrey VG. Distribution of NG, NG,-dimethylarginine in nuclear protein fractions. Biochem Biophys Res Commun. 1977;74(3):969–976. doi: 10.1016/0006-291x(77)91613-8. [DOI] [PubMed] [Google Scholar]
- 34.Cheng D, Cote J, Shaaban S, Bedford MT. The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol Cell. 2007;25(1):71–83. doi: 10.1016/j.molcel.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 35.Branscombe TL, Frankel A, Lee JH, Cook JR, Yang Z, Pestka S, Clarke S. PRMT5 (Janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J Biol Chem. 2001;276(35):32971–32976. doi: 10.1074/jbc.M105412200. [DOI] [PubMed] [Google Scholar]
- 36.Wolf SS. The protein arginine methyltransferase family: an update about function, new perspectives and the physiological role in humans. Cell Mol Life Sci. 2009;66(13):2109–2121. doi: 10.1007/s00018-009-0010-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gayatri S, Bedford MT. Readers of histone methylarginine marks. Biochim Biophys Acta. 2014;1839(8):702–710. doi: 10.1016/j.bbagrm.2014.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119(7):941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 39.Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437(7057):432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- 40.Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8(4):307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 41.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439(7078):811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 42.Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, Shi Y. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125(3):467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 43.Mosammaparast N, Shi Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu Rev Biochem. 2010;79:155–179. doi: 10.1146/annurev.biochem.78.070907.103946. [DOI] [PubMed] [Google Scholar]
- 44.Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22(9):1115–1140. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21(3):381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang B, Chen Y, Zhao Y, Bruick RK. JMJD6 is a histone arginine demethylase. Science. 2007;318(5849):444–447. doi: 10.1126/science.1145801. [DOI] [PubMed] [Google Scholar]
- 47.Liu W, Ma Q, Wong K, Li W, Ohgi K, Zhang J, Aggarwal AK, Rosenfeld MG. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell. 2013;155(7):1581–1595. doi: 10.1016/j.cell.2013.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Webby CJ, Wolf A, Gromak N, Dreger M, Kramer H, Kessler B, Nielsen ML, Schmitz C, Butler DS, Yates JR, 3rd, Delahunty CM, Hahn P, Lengeling A, Mann M, Proudfoot NJ, Schofield CJ, Bottger A. Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science. 2009;325(5936):90–93. doi: 10.1126/science.1175865. [DOI] [PubMed] [Google Scholar]
- 49.Feinberg AP, Oshimura M, Barrett JC. Epigenetic mechanisms in human disease. Cancer Res. 2002;62(22):6784–6787. [PubMed] [Google Scholar]
- 50.Handel AE, Ebers GC, Ramagopalan SV. Epigenetics: molecular mechanisms and implications for disease. Trends Mol Med. 2010;16(1):7–16. doi: 10.1016/j.molmed.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 51.Ghosh S, Karin M. Missing pieces in the NF-kappaB puzzle. Cell. 2002;109(Suppl):S81–96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- 52.Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18(18):2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 53.Bharti AC, Aggarwal BB. Nuclear factor-kappa B and cancer: its role in prevention and therapy. Biochem Pharmacol. 2002;64(5–6):883–888. doi: 10.1016/s0006-2952(02)01154-1. [DOI] [PubMed] [Google Scholar]
- 54.Basseres DS, Baldwin AS. Nuclear factor-kappaB and inhibitor of kappaB kinase pathways in oncogenic initiation and progression. Oncogene. 2006;25(51):6817–6830. doi: 10.1038/sj.onc.1209942. [DOI] [PubMed] [Google Scholar]
