Abstract
Anautogenous mosquitoes depend on vertebrate blood as nutrient source for their eggs. A highly efficient set of membrane transporters mediates the massive movement of nutrient amino acids between mosquito tissues after a blood meal. Here we report the characterization of the amino acid transporter Slimfast (Slif) from the yellow fever mosquito Aedes aegypti using codon-optimized heterologous expression. Slif is a well-known component of the target-of-rapamycin signaling pathway and fat body nutrient sensor, but its substrate specificity and transport mechanism were unknown. We found that Slif transports essential cationic and neutral amino acids with preference for arginine. It has an unusual dual-affinity mechanism with only the high affinity being Na+-dependent. Tissue-specific expression and blood meal-dependent regulation of Slif are consistent with conveyance of essential amino acids from gut to fat body. Slif represents a novel transport system and type of transceptor for sensing and transporting essential amino acids during mosquito reproduction.
Introduction
A principal feature in metazoan evolution is the loss of pathways for the biosynthesis of a group of nine essential amino acids (AAs)1. Concurrently, metazoans acquired effective mechanisms for the uptake of these and other AAs from their food, their distribution within the body, and the monitoring of intracellular and extracellular AA concentrations2, 3.
A blood meal (BM) taken from a vertebrate host is the source of essential AAs for egg development in anautogenous mosquitos4. Post blood meal (PBM) females mosquitos undergo complex changes in tissue-specific gene expression in order to transfer nutrient AAs from the digested meal to the developing oocytes, a process called vitellogenesis5, 6. Vitellogenesis is triggered upon the initial detection of a surge of free AAs in the mosquito circulation7, 8 and involves coordinated hormonal and nutritional regulation of enzymatic and transport processes in midgut, fat body, and ovaries8, 9, 10. The three subsequent key mechanisms of this process are: BM digestion in the midgut, synthesis and secretion of yolk protein precursors (YPPs) by the fat body, and receptor-mediated uptake and deposition of YPPs and other nutrient reserves in developing oocytes.
Up to 19% of the total ingested AAs are used for the synthesis of soluble yolk proteins while the rest is largely used for energy production11, 12, 13. The digested AAs are transported across the apical and the basolateral membranes of the gut epithelium into the hemolymph and subsequently across the plasma membrane of fat body cells. The yolk proteins synthesized by the fat body are secreted into the hemolymph and are absorbed by the developing oocytes via receptor-mediated endocytosis14, 15. Evidently, the membrane transport of free AAs is also necessary to supply a surge of protein synthesis in the ovaries PBM16. Hence, the rapid redistribution of nutrient AAs during mosquito vitellogenesis requires highly regulated and efficient transport mechanisms. However, the molecular properties and regulation of such mechanisms are only partly explored.
To date, only a few transporters that mediate uptake of nutrient AAs in mosquitoes have been characterized2, 17, 18. Most of these transporters are expressed in the larval stage and their role and significance in adult metabolism remains enigmatic. The aquatic larval stage of mosquitoes substantially differs from the terrestrial reproductive stage in terms of nutrient availability and metabolic demand2, 19.
In previous work, we have identified and characterized several AA transporters from the Solute Carrier Family 7 (SLC7) of Ae. aegypti the yellow fever mosquito. The SLC7 family includes two subgroups: the Cationic AA Transporters (CATs) and the Heterodimeric AA Transporters (HATs)20. RNAi-mediated knockdown of six of the eleven members of the SLC7 family reduced target of rapamycin (TOR)- mediated nutrient signaling in the mosquito fat body, which resulted in limited egg production21, 22. We cloned and characterized a first mosquito CAT, AaCAT1, and found that it is a Na+-independent transporter with a unique selectivity to L-histidine23. The AaCAT1 paralog, AaSlif (=CAT3) (GenBank accession #: XP_001662274) is closely related to fruit fly slimfast (slif). Characterization of slif homologs in mosquito is of high interest since it may function as a mechanism combining transporter and receptor properties (transceptor). Drosophila slif is involved in signaling AA-availability in the metabolic regulation of fly growth24 and behavioral control of feeding arousal25. Both these functions comprise appealing targets for vector control. However, transport function of slif was unknown and couldn’t be extrapolated from mammalian paralogs and insect CATs due to significant functional divergence of these transporters.
Here we describe the tissue-specific expression along with the biochemical-biophysical properties of AaSlif (= AaCAT3). Unexpectedly, our study revealed an unusual set of biophysical properties of this transporter including: dual-affinity for cationic and neutral AAs, lack of L-isomer selectivity, and a significant Na+- dependent component necessary for its high affinity transport mode. These findings expand our current knowledge of transport functions and biological significance of the CAT-SLC7 mechanisms. The present studies also describe properties of a new AA transceptor with critical roles in nutrition and reproduction of an important disease vector.
Results
Molecular cloning of AaSlif and genomic organization of a CAT cluster
To determine the AaSlif cDNA sequence we used 5’ RACE. We found an additional intron and an exon at the 5’-end of the transcript that was absent in the current annotation of the Ae. aegypti genome (VectorBase AAEL012131; see Supplementary Figure 1). The AaSlif gene consists of 5 exons and 4 introns, which are spliced to an 1881 nucleotide-long open reading frame (ORF) encoding a 626 amino acid protein. When compared to the published genome sequence26, the AaSlif ORF contains 21 synonymous SNPs, which likely represent differences between the Liverpool (Genome sequence in VectorBase) and the Rockefeller strains (this study) of Ae. aegypti. The gene is located on the reverse strand of the supercontig 1.658 (Supplementary Figure 1A). Two other genes of the AaCAT-SLC7 subfamily (AaCAT1 and AaCAT2) are localized in a proximal reverse cluster downstream from AaSlif. The fourth gene of the AaCAT-SLC7 subfamily AAEL012129 is found in the forward strand and encodes the complete ORF of AaCAT4 (Supplementary Figure 1A). AaCAT2 (AAEL012133) was only partly predicted in the current annotation of Aedes genome (Supplementary Figure 1, brown splicing pattern). Our in silico analysis revealed a splicing scheme that is also supported by the gene expression pattern found in studies of the mosquito transcriptome (Supplementary Figure 1, red + brown splicing pattern and magenta bars, respectively; based on the data from VectorBase).
Selection and conservation of the SBM in the Slif mechanism
In a previous phylogenetic study, we found that D. melanogaster slif (DmSilf) forms a phylogenetic cluster with two putative orthologs from Ae. aegypti, AaCAT1 and AaSlif23. Pairwise identity between DmSlif and AaCAT1 is 56.9%; between the DmSlif and AaSlif 58.6 %, with 337 and 367 identical sites, respectively. To identify structure-function conservation among members of this cluster we aligned the amino acid sequences of DmSlif orthologs from selected insect representatives against sequences and structural motifs of the recently crystalized prokaryotic relatives of the SLC7 family: the Methanocaldococcus jannaschii ApcT27 and Escherichia coli AdiC28 (Supplementary Figure 1B). In spite of only moderate pairwise identity, the prokaryotic and eukaryotic transporters aligned well in a region between the first and tenth transmembrane domain (TMD). The prokaryotic TMDs 11 and 12 aligned well to insect TMDs 11–12 as well as 13–14 with similar homology patterns (Supplementary Figure 1B). The possibility of an alternative alignment supports the idea that the two C-terminal TMDs were duplicated in an universal CAT ancestor23.