- 55.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441(7092):431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 56.Perkins ND. Post-translational modifications regulating the activity and function of the nuclear factor kappa B pathway. Oncogene. 2006;25(51):6717–6730. doi: 10.1038/sj.onc.1209937. [DOI] [PubMed] [Google Scholar]
- 57.Lu T, Jackson MW, Wang B, Yang M, Chance MR, Miyagi M, Gudkov AV, Stark GR. Regulation of NF-kappaB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc Natl Acad Sci U S A. 2010;107(1):46–51. doi: 10.1073/pnas.0912493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang T, Park KA, Li Y, Byun HS, Jeon J, Lee Y, Hong JH, Kim JM, Huang SM, Choi SW, Kim SH, Sohn KC, Ro H, Lee JH, Lu T, Stark GR, Shen HM, Liu ZG, Park J, Hur GM. PHF20 regulates NF-kappaB signalling by disrupting recruitment of PP2A to p65. Nat Commun. 2013;4:2062. doi: 10.1038/ncomms3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ea CK, Baltimore D. Regulation of NF-kappaB activity through lysine monomethylation of p65. Proc Natl Acad Sci U S A. 2009;106(45):18972–18977. doi: 10.1073/pnas.0910439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nishioka K, Chuikov S, Sarma K, Erdjument-Bromage H, Allis CD, Tempst P, Reinberg D. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 2002;16(4):479–489. doi: 10.1101/gad.967202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang H, Cao R, Xia L, Erdjument-Bromage H, Borchers C, Tempst P, Zhang Y. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol Cell. 2001;8(6):1207–1217. doi: 10.1016/s1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- 62.Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, McKinney K, Tempst P, Prives C, Gamblin SJ, Barlev NA, Reinberg D. Regulation of p53 activity through lysine methylation. Nature. 2004;432(7015):353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- 63.Lu T, Yang M, Huang DB, Wei H, Ozer GH, Ghosh G, Stark GR. Role of lysine methylation of NF-kappaB in differential gene regulation. Proc Natl Acad Sci U S A. 2013;110(33):13510–13515. doi: 10.1073/pnas.1311770110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang XD, Huang B, Li M, Lamb A, Kelleher NL, Chen LF. Negative regulation of NF-kappaB action by Set9-mediated lysine methylation of the RelA subunit. EMBO J. 2009;28(8):1055–1066. doi: 10.1038/emboj.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Levy D, Kuo AJ, Chang Y, Schaefer U, Kitson C, Cheung P, Espejo A, Zee BM, Liu CL, Tangsombatvisit S, Tennen RI, Kuo AY, Tanjing S, Cheung R, Chua KF, Utz PJ, Shi X, Prinjha RK, Lee K, Garcia BA, Bedford MT, Tarakhovsky A, Cheng X, Gozani O. Lysine methylation of the NF-kappaB subunit RelA by SETD6 couples activity of the histone methyltransferase GLP at chromatin to tonic repression of NF-kappaB signaling. Nat Immunol. 2011;12(1):29–36. doi: 10.1038/ni.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wei H, Wang B, Miyagi M, She Y, Gopalan B, Huang DB, Ghosh G, Stark GR, Lu T. PRMT5 dimethylates R30 of the p65 subunit to activate NF-kappaB. Proc Natl Acad Sci U S A. 2013;110(33):13516–13521. doi: 10.1073/pnas.1311784110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wei H, Mundade R, Lange KC, Lu T. Protein arginine methylation of non-histone proteins and its role in diseases. Cell Cycle. 2014;13(1):32–41. doi: 10.4161/cc.27353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang P, Guo L, Duan ZJ, Tepper CG, Xue L, Chen X, Kung HJ, Gao AC, Zou JX, Chen HW. Histone methyltransferase NSD2/MMSET mediates constitutive NF-kappaB signaling for cancer cell proliferation, survival, and tumor growth via a feed-forward loop. Mol Cell Biol. 2012;32(15):3121–3131. doi: 10.1128/MCB.00204-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee ST, Li Z, Wu Z, Aau M, Guan P, Karuturi RK, Liou YC, Yu Q. Context-specific regulation of NF-kappaB target gene expression by EZH2 in breast cancers. Mol Cell. 2011;43(5):798–810. doi: 10.1016/j.molcel.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 70.Chen X, El Gazzar M, Yoza BK, McCall CE. The NF-kappaB factor RelB and histone H3 lysine methyltransferase G9a directly interact to generate epigenetic silencing in endotoxin tolerance. J Biol Chem. 2009;284(41):27857–27865. doi: 10.1074/jbc.M109.000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dhillon AS, Hagan S, Rath O, Kolch W. MAP kinase signalling pathways in cancer. Oncogene. 2007;26(22):3279–3290. doi: 10.1038/sj.onc.1210421. [DOI] [PubMed] [Google Scholar]
- 72.Chen Z, Gibson TB, Robinson F, Silvestro L, Pearson G, Xu B, Wright A, Vanderbilt C, Cobb MH. MAP kinases. Chem Rev. 2001;101(8):2449–2476. doi: 10.1021/cr000241p. [DOI] [PubMed] [Google Scholar]
- 73.Kyriakis JM, Avruch J. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol Rev. 2001;81(2):807–869. doi: 10.1152/physrev.2001.81.2.807. [DOI] [PubMed] [Google Scholar]
- 74.Pearson G, Robinson F, Beers Gibson T, Xu BE, Karandikar M, Berman K, Cobb MH. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev. 2001;22(2):153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- 75.Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol Rev. 2011;75(1):50–83. doi: 10.1128/MMBR.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hsu JM, Chen CT, Chou CK, Kuo HP, Li LY, Lin CY, Lee HJ, Wang YN, Liu M, Liao HW, Shi B, Lai CC, Bedford MT, Tsai CH, Hung MC. Crosstalk between Arg 1175 methylation and Tyr 1173 phosphorylation negatively modulates EGFR-mediated ERK activation. Nat Cell Biol. 2011;13(2):174–181. doi: 10.1038/ncb2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Andreu-Perez P, Esteve-Puig R, de Torre-Minguela C, Lopez-Fauqued M, Bech-Serra JJ, Tenbaum S, Garcia-Trevijano ER, Canals F, Merlino G, Avila MA, Recio JA. Protein arginine methyltransferase 5 regulates ERK1/2 signal transduction amplitude and cell fate through CRAF. Sci Signal. 2011;4(190):ra58. doi: 10.1126/scisignal.2001936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wei TY, Juan CC, Hisa JY, Su LJ, Lee YC, Chou HY, Chen JM, Wu YC, Chiu SC, Hsu CP, Liu KL, Yu CT. Protein arginine methyltransferase 5 is a potential oncoprotein that upregulates G1 cyclins/cyclin-dependent kinases and the phosphoinositide 3-kinase/AKT signaling cascade. Cancer Sci. 2012;103(9):1640–1650. doi: 10.1111/j.1349-7006.2012.02367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang L, Pal S, Sif S. Protein arginine methyltransferase 5 suppresses the transcription of the RB family of tumor suppressors in leukemia and lymphoma cells. Mol Cell Biol. 2008;28(20):6262–6277. doi: 10.1128/MCB.00923-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Van Aller GS, Reynoird N, Barbash O, Huddleston M, Liu S, Zmoos AF, McDevitt P, Sinnamon R, Le B, Mas G, Annan R, Sage J, Garcia BA, Tummino PJ, Gozani O, Kruger RG. Smyd3 regulates cancer cell phenotypes and catalyzes histone H4 lysine 5 methylation. Epigenetics. 2012;7(4):340–343. doi: 10.4161/epi.19506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mazur PK, Reynoird N, Khatri P, Jansen PW, Wilkinson AW, Liu S, Barbash O, Van Aller GS, Huddleston M, Dhanak D, Tummino PJ, Kruger RG, Garcia BA, Butte AJ, Vermeulen M, Sage J, Gozani O. SMYD3 links lysine methylation of MAP3K2 to Ras-driven cancer. Nature. 2014;510(7504):283–287. doi: 10.1038/nature13320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wagner KW, Alam H, Dhar SS, Giri U, Li N, Wei Y, Giri D, Cascone T, Kim JH, Ye Y, Multani AS, Chan CH, Erez B, Saigal B, Chung J, Lin HK, Wu X, Hung MC, Heymach JV, Lee MG. KDM2A promotes lung tumorigenesis by epigenetically enhancing ERK1/2 signaling. J Clin Invest. 