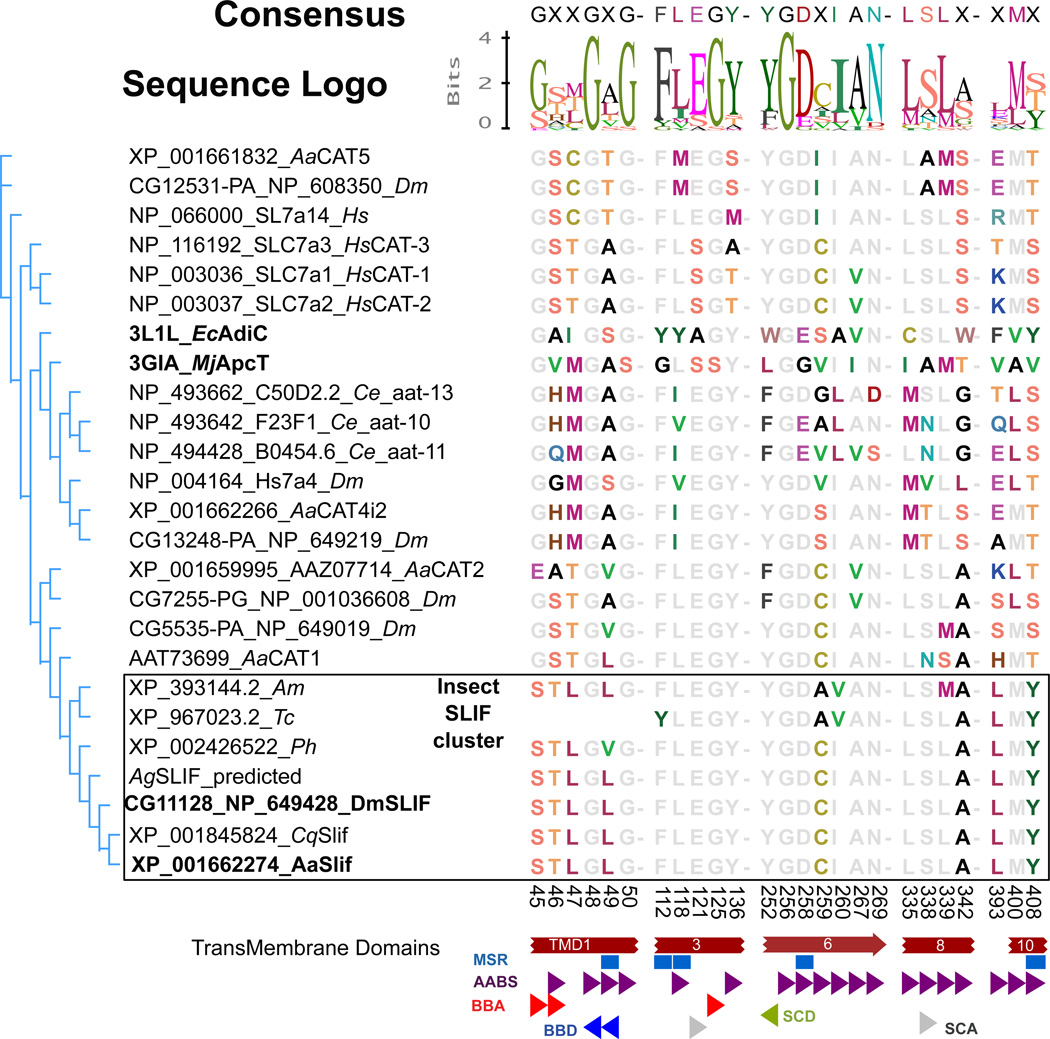
Fig. 1 shows the alignment pattern of a putative substrate-binding motif (SBM) interpolated from the EcAdiC 3D structure. It reveals both highly conserved and variable sites. Importantly, it defines that the predicted SBM residues are identical among dipteran slif orthologs and also are strongly conserved in the flour beetle (Tribolium castaneum), the body louse (Pediculus humanus corporis) and the honey bee (Apis mellifera) orthologs.
Fig 1. Bioinformatics analysis of the AaSlif SBM.
Shown are a Neighbor-Joining consensus tree and an alignment of the putative Substrate Binding Motif (SBM) residues of selected CAT-SLC7 members. Putative Slif orthologs (outlined box) form phylogenetic cluster with exactly one ortholog per selected insect genome and also show strong conservation of the SBM. The names of Drosophila and Aedes slif orthologs and their prokaryotic homologs used for the identification and alignment of SBMs are highlighted by bold font. Colored and gray fonts depict variable and significantly conserved residues, respectively. The aligned numbers indicate the amino acid positions in the AaSlif protein sequence. The partial SBM of the Tribolium Slif is due to its incomplete annotation. The colored shapes depict specific putative component of the SBMs abbreviated as: AABS, amino acid binding sites; BBA, backbone acceptor; BBD, backbone donor; MSR, mutation sensitive residues; SCA, side chain acceptor; SCD, side chain donor. Species abbreviations are: Aa, Aedes aegypti; Ce, Caenorhabditis elegans; Cq, Culex quinquefasciatus; Hs, Homo sapiens, Ph, Pediculus humanus, Tc, Tribolium castaneum.
Heterologous expression & electrophysiological characterization
The heterologous expression of the original AaSlif transcript (GenBank accession #: KM593906) in X. laevis oocyte resulted in a significant increase of Arg-induced currents compared to water-injected control oocytes (I = 10.2 ± 5.8 nA for 10 mM L-Arg; n > 3, p = 0.014; t-test). However, such a signal was considered too weak for an electrophysiological characterization of the transport mechanism.
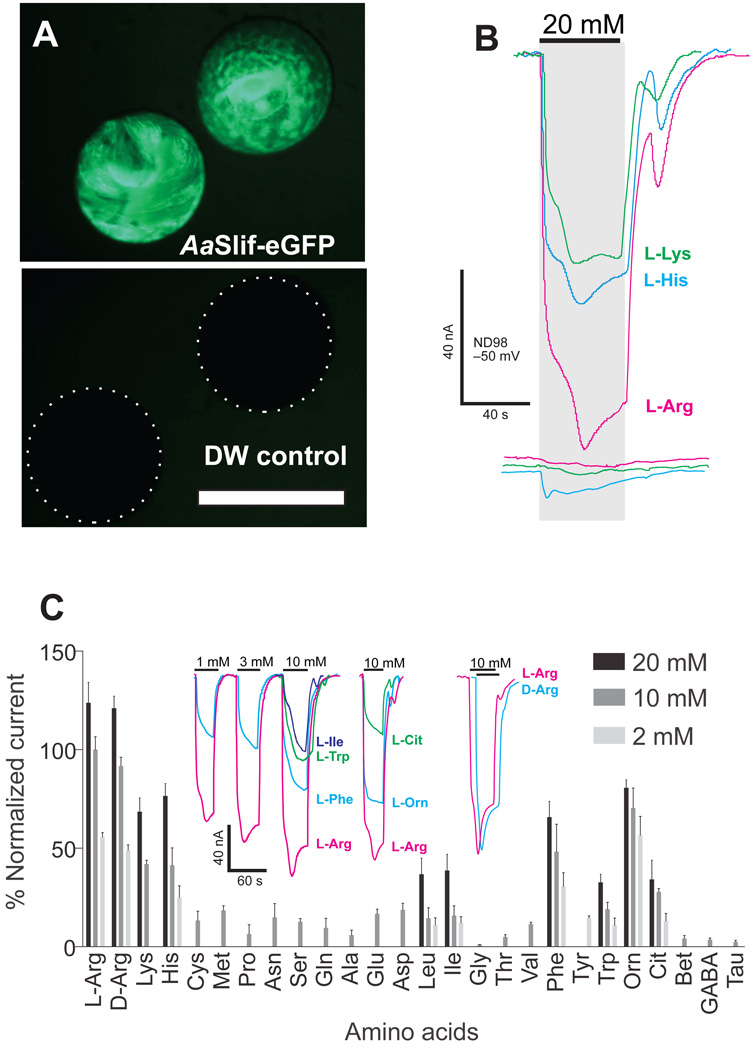
To improve AaSlif expression, we ordered synonymous optimized synthetic variants of AaSlif with a Xenopus-codon usage (synthesized by Genwiz® Inc., South Plainfield, NJ) and cloned them in the pXOOM expression vector. Two plasmids: AaSlifco-pXOOM (GenBank accession #: KM639700) and AaslifcoeGFP-pXOOM (GenBank accession #: KM593907) both with the AaSlif ORF and the latter also fused to eGFP at the C-terminus, were tested. The expression of AaSlifcoGFP induced bright GFP-specific fluorescence that was correlated with a large inward current upon application of cationic AAs in oocytes on days 4–10 after injection of cRNA (~10 nA at 1 µM and ~100 nA at 10 mM of L-Arg; Fig. 2A). Importantly, the expression of AaSlifco and AaSlifcoGFP resulted in no detectable differences in measured amplitude, saturable kinetics, and voltage dependency; therefore, both variants could be used interchangeably in the analysis (Supplementary Figure 2).
Fig. 2. Functional expression and AA-specificity profile of AaSlif.
A. Functional expression of codon-optimized AaSlif-eGFP fusion protein showed bright fluorescence compare to control deionized water injected oocytes (bottom panel, white dashed outlines indicate position of control oocytes that are almost invisible with identical setting of GFP-specific filter cube (Nikon 49583; EN GFP LP: Excitation Hq 470/40, Dichroic 495EM, Emission 500LP) and camera sensitivity; scale bar represents 1 mm. B. The expression of AaSlif correlates with large cationic amino acid-induced current responses. The colored lines represent currents induced in the AaSlif-eGFP expressing oocytes (top traces) versus control oocytes (bottom traces) after application 20 mM of L-enantiomer of Lysine (K), Histidine (H), and Arginine (R). C. AA specificity profile reconstructed from AA-induced currents. The bars are mean of percent normalized current + SD (n > 2 oocytes/samples per point) at 20, 10, and 2 mM concentrations of AAs. Currents were recorded using two-electrode voltage clamp. The colored traces show representative recordings of currents induced by 3 different concentration of L-Arg and L-Phe, as well as 10 mM of L-Trp, L-Ile, L-Arg, L-Orn, and L-Cit. Oocytes were clamped to –50 mV holding voltage (HV) in a constant flow micro-chamber perfused at ~ 2-chamber volume per second with ND98 or indicated solutions of AAs in 98 Na+ buffer. Currents were measured considering a steady-state current value and normalized to the mean of 10 mM L-Arg (= 100%) response in each oocyte for cross-oocyte comparison. Abbreviation: Orn, ornithine; Cit, citrulline; GABA, γ-aminobutyric acid; Tau, taurine.
AaSlif has an apparent preference for Arg (Fig. 2B; Arg > His ≈ Lys) without significant L-D enantiomer selectivity (Fig. 2C, current trace insert). AaSlif also generated significant inward currents on applications of L-citrulline (Cit) and L-ornithine (Orn), two metabolic AAs that we tested as intermediates of the Nitric Oxide (NO) synthesis and L-arginine recycling cascades (Supplementary Figure 2). In contrast, betaine, GABA, and taurine induced no significant responses. Notably, AaSlif also generates significant inward currents for neutral aliphatic (Ile, Leu, Met), aromatic (Phe, Trp, Tyr), and even acidic AAs (Supplementary Figure 2).
Substrate saturation kinetics, pH dependency, and substrate uptake
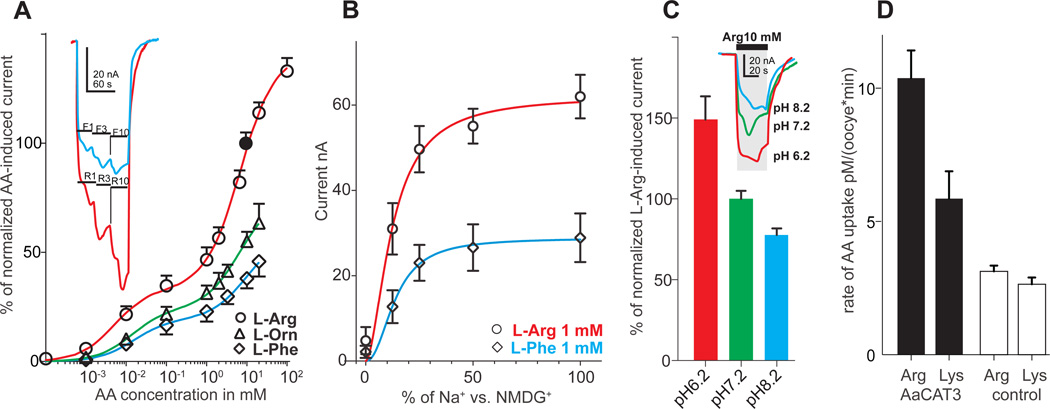
A substrate saturation assay revealed that AaSlif has two apparent saturation points: one at ~1 mM and second at ~20 mM of L-Arg (Fig. 3A, Supplementary Figure 3). The estimated KdHi and KdLo were 5.2 ± 1.4 µM and 6.9 ± 0.8 mM for L-Arg; 13.8 ± 6.2 µM and 6.8 ± 2.6 mM for L-Orn; and 15.6 ± 8.5 µM and 9.8 ± 6.5 mM for L-Phe. Our null hypothesis of a one point saturation model was rejected with p < 0.001, p = 0.018, and p = 0.034 for L-Arg, L-Phe and L-Orn, respectively (t-test). The amplitude of cationic and neutral amino acid-induced currents depended on the extracellular concentration of Na+, with apparent Na+ dissociation constant Kd = 12.4 ± 1.5 and 13.8 ± 2.2 mM and Hill constant η = 1.8 ± 0.6 and 2.2 ± 1.0 at 1 mM concentration of L-Arg and L-Phe, respectively (Fig. 3B). Acidification of the extracellular solution significantly increased the amplitudes of substrate-induced inward currents (~30–50% per pH unit; p < 0.001 for n > 3; t-test; Fig. 3C). To define the transport activity of AaSlif, we performed uptake assays with selected radioactive isotope-labeled substrates. The AaSlif expression increased the absorption of AAs in AaSlif-recombinant oocytes compared to deionized water-injected control oocytes. These results unequivocally verified that AaSlif functions as an AA transporter (Fig. 3D).
Fig. 3. AaSlif substrate saturation kinetics, pH dependency, and substrate specificity.
A. Saturation kinetic graphs for L-Arg, L-Phe and L-Orn induced currents. The insert show relative amplitudes of L-Arg and L-Phe-induced currents in AaSlif-expressing oocytes stimulated with 3 different concentrations of L-Arg and L-Phe. The data were normalized between different oocytes using mean value responses to 10 mM L-Arg (Filled circle point). Data are mean + or − SD for n > 3. B. Sodium saturation kinetics for 1 mM L-Arg and L-Phe induced currents. Data are mean ± SD for n = 3. C. pH dependency of 10 mM L-Arg induced currents. Bars are mean + SD for n = 3 oocyte/samples, that are significantly different p > 0.001; t-test. Insert shows a scaled superposition of typical L-Arg-induced currents induced upon sequential application of L-Arg at specified pH values. D. Uptake of radiolabeled cationic AAs in AaSlif-expressing oocytes (dark grey) vs. control oocytes (light grey). Data are mean + SD for n = 5 individual oocyte assays.
Cation-dependency of AA-induced currents in AaSlif oocytes
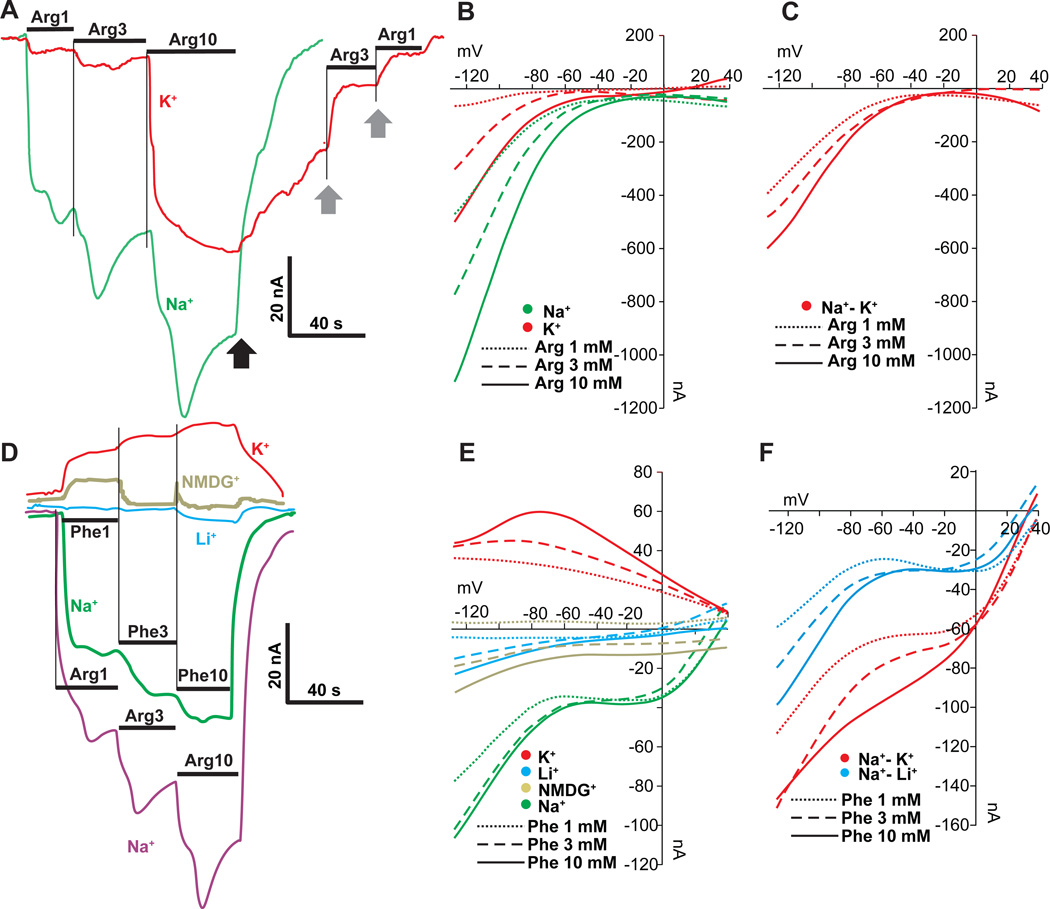
Substitution of extracellular Na+ by K+ significantly reduced the Arg-induced current (Fig. 4A) and modified voltage dependency of AaSlif, shifting the inflection point towards neutral (Fig. 4B). Surprisingly, the Na+- K+ substitution resulted in much lower current induced by Arg at concentrations below the KdLo (at 1 and 3 mM of L-Arg, reduction 75 ± 9.6% vs. same in Na+ media, n > 3) in comparison to the current induced by same substrate above the KdLo (at 10 mM of L-Arg, reduction 22 ± 4.6%, n > 3). It suggests that at low AA concentrations Arg-coupled current has large Na+-coupled component, while it may become largely Na+-independent at high concentrations of the organic substrate (Fig. 4A). The subtracted I/V plots showed additional details regarding interaction of the AaSlif mechanism with Na+ L-Arg+ ions (Fig. 4C). The data suggests that Na+ ions have a much greater contribution in the substrate-induced current compared to Arg+. This effect is especially profound at transmembrane voltages below − 60 mV.