2013;123(12):5231–5246. doi: 10.1172/JCI68642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen H, Kluz T, Zhang R, Costa M. Hypoxia and nickel inhibit histone demethylase JMJD1A and repress Spry2 expression in human bronchial epithelial BEAS-2B cells. Carcinogenesis. 2010;31(12):2136–2144. doi: 10.1093/carcin/bgq197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen H, Giri NC, Zhang R, Yamane K, Zhang Y, Maroney M, Costa M. Nickel ions inhibit histone demethylase JMJD1A and DNA repair enzyme ABH2 by replacing the ferrous iron in the catalytic centers. J Biol Chem. 2010;285(10):7374–7383. doi: 10.1074/jbc.M109.058503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Min J, Zaslavsky A, Fedele G, McLaughlin SK, Reczek EE, De Raedt T, Guney I, Strochlic DE, Macconaill LE, Beroukhim R, Bronson RT, Ryeom S, Hahn WC, Loda M, Cichowski K. An oncogene-tumor suppressor cascade drives metastatic prostate cancer by coordinately activating Ras and nuclear factor-kappaB. Nat Med. 2010;16(3):286–294. doi: 10.1038/nm.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Moore HM, Gonzalez ME, Toy KA, Cimino-Mathews A, Argani P, Kleer CG. EZH2 inhibition decreases p38 signaling and suppresses breast cancer motility and metastasis. Breast Cancer Res Treat. 2013;138(3):741–752. doi: 10.1007/s10549-013-2498-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2(7):489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 88.Hemmings BA, Restuccia DF. PI3K-PKB/Akt pathway. Cold Spring Harb Perspect Biol. 2012;4(9):a011189. doi: 10.1101/cshperspect.a011189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129(7):1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Testa JR, Tsichlis PN. AKT signaling in normal and malignant cells. Oncogene. 2005;24(50):7391–7393. doi: 10.1038/sj.onc.1209100. [DOI] [PubMed] [Google Scholar]
- 91.Yang WL, Wu CY, Wu J, Lin HK. Regulation of Akt signaling activation by ubiquitination. Cell Cycle. 2010;9(3):487–497. doi: 10.4161/cc.9.3.10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chan CH, Li CF, Yang WL, Gao Y, Lee SW, Feng Z, Huang HY, Tsai KK, Flores LG, Shao Y, Hazle JD, Yu D, Wei W, Sarbassov D, Hung MC, Nakayama KI, Lin HK. The Skp2-SCF E3 ligase regulates Akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis. Cell. 2012;149(5):1098–1111. doi: 10.1016/j.cell.2012.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.de la Cruz-Herrera CF, Campagna M, Lang V, Del Carmen Gonzalez-Santamaria J, Marcos-Villar L, Rodriguez MS, Vidal A, Collado M, Rivas C. SUMOylation regulates AKT1 activity. Oncogene. 2014 doi: 10.1038/onc.2014.48. [DOI] [PubMed] [Google Scholar]
- 94.Li R, Wei J, Jiang C, Liu D, Deng L, Zhang K, Wang P. Akt SUMOylation regulates cell proliferation and tumorigenesis. Cancer Res. 2013;73(18):5742–5753. doi: 10.1158/0008-5472.CAN-13-0538. [DOI] [PubMed] [Google Scholar]
- 95.Yamagata K, Daitoku H, Takahashi Y, Namiki K, Hisatake K, Kako K, Mukai H, Kasuya Y, Fukamizu A. Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol Cell. 2008;32(2):221–231. doi: 10.1016/j.molcel.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 96.Sakamaki J, Daitoku H, Ueno K, Hagiwara A, Yamagata K, Fukamizu A. Arginine methylation of BCL-2 antagonist of cell death (BAD) counteracts its phosphorylation and inactivation by Akt. Proc Natl Acad Sci U S A. 2011;108(15):6085–6090. doi: 10.1073/pnas.1015328108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Le Romancer M, Treilleux I, Leconte N, Robin-Lespinasse Y, Sentis S, Bouchekioua-Bouzaghou K, Goddard S, Gobert-Gosse S, Corbo L. Regulation of estrogen rapid signaling through arginine methylation by PRMT1. Mol Cell. 2008;31(2):212–221. doi: 10.1016/j.molcel.2008.05.025. [DOI] [PubMed] [Google Scholar]