Fig. 4. Cation dependency of AA-induced currents.
A. Example of substrate-induced currents in an AaSlif-expressing oocyte after application of 1, 3, and 10 mM of L-Arg in 98 Na+ and 98 K+ buffers (containing identical concentrations of Cl− and other ions). Black arrow indicates washing after L-Arg application with initial buffer saline. Grey arrows indicate accelerated recovery after re-application of 3 or 1 mM of L-Arg (for 98 K+ buffer only). B. Background-subtracted I/V plots of AaSlif-expressing oocytes in 98 Na+ and 98 K+ buffers with 3 different concentrations of L-Arg (insert). C. I/V plot calculated by subtraction of the current values measured in 98 K+ from those in 98 Na+. Results show no significant dependency of current on L-Arg concentration. D. L-Phe-induced currents in four different buffers. Results show that substitution of Na+ with Li+ and NMDG+ ions reduced substrate-induced currents, while the substitution with K+ reversed the current. E. I/V plots acquired from the same oocyte as in D with different cation compositions, as color-coded in insert. F. I/V plots calculated by subtraction of current values measured in 98 K+ and 98 Li+ buffers from those in 98 Na+ buffer.
We also noticed that Na+ - K+ substitution delayed the recovery of the transporter after washing with 10 mM L-Arg (Fig. 4A black arrow). The reduced recovery can be rescued by extracellular administration of 3 or 1 mM L-Arg (Fig. 4A gray arrows). This indicates that AaSlif can be locked in some ion conductive state upon interaction with high concentration of L-Arg at low Na+ or high K+ levels. However, a more detailed analysis of this phenomenon was beyond the scope of the characterization.
Phenylalanine induced currents in AaSlif-expressing oocytes
The L-Phe was tested as a representative of neutral aromatic substrates. Its application resulted in a substrate-induced inward current (Fig. 2C and Fig. 3 A and B). The Phe-induced current was strictly Na+ - dependent. Na+ ions cannot be substituted by NMDG+ (N-methyl D-gluconate) or Li+ (Fig. 4D–F). Interestingly, the substitution of Na+ with K+ resulted in a reversion of the Phe-induced current, which implies the presence of some cationic efflux or blockage of cationic influx, e.g. leak current (Fig. 4D and E). I/V plots for L-Phe with K+ display a characteristic nonlinearity with an inflection point close to − 70 mV (Fig. 4E).
AaSlif expression and regulation Post Blood Meal (PBM)
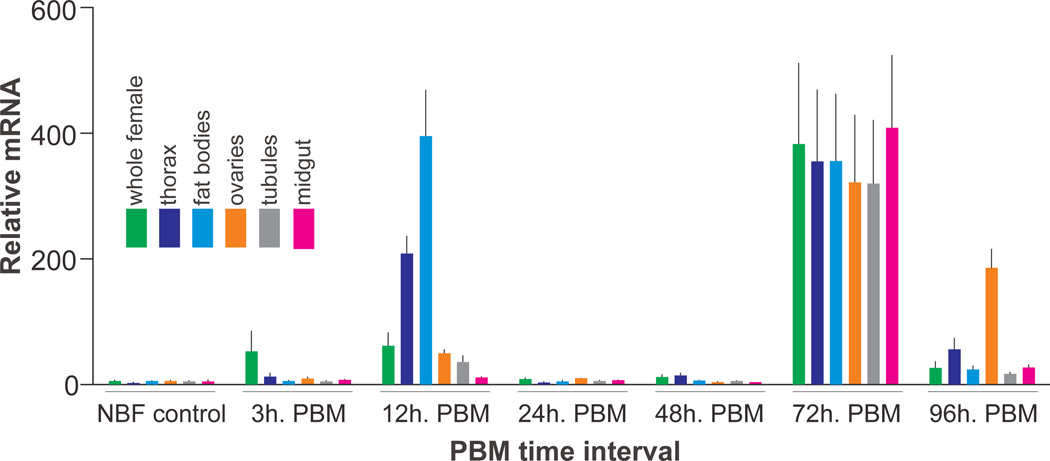
To reveal spatiotemporal expression and regulation of AaSlif we measured the accumulation/retention of its transcript in whole body, selected body parts, and organ samples isolated from non-blood fed (control) and blood fed mosquito females. The samples were isolated and tested after 3, 12, 24, 48, 72, and 96-hour PBM intervals. The AaSlif gene showed a varied and rapidly regulated expression profile (Fig. 5, Supplementary Figure 4). Specifically, it was significantly up regulated in whole females at 3, 12 and 72 h PBM (~10, 10, 60 times respectively; p<0.001; two-way ANOVA followed by Tukey’s HSD post hoc tests for n=3 samples). However, AaSlif expression declines in the whole body samples 24 and 48 h PBM close to the expression levels in the corresponding control samples. The elevated expression in 12 h PBM samples correlates with the intensified transcription of AaSlif in the fat bodies and thorax, as well as moderately elevated transcription in the ovaries and Malpighian tubules. In contrast, 72 h PBM samples showed strong over-expression across all selected body part and organs, including the mosquito gut. The level of transcript decreased in the 96h PBM samples, except for ovaries (Fig. 5).
Fig. 5. AaSlif expression and regulation in selected mosquito tissues and organs.
The transcript levels were determined using quantitative PCR (qPCR). The data represent relative quantities of AaSlif transcript that were normalized with qPCR levels of ribosomal protein S7 (rpS7) mRNA in the same tissue sample set. Bars are means + SEM for n = 3 replicates collected from different sets of mosquitoes (ANOVA followed by Tukey’s HSD post hoc test). The RNA samples were isolated from whole body and various body parts and organs of adult female mosquitoes grouped as shown by the color-coding insert. The horizontal scale indicates group-specific conditions: non-blood fed (NBF, control), 3, 12, 24, 48, 72, and 96 h post-blood meal (PBM).
Discussion
The requirement for acquisition of essential AAs through blood feeding makes anautogenous Ae. aegypti mosquitos an effective vector of important arboviral diseases, including dengue, yellow fever, and Chikungunya29. Blood meal-derived AAs are a major source of energy and building blocks during mosquito reproduction. Consequently, mosquitoes possess a highly efficient and tightly regulated system to distribute AAs between different tissues21. This transport system is only partly understood. In this study, we functionally expressed and characterized a mosquito ortholog of Drosophila slif, AaSlif. This transporter represents the first characterized member of the unique insect-specific cluster of the CAT-SLC7 subfamily of AA transporters23, 30 and the first characterized representative of a new transport system, SLIF.
The CAT acronym was originally coined for a group of mammalian AA transporters of the canonical y+ system, a Na+-independent transport mechanism selective for cationic L-AAs20, 31. The first CAT cloned and characterized via recombinant expression in Xenopus oocytes was serendipitously identified as a murine leukemia virus receptor with an explicit homology to yeast AA permeases and properties of the canonical y+ (CAT) system27, 28. Subsequent characterization of three from four existing mammalian members of the CAT-SLC7 subfamily showed similar AA selectivity patterns with some variations in affinity and tissue expression32. Our previous phylogenomic analysis of the CAT-SLC7 family showed that mammalian and putative insect CATs share a common ancestral root23. However, insect transporters formed independent clusters suggesting specificity of adaptations of CAT mechanisms in mammals and insects23. An important finding from the extended bioinformatics analysis of the present study is that there is only one slif orthologue per insect genome. Also, the putative SBMs of slif orthologs in fruit flies and mosquitos are identical even though these two insect groups diverged around 0.26 Bya33 (Fig. 1). These evidences strongly suggest that the SBM, transport mechanism, and biological functions of these transporters have undergone strong stabilizing (= purifying) selection. Another important finding is that SBMs of slif orthologs are distinct from the SBM of the previously characterized AaCAT123 and other mammalian and insect CATs (Fig. 1). These facts are consistent with the hypothesis that slif plays a unique role in insect metabolism and lacks genetic and functional redundancy.
Previously, three mosquito CATs: AaCAT1, AaCAT2, and AaSlif (= AaCAT3), were identified as components of the fat body nutrient sensor system that uses the TOR signaling pathway to activate reproductive processes in mosquito females after blood consumption21, 22. In the present study, we show that this trio plus AaCAT4 is physically co-localized in the mosquito genome (Supplementary Figure 1). We also found comparable transcripts of these CATs in the transcriptomic libraries at VectorBase34 (Supplementary Figure 1A, magenta pattern), supporting the notion that all four are functional genes. The tight clustering pattern suggest that CATs may be under a unified genetic control. However, considering the diverse SBMs we found in these transporters, the anticipated contributions in AA transport and signaling are clearly different. The genomic organization, together with the phylogenetic data, (Fig. 1) and functional data (this study and22) suggest that the CAT cluster in the Ae. aegypti genome is the result of gene duplications and subsequent functional adaptation of individual CATs for a specific physiological role, which may include a substrate specialization as found in AaCAT1 and AaSlif.
Using codon-optimized transcripts we achieved a significantly better expression of AaSlif in Xenopus oocytes compared to wild type transcript providing a unique opportunity for high-resolution electrophysiological characterization of this transporter. Chimeric fusion of AaSlif with eGFP did not modify expression and functional properties of this transporter (Fig. 2A) and could be used for the visual monitoring of CAT proteins in heterologous or naïve expression systems without functional artifacts.
Our functional characterization reveals that AaSlif is a high-efficiency carrier of cationic AAs with particular preference to Arg (Fig. 2 and 3). It also transports urea cycle metabolites that represent products of the NO synthesis pathway35. These characteristics of AaSlif are consistent with the generic properties of mammalian CAT-SLC7 transporters36. Nonetheless, the AaSlif mechanism differs from the y+ system of previously characterized mammalian CATs32 and the His-specific mosquito AaCAT123. AaSlif showed a substantially broader substrate spectrum compared to previously characterized CATs. Moreover, it equally transports L- and D-enantiomers of Arg (Fig. 2C); while, the mammalian CATs are strictly or significantly stereo-selective37, 38. It also generates significant inward currents for neutral aliphatic (Ile, Leu, Met), aromatic (Phe, Trp, Tyr), and even acidic AAs (Fig 2C). Such currents cannot be explained by facilitated diffusion of neutral AAs or cationic/neutral AA coupled exchange mechanisms found in mammalian CAT and HAT members of the SLC7 family23, 30. Such mechanisms should generate no outward currents in responses to neutral AAs. The ion substitution assay showed that the inward neutral AAs-induced current is due to involvement of inorganic ions (Fig. 4 D–F).
Unexpectedly, we observed complex saturation kinetics of the AaSlif consistent with an unusual dual-affinity mechanism and two dissociation constants: one in low-micromolar range and other in low-millimolar ranges (Fig. 3). Notably, its substrate-induced currents were pH-dependent and correlate with the uptake of radiolabeled substrates (Fig. 3C and D, respectively). Moreover, the high affinity mode of the AaSlif interaction with L-Arg was largely Na+-depended (Fig. 4 A–C). The Na+-coupling was also associated with the neutral AAs-induced inward current. This revealed a Na+- selectivity of the AaSlif mechanism that neither Li+ nor NMDG+ ions can substitute (Fig. 4). Interestingly, substitution of Na+ with K+ resulted in an inversion of the neutral AA-induced currents that was not observed upon Na+ substitution with Li+ or NMDG+ (Fig 4D–F). This indicates that K+ is a potent modulator of ion fluxes through AaSlif, a finding that requires further experimental analysis.
We propose that the identified properties represent adaptations of AaSlif for its action in an extended range of AA concentrations. At low AA availability, e.g. during the state-of-arrest before the mosquito takes a BM, it acts as secondary high affinity transporter using Na+ motive forces for the intracellular accumulation of AAs. In contrast, at high AA availability, e.g. after a BM, it acts as a low affinity, passive uniporter facilitating transmembrane diffusion of nutrient substrates without using the electrochemical energy of ion gradients. Dual-affinity mechanisms have never previously been reported among metazoan representatives of the Major Facilitator Superfamily (MSF). However, a dual affinities nitrate transporter (CHL1, also NRT1) has been recently reported among MFS representatives in plants39. When phosphorylated at Thr101, CHL1 acts as a high-affinity nitrate transporter, whereas, dephosphorylated transporter has low affinity. It has been suggested that switching involves dimerization, and that dual affinity CHL1 represents adaptation for managing highly variable concentrations of critical nutrients40.
AaSlif utilizes Na+- motive forces for the transport of neural AAs and also the high-affinity transport of cationic AAs. Such a coupling satisfies thermodynamic requirements for intracellular translocation of substrates from low concentration domains. The identification of the first CAT with explicit sodium dependency is truly remarkable for the field of transport biology. Although mammalian CATs generally do not use inorganic cations for translocation of cationic AAs, some Na+-dependency has been reported for representatives of the y+ system. For example, the first characterized murine CAT transports Cys and homoserine only in the presence of Na+ 41. Heterologous expressed human CAT-3 has been shown to mediate Na+ as well as K+-conductance42. Na+ dependency has also been reported in some HAT-SLC7 transporters resembling the y+L transport system32.
In this work, we have achieved unprecedented resolution for electrophysiological characterization of a CAT mechanism and tested a more comprehensive set of substrates compared to previous reports. Hence, it is possible that previous electrophysiological characterization of CAT mechanisms were simply incomplete. Alternatively, it is possible that a majority of vertebrate CATs substantially deviate from the insect slif type mechanism in such a way that these transporters either were never Na+ coupled, or secondarily lost the Na+ coupling. Such a possibility received indirect support from the comparative analysis of SBMs, which showed additional negatively charged glutamic acid residues in the AA position 121 (Fig.1). Such negatively charged SBMs are typical for Na+-coupled transport mechanisms. Additional analysis of the AaSlif mechanism could gain important insights about the molecular evolution and structural adaptations of CATs.
Amino acids play an important role as signaling molecules in mosquito vitellogenesis. Rising levels after a blood meal activate the TOR signaling pathway in midgut, fat body, and ovaries promoting tissue-specific reactions8, 10, 43, 44, 45, 46. TOR signaling in other key tissues involved in vitellogenesis like nervous system and Malpighian tubules has not yet been examined but it seems likely that these tissues are able to receive and process AA signals, too.
The AaSlif gene showed tissue-specific expression that was up-regulated by blood-meal with 12 and 72 h spikes in several of the tissues we examined (Fig. 5). The up-regulation of AaSlif at 12 h precedes the peak of digestive enzyme activities in the midgut and the peak in AA concentration in the Ae. aegypti hemolymph that coincide between 24 and 30 h PBM19. This suggests that female mosquitoes turn up the AaSlif mechanism to achieve rapid directed delivery of AAs to fat body and ovaries in times of abundance (PBM) and also to balance sufficient intracellular levels of AAs during limited accessibility of these substrates when hemolymph AA levels decline. The rather modest up-regulation of AaSlif in the midgut PBM is surprising and deserves additional studies. A possible explanation is that AaSlif may act through its Na+-independent high-throughput mode at this time in this tissue. Alternatively the midgut may utilize another group of AA transport systems than fat body or ovaries to shuttle the large amounts of amino acids into the hemolymph during this particular time point.
The high AaSlif expression in all tissues we found at 72 h PBM is surprising because at this time the digestion of the blood meal is already at its end and AA transport between tissues is thought to be declining. We have earlier shown similar transcription up-regulation during late vitellogenesis for several other SLC7-type AA transporters, specifically from the HAT family21. The gradual reduction of free AA levels at this time might trigger the over expression of these transporters. In the case of AaSlif this could compensate for a substrate concentration-dependent switch from its fast low-affinity transport mode to the slow high-affinity mode. An alternative explanation is that these proteins are synthesized in order to allow a quick response to the rapid changes in AA concentrations associated with a second blood meal. To fully understand the biological relevance of this late up-regulation phenomenon it would be interesting to determine the changes in the metabolomes and transportomes in these tissues over the cause of vitellogenesis with a comprehensive transcriptomics/metabolomics survey.
The AaSlif properties and expression profile also support its contribution to AAs signaling in the alimentary and reproductive tissues. Such a contribution is evident from our previous evaluation of the role of CATs in mosquito reproduction21, 22. This is also supported indirectly by data from studies of the slif mechanism in Drosophila. Fat body-specific suppression of DmSlif resulted in a global growth defect, similar to that seen in larvae grown under low nutrient conditions24, 48. Slif knock-down in the dopaminergic neurons of the Drosophila brain resulted in a strong inhibition of feeding25. This phenotype was associated with the General Control Nonderepressing 2 (GCN2)-mediated neuronal detection and behavioral rejection of AA-imbalanced meals by fly larvae. Therefore slif orthologs play an important role in the modulation of insect appetite and dietary source preferences.