- 98.Gonzalez ME, DuPrie ML, Krueger H, Merajver SD, Ventura AC, Toy KA, Kleer CG. Histone methyltransferase EZH2 induces Akt-dependent genomic instability and BRCA1 inhibition in breast cancer. Cancer Res. 2011;71(6):2360–2370. doi: 10.1158/0008-5472.CAN-10-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127(3):469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 100.Cadigan KM, Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11(24):3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 101.Polakis P. Drugging Wnt signalling in cancer. EMBO J. 2012;31(12):2737–2746. doi: 10.1038/emboj.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Arce L, Yokoyama NN, Waterman ML. Diversity of LEF/TCF action in development and disease. Oncogene. 2006;25(57):7492–7504. doi: 10.1038/sj.onc.1210056. [DOI] [PubMed] [Google Scholar]
- 103.Mosimann C, Hausmann G, Basler K. Beta-catenin hits chromatin: regulation of Wnt target gene activation. Nat Rev Mol Cell Biol. 2009;10(4):276–286. doi: 10.1038/nrm2654. [DOI] [PubMed] [Google Scholar]
- 104.Chang CJ, Yang JY, Xia W, Chen CT, Xie X, Chao CH, Woodward WA, Hsu JM, Hortobagyi GN, Hung MC. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-beta-catenin signaling. Cancer Cell. 2011;19(1):86–100. doi: 10.1016/j.ccr.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ding Q, Xia W, Liu JC, Yang JY, Lee DF, Xia J, Bartholomeusz G, Li Y, Pan Y, Li Z, Bargou RC, Qin J, Lai CC, Tsai FJ, Tsai CH, Hung MC. Erk associates with and primes GSK-3beta for its inactivation resulting in upregulation of beta-catenin. Mol Cell. 2005;19(2):159–170. doi: 10.1016/j.molcel.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 106.Cheng AS, Lau SS, Chen Y, Kondo Y, Li MS, Feng H, Ching AK, Cheung KF, Wong HK, Tong JH, Jin H, Choy KW, Yu J, To KF, Wong N, Huang TH, Sung JJ. EZH2-mediated concordant repression of Wnt antagonists promotes beta-catenin-dependent hepatocarcinogenesis. Cancer Res. 2011;71(11):4028–4039. doi: 10.1158/0008-5472.CAN-10-3342. [DOI] [PubMed] [Google Scholar]
- 107.Lu H, Sun J, Wang F, Feng L, Ma Y, Shen Q, Jiang Z, Sun X, Wang X, Jin H. Enhancer of zeste homolog 2 activates wnt signaling through downregulating CXXC finger protein 4. Cell Death Dis. 2013;4:e776. doi: 10.1038/cddis.2013.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hussain M, Rao M, Humphries AE, Hong JA, Liu F, Yang M, Caragacianu D, Schrump DS. Tobacco smoke induces polycomb-mediated repression of Dickkopf-1 in lung cancer cells. Cancer Res. 2009;69(8):3570–3578. doi: 10.1158/0008-5472.CAN-08-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shi B, Liang J, Yang X, Wang Y, Zhao Y, Wu H, Sun L, Zhang Y, Chen Y, Li R, Zhang Y, Hong M, Shang Y. Integration of estrogen and Wnt signaling circuits by the polycomb group protein EZH2 in breast cancer cells. Mol Cell Biol. 2007;27(14):5105–5119. doi: 10.1128/MCB.00162-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jung HY, Jun S, Lee M, Kim HC, Wang X, Ji H, McCrea PD, Park JI. PAF and EZH2 induce Wnt/beta-catenin signaling hyperactivation. Mol Cell. 2013;52(2):193–205. doi: 10.1016/j.molcel.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mohan M, Herz HM, Takahashi YH, Lin C, Lai KC, Zhang Y, Washburn MP, Florens L, Shilatifard A. Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom) Genes Dev. 2010;24(6):574–589. doi: 10.1101/gad.1898410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li Z, Nie F, Wang S, Li L. Histone H4 Lys 20 monomethylation by histone methylase SET8 mediates Wnt target gene activation. Proc Natl Acad Sci U S A. 2011;108(8):3116–3123. doi: 10.1073/pnas.1009353108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Toyokawa G, Cho HS, Masuda K, Yamane Y, Yoshimatsu M, Hayami S, Takawa M, Iwai Y, Daigo Y, Tsuchiya E, Tsunoda T, Field HI, Kelly JD, Neal DE, Maehara Y, Ponder BA, Nakamura Y, Hamamoto R. Histone lysine methyltransferase Wolf-Hirschhorn syndrome candidate 1 is involved in human carcinogenesis through regulation of the Wnt pathway. Neoplasia. 