An interesting question that warrants further research is to what degree AaSlif is involved in the nitric oxide (NO) signaling pathway in the mosquito fat body. NO is an important activator of the insect innate immune system and involved in the control of bacterial, and protozoan parasites49, 50. Nitric oxide synthase (NOS) catalyzes the synthesis of NO and the byproduct L-citrullin from its substrate L-Arg51. Both, L-citrullin and L-Arg are also substrates of AaSlif. Therefore it is possible if not likely that AaSlif plays a part in the control of NO synthesis by regulating intracellular L-Arg levels and thereby controlling NOS activity with consequences for the insect immune system and mosquito vectorial capacity.
AaSlif is the first characterized metazoan transporter with dual affinity for cationic and neutral AAs with a Na+-dependent component. Based on its unique properties and strong evolutionary conservation, we propose to define insect-specific orthologs of AaSlif as a new amino acid transport system, SLIF.
Expression profiles and transport kinetics suggest that AaSlif functions as a high-efficiency transporter genetically and biophysically adapted to act in diverse habitats where unautogenous mosquitos experience extremely variable availability of nutrient AAs, ranging from ‘very limited’ during a blood meal-preceding period to ‘over-saturated’ after a blood meal.
Arthropod-vectored infectious diseases put billions of people at risk worldwide. Novel means to control vector and pests populations are urgently needed. Our study reveals a new insect-specific transceptor for essential AAs, thereby opening new venues for the development of selective and environmentally-safe control methods targeting appetite, development, and reproduction in pest and vector insects.
Materials and Methods
Experimental animals
Aedes aegypti (Rockefeller strain) was used in all experiments. The mosquito population was maintained in the insectary of the Molecular Disease Vector Physiology Laboratory at New Mexico State University. Mosquitoes with high nutrient reserves were used for all experiments52. Larvae were reared using “Special Kitty” cat food (Walmart, Bentonville, AR). Adults were housed in cube-shaped cage (30 cm each side) with 20% sucrose solution at 26.5 °C, 70% RH and 16:8 hr (L:D) cycle. Chickens (Gallus gallus domesticus) were used to feed adult female mosquitoes, following the guidelines of Institutional Animal Care and Use Committee (IACUC) of the New Mexico State University under approved protocol (#2011-041).
Bioinformatic analysis
For combined phylogenomic-structural analysis the NCBI protein databases were screened for slif homologs using BLASTp. A set of protein sequences that were considered important and sufficient for comparative analysis of structural and evolutionary aspects of SLC7-CAT mechanisms was selected. Insect genomes were selected based on their taxonomic reciprocity, as well as quality and completes of an existed annotations. The protein sequences undergone multiple sequence alignment with MEGA53 implementation of the MUSCLE algorithm54 followed by manual correction. The transmembrane helices, known mutation sensitive site, and substrate interaction sites were interpolated from a consensus alignment of selected CATs with crystallized prokaryotic relative from Methanocaldococcus jannaschii, MjApcT27 and Escherichia coli, EcAdiC28 (Supplementary Figure 1B). The TMDs 11–14 were defined by using a TMD prediction algorithm and manual improvement. The coordinate file of bacterial proteins structure (3GIA, 3OB6, and 3L1L) were also used to build 3D models of AeCAT3 in L- arginine occlusion and outside open conformation that was used for docking of L- arginine and identification of additional substrate interaction residues integrated in the final figures. The Yasara55 structure software and MOE LigX56 algorithm were used for reconstruction of the 3D model and substrate interaction pattern, respectively. The identified residues position were trimmed from the complete aliment and used for preparing of substrate binding motifs diagram using Generous 6 software package57.
The evolutionary history was analyzed using the neighbor-joining, minimum evolution, maximum parsimony, and Bayesian methods that produced similar tree topologies. The neighbor-joining tree was selected as a more common representation of the evolutionary history of transport proteins. The tree was visualized using FigTree 1.3.1 software (A. Rambaut). It was drawn with estimated branch lengths used to infer the tree relative to a 3.7 billion-year-long scale from the root.
Molecular cloning and RACE verification
Total RNA was isolated from female mosquito 12 h after blood feeding using Trizol (Life Technologies, Carlsbad, CA). First-strand cDNA was synthesized on 1 µg of total RNA using Omniscript RT kit (QIAGEN, Valencia, CA). The AaCAT3 Open Reading Frame (ORF) was PCR amplified based on the predicted ORF of AAEL012131-RA gene (VectorBASE) using primers containing BamHI site for upstream primer or EcoRI site for downstream primer, cloned into pCR2.1 TOPO (Life Technologies, Carlsbad, CA) and sequenced. The Aaslif (AaCAT3) ORF was subcloned into an expression vector pXOOM. As we observed low expression signal with this construct we tested for a possibility of alternative 5’ extension of Aaslif ORF in mosquito genome using Rapid Amplification of cDNA Ends (5’ RACE). cDNA was synthesized using SMARTer RACE Amplification Kit (Clonetech, Mountain View, CA). 1 µg of RNA isolated from 12 h PBM females was used for the synthesis with 5’-CDS primer A and followed the product manual.
Gene-specific RACE PCR and nested PCR primers were designed using the 5’ sequence of AAEL012131-RA (VectorBase, for sequences see Supplementary Table 1). RACE PCR was performed with a gene-specific primer and Universal Primer A Mix or Nested Universal Primer A. RACE PCR products were separated by agarose gel electrophoresis, and bands were excised for gel extraction by QIAGEN Gel Extraction kit (QIAGEN, Valencia, CA). Gel-extracted PCR products were cloned into pCR2.1 TOPO vector using TOPO TA Cloning Kit (Life Technologies). Resulted 5’ end sequence was used as a query for local BLASTn against Ae. aegypti genome sequence (AaegL2, VectorBase) to find the genomic locus, and exon-intron boundary was manually adjusted so that the intron contains canonical splice site (GT-AG). As we did not identify alternatively spliced variant we concluded that the low expression was due to suboptimal translation in heterologous system and subsequently optimized AaSlif codon usage for expression in Xenopus laevis oocyte. Specifically, the Xenopus-optimized AaSlif was synthesized using GENEWIZ (South Plainfield, NJ) service. We also made a C-terminus fusion of AaSlif with eGFP reporter for monitoring the levels of expression and appropriate trafficking of the AaSlif in individual oocytes. The AaSlif (with or without eGFP) sequence was PCR amplified and cloned into pCR2.1 TOPO (Life Technologies, Carlsbad, CA) and restriction-digested insert was subcloned into the expression vector pXOOM using restriction sites BamHI (on 5’) and NotI (on 3’).
Heterologous expression and characterization
cRNA for heterologous expression was synthesized by in vitro transcription of XbaI-linearized Aaslifco-pXOOM and AaSlifcoeGFP-pXOOM plasmids using the mMessage-mMachine (Ambion Inc., Carlsbad, CA). The integrity and quantity of the RNA was confirmed by agarose gel electrophoresis. Surgically isolated, collagenase-treated, and defolliculated stage V-VI X. laevis oocytes were purchased from Ecocyte Bioscience US LLC (Austin, TX) and injected with ~35 ng of AaSlif cRNA using Nanoliter 2000 injector (WPI, Sarasota, FL). Oocytes were incubated for 2–6 days at 17 °C in sterile 98 mM Na+ oocyte media supplemented with 2.5 mM sodium pyruvate, 100 units/ml penicillin, 100 mg/ml streptomycin, and 5% horse serum. For ion dependency assay the 98 mM Na+ was substituted with the equimolar amount of K+, Li+ or NMDG+ (N-Methyl-D-glucamine). The exact recipes for solution preparation are summarized in the Supplementary Table 1. The substrate induced currents were recorded from voltage clamped oocytes at constant flow perfusion. The holding voltage was − 50 mV, except for ramp stimulation. The I/V data sets were acquired during ramp stimulation (step −130 mV; 200 ms, ramp −130 to +40 mV in 1 s; Supplementary Figure 2A). The I/V recording (1 kHz sampling) were filtered with digital 50 Hz low pass 8 pole Bessel filter, reduced by factor 30 (substitute average) and used to build I/V plots. The I/V before substrate application was subtracted to eliminate substrate independent current component. Additional corrections include subtraction of baseline drift current which reflects slow adaptation of ion pumps after ion substitution (typically < 5 nA), if required. The data (mean ± SD, n > 2 oocyte/sample for each point) were fit with a preferable two-site specific binding model: Site1 = BmaxHi*X/(KdHi+X), Site2 = BmaxLo*X/(KdLo+X), and Y = Site1+Site2 where: Bmax is apparent saturation current for High and Low affinity components, respectively; X are actual current values; and Kd are apparent dissociation constants. One site specific binding with Hill slope model was used as an Null hypothesis in evaluation of organic substrate binding kinetic and for approximation of Na+ binding data: Y=Bmax*Xη /(Kdη + Xη); were η is apparent Hill constant.
Radiolabeled amino acid uptake assay
AaSlif-expressing and control oocytes were conditioned in amino acid-free ND98 for 3–4 h and placed in 600 µL of 98 Na+ media containing 2 mM of final concentration of L-Arg and L-Lys with added 10 µL of 0.1 µCi/µL of L-[14C(U)]-Arg or 1 µCi/µL of L-[4,5–3H(N)]-Lys, respectively. After incubation for 20 min at room temperature oocytes were individually rinsed twice with excessive volume of ice-cold 98 Na+ media and lysed in 0.2 mL of 1% SDS solution with ultrasonic treatment. The individual samples were diluted with 2 mL of scintillation liquid and radioactivity was measured using a Beckman-Coulter LS6500 scintillation counter (Beckman, Indianapolis, IN). The amino acid uptake rate was extrapolated considering the isotope-labeled amounts, final concentrations, and specific activities of AAs (SA: L-Arg, 0.31 and L-Lys, 45 Ci/mmol, Moravek Biochemicals, Brea, CA). Disintegration per minute (DPM) data were converted to mmol/min using a constant (1 Ci = 2.22 × 1012 DPM): mmol/min = (DPM × molar ratio [all/isotope])/(2.2 × 1012 × SA × 20). Final data are shown in pmol/min.
Statistical analysis
Each experiment was repeated at least three times, using oocyte batches from different isolations. Statistical significance in the electrophysiology experiments was determined by unpaired Student’s t-test. All underlying assumptions of the test were met by the data. For quantitative PCR triplicate samples prepared from different mosquito individuals were analyzed. Two-way ANOVA followed by Tukey’s HSD post hoc test was used to validate significant differences of a spatio-temporal gene expression. The data met the assumptions of the test. The PRIZM5 (GraphPad Software, Inc. CA) and SigmaPlot/SigmaStat (Systat Software, Inc., CA) software was used for data collection, statistical evaluation, and final graph preparation. Statistical significance values and analysis-specific details are included in the result section and figure legends.
qPCR tissue expression analysis
RNA isolation, cDNA synthesis and qPCR analysis were completed in compliance with the MIQE guidelines58. Primer BLAST59 was used to develop gene-specific primers (Supplementary Table 2). Whole female (WF), fat body (FB), thorax (TX), Malpighian tubules (MT), ovaries (OV), and midgut (GT) were dissected from adult females non-blood fed, 3, 12, 24, 48, 72, and 96h post-blood meal. RNA was isolated using Trizol® (Invitrogen, Carlsbad, CA). First-strand cDNA synthesis was performed on 1 µg of total RNA in 20 µL reaction using Qiagen Omniscript® RT Kit (QIAGEN, Valencia, CA). Transcripts were normalized by qPCR analysis of ribosomal protein S7 (rpS7) levels(49) on an Eppendorf Mastercycler ep realplex® (Eppendorf, Hamburg, Germany) using iQ Supermix (Biorad, Hercules, CA) with 25 µl volume reactions. Each experiment was done with three independent biological replicates. PCR conditions were as follows: an initial incubation at 95 °C for 2 min; followed by 40 cycles of 95 °C for 15 sec, 55 °C for 15 sec, 72 °C for 20 sec.
Supplementary Material
Acknowledgements
This work was supported by grant # 1SC1AI109055 from the National Institute of Health.
Footnotes
Author contributions: D.Y.B., H.T. and I.A.H. designed the research; D.Y.B., S.D.R, R.M., H.T. and L.L.D. performed the research; D.Y.B., H.T., D.P.P., L.L.D. and I.A.H. analyzed the data; D.Y.B., H.T. and I.A.H. wrote the paper. D.Y.B. and H.T. contributed equally to this work.
The authors declare no conflict of interest.
References
- 1.Guedes R, Prosdocimi F, Fernandes G, Moura L, Ribeiro H, Ortega J. Amino acids biosynthesis and nitrogen assimilation pathways: a great genomic deletion during eukaryotes evolution. BMC genomics. 2011;12:S2. doi: 10.1186/1471-2164-12-S4-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boudko DY. Molecular basis of essential amino acid transport from studies of insect nutrient amino acid transporters of the SLC6 family (NAT-SLC6) Journal of insect physiology. 2012;58:433–449. doi: 10.1016/j.jinsphys.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boudko DY. Epithelial Transport Physiology. Springer: 2010. Molecular ontology of amino acid transport. (ed^(eds) [Google Scholar]
- 4.Attardo GM, Hansen IA, Raikhel AS. Nutritional regulation of vitellogenesis in mosquitoes: implications for anautogeny. Insect biochemistry and molecular biology. 2005;35:661–675. doi: 10.1016/j.ibmb.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Kokoza VA, Martin D, Mienaltowski MJ, Ahmed A, Morton CM, Raikhel AS. Transcriptional regulation of the mosquito vitellogenin gene via a blood meal-triggered cascade. Gene. 2001;274:47–65. doi: 10.1016/s0378-1119(01)00602-3. [DOI] [PubMed] [Google Scholar]
- 6.Price DP, et al. The Fat Body Transcriptomes of the Yellow Fever Mosquito Aedes aegypti, Pre- and Post- Blood Meal. PloS one. 2011;6 doi: 10.1371/journal.pone.0022573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uchida K, et al. Induction of oogenesis in mosquitoes (Diptera: Culicidae) by infusion of the hemocoel with amino acids. Journal of medical entomology. 2001;38:572–575. doi: 10.1603/0022-2585-38.4.572. [DOI] [PubMed] [Google Scholar]
- 8.Hansen IA, Attardo GM, Park JH, Peng Q, Raikhel AS. Target of rapamycin-mediated amino acid signaling in mosquito anautogeny. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:10626–10631. doi: 10.1073/pnas.0403460101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Badisco L, Van Wielendaele P, Vanden Broeck J. Eat to reproduce: a key role for the insulin signaling pathway in adult insects. Front Physiol. 2013;4:202. doi: 10.3389/fphys.2013.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansen IA, Attardo GM, Rodriguez SD, Drake LL. Four-way regulation of mosquito yolk protein precursor genes by juvenile hormone-, ecdysone-, nutrient-, and insulin-like peptide signaling pathways. Front Physiol. 2014;5:103. doi: 10.3389/fphys.2014.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.France KR, Judson C. Nitrogen partitioning and blood meal utilization by< i>Aedes aegypti</i>(Diptera Culicidae) Journal of insect physiology. 1979;25:841–846. [Google Scholar]
- 12.Marquardt WH. Biology of Disease Vectors. 2nd edn. Academic Press; 2004. [Google Scholar]
- 13.Briegel H. Mosquito reproduction: incomplete utilization of the blood meal protein for oögenesis. Journal of insect physiology. 1985;31:15–21. [Google Scholar]
- 14.Sappington TW, Hays AR, Raikhel AS. Mosquito Vitellogenin Receptor - Purification, Developmental and Biochemical-Characterization. Insect biochemistry and molecular biology. 1995;25:807–817. doi: 10.1016/0965-1748(95)00016-o. [DOI] [PubMed] [Google Scholar]
- 15.Sappington TW, Kokoza VA, Cho WL, Raikhel AS. Molecular characterization of the mosquito vitellogenin receptor reveals unexpected high homology to the Drosophila yolk protein receptor. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:8934–8939. doi: 10.1073/pnas.93.17.8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koller CN, Raikhel AS. Initiation of Vitellogenin Uptake and Protein-Synthesis in the Mosquito (Aedes-Aegypti) Ovary in Response to a Blood Meal. Journal of insect physiology. 1991;37:703–711. [Google Scholar]
- 17.Boudko D, Stevens B, Donly B, Harvey W. Nutrient amino acid and neurotransmitter transporters. Comprehensive Molecular Insect Science. 2005;4:255–309. [Google Scholar]
- 18.Gilbert LI, Gill SS. Insect pharmacology: channels, receptors, toxins and enzymes. Academic Press; 2010. [Google Scholar]
- 19.Clements AN. The Biology of Mosquitoes. Chapman & Hall; 1992. (ed^(eds). [Google Scholar]
- 20.Verrey F, Closs EI, Wagner CA, Palacin M, Endou H, Kanai Y. CATs and HATs: the SLC7 family of amino acid transporters. Pflugers Arch. 2004;447:532–542. doi: 10.1007/s00424-003-1086-z. [DOI] [PubMed] [Google Scholar]