2011;13(10):887–898. doi: 10.1593/neo.11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Blythe SA, Cha SW, Tadjuidje E, Heasman J, Klein PS. beta-Catenin primes organizer gene expression by recruiting a histone H3 arginine 8 methyltransferase, Prmt2. Dev Cell. 2010;19(2):220–231. doi: 10.1016/j.devcel.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cha B, Kim W, Kim YK, Hwang BN, Park SY, Yoon JW, Park WS, Cho JW, Bedford MT, Jho EH. Methylation by protein arginine methyltransferase 1 increases stability of Axin, a negative regulator of Wnt signaling. Oncogene. 2011;30(20):2379–2389. doi: 10.1038/onc.2010.610. [DOI] [PubMed] [Google Scholar]
- 116.Appella E, Anderson CW. Post-translational modifications and activation of p53 by genotoxic stresses. Eur J Biochem. 2001;268(10):2764–2772. doi: 10.1046/j.1432-1327.2001.02225.x. [DOI] [PubMed] [Google Scholar]
- 117.Lane DP. Exploiting the p53 pathway for cancer diagnosis and therapy. Br J Cancer. 1999;80(Suppl 1):1–5. [PubMed] [Google Scholar]
- 118.Prives C, Hall PA. The p53 pathway. J Pathol. 1999;187(1):112–126. doi: 10.1002/(SICI)1096-9896(199901)187:1<112::AID-PATH250>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 119.Brooks CL, Gu W. Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 regulation. Curr Opin Cell Biol. 2003;15(2):164–171. doi: 10.1016/s0955-0674(03)00003-6. [DOI] [PubMed] [Google Scholar]
- 120.Momand J, Wu HH, Dasgupta G. MDM2--master regulator of the p53 tumor suppressor protein. Gene. 2000;242(1–2):15–29. doi: 10.1016/s0378-1119(99)00487-4. [DOI] [PubMed] [Google Scholar]
- 121.Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91(3):325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 122.Zhang X, Wen H, Shi X. Lysine methylation: beyond histones. Acta Biochim Biophys Sin (Shanghai) 2012;44(1):14–27. doi: 10.1093/abbs/gmr100. [DOI] [PubMed] [Google Scholar]
- 123.Huang J, Berger SL. The emerging field of dynamic lysine methylation of non-histone proteins. Curr Opin Genet Dev. 2008;18(2):152–158. doi: 10.1016/j.gde.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 124.Lan F, Shi Y. Epigenetic regulation: methylation of histone and non-histone proteins. Sci China C Life Sci. 2009;52(4):311–322. doi: 10.1007/s11427-009-0054-z. [DOI] [PubMed] [Google Scholar]
- 125.West LE, Gozani O. Regulation of p53 function by lysine methylation. Epigenomics. 2011;3(3):361–369. doi: 10.2217/EPI.11.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ivanov GS, Ivanova T, Kurash J, Ivanov A, Chuikov S, Gizatullin F, Herrera-Medina EM, Rauscher F, 3rd, Reinberg D, Barlev NA. Methylation-acetylation interplay activates p53 in response to DNA damage. Mol Cell Biol. 2007;27(19):6756–6769. doi: 10.1128/MCB.00460-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA, Kubicek S, Opravil S, Jenuwein T, Berger SL. Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006;444(7119):629–632. doi: 10.1038/nature05287. [DOI] [PubMed] [Google Scholar]
- 128.Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M, Opravil S, Shiekhattar R, Bedford MT, Jenuwein T, Berger SL. p53 is regulated by the lysine demethylase LSD1. Nature. 2007;449(7158):105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- 129.Iwabuchi K, Bartel PL, Li B, Marraccino R, Fields S. Two cellular proteins that bind to wild-type but not mutant p53. Proc Natl Acad Sci U S A. 1994;91(13):6098–6102. doi: 10.1073/pnas.91.13.6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Iwabuchi K, Li B, Massa HF, Trask BJ, Date T, Fields S. Stimulation of p53-mediated transcriptional activation by the p53-binding proteins, 53BP1 and 53BP2. J Biol Chem. 1998;273(40):26061–26068. doi: 10.1074/jbc.273.40.26061. [DOI] [PubMed] [Google Scholar]
- 131.Shi X, Kachirskaia I, Yamaguchi H, West LE, Wen H, Wang EW, Dutta S, Appella E, Gozani O. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol Cell. 2007;27(4):636–646. doi: 10.1016/j.molcel.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Huang J, Dorsey J, Chuikov S, Perez-Burgos L, Zhang X, Jenuwein T, Reinberg D, Berger SL. G9a and Glp methylate lysine 373 in the tumor suppressor p53. J Biol Chem. 2010;285(13):9636–9641. doi: 10.1074/jbc.M109.062588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Collingwood TN, Urnov FD, Wolffe AP. Nuclear receptors: coactivators, corepressors and chromatin remodeling in the control of transcription. J Mol Endocrinol. 1999;23(3):255–275. doi: 10.1677/jme.0.0230255. [DOI] [PubMed] [Google Scholar]
- 134.McDonnell DP, Norris JD. Connections and regulation of the human estrogen receptor. Science. 2002;296(5573):1642–1644. doi: 10.1126/science.1071884. [DOI] [PubMed] [Google Scholar]
- 135.McKenna NJ, Lanz RB, O’Malley BW. Nuclear receptor coregulators: cellular and molecular biology. Endocr Rev. 1999;20(3):321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 136.Hervouet E, Cartron PF, Jouvenot M, Delage-Mourroux R. Epigenetic regulation of estrogen signaling in breast cancer. Epigenetics. 2013;8(3):237–245. doi: 10.4161/epi.23790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mann M, Cortez V, Vadlamudi RK. Epigenetics of estrogen receptor signaling: role in hormonal cancer progression and therapy. Cancers (Basel) 2011;3(3):1691–1707. doi: 10.3390/cancers3021691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hall JM, McDonnell DP. Coregulators in nuclear estrogen receptor action: from concept to therapeutic targeting. Mol Interv. 2005;5(6):343–357. doi: 10.1124/mi.5.6.7. [DOI] [PubMed] [Google Scholar]
- 139.Green KA, Carroll JS. Oestrogen-receptor-mediated transcription and the influence of co-factors and chromatin state. Nat Rev Cancer. 2007;7(9):713–722. doi: 10.1038/nrc2211. [DOI] [PubMed] [Google Scholar]
- 140.Heldring N, Pike A, Andersson S, Matthews J, Cheng G, Hartman J, Tujague M, Strom A, Treuter E, Warner M, Gustafsson JA. Estrogen receptors: how do they signal and what are their targets. Physiol Rev. 2007;87(3):905–931. doi: 10.1152/physrev.00026.2006. [DOI] [PubMed] [Google Scholar]
- 141.Teyssier C, Le Romancer M, Sentis S, Jalaguier S, Corbo L, Cavailles V. Protein arginine methylation in estrogen signaling and estrogen-related cancers. Trends Endocrinol Metab. 2010;21(3):181–189. doi: 10.1016/j.tem.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 142.Tsai MJ, O’Malley BW. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 143.Barnes CJ, Vadlamudi RK, Kumar R. Novel estrogen receptor coregulators and signaling molecules in human diseases. Cell Mol Life Sci. 2004;61(3):281–291. doi: 10.1007/s00018-003-3222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Magnani L, Lupien M. Chromatin and epigenetic determinants of estrogen receptor alpha (ESR1) signaling. Mol Cell Endocrinol. 2014;382(1):633–641. doi: 10.1016/j.mce.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 145.Dreijerink KM, Mulder KW, Winkler GS, Hoppener JW, Lips CJ, Timmers HT. Menin links estrogen receptor activation to histone H3K4 trimethylation. Cancer Res. 2006;66(9):4929–4935. doi: 10.1158/0008-5472.CAN-05-4461. [DOI] [PubMed] [Google Scholar]
- 146.Ansari KI, Kasiri S, Hussain I, Mandal SS. Mixed lineage leukemia histone methylases play critical roles in estrogen-mediated regulation of HOXC13. FEBS J. 2009;276(24):7400–7411. doi: 10.1111/j.1742-4658.2009.07453.x. [DOI] [PubMed] [Google Scholar]
- 147.Mo R, Rao SM, Zhu YJ. Identification of the MLL2 complex as a coactivator for estrogen receptor alpha. J Biol Chem. 2006;281(23):15714–15720. doi: 10.1074/jbc.M513245200. [DOI] [PubMed] [Google Scholar]