- 21.Carpenter VK, Drake LL, Aguirre SE, Price DP, Rodriguez SD, Hansen IA. SLC7 amino acid transporters of the yellow fever mosquito Aedes aegypti and their role in fat body TOR signaling and reproduction. Journal of insect physiology. 2012;58:513–522. doi: 10.1016/j.jinsphys.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Attardo GM, Hansen IA, Shiao SH, Raikhel AS. Identification of two cationic amino acid transporters required for nutritional signaling during mosquito reproduction. The Journal of experimental biology. 2006;209:3071–3078. doi: 10.1242/jeb.02349. [DOI] [PubMed] [Google Scholar]
- 23.Hansen IA, et al. AaCAT1 of the Yellow Fever Mosquito, Aedes aegypti A NOVEL HISTIDINE-SPECIFIC AMINO ACID TRANSPORTER FROM THE SLC7 FAMILY. Journal of Biological Chemistry. 2011;286:10803–10813. doi: 10.1074/jbc.M110.179739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colombani J, Raisin S, Pantalacci S, Radimerski T, Montagne J, Leopold P. A nutrient sensor mechanism controls Drosophila growth. Cell. 2003;114:739–749. doi: 10.1016/s0092-8674(03)00713-x. [DOI] [PubMed] [Google Scholar]
- 25.Bjordal M, Arquier N, Kniazeff J, Pin JP, Léopold P. Sensing of amino acids in a dopaminergic circuitry promotes rejection of an incomplete diet in Drosophila. Cell. 2014;156:510–521. doi: 10.1016/j.cell.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 26.Nene V, et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science (New York, NY. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaffer PL, Goehring A, Shankaranarayanan A, Gouaux E. Structure and mechanism of a Na+-independent amino acid transporter. Science (New York, NY. 2009;325:1010–1014. doi: 10.1126/science.1176088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao X, et al. Mechanism of substrate recognition and transport by an amino acid antiporter. Nature. 2010;463:828–832. doi: 10.1038/nature08741. [DOI] [PubMed] [Google Scholar]
- 29.Eisen L, Moore CG. Aedes (Stegomyia) aegypti in the continental United States: a vector at the cool margin of its geographic range. Journal of medical entomology. 2013;50:467–478. doi: 10.1603/me12245. [DOI] [PubMed] [Google Scholar]
- 30.Fotiadis D, Kanai Y, Palacin M. The SLC3 and SLC7 families of amino acid transporters. Mol Aspects Med. 2013;34:139–158. doi: 10.1016/j.mam.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 31.Hundal HS, Taylor PM. Amino acid transceptors: gate keepers of nutrient exchange and regulators of nutrient signaling. Am J Physiol Endocrinol Metab. 2009;296:E603–613. doi: 10.1152/ajpendo.91002.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Closs EI, Boissel JP, Habermeier A, Rotmann A. Structure and function of cationic amino acid transporters (CATs) J Membr Biol. 2006;213:67–77. doi: 10.1007/s00232-006-0875-7. [DOI] [PubMed] [Google Scholar]
- 33.Arensburger P, et al. Sequencing of Culex quinquefasciatus Establishes a Platform for Mosquito Comparative Genomics. Science (New York, NY. 2010;330:86–88. doi: 10.1126/science.1191864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lawson D, et al. VectorBase: a data resource for invertebrate vector genomics. Nucleic acids research. 2009;37:D583–D587. doi: 10.1093/nar/gkn857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boudko DY. Bioanalytical profile of the L-arginine/nitric oxide pathway and its evaluation by capillary electrophoresis. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;851:186–210. doi: 10.1016/j.jchromb.2007.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deves R, Boyd CA. Transporters for cationic amino acids in animal cells: discovery, structure, and function. Physiol Rev. 1998;78:487–545. doi: 10.1152/physrev.1998.78.2.487. [DOI] [PubMed] [Google Scholar]
- 37.Kim JW, Clos EIALCJ. Transport of cationic amino acids by the mouse ecotropic retrovirus receptor. Nature. 1991;532:725–728. doi: 10.1038/352725a0. [DOI] [PubMed] [Google Scholar]
- 38.Hosokawa Hea. Cloning and characterization of a brain-specific cationic amino acid transporter. The Journal of biological chemistry. 1997;272:8717–8722. doi: 10.1074/jbc.272.13.8717. [DOI] [PubMed] [Google Scholar]
- 39.YF LKT. Switching between the two action modes of the dual-affinity nitrate transporter CHL 1 by phosphorylation. the EMBO journall. 2003;22:1005–1013. doi: 10.1093/emboj/cdg118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.SUN Jea. Crystal structure of the plant dual-affinity nitrate transporter NRT1. Nature. 2014 doi: 10.1038/nature13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang HKMNR, Kabat D. Cell-surface receptor for ecotropic murine retroviruses is a basic amino-acid transporter. Nature. 1991;352:729–731. doi: 10.1038/352729a0. [DOI] [PubMed] [Google Scholar]
- 42.Gilles Wea. Monovalent cation conductance in Xenopus laevis oocytes expressing hCAT-3. Biochimica et Biophysica Acta (BBA)-Biomembranes. 2005;1668:234–239. doi: 10.1016/j.bbamem.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 43.Hansen IA, Attardo GM. Nutritional Signaling in Anautogenous Mosquitoes. In: Chandrasekar R, editor. Short Views on Insect Molecular Biology. South India: International Book Mission; 2009. (ed^(eds. [Google Scholar]
- 44.Dhara A, et al. Ovary ecdysteroidogenic hormone functions independently of the insulin receptor in the yellow fever mosquito, Aedes aegypti. Insect biochemistry and molecular biology. 2013;43:1100–1108. doi: 10.1016/j.ibmb.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brandon MC, Pennington JE, Isoe J, Zamora J, Schillinger A-S, Miesfeld RL. TOR signaling is required for amino acid stimulation of early trypsin protein synthesis in the midgut of Aedes aegypti mosquitoes. Insect biochemistry and molecular biology. 2008;38:916–922. doi: 10.1016/j.ibmb.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gulia-Nuss M, Robertson AE, Brown MR, Strand MR. Insulin-like peptides and the target of rapamycin pathway coordinately regulate blood digestion and egg maturation in the mosquito Aedes aegypti. PloS one. 2011;6:e20401. doi: 10.1371/journal.pone.0020401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Melikian HE. Neurotransmitter transporter trafficking: endocytosis, recycling, and regulation. Pharmacology & therapeutics. 2004;104:17–27. doi: 10.1016/j.pharmthera.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 48.Bradley GL, Leevers SJ. Amino acids and the humoral regulation of growth: fat bodies use slimfast. Cell. 2003;114:656–658. doi: 10.1016/s0092-8674(03)00721-9. [DOI] [PubMed] [Google Scholar]
- 49.Luckhart S, Vodovotz Y, Cui L, Rosenberg R. The mosquito Anopheles stephensi limits malaria parasite development with inducible synthesis of nitric oxide. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:5700–5705. doi: 10.1073/pnas.95.10.5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hillyer JF, Estevez-Lao TY. Nitric oxide is an essential component of the hemocyte-mediated mosquito immune response against bacteria. Developmental and comparative immunology. 2010;34:141–149. doi: 10.1016/j.dci.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 51.Stuehr DJ. Enzymes of the L-arginine to nitric oxide pathway. J Nutr. 2004;134:2748S–2751S. doi: 10.1093/jn/134.10.2748S. discussion 2765S–2767S. [DOI] [PubMed] [Google Scholar]
- 52.Price DP, Schilkey FD, Ulanov A, Hansen IA. Small mosquitoes, large implications: crowding and starvation affects gene expression and nutrient accumulation in Aedes aegypti. Parasit Vectors. 2015;8:252. doi: 10.1186/s13071-015-0863-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular biology and evolution. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krieger EKG, Vriend G. Increasing the precision of comparative models with YASARA NOVA- a self-parameterizing force field. Protiens. 2002;47:393–402. doi: 10.1002/prot.10104. [DOI] [PubMed] [Google Scholar]
- 56.Inc. CCG. Molecular Operating Environment (MOE), 2013.08. 2015 (ed^(eds) [Google Scholar]
- 57.Kearse M, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huggett JF, et al. The digital MIQE guidelines: Minimum Information for Publication of Quantitative Digital PCR Experiments. Clinical chemistry. 2013;59:892–902. doi: 10.1373/clinchem.2013.206375. [DOI] [PubMed] [Google Scholar]
- 59.Primer BLAST. 2010 Url: http://www.ncbi.nlm.nih.gov/tools/primer-blast/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.