- 148.Kim JH, Sharma A, Dhar SS, Lee SH, Gu B, Chan CH, Lin HK, Lee MG. UTX and MLL4 coordinately regulate transcriptional programs for cell proliferation and invasiveness in breast cancer cells. Cancer Res. 2014;74(6):1705–1717. doi: 10.1158/0008-5472.CAN-13-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kim H, Heo K, Kim JH, Kim K, Choi J, An W. Requirement of histone methyltransferase SMYD3 for estrogen receptor-mediated transcription. J Biol Chem. 2009;284(30):19867–19877. doi: 10.1074/jbc.M109.021485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhang X, Tanaka K, Yan J, Li J, Peng D, Jiang Y, Yang Z, Barton MC, Wen H, Shi X. Regulation of estrogen receptor alpha by histone methyltransferase SMYD2-mediated protein methylation. Proc Natl Acad Sci U S A. 2013;110(43):17284–17289. doi: 10.1073/pnas.1307959110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Garcia-Bassets I, Kwon YS, Telese F, Prefontaine GG, Hutt KR, Cheng CS, Ju BG, Ohgi KA, Wang J, Escoubet-Lozach L, Rose DW, Glass CK, Fu XD, Rosenfeld MG. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 2007;128(3):505–518. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C, Avvedimento EV. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319(5860):202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- 153.Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115(6):751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 154.Frietze S, Lupien M, Silver PA, Brown M. CARM1 regulates estrogen-stimulated breast cancer growth through up-regulation of E2F1. Cancer Res. 2008;68(1):301–306. doi: 10.1158/0008-5472.CAN-07-1983. [DOI] [PubMed] [Google Scholar]
- 155.Feng Q, Yi P, Wong J, O’Malley BW. Signaling within a coactivator complex: methylation of SRC-3/AIB1 is a molecular switch for complex disassembly. Mol Cell Biol. 2006;26(21):7846–7857. doi: 10.1128/MCB.00568-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wang H, Huang ZQ, Xia L, Feng Q, Erdjument-Bromage H, Strahl BD, Briggs SD, Allis CD, Wong J, Tempst P, Zhang Y. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 2001;293(5531):853–857. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- 157.Mostaqul Huq MD, Gupta P, Tsai NP, White R, Parker MG, Wei LN. Suppression of receptor interacting protein 140 repressive activity by protein arginine methylation. EMBO J. 2006;25(21):5094–5104. doi: 10.1038/sj.emboj.7601389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Teyssier C, Ma H, Emter R, Kralli A, Stallcup MR. Activation of nuclear receptor coactivator PGC-1alpha by arginine methylation. Genes Dev. 2005;19(12):1466–1473. doi: 10.1101/gad.1295005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ntziachristos P, Tsirigos A, Welstead GG, Trimarchi T, Bakogianni S, Xu L, Loizou E, Holmfeldt L, Strikoudis A, King B, Mullenders J, Becksfort J, Nedjic J, Paietta E, Tallman MS, Rowe JM, Tonon G, Satoh T, Kruidenier L, Prinjha R, Akira S, Van Vlierberghe P, Ferrando AA, Jaenisch R, Mullighan CG, Aifantis I. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature. 2014;514(7523):513–517. doi: 10.1038/nature13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hock H. A complex Polycomb issue: the two faces of EZH2 in cancer. Genes Dev. 2012;26(8):751–755. doi: 10.1101/gad.191163.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Simon C, Chagraoui J, Krosl J, Gendron P, Wilhelm B, Lemieux S, Boucher G, Chagnon P, Drouin S, Lambert R, Rondeau C, Bilodeau A, Lavallee S, Sauvageau M, Hebert J, Sauvageau G. A key role for EZH2 and associated genes in mouse and human adult T-cell acute leukemia. Genes Dev. 2012;26(7):651–656. doi: 10.1101/gad.186411.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.McGrath J, Trojer P. Targeting histone lysine methylation in cancer. Pharmacol Ther. 2015 doi: 10.1016/j.pharmthera.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 163.Lee YH, Coonrod SA, Kraus WL, Jelinek MA, Stallcup MR. Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proc Natl Acad Sci U S A. 2005;102(10):3611–3616. doi: 10.1073/pnas.0407159102. [DOI] [PMC free article] [PubMed] [Google Scholar]